Abstract
Objective
The purpose of this study was to investigate the mechanism of action of the Chinese herbal formula Buyang Huanwu Decoction (BYHWD), which is commonly used to treat nerve injuries, in the treatment of spinal cord injury (SCI) using a network pharmacology method.
Methods
BYHWD-related targets were obtained by mining the TCMSP and BATMAN-TCM databases, and SCI-related targets were obtained by mining the DisGeNET, TTD, CTD, GeneCards, and MalaCards databases. The overlapping targets of the abovementioned targets may be potential therapeutic targets for BYHWD anti-SCI. Subsequently, we performed protein-protein interaction (PPI) analysis, screened the hub genes using Cytoscape software, performed Gene Ontology (GO) annotation and KEGG pathway enrichment analysis, and finally achieved molecular docking between the hub proteins and key active compounds.
Results
The 189 potential therapeutic targets for BYHWD anti-SCI were overlapping targets of 744 BYHWD-related targets and 923 SCI-related targets. The top 10 genes obtained subsequently included AKT1, IL6, MAPK1, TNF, TP53, VEGFA, CASP3, ALB, MAPK8, and JUN. Fifteen signaling pathways were also screened out after enrichment analysis and literature search. The results of molecular docking of key active compounds and hub target proteins showed a good binding affinity for both.
Conclusion
This study shows that BYHWD anti-SCI is characterized by a multicomponent, multitarget, and multipathway synergy and provides new insights to explore the specific mechanisms of BYHWD against SCI.
1. Introduction
Spinal cord injury (SCI) is a group of disorders in which the integrity and continuity of the spinal cord are disrupted by trauma, tumor, hypoxia, inflammation, and other factors [1]. According to the World Health Organization (WHO) report, about 250,000 to 500,000 people worldwide suffer from SCI every year, and the population is mainly under 30 years old [2]. The Beijing SCI Epidemiology Survey Group reported in 2002 that the incidence of SCI in Beijing was about 60/106, an increase of nearly 10 times compared to the late 1980s, and that the per capita hospitalization cost was 27819.3 CNY [3]. The high disability rate and high consumption of SCI have imposed a heavy burden on the families of patients and society, while how to protect and restore injured neurons after SCI has been considered a great challenge in clinical and experimental research [4]. There is still a lack of effective treatment for SCI, but it is known that traditional Chinese medicine (TCM) has a long history and rich practical experience, which may be explored for the treatment of SCI [2].
Modern Chinese medicine scholars classify SCI as “Ti Duo” and “Wei Zheng” based on the TCM classics “Ling Shu Jing-Han Re Bing” and “Su Wen-Wei Lun” [3]. Based on the analysis of the etiology and pathogenesis of SCI by TCM, as well as the pathological changes of the damaged Governor Vessel and the symptoms corresponding to SCI, the main treatment of TCM is to unblock Governor Vessel and strengthen “Yang Qi,” supplemented by tonifying the kidney and filling the essence, strengthening the tendons and bones, activating blood circulation and removing blood stasis, and warming the meridians [3]. Buyang Huanwu Decoction (BYHWD), recorded in “Yi Lin Gai Cuo” by Wang Qingren in the Qing Dynasty, has been used in China for hundreds of years to treat stroke-related disabilities and improve the recovery of neurological function [5]. The whole formula of BYHWD consists of seven herbs: Chishao (Radix Paeoniae Rubra), Chuanxiong (Rhizoma Chuanxiong), Danggui (Radix Angelicae Sinensis), Dilong (Pheretima), Huangqi (Radix Astragali), Honghua (Flos Carthami), and Taoren (Semen Persicae) [5]. Studies have shown that BYHWD may promote the repair of SCI by upregulating the expression of Notch1 gene [6], and BYHWD combined with bone marrow mesenchymal stem cells (BMSCs) transplantation can promote the recovery after SCI by saving axotomized red nucleus neurons in rats [4]. Although some progress has been made in the study of BYHWD for SCI, the complexity of herbal prescriptions with multiple components, targets, and pathways makes the study of the specific mechanism of action of BYHWD for SCI difficult to clarify.
In the last decade, with the development of modern multiomics theories such as genomics, proteomics, and metabolomics, the introduction of a systems biology perspective, and the application of bioinformatics, the concept of network pharmacology has emerged. According to the network basis of drug action, network pharmacology methods, including protein-protein interaction (PPI) network, Gene Ontology (GO) annotation, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, are applied to infer the specific pathways of multimolecular drug action in the human body, which is consistent with the multicomponent, multitarget, and multipathway synergy of Chinese herbal medicine and helps to clarify the mechanism of action of BYHWD (Figure 1).
Figure 1.
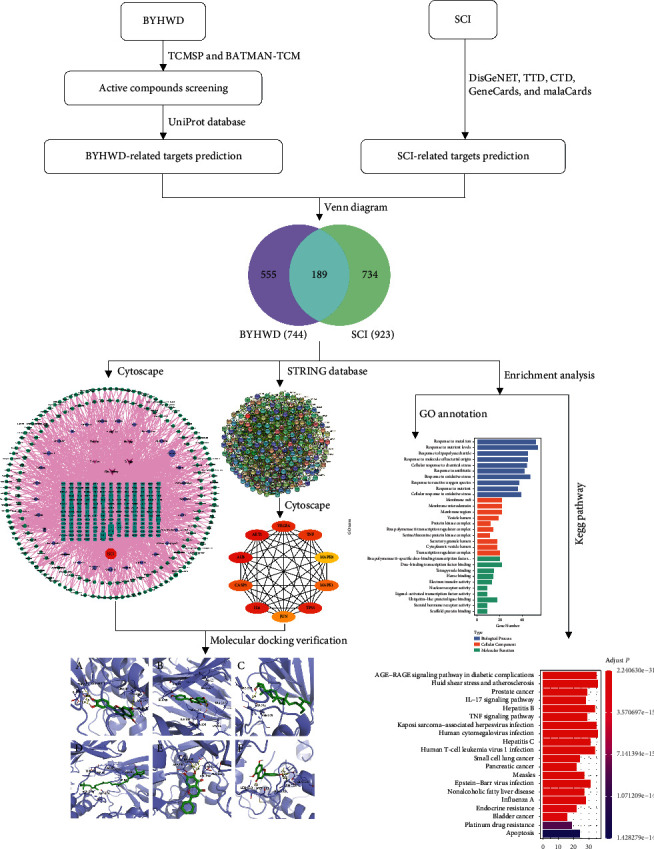
The overall flowchart of this study.
2. Materials and Methods
2.1. Acquisition of BYHWD-Related Targets and SCI-Related Targets
The compounds related to BYHWD were obtained by searching “Chishao or CHI SHAO or Radix Paeoniae Rubra,” “Chuanxiong or CHUAN XIONG or Rhizoma Chuanxiong,” “Danggui or DANG GUI or Radix Angelicae Sinensis,” “Dilong or DI LONG or Pheretima,” “Huangqi or HUANG QI or Radix Astragali,” “Honghua or HONG HUA or Flos Carthami,” and “Taoren or TAO REN or Semen Persicae” in Traditional Chinese Medicine Systems Pharmacology (TCMSP, version 2.3, https://tcmspw.com/tcmsp.php) database [7], and then the active compounds that met the criteria (oral bioavailability (OB) ≥ 30 and drug-likeness (DL) ≥ 0.18) were further screened by the absorption, distribution, metabolism, and excretion (ADME) principle [8, 9], and the corresponding target protein information of the active compounds was also obtained. Subsequently, by using the UniProt database (https://www.uniprot.org/) (species should be set to “Homo sapiens”), the target protein name is converted into the corresponding gene name and UniProt ID [10]. The compounds and targets related to BYHWD were also obtained directly by searching the above keywords in the BATMAN-TCM platform (http://bionet.ncpsb.org/batman-tcm/) according to the set criteria (score cutoff >25, adjusted P value < 0.05) [11]. Finally, the results of the two databases are summarized, integrated, and deduplicated to obtain BYHWD-related targets.
DisGeNet database (https://www.disgenet.org/, v7.0) (score ≥ 0.01) [12], GeneCards (https://www.genecards.org/) (relevance score ≥ 20) [13], MalaCards (https://www.malacards.org/) [14], Comparative Toxicogenomics Database (CTD) (http://ctdbase.org/, last update by June 2020) (inference score ≥ 20) [15], and Therapeutic Target Database (TTD) (http://db.idrblab.net/ttd/, last update by June 1, 2020) [16] were used to screen out potential targets related to SCI by searching for the keyword “Spinal cord injury or Spinal cord diseases.” The results of the five databases are summarized, integrated, and deduplicated to obtain SCI-related targets.
2.2. Acquisition of Potential Therapeutic Targets for BYHWD Anti-SCI
Potential therapeutic targets of BYHWD in the treatment of SCI should be derived from the overlapping targets of BYHWD-related targets and SCI-related targets obtained from the Venn online tool (http://www.bioinformatics.com.cn/). Subsequently, we used Cytoscape software (version 3.7.2) to construct a network diagram of “BYHWD-active compounds-target genes-SCI” [17].
2.3. Construction of PPI Network and Acquisition of Hub Genes
PPI is composed of proteins that interact with each other to participate in various aspects of life processes such as biological signaling, regulation of gene expression, energy and material metabolism, and cell cycle regulation [18]. The potential therapeutic targets of BYHWD against SCI were analyzed by a PPI network using the STRING database (http://string-db.org/; version 11) with the species set to “Homo sapiens” and a confidence level greater than 0.4 [19]. Subsequently, the PPI network was visualized by Cytoscape software (version 3.7.2).
To obtain the hub genes for BYHWD anti-SCI, we use the CytoHubba plug-in in Cytoscape software, which provides 12 topological analysis methods, including Degree, Maximal Clique Centrality (MCC), BottleNeck, Betweenness, ClusteringCoefficient, Closeness, Density of Maximum Neighborhood Component (DMNC), EcCentricity, Edge Percolated Component (EPC), Maximum Neighborhood Component (MNC), Radiality, and Stress [20, 21].
2.4. GO and KEGG Pathway Enrichment Analysis of Potential Therapeutic Genes for BYHWD Anti-SCI
R is commonly used for statistical analysis and plotting software, and its open-source and free features have been favored by many programming enthusiasts, resulting in the birth of many convenient and efficient packages, such as the ClusterProfiler package for GO annotation and KEGG pathway enrichment analysis [22–24]. In this study, we used ClusterProfiler package (adjusted P value < 0.05) to explore the biological processes (BP), cellular components (CC), molecular functions (MF), and signaling pathways associated with BYHWD anti-SCI.
2.5. Molecular Docking of Hub Genes and Key Active Compounds
Based on the previously mined BYHWD-related targets and related active compounds, the active compounds and herbs corresponding to the top 10 hub genes need to be sorted out. The interrelationship between “herbs in BYHWD-active compounds-top 10 hub genes” was visualized by a Sankey diagram (http://sankeymatic.com/). In the following, we will select key active compounds as small molecule ligands and corresponding large molecule receptors (proteins encoded by the top 10 hub genes) for molecular docking analysis. The raw files of key active compounds (MOL2 format) can be downloaded from PubChem database (https://pubchem.ncbi.nlm.nih.gov/), and the raw files of the top 10 core genes (PDB format) can be downloaded from the RCSB protein data (http://www.rcsb.org/), and after corresponding processing by AutoDock Tool [25], they are finally converted to PDBQT format for molecular docking in Pymol software (https://pymol.org/2/; version 2.4.1) [26]. The molecular docking score reflects the binding affinity, with smaller values representing higher affinity for the binding of small molecule ligands and large molecule receptors [27].
3. Results
3.1. Acquisition Results of BYHWD-Related Targets and SCI-Related Targets
A total of 775 compounds (BATMAN-TCM: 219; after deduplication: 189) in BYHWD were preliminarily retrieved from the TCMSP database and BATMAN-TCM platform (score cutoff >25; adjusted P value < 0.05), of which 119 (BATMAN-TCM: 8) were from Chishao, 189 (87) were from Chuanxiong, 125 (77) were from Danggui, 87 (23) were from Huangqi, 189 (22) were from Honghua, and 66 (2) were from Taoren. Relevant compounds and targets' information for Dilong was not retrieved in either database. After setting the ADME criteria (OB ≥ 30% and DL ≥ 0.18), a total of 103 active compounds (after removing duplicates and nontarget compounds: 60) were screened from the TCMSP database, of which 29 (After removing nontarget compounds: 14) were from Chishao, 7 (6) were from Chuanxiong, 2 (2) were from Danggui, 20 (17) were from Huangqi, 22 (17) were from Honghua, and 23 (19) were from Taoren. The BATMAN-TCM platform provides the gene names directly, while the TCMSP database first provides the target protein names of the active compounds and therefore needs to be converted to gene names via the UniProt database. Finally, after integrating the results of the two databases, we obtained 244 BYHWD-related active compounds and 744 BYHWD-related targets.
After deduplication, a total of 923 SCI-related targets were obtained by mining the DisGeNET (number: 84), TTD (number: 5), CTD (number: 553), GeneCards (number: 432), and MalaCards (number: 19) databases.
3.2. Acquisition Results of Potential Therapeutic Targets for BYHWD Anti-SCI
The overlapping targets of BYHWD-related targets and SCI-related targets were considered as potential therapeutic targets for BYHWD anti-SCI. As shown in Figure 2, we screened the overlapping targets of both by constructing a Venn diagram, and a total of 189 targets were obtained. Through multiple databases mining and construction of the Venn diagram, we obtained BYHWD-related compounds and potential therapeutic targets for BYHWD anti-SCI and finally used Cytoscape software to construct a “BYHWD-active compounds-target genes-SCI” network (Figure 3). In Figure 3, the lines between two nodes represent the existence of mutual relationships, and the larger the node is, the more relationships exist.
Figure 2.
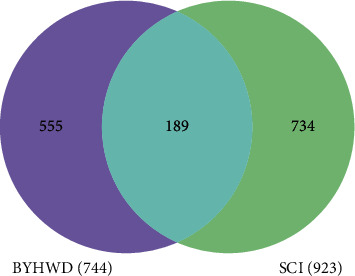
Venn diagram for BYHWD-related targets and SCI-related targets.
Figure 3.
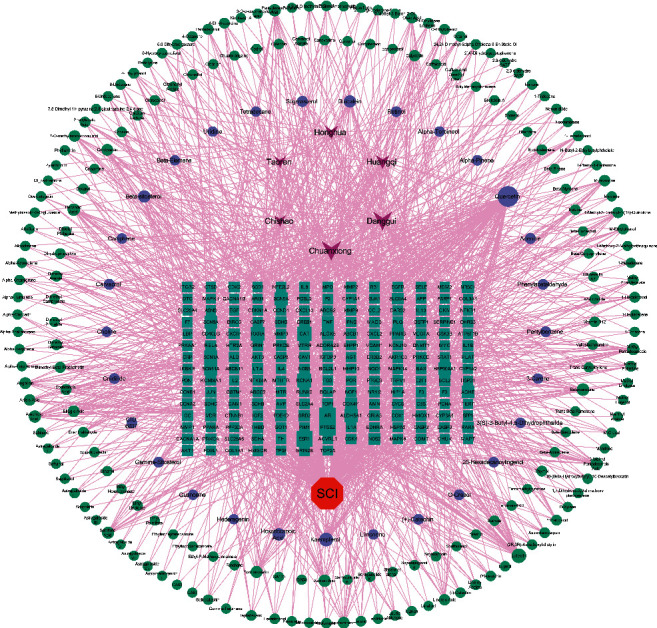
BYHWD-active compounds-target genes-SCI” network. The purple diamond represents the herbs contained in BYHWD; the red octagon represents SCI; the cyan rectangle represents the potential therapeutic targets for BYHWD anti-SCI; the light green ellipse represents the active compounds contained in BYHWD; the blue ellipse represents the common active compounds of the herb.
3.3. Acquisition Results of Hub Genes for BYHWD Anti-SCI and PPI Network Construction
In the STRING database, we first set the species to “Homo sapiens” and then entered 189 potential therapeutic targets for BYHWD anti-SCI to obtain the PPI network (Figure 4(a)), which involved 189 nodes and 3717 edges. The obtained TSV files were then imported into Cytoscape software (version 3.7.2) for further analysis and visualization (Figure 4(b)). To obtain the hub genes for BYHWD anti-SCI, based on the above the PPI network, we used the CytoHubba plug-in of Cytoscape software, which currently contains 12 topological analysis methods. We targeted the top 10 hub genes for BYHWD anti-SCI, and the results of the 12 algorithms each contained 10 genes. We integrated and analyzed the results to obtain a total of 45 different genes, which were then sorted by the number of algorithms to which they were attributed. Table 1 shows the basic information on hub genes for BYHWD anti-SCI. The top 10 hub genes (AKT1, IL6, MAPK1, TNF, TP53, VEGFA, CASP3, ALB, MAPK8, and JUN) sorted are completely consistent with some algorithms (Degree, Closeness, and MNC) (Figure 4(c)).
Figure 4.
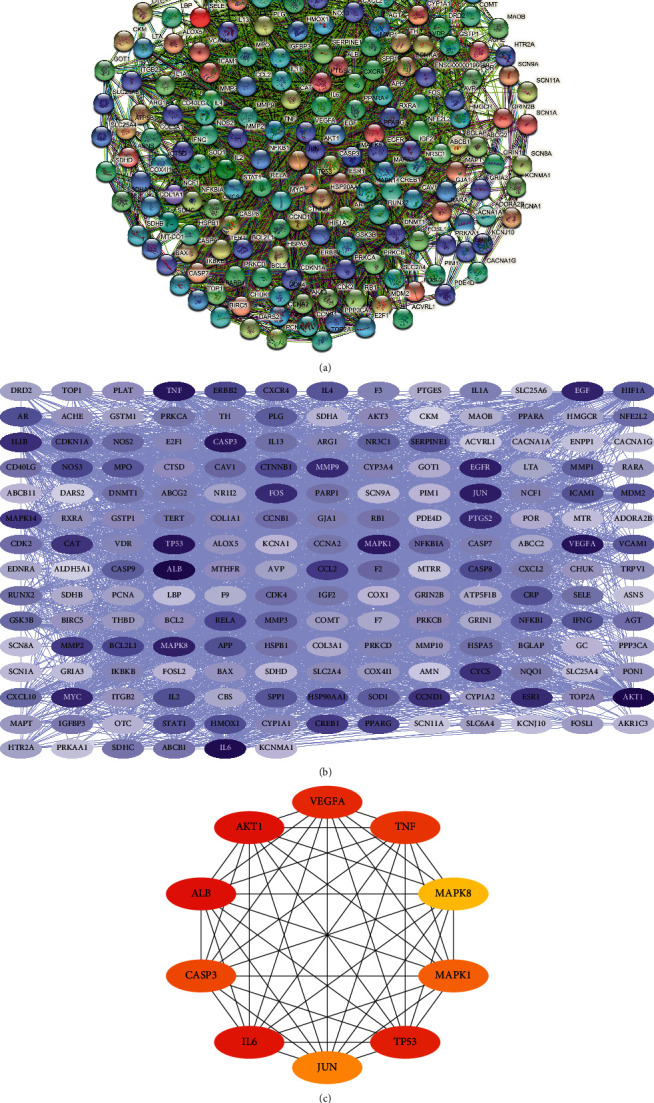
PPI network and top 10 hub genes for BYHWD anti-SCI. (a) PPI network constructed with STRING. (b) PPI network constructed with Cytoscape software (the darker the node color, the higher the number of connected proteins). (c) Top 10 hub genes for BYHWD anti-SCI were obtained by using the Degree algorithm.
Table 1.
Basic information of hub genes based on 12 topological algorithms.
| UniProt ID | Gene symbol | Protein names | Algorithms |
|---|---|---|---|
| P31749 | AKT1 | RAC-alpha serine/threonine-protein kinase | (1), (2), (3), (4), (5), (8), (10), (11), and (12) |
| P05231 | IL6 | Interleukin-6 | (1), (2), (3), (4), (5), (8), (10), (11), and (12) |
| P28482 | MAPK1 | Mitogen-activated protein kinase 1 | (1), (2), (3), (5), (8), (9), (10), (11), and (12) |
| P01375 | TNF | Tumor necrosis factor | (1), (2), (3), (4), (5), (8), (10), (11), and (12) |
| P04637 | TP53 | Cellular tumor antigen p53 | (1), (2), (3), (4), (5), (8), (10), (11), and (12) |
| P15692 | VEGFA | Vascular endothelial growth factor A | (1), (2), (3), (4), (5), (8), (10), (11), and (12) |
| P42574 | CASP3 | Caspase-3 | (1), (2), (3), (5), (8), (10), (11), and (12) |
| P02768 | ALB | Albumin | (1), (3), (5), (8), (10), (11), and (12) |
| P45983 | MAPK8 | Mitogen-activated protein kinase 8 | (1), (2), (4), (5), (8), and (10) |
| P05412 | JUN | Transcription factor AP-1 | (1), (2), (5), (10), and (11) |
| P04040 | CAT | Catalase | (3), (4), and (12) |
| P99999 | CYCS | Cytochrome c | (3) and (12) |
| P09238 | MMP10 | Stromelysin-2 | (6) and (7) |
| P07101 | TH | Tyrosine 3-monooxygenase | (4) and (9) |
| P37023 | ACVRL1 | Serine/threonine-protein kinase receptor R3 | (6) |
| P01019 | AGT | Angiotensinogen | (9) |
| P09917 | ALOX5 | Polyunsaturated fatty acid 5-lipoxygenase | (6) |
| P01185 | AVP | Vasopressin-neurophysin 2-copeptin | (9) |
| Q07812 | BAX | Apoptosis regulator BAX | (6) |
| O15392 | BIRC5 | Baculoviral IAP repeat-containing protein 5 | (6) |
| P55211 | CASP9 | Caspase-9 | (7) |
| P20248 | CCNA2 | Cyclin-A2 | (7) |
| P14635 | CCNB1 | G2/mitotic-specific cyclin-B1 | (7) |
| P13073 | COX4I1 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | (9) |
| P02741 | CRP | C-reactive protein | (4) |
| P02778 | CXCL10 | C-X-C motif chemokine 10 | (7) |
| P00533 | EGFR | Epidermal growth factor receptor | (11) |
| P15407 | FOSL1 | Fos-related antigen 1 | (6) |
| P15408 | FOSL2 | Fos-related antigen 2 | (6) |
| Q13224 | GRIN2B | Glutamate receptor ionotropic, NMDA 2B | (9) |
| Q16665 | HIF1A | Hypoxia-inducible factor 1-alpha | (9) |
| P04792 | HSPB1 | Heat shock protein beta-1 | (7) |
| P05362 | ICAM1 | Intercellular adhesion molecule 1 | (7) |
| P35225 | IL13 | Interleukin-13 | (7) |
| P05112 | IL4 | Interleukin-4 | (7) |
| P01374 | LTA | Lymphotoxin-alpha | (6) |
| P01106 | MYC | Myc proto-oncogene protein | (8) |
| P29474 | NOS3 | Nitric oxide synthase, endothelial | (9) |
| O14684 | PTGES | Prostaglandin E synthase | (6) |
| P35354 | PTGS2 | Prostaglandin G/H synthase 2 | (2) |
| Q15858 | SCN9A | Sodium channel protein type 9 subunit alpha | (4) |
| P16581 | SELE | E-selectin | (7) |
| P07204 | THBD | Thrombomodulin | (6) |
| Q8NER1 | TRPV1 | Transient receptor potential cation channel subfamily V member 1 | (9) |
| P11473 | VDR | Vitamin D3 receptor | (9) |
(1): Degree, (2): Maximal Clique Centrality (MCC), (3): Betweenness, (4): BottleNeck, (5): Closeness, (6): ClusteringCoefficient, (7): Density of Maximum Neighborhood Component (DMNC), (8): Edge Percolated Component (EPC), (9): EcCentricity, (10): Maximum Neighborhood Component (MNC), (11): Radiality, and (12): Stress
3.4. Results of GO and KEGG Pathway Enrichment Analysis
GO and KEGG pathway enrichment analysis are important tools to explore the biological processes and signal pathways involved in clinical drug therapy for diseases, as well as key genes. Through the ClusterProfiler package in R, we performed GO enrichment analysis on the 189 potential therapeutic targets for BYHWD anti-SCI and obtained 2842 GO items (after adjustment, P < 0.05), including 2567 BP items, 99 CC items, and 176 MF items. We combined the top 10 enrichment results for each of GO-BP, GO-CC, and GO-MF by a bar graph (Figure 5). The basic information of the top 10 GO enrichment items is shown in Table 2.
Figure 5.
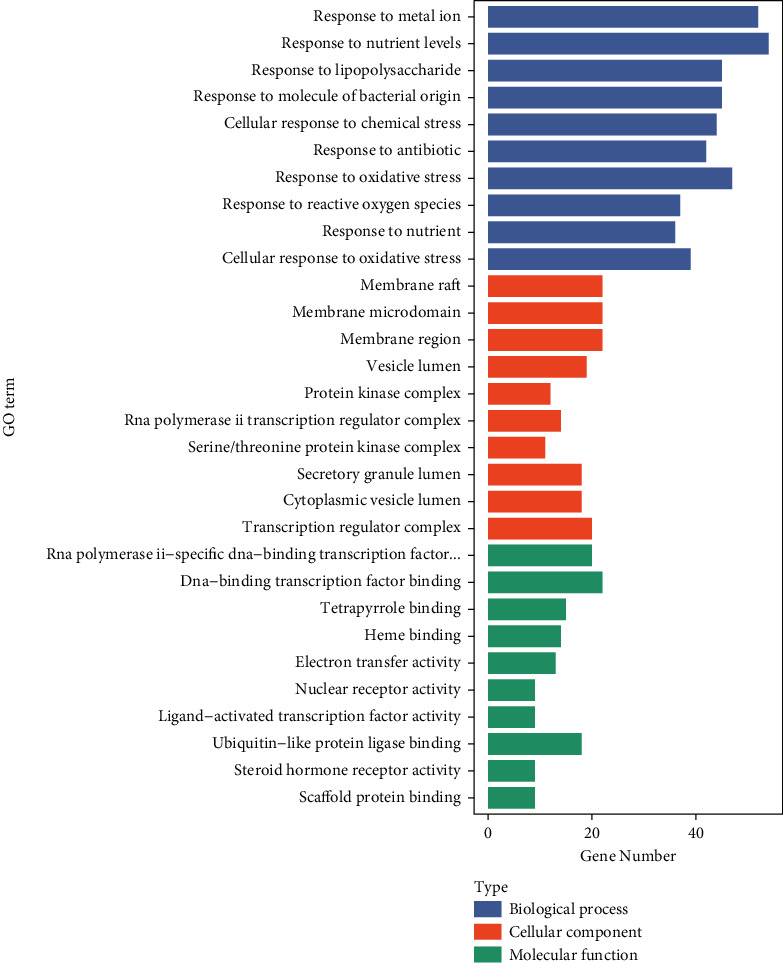
The bar chart of top 10 GO (BP, CC, and MF) enriched items.
Table 2.
Top 10 items of Gene Ontology (GO) enrichment analysis.
| GO items | ID | Description | P value | Adjusted P value | Gene number |
|---|---|---|---|---|---|
| Biological process | GO:0010038 | Response to metal ion | 2.68739E−45 | 1.20529E−41 | 52 |
| Biological process | GO:0031667 | Response to nutrient levels | 2.05974E−40 | 4.61896E−37 | 54 |
| Biological process | GO:0032496 | Response to lipopolysaccharide | 5.34106E−38 | 7.98488E−35 | 45 |
| Biological process | GO:0002237 | Response to molecule of bacterial origin | 3.1151E−37 | 3.4928E−34 | 45 |
| Biological process | GO:0062197 | Cellular response to chemical stress | 1.44121E−35 | 1.29277E−32 | 44 |
| Biological process | GO:0046677 | Response to antibiotic | 2.54411E−34 | 1.90172E−31 | 42 |
| Biological process | GO:0006979 | Response to oxidative stress | 3.05284E−34 | 1.956E−31 | 47 |
| Biological process | GO:0000302 | Response to reactive oxygen species | 9.13447E−34 | 5.12101E−31 | 37 |
| Biological process | GO:0007584 | Response to nutrient | 2.40193E−33 | 1.19696E−30 | 36 |
| Biological process | GO:0034599 | Cellular response to oxidative stress | 6.07803E−32 | 2.726E−29 | 39 |
| Cellular component | GO:0045121 | Membrane raft | 3.92204E−13 | 7.60946E−11 | 22 |
| Cellular component | GO:0098857 | Membrane microdomain | 4.18102E−13 | 7.60946E−11 | 22 |
| Cellular component | GO:0098589 | Membrane region | 8.84519E−13 | 1.07322E−10 | 22 |
| Cellular component | GO:0031983 | Vesicle lumen | 4.00266E−10 | 3.64242E−08 | 19 |
| Cellular component | GO:1902911 | Protein kinase complex | 5.90811E−10 | 4.10759E−08 | 12 |
| Cellular component | GO:0090575 | RNA polymerase II transcription regulator complex | 7.53142E−10 | 4.10759E−08 | 14 |
| Cellular component | GO:1902554 | Serine/threonine-protein kinase complex | 7.89922E−10 | 4.10759E−08 | 11 |
| Cellular component | GO:0034774 | Secretory granule lumen | 2.08462E−09 | 9.48503E−08 | 18 |
| Cellular component | GO:0060205 | Cytoplasmic vesicle lumen | 2.53536E−09 | 1.02541E−07 | 18 |
| Cellular component | GO:0005667 | Transcription regulator complex | 3.48128E−09 | 1.26718E−07 | 20 |
| Molecular function | GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | 1.80793E−11 | 7.91763E−09 | 20 |
| Molecular function | GO:0140297 | DNA-binding transcription factor binding | 2.70226E−11 | 7.91763E−09 | 22 |
| Molecular function | GO:0046906 | Tetrapyrrole binding | 4.34881E−11 | 8.49468E−09 | 15 |
| Molecular function | GO:0020037 | Heme binding | 1.91194E−10 | 2.801E−08 | 14 |
| Molecular function | GO:0009055 | Electron transfer activity | 2.62858E−10 | 3.08069E−08 | 13 |
| Molecular function | GO:0004879 | Nuclear receptor activity | 1.43492E−09 | 1.20123E−07 | 9 |
| Molecular function | GO:0098531 | Ligand-activated transcription factor activity | 1.43492E−09 | 1.20123E−07 | 9 |
| Molecular function | GO:0044389 | Ubiquitin-like protein ligase binding | 5.74415E−09 | 4.20759E−07 | 18 |
| Molecular function | GO:0003707 | Steroid hormone receptor activity | 7.34487E−09 | 4.78232E−07 | 9 |
| Molecular function | GO:0097110 | Scaffold protein binding | 1.18514E−08 | 5.67261E−07 | 9 |
In the same way, through the ClusterProfiler package in R, we performed KEGG pathway enrichment analysis on the 189 potential therapeutic targets for BYHWD anti-SCI and obtained 177 KEGG pathways (after adjustment, P < 0.05). The top 20 KEGG enrichment pathways are listed below, including AGE-RAGE signaling pathway in diabetic complications (hsa04933), Fluid shear stress and atherosclerosis (hsa05418), Prostate cancer (hsa05215), IL-17 signaling pathway (hsa04657), Hepatitis B (hsa05161), TNF signaling pathway (hsa04668), Kaposi sarcoma-associated herpesvirus infection (hsa05167), Human cytomegalovirus infection (hsa05163), Hepatitis C (hsa05160), Human T-cell leukemia virus 1 infection (hsa05166), Small cell lung cancer (hsa05222), Pancreatic cancer (hsa05212), Measles (hsa05162), Epstein-Barr virus infection (hsa05169), Nonalcoholic fatty liver disease (hsa04932), Influenza A (hsa05164), Endocrine resistance (hsa01522), Bladder cancer (hsa05219), Platinum drug resistance (hsa01524), and Apoptosis (hsa04210). The visualization of the abovementioned KEGG enrichment pathway is realized in Figure 6. More KEGG pathways were enriched, and another fine screening of the pathways was required. Therefore, the SCI-related literature was searched in the PubMed database, and then the retrieved potentially relevant pathways were compared with the enriched 177 pathways, and finally, 15 relatively relevant pathways for SCI were obtained. Subsequently, we used Cytoscape software to visualize the network relationship between potential therapeutic targets for BYHWD anti-SCI and pathways (Figure 7). Table 3 shows the basic information of 15 pathways that may be relevant for SCI.
Figure 6.
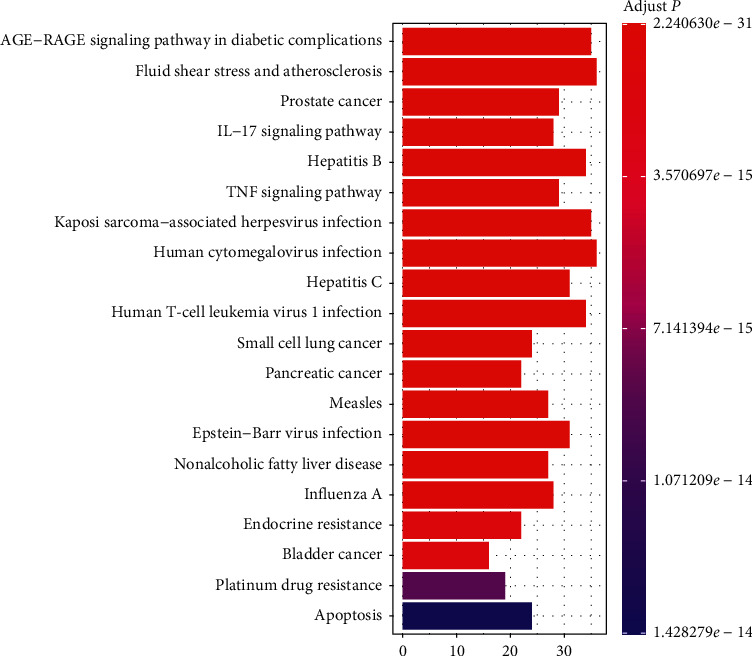
The bar chart of top 20 KEGG enriched pathways.
Figure 7.
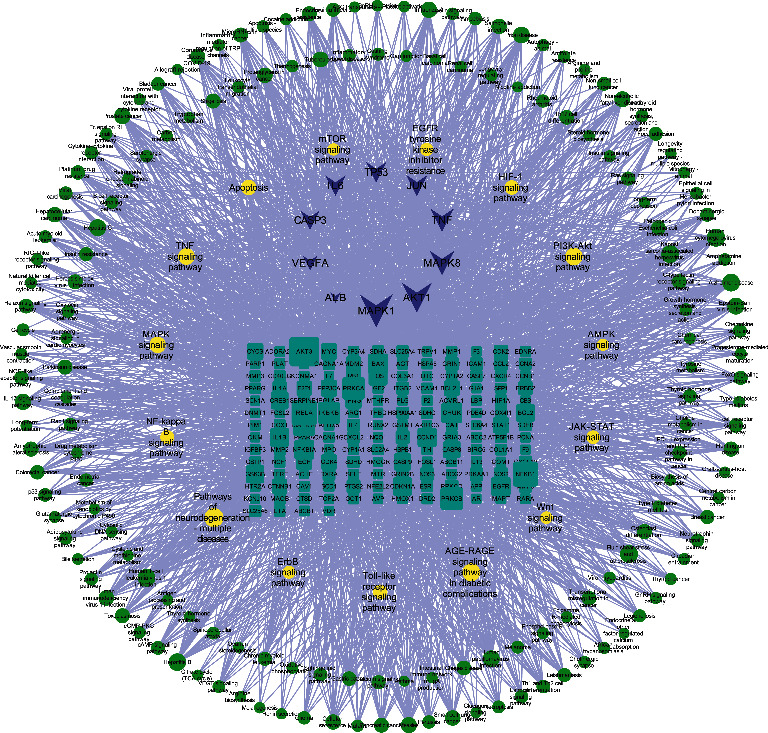
Potential therapeutic targets-pathways” network. The blue diamond represents the top 10 hub genes; the cyan rectangle represents the potential therapeutic targets for BYHWD anti-SCI; the yellow ellipse represents potential SCI-related pathways supported by literature; the green ellipse represents possible pathways related to SCI.
Table 3.
The enriched 15 possible related pathways for SCI.
| ID | Description | P value | Adjusted P value | Gene number |
|---|---|---|---|---|
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 8.58479E−34 | 2.24063E−31 | 35 |
| hsa04668 | TNF signaling pathway | 8.83261E−24 | 3.84219E−22 | 29 |
| hsa04210 | Apoptosis | 1.13129E−15 | 1.42828E−14 | 24 |
| hsa05022 | Pathways of neurodegeneration - multiple diseases | 1.12387E−14 | 1.0476E−13 | 41 |
| hsa04620 | Toll-like receptor signaling pathway | 5.71758E−14 | 4.52208E−13 | 20 |
| hsa04151 | PI3K-Akt signaling pathway | 1.43931E−13 | 1.00595E−12 | 34 |
| hsa04066 | HIF-1 signaling pathway | 1.4646E−13 | 1.00595E−12 | 20 |
| hsa04010 | MAPK signaling pathway | 5.84978E−12 | 3.39287E−11 | 29 |
| hsa04064 | NF-kappa B signaling pathway | 7.32616E−12 | 4.1568E−11 | 18 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 1.25843E−09 | 5.13202E−09 | 14 |
| hsa04012 | ErbB signaling pathway | 3.21508E−08 | 1.0622E−07 | 13 |
| hsa04630 | JAK-STAT signaling pathway | 4.40433E−07 | 1.27726E−06 | 16 |
| hsa04310 | Wnt signaling pathway | 0.000748937 | 0.001515292 | 11 |
| hsa04152 | AMPK signaling pathway | 0.001230137 | 0.002407294 | 9 |
| hsa04150 | mTOR signaling pathway | 0.002091493 | 0.003927191 | 10 |
3.5. Results of Molecular Docking of Hub Genes and Key Active Compounds
Sankey diagram was used to establish one-to-one correspondence between the top 10 hub genes (AKT1, IL6, MAPK1, TNF, TP53, VEGFA, CASP3, ALB, MAPK8, and JUN) for BYHWD anti-SCI and BYHWD-related compounds, providing the basis for the next molecular docking analysis. Among them, the gene that targets the most active compounds is TNF, and the key active compounds that target the most hub genes are quercetin and luteolin (Figure 8). As seen in the Sankey diagram, 6 of the 33 active compounds (quercetin, luteolin, beta-carotene, kaempferol, baicalein, and beta-sitosterol) targeted more hub genes, so we decided to perform the molecular docking between these 6 active compounds and top 10 hub genes. The results of the molecular docking score are presented as a heat map (Figure 9), with a range of −6.4 to −10.9 kcal·mol−1, representing the large molecule protein receptor (hub genes) that binds well to the small molecule ligand (key active compounds). As shown in Figure 10, the molecular docking process of each key active compound and its corresponding protein encoded by the hub gene with the best docking affinity was visualized using Pymol. Table 4 shows the basic information of active compounds targeting hub genes in BYHWD.
Figure 8.
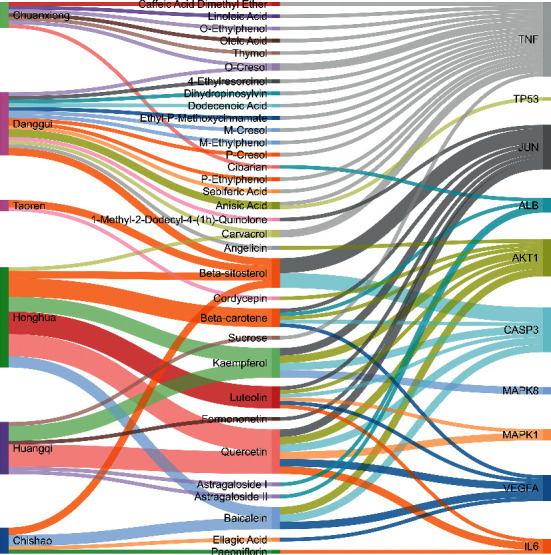
Sankey diagram of “herbs-active compounds-hub genes.” The band between the two bars represents a targeting relationship.
Figure 9.
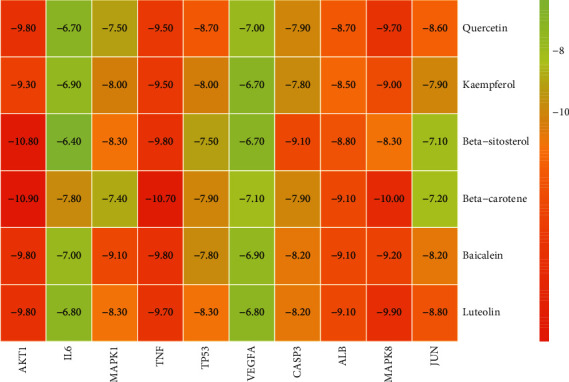
Heatmap of the scores of molecular docking in this study.
Figure 10.
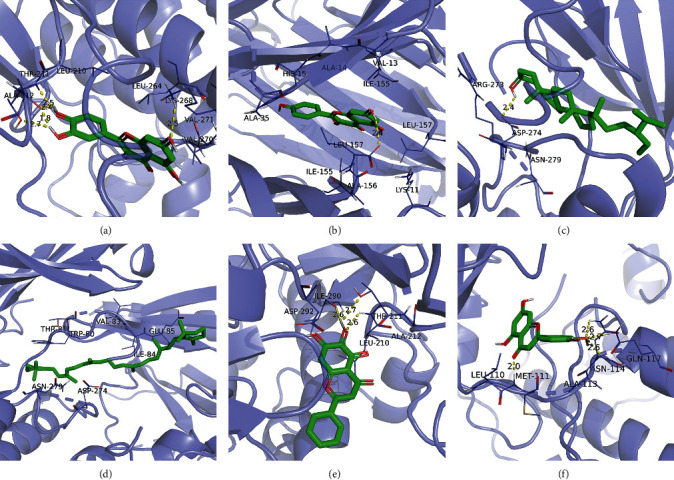
Results of molecular docking between hub genes and key active compounds. (a) Quercetin to AKT1. (b) Kaempferol to TNF. (c) Beta-sitosterol to AKT1. (d) Beta-carotene to AKT1. (e) Baicalein to AKT1. (f) Luteolin to MAPK8.
Table 4.
Basic information of active compounds targeting hub genes in BYHWD.
| Molecule ID | Molecule name | PubChem CID | OB (%) | DL | 2D Structure | Source database | Source | Targeted hub genes |
|---|---|---|---|---|---|---|---|---|
| MOL000098 | Quercetin | 5280343 | 46.43 | 0.28 |
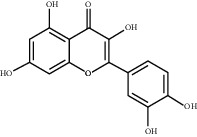
|
TCMSP | Honghua; Huangqi | AKT1, CASP3, IL6, JUN, MAPK1, and VEGFA |
| MOL000006 | Luteolin | 5280445 | 36.16 | 0.25 |
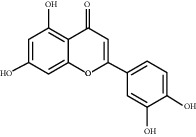
|
TCMSP | Honghua | AKT1, CASP3, IL6, JUN, MAPK1, and VEGFA |
| MOL002773 | Beta-carotene | 5280489 | 37.18 | 0.58 |
|
TCMSP | Honghua | AKT1, CASP3, ALB, JUN, and VEGFA |
| MOL000422 | Kaempferol | 5280863 | 41.88 | 0.24 |
|
TCMSP | Honghua; Huangqi | AKT1, CASP3, JUN, and MAPK8 |
| MOL002714 | Baicalein | 5281605 | 33.52 | 0.21 |
|
TCMSP | Chishao; Honghua | AKT1, CASP3, and VEGFA |
| MOL000358 | Beta-sitosterol | 222284 | 36.91 | 0.75 |
|
TCMSP | Chishao, Danggui, Honghua, and Taoren | CASP3 and JUN |
| MOL001002 | Ellagic acid | 5281855 | 43.06 | 0.43 |
|
TCMSP | Chishao | VEGFA |
| MOL001924 | Paeoniflorin | 442534 | 53.87 | 0.79 |
|
TCMSP | Chishao | IL6 |
| MOL000392 | Formononetin | 5280378 | 69.67 | 0.21 |
|
TCMSP | Huangqi | JUN |
| — | Anisic acid | 7478 | — | — |
|
BATMAN-TCM | Danggui | TNF and TP53 |
| — | Caffeic acid dimethyl ether | 717531 | — | — |
|
BATMAN-TCM | Chuanxiong | TNF |
| — | Cibarian | 100275 | — | — |
|
BATMAN-TCM | Chuanxiong | ALB |
| — | Linoleic acid | 5280450 | — | — |
|
BATMAN-TCM | Chuanxiong | TNF |
| — | O-Cresol | 335 | — | — |
|
BATMAN-TCM | Chuanxiong | TNF |
| — | O-Ethylphenol | 6997 | — | — |
|
BATMAN-TCM | Chuanxiong | TNF |
| — | Oleic acid | 445639 | — | — |
|
BATMAN-TCM | Chuanxiong | TNF |
| — | Thymol | 6989 | — | — |
|
BATMAN-TCM | Chuanxiong | TNF |
| — | 1-Methyl-2-dodecyl-4-(1h)-quinolone | 5319601 | — | — |
|
BATMAN-TCM | Danggui | JUN |
| — | 4-Ethylresorcinol | 17927 | — | — |
|
BATMAN-TCM | Danggui | TNF |
| — | Angelicin | 10658 | — | — |
|
BATMAN-TCM | Danggui | AKT1 |
| — | Carvacrol | 10364 | — | — |
|
BATMAN-TCM | Danggui | TNF |
| — | Dihydropinosylvin | 442700 | — | — |
|
BATMAN-TCM | Danggui | TNF |
| — | Dodecenoic acid | 96204 | — | — |
|
BATMAN-TCM | Danggui | TNF |
| — | Ethyl-P-methoxycinnamate | 5281783 | — | — |
|
BATMAN-TCM | Danggui | TNF |
| — | M-Cresol | 342 | — | — |
|
BATMAN-TCM | Danggui | TNF |
| — | M-Ethylphenol | 12101 | — | — |
|
BATMAN-TCM | Danggui | TNF |
| — | P-Cresol | 2879 | — | — |
|
BATMAN-TCM | Danggui | TNF |
| — | P-Ethylphenol | 31242 | — | — |
|
BATMAN-TCM | Danggui | TNF |
| — | Sebiferic acid | 5321206 | — | — |
|
BATMAN-TCM | Danggui | TNF |
| — | Astragaloside I | 13996685 | — | — |
|
BATMAN-TCM | Huangqi | ALB |
| — | Astragaloside II | 13996693 | — | — |
|
BATMAN-TCM | Huangqi | ALB |
| — | Sucrose | 5988 | — | — |
|
BATMAN-TCM | Huangqi | TNF |
| — | Cordycepin | 6303 | — | — |
|
BATMAN-TCM | Taoren | AKT1 |
OB, oral bioavailability; DL, drug-likeness.
4. Discussion
According to the report of the National SCI Statistics Center, there are about 17,000 new cases of SCI in the United States each year, and the annual incidence of SCI is about 54 cases per million people [1]. Over the past century, advances in understanding the mechanisms of SCI have changed the clinical management strategies for SCI, including surgical procedures, supportive measures, and rehabilitative training that have improved neurological outcomes and reduced morbidity in SCI patients [1]. However, there is still a lack of effective treatments for SCI and thus has been stimulating the desire of researchers to explore and try to find effective treatments, including drugs, surgical methods, and rehabilitation programs. As we all know, TCM has a long history of being used to treat various diseases in China and other Asian countries [28]. Among them, BYHWD, Jisuikang, single herb, and Governor Vessel electroacupuncture have all been shown in relevant studies to be used in the treatment of SCI [4, 5, 28–30].
BYHWD, which comes from Qing Dynasty medical classics, consists of seven herbs: Chishao, Chuanxiong, Danggui, Dilong, Huangqi, Honghua, and Taoren [5]. Based on the theory of TCM, Huangqi is the main ingredient in this formula, which has the function of tonifying Qi and nourishing blood, eliminating blood stasis without harming the proper function; Danggui is the secondary herb, which has the effect of invigorating blood and harmonizing blood; Chishao, Chuanxiong, Dilong, Honghua, and Taoren are the adjuvant herbs in this formula; all the five herbs have the function of activating blood circulation and removing blood stasis, promoting the flow of Qi and blood circulation; the combination of the five herbs not only activates blood circulation and removes blood stasis but also helps the Qi and blood that the main herbs benefit to reach the whole body [3, 31]. Studies have shown that BYHWD in combination with neural stem cells (NSCs) attenuates the process of demyelination or improves the recovery of myelin and exerts a synergistic effect on the neurological function recovery [5]. It has been shown that BYHWD restores hindlimb motor function in rats with SCI, and the neuroprotective effect is associated with the regulation of apoptosis-related protein expression [32]. BYHWD rescues axotomized neurons and promotes functional recovery after spinal cord injury in rats. A study published in 2008 showed that BYHWD protected the injured neurons and promoted functional recovery after SCI in rats [33]. However, since the specific mechanism of BYHWD for the treatment of SCI is still unclear due to its complex composition and wide range of effects, we decided to use a network pharmacology approach to investigate in this study and to lay the foundation for the next in-depth study.
In the first step, BYHWD-related compounds and related targets were obtained. We obtained relevant compounds and targets by mining two herbal-related databases, TCMSP and BATMAN-TCM. The mining methods of these two databases are different: TCMSP database (ADME principle: OB ≥ 30 and DL ≥ 0.18) first obtains BYHWD-related compounds, then obtains related target proteins, and finally needs to be converted into standard gene names by UniProt database; BATMAN-TCM platform (score cutoff >25, adjusted P value <0.05) directly obtains BYHWD-related compounds and target genes. The screening process of TCMSP database is divided into five steps: (1) the relevant compounds of each component of BYHWD are directly obtained (Chishao: 119, Chuanxiong: 189, Danggui: 125, Huangqi: 87, Honghua: 189, Taoren: 66, and Dilong: 0); (2) the relevant compounds of each component are screened by ADME criteria (Chishao: 29, Chuanxiong: 7, Danggui: 2, Huangqi: 20, Honghua: 22, Taoren: 23, and Dilong: 0); (3) the above compounds are screened again by removing the null targets (Chishao: 14, Chuanxiong: 6, Danggui: 2, Huangqi: 17, Honghua: 17, Taoren: 19, and Dilong: 0); (4) the above compounds are finally screened by removing duplicates (total number of active compounds: 60); and (5) the target proteins corresponding to the obtained active compounds are converted into gene names (total number of targets: 227). And the screening process of BATMAN-TCM platform is divided into two steps: (1) the relevant compounds and targets of each component of BYHWD are directly obtained (Chishao: 8, Chuanxiong: 87, Danggui: 77, Huangqi: 23, Honghua: 22, Taoren: 2, and Dilong: 0), and (2) the relevant compounds of each component are screened after deduplication (total number of active compounds: 189; the total number of targets: 586). The results of the two databases were integrated and deduplicated to finally obtain 244 BYHWD-related active compounds and 744 BYHWD-related targets. In the second step, a total of 923 SCI-related targets were acquired by mining 5 disease databases, including DisGeNET (number: 84), TTD (number: 5), CTD (number: 553), GeneCards (number: 432), and MalaCards (number: 19). Next, the Venn diagram of BYHWD-related targets and SCI-related targets was plotted by the website online tool to obtain 189 potential therapeutic targets of BYHWD against SCI.
In total, Cytoscape software was used four times in this study: (1) the “BYHWD-active compounds-target genes-SCI” network was constructed, involving 410 nodes and 1571 interactions; (2) the PPI network obtained from the STRING database was reconstructed here, involving 189 nodes and 3717 interactions; (3) the top 10 hub genes (AKT1, IL6, MAPK1, TNF, TP53, VEGFA, CASP3, ALB, MAPK8, and JUN) of BYHWD against SCI were obtained based on 12 topological analysis methods (Degree, MCC, Stress, ClusteringCoefficient, EcCentricity, BottleNeck, Closeness, Radiality, Betweenness, EPC, DMNC, and MNC) contained in the CytoHubba plug-in; (4) the “potential therapeutic targets-pathways” network was constructed, involving 347 nodes and 2650 interactions. Studies have shown that the protein encoded by TP53 and other factors regulate the regeneration, germination, and functional recovery of axons after central nervous system injury [34]; the rapid upregulation of CASP3 mRNA seen after SCI in rats may be associated with cell death in the spinal cord [35]; inflammatory mediators such as TNF-α and IL6 mediate the recruitment of inflammatory cells to the site of injury and by targeting these cytokines may be a potential strategy to reduce secondary injury in SCI [36, 37]; by targeting MAPK1, the overexpression of miRNA-433-5p protects motor dysfunction and inflammation after SCI [38].
The Sankey diagram was used once, mainly to show the correspondence between the top 10 hub genes and the corresponding active compounds contained in BYHWD (quercetin, luteolin, beta-carotene, kaempferol, baicalein, beta-sitosterol, carvacrol, O-cresol, anisic acid, 1-methyl-2-dodecyl-4-(1h)-quinolone, 4-ethylresorcinol, angelicin, astragaloside I, astragaloside II, caffeic acid dimethyl ether, cibarian, cordycepin, dihydropinosylvin, dodecenoic acid, ellagic acid, ethyl-P-methoxycinnamate, formononetin, linoleic acid, M-cresol, M-ethylphenol, O-ethylphenol, oleic acid, paeoniflorin, P-cresol, P-ethylphenol, sebiferic acid, sucrose, and thymol). Among them, quercetin targeted six hub genes, luteolin targeted six hub genes, beta-carotene targeted five hub genes, kaempferol targeted four hub genes, baicalein targeted three hub genes, beta-sitosterol targeted two hub genes, anisic acid targeted two hub genes, and the others targeted one hub gene. The experimental results showed that quercetin exerts a protective effect on the spinal cord by inhibiting the activation of the p38MAPK/iNOS signaling pathway in SCI rats and subsequently regulating secondary oxidative stress [39]; the combination of luteolin and palmitoylethanolamide reduced autophagy in SCI [40]; beta-carotene effectively reduced the course of secondary damage events after SCI by blocking NF-κB pathway activation [41]; baicalein may alleviate the harm caused by SCI by activating PI3K and inducing autophagy to reduce neuronal apoptosis [42].
Based on the results of the Sankey diagram, we performed molecular docking of the top 10 genes and the top 6 compounds in terms of the number of targeted genes (quercetin, luteolin, beta-carotene, kaempferol, baicalein, and beta-sitosterol). The results of molecular docking of 60 cohorts of hub target proteins and key active compounds showed a good binding affinity for both. We used R software twice, once to present the scores of the above molecular docking as a heat map, and another time to perform GO and KEGG pathway enrichment analysis using the ClusterProfiler package.
The top 10 results of the GO enrichment analysis of 189 potential therapeutic targets for BYHWD anti-SCI, based on the adjusted P value (from small to large), are as follows: response to metal ion (GO-BP:0010038), response to nutrient levels (GO-BP:0031667), response to lipopolysaccharide (GO-BP:0032496), response to molecule of bacterial origin (GO-BP:0002237), cellular response to chemical stress (GO-BP: 0062197), response to antibiotic (GO-BP:0046677), response to oxidative stress (GO-BP: 0006979), response to reactive oxygen species (GO-BP:0000302), response to nutrient (GO-BP:0007584), cellular response to oxidative stress (GO-BP: 0034599), membrane raft (GO-CC:0045121), membrane microdomain (GO-CC:0098857), membrane region (GO-CC:0098589), vesicle lumen (GO-CC: 0031983), protein kinase complex (GO-CC: 1902911), RNA polymerase II transcription regulator complex (GO-CC: 0090575), serine/threonine-protein kinase complex (GO-CC:1902554), secretory granule lumen (GO-CC: 0034774), cytoplasmic vesicle lumen (GO-CC: 0060205), transcription regulator complex (GO-CC: 0005667), RNA polymerase II-specific DNA-binding transcription factor binding (GO-MF:0061629), DNA-binding transcription factor binding (GO-MF:0140297), tetrapyrrole binding (GO-MF:0046906), heme binding (GO-MF:0020037), electron transfer activity (GO-MF: 0009055), nuclear receptor activity (GO-MF:0004879), ligand-activated transcription factor activity (GO-MF:0098531), ubiquitin-like protein ligase binding (GO-MF: 0044389), steroid hormone receptor activity (GO-MF:0003707), and scaffold protein binding (GO-MF:0097110). The first 20 results of the (adjusted P value based) KEGG pathway enrichment analysis of 189 potential therapeutic targets of BYHWD against SCI have been presented, and we screened the enriched pathways again to find possible relevant pathways for SCI. By searching the relevant literature, we screened a total of 15 pathways with strong relevance, as follows: AGE-RAGE signaling pathway in diabetic complications (hsa04933) [43], TNF signaling pathway (hsa04668), Apoptosis (hsa04210) [44], Pathways of neurodegeneration-multiple diseases (hsa05022), Toll-like receptor signaling pathway (hsa04620) [45], PI3K-Akt signaling pathway (hsa04151) [46], HIF-1 signaling pathway (hsa04066) [47], MAPK signaling pathway (hsa04010) [48], NF-kappa B signaling pathway (hsa04064) [49], EGFR tyrosine kinase inhibitor resistance (hsa01521) [50], JAK-STAT signaling pathway (hsa04630) [51], Wnt signaling pathway (hsa04310) [43], AMPK signaling pathway (hsa04152) [44], and mTOR signaling pathway (hsa04150) [46]. The above pathways and hub genes are not in a disconnected relationship; rather, many hub genes are important components of some of the pathways. Taking PI3K-Akt signaling pathway as an example, it contains AKT1, MAPK1, IL6, TP53, and VEGFA. Based on the results of the above network pharmacological analysis, we hypothesize that the key active compounds of BYHWD may exert anti-SCI effects through the above pathways and hub genes.
4.1. Limitations
This study still has some limitations. First, the corresponding experimental validation is lacking. Second, the compounds, targets, and pathways contained in these databases may not be exhaustive. Finally, compounds screened based on ADME principles may be missing other important compounds.
4.2. Future Perspectives
This study uncovered the multicompound, multitarget, and multipathway characteristics of BYHWD through a network pharmacology approach. Among them, the key compounds include quercetin, luteolin, beta-carotene, kaempferol, baicalein, and beta-sitosterol; the hub genes include AKT1, IL6, MAPK1, TNF, TP53, VEGFA, CASP3, ALB, MAPK8, and JUN; and the important pathways are the 15 related pathways mentioned above. These results clearly give a general direction for SCI-related researchers, and there is no doubt that the anti-SCI mechanism of BYHWD is much more than the above-mentioned research directions, and we will find more accurate targets and pathways while exploring them one by one.
5. Conclusion
Using a network pharmacology approach, we explored the potential mechanism of BYHWD as a classical herbal formula for treating nerve injury in the treatment of SCI. The top 10 core genes among the potential therapeutic targets for BYHWD anti-SCI include AKT1, IL6, MAPK1, TNF, TP53, VEGFA, CASP3, ALB, MAPK8, and JUN. Key active compounds of BYHWD against SCI include quercetin, luteolin, beta-carotene, kaempferol, baicalein, and beta-sitosterol. In addition, combining the KEGG pathway enrichment results and reviewing SCI-related literature, we obtained 15 SCI potentially relevant pathways, mainly including PI3K-Akt signaling pathway, MAPK signaling pathway, NF-kappa B signaling pathway, TNF signaling pathway, Apoptosis, Toll-like receptor signaling pathway, HIF-1 signaling pathway, JAK-STAT signaling pathway, Wnt signaling pathway, AMPK signaling pathway, and mTOR signaling pathway. The above analysis results demonstrated the multitarget, multicomponent, and multipathway characteristics of BYHWD in treating SCI, which laid the foundation for us to carry out specific experimental validation in the next step, and provided new ideas for researchers dedicated to the treatment of SCI with herbal extracts.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (no. 81873141).
Data Availability
The data used to support the findings of this study are included within the article.
Disclosure
Zhencheng Xiong and Feng Yang are joint first authors.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this article.
References
- 1.Courtine G., Sofroniew M. V. Spinal cord repair: advances in biology and technology. Nature Medicine. 2019;25(6):p. 898. doi: 10.1038/s41591-019-0475-6. [DOI] [PubMed] [Google Scholar]
- 2.Zheng Y., Qi S., Wu F., et al. Chinese herbal medicine in treatment of spinal cord injury: a systematic review and meta-analysis of randomized controlled trials. The American Journal of Chinese Medicine. 2020;48(7):p. 1593. doi: 10.1142/s0192415x20500792. [DOI] [PubMed] [Google Scholar]
- 3.Current status of traditional Chinese medicine research on spinal cord injury. World Journal of Integrated Traditional and Western Medicine. 2017;12(3):p. 440. [Google Scholar]
- 4.Yang P., Chen A., Qin Y., et al. Buyang huanwu decoction combined with BMSCs transplantation promotes recovery after spinal cord injury by rescuing axotomized red nucleus neurons. Journal of Ethnopharmacology. 2019;228:p. 123. doi: 10.1016/j.jep.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 5.Zhang M., Chai Y., Liu T., Xu N., Yang C. Synergistic effects of Buyang Huanwu decoction and embryonic neural stem cell transplantation on the recovery of neurological function in a rat model of spinal cord injury. Experimental and Therapeutic Medicine. 2015;9(4):p. 1141. doi: 10.3892/etm.2015.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Z. P., Huang M. N., Liu A. Q., Yuan Y. J., Zhao J. B., Mei X. F. Buyang Huanwu decoction up-regulates Notch1 gene expression in injured spinal cord. Neural Regeneration Research. 2015;10(8):p. 1321. doi: 10.4103/1673-5374.162767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6(13) doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsaioun K., Blaauboer B., Hartung T. Evidence-based absorption, distribution, metabolism, excretion (ADME) and its interplay with alternative toxicity methods. ALTEX. 2016;33(4):p. 343. doi: 10.14573/altex.1610101. [DOI] [PubMed] [Google Scholar]
- 9.Sheng S., Yang Z., Xu F., Huang Y. Network pharmacology-based exploration of synergistic mechanism of guanxin II formula (II) for coronary heart disease. Chinese Journal of Integrative Medicine. 2021;27(2):106–114. doi: 10.1007/s11655-020-3199-z. [DOI] [PubMed] [Google Scholar]
- 10.UniProt Consortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Research. 2018;46(5):p. 2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z., Guo F., Wang Y., et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Scientific Reports. 2016;6:p. 21146. doi: 10.1038/srep21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piñero J., Ramírez-Anguita J. M., Saüch-Pitarch J., et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Research. 2020;48(D1):p. D845. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stelzer G., Rosen N., Plaschkes I., et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Current Protocols in Bioinformatics. 2016;54:1–30. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 14.Rappaport N., Twik M., Plaschkes I., et al. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Research. 2017;45(D1):p. D877. doi: 10.1093/nar/gkw1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis A., Grondin C., Johnson R., et al. Comparative Toxicogenomics database (CTD): update 2021. Nucleic Acids Research. 2021;49(D1) doi: 10.1093/nar/gkaa891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Zhang S., Li F., et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Research. 2020;48(D1):p. D1031. doi: 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):p. 2498. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang K., Zeng L., Ge A., et al. Hedysarum multijugum MaximThe effect of .- compound on ischemic stroke: a research based on network and experimental pharmacology. Oxidative Medicine and Cellular Longevity. 2020;2020 doi: 10.1155/2020/6072380.6072380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D., Morris J. H., Cook H., et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research. 2017;45(D1):p. D362. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu C., Li R., Wu J. Effects of Yuanhu- Zhitong tablets on alcohol-induced conditioned place preference in mice. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie. 2020;133:p. 110962. doi: 10.1016/j.biopha.2020.110962. [DOI] [PubMed] [Google Scholar]
- 21.Chin C., Chen S., Wu H., Ho C., Ko M., Lin C. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Systems Biology. 2014;S11 doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium T. G. O. Gene Ontology consortium: going forward. Nucleic Acids Research. 2015;43:p. D1049. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M., Araki M., Goto S., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Research. 2008;36:p. D480. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012;16(5):p. 284. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trott O., Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31(2):p. 455. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu N., Hou J. Exploring the mechanism of action Xianlingubao Prescription in the treatment of osteoporosis by network pharmacology. Computational Biology and Chemistry. 2020;85:p. 107240. doi: 10.1016/j.compbiolchem.2020.107240. [DOI] [PubMed] [Google Scholar]
- 27.Chang J., Liu L., Wang Y., Sui Y., Li H., Feng L. Investigating the multitarget mechanism of traditional Chinese medicine prescription for cancer-related pain by using network pharmacology and molecular docking approach. Evidence-based complementary and alternative medicine. eCAM. 2020;7617261:p. 2020. doi: 10.1155/2020/7617261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y., Yang J., Wang X., et al. Research progress in use of traditional Chinese medicine for treatment of spinal cord injury. Biomedicine & Pharmacotherapy. 2020;127:p. 110136. doi: 10.1016/j.biopha.2020.110136. [DOI] [PubMed] [Google Scholar]
- 29.Wu C., Zhou Y., Tu P., et al. Jisuikang promotes the repair of spinal cord injury in rats by regulating NgR/RhoA/ROCK signal pathway. Evidence-based complementary and alternative medicine. eCAM. 2020;9542359:p. 2020. doi: 10.1155/2020/9542359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H., Yang Y., Deng Q., et al. Governor Vessel electro-acupuncture promotes the intrinsic growth ability of spinal neurons through activating calcitonin gene-related peptide/α-calcium/calmodulin-dependent protein kinase/neurotrophin-3 pathway after spinal cord injury. Journal of Neurotrauma. 2020;38(6):734–745. doi: 10.1089/neu.2020.7155. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y.-S., Woo S.-C., Kim S.-Y., Park J.-Y. Understanding the multi-herbal composition of Buyang Huanwu Decoction: a review for better clinical use. Journal of Ethnopharmacology. 2020;255 doi: 10.1016/j.jep.2020.112765.112765 [DOI] [PubMed] [Google Scholar]
- 32.Dong X., He X., Gao W. Neuroprotective effects of the Buyang Huanwu decoction on functional recovery in rats following spinal cord injury. The Journal of Spinal Cord Medicine. 2016;39(1):p. 85. doi: 10.1179/2045772314Y.0000000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen A., Wang H., Zhang J., et al. BYHWD rescues axotomized neurons and promotes functional recovery after spinal cord injury in rats. Journal of Ethnopharmacology. 2008;117(3):p. 451. doi: 10.1016/j.jep.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 34.Joshi Y., Sória M. G., Quadrato G., et al. The MDM4/MDM2-p53-IGF1 axis controls axonal regeneration, sprouting and functional recovery after CNS injury. Brain. 2015;138(7):p. 1843. doi: 10.1093/brain/awv125. [DOI] [PubMed] [Google Scholar]
- 35.Citron B. A., Arnold P. M., Sebastian C., et al. Rapid upregulation of caspase-3 in rat spinal cord after injury: mRNA, protein, and cellular localization correlates with apoptotic cell death. Experimental Neurology. 2000;166(2):p. 213. doi: 10.1006/exnr.2000.7523. [DOI] [PubMed] [Google Scholar]
- 36.Esposito E., Cuzzocrea S. Anti-TNF therapy in the injured spinal cord. Trends in Pharmacological Sciences. 2011;32(2):p. 107. doi: 10.1016/j.tips.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Kim J., Joshi H. P., Sheen S. H., et al. Resolvin D3 promotes inflammatory resolution, neuroprotection, and functional recovery after spinal cord injury. Molecular Neurobiology. 2021;58(1):p. 424. doi: 10.1007/s12035-020-02118-7. [DOI] [PubMed] [Google Scholar]
- 38.Zhou C. L., Li F., Wu X. W., et al. Overexpression of miRNA-433-5p protects acute spinal cord injury through activating MAPK1. European Review for Medical and Pharmacological Sciences. 2020;24(6):p. 2829. doi: 10.26355/eurrev_202003_20644. [DOI] [PubMed] [Google Scholar]
- 39.Song Y., Liu J., Zhang F., Zhang J., Shi T., Zeng Z. Antioxidant effect of quercetin against acute spinal cord injury in rats and its correlation with the p38MAPK/iNOS signaling pathway. Life Sciences. 2013;92(24-26):p. 1215. doi: 10.1016/j.lfs.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Siracusa R., Paterniti I., Bruschetta G., et al. The association of palmitoylethanolamide with Luteolin decreases autophagy in spinal cord injury. Molecular Neurobiology. 2016;53(6):p. 3783. doi: 10.1007/s12035-015-9328-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Zhou L., Ouyang L., Lin S., et al. Protective role of β-carotene against oxidative stress and neuroinflammation in a rat model of spinal cord injury. International Immunopharmacology. 2018;61:p. 92. doi: 10.1016/j.intimp.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 42.Li Y., Lin S., Xu C., Zhang P., Mei X. Triggering of autophagy by Baicalein in response to apoptosis after spinal cord injury: possible involvement of the PI3K activation. Biological and Pharmaceutical Bulletin. 2018;41(4):p. 478. doi: 10.1248/bpb.b17-00768. [DOI] [PubMed] [Google Scholar]
- 43.Wang H., Zhao Z., Liu C., et al. Receptor for advanced glycation end-products (RAGE) blockade do damage to neuronal survival via disrupting Wnt/β-catenin signaling in spinal cord injury. Neurochemical Research. 2018;43(7):p. 1405. doi: 10.1007/s11064-018-2555-2. [DOI] [PubMed] [Google Scholar]
- 44.Gao K., Niu J., Dang X. Neuroprotection of melatonin on spinal cord injury by activating autophagy and inhibiting apoptosis via SIRT1/AMPK signaling pathway. Biotechnology Letters. 2020;42(10):p. 2059. doi: 10.1007/s10529-020-02939-5. [DOI] [PubMed] [Google Scholar]
- 45.Xu S., Wang J., Jiang J., et al. TLR4 promotes microglial pyroptosis via lncRNA-F630028O10Rik by activating PI3K/AKT pathway after spinal cord injury. Cell Death & Disease. 2020;11(8):p. 693. doi: 10.1038/s41419-020-02824-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding Z., Dai C., Zhong L., et al. Neuregulin-1 converts reactive astrocytes toward oligodendrocyte lineage cells via upregulating the PI3K-AKT-mTOR pathway to repair spinal cord injury. Biomedicine & Pharmacotherapy. 2021;134:p. 111168. doi: 10.1016/j.biopha.2020.111168. [DOI] [PubMed] [Google Scholar]
- 47.Chen H., Li J., Liang S., et al. Effect of hypoxia-inducible factor-1/vascular endothelial growth factor signaling pathway on spinal cord injury in rats. Experimental and Therapeutic Medicine. 2017;13(3):p. 861. doi: 10.3892/etm.2017.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye J., Xue R., Ji Z. Y., et al. Effect of NT-3 on repair of spinal cord injury through the MAPK signaling pathway. European Review for Medical and Pharmacological Sciences. 2020;24(5):p. 2165. doi: 10.26355/eurrev_202003_20481. [DOI] [PubMed] [Google Scholar]
- 49.Wang C., Xu T., Lachance B., et al. Critical roles of sphingosine kinase 1 in the regulation of neuroinflammation and neuronal injury after spinal cord injury. Journal of Neuroinflammation. 2021;18(1):p. 50. doi: 10.1186/s12974-021-02092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue W., Zhao Y., Xiao Z., et al. Epidermal growth factor receptor-extracellular-regulated kinase blockade upregulates TRIM32 signaling cascade and promotes neurogenesis after spinal cord injury. Stem Cells. 2020;38(1):p. 118. doi: 10.1002/stem.3097. [DOI] [PubMed] [Google Scholar]
- 51.Alexander K., Tseng H., Fleming W., et al. Inhibition of JAK1/2 tyrosine kinases reduces neurogenic heterotopic ossification after spinal cord injury. Frontiers in Immunology. 2019;10:p. 377. doi: 10.3389/fimmu.2019.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.


