Abstract
Transcripts from noncoding repetitive elements (REs) in the genome may be involved in aging. However, they are often ignored in transcriptome studies on healthspan and lifespan, and their role in healthy aging interventions has not been characterized. Here, we analyze REs in RNA-seq datasets from mice subjected to robust healthspan- and lifespan-increasing interventions including calorie restriction, rapamycin, acarbose, 17-α-estradiol, and Protandim. We also examine RE transcripts in long-lived transgenic mice, and in mice subjected to a high-fat diet, and we use RNA-seq to investigate the influence of aerobic exercise on RE transcripts with aging in humans. We find that (a) healthy aging interventions/behaviors globally reduce RE transcripts, whereas aging and high-fat diet (an age-accelerating treatment) increase RE expression; and (b) reduced RE expression with healthy aging interventions is associated with biological/physiological processes mechanistically linked with aging. Our results suggest that RE transcript dysregulation and suppression are likely novel mechanisms underlying aging and healthy aging interventions, respectively.
Keywords: Healthspan, Life span, Noncoding RNA, RNA-seq, Transposable elements
Older age is the greatest risk factor for the development of most chronic diseases (1). Accordingly, recent large-scale “omics” studies have aimed to characterize novel genes and biological pathways that influence aging, and to identify related interventions (e.g., pharmaceutical compounds, exercise, nutrition) that increase longevity and healthspan (2). Indeed, advances in transcriptomics (e.g., RNA-seq) have led to important insight on many genes and pathways linked with “the hallmarks of aging” and broader health outcomes (3). However, most of these studies have focused on coding sequences—a small fraction of the genome. Noncoding, repetitive elements (REs, >60% of the genome) have been particularly neglected as “junk DNA,” despite growing evidence that they have many important biological functions (4).
REs include DNA transposons, retrotransposons, tandem repeats, satellites, and terminal repeats. A major fraction of REs, mainly DNA transposons and retrotransposons (e.g., LINEs, SINEs, LTRs), are transposable elements (TEs) with the ability to propagate, multiply, and change location in the genome (4). Most REs are in genomic regions that are chromatinized and suppressed (inactive), but recent reports show that certain TEs become active during aging, perhaps due to reduced chromatin architecture/stability (e.g., histone dysregulation) (5). Activation of these specific TEs may contribute to aging by causing genomic and/or cellular damage/stress (e.g., inflammation) (6). However, we have recently reported that aging is associated with a progressive, global increase in transcripts from most REs (not just specific TEs) in model organisms and humans (7). This global dysregulation of REs may have an important, more general role in aging, as RE transcripts have been linked with other key hallmarks of aging including oxidative stress and cellular senescence (8). In fact, it has been suggested that RE dysregulation itself may be an important hallmark of aging (9). If so, a logical prediction would be that interventions known to increase health/lifespan and reduce hallmarks of aging (e.g., calorie restriction [CR], select pharmacological agents, and exercise) should also generically suppress REs. Limited evidence suggests this may be true for CR and certain TEs in Drosophila (10), but global RE transcript expression has not been well studied in this context.
Results and Discussion
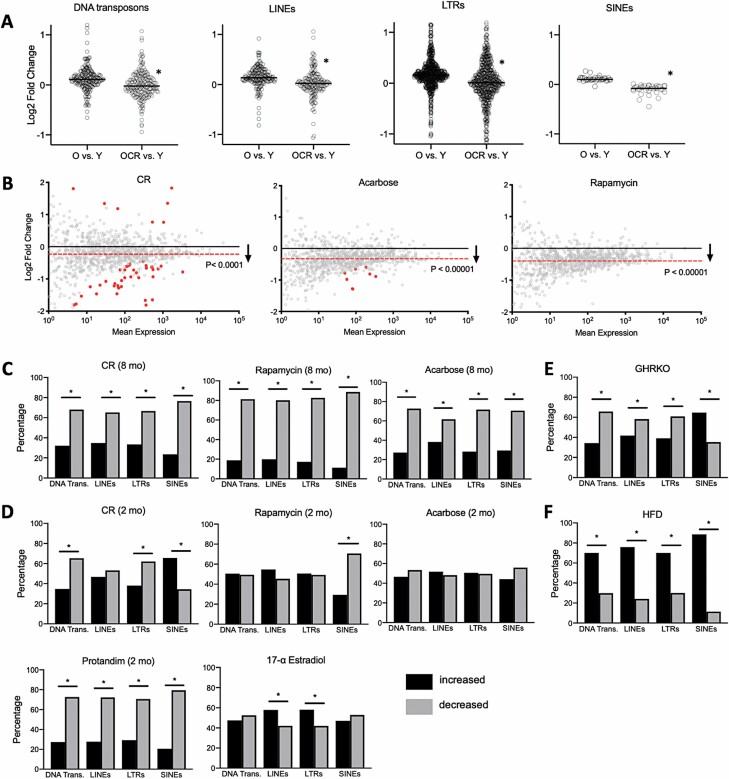
To determine if global RE transcript suppression might be a mechanism underlying healthy aging interventions, we first used the TEtranscripts pipeline to analyze REs in an RNA-seq dataset on livers (a key metabolic organ) from young and old mice, and old mice subjected to lifelong (24 months) CR (13). We found a small, but highly significant age-related increase in most major RE transcript types in this dataset, consistent with our previous work (7) and others’ (14). However, this effect was significantly attenuated with CR (Figure 1A). Based on this novel evidence of RE suppression by CR (arguably the strongest health/lifespan-enhancing intervention), we looked to confirm our results in an additional, pre-existing dataset including RNA-seq on livers) from mice subjected to different durations of CR and pharmacological interventions known to increase health/lifespan (rapamycin, acarbose, 17-α-estradiol [17aE], and Protandim) starting at 4 months of age, as well as data on transgenic, long-lived mice (2) (Figure 1). We focused on this large dataset, originally published as a “resource,” because it provides a unique opportunity to systematically interrogate transcriptome data on healthy aging interventions per se (without potential inter-study differences and/or age-related morbidity confounds). We also used an additional bioinformatics program, Repenrich2, to confirm similar findings with different analysis pipelines (because different programs use a variety of computational approaches to address challenges and potential artifacts in RE transcriptome analyses) (4), and we found similar results using both pipelines (Supplementary Data). In line with our initial results, we found that long-term (8 months) CR caused a significant, global reduction in RE transcripts, and that both long-term rapamycin and acarbose treatments were associated with a comparable, broad reduction in RE transcripts (Figure 1B), despite no global shifts in gene expression (Supplementary Figure 1A). These results suggest that a reduction in RE transcripts may be an important/conserved effect of healthy aging interventions, and they are consistent with the notion that acarbose and rapamycin may be “calorie restriction mimetics” (15). This effect was also clear when we examined intervention-associated RE reductions by major subtype (Figure 1C), all of which involved global, normally distributed shifts in RE expression (Supplementary Figure 1B−D).
Figure 1.
Healthy aging interventions reduce repetitive element (RE) transcripts. (A) Age-related increases in the major types of RE transcripts in old (O) or old calorie-restricted (OCR) vs young (Y) mice. N = 3/group, *p < .0001 (Shapiro–Wilk normality test followed by Mann–Whitney t test). All individual REs shown. (B) MA plots (M, log ratio; A, mean average) showing significant decreases in global RE transcripts with long-term (8-month) healthy aging interventions. Significantly reduced or increased REs (FDR < 0.1, identified using Deseq2) shown as solid/red points, and average transcript reduction indicated by dashed/red line. Likelihood of increased/decreased distribution calculated by chi-squared analysis. (C–F) Percentage of RE transcripts by type increased or decreased with long- or short-term (2-month) interventions (*p < .05, chi-squared analysis), and in growth hormone receptor knockout (GHRKO) and high-fat diet (HFD)-fed mice. Data were generated using both the TEtranscripts (A and B) and RepEnrich2 (C–F) pipelines. All relevant data/samples from datasets GSE92486, GSE131901, and GSE87565 were used for analyses (N = 3–6 mice/group), and raw data are provided in the Supplementary Data.
Short-term (2 months) interventions with other healthy aging compounds (some of which were not studied in longer-term experiments) influenced RE transcript levels to various degrees, although reductions were more pronounced with CR and Protandim, which is thought to activate endogenous antioxidant defenses (Figure 1D). Interestingly, the authors of the original study (2) observed similar variability in gene expression patterns, suggesting time/treatment-specific transcriptome effects. We also found a significant, largely inhibitory influence of growth hormone receptor knockout (GHRKO, a transgenic longevity model) on the main RE transcript types (Figure 1E). Moreover, in a separate dataset (16), we found that high-fat diet (HFD, a common “age-acclerating” intervention) significantly increased all major RE transcript types (Figure 1F), and many of the most increased REs with HFD were strongly reduced by most healthy aging interventions (Supplementary Data). Future studies are needed to determine if such RE transcript differences track with health/function (the datasets we analyzed did not include this information), but collectively, our results support the idea that global RE transcript levels are linked with healthspan/lifespan, as they are reduced by most “gold standard” anti-aging interventions and increased by one well-known pro-aging treatment (HFD, others should be investigated).
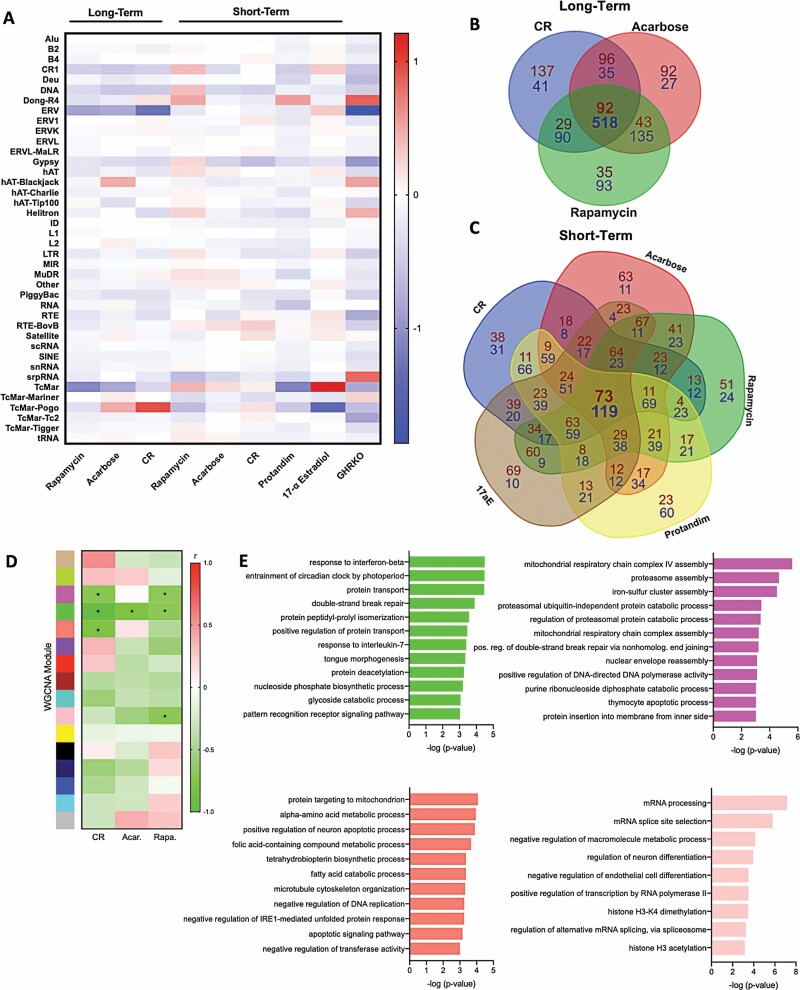
We did not note any particular patterns/specific REs that were the most reduced with different treatments (i.e., significant data points or largest fold-changes shown in Figure 1). Therefore, we next looked for similarities in the effects of healthy aging interventions on REs by subtype/family (Figure 2). With long-term treatment, CR and rapamycin influenced RE families most similarly, and most transcripts were decreased with all long-term treatments. Again, we observed variable patterns of RE expression by family with short-term treatments, and in GHRKO mice, most RE subtypes were decreased (Figure 2A). When we examined commonly modulated RE transcripts among all treatments, we found that long-term treatments decreased/increased a large number of common transcripts (518 and 92, respectively) (Figure 2B and Supplementary Data). Also, despite the variable RE/family expression patterns noted above, short-term treatments modulated many of the same transcripts (Figure 2C and Supplementary Data). Still, consistent with the idea that global RE modulation is linked with healthy aging interventions, we did not notice any particular enrichment for specific RE types/families in these common transcripts.
Figure 2.
Healthy aging interventions reduce most repetitive element (RE) transcript families and many similar individual transcripts. (A) Heatmap showing RE transcript families increased (red) and decreased (blue) by long- and short-term interventions, and in growth hormone receptor knockout (GHRKO) mice. (B and C) Venn diagrams showing the number of common decreased (blue) or increased (red) RE transcripts with long- and short-term interventions. (D) Weighted gene correlation network analysis (WGCNA) analysis heatmap of gene/RE modules influenced by long-term interventions (*modules that changed significantly with interventions, p < .001 in WGCNA). (E) Most specific biological processes (gene ontology terms) in each significant WGCNA module. Plots are based on the same data presented in Figure 1, including output from RepEnrich2 (A–C) and TEtranscripts (D and E). Exact p-values are noted in the Supplementary Data. All available samples/data from dataset GSE131901 were used for analyses (N = 6 mice/group).
Identifying potentially targetable biological mechanisms linking reduced RE expression with healthy aging interventions will require future experiments. However, to provide initial insight, we examined correlations among gene and RE expression patterns in mice subjected to long-term CR, rapamycin, and acarbose (treatments that reduced RE transcripts the most). To do this, we conducted a weighted gene correlationat and RE transcript counts (Figure 2). Although gene/RE signatures across interventions were not strikingly similar, we identified one WGCNA module (green) that decreased significantly with all interventions (Figure 2D). This module contained numerous DNA transposons and several LINE, ERV, and LTR transcripts (Supplementary Data). A GO analysis of the module also showed significant enrichment for many biological processes, including several linked with aging and disease (Figure 2E). In fact, the most specific GO terms included protein deacetylation, DNA repair, and immune activation/response pathways. The other gene/RE modules that decreased with CR and/or rapamycin were also enriched for specific GO terms including DNA repair, DNA/RNA processing, histone modifications, and stress responses (Figure 2E). These exploratory analyses do not definitively link reduced RE expression with such processes, but they fit with current thinking that age-related RE transcript accumulation could cause DNA damage (17) and immune activation/inflammation (6), and that RE dysregulation may be due to age-associated changes in chromatin/histones (18). Consistent with this latter possibility, we note that the TEtranscripts program can distinguish between unique RE reads (e.g., those embedded in genes) and multimapping reads, and that the age/treatment RE effects we observe occur in multimapped reads—suggesting underlying mechanisms like global chromatin dysregulation rather than age-related gene expression changes.
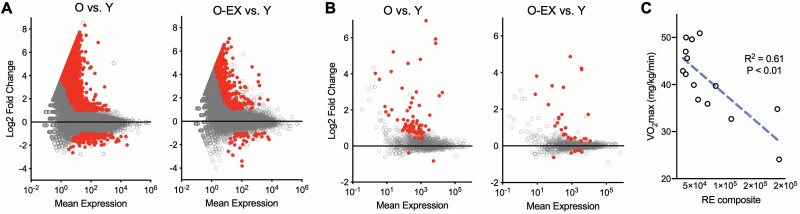
There is little or no RNA-seq data on true long-term CR or healthy aging compounds in older humans, as these are challenging clinical interventions to conduct (19). However, one well-studied intervention/behavior associated with increased healthspan and biological effects similar to CR is aerobic exercise (20). Others have studied the transcriptomic effects of clinical exercise interventions on select tissues, but proportionally, no studies have been nearly as long as an 8-month mouse intervention (~25% of the animal’s life). Therefore, as initial proof of concept, we conducted a cross-sectional study to determine if long-term exposure to this healthy aging behavior has the potential to reduce RE transcripts (Figure 3). We performed RNA-seq and gene/RE expression analyses on PBMCs from (a) young and older sedentary adults and (b) older habitually (≥5 years) exercising adults (Supplementary Data). Consistent with other reports (21), we found that older age was associated with altered PBMC gene expression, but these changes were attenuated in older exercising adults (Figure 3A). Moreover, in support of the idea that healthy aging interventions/behaviors may reduce RE expression in humans, we observed an increase in global RE transcript levels in older sedentary adult PBMCs, but this effect was strongly attenuated with exercise (Figure 3B). Similar to our findings in mice subjected to healthy aging interventions, we observed that many (~80%) of the transcripts reduced in older exercising adults versus their sedentary peers were also reduced in young versus older subjects, but we did not note any particular enrichment for specific RE types (Supplementary Figure 2). We did, however, find that maximal aerobic exercise capacity (VO2 max) was inversely related to a composite count of RE that are significantly increased with aging (Figure 3C), suggesting that greater aerobic fitness (and perhaps exposure to aerobic exercise) is directly linked with reduced RE expression. In fact, even when we performed multiple regression to account for other physiological variables related to health/aging (e.g., cholesterol, fasting glucose, etc.), VO2 max remained the strongest correlate of RE count (Supplemental Data). Interestingly, VO2 max is considered a key physiological predictor of longevity in humans (22), further demonstrating that REs may have an important role in human healthspan/lifespan.
Figure 3.
Long-term exercise is associated with reduced repetitive element (RE) transcript expression in humans. (A) MA plots showing gene expression differences in peripheral blood mononuclear cells of older (O) vs young (Y) sedentary, and older exercising (O-EX) vs Y adults. N = 4–5 matched samples per group. Note the ~1,200 transcripts significantly increased/decreased with aging but largely reversed with exercise (red data points, FDR < 0.1 and Log2 fold change >1, identified using Deseq2). (B) MA plots showing RE transcript levels in the same samples/subjects. Note general upward shift and numerous significantly increased RE transcripts with aging (red, FDR < 0.1, identified using Deseq2) that are largely reversed with exercise. (C) Correlation between maximal aerobic exercise capacity (VO2 max) and composite count of RE transcripts significantly increased with aging (O vs Y) in all subjects. All plots are based on data generated using the TEtranscripts pipeline (which includes gene expression counts), and human subjects characteristics are provided in the Supplementary Data.
Collectively, our results support the growing idea that global RE dysregulation may be an important mechanism of aging (and not simply an adverse effect of the process). Reversing age-related RE transcript accumulation may be necessary for healthy aging, as our present findings show that robust and reproducible health/lifespan-enhancing interventions consistently reduce RE expression. Indeed, we and others have reported age-associated increases in most RE transcript types (14,23), and this suggests a fundamental cellular mismanagement of REs with aging, which could have numerous deleterious causes and effects. For example, our current study suggests that histone modifications, DNA damage, and immune/inflammatory responses may be linked with RE dysregulation. Although our results are based solely on RNA-seq data and limited tissue types (liver and peripheral blood cells), we and others have demonstrated a link between RE transcripts and aging/healthy aging interventions using additional techniques (e.g., fluorescence in situ hybridization) and in other cells/tissues (e.g., fibroblasts, whole organisms) (7,24). Additional studies are required to determine if other anti-aging treatments (e.g., resveratrol, NAD+ activators) or pro-aging treatments (e.g., chemotherapy) influence REs. Future studies should also characterize RE dysregulation with aging in other metabolically active tissues (e.g., muscle) and key organs (e.g., the brain) and investigate underlying mechanisms, as this could lead to ideas for novel/potentially complementary healthy aging interventions.
Materials and Methods
RNA-seq Datasets and Availability
The data that support these findings can be found on the Gene Expression Omnibus website under accession numbers: GSE92486 (long-term CR), GSE131901 (8- and 2-month CR and pharmacological treatments), GSE87565 (HFD), and GSE153100 (human exercise).
Bioinformatics Analyses
RE transcripts were analyzed using the TEtranscripts and RepEnrich2 algorithms as previously described (7). Briefly, reads were trimmed, quality filtered with fastp, and then aligned to the genome (mm10 Mus musculus or Hg38 Homo sapiens) using Bowtie2 (RepEnrich2) or the STAR aligner (TEtranscripts). RE transcripts were then quantified using either TEtranscripts or RepEnrich2, which are pipelines to quantify RE transcripts by individual total counts, class, and family. Gene expression counts were extracted from bam alignment files produced during TEtranscripts analyses, and differential expression analyses of both REs and genes were performed using Deseq2 software. Weighted gene correlation network analysis (WGCNA) was performed according to standard procedures using normalized gene and RE counts for all samples, and a minimum module size of 300 to capture broader groups of REs that correlated with sample traits (specific interventions). All differential expression analyses and WGCNA calculations were performed using the same size factors, based on total library size normalized to the geometric mean of all libraries within each comparison. Gene ontology (GO) analyses of genes in the WGCNA modules were performed using the GOrilla algorithm, and specific GO modules were identified as terminal nodes in the directed acyclic graph produced by this program.
Human Subjects and RNA-seq Samples
RNA-seq was performed on peripheral blood mononuclear cells (PBMCs) from 12 healthy young (18–22 years) and older (62–74 years) adults. Subjects were non-obese, non-smokers, and healthy as assessed by medical history, physical examination, blood chemistries, and exercise ECG, and small groups were selected to match characteristics as closely as possible. Young (n = 5, 2 male) and older (n = 5, 2 male) sedentary subjects performed no regular exercise (<2 d/wk, < 30 min/d), whereas older exercising subjects (n = 4, 1 male) performed regular vigorous aerobic exercise (≥5 d/wk, >45 min/d) for the previous ≥5 years. The study conformed to the Declaration of Helsinki; all procedures were approved by the Institutional Review Board of the University of Colorado Boulder, and written informed consent was obtained from all subjects. Maximal oxygen consumption (VO2max) was assessed during treadmill exercise as previously described (11) and basic clinical measurements (eg, blood pressure) were performed using standard techniques. PBMCs were isolated from whole blood by traditional Ficoll gradient centrifugation, and RNA-seq and gene/RE expression analyses were performed using standard methods as previously described (12). Briefly, snap-frozen PBMC pellets were lysed in Trizol (Thermo), and RNA was recovered using a spin column kit (Direct-Zol, Zymo Research) that included a DNase I treatment to remove genomic DNA. Total RNA libraries were generated using Illumina Ribo-Zero kits to deplete ribosomal RNA, and libraries were sequenced on an Illumina NovaSeq 6000 platform to produce >40 M 150-bp single-end fastq reads per sample. Gene and RE expression analyses were performed as described above. RE composite scores for (stepwise) regression analyses were calculated as the total of all normalized RE reads that were increased with aging (i.e., in older vs young sedentary subjects), based on the idea that global rather than specific RE transcript accumulation may play a role in aging (although a composite RE score based on z-scores for each element yielded similar results).
Statistical Analyses
Differential expression of RE transcripts was quantified using Deseq2 software as previously described using size factors to account for library size differences among samples (12). Chi-squared analyses, normality and t tests, as well as heatmaps of increased/decreased RE and WGCNA modules were performed/constructed using GraphPad Prism software, and multiple regression analyses of RE predictors in human subjects were performed using JMP software.
Supplementary Material
Supplementary data are available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Supplementary Figure 1. Gene and RE expression changes with healthy aging interventions.
Supplementary Figure 2. Gene and RE expression in older exercising adults.
Acknowledgments
D.W. designed the study, wrote the paper, generated/analyzed data, and provided conceptual insight; A.N.C. analyzed data, provided conceptual insight, and edited the paper; M.S. edited the paper and provided conceptual insight; D.R.S. provided human PBMC samples, edited the paper, and provided conceptual insight; T.J.L. designed the study, wrote the paper, analyzed data, and provided conceptual insight.
Funding
This work was supported by the National Institute on Aging (grant number AG060302).
Conflict of Interest
None declared.
References
- 1. Niccoli T, Partridge L. Ageing as a risk factor for disease. Curr Biol. 2012;22:R741–R752. doi: 10.1016/j.cub.2012.07.024 [DOI] [PubMed] [Google Scholar]
- 2. Tyshkovskiy A, Bozaykut P, Borodinova AA, et al. Identification and application of gene expression signatures associated with lifespan extension. Cell Metab. 2019;30:573–593.e8. doi: 10.1016/j.cmet.2019.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bourque G, Burns KH, Gehring M, et al. Ten things you should know about transposable elements. Genome Biol. 2018;19:199. doi: 10.1186/s13059-018-1577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goodier JL. Restricting retrotransposons: a review. Mob DNA. 2016;7:16. doi: 10.1186/s13100-016-0070-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saleh A, Macia A, Muotri AR. Transposable elements, inflammation, and neurological disease. Front Neurol. 2019;10:894. doi: 10.3389/fneur.2019.00894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. LaRocca TJ, Cavalier AN, Wahl D. Repetitive elements as a transcriptomic marker of aging: evidence in multiple datasets and models. Aging Cell. 2020;19:e13167. doi: 10.1111/acel.13167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maxwell PH. What might retrotransposons teach us about aging? Curr Genet. 2016;62:277–282. doi: 10.1007/s00294-015-0538-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cardelli M. The epigenetic alterations of endogenous retroelements in aging. Mech Ageing Dev. 2018;174:30–46. doi: 10.1016/j.mad.2018.02.002 [DOI] [PubMed] [Google Scholar]
- 10. Wood JG, Jones BC, Jiang N, et al. Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila. Proc Natl Acad Sci USA. 2016;113:11277–11282. doi: 10.1073/pnas.1604621113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. LaRocca TJ, Seals DR, Pierce GL. Leukocyte telomere length is preserved with aging in endurance exercise-trained adults and related to maximal aerobic capacity. Mech Ageing Dev. 2010;131:165–167. doi: 10.1016/j.mad.2009.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. LaRocca TJ, Mariani A, Watkins LR, Link CD. TDP-43 knockdown causes innate immune activation via protein kinase R in astrocytes. Neurobiol Dis. 2019;132:104514. doi: 10.1016/j.nbd.2019.104514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hahn O, Grönke S, Stubbs TM, et al. Dietary restriction protects from age-associated DNA methylation and induces epigenetic reprogramming of lipid metabolism. Genome Biol. 2017;18:56. doi: 10.1186/s13059-017-1187-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Cecco M, Criscione SW, Peterson AL, Neretti N, Sedivy JM, Kreiling JA. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY). 2013;5:867–883. doi: 10.18632/aging.100621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim DH, Bang E, Jung HJ, et al. Anti-aging effects of calorie restriction (CR) and CR mimetics based on the senoinflammation concept. Nutrients. 2020;12(2). doi: 10.3390/nu12020422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siersbæk M, Varticovski L, Yang S, et al. High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss. Sci Rep. 2017;7:40220. doi: 10.1038/srep40220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hedges DJ, Deininger PL. Inviting instability: transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 2007;616:46–59. doi: 10.1016/j.mrfmmm.2006.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Booth LN, Brunet A. The aging epigenome. Mol Cell. 2016;62:728–744. doi: 10.1016/j.molcel.2016.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Di Francesco A, Di Germanio C, Bernier M, de Cabo R. A time to fast. Science. 2018;362:770–775. doi: 10.1126/science.aau2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seals DR, Justice JN, LaRocca TJ. Physiological geroscience: targeting function to increase healthspan and achieve optimal longevity. J Physiol. 2016;594:2001–2024. doi: 10.1113/jphysiol.2014.282665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gano LB, Donato AJ, Pierce GL, et al. Increased proinflammatory and oxidant gene expression in circulating mononuclear cells in older adults: amelioration by habitual exercise. Physiol Genomics. 2011;43:895–902. doi: 10.1152/physiolgenomics.00204.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Strasser B, Burtscher M. Survival of the fittest: VO2max, a key predictor of longevity? Front Biosci (Landmark Ed). 2018;23:1505–1516. doi: 10.2741/4657 [DOI] [PubMed] [Google Scholar]
- 23. Sedivy JM, Kreiling JA, Neretti N, et al. Death by transposition—the enemy within? Bioessays. 2013;35:1035–1043. doi: 10.1002/bies.201300097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Cecco M, Ito T, Petrashen AP, et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 2019;572:E5. doi: 10.1038/s41586-019-1350-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.