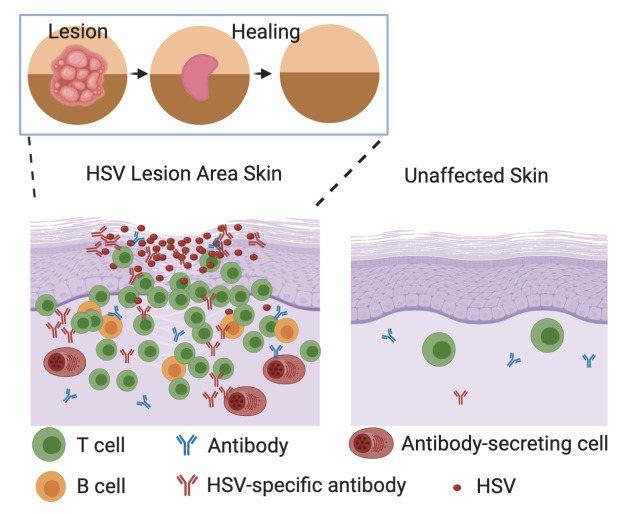
Abstract
Tissue-based T cells are important effectors in the prevention and control of mucosal viral infections; less is known about tissue-based B cells. We demonstrate that B cells and antibody-secreting cells (ASCs) are present in inflammatory infiltrates in skin biopsy specimens from study participants during symptomatic herpes simplex virus 2 (HSV-2) reactivation and early healing. Both CD20+ B cells, most of which are antigen inexperienced based on their coexpression of IgD, and ASCs — characterized by dense IgG RNA expression in combination with CD138, IRF4, and Blimp-1 RNA — were found to colocalize with T cells. ASCs clustered with CD4+ T cells, suggesting the potential for crosstalk. HSV-2–specific antibodies to virus surface antigens were also present in tissue and increased in concentration during HSV-2 reactivation and healing, unlike in serum, where concentrations remained static over time. B cells, ASCs, and HSV-specific antibody were rarely detected in biopsies of unaffected skin. Evaluation of samples from serial biopsies demonstrated that B cells and ASCs followed a more migratory than resident pattern of infiltration in HSV-affected genital skin, in contrast to T cells. Together, these observations suggest the presence of distinct phenotypes of B cells in HSV-affected tissue; dissecting their role in reactivation may reveal new therapeutic avenues to control these infections.
Keywords: Immunology, Infectious disease
Keywords: Adaptive immunity, Beta cells, Immunoglobulins
Introduction
In immunocompetent hosts, after initial exposure at the site of non-intact epithelialized skin or mucosa, herpes simplex virus 2 (HSV-2) establishes chronic latent infection of the associated dorsal root ganglion (1, 2). The spectrum of clinical disease among infected individuals is broad; some experience asymptomatic seroconversion, while others have 10 or more recurrences a year. While the mechanisms underlying viral reactivation and symptomatic disease are not known, the spectrum of clinical manifestations at the site of reactivation is thought to depend primarily on the host adaptive immune response, rather than intrinsic viral factors (3, 4). As suggested by human-based studies and supported by mathematical modeling and animal studies, cell-mediated immunity, by populations including tissue-resident memory CD8+ and infiltrating or resident memory CD4+ T cells, is an important factor in local viral control (5–10).
The role of humoral immunity in the human response to HSV-2 reactivation is less clear. Robust titers of binding and neutralizing antibodies are commonly detected in the blood but are not associated with the frequency of HSV-2 symptoms in natural infection (11, 12) or after vaccination (13–15). Vaccine-induced antibody to gD was not protective against primary infection in HSV-2–seronegative individuals (16). One proposed theory is that a dominant antigen, gD, leads to the production of antibodies that are binding and neutralizing but not protective (17). Other theories are that certain HSV-specific antibodies may function protectively in tissue near the site of viral release, but circulating antibody levels do not reflect this tissue-based event (18); and that cell-to-cell spread of virus is sufficient for propagation and development of symptomatic reactivation even in the presence of effective neutralizing antibodies, which are at a concentration below the threshold of efficacy (19).
Some evidence from animal models suggests that B cells are important for protection from HSV-2. B cell–deficient mice rapidly develop severe disease after HSV-2 genital challenge (6, 20), and this effect is limited if B cells are depleted after immunity has developed (21), suggesting that at least part of the role of B cells is in the primary immune response. Whether antiviral activity is related to systemic or local secretion of antibody is an area of active investigation. In a guinea pig HSV-2 infection model where recurrences have been observed, antibody-secreting cells (ASCs) have been seen only in secondary lymphoid and neuronal tissues (22, 23), and systemically infused antibody rescued control of HSV-2 shedding in B cell–depleted animals (21). In HSV-2–exposed mice, however, B cells (both CD20+ and CD138+IgG+) infiltrate into the vaginal mucosa in response to immunization and rechallenge with latency-deficient HSV-2. These B cells are associated with intraluminal vaginal secretion of HSV-specific IgG2 and IgA during viral challenge, but not at other times (24). In concert with the discovery of memory lymphoid clusters in mice infected with HSV-2, these recent observations have raised the possibility that B cells and ASCs may function outside their previously understood role in lymph tissue. In humans, the role of B cells during HSV-2 infection has not to our knowledge been studied. Single-cell dissociation studies have identified B cells in normal skin, but they are rare (25, 26). ASCs have been identified in tissue, extrinsic to blood vessels, in human foreskin (27). B cells have been observed, albeit infrequently, in inflammatory infiltrates related to HSV (28), and their presence in the skin has been confirmed by single-cell and RNA-Seq technology (26, 29, 30). Whether B cells and ASCs are actively involved during the immune response to HSV-2 reactivation in humans in nonmucosal skin, therefore, is unknown.
We utilized our repository of sequential genital skin biopsy specimens from persons with HSV-2 to explore whether B cells and ASCs are present during HSV-2 reactivation and clinical quiescence. HSV-2–specific antibody was present in tissue, and the concentration varied over time, suggesting the possibility of local antibody production. In a time of rapidly expanding information from single-cell dissociation studies, this histologic information confirms the importance of spatially informative methods, and suggests that further investigation into the role of B cells and ASCs in local inflammatory processes is warranted.
Results
Of 16 participants, 14 were women and 2 were men; the median age was 50 years (range 18–66). All 16 were seropositive for HSV-2, and 6 also had antibody to HSV-1. The most common site for biopsies was the buttock (n = 10; Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI142088DS1). Biopsies were performed when a clinically evident HSV-2 lesion was present and during healing, for a total of 5 or 6 biopsies per person. A control site biopsy of uninvolved skin was performed at the time of the lesion or 8 weeks after healing on either the arm (n = 7) or the contralateral genital skin (n = 9). There were no complications associated with serial biopsies (5, 8).
Histology of B cells in tissue by immunofluorescence.
B cells were identified by dual immunofluorescence (IF) staining for CD20 and CD79b in 47 of 65 punch biopsy specimens of active and early healing HSV-2 lesions from 11 participants from whom complete sets of 5 or 6 biopsy specimens taken from the period of active lesion to 8 weeks after healing were available. Per person, a median of 5 of 6 samples were found to have B cells (range 2–6); in most participants, B cells were present in all of the specimens from the HSV-2 lesion site. In 3 individuals, 10 or fewer B cells were seen in any of the samples, but CD20+CD79b+ B cells were seen in at least 2 of the specimens from each participant. CD20 and CD79b were chosen for IF due to their relative abundance on the B cell surface (CD20 is expressed at 6-fold-higher density than CD19; ref. 31) and consistent performance in this assay. To confirm the identity of these cells and minimize the risk of nonspecific antibody binding, we initially employed dual staining (Figure 1A). After quantification and confirmation of CD20 reliability by dual staining, CD20 was used independently to identify B cells in tissue biopsy specimens. The number of CD20+CD79b+ cells present in each biopsy section varied by person and time point. In general, B cells were more common during the lesion and early healing periods. They were infrequent in specimens taken during clinical quiescence (present in 3 of 11 specimens taken 8 weeks after healing) and were detected in only 3 of 16 control site samples from either uninvolved genital skin or arm (Figure 1A).
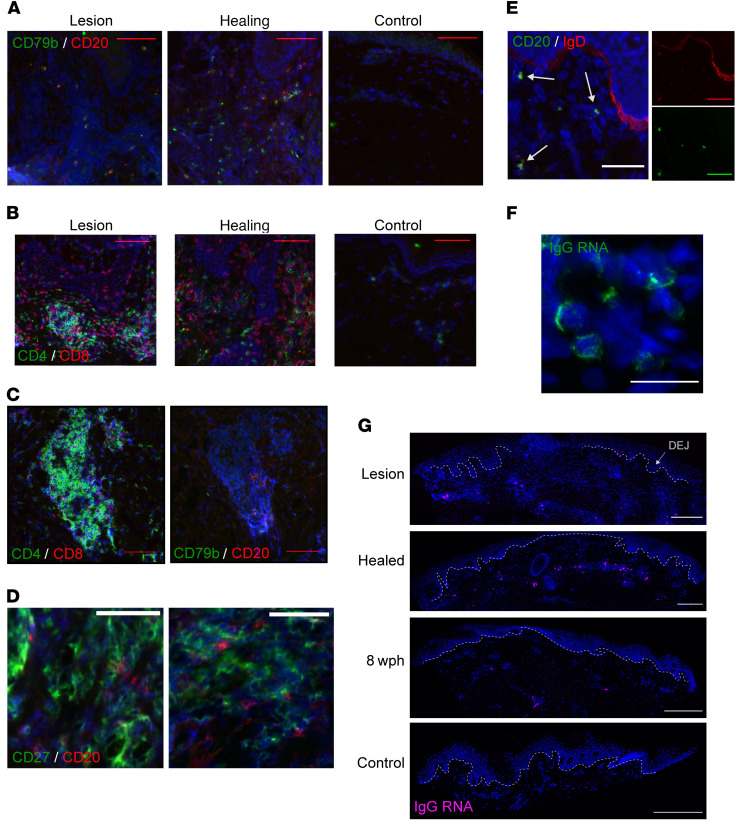
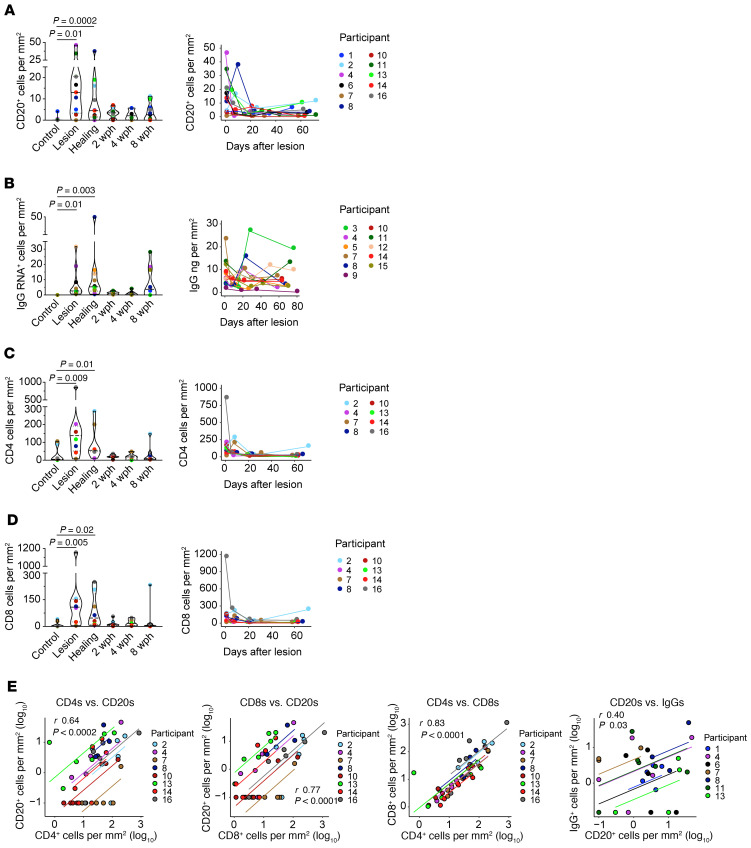
Figure 1. Antigen-naive CD20+ B cells and IgG RNA+ cells infiltrate lesion-area skin but not uninvolved skin during HSV-2 reactivation.
(A) B cells: CD20 (red) and CD79b (green) in serial genital skin biopsy samples taken during and after HSV-2 lesion from a single participant (results shown for 1 of 10 patients). No B cells were detected in the control sample. (B) T cells: CD4 (green) and CD8 (red) in serial specimens taken during and after HSV-2 reactivation (result shown for 1 of 8 patients). Scale bars: 100 μm. (C) Densely clustered CD4+CD8+ T cells (left) in this participant were accompanied by CD20+CD79b+ B cells (right); scale bars: 50 μm. (D) CD27 (green) and CD20 (red) in a lesion sample with no evidence of costaining (result shown for 1 of 6 patients); scale bars: 50 μm. (E) In a healing-skin specimen, B cells express CD20 (green) and IgD (red) (result shown for 1 of 5 patients). Arrows indicate sites of coexpression. Inset shows red and green channels separately. Scale bars: 50 μm. (F) IgG-producing cells are loaded with IgG RNA by FISH. IgG mRNA (green) in a healing specimen of HSV-2 reactivation (result shown for 1 of 9 patients). Original magnification, 40× by oil immersion. Scale bar: 25 μm. (G) IgG+ cells by FISH over time, showing distribution within the upper dermis and lymphocyte clusters. In this participant, the control specimen (from the arm) had no IgG+ cells. Dashed line indicates the DEJ. Images obtained at 20×; scale bars: 250 μm. Brightness in G was increased for consistency. wph, weeks after healing.
Histologically, B cells were identified by CD20 IF in the upper dermis (typically within 500 μm of the surface epithelium) and were not detected within the dermal-epidermal junction (DEJ; Figure 1A), where CD8+ T cells are often found (Figure 1B and ref. 5), or the multiple layers of the epidermis, consistent with histology in mice (32). CD20+CD79b+ cells were most often observed to be distributed within areas of immune cell infiltration (as in the healing biopsy in Figure 1A) but were identified with T cells in dense groups of lymphocytes by staining of serial sections (Figure 1C), suggesting in vivo interactions between the 2 subpopulations of cells. T cells are often observed to be present near hair follicles or skin-localized glandular structures, and this was likewise occasionally observed of B cells. CD27, a canonical marker of memory B cells, is more strongly expressed by activated T cells and thus is abundant in skin biopsy samples taken during HSV-2 reactivation and healing. Costaining of this marker may therefore be unreliable in samples with robust T cell infiltration, but in B cell–containing samples from 5 individuals, little to no overlap in staining of CD20 and CD27 was observed in B cells, suggesting that the majority of observed CD20+ cells were not memory B cells (Figure 1D).
To determine whether these CD20+CD27– cells could instead represent a naive or antigen-unexperienced population, we performed IgD costaining by IF in specimens from 5 participants. CD20+IgD+ cells were seen in all 5 individuals (representative image with dual-color images shown in Figure 1E). In 1 participant, nearly all CD20+ cells were also IgD+; however, in others there was more variability, and some CD20+ cells were observed to lack IgD+ costaining. We suspect that the majority of CD20+ cells observed were indeed antigen unexperienced, and there may also have been smaller populations of B cell of other phenotypes present.
To investigate the presence and distribution of ASCs, we performed FISH directed at IgG RNA (pooled IGHG1–4) to identify cells with dense expression of immunoglobulin transcripts. These cells were quantified in 57 specimens from 10 individuals. Both class-switched B cells and ASCs contain IgG mRNA; however, ASCs are known to express up to 1000-fold more immunoglobulin mRNA per cell than B cells (33, 34). High-intensity, cytoplasmic, punctate fluorescence indicative of ASCs was observed in 48 of the 57 tested specimens (Figure 1, F and G). Histologically, the distribution patterns of these IgG RNA+ cells followed those described above for CD20+ B cells. No IgG RNA+ cells were observed within the epithelial cell layers or in the DEJ in any sample. All IgG RNA+ cells observed were present in the dermis. No IgG RNA+ cells were identified by FISH in any of the control specimens.
To further confirm the phenotype of ASCs in our tissue samples, we evaluated the presence of the transcription factors Blimp-1 and IRF4 (35–37). Blimp-1 is encoded by the PRDM1 gene. Evidence of both PRDM1 and IgG RNA was detected in most but not all IgG RNA+ cells (Figure 2A). Further confirmation of the presence of ASCs was pursued by dual FISH with expression of the transcription factor IRF4, which is upregulated in ASCs by 5- to 10-fold compared with naive and memory B cells (38) and also induces Blimp-1 expression (39). Nuclear IRF4 production was frequently observed to be colocalized with IgG RNA transcripts by confocal microscopy (Figure 2B and Supplemental Figure 3). Taken together, these findings strongly indicate the presence of IgG-producing ASCs during HSV-2 reactivation and skin healing. To further confirm this phenotype, we also studied coexpression of MS4A1 (which encodes CD20) and SDC1 (syndecan-1, or CD138). SDC1 is a canonical marker in flow cytometry and histology of ASCs (40, 41); but it is also highly expressed on the surface of epithelial cells including keratinocytes, and less abundant on other types of skin cells including endothelial cells and fibroblasts (42). Transcripts of MS4A1 were visualized both with and without expression of IgG RNA, though more often these were seen independently (Figure 2C), and SDC1 signal was observed in some but not all cells expressing a high level of IgG RNA transcripts. Due to the abundance of SDC1 in skin, deconvolution microscopy was used to confirm that SDC1 and IgG RNA signals were concomitantly present in single cells. Figure 2D shows a single Z-stack image from these results, confirming that both SDC1 and IgG RNA signals were coming from the same cells. These constellations of findings provide evidence that ASCs are selectively increased in tissue during the time of lesion and early healing periods of genital HSV reactivation. They also show that ASCs of various stages of differentiation are present. The correlation between CD20+ and IgG+ cells by repeated-measure analysis was approximately 0.4 (P = 0.03; see below).
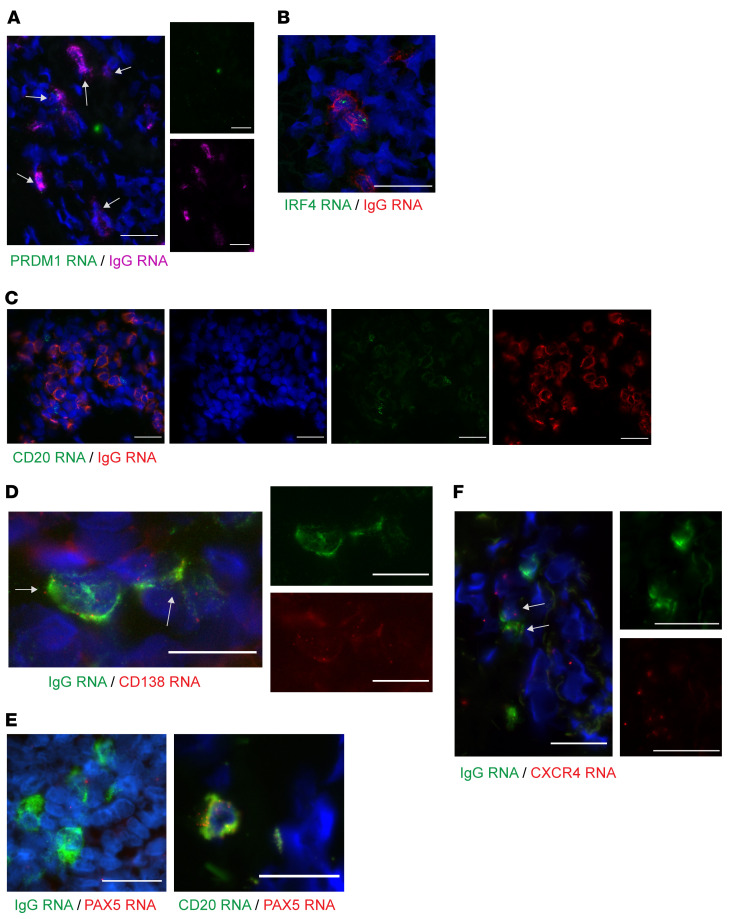
Figure 2. Cells expressing a high level of IgG RNA infiltrate in genital skin during the tissue-based immune response to HSV-2 reactivation.
(A) A healing lesion shows PRDM1 (Blimp-1) expression in many IgG RNA+ cells (40× immersion lens; scale bars: 25 μm). Result shown for 1 of 5 participants. Brightness increased for visualization. (B) IgG-producing cells, by FISH. IgG mRNA (red) and IRF4 (green) in a biopsy specimen taken during healing after HSV-2 reactivation. Note 2 cells with lower amplitude of IgG that are IRF4–. Image is 60× by confocal microscopy. Scale bar: 25 μm; result shown for 1 of 7 participants. (C) IgG RNA (red) and CD20 RNA (green) in adjacent cells without coexpression. Image is 40× by oil immersion microscopy; scale bars: 25 μm. Result shown for 1 of 5 participants. (D) IgG (green) and CD138 (red) coexpression by deconvolution microscopy (60×, single Z-stack image shown). Eight of 20 IgG RNA+ cells in this image also express CD138. Insets are single-color images of center cells. Result shown for 1 of 4 participants. (E) PAX5 RNA (red) and highly expressed IgG RNA (green) were not observed to colocalize, whereas PAX5 RNA and CD20 RNA were. Image is 40× by oil immersion microscopy. Scale bars: 25 μm; result shown for 1 of 5 participants. (F) In an active-lesion specimen, 2 cells show coexpression of CXCR4 and IgG (40× immersion lens; scale bars: 25 μm). Result shown for 1 of 3 persons. Arrows indicate cells with coexpression.
To investigate whether we could use FISH to identify rare memory B cells, we chose PAX5, or B cell–specific activator protein, a transcription factor that is expressed in naive and memory lineage B cells but is downregulated by 4- to 5-fold in ASCs (38). Colocalization of PAX5 at a high level with IgG RNA at a low level would indicate a memory or antigen-experienced B cell. In genital skin biopsy samples from 5 individuals, we did not observe robust PAX5 colocalization with IgG (Figure 2E). The overall frequency of PAX5 detection was low; in 4 participants studied, we did not observe any cells with dense PAX5/IgG RNA coexpression, and PAX5 signal was seen in cells not identified to have IgG transcription (potentially naive B cells). This was confirmed by consistently observed PAX5 costaining with MS4A1 in 2 individuals; all MS4A1+ cells also expressed PAX5 (Figure 2E). The finding of minimal or no PAX5 expression in cells containing a high level of IgG RNA transcripts supports that these cells were indeed ASCs.
While there are no validated markers to indicate tissue residence or migration of B cells, there is literature on B cell migration from animal models and studies of autoimmunity (30, 32, 43, 44). CXCR4 is a known marker of plasma cell migration and retention in the bone marrow, and is proposed to be important in plasmablast migration to sites of inflammation (44–47). Other chemokines, including CXCR3, CXCR5, and CCR7, have also been proposed (43). Here we saw evidence of CXCR4 RNA expression in IgG mRNA–containing cells, suggesting that CXCR4 may contribute to the migration of ASCs in skin during HSV reactivation (Figure 2F). Evidence of CXCR5 or CXCL13 expression was not seen. The primary ligand for CXCR4 (CXCL12) was observed in a gene expression study of HSV-infected tissue, but its expression was not significantly upregulated at healing or lesion time points (data accessible in the NCBI’s Gene Expression Omnibus database [refs. 48, 49], GEO GSE18527; Supplemental Figure 1).
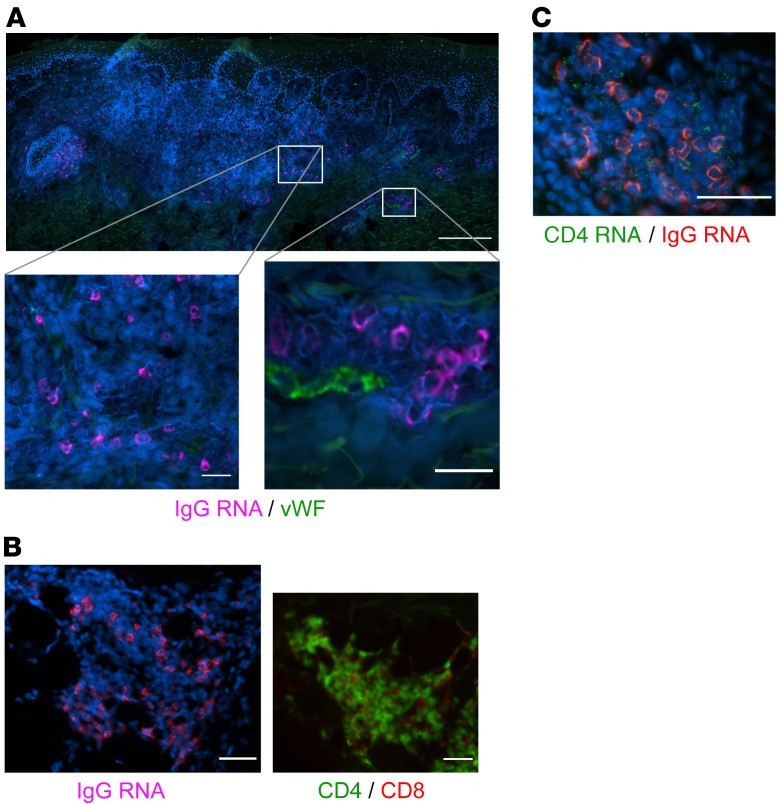
We performed detailed studies of the spatial localization of B cells to define their proximity to small capillaries. Costaining with vWF indicated that IgG RNA+ cells identified by FISH, while at times located near small capillaries, were not typically contained within vascular structures (Figure 3A). We observed many clusters of IgG RNA+ cells that did not have evidence of associated endothelial cells (Figure 1C and Figure 3, B and C). Some clusters appeared to contain primarily T cells, as in Figure 1D and as described previously (7, 8, 50), and others appeared to contain primarily ASCs, with a few T cells, as in Figure 3C. Clusters of IgG RNA+ cells were typically located in the dermis (not associated with the DEJ) and contained both CD8+ (Figure 3B, by FISH and IF over serial sections) and CD4+ (Figure 3C, by dual-probe FISH) T cells. Notably, these immune cell clusters were not detected in any of the 27 control specimens collected from either the arm or contralateral genital area. We also did not see any B cells or ASCs in vascular structures in control biopsies. Thus, dermis-infiltrating and capillary-associated B cells and ASCs are likely all related to HSV-2 reactivation in genital skin.
Figure 3. IgG RNA+ cells in relation to other tissue structures and cell types.
(A) ASCs are found infiltrating tissue distant from vascular structures, as well as adjacent to and within small capillaries. IgG-producing cells identified by FISH in a healing biopsy are found independently in tissue (lower left) as well as proximate to small capillaries (lower right) in biopsy specimens taken during healing after HSV-2 reactivation. Faint green signal is due to collagen autofluorescence. Blood vessel endothelial cells identified by vWF IF (green); IgG RNA+ cells, magenta. Scale bars: 200 μm; insets, 25 μm. vWF and IgG colocalization by combination FISH and IF was performed in specimens from 6 participants. While capillaries were identified in all samples, IgG cells were not present within these vascular structures in all samples. (B) IgG-producing cells by FISH (left) clustered in an area of dense T cell infiltration (right). CD4, green; CD8, red. Shown are serial sections of the same specimen. Scale bars: 50 μm. IgG FISH and T cell IF were performed in serial biopsy specimens from 4 participants. (C) Clustered IgG (red) and CD4 (green) by FISH; absence of vWF staining was confirmed. Scale bar: 50 μm. IgG and CD4 FISH was performed in specimens from 8 participants.
Kinetics of B cell response during HSV-2 reactivation and skin healing.
Overall, the magnitude and density of the B cell and ASC infiltrate (Figure 4, A and B) were greatest during the acute phases of reactivation; i.e., during clinical lesion and early healing. HSV-2 was detected by PCR at the time of lesion biopsy in 12 of 14 patients in whom this was tested, and at the first healing biopsy in patients 7 and 16. In most individuals, the highest density of CD20+ B cells was observed in samples when genital lesions were present (n = 8, median [range] density 18.4 [2.6–46.0] cells/mm2). In 3 patients, the density of CD20+ cells peaked at the time of healing, about 10 days after lesion onset (Figure 4A), with a median of 16.1 cells/mm2 (range 1.5–37.4 cells/mm2). To determine whether the peak density of CD20+ B cells was higher at either the active-lesion/early-healing or later healing time points, we compared the maximum levels in these specimens for each person. By Wilcoxon’s signed-rank test, the maximum CD20+ cell density of ulcerative or healing lesions was higher than the maximum of the 2-, 4-, or 8-weeks-post-healing specimens (P = 0.003). In 3 individuals (participants 1, 7, and 10), few B cells were seen in any samples. This overall pattern of higher infiltration at the time of symptomatic disease is identical to the pattern observed for T cell infiltration (see below).
Figure 4. Migration kinetics of B and T cells into genital skin during HSV-2 reactivation and tissue healing by type of biopsy and over time.
Density of (A) CD20+ cells by IF (n = 11), (B) IgG RNA+ cells by FISH (n = 10), (C) CD4+ T cells by IF (n = 8), and (D) CD8+ T cells by IF (n = 8) in tissue at the biopsy time points (left panels) and by participant over time (right panels). Statistical testing was by Friedman’s test with Dunn’s corrections for multiple comparisons. Each graph on the right presents cell counts from genital-area biopsies over time, as measured from the identified symptomatic lesion. (E) Correlation (r) between B and T cell subsets and between CD20+ and IgG RNA+ cells was calculated by repeated-measure correlation. Participants are labeled by color; results for multiple specimens from genital and control areas from each participant are included in each graph.
In 3 patients (participants 4, 11, and 15), we noted an increase in IgG RNA+ cells between the 4- and 8-weeks-post-healing biopsies. Similarly, CD20+ B cell density increased at 8 weeks after healing in patients 2 and 13. We speculated that this could be related to subclinical viral reactivation, although proof of this was not found in any participant by HSV PCR from tissue. In contrast to CD20+ B cells, by Wilcoxon’s signed-rank test, the maximum IgG RNA+ cell density of lesion or healing samples was no higher than the maximum of the 2-, 4-, or 8-weeks-post-healing samples (P = 0.2). In those 3 patients with a peak density of IgG RNA+ cells at 8 weeks after healing, the corresponding tissue antibody concentrations varied over time (participants 4, 11, and 15); 2 had a peak at the post-healing time, and 1 peaked during the lesion; none of these individuals had a corresponding peak in antibody titer at the 8-weeks-post-healing time point.
The density of CD20+ and IgG RNA+ cells was considerably lower than the density of T cells at the site of HSV-2 reactivation (Figure 4, C and D). As in past reports (8, 28), CD4+ and CD8+ T cells were found at highest density during the period of a symptomatic lesion in the majority of patients. As with CD20+ cells, by Wilcoxon’s signed-rank test, the maximum CD4+ and CD8+ cell density of lesion or healing samples was higher than the maximum of the 2-, 4-, or 8-weeks-post-healing specimens (P = 0.008, P = 0.04). We evaluated the relationship between B and T cell density in individual lesion specimens (Figure 4E). Specimens with the highest T cell density tended to be those with higher B cell density. By repeated-measure correlation, there was a correlation between the numbers of CD20+ B cells and CD4+ and CD8+ T cells, whereby biopsy samples with greater numbers of T cells also tended to have greater numbers of B cells when accounting for multiple biopsy results per person. The repeated-measure correlation coefficients (after log transformation) between CD20+ cells and CD4+ and CD8+ T cell subsets were 0.64 (P = 0.0002) and 0.77 (P < 0.0001). There was a small correlation between CD20 and IgG+ cells by repeated-measure correlation (r = 0.40, P = 0.03). None of these correlations were as strong as that between CD4+ and CD8+ T cells, which was 0.83 (P < 0.0001; Figure 4E). There was no correlation between IgG+ cells and CD4+ or CD8+ T cells (r = 0.28 and r = 0.31, P = 0.33 and P = 0.28, respectively) (Supplemental Figure 2). The simultaneous migration and spatial clustering of T and B cells in genital skin during HSV-2 reactivation and healing suggest that these lymphocyte subsets may be influenced by similar recruitment mechanisms.
Detection and kinetics of HSV-2–specific antibodies in genital tissue.
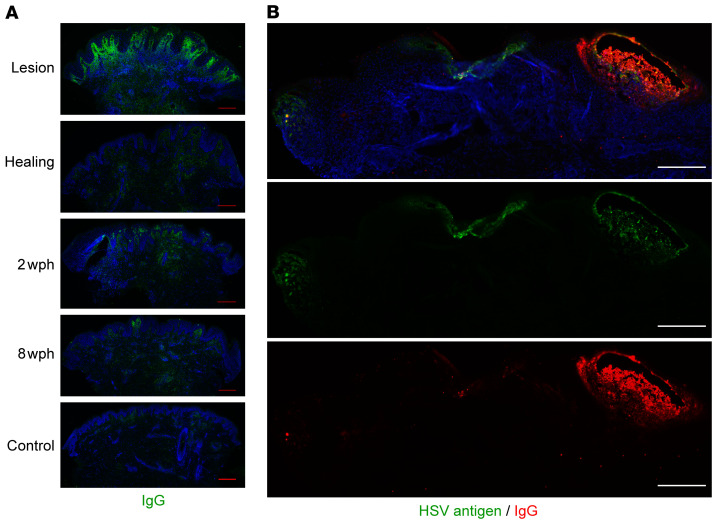
Antibody (IgG) was detectable by IF by using an antibody against human IgG in tissue. As with B cells, IgG was detected to the greatest extent in the upper dermis of lesion and early healing tissue biopsy sections, and was less detectable in specimens of uninvolved sites (Figure 5A). In a unique biopsy of an active HSV-2 vesicle and areas of HSV-2–associated epithelial cell necrosis, IgG was detectable colocalized with HSV antigen within the vesicle, and less prominent in the necrotic area (Figure 5B). Fluorescence was not detected with an isotype control, isolated secondary antibody, or if the tissue was blocked with 5% human serum prior to IF (Supplemental Figure 3). We therefore hypothesize that IgG detected in skin during HSV-2 reactivation (Figure 5) is produced by an antigen-specific process, such as by locally migrated B cells, and would thus be present at higher concentration than non–HSV-specific antibody.
Figure 5. Antibody is detectable during the tissue-based immune response to HSV-2 reactivation, and the amount is variable over time.
(A) Immunofluorescence detection of IgG (green) in genital skin biopsies in 1 participant. Scale bars: 250 μm. Brightness has been increased for visualization; results shown for 1 of 10 participants. (B) Spatial localization of in-tissue IgG versus HSV antigen by IF in biopsy of an active HSV-2 lesion (different subject). Antigen is seen in multiple locations, with the highest density of IgG at the site of vesicle formation. Scale bars: 250 μm. The distribution of IgG and HSV antigen across multiple sites of viral involvement was unique to this specimen.
We first investigated the trajectory of the systemic antibody response during HSV-2 reactivation and healing using sequential serum samples collected from 2 individuals (participants 1 and 6) concurrently with skin specimens (all other participants had a single blood draw at study entry, which may not have occurred within 6 months of their biopsy series). We used an antibody binding assay based on the Luminex platform to detect antibodies specific to the major HSV-2 surface glycoproteins gB2 and gD2. We included EBV gp350 and the stem of influenza hemagglutinin (Flu-HA) as highly prevalent, non-HSV targets. For all tested antigens, no variability in serum antibody level was detected over time, as evidenced by overlapping titration curves and the nearly identical AUC values calculated for these titration curves (Supplemental Figure 4 and Supplemental Table 2). This finding is consistent with earlier reports documenting the absence of a systemic antibody response during local HSV-2 reactivation (51–54).
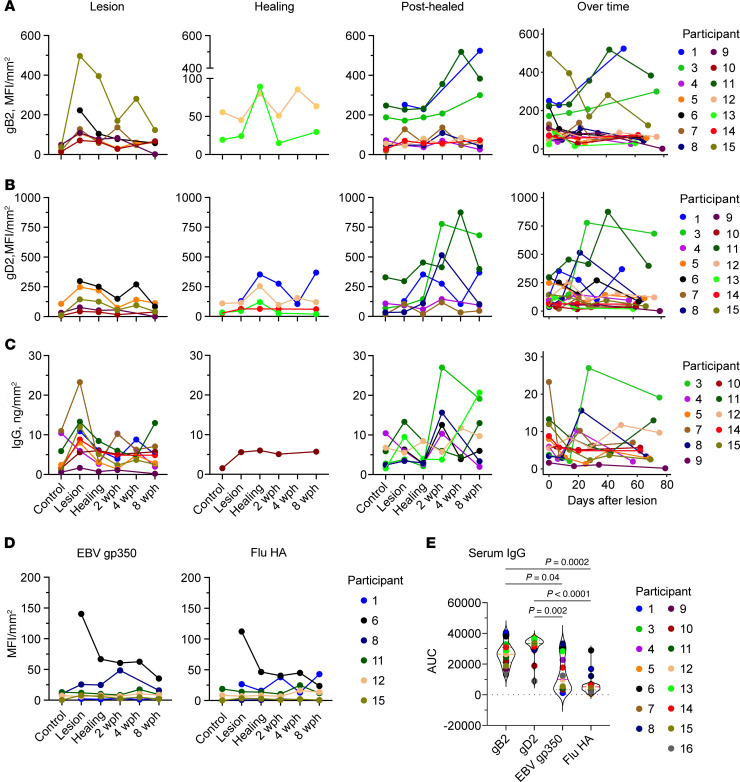
To investigate whether immunoglobulin observed in specimens by IF was specific to HSV-2, we extracted immunoglobulin from sequential biopsies of 14 participants to measure IgG reactive to gB2, gD2, and total IgG. IgG reactive to gB2 and gD2 proteins was detectable in the majority of the lesion area samples but was not detected in many specimens collected from the arm and contralateral skin (Figure 6, A and B). When followed over time, the pattern of shifts in concentration of HSV-specific IgG (Figure 6, A and B) and total IgG (Figure 6C) varied within the group and by individual, as was the case for B cells and ASCs. Across participants, the amount of HSV-specific antibody varied by up to 30-fold. Within participants, the differences in the amounts of HSV-specific antibody in different specimens likewise varied substantially (gB2 median 3.4-fold, range 1.7–57.8; and gD2 median 6.1-fold, range 2.3–94.7). Some participants had a peak HSV-2–specific IgG level during the lesion period; others had a peak during early healing; and the rest showed peak HSV-2–specific IgG during late healing. A higher concentration of total IgG was found at the lesion in samples from 6 participants and after healing in 4 participants; the total amount of IgG in the control biopsies was generally low (Figure 6C). Conversely, there was no major change observed over the course from the active-lesion to healing periods in the levels of IgG specific to control antigens in skin extracts, EBV gp350 and Flu-HA (Figure 6D), except in participant 6, for whom IgG at the lesion was substantially higher than for subsequent time points. Antibodies to control antigens were observed in skin extracts of only a few participants.
Figure 6. Levels of HSV-2–specific antibody and total IgG follow B cell and ASC density.
Concentration (expressed as MFI normalized to biopsy surface area [mm2]) of IgG to (A) gB2 (n = 14), (B) gD2 (n = 14), (C) total IgG (measured in ng/mm2; n = 13), and (D) control antigens EBV gp350 and Flu-HA (n = 6) in tissue over time and shown by participants, with antibody peak at lesion (first column) or healing (second and third columns) time points. If a person did not have a clear peak at one time point, they were included in multiple graphs. Levels of antibody over time (by number of days after lesion biopsy) are in the fourth column. Missing data points (such as the arm control in participant 6) correspond to specimens with insufficient tissue to run the test. (E) Area under the titration curve in a serum measurement of antibody specific to each of these antigens from a single sample taken from each person at study entry (n = 15). Serum titration curves are in Supplemental Figure 4, and AUC values are in Supplemental Table 2. Statistical testing was by Friedman’s test with Dunn’s corrections for multiple comparisons. All measurements were performed in duplicate.
We also used AUC of antibody titration curves to compare levels of circulating antibodies upon participant study intake. The amount of circulating antibody to gD2 was similar for the majority of the participants, while the amount of gB2-specific IgG was more varied among participants (Figure 6E, Supplemental Figure 5, and Supplemental Table 3). Both gB2- and gD2-specific IgGs were present at significantly higher levels than IgGs against non-HSV antigens, although the difference in levels between HSV-2 antigens and EBV gp350 was smaller.
This discordance between antibodies to HSV-specific and control antigens in skin and serum also suggests in-tissue antibody production. While some of the discrepancy between control and HSV-specific antibody in tissue may be accounted for by its greater abundance in serum, that was not consistently true. For example, the level of EBV gp350 IgG was similar to that of gB2 IgG in the serum of participant 13, whereas in skin EBV gp350 IgG was not detected, and gB2 IgG peaked at the healing time point (Figure 6D). In participant 8, EBV gp350 IgG in serum was present at a higher level than gD2- and gB2-specific antibodies, but in skin gD2 IgG peaked at 2 weeks after healing, while EBV gp350 IgG was increased at the lesion time point. Interestingly, Flu-HA but not EBV IgG mirrored the dynamics of HSV-specific antibody in this participant. Together, these findings support the hypothesis that there are multiple mechanisms of antibody infiltration into inflamed skin, including both transudation from serum and local production.
Discussion
Here we have shown that B cells of different developmental stages are present in biopsy tissue from individuals undergoing biopsies of active and healing skin lesions caused by HSV-2 reactivation. Our studies show B cells to be within the upper dermis and extrinsic to blood vessels in the weeks during and after HSV-2 reactivation. Spatially, we see 2 intriguing patterns of B cell infiltration. In the first, ASCs are present in the dermis at the site of HSV reactivation at variable density over time, and this coincides with the pattern of total immunoglobulin we see during genital lesions. The other pattern is the presence of largely antigen-inexperienced CD20+ B cells in T cell clusters and infiltrates. These CD20+ B cells primarily coexpress IgD but not memory markers and were not associated with Ig secretion by FISH staining — suggesting they are early-stage B cells that might participate in antigen processing or some as-yet-undefined function, such as regulation of inflammation. The robust correlation between B cells and CD4+ T cells corroborates this suggestion. Based on the finding of B cells in multiple stages of development in biopsy specimens and closely organized within T cell clusters, we propose that B cells may be involved in antigen presentation and regulation of inflammation (55), as well as local production of antibody. This diversity of tissue B cell subsets by spatiotemporal arrangement, phenotype, and a statistically significant but comparatively minor correlation between CD20 and IgG RNA also suggests that there is not a single mechanism of recruitment of different subsets of B cells. It is possible that tissue-infiltrating memory cells may give rise to ASCs during the course of HSV-2 reactivation and healing, but with the paucity of detected memory cells, it appears more likely that there is an independent mechanism by which ASCs enter inflamed tissue. The presence of B cells and ASCs at lower numbers than T cells in the skin, as is true in the blood, is consistent with multiple, diverse roles in immune activation, regulation, or antibody production.
We also found that HSV-2–specific antibody extracted from skin biopsy specimens varied in concentration over the course of reactivation and healing, and was present at a greater concentration than antibodies unrelated to HSV antigens, whereas no variability was seen in serum over the same time intervals. The change in tissue antibody concentration corresponds to the change seen in the density of ASCs, though they do not mirror each other directly. This is congruous with observations in other conditions where a peak in ASC density is offset in time from the peak in corresponding antibody production (56, 57). Overall, these findings are consistent with boosting of local antibody secretion by viral challenge, whereby tissue-based antibody concentration is increased during HSV-2 reactivation and healing, and much lower in uninvolved control tissues. The role of tissue-infiltrating antibody in control of viral replication during HSV-2 outbreaks has been suggested by mathematical and experimental models showing the importance of rapid suppression of cell-to-cell spread and neutralization of free virus during HSV-2 reactivation (4, 9). Further investigation to identify additional markers of tissue infiltration and recruitment of B cells is necessary; here we observed scant CXCR4 in IgG RNA+ cells but no CXCR5, CCR4, or CCR5 in B cells by IF in biopsy specimens (24). We suspect that there may be an antigen-ggdependent mechanism driving this process.
Mechanistic evaluation from human tissues is by necessity limited, and this study was limited in scope to those individuals available and willing to undergo serial biopsies and blood draws over the course of symptomatic HSV-2 infection. This typically excludes those with mild disease (<1–2 outbreaks/year) and by definition excludes any individuals without symptomatic HSV outbreaks, who may have a very different local immune response.
In summary, we have demonstrated that B cells and ASCs are present in the inflammatory infiltrate that results from HSV-2 reactivation in skin of the genital region. Phenotypic testing indicated that the ASCs were for the most part plasma cells, but this may include plasmablasts, and many, if not most, CD20+ cells seen were antigen inexperienced. The kinetics of tissue infiltration and egress of CD20+ cells, and to a lesser extent IgG+ cells, followed that seen in T cell studies, though it varied by patient. This apparent variability may be partly due to the limited sample size available for study; however, it paralleled the relative concentrations of HSV-2–specific antibody. That the concentration of HSV-2–specific antibody in tissue varied independently of the concentration of HSV-2–specific antibody in serum suggests that there are local mechanisms leading to either local production or recruitment of HSV-2–specific antibody. We propose an as-of-yet underappreciated role of B cells and ASCs in the human skin-based immune response to HSV-2 reactivation.
Methods
Study participants.
Healthy, HSV-2–seropositive adults with a history of symptomatic genital herpes were enrolled in a natural history biopsy protocol at the University of Washington Virology Research Clinic (UW-VRC) in Seattle. Participants had symptoms of genital herpes for longer than 1 year, and HSV-2 seropositivity was confirmed by HSV Western blot (58). Biopsies were performed by clinicians as described previously (5) at the time of an HSV-2 lesion; at lesion healing 10–14 days after onset; and in the same location 2, 4, and 8 weeks later. A control biopsy sample was obtained from the upper arm or contralateral genital region skin site. Blood for PBMC and plasma isolation was drawn either at enrollment or at each biopsy visit for some study participants.
Materials.
HSV-2 proteins gB2 and gD2 were provided by G.H. Cohen and R.J. Eisenberg (University of Pennsylvania, Philadelphia Pennsylvania, USA); gD2 was also provided by Immune Design. EBV gp350 was provided by A. McGuire (Fred Hutchinson Cancer Research Center). Stem of Flu-HA strain H1 1999 NC was expressed using vector VRC-3925 (59) in 293F cells and was provided by M. Gray and L. Stamatatos (Fred Hutchinson Cancer Research Center).
IF.
Biopsy specimens were flash frozen in optimum cutting temperature (OCT) compound (Sakura Finetek) and stored at −80°C. Slices of tissue (8 μm) perpendicular to the epidermal surface, including a cross section of the epidermis, DEJ, and dermis, were prepared by cryostat sectioning and mounted on glass slides. Slides were dehydrated at room temperature for 24 hours, frozen at –80°C, and fixed in acetone prior to use. To enumerate the relative density of B cells, T cells, ASCs, blood vessels, and/or antibody in tissue biopsy sections, the following primary antibodies were used; rabbit anti–human CD20 (1:100; Abcam ab78237), mouse anti-human CD79b (1:100; Novus Biologicals NBP1-28528), mouse anti-human IgG (1:100; eBioscience 05-4200), sheep anti–human vWF (FITC-conjugated, 1:5000; Abcam ab8822), mouse anti–human CD27 (1:100; BioLegend 356403), mouse anti–human CD20 (1:100; eBioScience 14-0202-82), mouse anti-human IgD (af647-conjugated, 1:50 or 1:100; BioLegend 348227), rabbit anti–HSV-2 (1:1000; Dako B0116), mouse anti-CD4 (1:1000; BioLegend 300502), and mouse anti-CD8 (af647-conjugated, 1:100; BD Pharmingen 557708). Prior to application of antibodies, nonspecific binding was blocked with 2% BSA, 10% casein, and 5% normal human sera, except for mouse anti-human IgG and IgD, for which human sera were omitted from the blocking buffer. TSA amplification (Invitrogen) was used for visualization of CD79b and IgG, and for CD20 in costaining with IgD. Tissue sections were mounted in Mowiol 40-88 containing 2.5% wt/vol DABCO (MilliporeSigma). Images were captured with a Nikon Eclipse Ti with NIS-Elements software using a Hamamatsu ORCA-Flash4.0 sCMOS camera and viewed in NIH ImageJ/Fiji (60). Cells were counted in ImageJ/Fiji with the Cell Counter plugin (61). The surface area of each biopsy was calculated in ImageJ.
FISH.
To determine the presence of IgG-producing cells in biopsy specimens and identify their spatial localization and interaction with other cell types, we performed FISH (RNAscope 2.0, ACD; ref. 62). Freshly sliced 8 μm frozen tissue sections were fixed at 4°C in 10% neutral buffered formalin, dehydrated, and treated with protease. Prepared sections were incubated with DNA-based probes (ACD) for pooled IGHG1–4 (IgG), CD4, MS4A1 (CD20), SDC1 (CD138), PAX5, CXCR4, PRDM1, and IRF4 transcripts, and fluorescently labeled for visualization. After nuclear staining with DAPI (Fluka), tissues were mounted with ProLong Gold Antifade (Thermo Fisher Scientific). Images were captured with a Nikon Eclipse Ti (as above) and viewed in ImageJ/Fiji. IgG RNA+ cells were counted manually over the entire slice, and the surface area of each biopsy slice was calculated in ImageJ (61) to determine B cell density. Some sections were then unmounted and stained by IF for vWF (Abcam) to determine the relative localization of B cells identified by FISH with respect to small capillaries. Colocalization of CD138 and IgG FISH probes was investigated by deconvolution (GE/Applied Precision DeltaVision Elite) and confocal (Zeiss LSM 780 NLO) microscopy at 60× magnification; confocal and deconvolution images were taken with a high-resolution cooled Photometrics HQ2 CCD camera.
Preparation of biopsy extract.
Five or 10 freshly sliced 10 μm biopsy cryosections were placed in a microcentrifuge tube on dry ice. To extract protein, tubes were brought to room temperature, and Tissue Extraction Reagent II (Invitrogen) was added (10 μL per biopsy slice). Tube content was mixed by pipetting, followed by sequential vortexing (30 seconds), sonication (30 seconds), and pulse centrifugation. This procedure was repeated twice, then samples were flash frozen on dry ice and kept until use at –20°C. Upon thawing, samples were vortexed, sonicated, and centrifuged for 5 minutes at 14,500g. The supernatant was collected to measure protein, total IgG, and HSV-specific antibodies via a Luminex binding antibody assay.
HSV-2 Luminex binding antibody assay.
HSV-2 proteins (gB2, gD2) and control antigens (EBV gp350, Flu-HA) were coupled to MagPlex beads using an antibody coupling kit (Luminex) and stored at 4°C. To measure HSV-2–specific antibody, MagPlex beads were incubated with blocking buffer (5% Blotting-Grade Blocker [Bio-Rad], 0.05% Tween-20 [MilliporeSigma], PBS) to minimize nonspecific binding. Beads were then washed and mixed with serial dilutions of biopsy extracts or serum samples in assay buffer (Pierce Protein-Free [PBS] Blocking Buffer, 1% Blotting-Grade Blocker, and 0.05% Tween-20). Sera pooled from 10 HSV-2–seropositive and 10 HSV-2–seronegative donors were used as positive and negative controls, respectively. After incubation with biopsy extracts or serum samples, MagPlex beads were washed with PBST (PBS, 0.05% Tween-20) and incubated with anti-human IgG Fc-PE (SouthernBiotech). Finally, beads were washed 3 times and resuspended in PBS containing 1% BSA and 0.05% Tween-20. Median fluorescence intensity (MFI) by PE fluorescence for each bead type was collected on Luminex 200 instruments operated by MagPlex software (Hitachi) or xPonent Software (Luminex). The level of background was assigned by the MFI of antigen-conjugated beads incubated first with buffer (in place of serum), then with secondary antibody. Background MFI values for each antigen were subtracted from experimental measurements. The MFI value of the biopsy extracts was normalized to the surface area of each biopsy slice (as measured in adjacent slices) to account for variation in size and protein composition of biopsy specimens. Values from the dilution curves were used to determine AUC. Serologic data were analyzed in GraphPad Prism.
HSV-2 quantitative PCR.
DNA from biopsies and genital swabs was extracted using tissue kits (EZ1; QIAGEN) from 60 to 80 μm cross-sectioned samples from each biopsy specimen, as previously described (63, 64), including subtype-specific typing by real-time PCR (65). HSV-2 DNA copy numbers were normalized to 1 × 106 cells using β-globin copy numbers. Primers used were as follows: 5′-TGAAGGCTCATGGCAAGAAA-3′ and 5′-GCTCACTCAGTGTGGCAAAGG-3′, with 5′-TCCAGGTGAGCCAGGCCATCACTA-3′ as a probe. Detection limits of HSV-2 DNA quantification were 1 copy per 50,000 cells in tissue, as previously published (50, 63).
Statistics.
All statistical and data analysis was performed using R Studio (R 3.4.1; ref. 66) or GraphPad Prism version 8.2.0 for Mac (GraphPad Software). To compare cell density and antibody concentration between biopsy time points, Friedman’s test with Dunn’s correction for multiple comparisons was used. To test the observation of greatest B cell and ASC density, the maximum for lesion or newly healed specimens was compared with the maximum at 2, 4, or 8 weeks after healing by paired Wilcoxon’s test. We employed repeated-measure correlation to account for the presence of multiple biopsies from each participant (67). The repeated-measure correlation coefficient between the density of cell types (r) and statistical test value (P) is shown in the scatter plots. To avoid taking log of 0, a small pseudo-count (50% of the lowest value) was added to each measurement prior to log transformation. Comparison of AUC calculated for sera collected at different time points was performed with Friedman’s test with Dunn’s correction for multiple comparisons. Statistical significance was defined as adjusted, 2-sided P value less than 0.05.
Study approval.
The natural history biopsy protocol was reviewed and approved by the University of Washington Human Subjects Review Committee, and written, informed consent was obtained from each participant.
Author contributions
ESF, AMS, JZ, and LC designed the study. ESF, AMS, AK, KP, RMB, and SSC performed tissue biopsy sample analysis. CJ and AW recruited participants and provided clinical guidance. ESF, AMS, RMB, DS, RR, PTE, JZ, and TP performed data analysis. ESF, AMS, JZ, and LC prepared the manuscript. All authors reviewed and approved the manuscript. ESF is listed first as co–first author due to contributing to the revision of the manuscript; ESF and AMS contributed equally to the design and undertaking of experiments and the analysis presented here.
Supplementary Material
Acknowledgments
The authors would like to express their appreciation to Mindy Miner for assistance in editing and formatting the final manuscript and to David McDonald for assistance in confocal and deconvolution microscopy. Funding sources include NIH grants K08 AI148588 (to ESF), U19 AI113173 (ESF), P01 AI030731 (JZ, LC, and AW), R01 AI042528 (JZ and LC), R01 AI134878 (to JZ and LC), and P30 CA015704 (core facilities); and a Fred Hutchinson Cancer Research Center Vaccine and Infectious Diseases Division (VIDD) Initiative grant (to AMS).
Version 1. 03/30/2021
In-Press Preview
Version 2. 05/03/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(9):e142088.https://doi.org/10.1172/JCI142088.
See the related Commentary at B cells join T cell clusters in the host response to recurrent herpes simplex virus 2 infection.
Contributor Information
Emily S. Ford, Email: esford3@fredhutch.org.
Anton M. Sholukh, Email: asholukh@fredhutch.org.
RuthMabel Boytz, Email: rmboytz@gmail.com.
Savanna S. Carmack, Email: savannacarmack@hotmail.com.
Alexis Klock, Email: alexismm@u.washington.edu.
Khamsone Phasouk, Email: kphasouk@fhcrc.org.
Danica Shao, Email: jshao@fredhutch.org.
Raabya Rossenkhan, Email: rrossenk@fredhutch.org.
Paul T. Edlefsen, Email: pedlefse@fhcrc.org.
Tao Peng, Email: tpeng@fhcrc.org.
Christine Johnston, Email: cjohnsto@u.washington.edu.
Anna Wald, Email: annawald@uw.edu.
Jia Zhu, Email: jiazhu@uw.edu.
Lawrence Corey, Email: lcorey@fredhutch.org.
References
- 1.Wald A, et al. Virologic characteristics of subclinical and symptomatic genital herpes infections. N Engl J Med. 1995;333(12):770–775. doi: 10.1056/NEJM199509213331205. [DOI] [PubMed] [Google Scholar]
- 2.Benedetti J, et al. Recurrence rates in genital herpes after symptomatic first-episode infection. Ann Intern Med. 1994;121(11):847–854. doi: 10.7326/0003-4819-121-11-199412010-00004. [DOI] [PubMed] [Google Scholar]
- 3.Schiffer JT, et al. Mathematical modeling predicts that increased HSV-2 shedding in HIV-1 infected persons is due to poor immunologic control in ganglia and genital mucosa. PLoS One. 2016;11(6):e0155124. doi: 10.1371/journal.pone.0155124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiffer JT, et al. Mucosal host immune response predicts the severity and duration of herpes simplex virus-2 genital tract shedding episodes. Proc Natl Acad Sci U S A. 2010;107(44):18973–18978. doi: 10.1073/pnas.1006614107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, et al. Virus-specific CD8+ T cells accumulate near sensory nerve endings in genital skin during subclinical HSV-2 reactivation. J Exp Med. 2007;204(3):595–603. doi: 10.1084/jem.20061792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iijima N, et al. Dendritic cells and B cells maximize mucosal Th1 memory response to herpes simplex virus. J Exp Med. 2008;205(13):3041–3052. doi: 10.1084/jem.20082039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J, et al. Persistence of HIV-1 receptor-positive cells after HSV-2 reactivation is a potential mechanism for increased HIV-1 acquisition. Nat Med. 2009;15(8):886–892. doi: 10.1038/nm.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu J, et al. Immune surveillance by CD8αα+ skin-resident T cells in human herpes virus infection. Nature. 2013;497(7450):494–497. doi: 10.1038/nature12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiffer JT, et al. Rapid localized spread and immunologic containment define Herpes simplex virus-2 reactivation in the human genital tract. Elife. 2013;2:e00288. doi: 10.7554/eLife.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iijima N, Iwasaki A. T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science. 2014;346(6205):93–98. doi: 10.1126/science.1257530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mertz GJ, et al. Frequency of acquisition of first-episode genital infection with herpes simplex virus from symptomatic and asymptomatic source contacts. Sex Transm Dis. 1985;12(1):33–39. doi: 10.1097/00007435-198501000-00007. [DOI] [PubMed] [Google Scholar]
- 12.Cairns TM, et al. Dissection of the antibody response against herpes simplex virus glycoproteins in naturally infected humans. J Virol. 2014;88(21):12612–12622. doi: 10.1128/JVI.01930-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flechtner JB, et al. Immune responses elicited by the GEN-003 candidate HSV-2 therapeutic vaccine in a randomized controlled dose-ranging phase 1/2a trial. Vaccine. 2016;34(44):5314–5320. doi: 10.1016/j.vaccine.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein DI, et al. Therapeutic vaccine for genital herpes simplex virus-2 infection: findings from a randomized trial. J Infect Dis. 2017;215(6):856–864. doi: 10.1093/infdis/jix004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wald A, et al. Therapeutic HSV-2 vaccine (GEN003) results in durable reduction in genital lesions at 1 year. Open Forum Infect Dis. 2014;1(Suppl 1):S55–S56. doi: 10.1093/ofid/ofu051.150. [DOI] [Google Scholar]
- 16.Gilbert PB, et al. Antibody to HSV gD peptide induced by vaccination does not protect against HSV-2 infection in HSV-2 seronegative women. PLoS One. 2017;12(5):e0176428. doi: 10.1371/journal.pone.0176428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burn C, et al. A herpes simplex virus (HSV)-2 single-cycle candidate vaccine deleted in glycoprotein D protects male mice from lethal skin challenge with clinical isolates of HSV-1 and HSV-2. J Infect Dis. 2017;217(5):754–758. doi: 10.1093/infdis/jix628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y, et al. Maternal antiviral immunoglobulin accumulates in neural tissue of neonates to prevent HSV neurological disease. mBio. 2017;8(4):1–14. doi: 10.1128/mBio.00678-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Criscuolo E, et al. Cell-to-cell spread blocking activity is extremely limited in the sera of herpes simplex virus 1 (HSV-1)- and HSV-2-infected subjects. J Virol. 2019;93(11):e00070–19. doi: 10.1128/JVI.00070-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harandi AM, et al. Differential roles of B cells and IFN-gamma-secreting CD4(+) T cells in innate and adaptive immune control of genital herpes simplex virus type 2 infection in mice. J Gen Virol. 2001;82(4):845–853. doi: 10.1099/0022-1317-82-4-845. [DOI] [PubMed] [Google Scholar]
- 21.Dudley KL, et al. Immune protection against HSV-2 in B-cell-deficient mice. Virology. 2000;270(2):454–463. doi: 10.1006/viro.2000.0298. [DOI] [PubMed] [Google Scholar]
- 22.Milligan GN, Bernstein DI. Generation of humoral immune responses against herpes simplex virus type 2 in the murine female genital tract. Virology. 1995;206(1):234–241. doi: 10.1016/S0042-6822(95)80038-7. [DOI] [PubMed] [Google Scholar]
- 23.Bourne N, et al. Increased frequency of virus shedding by herpes simplex virus 2-infected guinea pigs in the absence of CD4+ T lymphocytes. J Virol. 2018;93(4):e01721–18. doi: 10.1128/JVI.01721-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh JE, et al. Migrant memory B cells secrete luminal antibody in the vagina. Nature. 2019;571(7763):122–126. doi: 10.1038/s41586-019-1285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandal A, et al. Cell and fluid sampling microneedle patches for monitoring skin-resident immunity. Sci Transl Med. 2018;10(467):eaar2227. doi: 10.1126/scitranslmed.aar2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabib T, et al. SFRP2/DPP4 and FMO1/LSP1 define major fibroblast populations in human skin. J Invest Dermatol. 2018;138(4):802–810. doi: 10.1016/j.jid.2017.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemos MP, et al. In men at risk of HIV infection, IgM, IgG1, IgG3, and IgA reach the human foreskin epidermis. Mucosal Immunol. 2016;9(3):798–808. doi: 10.1038/mi.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cunningham AL, et al. Evolution of recurrent herpes simplex lesions. An immunohistologic study. J Clin Invest. 1985;75(1):226–233. doi: 10.1172/JCI111678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egbuniwe IU, et al. Revisiting the role of B cells in skin immune surveillance. Trends Immunol. 2015;36(2):102–111. doi: 10.1016/j.it.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Geherin S, et al. The skin, a novel niche for recirculating B cells. J Immunol. 2012;188(12):6027–6035. doi: 10.4049/jimmunol.1102639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginaldi L, et al. Changes in antigen expression on B lymphocytes during HIV infection. Pathobiology. 1998;66(1):17–23. doi: 10.1159/000027990. [DOI] [PubMed] [Google Scholar]
- 32.Wilson RP, et al. IgM plasma cells reside in healthy skin and accumulate with chronic inflammation. J Invest Dermatol. 2019;139(12):2477–2487. doi: 10.1016/j.jid.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens RH, et al. Immunoglobulin heavy chain mRNA in mitogen-stimulated B cells. Eur J Immunol. 1975;5(1):47–53. doi: 10.1002/eji.1830050111. [DOI] [PubMed] [Google Scholar]
- 34.Coronella JA, et al. Amplification of IgG VH and VL (Fab) from single human plasma cells and B cells. Nucleic Acids Res. 2000;28(20):e85. doi: 10.1093/nar/28.20.e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nutt SL, et al. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15(3):160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 36.Tellier J, et al. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat Immunol. 2016;17(3):323–330. doi: 10.1038/ni.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nutt SL, et al. The genetic network controlling plasma cell differentiation. Semin Immunol. 2011;23(5):341–349. doi: 10.1016/j.smim.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Ellebedy AH, et al. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat Immunol. 2016;17(10):1226–1234. doi: 10.1038/ni.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minnich M, et al. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat Immunol. 2016;17(3):331–343. doi: 10.1038/ni.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chilosi M, et al. CD138/syndecan-1: a useful immunohistochemical marker of normal and neoplastic plasma cells on routine trephine bone marrow biopsies. Mod Pathol. 1999;12(12):1101–1106. [PubMed] [Google Scholar]
- 41.O’Connell FP, et al. CD138 (syndecan-1), a plasma cell marker immunohistochemical profile in hematopoietic and nonhematopoietic neoplasms. Am J Clin Pathol. 2004;121(2):254–263. doi: 10.1309/617DWB5GNFWXHW4L. [DOI] [PubMed] [Google Scholar]
- 42.Kind S, et al. Prevalence of syndecan-1 (CD138) expression in different kinds of human tumors and normal tissues. Dis Markers. 2019;2019:4928315. doi: 10.1155/2019/4928315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bugatti S, et al. B cells in rheumatoid arthritis: from pathogenic players to disease biomarkers. Biomed Res Int. 2014;681678:1–14. doi: 10.1155/2014/681678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hauser AE, et al. Chemotactic responsiveness toward ligands for CXCR3 and CXCR4 is regulated on plasma blasts during the time course of a memory immune response. J Immunol. 2002;169(3):1277–1282. doi: 10.4049/jimmunol.169.3.1277. [DOI] [PubMed] [Google Scholar]
- 45.Nie Y, et al. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J Exp Med. 2004;200(9):1145–1156. doi: 10.1084/jem.20041185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caraux A, et al. Circulating human B and plasma cells. Age-associated changes in counts and detailed characterization of circulating normal CD138- and CD138+ plasma cells. Haematologica. 2010;95(6):1016–1020. doi: 10.3324/haematol.2009.018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nance CL, Shearer WT. SDF-1α regulates HIV-1-gp120-induced changes in CD79b surface expression and Ig production in activated human B cells. Clin Immunol. 2002;105(2):208–214. doi: 10.1006/clim.2002.5284. [DOI] [PubMed] [Google Scholar]
- 48.Peng T, et al. Evasion of the mucosal innate immune system by herpes simplex virus type 2. J Virol. 2009;83(23):12559–12568. doi: 10.1128/JVI.00939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hensel MT, et al. Selective expression of CCR10 and CXCR3 by circulating human herpes simplex virus-specific CD8 T cells. J Virol. 2017;91(19):e00810-17. doi: 10.1128/JVI.00810-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng T, et al. An effector phenotype of CD8+ T cells at the junction epithelium during clinical quiescence of herpes simplex virus 2 infection. J Virol. 2012;86(19):10587–10596. doi: 10.1128/JVI.01237-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Douglas RG, Couch RB. A prospective study of chronic herpes simplex virus infection and recurrent herpes labialis in humans. J Immunol. 1970;2(2):289–295. [PubMed] [Google Scholar]
- 52.Friedman MG, Kimmel N. Herpes simplex virus-specific serum immunoglobulin A: detection in patients with primary or recurrent herpes infections and in healthy adults. Infect Immun. 1982;37(1):374–377. doi: 10.1128/IAI.37.1.374-377.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juto P, Settergren B. Specific serum IgA, IgG and IgM antibody determination by a modified indirect ELISA-technique in primary and recurrent herpes simplex virus infection. J Virol Methods. 1988;20(1):45–55. doi: 10.1016/0166-0934(88)90039-0. [DOI] [PubMed] [Google Scholar]
- 54.Woodman CB, et al. The relative infrequency and low levels of neutralising and immunoprecipitating antibody to herpes simplex viruses types 1 and 2 in patients with a history of recurrent herpes genitalis. Med Microbiol Immunol. 1983;171(4):243–250. doi: 10.1007/BF02123498. [DOI] [PubMed] [Google Scholar]
- 55.Griss J, et al. B cells sustain inflammation and predict response to immune checkpoint blockade in human melanoma. Nat Commun. 2019;10(1):4186. doi: 10.1038/s41467-019-12160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanchard-Rohner G, et al. Appearance of peripheral blood plasma cells and memory B cells in a primary and secondary immune response in humans. Blood. 2009;114(24):4998–5002. doi: 10.1182/blood-2009-03-211052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He XS, et al. Plasmablast-derived polyclonal antibody response after influenza vaccination. J Immunol Methods. 2011;365(1–2):67–75. doi: 10.1016/j.jim.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ashley RL, et al. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunodot enzyme assay for detecting antibodies to herpes simplex virus types 1 and 2 in human sera. J Clin Microbiol. 1988;26(4):662–667. doi: 10.1128/JCM.26.4.662-667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lingwood D, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489(7417):566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Diem K, et al. Image analysis for accurately counting CD4+ and CD8+ T cells in human tissue. J Virol Methods. 2015;222:117–121. doi: 10.1016/j.jviromet.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagnostics. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Magaret AS, et al. Optimizing PCR positivity criterion for detection of herpes simplex virus DNA on skin and mucosa. J Clin Microbiol. 2007;45(5):1618–1620. doi: 10.1128/JCM.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wald A, et al. Polymerase chain reaction for detection of herpes simplex virus (HSV) DNA on mucosal surfaces: comparison with HSV isolation in cell culture. J Infect Dis. 2003;188(9):1345–1351. doi: 10.1086/379043. [DOI] [PubMed] [Google Scholar]
- 65.Corey L, et al. Differentiation of herpes simplex virus types 1 and 2 in clinical samples by a real-time taqman PCR assay. J Med Virol. 2005;76(3):350–355. doi: 10.1002/jmv.20365. [DOI] [PubMed] [Google Scholar]
- 66. R Foundation for Statistical Computing. R: a language and environment for statistical computing. https://www.R-project.org/ Accessed March 24, 2021.
- 67.Bakdash JZ, Marusich LR. Repeated measures correlation. Front Psychol. 2017;8:1–13. doi: 10.3389/fpsyg.2017.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.