Abstract
Background
DNA damage and inflammation are common in end-stage renal disease (ESRD). Our aim was to evaluate the levels of circulating cell-free DNA (cfDNA) and the relationship with inflammation, anaemia, oxidative stress and haemostatic disturbances in ESRD patients on dialysis. By performing a 1-year follow-up study, we also aimed to evaluate the predictive value of cfDNA for the outcome of ESRD patients.
Methods
A total of 289 ESRD patients on dialysis were enrolled in the study: we evaluated cfDNA, haemogram, serum iron, hepcidin, inflammatory and oxidative stress markers, and haemostasis. Events and causes of deaths were recorded throughout the follow-up period.
Results
ESRD patients, as compared with controls, presented significantly higher levels of cfDNA, hepcidin, and inflammatory and oxidative stress markers, and significantly lower values of iron and anaemia-related haemogram parameters. The all-cause mortality rate was 9.7%; compared with alive patients, deceased patients (n = 28) were older and presented significantly higher values of inflammatory markers and of cfDNA, which was almost 2-fold higher. Furthermore, cfDNA was the best predictor of all-cause mortality and cardiovascular mortality in ESRD patients, in both unadjusted and adjusted models for basic confounding factors in dialysis.
Conclusions
Our data show cfDNA to be a valuable predictive marker of prognosis in ESRD patients on dialysis treatment; high levels of cfDNA were associated with a poor outcome.
Keywords: cell-free DNA, CKD outcome, end-stage renal disease, inflammation
INTRODUCTION
Morbidity and mortality rates are still very high in chronic kidney disease (CKD) patients, as compared with the general population. The identification of predictive biomarkers of morbidity and mortality is an emergent area of interest in facing this global health problem.
Inflammation is a common feature in CKD, and it is particularly enhanced in end-stage renal disease (ESRD) patients on haemodialysis [1]. This inflammatory process has been associated with endothelial dysfunction, atherosclerotic modifications [2, 3] and haemostatic disturbances [4, 5], increasing the risk for thrombotic events.
The reduced erythropoietin production by failing kidneys and the erythropoietic suppression by uraemic toxins lead to the development of anaemia. The enhanced release of interleukin (IL)-6 by inflammatory cells triggers the synthesis of hepcidin, the main regulator of iron metabolism, contributing to worsening of anaemia by reducing iron absorption and mobilization from iron storage [6].
The enhanced inflammatory process in CKD patients appears to contribute to increase cell-free DNA (cfDNA) [7, 8]. Circulating cfDNA is composed of genomic DNA that circulates in a cell-free form (mRNAs and miRNAs) and as mitochondrial DNA (mtDNA); cfDNA seems to result from apoptosis, NETosis and necrosis of blood cells [9, 10]. Netosis has been associated with common comorbidities in ESRD patients [11, 12]. Increased cfDNA has been reported in inflammatory conditions, such as psoriasis, cancer, diabetes and CKD [8, 13–15]. It appears that cfDNA is able to selectively induce in vitro IL-6 production by monocytes [16]; the development of systemic inflammation has been related to the release of cfDNA (mainly) by inflammatory cells [17], explaining the positive correlation with inflammatory response, and suggesting a two-way relationship between cfDNA and inflammation. Increased levels of cfDNA have been also associated with higher mortality risk in CKD [7, 18]. Inaccurately repaired or unrepaired nuclear or mtDNA may have pathophysiological relevance for cancer development and cardiovascular complications in CKD patients [19].
Our aim was to evaluate cfDNA and study its relationship with inflammation, anaemia, oxidative stress and haemostatic markers in a large cohort of ESRD patients on dialysis. Moreover, by performing a 1-year follow-up study to record causes and events of death, we aimed to evaluate the predictive value of cfDNA for the outcome of ESRD patients.
MATERIALS AND METHODS
Subjects
Patients were recruited in five dialysis clinics from the North of Portugal. The Committee on Ethics, from the Faculty of Pharmacy, Porto University, approved the study protocol, as well as the Directors of Dialysis Clinics.
As a control group, we selected apparently healthy volunteers matched with patients, as far as possible, for gender and age (n = 22); subjects with history of renal, inflammatory, neoplasia or cardiovascular diseases were excluded. Creatinine and urea were evaluated in these controls to assure normal renal function.
Patients and controls participated in the study after providing informed and written consent, and their privacy rights were respected. ESRD patients presenting neoplasia, infectious and/or inflammatory conditions were excluded from the study.
The study included 289 ESRD patients under dialysis therapy for at least 90 days; dialysis was performed using FX-class® high-flux polysulphone dialysers (Fresenius, Germany); 41 patients were under high-flux haemodialysis, while 248 were under on-line haemodiafiltration. Dialysis was performed three times/week for 3–5 h; patients were on dialysis for a median (interquartile range) period of 3.74 (1.65–7.34) years. Dialysis clearance of urea was expressed as eKt/V. The aetiologies for CKD were diabetic nephropathy (n = 101), hypertensive nephrosclerosis (n = 36), chronic glomerulonephritis (n = 23), polycystic kidney disease (n = 19), other diseases (n = 44) and uncertain aetiology (n = 66). Considering major ESRD comorbidities, 61.9% presented arterial hypertension and 42.9% presented diabetes mellitus. Concerning vascular access for dialysis procedure, 42 patients used central venous catheter, 233 arteriovenous fistula and 14 patients used arteriovenous graft.
A 1-year follow-up was performed, identifying causes and events of death, in order to evaluate the predictive risk of mortality for the several parameters under study.
Analytical assays
Blood was collected immediately before a mid-week dialysis session, into tubes with and without anticoagulant (ethylenediaminetetraacetic acid), to obtain whole blood, plasma and serum. Samples were processed within 2 h of collection; aliquots of plasma and serum were prepared and stored at −80°C until assayed.
For blood cell counts, we used an automatic blood cell counter (Sysmex K1000; Sysmex, Hamburg, Germany). Iron concentration was determined, using a colorimetric method (Iron, Randox Laboratories Ltd, UK).
Hepcidin, IL-6, tumour necrosis factor (TNF)-α, soluble TNF receptor 2 (sTNFR2), pentraxin (PTX) 3, elastase, tissue inhibitor of metalloproteinases (TIMP)-1, tissue plasminogen activator (tPA), plasminogen activator inhibitor (PAI)-1 and D-dimer were determined using enzyme-linked immunosorbent assays (Human Hepcidin Quantikine ELISA Kit, Human IL-6 Quantikine HS ELISA Kit, Human TNF-α Quantikine HS ELISA Kit, Human TNF RII Immunoassay and PTX3/TSG-14 Quantikine ELISA Kit, R&D Systems, Minneapolis, MN, USA; Human TIMP1 ELISA kit, PMN Elastase Human ELISA Kit, Human Tissue Plasminogen Activator ELISA kit, Human PAI1 ELISA kit and D-Dimer ELISA kit, Abcam, Cambridge, UK); high-sensitivity C-reactive protein (hsCRP) was determined by immunoturbidimetry [CRP (Latex) High-Sensitivity, Roche Diagnostics, Basel, Switzerland].
Total antioxidant status (TAS) was evaluated using the colorimetric ferric reducing ability of plasma (FRAP) assay, according to Benzie and Strain [20]; briefly, the reduction of ferric tripyridyltriazine (Fe3+-TPTZ) complex to the ferrous form (Fe2+-TPTZ), at low pH, causes a colorimetric change; the changes in the absorbance of the samples, at 593 nm, are compared with standard reaction mixtures of ferrous ions [0.1–1.0 mM Iron(II) sulphate heptahydrate], to obtain TAS values. This method is easy to setup and perform, and valuable for clinical studies, as shown in other studies [21, 22].
Lipid peroxidation (LPO) was evaluated according to Mihara et al. [23], with some modifications. Briefly, 25.0 µL of plasma was mixed with 180.0 µL of 1% H3PO4 (v/v) and 60.0 µL of 0.6% thiobarbituric acid (TBA) (w/v) and heat denatured for 45 min. After immersion in ice for 10 min, TBA reactive substances were extracted with butanol (240.0 µL) and measured by spectrophotometry (535 nm); the ratio TAS/LPO was calculated and presented as a measure of oxidative stress.
For the evaluation of serum cfDNA levels, we used a rapid and direct fluorescent assay, according to Goldshtein et al. [24]. In this assay, SYBR Gold Nucleic Acid Gel Stain (Invitrogen, Paisley, UK; diluted 1:10 000) was mixed with serum and with standard samples (10.0 µL); the fluorescence was measured at an emission wavelength of 535 nm and an excitation wavelength of 485 nm. Salmon sperm DNA and 10% (m/v) bovine serum albumin, in phosphate-buffered saline pH 7.4, were used to prepare standards (156.25–10 000 ng/mL), in order to mimic the fluorescence background of serum; a set of four serum samples were used as internal controls in all assays, to ensure reproducibility.
Urea and creatinine were determined by colorimetric methods (Roche Diagnostics, Basel, Switzerland).
Statistical analysis
We used the Statistical Package for Social Sciences (SPSS, version 24.0, Chicago, IL, USA), for Windows. Data distribution was evaluated by Kolmogorov–Smirnov analysis. For comparison between groups, we used, for continuous variables, the Mann–Whitney U test and the unpaired Student’s t-test, in accordance with the Gaussian distribution of the variables; for categorical variables, chi-squared test and Fisher’s exact test were employed. Results are presented as mean ± standard deviation or as median (interquartile range) for variables with normal or non-normal distribution, respectively. Adjustment for confounding factors (e.g. age) was performed using analysis of covariance, followed by Bonferroni correction [variable(s) data respected a normal distribution]. Data presenting a non-Gaussian distribution were transformed to data with normal distribution, using the Templeton method [25]. Strength of the correlations between variables was determined through Spearman’s rank correlation coefficient. To estimate (all-cause, cardiovascular and non-cardiovascular) mortality hazard ratio (HR), we performed a Cox regression analysis. A P-value <0.05 was considered as statistically significant.
RESULTS
Data for controls and ESRD patients are presented in Table 1. As compared with controls, ESRD patients were older. Haemoglobin (Hb) concentration, haematocrit, red blood cell count, mean corpuscular Hb concentration (MCHC), lymphocytes and platelets were significantly lower, while mean corpuscular volume, white blood cells (WBCs) and neutrophils were significantly higher in ESRD patients. IL-6, hsCRP, TNF-α, sTNFR2, PTX3, TIMP-1, LPO, TAS, TAS/LPO, D-dimer, tPA/PAI-1, hepcidin, ferritin, urea and creatinine were significantly higher, and iron, PAI-1, and TNF-α/sTNFR2 and elastase/neutrophil ratios were significantly lower. The cfDNA levels were significantly higher in ESRD (Figure 1). The significant differences found for WBC, TNF-α, sTNFR2 and TIMP-1 were lost after statistical adjustment for age, suggesting that their changes are also associated with aging.
Table 1.
Demographic, clinical and analytical data for controls and ESRD patients and for those who remained alive or were deceased during a 1-year follow-up period
| ESRD patients (n = 289) |
||||
|---|---|---|---|---|
| Parameters | Controls (n = 22) | ESRD patients (n = 289) | Alive (n = 261) | Deceased (n = 28) |
| Age (years) | 57 (52–60) | 71 (60–79)*** | 71 (59–79) | 77 (67–82)† |
| Gender (F/M) | 14/8 | 131/158 | 117/144 | 14/14 |
| BMI (kg/m2) | 24.3 ± 3.4 | 25.8 ± 4.6 | 25.8 ± 4.5 | 25.0 ± 5.2 |
| Dialysis vintage (years) | – | 3.74 (1.65–7.36) | 3.68 (1.65–7.17) | 4.64 (1.51–8.78) |
| URR (%) | – | 79 (76–83) | 79 (76–83) | 79 (75–82) |
| eKt/V | – | 1.62 (1.44–1.81) | 1.62 (1.45–1.81) | 1.66 (1.42–1.79) |
| Ultrafiltration volume (L) | – | 2.3 (1.7–3.0) | 2.4 (1.8–3.0) | 2.1 (1.3–2.7)a,† |
| Calcium (mg/dL) | – | 9.14 (8.74–9.46) | 9.11 (8.70–9.41) | 9.43 (9.01–9.83)†† |
| Phosphorus (mg/dL) | – | 4.18 (3.35–5.00) | 4.18 (3.41–5.00) | 3.90 (3.15–5.17) |
| PTH (pg/mL) | – | 338 (203–502) | 340 (214–513) | 262 (140–479) |
| Albumin (g/dL) | – | 3.80 (3.60–4.10) | 3.80 (3.60–4.10) | 3.55 (3.30–3.88)††† |
| Urea (mg/dL) | 32 (27–36) | 116 (97–145)a,*** | 118 (98–147) | 105 (77–134)a,† |
| Creatinine (mg/dL) | 0.8 (0.7–0.9) | 7.9 (6.5–9.6)a,*** | 8.0 (6.6–9.7) | 7.6 (5.2–9.1) |
| Hb (g/dL) | 13.6 (13.1–15.4) | 11.4 (10.7–12.2)*** | 11.4 (10.7–12.1) | 11.5 (10.2–12.5) |
| Haematocrit (%) | 41 (39–45) | 35 (33–37)*** | 35 (33–37) | 36 (32–39) |
| RBC (x1012/L) | 4.54 (4.27–4.95) | 3.72 (3.45–4.01)*** | 3.71 (3.46–3.99) | 3.80 (3.41–4.07) |
| MCV (fL) | 91 (88–93) | 95 (92–98)*** | 95 (92–98) | 95 (91–98) |
| MCHC (g/dL) | 33.6 (33.2–33.9) | 32.5 (31.8–33.1)*** | 32.6 (31.8–33.2) | 32.2 (31.4–32.7)a,† |
| WBC (×109/L) | 5.30 (4.70–6.53) | 6.31 (5.29–7.62)a,* | 6.30 (5.25–7.60) | 6.80 (5.69–8.54) |
| Neutrophils (×109/L) | 2.95 (2.18–3.85) | 3.90 (3.25–5.00)*** | 3.90 (3.20–4.90) | 4.50 (3.62–5.88)a,† |
| Lymphocytes (×109/L) | 1.95 (1.78–2.53) | 1.50 (1.20–1.90)*** | 1.50 (1.20–1.90) | 1.40 (1.00–1.70) |
| Monocytes (×109/L) | 0.34 (0.30–0.45) | 0.41 (0.32–0.52) | 0.41 (0.32–0.52) | 0.46 (0.39–0.52) |
| Platelets (×109/L) | 289 (225–355) | 197 (161–233)*** | 197 (161–233) | 200 (149–243) |
| Iron (μg/dL) | 106 (91–133) | 55 (45–74)*** | 56 (46–75) | 50 (36–62) |
| Ferritin (ng/mL) | 69 (37–146) | 302 (178–454)*** | 302 (177–456) | 310 (187–448) |
| Hepcidin (ng/mL) | 17 (8–37) | 78 (41–132)*** | 79 (41–131) | 67 (29–135) |
| LPO (nmol/mL) | 9.68 (8.44–12.0) | 11.1 (9.44–13.2)** | 11.1 (9.68–13.2) | 10.7 (9.12–12.3) |
| TAS (μM) | 352 (272–417) | 505 (428–609)*** | 514 (434–619) | 451 (368–526)† |
| TAS/LPO | 33.5 (24.8–51.0) | 44.8 (36.5–57.0)** | 45.2 (37.2–57.0) | 42.0 (32.2–57.0) |
| tPA (ng/mL) | 5.0 (4.5–5.7) | 4.2 (3.0–6.1) | 4.1 (2.9–5.9) | 4.6 (3.7–7.0) |
| PAI-1 (ng/mL) | 26 (16–47) | 7.6 (4.7–11.6)*** | 7.6 (4.7–11.4) | 7.3 (4.8–12.2) |
| tPA/PAI-1 | 0.20 (0.14–0.32) | 0.56 (0.39–0.80)*** | 0.56 (0.38–0.79) | 0.68 (0.40–1.28) |
| D-dimer (ng/mL) | 153 (123–267) | 518 (364–930)*** | 506 (357–902) | 838 (472–1193)a,† |
| IL-6 (pg/mL) | 1.05 (0.68–1.54) | 4.10 (2.69–7.33)*** | 3.89 (2.62–6.84) | 7.56 (4.15–13.1)††† |
| hsCRP (mg/dL) | 0.10 (0.04–0.21) | 0.37 (0.16–0.81)*** | 0.35 (0.15–0.77) | 0.70 (0.31–1.37)†† |
| TNF-α (pg/mL) | 0.83 (0.71–1.09) | 3.38 (2.66–4.59)a,*** | 3.38 (2.66–4.58) | 3.28 (2.63–4.69) |
| sTNFR2 (ng/mL) | 2.0 (1.7–2.3) | 14.7 (11.9–17.5)a,*** | 14.6 (11.7–17.3) | 17.1 (14.1–20.3)†† |
| TNF-α/sTNFR2 (×10−3) | 0.42 (0.36–0.54) | 0.24 (0.18–0.32)*** | 0.24 (0.19–0.32) | 0.20 (0.14–0.25)†† |
| PTX3 (ng/mL) | 0.62 (0.50–0.75) | 1.40 (0.99–2.05)*** | 1.32 (0.96–1.86) | 2.06 (1.41–2.44)†† |
| Elastase (ng/mL) | 48 (39–57) | 43 (33–55) | 41 (33–55) | 50 (40–69)† |
| Elastase/neutrophil | 16 (12–20) | 11 (8–13)*** | 11 (8–13) | 11 (8–14) |
| TIMP-1 (ng/mL) | 243 (212–325) | 535 (469–620)a,*** | 530 (466–617) | 580 (499–725) |
| cfDNA (μg/mL) | 0.53 (0.29–0.75) | 0.84 (0.56–1.22)*** | 0.80 (0.54–1.17) | 1.22 (0.74–1.72)†† |
Data are presented as mean ± standard deviation or median (interquartile range), except for gender (n/n).
Loss of significance after statistical adjustment (ANCOVA) for age.
P versus control <0.05.
P versus control <0.01.
P versus control ≤0.001.
P deceased versus alive <0.05.
P deceased versus alive <0.01.
P deceased versus alive ≤0.001.
F, female; M, male; BMI, body mass index; URR, urea reduction ratio; PTH, parathyroid hormone; RBC, red blood cell count, MCV, mean corpuscular volume; MCHC, mean corpuscular hemoglobin concentration; WBC, white blood cell count; LPO, lipid-peroxidation; TAS, total antioxidant status; tPA, tissue plasminogen activator; PAI, plasminogen activator inhibitor; IL, interleukin; hsCRP, high-sensitivity C-reactive protein; TNF, tumor necrosis factor; sTNFR, soluble TNF receptor; PTX, pentraxin; TIMP, tissue inhibitor of metalloproteinases; cfDNA, cell-free DNA.
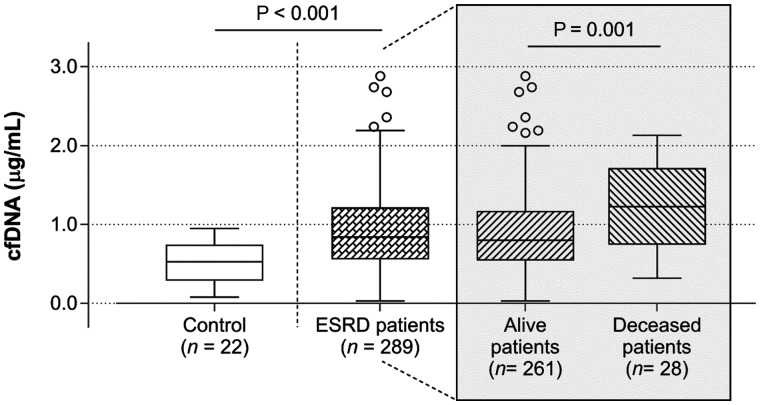
FIGURE 1:
cfDNA levels for controls and ESRD patients under dialysis therapy and for those who remained alive or were deceased during a 1-year following up period.
The mortality rate for ESRD patients was 9.7% and mean time of survival was 199 (range: 77–293) days. The causes of mortality were cardiovascular events (46.4%), infections (10.7%), cachexia (7.1%), miscellaneous (14.3%) or unknown (21.4%).
ESRD patients deceased (n = 28) during the 1-year follow-up, as compared with alive patients (n = 261), presented significantly lower albumin and higher calcium, elastase and inflammatory markers (IL-6, hsCRP, sTNFR2 and PTX3), and were older; the changes in ultrafiltration volume, urea, MCHC, neutrophils and D-dimer lost significance after adjusting for age. cfDNA values were almost 2-fold higher than those presented by alive patients (Figure 1).
We estimated the HR for the parameters that were significantly different between alive and deceased ESRD patients. Both in the unadjusted and the adjusted model for basic confounding factors in dialysis patients (age, dialysis vintage, vascular access, Kt/V, and the comorbidities, diabetes mellitus and history of cardiovascular disease), we found that cfDNA, PTX3, neutrophils, sTNFR2, IL-6, age, elastase, TAS, urea and ultrafiltration volume appeared as independent predictors of all-cause mortality, in decreasing order of HR (Table 2). The cfDNA emerged as the most important determinant of mortality in this cohort: the risk of death increases 2.651-fold in the unadjusted model and 2.146-fold in the adjusted model, for each increment of 1 μg/mL of cfDNA. The patient group was divided into quartiles (Q) by their cfDNA values (n = 72 in each Q1, Q2, Q4; n = 73 in Q3). The numbers of deceased patients for Q1–Q4 were 3 (10.7% of total mortality), 4 (14.3%), 7 (25.0%) and 14 (50.0%), respectively. In other words, patients with cfDNA above the median concentration of 0.84 μg/mL had a 1-year mortality of 15%, while patients with cfDNA below the median had a 1-year mortality of 4.9%. By performing a univariate Cox regression and a multivariate Cox regression (adjusted for age, dialysis vintage, vascular access, Kt/V, and the comorbidities diabetes mellitus and history of cardiovascular disease) for ESRD patients deceased from cardiovascular events, we found that the HR for cfDNA was higher, when compared with the HR of all-cause mortality (Table 2), both in the unadjusted (HR 3.057; P = 0.007) and in the adjusted (HR 2.882; P = 0.007) analysis. The same analysis for patients deceased from non-cardiovascular-related causes, in unadjusted (HR 2.296; P = 0.044) and adjusted (HR 1.771; P = 0.197) analyses, showed loss of cfDNA predictive power.
Table 2.
Univariate and multivariate Cox regression analysis for all-cause mortality in ESRD patients (n = 289)
| Univariate Cox regressiona |
Multivariate Cox regressionb |
|||||||
|---|---|---|---|---|---|---|---|---|
| Parameters | P-value | HR | 95% CI for HR |
P-value | HR | 95% CI for HR |
||
| Lower | Upper | Lower | Upper | |||||
| Age (years) | 0.026 | 1.037 | 1.004 | 1.071 | 0.040 | 1.036 | 1.002 | 1.072 |
| Ultrafiltration volume (L) | 0.035 | 0.613 | 0.389 | 0.967 | 0.040 | 0.606 | 0.375 | 0.977 |
| Urea (mg/dL) | 0.027 | 0.987 | 0.975 | 0.998 | 0.033 | 0.986 | 0.974 | 0.999 |
| MCHC (g/dL) | 0.035 | 0.727 | 0.541 | 0.978 | 0.077 | 0.747 | 0.540 | 1.033 |
| Neutrophils (×109/L) | 0.001 | 1.270 | 1.098 | 1.470 | 0.007 | 1.251 | 1.162 | 1.474 |
| TAS (μM) | 0.019 | 0.997 | 0.994 | 0.999 | 0.049 | 0.997 | 0.994 | 1.000 |
| D-dimer (ng/mL) | 0.434 | 1.000 | 1.000 | 1.000 | 0.927 | 1.000 | 1.000 | 1.000 |
| IL-6 (pg/mL) | 0.005 | 1.032 | 1.009 | 1.055 | 0.003 | 1.038 | 1.012 | 1.064 |
| hsCRP (mg/dL) | 0.085 | 1.200 | 0.975 | 1.476 | 0.100 | 1.210 | 0.946 | 1.518 |
| sTNFR2 (ng/mL) | 0.003 | 1.077 | 1.026 | 1.132 | 0.008 | 1.075 | 1.019 | 1.134 |
| TNF-α/sTNFR2 (×10−3) | 0.116 | 0.000 | 0.000 | – | 0.160 | 0.000 | 0.000 | – |
| PTX3 (ng/mL) | 0.000 | 1.487 | 1.228 | 1.801 | 0.000 | 1.488 | 1.198 | 1.848 |
| Elastase (ng/mL) | 0.000 | 1.002 | 1.001 | 1.001 | 0.000 | 1.003 | 1.002 | 1.004 |
| cfDNA (μg/mL) | 0.001 | 2.651 | 1.486 | 4.769 | 0.013 | 2.146 | 1.177 | 3.912 |
Unadjusted HRs for each variable.
HR for each variable adjusted for age, dialysis vintage, vascular access, Kt/V, and the comorbidities, diabetes mellitus and history of cardiovascular disease (multivariate analysis of age as an independent covariate was adjusted for dialysis, vascular access, Kt/V, and the comorbidities diabetes mellitus and history of cardiovascular disease).
CI, confidence interval;
MCHC, mean corpuscular hemoglobin concentration; TAS, total antioxidant status; IL, interleukin; hsCRP, high sensitivity C-reactive protein; sTNFR, soluble tumor necrosis factor receptor; PTX, pentraxin; cfDNA, cell-free DNA.
HR values that present p values < 0.05 (i.e. significant predictive value) are highlighted in bold.
Our data also showed that cfDNA correlated positively with age (rS= 0.144; P = 0.015), WBC (rS= 0.211; P < 0.001), neutrophils (rS= 0.225; P < 0.001), monocytes (rS= 0.167; P = 0.004), TIMP-1 (rS= 0.186; P = 0.001), elastase (rS= 0.330; P < 0.001), IL-6 (rS= 0.435; P < 0.001), hsCRP (rS= 0.440; P < 0.001), sTNFR2 (rS= 0.228; P < 0.001), LPO (rS= 0.180; P = 0.002), tPA (rS= 0.264; P < 0.001), PAI-1 (rS= 0.156; P = 0.008) and D-dimer (rS= 0.235; P < 0.001). cfDNA correlated negatively with TAS/LPO (rS= −0.207; P < 0.001), iron (rS= −0.155; P = 0.008) and urea reduction ratio (URR; rS= −0.143; P = 0.015).
DISCUSSION
In line with previous studies, we found that ESRD patients on dialysis treatment presented increased cfDNA levels, inflammation, oxidative stress, hypercoagulable state and anaemia, which are apparently due to a functional iron deficiency [1, 4–6, 26].
In ESRD patients on dialysis, the uraemic milieu and the dialysis procedure appear to lead to activation of inflammatory cells, creating a pro-inflammatory milieu [16, 27–30]. We found that cfDNA correlated positively with leucocyte, neutrophil and monocyte counts, as well as with their activation/inhibitors products, elastase and TIMP-1, strengthening the interplay between inflammatory cells and cfDNA. Our data also strengthen the linkage between pro-inflammatory cytokines/receptors with cfDNA, as showed by the positive correlations of cfDNA with IL-6, CRP and sTNFR2. cfDNA released by apoptotic leucocytes may selectively induce activation of monocytes, with release of different pro-inflammatory products, such as IL-6 [16], which is able to stimulate hepatic production of CRP and hepcidin, contributing to the enhancement of inflammation and to disturbances in iron metabolism. In fact, cfDNA correlated inversely with serum iron and positively with IL-6. The enhanced hepcidin production, by triggering endocytosis and proteolysis of ferroportin on membrane surface of duodenal enterocytes, iron-recycling macrophages and of iron-storing hepatocytes, inhibits iron absorption and mobilization [31], contributing to the anaemia observed in dialysis patients.
High blood levels of sTNFRs have been associated with kidney dysfunction in elderly [32], to worsening of renal dysfunction in diabetic CKD patients [33, 34] and to an increase in all-cause mortality and in the risk for cardiovascular events in advanced CKD [35]. In accordance with the literature, we found an enhancement in TNF-system activity (TNF-α and sTNFR2) and a positive correlation of sTNFR2 with cfDNA.
It has been reported that cfDNA correlates positively with aging and with age-associated inflammation [36]. We also found that cfDNA correlated positively with both age and inflammatory markers, although it appears to be mostly associated with the inflammatory milieu.
The ESRD patients on dialysis presented a hypercoagulable state and altered fibrinolysis, as shown by a decrease in PAI-1 and an increase in D-dimer and tPA/PAI-1 ratio. The cfDNA may induce coagulofibrinolytic changes by activating the intrinsic coagulation pathway [37]; in fibrinolysis, cfDNA appears to have a dual opposite role—it enhances plasminogen activation by tPA, potentiating fibrinolysis, and concomitantly, attenuates fibrinolysis by triggering the inactivation of tPA by PAI-1 [38]. In sepsis, cfDNA modulates clot structure and impairs fibrinolysis by inhibiting plasmin-mediated fibrin degradation [39]. cfDNA correlated positively with tPA, PAI-1 and D-dimer, probably as a result of opposing roles in fibrinolysis; thus, our data strengthen the association of cfDNA with coagulofibrinolytic disturbances in ESRD patients on dialysis.
Like lipids and proteins, DNA is susceptible to oxidative peroxidation by reactive oxygen species released by activated inflammatory cells [40]. We found a positive correlation of cfDNA with LPO, and a negative correlation with TAS/LPO, strengthening the association of cfDNA with a pro-oxidative environment in ESRD. In accordance with other reports [41–43], we found increased TAS levels, probably resulting from a continuous antioxidant upregulation to overwhelm the development of oxidative stress in these patients on dialysis. To face the development/enhancing of oxidative stress and other associated complications, complex B vitamins (including folic acid) are routinely administered to these patients after each dialysis session, which may also contribute to this increase. Actually, folate supplementation seems to improve serum total antioxidant capacity [44]. Furthermore, uric acid is one of the antioxidants evaluated by FRAP assay and, therefore, may explain the increase in TAS, as uric acid is known to increase in pre-haemodialysis CKD patients. We must keep in mind that we collected blood samples immediately before dialysis session, and therefore many compounds with antioxidant capacity (such as uric acid) are present in high concentrations. Actually, it was reported that FRAP decreases after haemodialysis [42].
cfDNA correlated inversely with URR, a measure of dialysis adequacy, suggesting that a decrease in removal of waste products, like urea, creates a favourable milieu for DNA damage.
To evaluate the predictive value of the biomarkers under study, for the outcome of patients, we performed a 1-year follow-up study recording events and causes of deaths.
Deceased ESRD patients were older and presented higher levels of cfDNA, inflammatory markers (IL-6, CRP, PTX3 and elastase) and reduced antioxidant defences (TAS), as compared with alive patients.
From the evaluation of HRs for the parameters that were significantly different in deceased patients, when compared with alive patients, with and without adjustment for confounding factors (age, dialysis vintage, vascular access, Kt/V, and the comorbidities, diabetes mellitus and history of cardiovascular disease), cfDNA emerged as the best independent predictor for all-cause mortality. Mortality risk increases 2.651-fold in the unadjusted model and 2.146-fold in the adjusted model, for each increase of 1 μg/mL in cfDNA levels. The distribution of deceased patients per quartile of cfDNA was expressively different; the number of deceased patients for Q1–Q4 was 3 (10.7% of total mortality), 4 (14.3%), 7 (25.0%) and 14 (50.0%), respectively. Twenty-one out of the 28 deceased patients were in Q3 and Q4 of cfDNA values, presenting a cfDNA >0.84 μg/mL.
Furthermore, the evaluation of HR in patients deceased from cardiovascular-related causes, unadjusted and adjusted for confounding factors, showed that the HR for cfDNA was even higher than the value obtained in the analysis for all-cause mortality (Table 2), both in unadjusted (HR 3.057; P = 0.007) and in adjusted (HR 2.882; P = 0.007) analysis. Conversely, the same analysis, for patients deceased from non-cardiovascular disease-related causes, both in unadjusted (HR 2.296; P = 0.044) and adjusted (HR 1.771; P = 0.197) analyses, showed a clear loss of cfDNA predictive power. This suggests that cfDNA is more valuable as an independent predictor of cardiovascular mortality, further strengthening the link between the levels of cfDNA and vascular inflammation.
Our data strongly suggest the value of circulating cfDNA as an accurate predictor of mortality in ESRD patients, validating data obtained in smaller cohorts [7, 18]. It might be also a good marker for the outcome of ESRD patients with other inflammatory-associated diseases. A biological role was also proposed for cfDNA in tumour progression by oncogenesis of host cells [45]. In addition, markers of NETosis, like cfDNA and DNA–histone complexes, were reported as important factors for major adverse cardiovascular events in haemodialysis patients [11, 12].
With an HR >1.488, the inflammatory marker PTX3 appeared as the second most prominent independent predictor of mortality risk. We recently reported that PTX3 can be used as broad inflammatory biomarker, presenting a close association with inflammation, malnutrition, cardiovascular disease and renal fibrosis, and, in accordance with the present work, a great potential to predict mortality risk in dialysis patients [46].
The complexity of comorbid conditions and multidrug therapy (e.g. statins, diuretics, antihypertensives, erythropoiesis-stimulating agents, iron) represents a limitation when interpreting data from ESRD patients. The follow-up period was relatively short, which may limit mortality analysis, due to the sample size analysed. Being a cross-sectional study, we were not able to study the temporal changes of cfDNA and its correlation with morbidity and mortality. Nonetheless, it involved a large population (and adjustment for confounding factors was performed), which allows more accurate associations to be established and includes several outcomes that allow reinforcing conclusions.
Our study suggests that cfDNA can be predictive of prognosis in ESRD patients on dialysis, with increased cfDNA levels being an indication of a poor prognosis.
FUNDING
The work was supported by UIDB/04378/2020 with funding from FCT/MCTES through national funds, by North Portugal Regional Coordination and Development Commission (CCDR-N)/NORTE2020/Portugal 2020 (Norte-01-0145-FEDER-000024) and by REQUIMTE-Rede de Química e Tecnologia-Associação in the form of a researcher (S.R.)—project Dial4Life co-financed by FCT/MCTES (PTDC/MEC-CAR/31322/2017) and FEDER/COMPETE 2020 (POCI-01-0145-FEDER-031322).
CONFLICT OF INTEREST STATEMENT
None declared. Data in the manuscript are original, and this manuscript has not been previously published and is not under consideration for publication elsewhere.
REFERENCES
- 1. Amdur RL, Feldman HI, Gupta J. et al.; the CRIC Study Investigators. Inflammation and progression of CKD: the CRIC study. Clin J Am Soc Nephrol 2016; 11: 1546–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Recio-Mayoral A, Banerjee D, Streather C. et al. Endothelial dysfunction, inflammation and atherosclerosis in chronic kidney disease–a cross-sectional study of predialysis, dialysis and kidney-transplantation patients. Atherosclerosis 2011; 216: 446–451 [DOI] [PubMed] [Google Scholar]
- 3. Ioannou K, Stel VS, Dounousi E. et al. Inflammation, endothelial dysfunction and increased left ventricular mass in chronic kidney disease (CKD) patients: a longitudinal study. PLoS One 2015; 10: e0138461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adams MJ, Irish AB, Watts GF. et al. Hypercoagulability in chronic kidney disease is associated with coagulation activation but not endothelial function. Thromb Res 2008; 123: 374–380 [DOI] [PubMed] [Google Scholar]
- 5. Huang MJ, Wei RB, Wang Y. et al. Blood coagulation system in patients with chronic kidney disease: a prospective observational study. BMJ Open 2017; 7: e014294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wagner M, Ashby DR, Kurtz C. et al. Hepcidin-25 in diabetic chronic kidney disease is predictive for mortality and progression to end stage renal disease. PLoS One 2015; 10: e0123072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coimbra S, Santos-Silva A, Costa E. et al. DNA damage in end-stage renal disease patients. assessment by in vitro comet assay and by cell-free DNA quantification. In: Larramendy ML, Soloneski S (eds). Genotoxicity: A Predictable Risk to Our Actual World. London: IntechOpen, ; 2017, 19–41 [Google Scholar]
- 8. Kohlova M, Ribeiro S, do Sameiro-Faria M. et al. Circulating cell-free DNA levels in hemodialysis patients and its association with inflammation, iron metabolism, and rhEPO doses. Hemodial Int 2013; 17: 664–667 [DOI] [PubMed] [Google Scholar]
- 9. Lichtenstein AV, Melkonyan HS, Tomei LD. et al. Circulating nucleic acids and apoptosis. Ann N Y Acad Sci 2006; 945: 239–249 [DOI] [PubMed] [Google Scholar]
- 10. Frank MO. Circulating cell-free DNA differentiates severity of inflammation. Biol Res Nurs 2016; 18: 477–488 [DOI] [PubMed] [Google Scholar]
- 11. Korabecna M, Tesar V.. NETosis provides the link between activation of neutrophils on hemodialysis membrane and comorbidities in dialyzed patients. Inflamm Res 2017; 66: 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jeong JC, Kim J-E, Gu J-Y. et al. Significance of the DNA-histone complex level as a predictor of major adverse cardiovascular events in hemodialysis patients: the effect of uremic toxin on DNA-histone complex formation. Blood Purif 2016; 41: 64–71 [DOI] [PubMed] [Google Scholar]
- 13. Aarthy R, Mani S, Velusami S. et al. Role of circulating cell-free DNA in cancers. Mol Diagn Ther 2015; 19: 339–350 [DOI] [PubMed] [Google Scholar]
- 14. Coimbra S, Catarino C, Costa E. et al. Circulating cell-free DNA levels in Portuguese patients with psoriasis vulgaris according to severity and therapy. Br J Dermatol 2014; 170: 939–942 [DOI] [PubMed] [Google Scholar]
- 15. El Tarhouny SA, Hadhoud KM, Ebrahem MM. et al. Assessment of cell-free DNA with microvascular complication of type II diabetes mellitus, using PCR and ELISA. Nucleosides Nucleotides Nucleic Acids 2010; 29: 228–236 [DOI] [PubMed] [Google Scholar]
- 16. Atamaniuk J, Kopecky C, Skoupy S. et al. Apoptotic cell-free DNA promotes inflammation in haemodialysis patients. Nephrol Dial Transplant 2012; 27: 902–905 [DOI] [PubMed] [Google Scholar]
- 17. van der Meer AJ, Kroeze A, Hoogendijk AJ. et al. Systemic inflammation induces release of cell-free DNA from hematopoietic and parenchymal cells in mice and humans. Blood Adv 2019; 3: 724–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tovbin D, Novack V, Wiessman MP. et al. Circulating cell-free DNA in hemodialysis patients predicts mortality. Nephrol Dial Transplant 2012; 27: 3929–3935 [DOI] [PubMed] [Google Scholar]
- 19. Schupp N, Stopper H, Heidland A.. DNA damage in chronic kidney disease: evaluation of clinical biomarkers. Oxid Med Cell Longev 2016; 2016: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benzie IF, Strain JJ.. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: the FRAP assay. Anal Biochem 1996; 239: 70–76 [DOI] [PubMed] [Google Scholar]
- 21. Ruskovska T, Jansen EHJM, Antarorov R.. Evaluation of assays for measurement of serum (anti)oxidants in hemodialysis patients. Biomed Res Int 2014; 2014: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jansen EH, Ruskovska T.. Comparative analysis of serum (anti)oxidative status parsmall a, cyrillicmeters in healthy persons. Int J Mol Sci 2013; 14: 6106–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mihara M, Uchiyama M.. Determination of malonaldehyde precursor in tissues by thiobarbituric acid test. Anal Biochem 1978; 86: 271–278 [DOI] [PubMed] [Google Scholar]
- 24. Goldshtein H, Hausmann MJ, Douvdevani A.. A rapid direct fluorescent assay for cell-free DNA quantification in biological fluids. Ann Clin Biochem 2009; 46: 488–494 [DOI] [PubMed] [Google Scholar]
- 25. Templeton GF. A two-step approach for transforming continuous variables to normal: implications and recommendations for IS research. Commun Assoc Inf Syst 2011; 28: 41–58 [Google Scholar]
- 26. Locatelli F, Canaud B, Eckardt KU. et al. Oxidative stress in end-stage renal disease: an emerging threat to patient outcome. Nephrol Dial Transplant 2003; 18: 1272–1280 [DOI] [PubMed] [Google Scholar]
- 27. Andrikos E, Buoncristiani E, D’Intini V. et al. Effect of daily hemodialysis on monocytes apoptosis. Blood Purif 2005; 23: 79–82 [DOI] [PubMed] [Google Scholar]
- 28. Cendoroglo M, Jaber BL, Balakrishnan VS. et al. Neutrophil apoptosis and dysfunction in uremia. J Am Soc Nephrol 1999; 10: 93–100 [DOI] [PubMed] [Google Scholar]
- 29. Koller H, Hochegger K, Zlabinger GJ. et al. Apoptosis of human polymorphonuclear neutrophils accelerated by dialysis membranes via the activation of the complement system. Nephrol Dial Transplant 2004; 19: 3104–3111 [DOI] [PubMed] [Google Scholar]
- 30. Opatrna S, Wirth J, Korabecna M. et al. Cell-free plasma DNA during hemodialysis. Ren Fail 2009; 31: 475–480 [DOI] [PubMed] [Google Scholar]
- 31. Coimbra S, Catarino C, Santos-Silva A.. The role of adipocytes in the modulation of iron metabolism in obesity. Obes Rev 2013; 14: 771–779 [DOI] [PubMed] [Google Scholar]
- 32. Carlsson AC, Larsson TE, Helmersson-Karlqvist J. et al. Soluble TNF receptors and kidney dysfunction in the elderly. J Am Soc Nephrol 2014; 25: 1313–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lopes-Virella MF, Baker NL, Hunt KJ . et al. the DCCT/EDIC Research Group. Baseline markers of inflammation are associated with progression to macroalbuminuria in type 1 diabetic subjects. Diabetes Care 2013; 36: 2317–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carlsson AC, Carrero J-JS, Stenvinkel P. et al. High levels of soluble tumor necrosis factor receptors 1 and 2 and their association with mortality in patients undergoing hemodialysis. Cardiorenal Med 2015; 5: 89–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Neirynck N, Glorieux G, Schepers E. et al. Soluble tumor necrosis factor receptor 1 and 2 predict outcomes in advanced chronic kidney disease: a prospective cohort study. PLoS One 2015; 10: e0122073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jylhava J, Nevalainen T, Marttila S. et al. Characterization of the role of distinct plasma cell-free DNA species in age-associated inflammation and frailty. Aging Cell 2013; 12: 388–397 [DOI] [PubMed] [Google Scholar]
- 37. Gould TJ, Vu TT, Swystun LL. et al. Neutrophil extracellular traps promote thrombin generation through platelet-dependent and platelet-independent mechanisms. Arterioscler Thromb Vasc Biol 2014; 34: 1977–1984 [DOI] [PubMed] [Google Scholar]
- 38. Komissarov AA, Florova G, Idell S.. Effects of extracellular DNA on plasminogen activation and fibrinolysis. J Biol Chem 2011; 286: 41949–41962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gould TJ, Vu TT, Stafford AR. et al. Cell-free DNA modulates clot structure and impairs fibrinolysis in sepsis. Arterioscler Thromb Vasc Biol 2015; 35: 2544–2553 [DOI] [PubMed] [Google Scholar]
- 40. Corredor Z, Stoyanova E, Rodriguez-Ribera L. et al. Genomic damage as a biomarker of chronic kidney disease status. Environ Mol Mutagen 2015; 56: 301–312 [DOI] [PubMed] [Google Scholar]
- 41. Maciejczyk M, Szulimowska J, Taranta-Janusz K. et al. Salivary FRAP as a marker of chronic kidney disease progression in children. Antioxidants 2019; 8: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sangeetha Lakshmi B, Harini Devi N, Suchitra MM. et al. Changes in the inflammatory and oxidative stress markers during a single hemodialysis session in patients with chronic kidney disease. Ren Fail 2018; 40: 534–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sridhar A, Srinivasa Rao P, Sivakumar V. et al. Study of oxidant and anti-oxidant status in patients with chronic kidney disease. J Clin Sci Res 2018; 7: 124–130 [Google Scholar]
- 44. Aghamohammadi V, Gargari BP, Aliasgharzadeh A.. Effect of folic acid supplementation on homocysteine, serum total antioxidant capacity, and malondialdehyde in patients with type 2 diabetes mellitus. J Am Coll Nutr 2011; 30: 210–215 [DOI] [PubMed] [Google Scholar]
- 45. Garcia-Olmo DC, Garcia-Olmo D.. Biological role of cell-free nucleic acids in cancer: the theory of genometastasis. Crit Rev Oncog 2013; 18: 153–161 [DOI] [PubMed] [Google Scholar]
- 46. Valente MJ, Rocha S, Coimbra S. et al. Long pentraxin 3 as a broader biomarker for multiple risk factors in end-stage renal disease: association with all-cause mortality. Mediators Inflamm 2019; 2019: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]