Abstract
Purpose
Noninvasive prenatal testing (NIPT) is a highly sensitive and specific method for detection of fetal chromosomal aneuploidies from maternal plasma. The objective of this study was to determine the performance of a new paired-end sequencing-based NIPT assay in 13,607 pregnancies from a single center in Germany.
Methods
Samples from 13,607 pregnant women who previously underwent NIPT were analyzed using VeriSeq NIPT Solution v2 assay for presence of common fetal trisomies and monosomy X. Follow-up to determine clinical truth was carried out.
Results
Of the 13,607 cases, 13,509 received a NIPT call resulting in a low study failure rate of 0.72%. There were 188 (1.4%) high-risk calls: 117 trisomy 21, 34 trisomy 18, 23 trisomy 13, one trisomy 21 + 13, and 13 monosomy X. High sensitivities and specificities of ≥ 98.89% were reported for all four aneuploidy conditions. Of the high-risk cases, clinical follow-up data were available for 77.1% (145/188). Clinical follow-up of high-risk calls revealed an overall positive predictive value of 84.8% (potential range 65.4–88.3%). NIPT results were provided for samples across a range of fetal fractions, down to 2% fetal fraction.
Conclusion
The VeriSeq NIPT Solution v2 assay detected fetal chromosomal aneuploidies across a range of fetal fractions with high sensitivities and specificities observed based on known clinical outcomes, a high overall PPV, and a low failure rate.
Electronic supplementary material
The online version of this article (10.1007/s00404-020-05856-0) contains supplementary material, which is available to authorized users.
Keywords: Noninvasive prenatal testing, Fetal chromosomal aneuploidies, Fetal fraction, Positive predictive value, VeriSeq NIPT Solution
Introduction
The discovery of fetal cell-free DNA (cfDNA) in maternal circulation in 1997 [1] facilitated the development and commercial availability of noninvasive prenatal testing (NIPT) assays to screen for the presence of fetal chromosomal anomalies. These NIPT assays have been shown to have superior performance over traditional serum screening methods [2]; a recent meta-analysis found that NIPT could detect at least 98% of common fetal trisomies in singleton pregnancies with a combined false-positive rate of 0.13% [3]. The use of NIPT in the general pregnancy population has been endorsed by a number of professional societies, including the German Society of Human Genetics [4–7], and recommendations from German Society for Ultrasound in Medicine (DEGUM), Austrian Society for Ultrasound in Medicine (ÖGUM), Swiss Society for Ultrasound in Medicine (SGUM), and Fetal Medicine Foundation (FMF) Germany regarding prenatal screening including cfDNA screening were recently published [8]. The 2015 Austria–Swiss–German guidelines for NIPT endorse the use of NIPT to screen for fetal trisomy 21 in all pregnant women, but do not recommend the screening of sex chromosome aneuploidies or microdeletions without reservation [9]. The guidelines note that the performance of cfDNA screening for trisomy 13 and 18 is lower than that observed for trisomy 21. Germany’s publicly funded insurance system plans to cover NIPT for common fetal trisomies in pregnancies with increased need of surveillance and pregnancies with special risks in the near future.
The range of conditions that NIPT assays can screen for has expanded since their commercial availability in 2011. Initially, NIPT assays screened for the presence of common fetal trisomies only, i.e., trisomy 21, trisomy 18, and trisomy 13 [10, 11]. This was then expanded to include optional testing for fetal sex and sex chromosome aneuploidies [12, 13], and later to include the option of screening for common microdeletions [14, 15], rare autosomal aneuploidies, and partial deletions and duplications [16–18]. The additional fetal anomalies that are captured following genome-wide testing can impact patient management as they are often associated with serious pregnancy complications [16, 19]. The ability of commercially available NIPT assays to detect additional fetal anomalies is dependent on the technology used by the assay. The primary NIPT technologies at present include massively parallel whole-genome next-generation sequencing (NGS) [20–22], single-nucleotide polymorphism (SNP)-based targeted sequencing [23, 24], and microarray sequencing [25], of which only some NGS-based assays are currently validated to detect rare autosomal aneuploidies and partial deletions and duplications. Massively parallel sequencing techniques can use either single-end or paired-end sequencing; the use of paired-end sequencing is advantageous as it provides information on both the DNA fragment size and location. Several studies have shown the use of this type of sequencing in the noninvasive screening of fetal chromosomal aneuploidies [26, 27], and the first paper on clinical use of paired-end sequencing was published in 2017 [28].
The fetal fraction (FF) of a pregnancy plasma sample has been shown to play a key role in NIPT [29]. Patient characteristics can impact the fetal fraction level in NIPT samples; a positive correlation between gestational age and fetal fraction and a negative correlation between maternal weight and fetal fraction have been shown [24, 30, 31]. Low fetal fraction has been found to be one of the main reasons for NIPT assay failures. Studies have also shown that samples with very low fetal fraction have an increased risk of aneuploidy, particularly trisomy 18 and trisomy 13 [23, 32–34]; this is important as some NIPT assays apply a fetal fraction cut-off and, thus, may miss a number of affected pregnancies.
Our laboratory previously reported on the performance of a SNP-based NIPT assay in the analysis of cfDNA in 3000 cases [35]. The objective of the current study was to determine the performance of the VeriSeq™ NIPT Solution v2 assay, a paired-end sequencing-based NGS assay, in testing for fetal chromosomal aneuploidies in 13,607 general pregnancy samples. We also wanted to investigate the role of fetal fraction in our study, and to determine if patient characteristics had an impact on study outcomes.
Materials and methods
Study patient/sample details
This retrospective analysis study involved 13,607 consecutive patients undergoing NIPT from the general pregnancy population collected over a 17-month period between December 2017 and April 2019. Both singleton and twin pregnancy samples of at least 10 weeks’ gestation were included in the study; exclusion criteria included a known vanishing twin or a higher-grade multiple pregnancy. For this study, the existing sequencing files from the original sample analysis were reanalyzed using the bioinformatic pipeline of the new NIPT assay, VeriSeq NIPT Solution v2, being implemented in the laboratory. Results from the reanalysis were not communicated to the patients or their physicians. Amedes observes the provisions of the Federal Data Protection Act. Patient consent was obtained from all patients involved in the study for use of their data for appropriate quality control and improvements for NIPT assays. In addition, all data were deidentified before being included in the study.
Indications for NIPT included advanced maternal age, a positive screening test result (ultrasound, serum marker), other medical reasons, or patient anxiety. Other medical reasons included an abnormal ultrasound, known disease such as diabetes, epilepsy, and carcinoma, medication such as chemotherapy, a previous history of pregnancy complications including miscarriage or a previously affected pregnancy (e.g., trisomy 21, 18, 13, monosomy X), a genetic aberration in the family (e.g., trisomy 21), or consanguinity. If no information about the indication was provided, patient anxiety or advanced maternal age (if the patent was ≥ 35 years) was taken as the indication. If several of the above mentioned indications were provided, the cases were assigned to the indication groups with the following priority to assign only one indication group to each patient: (1) positive screening test result, (2) advanced maternal age, (3) other medical reasons, and (4) patient anxiety. The correctness of the classification of the cases to the different indication groups, as well as all other information of the patient anamnesis and the feedback regarding the clinical outcome, was dependent on the correctness of the information received.
Clinical outcomes, i.e., clinical truth, for study cases were determined by invasive prenatal diagnostic techniques (cytogenetic analysis after chorionic villus sampling (CVS) and/or amniocentesis), products of conception analysis (cytogenetic analysis of the abortion tissue or placenta samples, post-mortem examination such as autopsy or macroscopic assessment of the abortion), and postnatal cytogenetic analysis, as well as by ultrasound and newborn physical exam. NIPT results positive for fetal aneuploidy were considered confirmed when validated by either invasive prenatal diagnostics or an anomaly observed on ultrasound that matched the high-risk NIPT call. Low-risk NIPT results were considered confirmed if the attending physician provided feedback on a healthy born child with no or minor anomalies that did not clearly fit trisomy 21, 18, 13 or monosomy X (e.g., clubfeet, renal pelvic dilatation, heart defects such as ventricular septal defect, or intrauterine growth restriction due to placental insufficiency).
VeriSeq NIPT Solution v2 assay
NIPT analysis was carried out using the VeriSeq NIPT Solution v2 bioinformatic pipeline (Illumina Inc., San Diego, CA, USA). This assay uses a paired-end sequencing technique and has two reporting modes: Basic, with reporting for common trisomies and sex chromosomes (if selected), and Genome-wide analysis for detection of the presence of genome-wide fetal anomalies (including rare autosomal aneuploidies and partial deletions and duplications ≥ 7 Mb). As study patients originally consented to the analysis of their samples for common trisomies and sex chromosomes (if selected), results for anomalies other than those screened for using basic mode are not reported in this study. Following analysis of existing sequencing files by the VeriSeq NIPT Assay Software v2, samples were called as Anomaly Detected or No Anomaly Detected for common fetal trisomies (trisomy 21, trisomy 18, and trisomy 13) as well as monosomy X (singletons only). A fetal fraction estimate of the sample in question was also provided.
Fetal fraction was estimated using observed coverage data and fragment size distribution. The coverage-based estimate was obtained using methodology similar to a previously described method [36]. Size-based FF estimates were based on the fact that fetal cfDNA fragments are, on average, shorter than maternal fragments [27, 36, 37].
Following data analysis, a log likelihood ratio (LLR) score was provided for each sample. This is the probability of a sample being affected given the sample’s estimated FF and observed coverage. The assay software also uses a dynamic threshold metric known as the individualized Fetal Aneuploidy Confidence Test (iFACT), which determines whether there is sufficient sequencing coverage for each individual sample given the FF estimate for that sample; samples that did not meet this threshold were reported as QC failures. Other QC metrics such as uniformity of coverage were also taken into account. The assay also provided a T-Statistics value, which was used to help differentiate between low-risk and high-risk samples.
Statistics
Statistical data analysis was performed using Microsoft Excel 2010. Where applicable, statistical significance of differences was assessed by Student’s t test; a p value < 0.05 was considered to be significant. Binomial 95% confidence intervals (CI) were calculated for sensitivity and specificity estimates.
In case of an inconclusive NIPT result of the first blood sample a second blood sample was taken for re-analysis. In these cases, the result of the first analysis was excluded in the statistical analysis, so that only one result per patient was included in the statistical evaluation. In addition, in some patients where a NIPT result could be obtained in the analysis of a first blood sample, but a low FF of less than 4% (3% FF or 2% FF) was present, another blood sample was taken for repeat analysis, which was processed as a gesture of goodwill. Again, in these cases, the result of the first analysis was excluded from the statistical analysis, with the exception of cases in which the repeat analysis did not yield a NIPT result; in the latter cases, the result of the first analysis was included in the statistics with a FF of 2% or 3%, and the unsuccessful repeat analysis was excluded.
Results
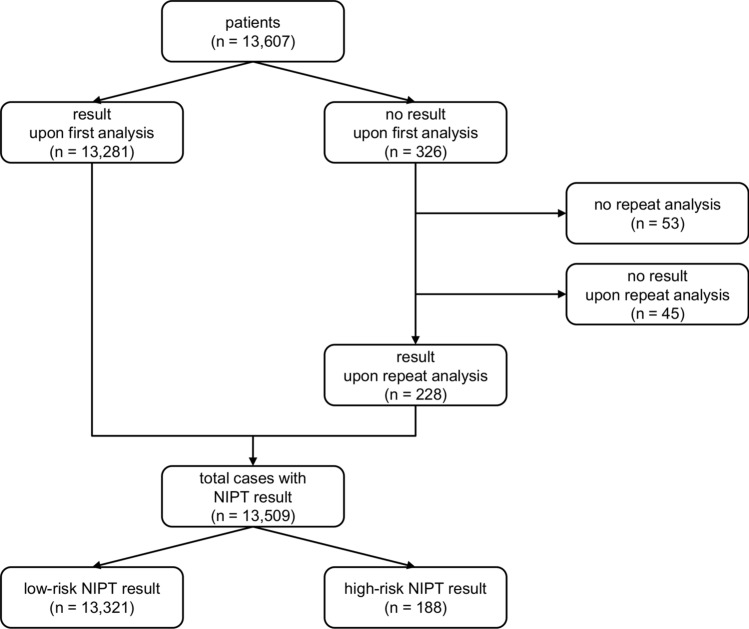
A total of 13,607 pregnancy cases were included in the study, which included 13,333 singleton and 274 twin pregnancy samples. Of these, 13,281 (97.6%) got an NIPT result upon analysis of the first blood sample. Of the 326 samples that did not get a result using the initial blood sample, 273 underwent a repeat analysis on a second blood sample, with 228 (83.5%) cases getting a result. Overall, a total of 13,509 patients obtained an NIPT result (Fig. 1); the study failure rate was 0.72% (98/13,607). Clinical outcomes were available for 2623 cases; outcomes were based on invasive diagnostic techniques, ultrasound, or new-born physical exam (Supplementary Table 1; Online Resource 1).
Fig. 1.
Flowchart of study samples
Patient demographics for the 13,607 patients included in the overall study are shown in Table 1. Almost half of all patients initially chose to undergo NIPT due to parental anxiety. The mean fetal fraction of the study samples was 9.62%, with a range of 1.20–33.94%. The relationship between fetal fraction and patient BMI is shown in Fig. 2. Overall, 86.6% of patients presented for testing in their first trimester; the relationship between gestational age and other patient characteristics (maternal age, BMI, and fetal fraction) is shown in Supplementary Fig. 1 (Online Resource 2). As expected, the BMI increased with increasing gestational age as did the fetal fraction; a marked increase of fetal fraction was observed only from > 20 weeks of gestation, whereas in the previous weeks of gestation a significant but only slight increase in the fetal fraction was observed. The relationships between indications for screening and maternal age, BMI, and fetal fraction were also investigated (Supplementary Fig. 2; Online Resource 2).
Table 1.
Patient demographics for the study cohort
| Study cohort (n = 13,607) | |
|---|---|
| Maternal age (year) | |
| Mean ± SEM | 33.68 ± 0.04 |
| Range | 17.45–55.83 |
| Gestational age (week) | |
| Mean ± SEM | 12.48 ± 0.02 |
| Range | 10.00–36.57 |
| BMI | |
| Mean ± SEM | 24.87 ± 0.05 |
| Range | 15.05–60.96 |
| Indication for screening [n (%)] | |
| Advanced maternal age | 5755 (42.3) |
| Positive screening test resulta | 819 (6.0) |
| Other medical reasonsb | 748 (5.5) |
| Patient anxiety | 6285 (46.2) |
Yr year, wk week, SEM standard error of the mean, BMI body mass index
aPositive screening test result includes ultrasound or serum marker screening
bOther medical reasons include e.g., abnormal ultrasound, known diseases of the patient (e.g., diabetes, epilepsy, and carcinoma), medication (e.g., chemotherapy), or a high-risk family medical history such as a previous miscarriage, a genetic aberration in a previous pregnancy (e.g., trisomy 21, 18, 13, monosomy X), a genetic aberration in the family (e.g., trisomy 21), or consanguinity
Fig. 2.
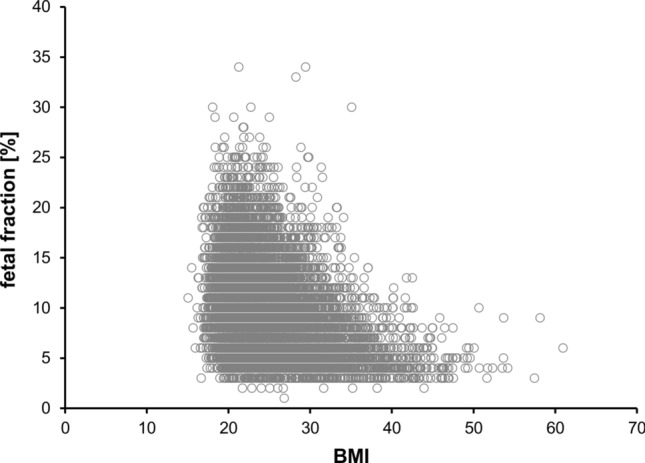
Relationship between fetal fraction and patient BMI
Of the 13,509 patients that received a NIPT result, 98.6% (13,321) were called as low risk, and 1.4% (188) as high risk for presence of a fetal chromosomal aneuploidy (Fig. 1). Clinical outcomes were available for 2623 cases (Supplementary Table 1; Online Resource 1). Of the 13,321 cases reported as low-risk for fetal aneuploidy, clinical outcomes were available for 2478 cases (18.6%). Of these, there was one false-negative call that was found to be a trisomy 21 case; the fetal fraction of this false-negative case was 3%. Of the 188 high-risk calls, 117 were trisomy 21, 34 were trisomy 18, 23 were trisomy 13, one was a trisomy 21 + 13, and 13 were monosomy X calls; 1.3% of patients in this study received a positive NIPT call for presence of a classic trisomy. Follow-up was available for 77.1% (145/188) of these high-risk cases. Based on 123 true-positive and 22 false-positive cases, sensitivities of ≥ 98.89% and specificities of ≥ 99.73% were observed for the high-risk cases as shown in Table 2; results for the one trisomy 21 + 13 case are not shown but this case was found to be a true positive. An overall PPV of 84.8% (123/145) for high-risk cases was observed; PPV results for trisomy 21, trisomy 18, trisomy 13 and monosomy X cases are shown in Table 2. A theoretical PPV range was calculated based on the assumption that high-risk cases without follow-up were either all true positives or all true negatives; the theoretical PPV range for high-risk cases was 65.4% (123/188)–88.3% (166/188). Stratification of NIPT results by maternal age, BMI, and FF is shown in Supplementary Fig. 3 (Online Resource 2); trisomy 18 and trisomy 13 were found to be associated with a significantly lower fetal fraction (p < 0.01 and p < 0.001, respectively, compared to low-risk NIPT results). Fetal fractions of all low-risk and high-risk NIPT cases stratified by patient BMI are shown in Supplementary Fig. 4 (Online Resource 2). The LLR scores and T-Statistics values for the true-positive and false-positive high-risk cases are shown in Supplementary Table 2 (Online Resource 1). As can be seen, both the LLR score and the T-Statistics value were much lower for the false-positive trisomy 21 cases than for the true-positive trisomy 21 cases.
Table 2.
Sensitivities, specificities, and positive predictive values for the high-risk NIPT cases
| Trisomy 21 | Trisomy 18 | Trisomy 13 | Monosomy X | |
|---|---|---|---|---|
| Cases (n) | 117 | 34 | 23 | 13 |
| Cases with follow-up [n (%)] | 96 (82.1) | 23 (67.6) | 15 (65.2) | 10 (76.9) |
| Sensitivity [% (n/N; 95% CI)] | 98.89 (89/90; 93.96–99.97) | > 99.99 (19/19; 82.35–100) | > 99.99 (9/9; 66.37–100) | > 99.99 (5/5; 47.82–100) |
| Specificity [% (n/N; 95% CI)] | 99.73 (2566/2573; 99.44–99.89) | 99.84 (2496/2500; 99.59–99.96) | 99.76 (2486/2492; 99.48–99.91) | 99.80 (2482/2487; 99.53–99.93) |
| PPV [% (n/N)] | 92.7 (89/96) | 82.6 (19/23) | 60.0 (9/15) | 50.0 (5/10) |
| Theoretical lower PPV [% (n/N)] | 76.1 (89/117) | 55.9 (19/34) | 39.1 (9/23) | 38.5 (5/13) |
| Theoretical upper PPV [% (n/N)] | 94.0 (110/117) | 88.2 (30/34) | 73.9 (17/23) | 61.5 (8/13) |
Overall, 2.0% (264/13,509) of samples had a fetal fraction less than 4%, 43.9% (5933/13,509) of samples had a fetal fraction between 4 and 8%, and 54.1% (7312/13,509) of samples had a fetal fraction greater than 8%. The breakdown of high-risk NIPT results by fetal fraction is shown in Fig. 3. As can be seen, the majority of high-risk cases had a fetal fraction between 4 and 8% (49.5%; 93/188). Of the 264 samples with a fetal fraction < 4%, 21 were classified as high-risk by NIPT—six trisomy 21, six trisomy 18, and nine trisomy 13. Outcomes were available for nine of these 21 cases resulting in a PPV of 66.7% (6/9). For low-risk cases, an NPV of 98.1% (52/53) for cases with a fetal fraction of less than 4% was observed. An NPV of > 99.9% was observed for cases with a fetal fraction between 4 and 8% (1198/1198), and for cases with a fetal fraction of ≥ 8% (1227/1227).
Fig. 3.
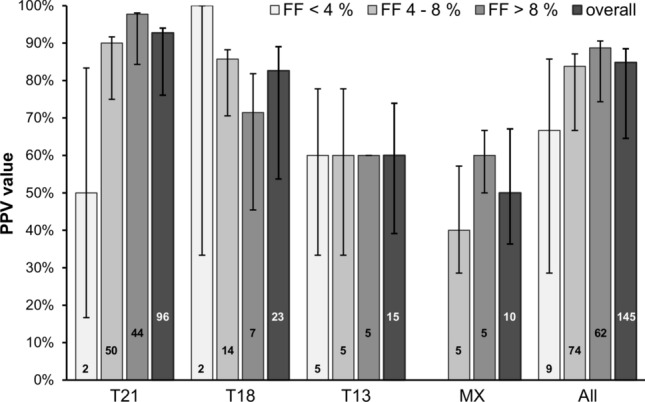
PPV values for high-risk cases for cases with fetal fractions < 4%, 4–8%, and > 8%
Discussion
Here, we show that the VeriSeq NIPT Solution v2 assay was able to screen for fetal chromosomal aneuploidies with high sensitivities and specificities observed based on known clinical outcomes. The sensitivities and specificities reported in our study are in line with those found in a recent meta-analysis by Gil et al. [3] Our study also had a low overall failure rate of less than 1%. For clinical studies, the positive predictive value is a key metric that should be reported along with the assay sensitivity and specificity, provided that clinical follow-up is possible; here, clinical truth was available for the majority of high-risk cases. High PPVs were observed in our study, particularly for trisomy 21 and trisomy 18 cases, at levels similar to those shown in previous studies [22, 24, 38]. The limited follow-up of the low-risk cases in our study found one false-negative case that was determined to be a trisomy 21 case.
We also looked at the impact of patient characteristics on study outcomes. We observed a positive correlation between gestational age and fetal fraction, which has been noted in several previous studies [24, 30, 31]. As expected, a positive correlation was seen between gestational age and BMI. We also noted that there was a significantly lower fetal fraction in cases that screened positive for trisomy 18 and trisomy 13. As the cfDNA that is analyzed in NIPT assays is derived from the placenta, the smaller placental volume that has previously been reported in pregnancies affected with either trisomy 18 or trisomy 13 [39] is likely the cause of the lower FF levels observed in our study for these trisomic conditions.
One of the strengths of our study is that it involves a large cohort of patients from a general pregnancy population, providing further evidence for the utility of NIPT in that patient population. We demonstrate the high test performance of this NIPT assay, with high sensitivities, specificities, and PPVs reported for the NIPT-positive cases. We also report a low assay failure rate of less than 1% following retesting of a second blood sample for any samples that did not get a result with the initial blood sample. Having a low failure rate is an important feature for any NIPT assay as it allows patients to avoid having to decide whether or not to undergo an invasive diagnostic test without an NIPT result which can add to parental anxiety; professional societies often recommend that failed NIPT samples should be considered as high risk [4, 40, 41]. A limitation of our study was the inability to report on the additional findings of the genome-wide assay due to the retrospective nature of the study. This meant that we were unable to provide any performance data relating to the screening of rare autosomal aneuploidies and partial deletions and duplications. The ability of this NIPT assay to screen for fetal chromosomal aneuploidies even at very low fetal fractions is important, as cases with low fetal fraction have been shown to be associated with adverse outcomes [42]. However, the small number of samples in our study that had a fetal fraction less than 4% precluded any meaningful analysis of FF performance at these low levels. Another limitation of this study was that outcome information was limited for cases with a negative NIPT result, which is consistent with many other published NIPT studies [22, 24].
Currently, there is no insurance coverage for NIPT in Germany and so patients need to pay for NIPT out-of-pocket which can be prohibitive in many cases. However, Germany’s publicly funded health insurance system plans to cover NIPT for trisomies 21, 13, and 18 for particular pregnancies with increased need of surveillance and pregnancies with special risks. Introduction of nationwide health coverage for NIPT has been shown to result in a considerable increase in update of this type of prenatal screening for women with increased risk of fetal aneuploidy [43, 44]. It is important that patients are informed that NIPT is not a diagnostic test, and that high-risk NIPT calls should be confirmed by invasive diagnostic procedures such as CVS or amniocentesis. Patients who are found to be at high-risk following an abnormal ultrasound should be recommended to undergo an invasive diagnostic procedure to avoid any incorrect NIPT results that can occur due to reasons such as confined placental mosaicism [45].
In conclusion, the VeriSeq NIPT Solution v2 assay showed strong performance for the detection of fetal chromosomal aneuploidies in our large population of general pregnancy patients across a wide range of fetal fractions and can help to avoid unnecessary invasive procedures that risk harming pregnant woman.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Cosmin Deciu and Sarah Kinnings (Illumina, Inc.) for their assistance with this project, as well as Kristine Jinnett and Kirsten Curnow (Illumina, Inc.) for their assistance in the preparation of the manuscript. Wolf Kupatt and Christoph Keck (amedes) have always supported and advanced the NIPT project excellently. Also, we want to thank the many gynaecologists who gave us valuable information on the fetal outcome.
Author contributions
Data analysis and paper preparation were carried out by HB and BE; Assistance with data analysis was provided by NK, AT and SK; paper preparation and discussion was assisted by RG and TW.
Funding
Assistance with data analysis and paper preparation was provided by Illumina, Inc. but no financial renumeration was provided directly to the authors.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Amedes observes the provisions of the Federal Data Protection Act.
Consent to participate
Patient consent was obtained from all patients involved in the study for use of their data for appropriate quality control and improvements for NIPT assays. In addition, all data were deidentified before being included in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–487. doi: 10.1016/s0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 2.Norton ME, Baer RJ, Wapner RJ, Kuppermann M, Jelliffe-Pawlowski LL, Currier RJ. Cell-free DNA vs sequential screening for the detection of fetal chromosomal abnormalities. Am J Obstet Gynecol. 2016;214(6):727–726. doi: 10.1016/j.ajog.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 3.Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol. 2017;50(3):302–314. doi: 10.1002/uog.17484. [DOI] [PubMed] [Google Scholar]
- 4.Benn P, Borrell A, Chiu RW, Cuckle H, Dugoff L, Faas B, Gross S, Huang T, Johnson J, Maymon R, Norton M, Odibo A, Schielen P, Spencer K, Wright D, Yaron Y. Position statement from the Chromosome Abnormality Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat Diagn. 2015;35(8):725–734. doi: 10.1002/pd.4608. [DOI] [PubMed] [Google Scholar]
- 5.Gregg AR, Skotko BG, Benkendorf JL, Monaghan KG, Bajaj K, Best RG, Klugman S, Watson MS. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2016;18(10):1056–1065. doi: 10.1038/gim.2016.97. [DOI] [PubMed] [Google Scholar]
- 6.Dondorp W, de Wert G, Bombard Y, Bianchi DW, Bergmann C, Borry P, Chitty LS, Fellmann F, Forzano F, Hall A, Henneman L, Howard HC, Lucassen A, Ormond K, Peterlin B, Radojkovic D, Rogowski W, Soller M, Tibben A, Tranebjaerg L, van El CG, Cornel MC, European Society of Human G, American Society of Human G Non-invasive prenatal testing for aneuploidy and beyond: challenges of responsible innovation in prenatal screening. Eur J Hum Genet. 2015;23(11):1438–1450. doi: 10.1038/ejhg.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stellungnahme der Deutschen Gesellschaft für Humangenetik zur Analyse fetaler DNA aus dem mütterlichen Blut. https://www.gfhev.de/leitlinien. 2013.
- 8.Kozlowski P, Burkhardt T, Gembruch U, Gonser M, Kahler C, Kagan KO, von Kaisenberg C, Klaritsch P, Merz E, Steiner H, Tercanli S, Vetter K, Schramm T. DEGUM, OGUM, SGUM and FMF Germany recommendations for the implementation of first-trimester screening, detailed ultrasound, cell-Free DNA screening and diagnostic procedures. Ultraschall Med. 2019;40(2):176–193. doi: 10.1055/a-0631-8898. [DOI] [PubMed] [Google Scholar]
- 9.Schmid M, Klaritsch P, Arzt W, Burkhardt T, Duba HC, Hausler M, Hafner E, Lang U, Pertl B, Speicher M, Steiner H, Tercanli S, Merz E, Heling KS, Eiben B. Cell-free DNA testing for fetal chromosomal anomalies in clinical practice: Austrian–German–Swiss recommendations for non-invasive prenatal tests (NIPT) Ultraschall Med. 2015;36(5):507–510. doi: 10.1055/s-0035-1553804. [DOI] [PubMed] [Google Scholar]
- 10.Nicolaides KH, Syngelaki A, Ashoor G, Birdir C, Touzet G. Noninvasive prenatal testing for fetal trisomies in a routinely screened first-trimester population. Am J Obstet Gynecol. 2012;207(5):374.e371–376. doi: 10.1016/j.ajog.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 11.Palomaki GE, Deciu C, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, van den Boom D, Bombard AT, Grody WW, Nelson SF, Canick JA. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet Med. 2012;14(3):296–305. doi: 10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samango-Sprouse C, Banjevic M, Ryan A, Sigurjonsson S, Zimmermann B, Hill M, Hall MP, Westemeyer M, Saucier J, Demko Z, Rabinowitz M. SNP-based non-invasive prenatal testing detects sex chromosome aneuploidies with high accuracy. Prenat Diagn. 2013;33(7):643–649. doi: 10.1002/pd.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazloom AR, Dzakula Z, Oeth P, Wang H, Jensen T, Tynan J, McCullough R, Saldivar JS, Ehrich M, van den Boom D, Bombard AT, Maeder M, McLennan G, Meschino W, Palomaki GE, Canick JA, Deciu C. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell-free DNA from maternal plasma. Prenat Diagn. 2013;33(6):591–597. doi: 10.1002/pd.4127. [DOI] [PubMed] [Google Scholar]
- 14.Wapner RJ, Babiarz JE, Levy B, Stosic M, Zimmermann B, Sigurjonsson S, Wayham N, Ryan A, Banjevic M, Lacroute P, Hu J, Hall MP, Demko Z, Siddiqui A, Rabinowitz M, Gross SJ, Hill M, Benn P. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am J Obstet Gynecol. 2015;212(3):332–339. doi: 10.1016/j.ajog.2014.11.041. [DOI] [PubMed] [Google Scholar]
- 15.Gross SJ, Stosic M, McDonald-McGinn DM, Bassett AS, Norvez A, Dhamankar R, Kobara K, Kirkizlar E, Zimmermann B, Wayham N, Babiarz JE, Ryan A, Jinnett KN, Demko Z, Benn P. Clinical experience with single-nucleotide polymorphism-based non-invasive prenatal screening for 22q11.2 deletion syndrome. Ultrasound Obstet Gynecol. 2016;47(2):177–183. doi: 10.1002/uog.15754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pertile MD, Halks-Miller M, Flowers N, Barbacioru C, Kinnings SL, Vavrek D, Seltzer WK, Bianchi DW. Rare autosomal trisomies, revealed by maternal plasma DNA sequencing, suggest increased risk of feto-placental disease. Sci Transl Med. 2017 doi: 10.1126/scitranslmed.aan1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiorentino F, Bono S, Pizzuti F, Duca S, Polverari A, Faieta M, Baldi M, Diano L, Spinella F. The clinical utility of genome-wide non invasive prenatal screening. Prenat Diagn. 2017;37(6):593–601. doi: 10.1002/pd.5053. [DOI] [PubMed] [Google Scholar]
- 18.Liang D, Lin Y, Qiao F, Li H, Wang Y, Zhang J, Liu A, Ji X, Ma D, Jiang T, Hu P, Xu Z. Perinatal outcomes following cell-free DNA screening in > 32,000 women: clinical follow-up data from a single tertiary center. Prenat Diagn. 2018;38(10):755–764. doi: 10.1002/pd.5328. [DOI] [PubMed] [Google Scholar]
- 19.Scott F, Bonifacio M, Sandow R, Ellis K, Smet ME, McLennan A. Rare autosomal trisomies: important and not so rare. Prenat Diagn. 2018;38(10):765–771. doi: 10.1002/pd.5325. [DOI] [PubMed] [Google Scholar]
- 20.Sehnert AJ, Rhees B, Comstock D, de Feo E, Heilek G, Burke J, Rava RP. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin Chem. 2011;57(7):1042–1049. doi: 10.1373/clinchem.2011.165910. [DOI] [PubMed] [Google Scholar]
- 21.Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, Tim R, Lu V, McCullough R, McCarthy E, Nygren AO, Dean J, Tang L, Hutchison D, Lu T, Wang H, Angkachatchai V, Oeth P, Cantor CR, Bombard A, van den Boom D. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011;204(3):205–211. doi: 10.1016/j.ajog.2010.12.060. [DOI] [PubMed] [Google Scholar]
- 22.Taneja PA, Snyder HL, de Feo E, Kruglyak KM, Halks-Miller M, Curnow KJ, Bhatt S. Noninvasive prenatal testing in the general obstetric population: clinical performance and counseling considerations in over 85,000 cases. Prenat Diagn. 2016;36(3):237–243. doi: 10.1002/pd.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pergament E, Cuckle H, Zimmermann B, Banjevic M, Sigurjonsson S, Ryan A, Hall MP, Dodd M, Lacroute P, Stosic M, Chopra N, Hunkapiller N, Prosen DE, McAdoo S, Demko Z, Siddiqui A, Hill M, Rabinowitz M. Single-nucleotide polymorphism-based noninvasive prenatal screening in a high-risk and low-risk cohort. Obstet Gynecol. 2014;124(2 Pt 1):210–218. doi: 10.1097/aog.0000000000000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dar P, Curnow KJ, Gross SJ, Hall MP, Stosic M, Demko Z, Zimmermann B, Hill M, Sigurjonsson S, Ryan A, Banjevic M, Kolacki PL, Koch SW, Strom CM, Rabinowitz M, Benn P. Clinical experience and follow-up with large scale single-nucleotide polymorphism-based non-invasive prenatal aneuploidy testing. Am J Obstet Gynecol. 2014;211(5):527.e521–517. doi: 10.1016/j.ajog.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Juneau K, Bogard PE, Huang S, Mohseni M, Wang ET, Ryvkin P, Kingsley C, Struble CA, Oliphant A, Zahn JM. Microarray-based cell-free DNA analysis improves noninvasive prenatal testing. Fetal Diagn Ther. 2014;36(4):282–286. doi: 10.1159/000367626. [DOI] [PubMed] [Google Scholar]
- 26.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin Chem. 2010;56(8):1279–1286. doi: 10.1373/clinchem.2010.144188. [DOI] [PubMed] [Google Scholar]
- 27.Yu SC, Chan KC, Zheng YW, Jiang P, Liao GJ, Sun H, Akolekar R, Leung TY, Go AT, van Vugt JM, Minekawa R, Oudejans CB, Nicolaides KH, Chiu RW, Lo YM. Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc Natl Acad Sci USA. 2014;111(23):8583–8588. doi: 10.1073/pnas.1406103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cirigliano V, Ordonez E, Rueda L, Syngelaki A, Nicolaides KH. Performance of the neoBona test: a new paired-end massively parallel shotgun sequencing approach for cell-free DNA-based aneuploidy screening. Ultrasound Obstet Gynecol. 2017;49(4):460–464. doi: 10.1002/uog.17386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hui L, Bianchi DW. Fetal fraction and noninvasive prenatal testing: what clinicians need to know. Prenat Diagn. 2020;40(2):155–163. doi: 10.1002/pd.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat Diagn. 2013;33(7):662–666. doi: 10.1002/pd.4119. [DOI] [PubMed] [Google Scholar]
- 31.Scott FP, Menezes M, Palma-Dias R, Nisbet D, Schluter P, da Silva CF, McLennan AC. Factors affecting cell-free DNA fetal fraction and the consequences for test accuracy. J Matern Fetal Neonatal Med. 2017 doi: 10.1080/14767058.2017.1330881. [DOI] [PubMed] [Google Scholar]
- 32.Ryan A, Hunkapiller N, Banjevic M, Vankayalapati N, Fong N, Jinnett KN, Demko Z, Zimmermann B, Sigurjonsson S, Gross SJ, Hill M. Validation of an enhanced version of a single-nucleotide polymorphism-based noninvasive prenatal test for detection of fetal aneuploidies. Fetal Diagn Ther. 2016;40(3):219–223. doi: 10.1159/000442931. [DOI] [PubMed] [Google Scholar]
- 33.Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, Brar H, Tomlinson MW, Pereira L, Spitz JL, Hollemon D, Cuckle H, Musci TJ, Wapner RJ. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015;372(17):1589–1597. doi: 10.1056/NEJMoa1407349. [DOI] [PubMed] [Google Scholar]
- 34.Fiorentino F, Bono S, Pizzuti F, Mariano M, Polverari A, Duca S, Sessa M, Baldi M, Diano L, Spinella F. The importance of determining the limit of detection of non-invasive prenatal testing methods. Prenat Diagn. 2016;36(4):304–311. doi: 10.1002/pd.4780. [DOI] [PubMed] [Google Scholar]
- 35.Eiben B, Krapp M, Borth H, Kutur N, Kreiselmaier P, Glaubitz R, Deutinger J, Merz E. Single nucleotide polymorphism-based analysis of cell-free fetal DNA in 3000 cases from Germany and Austria. Ultrasound Int Open. 2015;1(1):E8–E11. doi: 10.1055/s-0035-1555765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SK, Hannum G, Geis J, Tynan J, Hogg G, Zhao C, Jensen TJ, Mazloom AR, Oeth P, Ehrich M, van den Boom D, Deciu C. Determination of fetal DNA fraction from the plasma of pregnant women using sequence read counts. Prenat Diagn. 2015;35(8):810–815. doi: 10.1002/pd.4615. [DOI] [PubMed] [Google Scholar]
- 37.Chan KC, Zhang J, Hui AB, Wong N, Lau TK, Leung TN, Lo KW, Huang DW, Lo YM. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem. 2004;50(1):88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- 38.Xu L, Huang H, Lin N, Wang Y, He D, Zhang M, Chen M, Chen L, Lin Y. Non-invasive cell-free fetal DNA testing: a multicenter follow-up study of 31,515 singleton pregnancies in southeastern China. Ultrasound Obstet Gynecol. 2019 doi: 10.1002/uog.20416. [DOI] [PubMed] [Google Scholar]
- 39.Wegrzyn P, Faro C, Falcon O, Peralta CF, Nicolaides KH. Placental volume measured by three-dimensional ultrasound at 11–13 + 6 weeks of gestation: relation to chromosomal defects. Ultrasound Obstet Gynecol. 2005;26(1):28–32. doi: 10.1002/uog.1923. [DOI] [PubMed] [Google Scholar]
- 40.The American College of Obstetricians and Gynecologists, Society for Maternal Fetal Medicine Practice Bulletin No. 163 summary: screening for fetal aneuploidy. Obstet Gynecol. 2016;127(5):979–981. doi: 10.1097/aog.0000000000001439. [DOI] [PubMed] [Google Scholar]
- 41.Salomon LJ, Alfirevic Z, Audibert F, Kagan KO, Paladini D, Yeo G, Raine-Fenning N. ISUOG updated consensus statement on the impact of cfDNA aneuploidy testing on screening policies and prenatal ultrasound practice. Ultrasound Obstet Gynecol. 2017;49(6):815–816. doi: 10.1002/uog.17483. [DOI] [PubMed] [Google Scholar]
- 42.Krishna I, Badell M, Loucks TL, Lindsay M, Samuel A. Adverse perinatal outcomes are more frequent in pregnancies with a low fetal fraction result on noninvasive prenatal testing. Prenat Diagn. 2016;36(3):210–215. doi: 10.1002/pd.4779. [DOI] [PubMed] [Google Scholar]
- 43.Vinante V, Keller B, Huhn EA, Huang D, Lapaire O, Manegold-Brauer G. Impact of nationwide health insurance coverage for non-invasive prenatal testing. Int J Gynaecol Obstet. 2018;141(2):189–193. doi: 10.1002/ijgo.12422. [DOI] [PubMed] [Google Scholar]
- 44.Huang T, Dougan S, Walker M, Armour CM, Okun N. Trends in the use of prenatal testing services for fetal aneuploidy in Ontario: a descriptive study. CMAJ Open. 2018;6(4):E436–E444. doi: 10.9778/cmajo.20180046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eiben B. Fallstricke beim NIPT—eine kritische Betrachtung. Frauenarzt. 2016;57:567. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.