Summary
Exome and genome sequencing have proven to be effective tools for the diagnosis of neurodevelopmental disorders (NDDs), but large fractions of NDDs cannot be attributed to currently detectable genetic variation. This is likely, at least in part, a result of the fact that many genetic variants are difficult or impossible to detect through typical short-read sequencing approaches. Here, we describe a genomic analysis using Pacific Biosciences circular consensus sequencing (CCS) reads, which are both long (>10 kb) and accurate (>99% bp accuracy). We used CCS on six proband-parent trios with NDDs that were unexplained despite extensive testing, including genome sequencing with short reads. We identified variants and created de novo assemblies in each trio, with global metrics indicating these datasets are more accurate and comprehensive than those provided by short-read data. In one proband, we identified a likely pathogenic (LP), de novo L1-mediated insertion in CDKL5 that results in duplication of exon 3, leading to a frameshift. In a second proband, we identified multiple large de novo structural variants, including insertion-translocations affecting DGKB and MLLT3, which we show disrupt MLLT3 transcript levels. We consider this extensive structural variation likely pathogenic. The breadth and quality of variant detection, coupled to finding variants of clinical and research interest in two of six probands with unexplained NDDs, support the hypothesis that long-read genome sequencing can substantially improve rare disease genetic discovery rates.
Keywords: long read sequencing, clinical sequencing, neurodevelopmental disorder, structural variation, mobile element insertion
Here we apply “long-read” genome sequencing technology on six children with an undiagnosed neurodevelopmental disorder. In two of those six, we detected clinically relevant genetic variation that had previously been missed. Our data suggest that long reads will substantially improve yield of genomic testing in individuals with rare disease.
Introduction
Neurodevelopmental disorders (NDDS) are a heterogeneous group of conditions that lead to a range of physical and intellectual disabilities and collectively affect 1%–3% of children.1 Many NDDs result from large-effect genetic variation, which often occurs de novo,2 with hundreds of genes known to associate with disease.3 Owing to this combination of factors, exome and genome sequencing (ES/GS) have proven to be powerful tools for both clinical diagnostics and research on the genetic causes of NDDs. However, while discovery power and diagnostic yield of genomic testing have consistently improved over time,4 most NDDs cannot be attributed to currently detectable genetic variation.5
There are a variety of hypotheses that might explain the fact that most NDDs cannot be traced to a causal genetic variant after ES/GS, including potential environmental causes and complex genetic effects driven by small-effect variants.6 However, one likely possibility is that at least some NDDs result from highly penetrant variants that are missed by typical genomic testing. ES/GS are generally performed by generating millions of “short” sequencing reads, often paired-end 150 bp reads, followed by alignment of those reads to the human reference assembly and detection of variation from the reference. Various limitations of this process, such as confident alignment of variant reads to a unique genomic location, make it difficult to detect many variants, including some known to be highly penetrant contributors to disease. Examples of NDD-associated variation that might be missed include low-complexity repeat variants,7 small to moderately sized structural variants (SVs),4,8 and mobile element insertions (MEIs).9,10 Indeed, despite extensive effort from many groups, detection of such variation remains plagued by high error rates, both false positives (FPs) and false negatives (FNs), and it is likely that many such variants are simply invisible to short-read analysis.11
One potential approach to overcome variant detection limitations in ES/GS is to use sequencing platforms that provide longer reads. Long reads allow for more comprehensive and accurate read alignment to the reference assembly, including within and near to repetitive regions, and de novo assembly.12 However, to date, the utility of these long reads has been limited for several reasons, including cost, requirements on size, quantity and quality of input DNA, and high base-pair-level error rates. Recently, Pacific Biosciences released an approach, called circular consensus sequencing (CCS), or “HiFi,” in which fragments of DNA are circularized and then sequenced repeatedly.13 This leads to sequence reads that are both long (>10 kb) and accurate at the base pair level (>99%). In principle, such an approach holds great potential for more comprehensive and accurate detection of human genetic variation, especially in the context of rare genetic disease.
We have used CCS to analyze six proband-parent trios affected with NDDs that we previously sequenced using a typical Illumina genome sequencing (IGS) approach but in whom no causal or even potentially causal genetic variant was found. The CCS data were used to detect variation within each trio and generate de novo genome assemblies, with a variety of metrics indicating that the results are more comprehensive and accurate, especially for complex variation, than those seen in short-read datasets. In one proband, we identified an L1-mediated de novo insertion within CDKL5 that leads to a duplicated coding exon and is predicted to lead to a frameshift and loss of function. Transcript analyses confirm that the duplicated exon is spliced into mRNA in the proband. We have classified this variant as likely pathogenic (LP) using American College of Medical Genetics (ACMG) standards.14 In a second proband, we found multiple large SVs that together likely disrupt at least seven protein-coding genes. Our observations support the hypothesis that long-read genome analysis can substantially improve success rates for the detection of variation associated with rare genetic conditions.
Material and methods
Illumina sequencing, variant calling, and analysis
Six probands and their unaffected parents were enrolled in a research study aimed at identifying genetic causes of NDDs,15 which was monitored by the Western Institutional Review Board (IRB) (20130675). All six of these families underwent trio IGS between 4 and 5 years ago, which was performed as described.15 Briefly, whole-blood genomic DNA was isolated using the QIAsymphony (QIAGEN), and sequencing libraries were constructed by the HudsonAlpha Genomic Services Lab, using a standard protocol that included PCR amplification. Sequencing was performed on the Illumina HiSeqX using paired-end reads with a read length of 150 bp. Each genome was sequenced at an approximate mean depth of 30×, with at least 80% of base positions reaching 20× coverage. While originally analyzed using hg37, for this study reads were aligned to hg38 using DRAGEN version 07.011.352.3.2.8b. Variants were discovered (in gvcf mode) with DRAGEN, and joint genotyping was performed across six trios using GATK version 3.8-1-0-gf15c1c3ef. SVs were called using a combination of Delly (v0.6.01),16 CNVnator (v0.3.2),17 ERDS (v1.1),18 and Manta (v1.1.1.).19 Individual SVs were then annotated with gene features and allele frequencies from 1000 Genomes,20 gnomAD,21 NDD publications,22,23 and an internal SV database. We merged SVs from the various callers when they were of the same SV type and exhibited at least 50% reciprocal overlap. SVs that were only called by one caller were discarded unless they were >400 kb. MEIs were called using MELT (v2.02)24 run in MELT-SINGLE mode. Variant analysis and interpretation were performed using ACMG guidelines,14 similar to that which we previously performed.4,15 None of the probands had a pathogenic (P), likely pathogenic, or variant of uncertain significance (VUS) identified by IGS, either at the time of original analysis or after a reanalysis performed at the time of generation of long-read data. In all trios, expected relatedness was confirmed.25 IGS data for probands 1–5 are available via dbGAP (project accession number dbGAP: phs001089). Complete IGS data for proband 6 is not available due to consent restrictions.
Long-read sequencing, variant calling, analysis, and de novo assemblies
Long-read sequencing was performed using CCS mode on a PacBio Sequel II instrument (Pacific Biosciences of California). Libraries were constructed using a SMRTbell Template Prep Kit 1.0 and tightly sized on a SageELF instrument (Sage Science, Beverly, MA, USA). Sequencing was performed using a 30 h movie time with 2 h pre-extension, and the resulting raw data were processed using either the CCS3.4 or CCS4 algorithm, as the latter was released during the course of the study. Comparison of the number of high-quality insertion or deletion (indel) events in a read versus the number of passes confirmed that these algorithms produced comparable results. Probands were sequenced to an average CCS depth of 32× (range, 25× to 44×), while parents were covered at an average depth of 16× (range, 10× to 22×; Table 1). CCS reads were aligned to the complete GRCh38.p13 human reference. For single-nucleotide variants (SNVs) and indels, CCS reads were aligned using the Sentieon v.201808.07 implementation of the BWA-MEM aligner,26 and variants were called using DeepVariant v0.1027 and joint-genotyped using GLNexus v1.2.6.28 For SVs, reads were aligned using pbmm2 1.0.0, and SVs were called using pbsv v2.2.2. Candidate de novo SVs required a proband genotype of 0/1 and parent genotypes of 0/0, with ≥6 alternate reads in the proband and 0 alternate reads, and ≥5 reference reads in the parents.
Table 1.
Probands selected for PacBio sequencing
| Family ID | Proband gender | Race | Major phenotypic features | Previous genetic testing |
PacBio CCS coverage (P/D/M) | Average insert size (bp) (P/D/M) | |||
|---|---|---|---|---|---|---|---|---|---|
| Array | Single gene test(s) or panel(s)a | ES/GS | Other normal test results | ||||||
| 1 | F | C | seizures, facial dysmorphism, hypotonia | normal | normal ×2 | no findings (both) | karyotype | 25×/10×/11× | 12,655/12,238/12,884 |
| 2 | F | AA | ID, seizures, hypotonia | normal | normal ×7 | no findings (both) | mito | 26×/16×/12× | 12,651/12,865/12,600 |
| 3 | M | C | ID, seizures | VUS dup | normal ×3 | no findings (GS) | fragile X | 35×/19×/22× | 14,393/16,604/16,344 |
| 4 | F | C/AA | ID, facial dysmorphism, hypotonia | normal | normal ×1 | no findings (GS) | fragile X | 44×/14×/20× | 11,420/11,555/11,197 |
| 5 | M | C | ID, seizures, speech delay, brain MRI abnormalities | normal | normal ×4 | no findings (GS) | mito | 30×/16×/20× | 21,145/19,264/21,568 |
| 6 | F | C | ID, seizures, speech delay | normal | NP | no findings (GS) | NP | 33×/19×/14× | 12,452/12,183/13,641 |
ES/GS, exome sequencing/genome sequencing; P, proband; D, dad; M, mom; F, female; M, male; C, Caucasian; AA, African American; ID, intellectual disability; NP, not performed.
Some VUS SNVs have been reported in these probands.
For one proband (proband 4), we used several strategies to create de novo assemblies using 44× CCS data. Assemblies were generated using canu (v1.8),29 Falcon unzip (falcon-kit 1.8.1),30 HiCanu (hicanu_rc +325 changes [r9818 86bb2e221546c76437887d3a0ff5ab9546f85317]),31 and hifiasm (v 0.5-dirty-r247).32 Hifiasm was used to create two assemblies. First, the default parameters were used, followed by two rounds of Racon (v1.4.10) polishing of contigs. Second, trio-binned assemblies were built using the same input CCS reads, in addition to kmers generated from a 36× paternal Illumina library and a 37× maternal Illumina library (singletons were excluded). The kmers were generated using yak(r55) using the suggested parameters for running a hifiasm trio assembly (kmer size = 31 and Bloom filter size of 2∗∗37). Maternal and paternal contigs went through two rounds of Racon (v1.4.10) polishing. Trio-binned assemblies were built for the remaining probands in the same way. Individual parent assemblies were also built with hifiasm (v0.5-dirty-r247) using default parameters. The resulting contigs went through two rounds of Racon (v1.4.10) polishing.
Coordinates of breakpoints were defined by a combination of assembly-assembly alignments using minimap233 (followed by use of bedtools bamToBed), visual inspection of CCS read alignments, and BLAT. Rearranged segments in the chromosome 6 region were restricted to those >4 kb. Dot plots illustrating sequence differences were created using Gepard.34
QC statistics
SNV and indel concordance and de novo variant counts were calculated using bcftools v1.9 and rtg-tools vcfeval v3.9.1. “High-quality de novo” variants were defined as PASS variants (IGS/GATK only) on autosomes (on primary contigs only) that were biallelic with total alelle depth (DP) ≥ 7 and genotype quality (GQ) ≥ 35. Additional requirements were a proband genotype of 0/1, with ≥2 alternate reads and an allele balance ≥0.3 and ≤0.7. Required parent genotypes were 0/0, with alternate allele depth of 0. Mendelian error rates were also calculated using bcftools. “Rigorous” error rates were restricted to PASS variants (IGS/GATK only) on autosomes with GQ > 20, and DP > 5. Total variant counts per trio were calculated using Variant Effect Predictor (VEP, v98), counting multi-allelic sites as one variant. SV counts were calculated using bcftools and R. Counts were restricted to calls designated as “PASS,” with an alternate allele depth (AD) ≥ 2. Candidate SV de novos required proband genotype of 0/1 and parent genotypes of 0/0, with ≥6 alternate reads in the proband and 0 alternate reads and ≥5 reference reads in the parents. De novo MELT calls in IGS data were defined as isolated proband calls where the parent did not have the same type (ALU, L1, or SVA) of call within 1 kb as calculated by bedtools closest v.2.25.0. These calls were then filtered (using bcftools) for “PASS” calls and varying depths, defined as the number of read pairs supporting both sides of the breakpoint (left read pairs, LP; right read pairs, RP). To create a comparable set of de novo mobile element calls in CCS data, individual calls were extracted from the pbsv joint-called VCF using bcftools and awk and isolated proband calls were defined as they were for the IGS data and filtered (using bcftools) for PASS calls and varying depths, defined as the proband alternate allele depth (AD[1]).
Simple repeat and low-mappability regions
We generated a bed file of disease-related low-complexity repeat regions in 35 genes from previous studies.7,35 Most regions (25) include triplet nucleotide repeats, while the remainder include repeat units of 4–12 bp. Reads aligning to these regions were extracted from bwa-mem-aligned bams and visualized using the Integrated Genomics Viewer (IGV36). Proband depths of MAPQ60 reads spanning each region were calculated using bedtools multicov v2.28.0. For the depth calculations, regions were expanded by 15 bp on either side (using bedtools slop) to count reads anchored into non-repeat sequence. The mean length of these regions was 83 bp, with a max of 133 bp.
Low-mappability regions were defined as the regions of the genome that do not lie in Umap k100 mappable regions.37 Regions ≥100,000 nt long and those on non-primary contigs were removed, leaving a total of 242,222 difficult-to-map regions with average length of 411 bp. Proband depths of MAPQ60 reads spanning each region were calculated using bedtools multicov v2.28.0. High-quality protein-altering variants in probands were defined using VEP annotations and counted using bcftools v1.9. Requirements included a heterozygous or homozygous genotype in the proband, with ≥4 alternate reads, an allele balance ≥0.3 and ≤0.7, GQ > 20, and DP > 5. Reads supporting 57 loss-of-function variants (high quality and low quality) in proband 5 were visualized with IGV and semiquantitatively scored to assess call accuracy. Approximate counts of reads were recorded and grouped by mapping quality (MapQ = 0 and MapQ ≥ 1), along with subjective descriptions of the reads. The total evidence across CCS and IGS reads was used to estimate truth and score each variant call as true positive (TP), FP, true negative (TN), FN, or undetermined (UN).
CDKL5 cDNA amplicon sequencing
Total RNA was extracted from whole blood in PAXgene tubes using a PAXgene Blood RNA Kit version 2 (PreAnalytiX, #762164) according to the manufacturer’s protocol. cDNA was generated with a High-Capacity Reverse Transcription Kit (Applied Biosystems, #4368814) using 500 ng of extracted RNA from each individual as input. Primers were designed to CDKL5 exons 2, 5, and 6 to generate two amplicons spanning the potentially disrupted region of CDKL5 mRNA. Select amplicons were purified and sent to MCLAB (Molecular Cloning Laboratories, South San Francisco, CA, USA) for Sanger sequencing. See Supplemental methods for additional details, including primers.
Genomic DNA PCR to confirm relevant breakpoints in probands 4 and 6 and Alu insertions
We performed PCR to amplify products spanning junctions of various insertions and breakpoints, using the genomic DNA (gDNA) of the probands and parents as template. Select amplicons were purified and sent to MCLAB (Molecular Cloning Laboratories, South San Francisco, CA, USA) for Sanger sequencing. See Supplemental methods for additional details, including primers.
DGKB/MLLT3 qPCR
Total RNA was extracted from whole blood using a PAXgene Blood RNA Kit version 2 (PreAnalytiX, #762164), and cDNA was generated with a High-Capacity Reverse Transcription Kit (Applied Biosystems, #4368814) in an identical fashion as described for CDKL5 cDNA amplicon sequencing. For qPCR, Two TaqMan probes targeting the MLLT3 exon 3–4 and exon 9–10 splice junctions (ThermoFisher, Hs00971092_m1 and Hs00971099_m1) were used with cDNA diluted 1:5 in dH2O to perform qPCR for six replicates per sample on an Applied Biosystems Quant Studio 6 Flex. Differences in CT values from the median CT values for either an unrelated family or the proband’s parents were used to compute relative expression levels. See Supplemental methods for additional details, including primers.
Results
Affected probands and their unaffected parents were enrolled in a research study aimed at identifying genetic causes of NDDs.15 All trios were originally subject to IGS and analysis using ACMG standards14 to find pathogenic or likely pathogenic variants, or VUSs. Within the subset of probands for which no variants of interest (pathogenic, likely pathogenic, VUS) were identified either originally or after subsequent reanalyses,4,15 six trios were selected for sequencing using the PacBio Sequel II CCS approach (Table 1). These trios were selected for those with a strong suspicion of a genetic disorder, in addition to diversifying with respect to gender and ethnicity. Parents were sequenced, at a relatively reduced depth, to facilitate identification of de novo variation.
QC of CCS data
Variant calls from CCS data and IGS data were largely concordant (Table S1A). When comparing each individual’s variant calls in the Genome in a Bottle (GIAB) high-confidence regions38 between CCS and IGS, concordance was 94.63%, with higher concordance for SNVs (96.88%) than indels (75.96%). Concordance was slightly higher for probands only, likely due to the lower CCS read-depth coverage in parents. While CCS data showed a consistently lower number of SNV calls than IGS (mean = 7.0 M versus 7.45 M, per trio), more de novo SNVs at high QC stringency were produced in CCS data than IGS (mean SNVs = 89 versus 38; Tables S1B and S1C). CCS yielded far fewer de novo indels at these same thresholds (mean indels, 11 versus 148), with the IGS de novo indel count being much higher than biological expectation39 and likely mostly FP calls (Table S1C). In examining reads supporting variation that was uniquely called in each set, we found that CCS FP de novos were usually FN calls in the parent, due to lower genome-wide coverage in the parent and the effects of random sampling (i.e., sites at which there were 7 or more CCS reads in a parent that randomly happened to all derive from the same allele; Table S1C). Mendelian error rates in autosomes were lower in CCS data relative to IGS (harmonic mean of high-quality calls, 0.18% versus 0.34%; Table S1D), suggesting the CCS SNV calls are of higher accuracy, consistent with previously published data.13
Each trio had an average of ∼56,000 SVs among all three members, including an average of 59 candidate de novo SVs per proband (Table S1E). Trio SVs mainly represent insertions (48%) and deletions (43%), followed by duplications (6%), single breakends (BND) (3%), and inversions (<1%).
Trio-binned hifiasm de novo assemblies were built for each proband. The average N50 for proband trio-based assemblies was 35.4 Mb (Table S2A). Several assemblers were used to build de novo assemblies for one proband (proband 4). Canu, Falcon, and HiCanu all produced high-quality assemblies, but hifiasm assemblies were of highest quality (Table S2B). Use of trio-binned hifiasm allowed assembly of high-quality maternal- and paternal-specific contigs with an average N50 of 45.65 Mb, approaching that of hg38.
Variation in simple repeat regions
Accurate genotyping of simple repeat regions like trinucleotide repeat expansions presents a challenge in short-read data where the reads are often not long enough to span variant alleles. We assessed the ability of CCS to detect variation in these genomic regions and compared that to IGS, which in this case was produced from libraries produced with a PCR amplification step. We first examined variation in FMR1 (MIM: 309550). Expansion of a trinucleotide repeat in the 5′ UTR of FMR1 is associated with fragile X syndrome (MIM: 300624), the second-most common genetic cause of intellectual disability.40 Visualization of this region in all 18 individuals indicated insertions in all but two samples in the CGG repeat region of FMR1 relative to hg38, with a range of insertion sizes from 6–105 bp (Table S3; Figure S1). When manually inspecting these regions, while one or two major alternative alleles are clearly visible, there are often minor discrepancies in insertion lengths, often by multiples of 3. It is unclear if this represents true somatic variation or if this represents inaccuracy of consensus generation in CCS processing.
Like that for FMR1, manual curation of 34 other disease-causal repeat regions in each proband indicated that alignment of CCS reads provides a more accurate assessment of variation in these regions compared to IGS. When looking at region-spanning reads with high-quality alignment (mapQ = 60), 97% (34 of 35) of the regions were covered by at least 10 CCS reads in all six probands, as compared to 11% (4 of 35) of regions with high-quality IGS reads (Table S4A). While all query regions measured ≤144 bp (which includes an extension of 15 bp on either end of the repeat region), seven query regions were ≥100 bp. When considering only regions of interest <100 bp, 14% (4 of 28 regions) are covered by at least 10 high-quality IGS reads in each proband. Mean coverage of high-quality, region-spanning reads across probands was higher in CCS data than in IGS (29 versus 11; Table S4A). Of all repeat regions studied, none harbored variation classified as pathogenic/likely pathogenic/VUS.
We also compared coverage of high-quality CCS and IGS reads in low-mappability regions of the genome, specifically those that cannot be uniquely mapped by 100 bp kmers.37 While over half of these regions (62.5%) were fully covered by at least 10 high-quality CCS reads (mapQ = 60) in all six probands, only 19.3% of the regions met the same coverage metrics in the IGS data (Table S4B). The average CCS read depth in these regions was 26 reads, versus 8 reads in IGS. Within these regions, CCS yielded twice as many high-quality, protein-altering variants in each proband when compared to IGS (182 in CCS versus 85 in IGS) (Table S4C). Outside of the low-mappability regions, counts of protein-altering variants were similar (6,627 in CCS versus 6,759 in IGS).
To assess the accuracy of the protein-altering variant calls in low-mappability regions, we visualized reads for 57 loss-of-function variants detected by CCS, IGS, or both in proband 5 and used the totality of read evidence to score each variant as TP, FP, TN, FN, or UN. Six of these were “high-quality” calls (see Material and methods), and all of these were correctly called in CCS (TPs, 100%); in IGS, two were correctly called (TPs, 33%) and four were undetected (FNs, 67%) (Table S4D). Among all 57 unfiltered variant calls, most CCS calls were correct (29 TP, 15 TN, total 77%), while most IGS calls were incorrect (16 FP, 22 FN, total 67%) (Table S4E).
MEIs
We searched for MEIs in these six probands within the IGS data using MELT (Tables S5A and S5B)24 and within CCS data using pbsv (Tables S1E, S5C, and S5D; see also Material and methods). Our results suggest that CCS detection of MEIs is far more accurate. For example, it has been estimated that there exists a de novo Alu insertion in ∼1 in every 20 live births (mean of 0.05 per individual).41,42 However, at stringent QC filters (i.e., ≥5 read-pairs at both breakpoints, PASS, and no parental calls of the same MEI type within 1 kb), a total of 82 candidate de novo Alu insertions (average of 13.7) were called across the six probands using the IGS data (Table S5B), a number far larger than expected. Inspection of these calls indicated that most were bona fide heterozygous Alu insertions in the proband that were inherited but undetected in the parents. Filtering changes to improve sensitivity comes at a cost of elevated FP rates; for example, requiring only 2 supporting read pairs at each breakpoint leads to an average of ∼55 candidate de novo Alu insertions per proband (Table S5B). In contrast, using the CCS data and stringent QC filters (≥5 alternate reads, PASS, and no parental calls within 1 kb), we identified a total of only 6 candidate de novo Alu MEIs among the 6 probands (Table S5D), an observation that is far closer to biological expectation. We retained 4 candidate de novo Alu MEIs after further inspection of genotype and parental reference read depth (Table S1E). One of these 4 appears genuine, while the other three appear to be correctly called in the proband but missed in the parents owing to low read-depth, such that the Alu insertion-bearing allele was not covered by any CCS reads (Figure S2). Three of these four were confirmed by PCR, with PCR at the fourth yielding unclear results, and amplification and results were consistent with observations in IGV (Figure S3; Supplemental methods).
A likely pathogenic de novo SV in CDKL5
Analysis of SV calls and visual inspection of CCS data in proband 6 indicated a de novo SV within the CDKL5 gene (MIM: 300203; Figure 1). Given the de novo status of this event, the association of CDKL5 with early infantile epileptic encephalopathy 2 (EIEE2, MIM: 300672), and the overlap of disease with the proband’s phenotype (see Supplemental note), which includes intellectual disability, developmental delay, and seizures, we prioritized this event as the most compelling candidate variant in this proband.
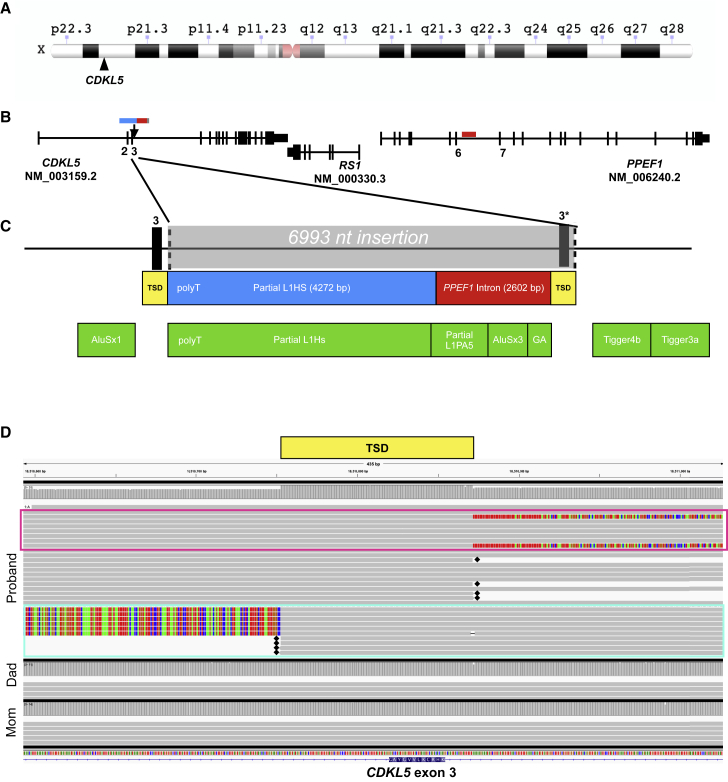
Figure 1.
Proband 6 has a de novo insertion resulting in duplication of exon 3 of CDKL5
(A) Ideogram showing location of CDKL5 on chromosome X. Ideogram is from the NCBI Genome Decoration Page.
(B) Gene structure of CDKL5, RS1, and PPEF1, indicating the location of the 6,993 bp insertion in CDKL5 (blue/red/gray bars) and location of the origin of the duplicated PPEF1 intronic sequence (red).
(C) Zoomed-in view of the insertion. The gray box indicates the entire 6,993 nt insertion, which consists of a partial L1HS retrotransposon (blue box), duplicated PPEF1 intronic sequence (red box), and target site duplication (TSD, yellow box) with duplicated exon 3 (3∗). Green boxes indicate RepeatMasker annotation of the proband’s insertion-bearing, contig sequence.
(D) Alignment of CCS reads near exon 3 of CDKL5 in IGV in proband 6 and her parents. Gray reads represent alignment to reference, and multicolor alignments represent unaligned ends of reads. The TSD is indicated by a yellow box. Reads highlighted by the pink box include examples of reads that align to reference upstream of the insertion, contain the TSD, and then have inserted sequence at their 3′ end. Those highlighted in the turquoise box represent inserted sequence, TSD, and reference sequence downstream of the insertion. Note that some reads have hard-clipped bases, which are designated with a black diamond.
A trio-based de novo assembly in this proband identified a 45.3 Mb paternal contig and a 50.6 Mb maternal contig in the region surrounding CDKL5. While these contigs align linearly across the majority of the p arm of chromosome X (Figure S4), alignment of the paternal contig to GRCh38 revealed a heterozygous 6,993 bp insertion in an intron of CDKL5 (chrX: 18,510,871–18,510,872_ins6993 [GenBank: GRCh38]; Figure 1; Figure S5). Analysis of SNVs in the region surrounding the insertion confirm that it lies on the proband’s paternal allele. However, mosaicism is suspected, as there exist paternal haplotype reads within the proband that do not harbor the insertion (5 of 8 paternal reads without the insertion at the 5′ end of the event, and 7 of 16 paternal reads without insertion at the 3′ end of the event; Figure S6).
Annotation of the insertion indicated that it contains three distinct segments: 4,272 bp of a retrotransposed, 5′ truncated L1HS mobile element (including a poly[A] tail), 2,602 bp of sequence identical to an intron of the nearby PPEF1 gene (g.18738310_18740911 [GenBank: NC_000023.11]; [c.235+4502_235+7103 (GenBank: NM_006240.2)]), and a 119 bp target-site duplication (TSD) that includes a duplicated exon 3 of CDKL5 (35 bp) and surrounding intronic sequence (chrX: 18510753–18510871 [GenBank: GRCh38]; [c.65–67 (GenBank: NM_003159.2) to c.99+17 (GenBank: NM_003159.2)]; 119 bp total) (Figure 1; Figure S7). The 2,602 bp copy of PPEF1 intronic sequence includes the 5′ end (1,953 bp) of an L1PA5 element that is ∼6.5% divergent from its consensus L1, an AluSx element, and additional repetitive and non-repetitive intronic sequence. The size and identity of this insert in the proband, and absence in both parents, was confirmed by PCR amplification and partially confirmed by Sanger sequencing (see Supplemental methods; Figure S7).
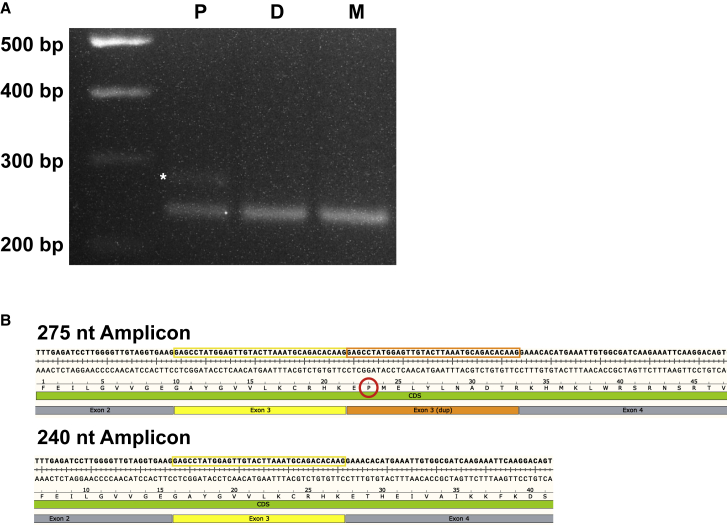
Exon 3 of CDKL5, which lies within the target-site duplication of the L1-mediated insertion, is a coding exon that is 35 bp long; inclusion of a second copy of exon 3 into CDKL5 mRNA is predicted to lead to a frameshift (Thr35ProfsTer52; Figure 2). To determine the effect of this insertion on CDKL5 transcripts, we performed RT-PCR from RNA isolated from each member of the trio. Using primers designed to span from exon 2 to exon 5, all three members of the trio had an expected amplicon of 240 bp. However, the proband had an additional amplicon of 275 bp (Figure 2A). Sanger sequencing of this amplicon indicated that a duplicate exon 3 was spliced into this transcript (Figure 2B). The presence of transcripts with a second copy of exon 3 strongly supports the hypothesis that the variant leads to a CDKL5 loss-of-function effect in the proband.
Figure 2.
The duplicated CDKL5 exon 3 is present in a subset of the proband’s CDKL5 transcripts
(A) RT-PCR using primers specific to exons 2–5 of CDKL5 cDNA results in a 240 bp amplicon in proband (P), dad (D), and mom (M). An additional 275 bp amplicon is present only in the proband (asterisk).
(B) Sanger sequencing of both amplicons from the proband confirmed that the 240 bp amplicon includes the normal, expected sequencing and inclusion of a duplicated exon 3 in the upper, 275 bp band. This is predicted to lead to a frameshift (red circle) and downstream stop, p.Thr35ProfsTer52. Yellow outlined box, exon 3 sequence; orange outlined box, duplicated exon 3 sequence.
Multiple large de novo SVs in proband 4
Analysis of SV calls in proband 4 indicated several large, complex, de novo events affecting multiple chromosomes (6, 7, and 9). To assess the structure of the proband’s derived chromosomes, we inspected the trio-binned de novo assembly for this proband.
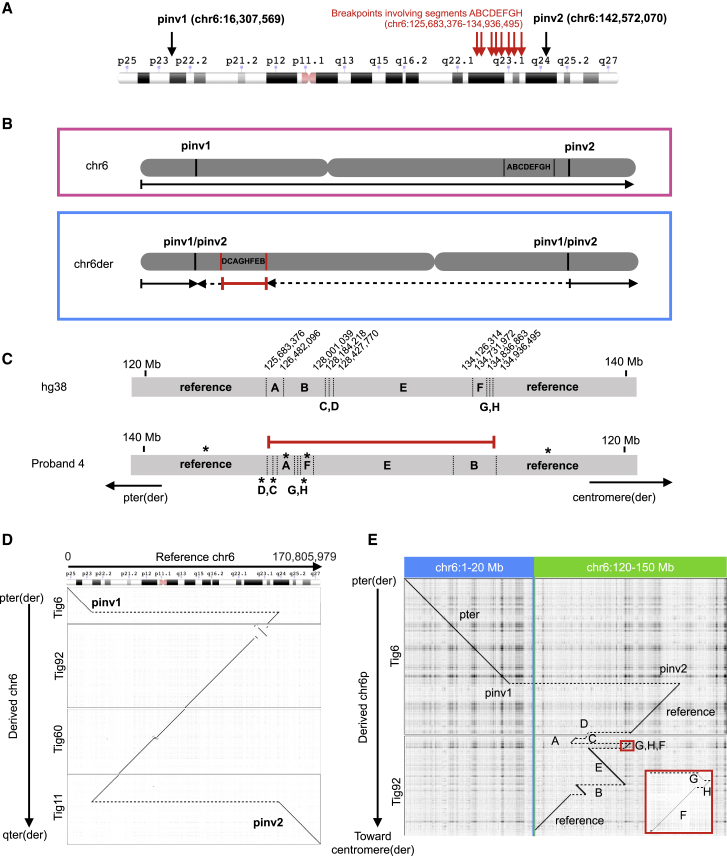
Four paternal contigs were assembled for chromosome 6, which showed many structural changes compared to reference chromosome 6 (Figure 3). The proband harbors a pericentric inversion, with breakpoints at chr6: 16,307,569 (6p22.3) and chr6: 142,572,070 (6q24.2; Figures 3A and 3B; Table S6A). In addition, a 9.3 Mb region near 6q22.31–6q23.3 contained at least eight additional breakpoints, with local rearrangement of eight segments, some of which are inverted (ABCDEFGH in reference versus DCAGHFEB; Figure 3C; Table S6B). The median fragment size is just over 400 kb (range, 99 kb to 5.7 Mb; Table S6B). While the ends of several fragments do overlap annotated repeats, many do not. We were not able to identify microhomology at the junctions of these eight segments, the majority of which (7/8) were PCR confirmed in the proband (Table S6B; Figures S8 and S9; Supplemental methods). Together, the 10 breakpoints identified across chromosome 6 are predicted to disrupt at least six genes, five of which are annotated as protein coding (Table S6A). None of these have been associated with neurodevelopmental disease.
Figure 3.
Proband 4 has several large structural changes on chromosome 6
(A) Ideogram with annotation of chromosome 6 breakpoints identified in proband 4, including pericentric inversion breakpoints (pinv1, pinv2) and multiple breakpoints of a complex genomic rearrangement (red arrows). Ideogram is from the NCBI Genome Decoration Page.
(B) Schematic of proband 4’s maternal (pink box) and paternal (blue box) chromosome 6 structures. The maternal structure matches reference, while the paternally inherited derived chromosome 6 has pericentric inversion breakpoints (pinv1/pinv2) and a complex cluster of rearranged fragments (DCAGHFEB).
(C) Zoomed-in view of (B), showing the schematic of additional fragmentation near 6q22.31–6q23.3 (vertical dashed lines). Asterisks indicate inverted sequence as compared to hg38 reference. See Table S6 for additional breakpoint coordinates and details.
(D) Alignment of four sequential paternal contigs to reference chromosome 6 identified a pericentric inversion spanning 6p22.3 to 6q24.2 and a 9.3 Mb region near 6q22.31–6q23.3 with several additional breaks.
(E) Zoomed-in view of (D), showing additional fragmentation near 6q22.31–6q23.3.
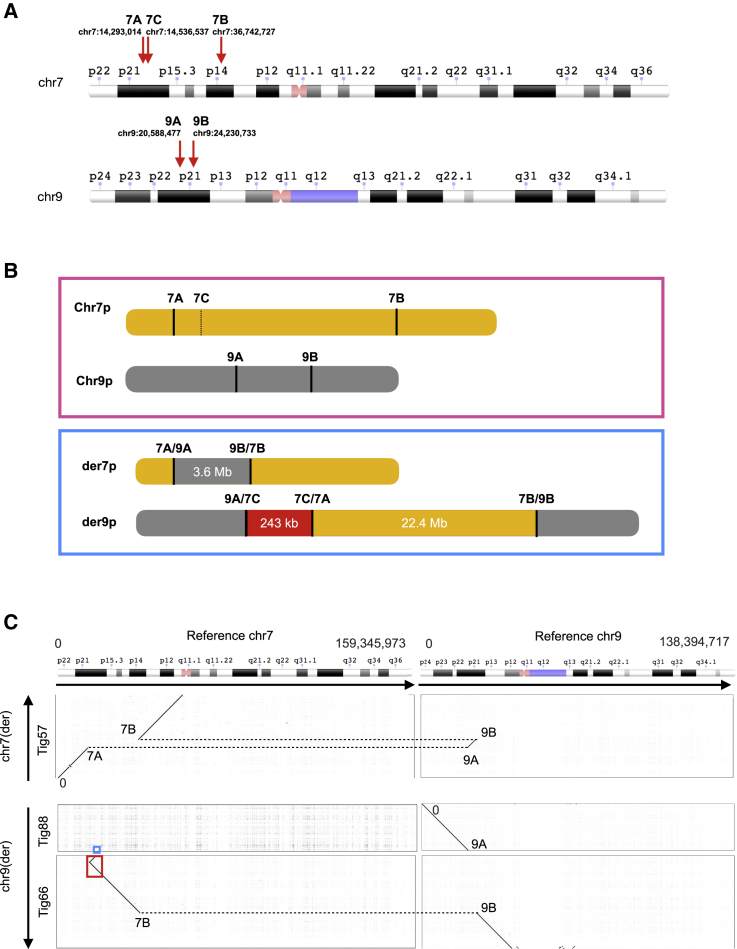
CCS reads and contigs from the de novo paternal assembly of proband 4 also support structural variation involving chromosomes 7 and 9, with five breakpoints (Figure 4). The proband has two insertional translocations in addition to an inversion at the 5′ end of the chromosome 7 sequence within the derived 9p arm. Manual curation of SNVs surrounding all breakpoints confirmed that all variation lies on the paternal allele, and no mosaicism is suspected. Manual curation of the proband’s de novo assembly (specifically tig66) was required to resolve an assembly artifact (Figure S10; Supplemental methods).
Figure 4.
Proband 4 has two insertional translocations between chromosomes 7 and 9 and an inversion
(A) Ideogram with annotation of chromosome 7 and 9 breakpoints identified in proband 4. Ideograms are from the NCBI Genome Decoration Page.
(B) Schematic of the proband’s maternal (pink box) and paternal (blue box) p arms of chromosomes 7 and 9. The proband’s maternal alleles match reference. The paternal sequences represent the outcome of translocations (7A;9A and 7B;9B) and inversion (7A;7C), with fragment sizes shown. The red fragment in paternal der9p is inverted with respect to hg38 reference.
(C) Alignment of three paternal contigs to reference chromosomes 7 and 9 identified two insertional translocations. See Figure S6 and Supplemental methods regarding blue and red boxed areas.
The net effect of the translocations and inversion is likely disruption of two protein-coding genes: DGKB (MIM: 604070) on chromosome 7 and MLLT3 (MIM: 159558) on chromosome 9, neither of which has been associated with disease (Table S6A). To determine if MLLT3 transcripts are disrupted in this proband, we performed qPCR using RNA from each member of the trio, in addition to three unrelated individuals (family 3). Using two validated TaqMan probes near the region of interest (exons 3–4 and exons 9–10), we found that proband 4 showed a ∼35%–39% decrease in MLLT3 compared to her parents and a 38%–45% decrease relative to unrelated individuals (Figure S11; Table S7). Expression of DGKB was not examined, as the gene is not expressed at appreciable levels in blood.43
Analysis of CCS-detected SVs in IGS reads
None of the disease-associated variation described here and detected by CCS analysis was identified in our IGS analyses. We analyzed raw variant calls and IGS reads at each of the relevant breakpoints to determine why such variants were not detected (Figures S12–S15).
In the case of CDKL5, MELT did not call any L1, SVA, or Alu-mediated insertions with 1 Mb of CDKL5. This is likely, at least in part, because the insertion is L1-mediated but has a non-L1 sequence at one breakpoint. However, in retrospectively searching for structural variation near CDKL5 from our standard SV pipeline, we found that Delly and Manta both called a 230 kb duplication event in CDKL5. The call passed our frequency filters and was flagged as de novo. However, upon inspection, read depth and allele ratios clearly did not support a duplication event (Figure S16). Retrospectively, it is clear that this “230 kb duplication” call resulted from the duplication and insertion of a segment of PPEF1 intronic sequence into the CDKL5 intron. However, the Delly and Manta calls are plainly not correct and at the time of initial IGS analysis were disregarded.
In the case of the multiple complex breakpoints identified in proband 4, most of the breakpoints were in fact called as BND or inversions by Manta (Table S6). However, Manta is the only tool capable of detecting such variation, and our pipeline requires concordance from at least two callers for small SVs (see Material and methods); thus, these events were disregarded. Furthermore, it is important to note that the proband had 814 potentially de novo BND/inversion calls from Manta, a number that is indicative of an untenably high number of false de novo calls (be they inherited or simply FP variants). In addition, typical strategies to curate and interpret candidate variation, including filtration using population frequencies, are unavailable for these categories of variation. The net result is that these variants were not evaluated in our routine analysis process. Lastly, even to the extent that individual breakpoints were flagged in IGS analysis, the lack of a coherent assembly of how the individual breakpoints and fragments relate to one another would have precluded meaningful evaluation.
Discussion
Here we describe CCS long-read sequencing of six probands with NDDs who had previously undergone extensive genetic testing with no variants found to be relevant to disease. Generally, the CCS genomes appeared to be highly comprehensive and accurate in terms of variant detection, facilitating detection of a diversity of variant types across many loci, including those that prove challenging to analysis with short reads. Detection of simple-repeat expansions and variants within low-mappability regions, for example, was more accurate and comprehensive in CCS data than that seen in IGS, and many complex SVs were plainly visible in CCS data but missed by IGS.
Given the importance of de novo variation in rare disease diagnostics, especially for NDDs, it is also important to note the qualities of discrepant de novo calls between the two technologies. We found that most of the erroneously called de novo variants in the CCS data were correctly called as heterozygous in the proband but missed in the parents due to lower coverage and random sampling effects such that the variant allele was simply not covered by any reads in the transmitting parent. Such errors could be mitigated by sequencing parents more deeply. In contrast, de novo variants unique to IGS were enriched for systematic artifacts that cannot be corrected for with higher read-depth. Indels, for example, are a well-known source of error and heavily enriched among IGS de novo variant calls.
In one proband we identified a likely pathogenic, de novo L1-mediated insertion in CDKL5. CDKL5 encodes cyclin-dependent kinase-like 5, a serine-threonine protein kinase that plays a role in neuronal morphology, possibly via regulation of microtubule dynamics.44 Variation in CDKL5 has been associated with EIEE2 (MIM: 300672), an X-linked dominant syndrome characterized by infantile spasms, early-onset intractable epilepsy, hypotonia, and variable additional Rett-like features.45,46 CDKL5 is one of the most commonly implicated genes identified by ES/GS in individuals with epilepsy.47 SNVs, small insertions and deletions, copy-number variants (CNVs), and balanced translocations have all been identified in affected individuals, each supporting a haploinsufficiency model of disease.48 We also note that de novo SVs, including deletions and at least one translocation, have been reported with a breakpoint in intron 3, near the breakpoint identified here48, 49, 50, 51 (Table S8; Figure S17). The variant observed here appears to be mosaic, and we note that a recent study found that 8.8% of previously reported CDKL5 mutations are also mosaic.52 While most such mutations have been identified in males rather than females, noting that pathogenic CDKL5 variation is often lethal in males, there is not an obvious relationship between phenotypic severity, gender, variant type, and mosaicism.53
The variant harbors two classic marks of an L1HS insertion, including the preferred L1 EN consensus cleavage site (5′-TTTT/G-3′), and a 119-bp TSD, which, in this case, includes exon 3 of CDKL5. Although TSDs are often fewer than 50 bp long, TSDs up to 323 bp have been detected.54 The variant appears to be a chimeric L1 insertion. The 3′ end of the insertion represents retrotransposition of an active L1HS mobile element, with a signature poly(A) tail. However, the 5′ portion of the L1 sequence has greater identity to an L1 sequence within an intron of PPEF1, which lies about 230 kb downstream of CDKL5. Additional non-L1 sequence at the 5′ end of the insertion is identical to an intronic segment of PPEF1. While transduction of sequences at the 3′ end of L1 sequence has been described,55 the PPEF1 intronic sequence here lies at the 5′ end of the L1. A chimeric insertion similar to that observed here has been described previously and has been proposed to result from a combination of retrotransposition and a synthesis-dependent strand annealing (SDSA)-like mechanism.54
Using ACMG variant classification guidelines, we classified this variant as likely pathogenic. The variant was experimentally confirmed to result in frameshifted transcripts due to exon duplication and was shown to be de novo, allowing for use of both the PVS1 (loss of function)56 and PM2 (de novo)57 evidence codes. Use of likely pathogenic, as opposed to pathogenic, reflects the uncertainty resulting from the intrinsically unusual nature of the variant and its potential somatic mosaicism, in addition to the fact that its absence from population variant databases is not in principle a reliable indicator of true rarity. Identification of additional MEIs and other complex SVs in other individuals will likely aid in disease interpretation by both facilitating more accurate allele frequency estimation and by improving interpretation guidelines.
More generally, MEIs have been previously described as a pathogenic mechanism of gene disruption, but their contribution to developmental disorders has been limited to a modest number of individuals in a few studies, each of which report pathogenic/likely pathogenic variation lying within coding exons.9,10 However, the MEI observed here in CDKL5 would likely be missed by exome sequencing as the breakpoints are intronic, and in fact it was also missed in our previous short-read genome sequencing analysis.15 Global analyses of MEIs, such as our assessment of de novo Alu insertion rates (Table S5), also support the conclusion that MEI events are far more effectively detected within CCS data compared to that seen in short-read genomes. We find it likely that long-read sequencing will uncover MEIs that disrupt gene function and lead to NDDs in many currently unexplained cases.
CCS data also led to the detection of multiple large, complex, de novo SVs in proband 4, affecting at least three chromosomes. Both complex chromosomal rearrangements (CCRs), which involve at least three cytogenetically visible breakpoints on two or more chromosomes, and complex genomic rearrangements (CGRs), which are often on a smaller scale but more complex, have been reported in individuals with NDDs or other congenital anomalies.58, 59, 60, 61 Proband 4 appears to have both a CGR and a CCR, the latter of which includes insertional translocations and an inversion on chromosomes 7 and 9. The CGR consists of local rearrangement of eight segments near 6q22.31–6q23.3 and appears to represent chromothripsis, as the segments are localized, do not have microhomology at their breaks, and show no significant copy gain or loss in the region (Figure S18), all of which are characteristics of chromothripsis.62 The location of this cluster near one of the breakpoints of the pericentric inversion is consistent with observations that missegregated chromosomes can undergo micronucleus formation and shattering.63 However, we cannot rule out other related mechanisms under the umbrella term of chromoanagenesis.64
One of the most compelling disease causal candidate genes affected in proband 4 is MLLT3, which is predicted to be moderately intolerant to loss-of-function variation (pLI = 1, o/e = 0 [0–0.13];21 RVIS = 21.1%65). MLLT3, also known as AF9, undergoes somatic translocation with the MLL gene, also known as KMT2A (MIM: 159555), in individuals with acute leukemia; pathogenicity in these cases results from expression of an in-frame KMT2A-MLLT3 fusion protein and subsequent deregulation of target HOX genes.66 Balanced translocations between chromosome 4 and chromosome 9, resulting in disruption of MLLT3, have been previously reported in two individuals, each with NDDs including intractable seizures.67,68 Although proband 4 does not exhibit seizures, she does have features that overlap the described probands, including speech delay, hypotonia, and fifth-finger clinodactyly.
While we cannot be certain of the pathogenic contribution of any one SV in proband 4, we consider the number, size, and extent of de novo structural variation to be likely pathogenic. ACMG recommendations on the interpretation of copy number variation were recently published, and although the events in proband 4 appear to be copy neutral, we attempted to apply modifications of these guidelines to these events.69 The most compelling evidence for pathogenicity of these events is their de novo status (evidence code 5A); disruption of at least six protein-coding genes at the breakpoints (3A), at least one of which is predicted to be haploinsufficient (2H); and the total number and genomic extent of large SVs. While several of these can be captured by current evidence codes, they are weakened by the lack of affected disease-associated genes and the lack of a highly specific phenotype in the proband. Further, although the SVs are large events, including a shattering of a >9 Mb region of the genome, we do not know the molecular effect on genes that are nearby but not spanning the breakpoints. Identification of additional complex structural variation like that in this proband will aid in development of additional guidelines for classification of these events.
Retrospective analysis of the disease-associated events described here did identify reads in the IGS data that support the majority of the breakpoints (Figures S12–S15; Table S6). However, there are multiple reasons why these events were not originally identified by our standard IGS analyses, including discrepancies among calling algorithms, incorrect or incomplete descriptions of the sizes and natures of the events, and filtration steps that are required to make IGS interpretation pipelines effective and sustainable.
We note that our sample size, with only six total trios and two individuals with clinically relevant discoveries, is clearly too small to make precise predictions about the diagnostic yield of long-read sequencing. However, we believe the yield will be substantial. As a baseline, it is likely to be at least as high as that from short reads, given that there is no evidence of a sensitivity loss for short-read-detectable variation (e.g., SNVs and short indels). The key unknown is thus the additional yield from long-read sequencing in cases that harbor no clinically relevant variation detected by short-read sequencing. In that light, our observations are inconsistent with a very low yield. If we were to assume, as an example, that the true yield for long reads in unsolved cases is only 1%, it is unlikely that we would have observed 2 successes in 6 individuals (p = 0.0015, binomial test). Of course, the 6 unsolved probands were not randomly sampled from the set of all unsolved probands, and small counts are always intrinsically uncertain. Thus, studies of larger cohorts are necessary to estimate the magnitude of increased diagnostic yield from long-read genome sequencing.
In addition to the need for larger studies, it is also important to consider factors like costs and DNA input requirements, which remain obstacles to widespread adoption of long-read genome sequencing. Additionally, refining and optimizing computational pipelines and establishing benchmarks and quality-control metrics will also be necessary. That said, there have been considerable improvements, especially recently, on cost and DNA input requirements,70 and the computational and analytical challenges, while non-trivial, are tractable.
Considering the evidence supporting the superior variant detection ability of long reads presented here and elsewhere,70,71 we believe that the overall diagnostic yield for long reads will prove to be substantially better than current yields and that long-read genome analysis will supplant short-read analysis of individuals with rare disease in the coming years.
Acknowledgments
This work was supported by a grant from the National Human Genome Research Institute (UM1HG007301). Some reagents were provided by PacBio as part of an early-access testing program. We thank our colleagues at HudsonAlpha who provided advice and general support, including Amy Nesmith Cox, Greg Barsh, Kelly East, Whitley Kelley, David Bick, and Elaine Lyon, in addition to the HudsonAlpha Genomic Services Laboratory and Clinical Services Laboratory. We also thank the clinical team at North Alabama Children’s Specialists. Finally, we are grateful to the families who participated in this study.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2021.100023.
Data and code availability
All relevant variant data are supplied within the paper or in supporting files. Complete IGS data for probands 1–5 are available via dbGAP (dbGAP: phs001089.v3.p1). CCS data for these proband will also be available via dbGAP under the same project. Complete IGS and CCS data for proband 6 are not available due to privacy and IRB reasons.
Web resources
Bcftools, https://samtools.github.io/bcftools/
Bedtools2, https://github.com/arq5x/bedtools2
Burrows-Wheeler Aligner (bwa), http://bio-bwa.sourceforge.net/
CNVnator, https://github.com/abyzovlab/CNVnator
dbGAP, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001089.v3.p1
DeepVariant, https://github.com/google/deepvariant
Ensembl Variant Effect Predictor (VEP), https://useast.ensembl.org/info/docs/tools/vep/index.html
Genome Analysis Toolkit (GATK), https://gatk.broadinstitute.org/hc/en-us
Genome Pair Rapid Dotter (Gepard), https://github.com/univieCUBE/gepard
gnomAD Genome Aggregation Database, https://gnomad.broadinstitute.org/
Integrative Genomics Viewer (IGV), http://software.broadinstitute.org/software/igv/
Mobile element locator tool (MELT), https://melt.igs.umaryland.edu/
NCBI Genome Decoration Page, https://www.ncbi.nlm.nih.gov/genome/tools/gdp
OMIM, https://www.omim.org
Pacific Biosciences/CCS, https://github.com/PacificBiosciences/ccs
Pacific Biosciences/pbmm2, https://github.com/PacificBiosciences/pbmm2
Pacific Biosciences/pbsv, https://github.com/PacificBiosciences/pbsv
RealTimeGenomics/rtg-tools, https://github.com/RealTimeGenomics/rtg-tools
Sentieon, https://github.com/Sentieon
The R Project for Statistical Computing (R), http://www.r-project.org
Umap k100 mappable regions, https://bismap.hoffmanlab.org/
Supplemental information
References
- 1.Ropers H.H. Genetics of intellectual disability. Curr. Opin. Genet. Dev. 2008;18:241–250. doi: 10.1016/j.gde.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Vissers L.E., de Ligt J., Gilissen C., Janssen I., Steehouwer M., de Vries P., van Lier B., Arts P., Wieskamp N., del Rosario M., et al. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 3.Wellcome Sanger Institute. D.D.D. Development Disorder Genotype - Phenotype Database. https://decipher.sanger.ac.uk/ddd/ddgenes
- 4.Hiatt S.M., Amaral M.D., Bowling K.M., Finnila C.R., Thompson M.L., Gray D.E., Lawlor J.M.J., Cochran J.N., Bebin E.M., Brothers K.B., et al. Systematic reanalysis of genomic data improves quality of variant interpretation. Clin. Genet. 2018;94:174–178. doi: 10.1111/cge.13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark M.M., Stark Z., Farnaes L., Tan T.Y., White S.M., Dimmock D., Kingsmore S.F. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom. Med. 2018;3:16. doi: 10.1038/s41525-018-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niemi M.E.K., Martin H.C., Rice D.L., Gallone G., Gordon S., Kelemen M., McAloney K., McRae J., Radford E.J., Yu S., et al. Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature. 2018;562:268–271. doi: 10.1038/s41586-018-0566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMurray C.T. Expansions in simple DNA repeats underlie ∼20 severe neuromuscular and neurodegenerative disorders. Nat. Publ. Gr. 2010;11:786–799. [Google Scholar]
- 8.Asadollahi R., Oneda B., Joset P., Azzarello-Burri S., Bartholdi D., Steindl K., Vincent M., Cobilanschi J., Sticht H., Baldinger R., et al. The clinical significance of small copy number variants in neurodevelopmental disorders. J. Med. Genet. 2014;51:677–688. doi: 10.1136/jmedgenet-2014-102588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torene R.I., Galens K., Liu S., Arvai K., Borroto C., Scuffins J., Zhang Z., Friedman B., Sroka H., Heeley J., et al. Mobile element insertion detection in 89,874 clinical exomes. Genet. Med. 2020;22:974–978. doi: 10.1038/s41436-020-0749-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner E.J., Prigmore E., Gallone G., Danecek P., Samocha K.E., Handsaker J., Gerety S.S., Ironfield H., Short P.J., Sifrim A., et al. Contribution of retrotransposition to developmental disorders. Nat. Commun. 2019;10:4630. doi: 10.1038/s41467-019-12520-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahmoud M., Gobet N., Cruz-Dávalos D.I., Mounier N., Dessimoz C., Sedlazeck F.J. Structural variant calling: the long and the short of it. Genome Biol. 2019;20:246. doi: 10.1186/s13059-019-1828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mantere T., Kersten S., Hoischen A. Long-Read Sequencing Emerging in Medical Genetics. Front. Genet. 2019;10:426. doi: 10.3389/fgene.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wenger A.M., Peluso P., Rowell W.J., Chang P.C., Hall R.J., Concepcion G.T., Ebler J., Fungtammasan A., Kolesnikov A., Olson N.D., et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019;37:1155–1162. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowling K.M., Thompson M.L., Amaral M.D., Finnila C.R., Hiatt S.M., Engel K.L., Cochran J.N., Brothers K.B., East K.M., Gray D.E., et al. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med. 2017;9:43. doi: 10.1186/s13073-017-0433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rausch T., Zichner T., Schlattl A., Stütz A.M., Benes V., Korbel J.O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abyzov A., Urban A.E., Snyder M., Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu M., Need A.C., Han Y., Ge D., Maia J.M., Zhu Q., Heinzen E.L., Cirulli E.T., Pelak K., He M., et al. Using ERDS to infer copy-number variants in high-coverage genomes. Am. J. Hum. Genet. 2012;91:408–421. doi: 10.1016/j.ajhg.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 20.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coe B.P., Witherspoon K., Rosenfeld J.A., van Bon B.W.M., Vulto-van Silfhout A.T., Bosco P., Friend K.L., Baker C., Buono S., Vissers L.E.L.M., et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper G.M., Coe B.P., Girirajan S., Rosenfeld J.A., Vu T.H., Baker C., Williams C., Stalker H., Hamid R., Hannig V., et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner E.J., Lam V.K., Harris D.N., Chuang N.T., Scott E.C., Pittard W.S., Mills R.E., Devine S.E., 1000 Genomes Project Consortium The Mobile Element Locator Tool (MELT): population-scale mobile element discovery and biology. Genome Res. 2017;27:1916–1929. doi: 10.1101/gr.218032.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kendig K.I., Baheti S., Bockol M.A., Drucker T.M., Hart S.N., Heldenbrand J.R., Hernaez M., Hudson M.E., Kalmbach M.T., Klee E.W., et al. 2018. Computational performance and accuracy of Sentieon DNASeq variant calling workflow. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poplin R., Chang P.C., Alexander D., Schwartz S., Colthurst T., Ku A., Newburger D., Dijamco J., Nguyen N., Afshar P.T., et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018;36:983–987. doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 28.Lin M.F., Rodeh O., Penn J., Bai X., Reid J.G., Krasheninina O., Salerno W.J. 2018. GLnexus: joint variant calling for large cohort sequencing. bioRxiv. [DOI] [Google Scholar]
- 29.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin C.S., Peluso P., Sedlazeck F.J., Nattestad M., Concepcion G.T., Clum A., Dunn C., O’Malley R., Figueroa-Balderas R., Morales-Cruz A., et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nurk S., Walenz B., Rhie A., Vollger M., Logsdon G., Grothe R., Miga K., Eichler E., Phillippy A., Koren S. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. bioRxiv. 2020 doi: 10.1101/gr.263566.120. 2020.03.14.992248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng H., Concepcion G.T., Feng X., Zhang H., Li H. 2020. Haplotype-resolved de novo assembly with phased assembly graphs. arXiv, 2008.01237v1.https://arxiv.org/abs/2008.01237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krumsiek J., Arnold R., Rattei T. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 35.Khristich A.N., Mirkin S.M. On the wrong DNA track: Molecular mechanisms of repeat-mediated genome instability. J. Biol. Chem. 2020;295:4134–4170. doi: 10.1074/jbc.REV119.007678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson J.T., Thorvaldsdóttir H., Wenger A.M., Zehir A., Mesirov J.P. Variant review with the integrative genomics viewer. Cancer Res. 2017;77:e31–e34. doi: 10.1158/0008-5472.CAN-17-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karimzadeh M., Ernst C., Kundaje A., Hoffman M.M. Umap and Bismap: quantifying genome and methylome mappability. Nucleic Acids Res. 2018;46:e120. doi: 10.1093/nar/gky677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zook J.M., McDaniel J., Olson N.D., Wagner J., Parikh H., Heaton H., Irvine S.A., Trigg L., Truty R., McLean C.Y., et al. An open resource for accurately benchmarking small variant and reference calls. Nat. Biotechnol. 2019;37:561–566. doi: 10.1038/s41587-019-0074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A., et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rousseau F., Rouillard P., Morel M.L., Khandjian E.W., Morgan K. Prevalence of carriers of premutation-size alleles of the FMRI gene--and implications for the population genetics of the fragile X syndrome. Am. J. Hum. Genet. 1995;57:1006–1018. [PMC free article] [PubMed] [Google Scholar]
- 41.Xing J., Zhang Y., Han K., Salem A.H., Sen S.K., Huff C.D., Zhou Q., Kirkness E.F., Levy S., Batzer M.A., Jorde L.B. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feusier J., Watkins W.S., Thomas J., Farrell A., Witherspoon D.J., Baird L., Ha H., Xing J., Jorde L.B. Pedigree-based estimation of human mobile element retrotransposition rates. Genome Res. 2019;29:1567–1577. doi: 10.1101/gr.247965.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., et al. GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbiero I., Peroni D., Siniscalchi P., Rusconi L., Tramarin M., De Rosa R., Motta P., Bianchi M., Kilstrup-Nielsen C. Pregnenolone and pregnenolone-methyl-ether rescue neuronal defects caused by dysfunctional CLIP170 in a neuronal model of CDKL5 Deficiency Disorder. Neuropharmacology. 2020;164:107897. doi: 10.1016/j.neuropharm.2019.107897. [DOI] [PubMed] [Google Scholar]
- 45.Bahi-Buisson N., Nectoux J., Rosas-Vargos H., Milh M., Boddaert N., Girard B., Cances C., Ville D., Afenjar A., Rio M., et al. Key clinical features to identify girls with CDKL5 mutations. Brain. 2008;131:2647–2661. doi: 10.1093/brain/awn197. [DOI] [PubMed] [Google Scholar]
- 46.Kadam S.D., Sullivan B.J., Goyal A., Blue M.E., Smith-Hicks C. Rett syndrome and CDKL5 deficiency disorder: From bench to clinic. Int. J. Mol. Sci. 2019;20:5098. doi: 10.3390/ijms20205098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Symonds J.D., McTague A. Epilepsy and developmental disorders: Next generation sequencing in the clinic. Eur. J. Paediatr. Neurol. 2020;24:15–23. doi: 10.1016/j.ejpn.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 48.Erez A., Patel A.J., Wang X., Xia Z., Bhatt S.S., Craigen W., Cheung S.W., Lewis R.A., Fang P., Davenport S.L.H., et al. Alu-specific microhomology-mediated deletions in CDKL5 in females with early-onset seizure disorder. Neurogenetics. 2009;10:363–369. doi: 10.1007/s10048-009-0195-z. [DOI] [PubMed] [Google Scholar]
- 49.Bartnik M., Derwińska K., Gos M., Obersztyn E., Kołodziejska K.E., Erez A., Szpecht-Potocka A., Fang P., Terczyńska I., Mierzewska H., et al. Early-onset seizures due to mosaic exonic deletions of CDKL5 in a male and two females. Genet. Med. 2011;13:447–452. doi: 10.1097/GIM.0b013e31820605f5. [DOI] [PubMed] [Google Scholar]
- 50.Córdova-Fletes C., Rademacher N., Müller I., Mundo-Ayala J.N., Morales-Jeanhs E.A., García-Ortiz J.E., León-Gil A., Rivera H., Domínguez M.G., Kalscheuer V.M. CDKL5 truncation due to a t(X;2)(p22.1;p25.3) in a girl with X-linked infantile spasm syndrome. Clin. Genet. 2010;77:92–96. doi: 10.1111/j.1399-0004.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- 51.Sanchis-Juan A., Stephens J., French C.E., Gleadall N., Mégy K., Penkett C., Shamardina O., Stirrups K., Delon I., Dewhurst E., et al. Complex structural variants in Mendelian disorders: identification and breakpoint resolution using short- and long-read genome sequencing. Genome Med. 2018;10:95. doi: 10.1186/s13073-018-0606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stosser M.B., Lindy A.S., Butler E., Retterer K., Piccirillo-Stosser C.M., Richard G., McKnight D.A. High frequency of mosaic pathogenic variants in genes causing epilepsy-related neurodevelopmental disorders. Genet. Med. 2018;20:403–410. doi: 10.1038/gim.2017.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Demarest S.T., Olson H.E., Moss A., Pestana-Knight E., Zhang X., Parikh S., Swanson L.C., Riley K.D., Bazin G.A., Angione K., et al. CDKL5 deficiency disorder: Relationship between genotype, epilepsy, cortical visual impairment, and development. Epilepsia. 2019;60:1733–1742. doi: 10.1111/epi.16285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gilbert N., Lutz S., Morrish T.A., Moran J.V. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodier J.L., Ostertag E.M., Kazazian H.H., Jr. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 56.Abou Tayoun A.N., Pesaran T., DiStefano M.T., Oza A., Rehm H.L., Biesecker L.G., Harrison S.M., ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI) Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum. Mutat. 2018;39:1517–1524. doi: 10.1002/humu.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sequence Variant Interpretation Working Group . 2018. ClinGen Sequence Variant Interpretation Recommendation for de novo Criteria (PS2/PM6)-Version 1.0.https://clinicalgenome.org/working-groups/sequence-variant-interpretation/ [Google Scholar]
- 58.Middelkamp S., Vlaar J.M., Giltay J., Korzelius J., Besselink N., Boymans S., Janssen R., de la Fonteijne L., van Binsbergen E., van Roosmalen M.J., et al. Prioritization of genes driving congenital phenotypes of patients with de novo genomic structural variants. Genome Med. 2019;11:79. doi: 10.1186/s13073-019-0692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plesser Duvdevani M., Pettersson M., Eisfeldt J., Avraham O., Dagan J., Frumkin A., Lupski J.R., Lindstrand A., Harel T. Whole-genome sequencing reveals complex chromosome rearrangement disrupting NIPBL in infant with Cornelia de Lange syndrome. Am. J. Med. Genet. A. 2020;182:1143–1151. doi: 10.1002/ajmg.a.61539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lei M., Liang D., Yang Y., Mitsuhashi S., Katoh K., Miyake N., Frith M.C., Wu L., Matsumoto N. Long-read DNA sequencing fully characterized chromothripsis in a patient with Langer-Giedion syndrome and Cornelia de Lange syndrome-4. J. Hum. Genet. 2020;65:667–674. doi: 10.1038/s10038-020-0754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang F., Carvalho C.M.B., Lupski J.R. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hattori A., Fukami M. Established and Novel Mechanisms Leading to de novo Genomic Rearrangements in the Human Germline. Cytogenet. Genome Res. 2020;160:167–176. doi: 10.1159/000507837. [DOI] [PubMed] [Google Scholar]
- 63.Ly P., Teitz L.S., Kim D.H., Shoshani O., Skaletsky H., Fachinetti D., Page D.C., Cleveland D.W. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 2017;19:68–75. doi: 10.1038/ncb3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang C.Z., Leibowitz M.L., Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27:2513–2530. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krivtsov A.V., Armstrong S.A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 67.Pramparo T., Grosso S., Messa J., Zatterale A., Bonaglia M.C., Chessa L., Balestri P., Rocchi M., Zuffardi O., Giorda R. Loss-of-function mutation of the AF9/MLLT3 gene in a girl with neuromotor development delay, cerebellar ataxia, and epilepsy. Hum. Genet. 2005;118:76–81. doi: 10.1007/s00439-005-0004-1. [DOI] [PubMed] [Google Scholar]
- 68.Striano P., Elia M., Castiglia L., Galesi O., Pelligra S., Striano S. A t(4;9)(q34;p22) translocation associated with partial epilepsy, mental retardation, and dysmorphism. Epilepsia. 2005;46:1322–1324. doi: 10.1111/j.1528-1167.2005.64304.x. [DOI] [PubMed] [Google Scholar]
- 69.Riggs E.R., Andersen E.F., Cherry A.M., Kantarci S., Kearney H., Patel A., Raca G., Ritter D.I., South S.T., Thorland E.C., et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) Genet. Med. 2020;22:245–257. doi: 10.1038/s41436-019-0686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Logsdon G.A., Vollger M.R., Eichler E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020;21:597–614. doi: 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cretu Stancu M., van Roosmalen M.J., Renkens I., Nieboer M.M., Middelkamp S., de Ligt J., Pregno G., Giachino D., Mandrile G., Espejo Valle-Inclan J., et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 2017;8:1326. doi: 10.1038/s41467-017-01343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant variant data are supplied within the paper or in supporting files. Complete IGS data for probands 1–5 are available via dbGAP (dbGAP: phs001089.v3.p1). CCS data for these proband will also be available via dbGAP under the same project. Complete IGS and CCS data for proband 6 are not available due to privacy and IRB reasons.