Abstract
The aging process is characterized by the presence of high interindividual variation between individuals of the same chronical age prompting a search for biomarkers that capture this heterogeneity. Epigenetic clocks measure changes in DNA methylation levels at specific CpG sites that are highly correlated with calendar age. The discrepancy resulting from the regression of DNA methylation age on calendar age is hypothesized to represent a measure of biological aging with a positive/negative residual signifying age acceleration (AA)/deceleration, respectively. The present study examines the associations of 4 epigenetic clocks—Horvath, Hannum, PhenoAge, GrimAge—with a wide range of clinical phenotypes (walking speed, grip strength, Fried frailty, polypharmacy, Mini-Mental State Examination (MMSE), Montreal Cognitive Assessment (MOCA), Sustained Attention Reaction Time, 2-choice reaction time), and with all-cause mortality at up to 10-year follow-up, in a sample of 490 participants in the Irish Longitudinal Study on Ageing (TILDA). HorvathAA and HannumAA were not predictive of health; PhenoAgeAA was associated with 4/9 outcomes (walking speed, frailty MOCA, MMSE) in minimally adjusted models, but not when adjusted for other social and lifestyle factors. GrimAgeAA by contrast was associated with 8/9 outcomes (all except grip strength) in minimally adjusted models, and remained a significant predictor of walking speed, .polypharmacy, frailty, and mortality in fully adjusted models. Results indicate that the GrimAge clock represents a step-improvement in the predictive utility of the epigenetic clocks for identifying age-related decline in an array of clinical phenotypes promising to advance precision medicine.
Keywords: Epigenetics, DNA methylation, Health, Mortality
Individuals of the same calendar age exhibit marked divergence in their biological aging rate, indicating that the progression of time represents an incomplete account of the aging process (1). This simple observation has spawned a search for biomarkers that are sensitive to individual differences in the pace of aging. A number of candidate biomarkers have been mooted (2), but among these, the epigenetic clocks hold arguably the most promise as a sensitive biomarker of aging. The development of the epigenetic clocks followed from the observation that patterns of DNA methylation (DNAm)—which refers to the addition or removal of methyl (–CH3) groups to Cytosine-phospho-Guanine (CpG) sites—change with age. The advent of DNAm microarray technology enabled the identification of the specific genomic locations of CpG sites that were differentially methylated according to age. The most commonly used of what is now referred to as the first-generation clocks are Hannum’s blood-specific clock (3), and Horvath’s pan-tissue clock (4), which are based on levels of DNAm at 71 and 353 CpG sites, respectively. These clocks are highly correlated with chronological age (5), and the discrepancy resulting from the regression of DNAm age on calendar age—a putative measure of biological age acceleration (AA)—is associated with increased risk of all-cause mortality (6,7). Hereinafter we refer to the AA estimates derived from the epigenetic clocks with the suffix AA.
In a wide-ranging review describing the development and evolution of the epigenetic clocks, Horvath and Raj (5) acknowledged that the first-generation clocks exhibited only weak associations with clinical measures of physiological dysregulation such as blood pressure or glucose metabolism that anticipate hard disease endpoints. Recently, Zhang et al (8) have shown that, in principle, a near-perfect predictor of calendar age can be estimated from DNAm levels, but its association with mortality attenuates with increased accuracy in predicting calendar age; the logical corollary of which is that the training sets being used for deriving accurate biological age estimators require more for their calibration than simply calendar age. The second-generation clocks, PhenoAge (9) and GrimAge (10) were designed to overcome these limitations by including DNAm correlates of morbidity and mortality. The PhenoAge clock was developed using a 2-stage approach. In the first stage, a weighted composite of 10 clinical characteristics (calendar age, albumin, creatinine, glucose, C-reactive protein, lymphocyte percentage, mean cell volume, red blood cell distribution weight, alkaline phosphatase, and white blood cell [WBC] count) was used to develop a phenotypic age estimator. In the second stage, the phenotypic age estimator was regressed on DNAm levels resulting in the identification of 513 CpG sites that exhibited marked differences in disease and mortality among individuals of the same calendar age. Initial validation work by Levine and colleagues confirmed that PhenoAgeAA outperformed the first-generation clocks in the prediction of many age-related diseases and life span. Lu and colleagues (10) adopted a slightly different strategy in the development of the latest clock—GrimAge. In the first stage, they identified DNAm-based surrogates of 12 plasma proteins as well as smoking pack-years. In the second stage, they regressed time-to-death due to all-cause mortality on the DNAm-based markers of plasma protein levels and smoking pack-years, identifying 1030 CpG sites that jointly predicted mortality risk.
Given the novelty of these measures, the validation of the second-generation clocks has only really begun in earnest, but early studies suggest that they represent an important step forward in our ability to predict a range of important age-related health outcomes and life span. While not the primary focus of the paper, McCrory et al (11) reported that PhenoAgeAA was associated with slower performance on the timed-up-and-go task, and with higher risk of prefrailty/frailty, but not with activity limitations in a paper comparing the performance of allostatic load and the epigenetic clocks for advancing our understanding of how low socioeconomic position (SEP) gets biologically embedded. A separate paper by the same group (12) noted that PhenoAgeAA correlated 0.21 with allostatic load—a compositive measure of physiological dysregulation—while the correlation of HorvathAA and HannumAA with allostatic load was close to zero in the overall sample, and higher for men compared with women.
Maddock et al (13) examined associations of HorvathAA, HannumAA, PhenoAgeAA, and GrimAgeAA with 3 measures of functional health (grip strength, chair-rise speed, and lung function) and 2 cognitive performance measures (word recall and mental speed) between the ages of 45–87 using data for 3 British cohorts: the 1946 National Survey of Health and Development (NSHD), 1958 National Child Development Study (NCDS), and Twins UK. They found that the first-generation clocks were not significantly associated with any of the 5 outcome measures in the meta-analysis of these cohorts, but that the second-generation clocks were. Specifically, PhenoAgeAA was associated with lower grip strength, worse lung function, and slower mental speed; while GrimAgeAA was associated with worse lung function, poorer word recall, and slower mental speed. Importantly, GrimAgeAA as assessed at baseline was associated with a faster rate of decline in grip strength and lung function between 53 and 69 years of age in the NSHD cohort.
In one of the most comprehensive investigations to date, Hillary and collaborators (14) present further evidence that the GrimAge clock exhibits remarkable promise as a biomarker of aging. Using data for some 709 individuals (mean age = 73) participating in the 1936 Lothian Birth Cohort (LBC), they reported that GrimAgeAA was inversely associated with lung function, levels of iron, low density lipoprotein, and total cholesterol; and positively associated with measured weight, body mass index (BMI), C-reactive protein, creatinine, and interleukin 6. GrimAgeAA was also found to be negatively associated with performance on a battery of cognitive ability measures including, choice reaction time (CRT), digit symbol-coding, symbol search, matrix reasoning, and general intelligence, decreased brain region volume, and increased white matter hyperintensities. It also predicted increased risk of all-cause mortality with an 81% increase in hazard per standard deviation (SD) increase in GrimAgeAA.
Nevertheless, Dugué (15) in a recent critique of the epigenetic clocks cautioned that early studies generally report stronger associations than later studies and are more likely to be affected by publication bias. This novel study provides important new evidence concerning the relevance of the epigenetic clocks for clinical medicine by examining the utility of 4 epigenetic AA indices—HorvathAA, HannumAA, PhenoAgeAA, and GrimAgeAA—for predicting functional decline, disability, comorbidity, and cognitive impairment/dementia using a series of measures that are well validated in the clinical literature as markers of age-related decline. We also examine the utility of epigenetic AA for predicting time to event due to all-cause mortality at up to 10-year follow-up.
Experimental Procedures
Ethical Statement
Ethical approval for the Irish Longitudinal Study on Ageing (TILDA) study was obtained from the Trinity College Dublin Research Ethics Committee and signed informed consent was obtained from all participants.
Participants
The TILDA study design and sample selection is described in detail elsewhere (16,17). Briefly, the study involves a nationally representative sample of 8175 community-dwelling older persons aged 50 years and older at baseline. Respondents complete a 90-minute computer-assisted personal interview in the home and a separate self-completion questionnaire containing more sensitive information that is returned via post. All participating respondents are invited to attend for a detailed clinic-based health assessment administered by trained nursing staff in either a center-based or home-based setting. The present study uses a subsample (n = 490) of the baseline TILDA cohort who were selected into the epigenetic study based on their life course social class trajectory into 4 groups: stable high, stable low, upwardly mobile, downwardly mobile as described in (11).
Epigenetic AA Measures
DNAm levels were assessed using the Infinium Human Methylation 850k Beadchip (Illumina Inc., San Diego, CA). Horvath DNAm Age, Hannum DNAm Age, DNAm PhenoAge, and DNAm GrimAge were estimated from whole blood according to procedures described in Fiorito et al (18) and Lu et al (10). Since the difference between DNAm Age and chronological age could be correlated with chronological age and WBC percentage, Chen and colleagues introduced the “extrinsic epigenetic AA” as the residuals from the regression of DNAm Age on chronological age, and the “intrinsic epigenetic AA” as the residuals from the regression of DNAm Age on chronological age and WBC percentage (6). The latter were estimated using the Houseman et al (19) algorithm. For all 4 epigenetic clocks, we utilized the “intrinsic” epigenetic AA in the main analyses to take account of possible differences in WBC by age and other risk factors, and we used the “extrinsic” epigenetic AA in sensitivity analyses.
TILDA Health Assessment
TILDA participants were invited to undergo a detailed health assessment described in (20). If a respondent could not attend the health center but was agreeable to completing a health assessment, a trained nurse administered a subset of the tests in the respondent’s home. Walking speed was only measured in the health center but all other measures were administered at both center-based and home-based assessments.
Physical Health Outcome Measures
Walking speed
Walking speed was assessed using a 4.88 meter (m) computerized walkway with embedded pressure sensors (GAITRite, CIR Systems Inc., New York, NY). Participants completed 2 walks along the mat at their normal walking speed. Each trial started 2.5 m before and ended 2 m after the walkway in order to allow room for acceleration and deceleration. The average of the 2 readings represented the overall walking speed measure expressed in centimeters traveled per second (cm/s).
Grip strength
Grip strength was measured using a Baseline Hydraulic Hand Dynamometer, which consisted of a gripping handle with a strain gauge and an analogue reading scale that increased in 2-kilogram (kg) increments. Respondents were instructed to hold the device in their hand with the forearm at a right angle to their upper arm. The participant was then instructed to squeeze as hard as they could for a few seconds. Results were rounded to the nearest whole number (ie, if the dial fell between 22 and 24 kg, grip strength was recorded as 23 kg). This procedure was repeated twice in each of the dominant and nondominant hands and the average of the 4 measurements was used to indicate grip strength.
Polypharmacy
Doctor prescribed medication usage was captured as part of the household interview and confirmed by cross-checking the labels on the medicinal packaging. The international nonproprietary name for any regularly taken medications was assigned and coded using Anatomic Therapeutic Classification codes. Polypharmacy was defined as currently taking 5+ doctor prescribed medications and used as a binary variable in the analysis. Previous studies have documented strong concordance between self-reported interview data and pharmacy records (21,22).
Fried frailty score
Frailty was operationalized using TILDA population-specific cut points following the methodology of Fried and colleagues (23,24). Weakness: Mean grip strength was calculated from an average of 2 measurements on the dominant hand using baseline dynamometer. Weight and height were measured using standardized procedures during the health center assessments. Participants scoring in the 20th percentile of sex- and BMI-adjusted grip strength were given a score of 1, those above the 20th percentile scored 0. Physical activity: Kilocalories (kcals) of energy expended were calculated from the International Physical Activity Questionnaire-Short Form (IPAQ-SF). Participants scoring in the 20th percentile of sex-adjusted kcals were given a score of 1, those above the 20th percentile scored 0. Slow walking speed: recorded time taken in seconds (s) to perform the timed-up-and-go task. Participants scoring in the slowest 20th percentile of sex- and height-adjusted timed-up-and-go time were given a score of 1, those above the slowest 20th percentile scored 0. Unintended weight loss was ascertained by the question “In the past year have you lost 10 pounds (4.5 kg) or more in weight when you were not trying to.” Participants who responded “yes” were given a score of 1, those who responded “no” scored 0. Exhaustion was captured using 2 items from the 20-item Center for Epidemiological Studies-Depression (CES-D) scale. Participants were asked how often they felt that “I could not get going” and “I felt that everything I did was an effort.” Participants who responded “a moderate amount/all of the time” to either question were given a score of 1, those who responded “none/some of the time” to both questions scored 0. The total score (range 0–5) was modeled as the response variable.
Global Cognitive/Neuropsychological Outcome Measures
Mini-Mental State Examination
The Mini-Mental State Examination (MMSE) (25) was originally designed to provide a brief, standardized assessment of mental status in psychiatric patients, although it is now used widely in clinical and epidemiological settings as a screening tool for the assessment of global cognitive impairment/dementia. It takes approximately 5–10 minutes to administer and assesses attention and concentration, memory, language, visuoconstructional skills, calculation, and orientation. Because the instrument suffers from ceiling effects, we used the count of errors in the analysis (ie, 30 − number of items correct) as the dependent variable.
Montreal Cognitive Assessment
Global cognitive status was also measured using the Montreal Cognitive Assessment (MOCA) (26). It designed as a brief cognitive screening tool to detect mild cognitive impairment among community- and clinic-based samples. The test takes approximately 10 minutes to administer and assesses performance across a number of cognitive domains including: executive functioning/visuospatial abilities, attention/concentration, language; animal naming, short-term memory, abstraction, and orientation. The instrument yields a sum score ranging from 0 to 30. Recent studies involving clinical samples suggest the MOCA performs better than the MMSE in detecting early signs of cognitive decline 15 and hence servers as a useful epidemiological screening tool for identifying the transitional state between normal cognitive functioning and more severe cognitive impairment. Again, we modeled the count of errors as the dependent variable (ie, 30 − number of items correct).
Sustained Attention Reaction Time Task
The ability to sustain attention is an important component of normal executive functioning, and normal cognitive aging is associated with declines in selective (ie, ability to attend) and divided (ie, ability to focus on multiple tasks simultaneously) attention (27). The Sustained Attention Reaction Time (SART) is a computerized performance reaction time (RT) task (28). It requires participants to attend to a repeating stream of digits (1–9) and to press the response key for each number that they see (GO trials), except number “3” (NO–GO trials). Each digit from 1 through 9 appears for 300 milliseconds (ms), with an interval of 800 ms between digits, and the cycle is repeated 23 times giving a total of 207 trials. The task takes approximately 4 minutes to complete. Commission errors (ie, responding to NO–GO trials) reflect lapses of sustained attention, and omission errors (ie, failure to respond to GO trials) reflect a break from task engagement, also corresponding to lapsing attention. The total count of commission errors made on the task was modeled as the response variable.
Choice reaction time task
Processing speed slows in aging leading some to speculate that it may represent a biomarker of cognitive aging (29). Participants completed a standard 2-CRT task as part of a detailed neuropsychological battery. They were instructed to rest their finger on a “home” plate and wait for a stimulus to appear on screen stating “Yes” or “No.” They then had to move their finger from the home plate to the corresponding “Yes” or “No” button. Participants completed several practice trials in the presence of the clinical nurse to ensure familiarity with the test procedure. Respondents were only permitted to enter the test phase if they demonstrated sufficient understanding of the task during the test phase. The trial phase consisted of a series of 100 trials with the stimulus prompt alternating in a randomized sequence and appearing approximately 50% of the time for each stimulus response. RT can be decomposed into a decision time (interlude between stimulus onset and movement from the home plate) and movement time component (interlude between movement from home plate to the relevant response key). As the CRT data exhibited marked positive skew, it was log-transformed prior to analysis.
All-Cause Mortality
In total, 35/490 participants or 7.2% of the epigenetic subsample were confirmed as deceased at up to 10-year follow-up (ie, by end of Wave 5 sweep of data collection on December 20, 2018). Mortality status was available from 2 sources: (i) through data linkage to the General Registrar’s Office (GRO) National Death Registry which was done in March 2017; and (ii) via an End of Life (EOL) interview for deaths occurring subsequent to administrative record linkage, which was conducted with the respondent’s next of kin (if available). Date of death was available for 17/35 respondents from the GRO and for a further 7/35 via the EOL interview. Time of death was unavailable for the remaining 11 cases because no EOL was completed or because TILDA allows a period of 6 months to elapse before attempting to conduct an EOL interview with the spouse/family of recently deceased cohort members. In these instances, we imputed date of death as half of the mean between-wave interval after the “date of last contact.”
Covariates
In addition to age (years) and sex (male, female), we controlled for other socioeconomic characteristics and lifestyle factors that could potentially confound the association of the putative biological aging measures with the various outcome measures. Life course social class trajectory was measured using the cross-classification of participants’ childhood (ie, father’s) and adulthood (ie, own contemporaneous) social class into 4 categories as follows: stable professional, upwardly mobile, stable unskilled, and downwardly mobile. Smoking history is represented using a 3-level variable: Never Smoked, Past Smoker, Current Smoker. Physical activity was assessed using the 8-item short form of the International Physical Activity Questionnaire (IPAQ) (30). It measures the amount of time (minutes) spent walking and engaged in moderate and vigorous physical activity, and the amount of time spent sedentary and categorized into: Low, Medium, and High levels of physical activity as per the IPAQ protocol (www.ipaq.ki.se). Hazardous drinking was assessed using the CAGE alcohol screening test (31). The scale comprises 4 items and follows a dichotomous yes/no response format. Answering yes to 2 or more questions indicates a clinically significant profile and constitutes potentially hazardous drinking. BMI was calculated from measured height and weight. Height was measured using a SECA 240 wall mounted measuring rod and weight was measured using a SECA electronic floor scales.
Treatment of Missing Cases
The overall level of missing data for the covariates was small. Six individuals were missing on the physical activity measure and 42 individuals were missing on the alcohol consumption measure; so, we coded these as “missing” using dummy variables so that they would not be lost to the analysis. One person was missing data for height and weight. The amount of missing data on the outcome measures was small overall and generally due to nonsystematic sources of variance (eg, technical problems).
Statistical Analysis
All analyses were conducted using Stata 15.0 (StataCorp, College Station, TX). The outcome measures were regressed separately on each of the 4 epigenetic AA measures adjusting for chronological age (years), sex, and WBC counts using ordinary least squares regression (walking speed, grip strength, reaction time), Poisson regression (frailty scores), negative binomial regression (MOCA errors, MMSE errors, SART commission errors), logistic regression (polypharmacy), and Cox regression (mortality) as appropriate. We report the change in each of the outcome measures associated with a standard unit increase in AA in each of the 4 biological aging measures. In the analyses involving walking speed and grip strength, we adjusted additionally for measured height (cm) which differs between men and women and is strongly associated with stride length and strength. In secondary models we adjusted additionally for participants’ life course socioeconomic trajectory in order to take account of characteristics associated with selection into the sample, as well as smoking, hazardous alcohol consumption, physical activity levels, and BMI to determine whether any putative association of the AA measures with health survived adjustment for lifestyle factors. We tested for effect modification by sex by fitting separate sex * AA interaction terms with respect to each outcome in the minimally adjusted models. Only 2 of the 32 contrasts were significant—both on the SART task, but in opposite directions. Women made a significantly higher number of commission errors on the SART task per 1-year increase in GrimAgeAA, but significantly fewer errors per 1-year increase in HannumAA. We report results therefore for the overall sample.
Results
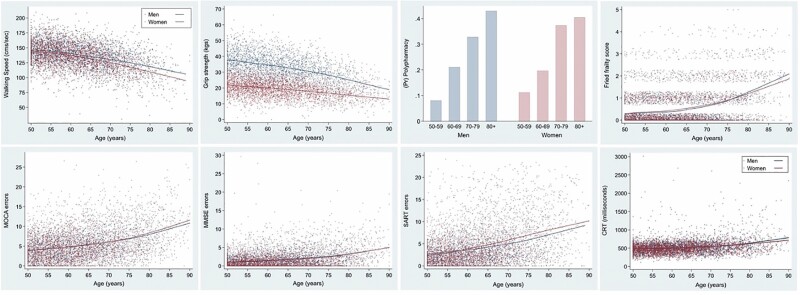
Figure 1 depicts scatterplots for each of the physical health and cognitive functioning measures using locally weighted scatterplot smoothing (lowess) to describe the functional form of change in each of the dependent variables by age and sex. It demonstrates clearly that the various outcome measures are highly sensitive to age-related decline. Walking speed and grip strength decline steadily with age, Fried frailty scores increase with age, as does the prevalence of polypharmacy. Likewise, the number of errors on the MOCA, MMSE, and SART task increases sharply with age, as does performance on the CRT task. Normative values and age gradients for many of these measures derived using the TILDA data set have been published elsewhere (32). Table 1 describes the characteristics of the sample and reports mean scores (SD) as well as the range for each of the functional health and cognitive performance measures. The mean age of the sample was 62.2 years and 50.2% were female. This compared with a mean clock age of 61.3, 60.9, 61.2, and 60.8 years for Horvath, Hannum, PhenoAge, and GrimAge clocks, respectively. Scatterplots of DNAm age by chronological age for Horvath (r = 0.74), Hannum (r = 0.74), PhenoAge (r = 0.85), and GrimAge (r = 0.81) are shown in Supplementary Figure 1. The magnitude of the Pearson intercorrelations between the various epigenetic AA measures is as follows: HorvathAA correlated 0.83, 0.06, and −0.02 with HannumAA, PhenoAgeAA, and GrimAgeAA, respectively. HannumAA correlated 0.03 and 0.00 with PhenoAgeAA and GrimAgeAA, respectively, and PhenoAgeAA correlated 0.40 with GrimAgeAA.
Figure 1.
Plots describing the relationship of the physical and cognitive outcome measures by age and sex. Fitted regression lines were estimated using locally weighted scatterplot smoothing. As polypharmacy was a binary outcome, we plotted the proportion of cases taking 5+ medications in 10-year age intervals.
Table 1.
Baseline Characteristics of the TILDA Epigenetic Subsample
| Mean (SD) or n (%) | |
|---|---|
| Horvath DNAm age | 61.3 (11.0) |
| Hannum DNAm age | 60.9 (11.0) |
| PhenoAge DNAm age | 61.2 (9.6) |
| GrimAge DNAm age | 60.8 (7.6) |
| Horvath age acceleration (AA) | 0.00 (7.4) |
| Hannum age acceleration (AA) | 0.00 (7.3) |
| PhenoAge age acceleration (AA) | 0.00 (5.0) |
| GrimAge age acceleration (AA) | 0.00 (4.3) |
| Age | 62.2 (8.3) |
| Female sex | 246 (50.2%) |
| Social class trajectory | |
| -Stable high | 123 (25.1%) |
| -Upwardly mobile | 125 (25.5%) |
| -Downwardly mobile | 121 (24.7%) |
| -Stable low | 121 (24.7%) |
| Smoking | |
| -Never smoked | 193 (39.4) |
| -Past smoker | 211 (43.1) |
| -Current smoker | 86 (17.6) |
| Hazardous drinking | |
| -No | 371 (75.7%) |
| -Yes | 77 (15.7%) |
| -Missing | 42 (8.6%) |
| Physical Activity | |
| -Low | 142 (29.0%) |
| -Moderate | 175 (35.7%) |
| -High | 167 (34.1%) |
| -Missing | 6 (1.2%) |
| BMI | 28.8 (5.1) |
| Walking speed (cm/s) | 136.1 (21.6) |
| Grip strength (kg) | 26.5 (9.4) |
| Polypharmacy | 94 (19.2%) |
| Fried frailty score | 0.43 (0.74) |
| MOCA errors | 4.9 (3.4) |
| MMSE errors | 1.5 (1.9) |
| SART errors | 3.7 (4.0) |
| Raw CRT (ms) | 505.7 (139.2) |
Notes: BMI = body mass index; CRT = choice reaction time; DNAm = DNA methylation; MMSE = Mini-Mental State Examination; MOCA = Montreal Cognitive Assessment; SART = Sustained Attention Reaction Time; TILDA = Irish Longitudinal Study on Ageing.
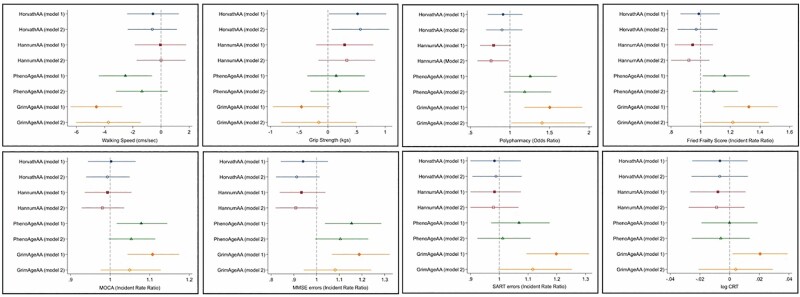
Figure 2 shows the change in each of the physical health and cognitive performance measures associated with a standard unit (z-score) increase in epigenetic AA according to each of the 4 clocks while adjusting for calendar age and sex in the minimally adjusted models (Model 1), and additionally for life course socioeconomic characteristics, smoking, hazardous levels of alcohol consumption, physical activity levels, and BMI in the fully adjusted models (Model 2). The unstandardized beta coefficients (B), odds ratios (ORs), and incident rate ratios (IRRs) and associated 95% confidence intervals (CIs) are reported in Supplementary Table 1. There were only 2 significant associations of the first-generation clocks with any of the outcome measures under investigation. HorvathAA was associated with higher grip strength in both the minimally and full multivariable-adjusted models, while HannumAA was associated with reduced risk of polypharmacy in the fully adjusted models.
Figure 2.
Association of z-score standardized cell intrinsic epigenetic age acceleration measures with physical and cognitive functioning in the minimally (Model 1) and full multivariable-adjusted models (Model 2). Model 1: adjusted for age, sex, white blood cell counts (+height for walking speed and grip strength). Model 2: Model 1 + life course social class trajectory, smoking, physical activity, body mass index (BMI).
PhenoAgeAA was associated with 4/8 physical and cognitive outcomes in the minimally adjusted models including, slower walking speed (B = −2.53, 95% CI = −4.41, −0.65; p = .009), higher Fried frailty (IRR = 1.16, 95% CI = 1.01, 1.33; p = .030), MOCA errors (IRR = 1.08, 95% CI = 1.02, 1.14; p = .012), and MMSE errors (IRR = 1.15, 95% CI = 1.04, 1.29; p = .009), but none of these associations survived multivariable adjustment. GrimAgeAA was associated with 7/8 physical and cognitive health outcomes in the minimally adjusted models: slower walking speed (B = −4.59, 95% CI = −6.41, −2.76; p < .001), increased polypharmacy (OR = 1.50, 95% CI = 1.18, 1.91; p < .001), higher Fried frailty score (IRR = 1.33, 95% CI = 1.16, 1.52; p < .001), MOCA errors (IRR = 1.11, 95% CI = 1.04, 1.17; p < .001), MMSE errors (IRR = 1.19, 95% CI = 1.07, 1.32; p = .002), SART errors (IRR = 1.20, 95% CI = 1.09, 1.31; p < .001), log CRT * 100 (B = 2.05, 95% CI = 0.20, 3.91; p = .030); and continued to be associated with 3/8 outcomes (walking speed, frailty score, and polypharmacy) in the full multivariable-adjusted models.
Table 2 (Model 1) shows that a standard unit increase in GrimAgeAA was associated with a 2-fold increase (hazard ratio [HR] = 2.05, 95% CI = 1.45, 2.90; p < .001) in the hazard of all-cause mortality at up to 10-year follow-up, and these estimates were only marginally affected when adjusted for socioeconomic characteristics and lifestyle factors (HR = 1.91, 95% CI = 1.23, 2.96; p = .004) (Table 2, Model 2). None of the other epigenetic AA measures significantly predicted mortality.
Table 2.
Hazard Ratios (HRs) for All-Cause Mortality at up to 10-Year Follow-Up Associated With a Standard Unit (z-score) Increase in Cell Intrinsic Epigenetic Age Acceleration in the Minimally and Full Multivariable-Adjusted Models (n = 489)
| Model 1 | Model 2 | |
|---|---|---|
| HR (95% CI) | HR (95% CI) | |
| HorvathAA | 0.97 (0.70, 1.36) | 1.03 (0.74, 1.44) |
| HannumAA | 0.89 (0.64, 1.24) | 0.92 (0.67, 1.28) |
| PhenoAgeAA | 1.26 (0.92, 1.74) | 1.13 (0.81, 1.57) |
| GrimAgeAA | 2.05 (1.45, 2.90)*** | 1.91 (1.23, 2.96)** |
Notes: AA = age acceleration; CI = confidence interval.
***significant at the p < .001 level; **significant at the .01 level.
Sensitivity Checks
As a sensitivity check, we also calculated the relationship between epigenetic AA adjusted for calendar age but not WBC percentages (ie, cell extrinsic epigenetic age) (Supplementary Table 2). In general, results were consistent with what we found for the WBC adjusted measures so here we discuss only the differences. PhenoAgeAA was now additionally associated with polypharmacy (OR = 1.32, 95% CI = 1.04, 1.67; p = .024), and GrimAgeAA was now additionally associated with lower grip strength (B = −0.60, 95% CI = −1.10, −0.10; p = .019) in the minimally adjusted, but not full multivariable-adjusted models. Adjusting for socioeconomic characteristics and other lifestyle factors, GrimAgeAA remained associated with 4/8 physical and cognitive outcomes (ie, slower walking speed, polypharmacy, Fried frailty score, MOCA errors) as well as all-cause mortality. In response to a reviewer query, we also investigated whether the DNAm surrogate components of the GrimAge clock were more predictive of outcome than the overall measure (Supplementary Table 3). DNAm smoking pack-years was strongly predictive of an adverse outcome (9 outcomes), as was DNAm tissue inhibitor metalloproteinaise (TIMP1) (7 outcomes), and DNAm adrenomedullin (ADM) (5 outcomes), but each of the components was found to relate to each of the outcomes to a varying degree. This is not surprising, GrimAge was developed as a linear combination of these DNAm-based biomarkers, which in turn were significantly associated with mortality in a Cox regression model. Nevertheless, this supplementary analysis may generate some interesting new leads as DNAm TIMP1 was more strongly associated with walking speed, polypharmacy, frailty, MOCA, MMSE, and SART errors than any of the other DNAm-based surrogate biomarkers, or indeed, GrimAge, making it a candidate worthy of further study in the context of aging.
Discussion
This study compared the utility of the so-called first- and second-generation epigenetic clocks for predicting functional health, cognitive/neuropsychological performance, and all-cause mortality in a subsample of TILDA participants confirming that the new GrimAge clock represents a strong marker of premature aging with huge implications for clinical medicine. The results of this study confirm Horvath and Raj’s (5) previous observation that the first-generation clocks are not sensitive predictors of age-related decline in clinical health measures. Few of the associations were significant, and even among those that were, the observed relationships were in the opposite direction to what one would hypothesize. A standard unit increase in HorvathAA was paradoxically associated with higher grip strength, and a standard unit increase in HannumAA was associated with reduced risk of polypharmacy. The mostly null associations for the first-generation clocks are consistent with the results obtained by Maddock et al (13) in a recent study involving the British cohort studies. A second interesting observation is the difference in the direction of effects across the clocks. The reasons for these differences are not immediately apparent, although McCrory et al (12) previously speculated that the first-generation clocks may be less well calibrated for detecting age-related decline as they did not incorporate clinical biomarkers in their derivation and hence may be less sensitive to capturing the biological AA that results from extrinsic life course stressors.
The second-generation clocks by contrast markedly outperformed the first-generation clocks in the prediction of many functional health and cognitive performance measures, and GrimAgeAA-estimated risks were about twice those estimated for PhenoAgeAA. PhenoAgeAA was associated with slower walking speed, higher Fried frailty, MOCA errors, and MMSE errors, but these relationships no longer held when adjusted for socioeconomic characteristics and other lifestyle-related factors. GrimAgeAA by contrast was associated with all outcome variables under investigation, except grip strength, and continued to be associated with 4 of them (walking speed, polypharmacy, Fried frailty score, and all-cause mortality) even in the full multivariable-adjusted models; a finding which implies that the GrimAge clock is tapping variation in the pace of aging that is not simply due to SEP and other lifestyle-related factors. This, despite the fact that GrimAgeAA is more strongly associated with SEP than the other clocks.
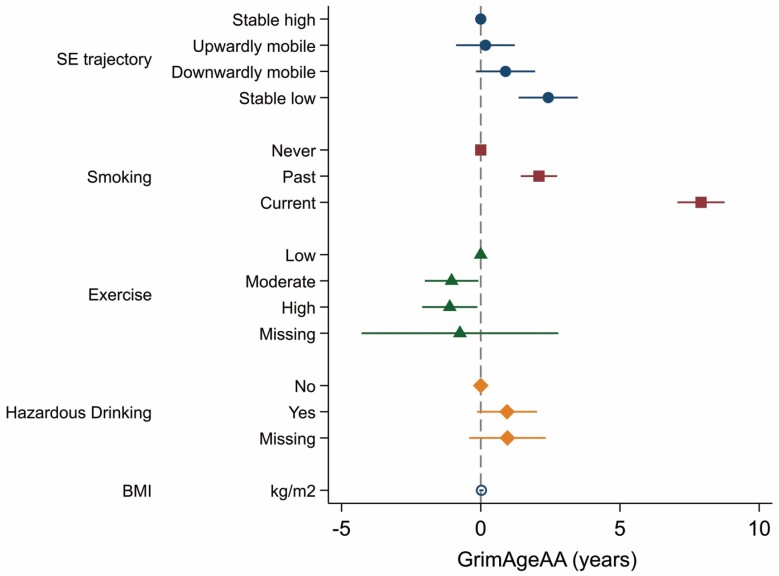
McCrory et al (11) have previously shown that HorvathAA, HannumAA, and PhenoAgeAA are not strongly related to socioeconomic characteristics in the TILDA epigenetic sample, but in this study, having a stable low SEP across the life course (ie, low childhood social class/low adulthood social class) was associated with 2.42 years (95% CI = 1.36, 3.49; p < .001) of epigenetic AA according to the GrimAge clock compared with those who were stable high (ie, high childhood social class/high adulthood social class) in SEP. Being a current smoker was associated with 7.91 years (95% CI = 7.06, 8.76; p < .001) of GrimAgeAA, being a hazardous drinker according to the CAGE questionnaire was associated with 0.94 years (−0.13, 2.02, p = .086) of GrimAgeAA, while levels of physical activity were inversely associated with GrimAgeAA. Finally, BMI was found to be unrelated to GrimAgeAA (Figure 3).
Figure 3.
Age, sex, and white blood cell adjusted independent association of GrimAgeAA with socioeconomic characteristics and other lifestyle factors.
Other factors which we did not adjust for in this analysis but may mediate the association of GrimAgeAA with our broad array of clinical outcomes includes: (i) genetics, as the GrimAge clock was calibrated by regressing time to death due to all-cause mortality on DNAm levels; (ii) life course psychosocial stressors, which have been linked with accelerated epigenetic aging (33) and physiological wear and tear in tissues and organ systems (34); (iii) nutritional deficiencies, particularly folate insufficiency, which is a key nutritional factor in 1-carbon metabolism (35); and (iv) exposure to environmental toxins (36), which may elicit epigenetic modifications (37), and are known to be deleterious to health. Future research should be designed to address these possibilities.
Shiels and colleagues (38) contend that a sequitur for any valid biomarker of aging is that it shows a statistically significant association with a measure of health or organ functional capacity over and above calendar age. We went a step further to examine whether epigenetic AA measures are clinically relevant as well as statistically relevant. On the basis of previous exploratory work involving HorvathAA, HannumAA, and PhenoAgeAA, McCrory and collaborators (12) questioned whether the residual from the regression of DNAm age on chronological age represented the sine qua non of biological aging. The new GrimAge clock seems to allay many of these concerns, reaffirming the belief that DNAm levels at very few CpG sites across the human methylome may be informative about physical health and brain health independently of calendar age.
Nevertheless, what remains to be established is whether epigenetic AA predicts changes in health and cognition prospectively, and perhaps more pertinently, whether change in the epigenetic AA measures is associated with change in physical health and cognitive performance. This is important in order to help establish whether DNAm is a cause or a consequence of aging. To date, the evidence is inconclusive (13). Unfortunately, few studies have measured change in both health and patterns of DNAm over time, even though these studies are urgently needed to help disambiguate the temporal ordering of the observed relationships. Levine et al. (39) recently suggested that the reason the second-generation clocks outperform their progenitors is because they were trained on longitudinal data. Although speculative, it is highly likely that the third-generation clocks will be trained on data that assess changes in DNAm levels and change in the type of age-related phenotypes measured so comprehensively in the TILDA study prospectively. There is also a concomitant need to explore the functional significance of genes that are regulated by the CpG sites that are most informative of biological aging, as this may will help inform efforts to compress disease and morbidity and advance precision medicine.
The study benefits from the use of a deeply phenotyped community-dwelling older cohort for whom we have a rich array of clinical health and cognitive performance measures administered in a health center conducted by trained nurses using standardized operating protocols. We also capture older, frailer individuals as a nurse also administers a home health assessment to those who cannot attend the center and who may be lost to follow-up in other studies. Our study also has a number of weaknesses, perhaps the most notable of which is the relatively small (by epidemiological standards) sample size, and the selective nature of the sample which was originally designed to look at the impact of life course socioeconomic trajectories on epigenetic aging rates.
This study provides important new evidence concerning the utility of the new GrimAge epigenetic clock for predicting age-related decline in a number of important health and cognitive domains relevant to clinical medicine. The fact that GrimAgeAA was so strongly associated with health span and life span is potentially exciting as it holds out the tantalizing prospect that we may have identified a biological aging surrogate with strong theoretical underpinnings that has prognostic utility as an indicator of a person’s state of health. Recent work suggests that epigenetic aging is potentially reversible (40), so the epigenetic clocks may also represent a means for quantifying the efficacy of interventions designed to retard or reverse the aging process—the holy grail of Geroscience. In summary, evidence is accumulating that DNAm plays a central role in the aging process, and the epigenetic clocks may represent a highly accurate means for quantifying the rate of biological decay in what has been heralded the new era of precision medicine.
Funding
This work was supported by the Health Research Board (HRB) of Ireland under an Emerging Investigator Award (EIA-2017-012) to C.M.C. This work was also supported by the LIFEPATH grant to P.V. at Imperial College London (European Commission H2020 grant, grant number: 633666). Funding for the TILDA project was supported by the Irish Government, the Atlantic Philanthropies, and Irish Life plc. S.H. and A.T.L. were supported by NIH/NIA IU01AG060908. The funders had no involvement in the study design, collection, analysis and interpretation of data, or authorship of the submitted work.
Conflict of Interest
S.H. and A.T.L. are named as inventors on a patent application surrounding GrimAge that was filed by their employers, UC Regents.
Author Contributions
C.M.C., G.F., P.V., and R.A.K. conceived and designed the study. C.M.C. conducted the statistical analysis and wrote the preliminary draft of the manuscript. G.F., B.H., S.P., A.M.O.H., A.H., C.N.C., A.T.L., and S.H. contributed to the development of methods used in the paper. All authors critically revised the manuscript through 2 iterations of internal review and contributed important intellectual content. C.M.C. holds all data files and analysis files to enable replication of the findings.
Supplementary Material
References
- 1. Hägg S, Belsky DW, Cohen AA. Developments in molecular epidemiology of aging. Emerg Top Life Sci. 2019;3:411–442. doi: 10.1042/ETLS20180173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. EBioMedicine. 2017;21:29–36. doi: 10.1016/j.ebiom.2017.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3 [DOI] [PubMed] [Google Scholar]
- 6. Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY). 2016;8:1844–1865. doi: 10.18632/aging.101020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marioni RE, Shah S, McRae AF, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi: 10.1186/s13059-015-0584-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Q, Vallerga CL, Walker RM, et al. Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med. 2019;11:54. doi: 10.1186/s13073-019-0667-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10:573–591. doi: 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019;11:303–327. doi: 10.18632/aging.101684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCrory C, Fiorito G, Ni Cheallaigh C, et al. How does socio-economic position (SEP) get biologically embedded? A comparison of allostatic load and the epigenetic clock(s). Psychoneuroendocrinology. 2019;104:64–73. doi: 10.1016/j.psyneuen.2019.02.018 [DOI] [PubMed] [Google Scholar]
- 12. McCrory C, Fiorito G, McLoughlin S, et al. Epigenetic clocks and allostatic load reveal potential sex-specific drivers of biological aging. J Gerontol A Biol Sci Med Sci. 2020;75:495–503. doi: 10.1093/gerona/glz241 [DOI] [PubMed] [Google Scholar]
- 13. Maddock J, Castillo-Fernandez J, Wong A, et al. DNA methylation age and physical and cognitive aging. J Gerontol A Biol Sci Med Sci. 2020;75:504–511. doi: 10.1093/gerona/glz246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hillary RF, Stevenson AJ, Cox, SR, et al. An epigenetic predictor of death captures multi-modal measures of brain health. Mol Psychiatr. 2019. doi: 10.1038/s41380-019-0616-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dugué PA, Li S, Hopper JL, Milne RL. Chapter 3 - DNA methylation-based measures of biological aging. In: Tollefsbol TO, ed. Epigenetics in Human Disease. 2nd ed. Vol. 6. Academic Press; 2018:39–64. doi: 10.1016/B978-0-12-812215-0.00003-0 [DOI] [Google Scholar]
- 16. Kearney PM, Cronin H, O’Regan C, et al. Cohort profile: the Irish Longitudinal Study on Ageing. Int J Epidemiol. 2011;40:877–884. doi: 10.1093/ije/dyr116 [DOI] [PubMed] [Google Scholar]
- 17. Whelan BJ, Savva GM. Design and methodology of the Irish Longitudinal Study on Ageing. J Am Geriatr Soc. 2013;61(suppl 2):S265–S268. doi: 10.1111/jgs.12199 [DOI] [PubMed] [Google Scholar]
- 18. Fiorito G, Polidoro S, Dugué PA, et al. Social adversity and epigenetic aging: a multi-cohort study on socioeconomic differences in peripheral blood DNA methylation. Sci Rep. 2017;7:16266. doi: 10.1038/s41598-017-16391-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cronin H, O’Regan C, Finucane C, Kearney P, Kenny RA. Health and aging: development of the Irish Longitudinal Study on Ageing health assessment. J Am Geriatr Soc. 2013;61(suppl 2):S269–S278. doi: 10.1111/jgs.12197 [DOI] [PubMed] [Google Scholar]
- 21. Drieling RL, LaCroix AZ, Beresford SA, Boudreau DM, Kooperberg C, Heckbert SR. Validity of self-reported medication use compared with pharmacy records in a cohort of older women: findings from the women’s health initiative. Am J Epidemiol. 2016;184:233–238. doi: 10.1093/aje/kwv446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richardson K, Kenny RA, Peklar J, Bennett K. Agreement between patient interview data on prescription medication use and pharmacy records in those aged older than 50 years varied by therapeutic group and reporting of indicated health conditions. J Clin Epidemiol. 2013;66:1308–1316. doi: 10.1016/j.jclinepi.2013.02.016 [DOI] [PubMed] [Google Scholar]
- 23. Fried LP, Ferrucci L, Darer J, Williamson JD, Anderson G. Untangling the concepts of disability, frailty, and comorbidity: implications for improved targeting and care. J Gerontol A Biol Sci Med Sci. 2004;59:255–263. doi: 10.1093/gerona/59.3.m255 [DOI] [PubMed] [Google Scholar]
- 24. Fried LP, Tangen CM, Walston J, et al. ; Cardiovascular Health Study Collaborative Research Group . Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci. 2001;56:M146–M156. doi: 10.1093/gerona/56.3.m146 [DOI] [PubMed] [Google Scholar]
- 25. Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6 [DOI] [PubMed] [Google Scholar]
- 26. Nasreddine ZS, Phillips NA, Bédirian V, et al. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 2005;53:695–699. doi: 10.1111/j.1532-5415.2005.53221.x [DOI] [PubMed] [Google Scholar]
- 27. Murman DL. The impact of age on cognition. Semin Hear. 2015;36:111–121. doi: 10.1055/s-0035-1555115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roalf DR, Moberg PJ, Xie SX, et al. Comparative accuracies of two common screening instruments for classification of Alzheimer’s disease, mild cognitive impairment, and healthy aging. Alzheimers Dement. 2013;9:529–537. doi: 10.1016/j.jalz.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deary IJ, Johnson W, Starr JM. Are processing speed tasks biomarkers of cognitive aging? Psychol Aging. 2010;25:219–228. doi: 10.1037/a0017750 [DOI] [PubMed] [Google Scholar]
- 30. Craig CL, Marshall AL, Sjöström M, et al. International physical activity questionnaire: 12-country reliability and validity. Med Sci Sports Exerc. 2003;35:1381–1395. doi: 10.1249/01.MSS.0000078924.61453.FB [DOI] [PubMed] [Google Scholar]
- 31. Ewing JA. Detecting alcoholism. The CAGE questionnaire. JAMA. 1984;252:1905–1907. doi: 10.1001/jama.252.14.1905 [DOI] [PubMed] [Google Scholar]
- 32. Kenny RA, Coen RF, Frewen J, et al. Normative values of cognitive and physical function in older adults: findings from the Irish Longitudinal Study on Ageing. J Am Geriatr Soc. 2013;61(suppl 2):S279–S290. doi: 10.1111/jgs.12195 [DOI] [PubMed] [Google Scholar]
- 33. Zannas AS. Editorial perspective: psychological stress and epigenetic aging – what can we learn and how can we prevent? J Child Psychol Psychiatr. 2016;57:674–675. doi: 10.1111/jcpp.12535 [DOI] [PubMed] [Google Scholar]
- 34. Castagné R, Gares V, Karimi M, et al. Allostatic load and subsequent all-cause mortality: which biological markers drive the relationship? Findings from a UK birth cohort. Eur J Epidemiol. 2018;33:441–458. doi: 10.1007/s10654-018-0364-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol. 2012;863:359–376. doi: 10.1007/978-1-61779-612-8_23 [DOI] [PubMed] [Google Scholar]
- 36. Vineis P. From John Snow to omics: the long journey of environmental epidemiology. Eur J Epidemiol. 2018;33:355–363. doi: 10.1007/s10654-018-0398-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Plusquin M, Chadeau-Hyam M, Ghantous A, et al. DNA methylome marks of exposure to particulate matter at three time points in early life. Environ Sci Technol. 2018;52:5427–5437. doi: 10.1021/acs.est.7b06447 [DOI] [PubMed] [Google Scholar]
- 38. Shiels PG, Stenvinkel P, Kooman JP, McGuinness D. Circulating markers of ageing and allostatic load: a slow train coming. Pract Lab Med. 2017;7:49–54. doi: 10.1016/j.plabm.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Levine ME. Assessment of epigenetic clocks as biomarkers of aging in basic and population research. J Gerontol A Biol Sci Med Sci. 2020;75:463–465. doi: 10.1093/gerona/glaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fahy GM, Brooke RT, Watson JP, et al. Reversal of epigenetic aging and immunosenescent trends in humans. Aging Cell. 2019;18:e13028. doi: 10.1111/acel.13028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.