Abstract
Objectives
To compare detection patterns of 80 cephalometric landmarks identified by an automated identification system (AI) based on a recently proposed deep-learning method, the You-Only-Look-Once version 3 (YOLOv3), with those identified by human examiners.
Materials and Methods
The YOLOv3 algorithm was implemented with custom modifications and trained on 1028 cephalograms. A total of 80 landmarks comprising two vertical reference points and 46 hard tissue and 32 soft tissue landmarks were identified. On the 283 test images, the same 80 landmarks were identified by AI and human examiners twice. Statistical analyses were conducted to detect whether any significant differences between AI and human examiners existed. Influence of image factors on those differences was also investigated.
Results
Upon repeated trials, AI always detected identical positions on each landmark, while the human intraexaminer variability of repeated manual detections demonstrated a detection error of 0.97 ± 1.03 mm. The mean detection error between AI and human was 1.46 ± 2.97 mm. The mean difference between human examiners was 1.50 ± 1.48 mm. In general, comparisons in the detection errors between AI and human examiners were less than 0.9 mm, which did not seem to be clinically significant.
Conclusions
AI showed as accurate an identification of cephalometric landmarks as did human examiners. AI might be a viable option for repeatedly identifying multiple cephalometric landmarks.
Keywords: Automated identification, Cephalometric landmarks, Artificial intelligence, Machine learning, Deep learning
INTRODUCTION
Recently, in the field of automated identification of cephalometric landmarks, the latest deep learning method based on the You-Only-Look-Once version 3 algorithm (YOLOv3)1,2 detected 80 landmarks and resulted in not only more accurate but also faster detecting performance.3 The performance of an automated identification system (AI) has traditionally been compared by the successful detection rates of 19 skeletal landmarks with a 2-mm range, which has conventionally been accepted as a clinical error range at AI performance competitions.4–6 Rather than again comparing certain AI techniques to other AI techniques to determine which were more accurate, the present study proposed a new automatic identification method and tested whether this new AI method was better and more reliable than clinically experienced human experts. This could be more interesting and actually applicable to clinicians. However, when it comes to a reliability measure when identifying a certain cephalometric landmark, there is no firm “ground truth” or gold standard that can provide validation as to where the true location of the landmark is.7–9 Consequently, a study design to answer questions as to (1) whether differences between AI and human examiners would be smaller than those between human examiners (better accuracy in finding landmarks) and (2) whether AI might result in smaller differences upon repeated detection trials than those resulted by humans (better reproducibility of landmarks) would be helpful and appropriate. These results could indicate if AI could be safely proposed for use in clinical practice.
The purpose of this study was to compare detection patterns of 80 cephalometric landmarks identified by a recently proposed deep-learning method, YOLOv3, with those identified by human examiners. The pattern of differences according to image quality and metallic artifacts on images was also investigated. The null hypothesis was that there would be no significant difference between AI and human examiners regarding (1) accuracy in finding landmarks and (2) reproducibility of landmarks.
MATERIALS AND METHODS
The institutional review board for the protection of human subjects reviewed and approved the research protocol (S-D 2018010 and ERI 19007).
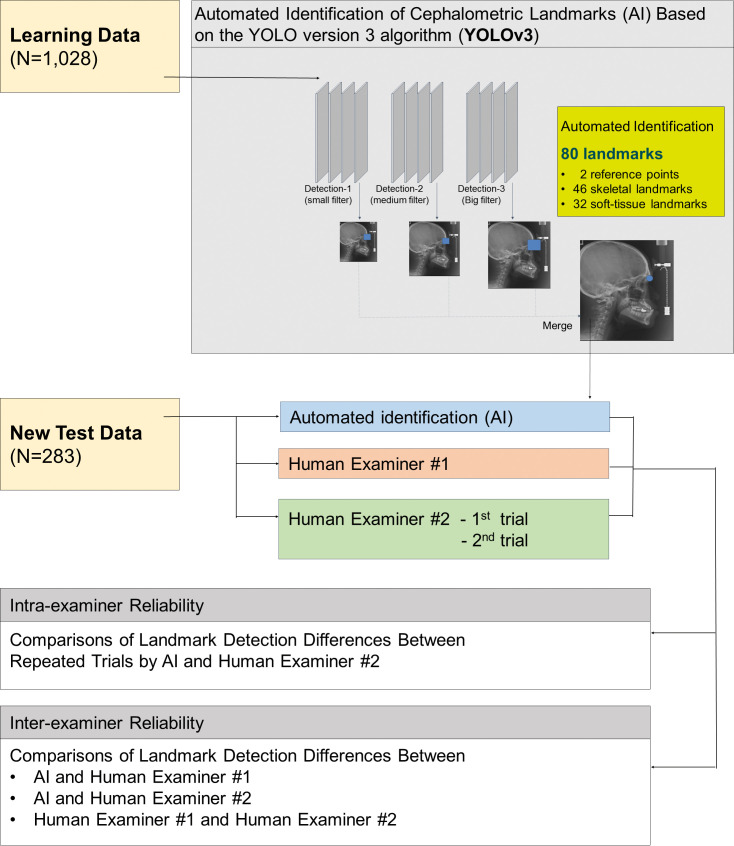
Figure 1 summarizes the experimental design used in the present study. The x-ray image characteristics of data are listed in Table 1. As a result of ethical concerns, the authors' institution has not permitted researchers to use a high-quality electronic medical image in the DICOM format. All of the learning data images were downloaded in the .jpg format with a resolution of 150 and 300 DPI. When digitizing the test data images, a minimum resolution of 150 DPI was maintained.
Figure 1.
Flow diagram summarizing the experimental design.
Table 1.
Descriptive Summary of Study Data
| Study Variables |
N (%) |
|
| Learning data | 1028 (100) | |
| Gender | Female | 507 (49.3) |
| Skeletal classification | Class II | 178 (17.3) |
| Class III | 719 (70.0) | |
| Test data | 283 (100) | |
| Gender | Female | 146 (51.6) |
| Skeletal classification | Class II | 32 (11.3) |
| Class III | 251 (88.7) | |
| Image quality | Good | 248 (87.6) |
| Fair | 13 (4.6) | |
| Poor | 22 (7.8) | |
| Metallic artifactsa | Yes | 140 (49.5) |
Metallic artifacts included full-mouth fixed orthodontic appliances, massive prostheses, and/or surgical bone plates.
The same 1311 lateral cephalometric radiograph images that consisted of 1028 learning and 283 test data were applied in the development stage of the YOLOv3-based AI.3 A total of 80 landmarks comprising two vertical reference points and 46 hard tissue and 32 soft tissue landmarks3 were manually identified by a single examiner (examiner 1) who has 28 years of clinical orthodontic practice experience.
The test set of 283 images were also manually identified for the same 80 landmarks by examiner 2 twice within 3-month intervals. Examiner 2 was a third-year resident at the same institution as examiner 1.
The test data were film images with varying degrees of image quality. Test images were classified according to gender, skeletal classification, and presence of metallic artifacts. These x-ray images displayed varying degrees of image quality, which were subjectively classified as “good,” “fair,” or “poor” (Table 1).
The AI method based on the YOLOv3 algorithm applied in the present study was described in detail in part 1 of this AI project.3 The deep-learning was processed by a workstation running Ubuntu 18.04.1 LTS with NVIDIA Tesla V100 GPU (NVIDIA Corporation, Santa Clara, Calif). After the deep-learning procedure on 80 landmark locations of 1028 images was conducted, the trained AI automatically found each landmark on the 283 test images.
Differences between AI and examiner 1, differences between examiners 1 and 2, and differences between examiner 2's first and second trials were calculated in terms of distance measured in millimeter scales. To compare the detection accuracy between AI and human examiners controlling for multiplicity problems, t-tests with the Bonferroni correction of alpha errors were performed. To investigate which image factors might have influence on significant differences in the landmark identification, multiple linear regression analyses were conducted.
To visualize and evaluate the error pattern in two-dimensional space, scattergrams with 95% confidence ellipses7,8 were depicted. Language R (Vienna, Austria)10 was used throughout all of the statistical analyses.
RESULTS
Accuracy in Finding Landmarks
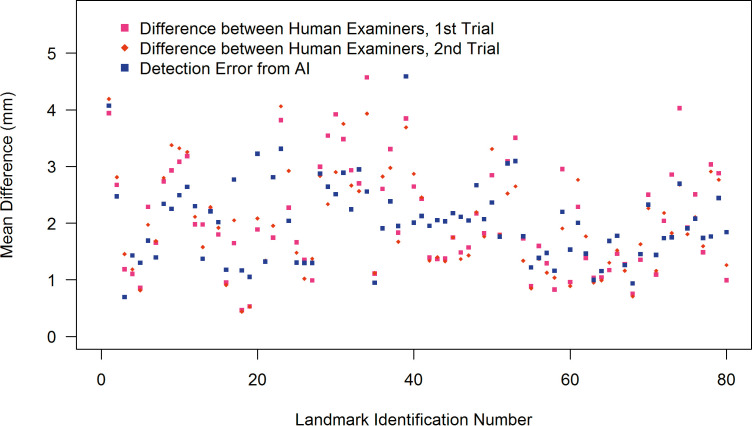
When identifying 46 skeletal landmarks, AI showed better accuracy in 14 out of 46 landmarks, the human examiner did better in 14 out of 46 landmarks, and the remaining 18 out of 46 did not show statistically significant differences. Regarding the 32 soft tissue landmarks, AI showed a better accuracy in 5 out of 32, the human examiner did better in 7 out of 32, and the remaining 20 out of 32 did not show statistically significant differences (Figure 2; Table 2).
Figure 2.
Point plots summarizing the mean differences between human examiners and between AI vs humans.
Table 2.
Comparisons Between Differences between Automated Identification System (AI) and Human Examiners and between Human Examiners. Values Are Point-to-Point Errors in Millimeter Units. More Accurate Results Are Marked with a √ Symbol
| Detection Error from AI |
Difference Between Human Examiners |
Mean Difference | More Accurate Result from |
|||||||||
| Landmarka |
Mean |
SD |
Min |
Max |
Mean |
SD |
Min |
Max |
Between AI and Human Examiner |
AI |
Human Examiner |
P-Valueb |
| Sella | 0.7 | 0.9 | 0.0 | 10.9 | 1.3 | 0.5 | 0.2 | 3.5 | −0.6 | √ | <.0001 | |
| Nasion | 1.4 | 1.2 | 0.0 | 12.6 | 1.1 | 0.9 | 0.1 | 5.6 | 0.3 | .1037 | ||
| Nasal tip | 1.3 | 0.8 | 0.0 | 6.0 | 0.8 | 0.5 | 0.1 | 3.1 | 0.5 | √ | <.0001 | |
| Porion | 1.7 | 1.4 | 0.0 | 13.1 | 2.1 | 1.3 | 0.3 | 8.9 | −0.4 | √ | .0157 | |
| Orbitale | 1.4 | 0.9 | 0.0 | 7.5 | 1.7 | 1.1 | 0.1 | 9.2 | −0.3 | .0886 | ||
| Anterior nasal spine | 2.3 | 1.9 | 0.1 | 17.5 | 2.0 | 1.6 | 0.2 | 10.0 | 0.3 | 1.0000 | ||
| Posterior nasal spine | 1.4 | 1.1 | 0.0 | 9.2 | 1.8 | 1.3 | 0.1 | 7.6 | −0.4 | √ | .0038 | |
| Point A | 2.2 | 1.5 | 0.1 | 8.9 | 2.2 | 1.5 | 0.1 | 7.7 | 0.0 | 1.0000 | ||
| U1 root tip | 2.8 | 1.7 | 0.0 | 10.2 | 1.8 | 1.2 | 0.0 | 7.0 | 1.0 | √ | <.0001 | |
| U1 incisal edge | 1.2 | 0.7 | 0.0 | 3.6 | 0.5 | 0.3 | 0.0 | 2.8 | 0.7 | √ | <.0001 | |
| L1 incisal edge | 1.1 | 0.7 | 0.0 | 4.2 | 0.5 | 0.4 | 0.0 | 4.0 | 0.6 | √ | <.0001 | |
| L1 root tip | 3.2 | 1.7 | 0.1 | 10.2 | 2.0 | 1.3 | 0.3 | 7.9 | 1.2 | √ | <.0001 | |
| Point B | 3.3 | 2.0 | 0.1 | 10.3 | 3.9 | 2.2 | 0.1 | 11.0 | −0.6 | √ | .0264 | |
| Protuberance menti | 2.0 | 1.6 | 0.0 | 10.3 | 2.6 | 1.7 | 0.2 | 11.9 | −0.6 | √ | .0063 | |
| Pogonion | 1.3 | 1.2 | 0.0 | 11.3 | 1.6 | 1.3 | 0.1 | 8.4 | −0.3 | .9907 | ||
| Gnathion | 1.3 | 0.8 | 0.0 | 5.4 | 1.2 | 0.7 | 0.2 | 4.6 | 0.1 | 1.0000 | ||
| Menton | 1.3 | 0.7 | 0.1 | 6.7 | 1.2 | 0.7 | 0.1 | 4.8 | 0.1 | 1.0000 | ||
| Gonion, constructed | 2.9 | 1.6 | 0.0 | 16.1 | 2.9 | 1.9 | 0.5 | 13.4 | 0.0 | 1.0000 | ||
| Gonion, anatomic | 2.2 | 1.6 | 0.1 | 11.1 | 2.8 | 2.0 | 0.2 | 13.1 | −0.6 | √ | .0237 | |
| Articulare | 0.9 | 0.6 | 0.0 | 4.2 | 1.1 | 0.8 | 0.1 | 5.1 | −0.2 | .3024 | ||
| Condylion | 1.9 | 1.5 | 0.2 | 15.6 | 1.8 | 1.1 | 0.2 | 8.1 | 0.1 | 1.0000 | ||
| Pterygoid | 2.1 | 5.8 | 0.0 | 96.9 | 2.4 | 1.9 | 0.3 | 10.1 | −0.3 | 1.0000 | ||
| Basion | 2.0 | 1.5 | 0.0 | 10.3 | 1.4 | 1.1 | 0.1 | 6.9 | 0.6 | √ | <.0001 | |
| Glabella | 2.1 | 4.1 | 0.1 | 65.4 | 1.8 | 1.4 | 0.2 | 11.6 | 0.3 | 1.0000 | ||
| nasion | 1.8 | 1.3 | 0.0 | 7.5 | 1.8 | 1.3 | 0.1 | 6.8 | 0.0 | 1.0000 | ||
| supranasal tip | 1.8 | 1.9 | 0.0 | 18.4 | 1.5 | 1.0 | 0.1 | 5.5 | 0.3 | 1.0000 | ||
| pronasale | 1.2 | 1.3 | 0.0 | 11.0 | 0.9 | 0.6 | 0.1 | 3.9 | 0.3 | √ | .0037 | |
| columella | 1.4 | 1.1 | 0.0 | 9.7 | 1.5 | 0.8 | 0.2 | 6.5 | −0.1 | 1.0000 | ||
| subnasale | 1.2 | 0.8 | 0.0 | 4.0 | 0.9 | 0.5 | 0.2 | 3.6 | 0.3 | √ | .0033 | |
| point A | 1.5 | 1.0 | 0.0 | 5.9 | 0.9 | 0.7 | 0.1 | 4.1 | 0.6 | √ | <.0001 | |
| superior labial sulcus | 2.0 | 1.6 | 0.0 | 9.3 | 2.5 | 1.6 | 0.1 | 8.5 | −0.5 | √ | .0074 | |
| labiale superius | 1.5 | 1.1 | 0.0 | 6.9 | 1.6 | 1.2 | 0.1 | 6.5 | −0.1 | 1.0000 | ||
| upper lip | 1.0 | 0.9 | 0.0 | 9.6 | 1.0 | 0.7 | 0.0 | 4.3 | 0.0 | 1.0000 | ||
| stomion superius | 1.7 | 1.2 | 0.1 | 6.3 | 1.2 | 1.1 | 0.1 | 8.3 | 0.5 | √ | .0002 | |
| stomion inferius | 1.8 | 1.6 | 0.1 | 14.7 | 1.5 | 1.6 | 0.1 | 15.9 | 0.3 | 1.0000 | ||
| lower lip | 0.9 | 0.6 | 0.0 | 5.0 | 0.7 | 0.5 | 0.1 | 2.7 | 0.2 | √ | .0022 | |
| labiale inferius | 1.5 | 1.0 | 0.0 | 5.9 | 1.5 | 1.0 | 0.1 | 7.5 | 0.0 | 1.0000 | ||
| point B | 1.4 | 1.3 | 0.1 | 12.3 | 1.1 | 0.8 | 0.0 | 5.6 | 0.3 | √ | .0438 | |
| protuberance menti | 1.7 | 1.4 | 0.1 | 9.6 | 2.1 | 1.4 | 0.2 | 9.0 | −0.4 | .0896 | ||
| pogonion | 1.7 | 1.9 | 0.0 | 16.8 | 2.3 | 1.9 | 0.2 | 11.5 | −0.6 | √ | .0197 | |
| gnathion | 2.7 | 2.3 | 0.1 | 19.1 | 3.4 | 2.2 | 0.5 | 14.9 | −0.7 | √ | .0307 | |
| menton | 1.9 | 1.6 | 0.1 | 11.0 | 1.9 | 1.5 | 0.2 | 10.6 | 0.0 | 1.0000 | ||
The landmarks and data included in this table were chosen to concisely describe the results. Uppercase letters were used to indicate skeletal landmarks, and lowercase letters were used to indicate soft tissue landmarks.
Results from t-tests with Bonferroni correction. AI, automatic identification of cephalometric landmarks based on a deep learning method (YOLOv3). SD indicates standard deviation; Min, minimum; and Max, maximum.
The mean detection error between AI and the human was 1.46 ± 2.97 mm. The mean difference between human examiners was 1.50 ± 1.48 mm.
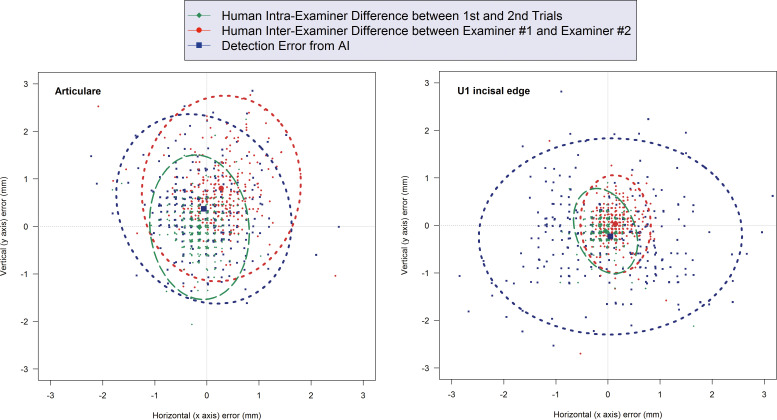
Figure 3 illustrates representative cases in which there was no statistically significant difference between AI and the human examiners (ie, Articulare) and in which the human demonstrated a more accurate detection (ie, upper incisal edge). In either case, however, comparisons in the mean detection errors between AI and the human were less than 0.9 mm. The only exception was the landmark for the lower incisor root tip that showed a 1.2-mm greater error from AI than did the human examiner (Table 2).
Figure 3.
Scattergrams and 95% confidence ellipses illustrating representative cases. Left: when there is no statistically significant difference between AI and human examiners (Articulare); Right: when human examiners demonstrate a more accurate detection (upper incisal edge).
Reproducibility of Landmarks Upon Repeated Trials
Upon repeated trials, AI always detected identical positions on each landmark, while the human intraexaminer variability from repeated detection trials was 0.97 ± 1.03 mm.
Comparisons According to Image Variables
The results of the multiple linear regression analysis indicated that AI's accuracy in finding landmarks was not meaningfully affected by image variables such as gender, skeletal classification, image quality, and presence of metallic artifacts.
DISCUSSION
The present study was formulated to investigate whether AI might be a viable option for the repetitive and arduous task of identifying multiple cephalometric landmarks for use in clinical orthodontic practice. The null hypothesis that there would be no difference between AI and human examiners regarding accuracy in finding landmarks could not be rejected. The mean detection errors between AI and the human did not exceed 0.9 mm, except at only one landmark for the lower incisor root tip, which showed a 1.2-mm difference. In all landmarks, AI demonstrated as accurate identification as did trained orthodontists. In general, all of those mean differences showing less than 2 mm would not seem to be clinically significant errors. However, since AI always detected identical positions, the reproducibility by AI upon repeated detection trials was definitely better than that associated with human examiners.
Among the machine learning methods, deep-learning methods have demonstrated superiority in automatically recognizing anatomical landmarks on diagnostic images. Studies on related topics in various fields have also been gaining more popularity.3,11–14 Although three-dimensional images have gained popularity these days,15–19 two-dimensional cephalometric analysis is still a vital tool in orthodontic diagnosis and treatment planning since it provides information regarding a patient's skeletal and soft tissue. Currently, computer-assisted cephalometric analysis eliminates human-induced mechanical errors. Fully automatic cephalometric analysis has long been attempted with the intention of reducing the time required to obtain a cephalometric analysis, improving the accuracy of landmark identification, and reducing the errors caused by a clinician's subjectivity. However, previous studies detected a limited number of landmarks, less than 20, and the accuracy results were not satisfactory for use in clinical orthodontic practice. For example, in 2009, 10 landmarks on 41 digital images were identified.20 In 2013, 16 landmarks were identified on 40 cephalometric radiographs, and the mean error from automatically identified landmarks was 2.59 mm.21 The accuracy of those automated methods was not as good as that associated with manual identification. In addition, cephalometric landmarks need not be limited to simply obtaining the skeletal characteristics of patients but could be also be applied to plan treatment and to predict treatment outcomes, including soft tissue drape changes. For those purposes, an expanded number, even hundreds, of variables of anatomic landmarks is necessary.14,22–25
In the present study, unlike the learning data that included images from a variety of malocclusion patients, the test images were selected from patients who had a severe type of mandibular deficiency, prognathism, or facial asymmetry. They eventually had orthognathic surgeries performed. From the first formulation of the current study, the selection of these types of patients was intended to test the performance of AI in a more difficult condition, rather than identifying landmarks on images from good-looking subjects. The descriptive summary in Table 1 reflects and matches well with the current trend of patients seeking a university-affiliated dental healthcare institution that has a high proportion of orthodontic patients with severe skeletal discrepancies.26
The cephalometric landmarks identified could potentially result in errors on both the x and y axes. There are several advantages when visualizing results with scattergrams and the 95% confidence ellipse that was a two-dimensional expansion of the Bland-Altman plot.7,8 One of them is to observe the correlation between the x- and y-axis errors in the shape of the ellipse. Closer to an isometric circle indicates more independence between the x- and y-axis errors. The greater the degree of deformation of the ellipse, the greater the indication of the correlation between the x- and y-axis errors.7,8
In general, the pattern of differences between AI and human examiners demonstrated that AI acted like a human examiner. For example, when human examiners had difficulties in identifying landmarks on poor-quality images, so did AI. This might be the reason why image factors did not meaningfully affect the accuracy of AI in finding landmarks. In those subjects with fixed orthodontic appliances, massive prostheses, and/or surgical bone plates, it was initially anticipated that there would be difficulties in identifying the landmarks because of the multiple metallic artifacts. However, metal artifacts did not appear to have a clinically significant impact on the identification of landmarks either.
One strength of the present study might be that it included the largest number of both learning and test data sets when compared to previous studies. The number of cephalometric landmarks was also the greatest: 80 landmarks including soft tissue glabella to the terminal point on the neck. Conventional key landmarks that have previously been required for cephalometric analysis as well as a large number of other landmarks are essential for accurately predicting posttreatment changes.22–25
As a limitation of the present study, the way AI learned during the training session and how it identified landmarks later in the test step are not explainable without describing computer science jargon. Although some technical details have been necessary, this present study intended to focus on showcasing the results from AI. Further details of the modification algorithms appear elsewhere.1,2 Upon repeated trials, AI always found identical positions. However, during preceding pilot studies, when the quantity of learning data was less than 500 images, AI did not identify an identical point. In this regard, how much learning data might be sufficient enough to teach AI is currently unknown. Furthermore, it could be conjectured that the number of target landmarks might also be a contributing factor in deciding a sufficient number for learning data. A study to elucidate the sufficient quantity of data for deep-learning of AI might be necessary in the future.
From the clinical perspective, however, AI would never replace trained specialists in orthodontics, nor might AI intend to replace a comprehensive orthodontic training program. Rather it could supplement, augment, and amplify diagnostic performance by objectively evaluating each patient seeking orthodontic treatment. The AI proposed in the present study can be compatible with the current clinical environment and would retain its validity under the constant supervision of experts in orthodontics.
CONCLUSIONS
In general, the pattern of differences between AI and human examiners demonstrated that AI acted like human examiners. AI showed as accurate an identification of cephalometric landmarks as did human examiners.
Upon repeated trials, AI detected always identical positions, which implies that AI might be a more reliable option for repeatedly identifying multiple cephalometric landmarks.
ACKNOWLEDGMENTS
This study was partly supported by grant 05-2018-0018 from the Seoul National University Dental Hospital Research Fund and the Seoul R&BD program (grant number IC 170010) funded by the Seoul Metropolitan Government.
Disclosure
The final form of the machine learning system was developed by DDH Inc (Seoul, Korea), which is expected to own the patent in the future. Among the coauthors, Hansuk Kim and Soo-Bok Her are shareholders of DDH Inc. Youngsung Yu and Girish Srinivasan are employees there. Other authors do not have a conflict of interest.
REFERENCES
- 1.Redmon J, Farhadi A. Yolov3: an incremental improvement. arXiv preprint arXiv180402767. 2018 Available at: https://arxiv.org/pdf/1804.02767.pdf.
- 2.Redmon J, Divvala S, Girshick R, Farhadi A. You Only Look Once Unified RealTime Object Detection. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016. pp. 779–788. [Google Scholar]
- 3.Park JH, Hwang HW, Moon JH, et al. Automated identification of cephalometric landmarks—part 1. Comparisons between the latest deep learning methods YOLOv3 and SSD. Angle Orthod. 2019;89:903–909. doi: 10.2319/022019-127.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arik SÖ, Ibragimov B, Xing L. Fully automated quantitative cephalometry using convolutional neural networks. J Med Imag. 2017;4:014501. doi: 10.1117/1.JMI.4.1.014501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang CW, Huang CT, Hsieh MC, et al. Evaluation and comparison of anatomical landmark detection methods for cephalometric x-ray images: a grand challenge. IEEE Trans Med Imaging. 2015;34:1890–1900. doi: 10.1109/TMI.2015.2412951. [DOI] [PubMed] [Google Scholar]
- 6.Wang CW, Huang CT, Lee JH, et al. A benchmark for comparison of dental radiography analysis algorithms. Med Image Anal. 2016;31:63–76. doi: 10.1016/j.media.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Donatelli RE, Lee SJ. How to report reliability in orthodontic research: part 1. Am J Orthod Dentofacial Orthop. 2013;144:156–161. doi: 10.1016/j.ajodo.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Donatelli RE, Lee SJ. How to report reliability in orthodontic research: part 2. Am J Orthod Dentofacial Orthop. 2013;144:315–318. doi: 10.1016/j.ajodo.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 9.Donatelli RE, Lee SJ. How to test validity in orthodontic research: a mixed dentition analysis example. Am J Orthod Dentofacial Orthop. 2015;147:272–279. doi: 10.1016/j.ajodo.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 10.R Development Core Team. R A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 11.Montúfar J, Romero M, Scougall-Vilchis RJ. Automatic 3-dimensional cephalometric landmarking based on active shape models in related projections. Am J Orthod Dentofacial Orthop. 2018;153:449–458. doi: 10.1016/j.ajodo.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 12.Montúfar J, Romero M, Scougall-Vilchis RJ. Hybrid approach for automatic cephalometric landmark annotation on cone-beam computed tomography volumes. Am J Orthod Dentofacial Orthop. 2018;154:140–150. doi: 10.1016/j.ajodo.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 13.Ponce-Garcia C, Lagravere-Vich M, Cevidanes LHS, Ruellas ACdO, Carey J, Flores-Mir C. Reliability of three-dimensional anterior cranial base superimposition methods for assessment of overall hard tissue changes: a systematic review. Angle Orthod. 2018;88:233–245. doi: 10.2319/071217-468.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang TJ, Eo SH, Cho HJ, Donatelli RE, Lee SJ. A sparse principal component analysis of Class III malocclusions. Angle Orthod. 2019;89:768–774. doi: 10.2319/100518-717.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sam A, Currie K, Oh H, Flores-Mir C, Lagravere-Vich M. Reliability of different three-dimensional cephalometric landmarks in cone-beam computed tomography: a systematic review. Angle Orthod. 2019;89:317–332. doi: 10.2319/042018-302.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castillo JC, Gianneschi G, Azer D, et al. The relationship between 3D dentofacial photogrammetry measurements and traditional cephalometric measurements. Angle Orthod. 2019;89:275–283. doi: 10.2319/120317-825.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isidor S, Carlo GD, Cornelis MA, Isidor F, Cattaneo PM. Three-dimensional evaluation of changes in upper airway volume in growing skeletal Class II patients following mandibular advancement treatment with functional orthopedic appliances. Angle Orthod. 2018;88:552–559. doi: 10.2319/083117-587.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanikawa C, Takada K. Test-retest reliability of smile tasks using three-dimensional facial topography. Angle Orthod. 2018;88:319–328. doi: 10.2319/062617-425.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng J, Yu H, Yin Y, et al. Esthetic evaluation of facial cheek volume: a study using 3D stereophotogrammetry. Angle Orthod. 2019;89:129–137. doi: 10.2319/020418-97.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonardi R, Giordano D, Maiorana F. An evaluation of cellular neural networks for the automatic identification of cephalometric landmarks on digital images. J Biomed Biotechnol. 2009. p. 717102. [DOI] [PMC free article] [PubMed]
- 21.Shahidi S, Oshagh M, Gozin F, Salehi P, Danaei SM. Accuracy of computerized automatic identification of cephalometric landmarks by a designed software. Dentomaxillofac Radiol. 2013;42:20110187. doi: 10.1259/dmfr.20110187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HJ, Suh HY, Lee YS, et al. A better statistical method of predicting postsurgery soft tissue response in Class II patients. Angle Orthod. 2014;84:322–328. doi: 10.2319/050313-338.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YS, Suh HY, Lee SJ, Donatelli RE. A more accurate soft-tissue prediction model for Class III 2-jaw surgeries. Am J Orthod Dentofacial Orthop. 2014;146:724–733. doi: 10.1016/j.ajodo.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Suh HY, Lee HJ, Lee YS, Eo SH, Donatelli RE, Lee SJ. Predicting soft tissue changes after orthognathic surgery: the sparse partial least squares method. Angle Orthod. 2019;89:910–916. doi: 10.2319/120518-851.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon KS, Lee HJ, Lee SJ, Donatelli RE. Testing a better method of predicting postsurgery soft tissue response in Class II patients: a prospective study and validity assessment. Angle Orthod. 2015;85:597–603. doi: 10.2319/052514-370.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CH, Park HH, Seo BM, Lee SJ. Modern trends in Class III orthognathic treatment: a time series analysis. Angle Orthod. 2017;87:269–278. doi: 10.2319/043016-349.1. [DOI] [PMC free article] [PubMed] [Google Scholar]