Abstract
Recombinant adeno-associated viruses (rAAVs) are well-established vectors for delivering therapeutic genes. However, previous reports have suggested that wild-type AAV is linked to hepatocellular carcinoma, raising concern with the safety of rAAVs. In addition, a recent long-term follow-up study in canines, which received rAAVs for factor VIII gene therapy, demonstrated vector integration into the genome of liver cells, reviving the uncertainty between AAV and cancer. To further explore this relationship, we performed large-scale molecular epidemiology of AAV in resected tumor samples and non-lesion tissues collected from 413 patients, reflecting nine carcinoma types: breast carcinoma, rectal cancer, pancreas carcinoma, brain tumor, hepatoid adenocarcinoma, hepatocellular carcinoma, gastric carcinoma, lung squamous, and adenocarcinoma. We found that over 80% of patients were AAV-positive among all nine types of carcinoma examined. Importantly, the AAV sequences detected in patient-matched tumor and adjacent non-lesion tissues showed no significant difference in incidence, abundance, and variation. Additionally, no specific AAV sequences predominated in tumor samples. Our data shows that AAV genomes are equally abundant in tumors and adjacent normal tissues, but lack clonality. The finding critically adds to the epidemiological profile of AAV in humans, and provides insights that may assist rAAV-based clinical studies and gene therapy strategies.
Introduction
Adeno-associated virus (AAV) is a ~26 nm-wide, icosahedral 60-mer belonging to the dependoparvovirus family of single-strand DNA viruses (1). The AAV genome consists of four known open reading frames (Figure 1), 1) rep, which encodes for the four replication proteins Rep78, Rep68, Rep52, and Rep40; 2) cap, which encodes for the three capsid proteins, VP1, VP2, and VP3; 3) assembly activating protein (AAP), which recruits capsid monomers to nucleoli and drives capsid assembly; and 4) the recently discovered membrane-associated accessory protein (MAAP), whose function is not currently known (1, 2). AAV’s non-pathogenic nature, relatively low immunological profile, and its dependence on adenovirus or herpesvirus to complete its lifecycle, have made AAV an ideal gene transfer vector in vivo and in vitro (1). Vectorized AAVs are now recognized as the preeminent vehicle to deliver therapeutic genes to treat human genetic diseases. Recombinant AAVs (rAAVs) employed as gene therapy vectors are void of all viral genes. The only remaining element is the inverted terminal repeat (ITR) that reside at the 5’ and 3’ ends of the genome and is vital for vector genome replication and packaging during production (1). The ITRs are also critical for conversion of the single-strand genome into the double-stranded species required for gene transcription in the host cell. They are also crucial to the formation of the circular episomal forms of AAV, which were first discovered to persist in tissues for rAAVs in non-dividing cells (3), and were later demonstrated as the predominant form for natural viral genomes in the nuclei of infected cells (4). The ITR is also a vital structure for low-frequency integration into the host cell genome in the presence of Rep proteins (5, 6).
Figure 1. Schematic of the AAV genome and sites used for PCR screening.
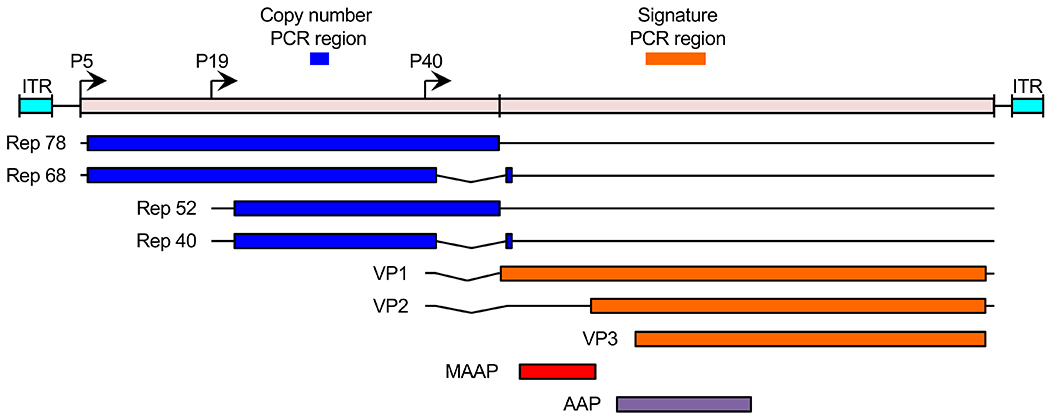
The AAV genome is comprised of four known open reading frames, rep (blue), cap (orange), MAAP (red), and AAP (purple). The rep and cap ORFs encode four and three isoforms, respectively. Transcription is driven by the viral P5, P19, and P40 promoters (arrows). The genome is flanked by inverted terminal repeat (ITR, cyan) sequences. Copy number per host cell genome was quantified by qPCR using primers spanning the “copy number PCR region”. Molecular identification of serotype and variation was determined by high-throughput sequencing of the “signature PCR region”.
Early in vitro evidence showed that AAV has the capacity to preferentially integrate into the human genome at the AAVS1 locus on chromosome 19 (7). Several subsequent in vivo studies have found that AAV vectors can integrate throughout the host cell genome in a variety of rodent tissues (8–12). However, the most striking body of work, relates to the manifestation of hepatocellular carcinoma by vectors that confer high liver tropism and subsequent vector genome integration (8, 9, 11, 13, 14). The disease outcome observed in mice is due to specific integration of rAAV genomes into the Rian locus, a position enriched with cancer-driving miRNAs. Fortunately, evidence of preferential integration into the Rian locus homolog in humans (Dlk1-Dio3 cluster) is lacking (15); and thus, this outcome was assumed to be unique to mice. However, studies reporting an association between HCC in humans and wild-type AAV integration into cancer driver gene promoters (16, 17) have re-raised concerns. In response to these reports, the gene therapy field came to a consensus that the evidence for wild-type AAV integration does not inform whether vectored AAV integration can trigger cancer development in humans (18–20). Nonetheless, the report continues to draw controversy, as it serves to raise the possibility of integration events leading to unexpected outcomes. Furthermore, recombinant AAV (rAAV) and wild-type AAVs still share in common the ITR sequence, the essential element that drives genome recombination and integration (21, 22). Most recently, a report on a ten-year follow-up study in six dogs receiving gene therapy vector for hemophilia A (factor VIII), revealed integration of vector genomes in liver tissues (23). Although the dogs did not show signs of malignancy and tumor development, cells with integration events were clonally expanded, re-raising concerns over genotoxicity related to vector integration.
In light of these new reports, we aimed to further investigate the prevalence of AAV in cancer patient tissues to provide further insight into the occurrence of clonally expanded AAV genomes in cancer patients. We profiled 413 patients receiving cancer treatment at West China Hospital by molecular analysis of tumor resections and adjacent non-lesion tissues. Our data shows that both tumor mass and normal tissues showed a high degree of AAV positivity, similar to those observed in other epidemiological studies (24–28). Interestingly, among these tissues, we saw a high diversity of sequences that lacked clonal representation, demonstrating that the cancer patients queried in this study lack evidence of AAV integration and subsequent clonal expansion, which was expected to be a hallmark of events that would lead to tumorigenesis.
Materials/Subjects and Methods:
Tissue collection and tumor grading
Under approval of West China Hospital Institution Ethics Committee, all tissue samples were acquired from patients who were diagnosed by radiological and biopsy examination, seeking medical treatment, and receiving tumorectomies (Department of Oncology, West China Hospital of Sichuan University, Chengdu, China). Fresh and frozen specimens were collected along with patients’ ages, genders, and tumor classifications defined by the hereditary disease resource center of West China Hospital. Adjacent non-tumor tissues defined as 3 cm from lesions, were also obtained as indicated. Tumor grading was performed by frozen section examination and intraoperative frozen section diagnosis. All samples were labeled and stored in liquid nitrogen until DNA extraction. Downstream analyses was blinded to group allocation.
DNA extraction
Frozen tissues were thawed to room temperature and about 25 mg of tissue was obtained with disposable scalpels. Extraction of DNA from tissues were performed using the QIAampDNA Mini Kit (Shanghai, China; Qiagen, #51306) according to manufacturer’s recommendations. Purified DNA samples were stored at −20°C. To avoid cross-contamination by environmental AAV genomes, extractions and subsequent PCR procedures were all performed in a sterile UV-irradiated PCR cabinet (Singapore, Airstream® ESCO). All surfaces and equipment were sprayed with DNA-Exitus Plus (Cenghdu, China; Applichem, Cat No: A7089) and wiped clean with Milli-Q water after 15 min.
Signature and quantitative PCR
AAV positivity was determined by detection of amplicons spanning a “signature” region of the cap ORF encompassing variable region I within VP3 (29) generated by polymerase chain reaction (PCR) with 2x GoldStar Taq MasterMix (Beijing, China; ComWin Biotech CW0960 1ML) and PCR primers that are complementary to semi-conserved regions flanking the signature region:
5’-GGTAATGCCTCAGGAAATTGGCATT-3’
5’-GAATCCCCAGTTGTTGTTGATGAGTC-3’
AAV-positive samples were determined by production of a 255-bp band on 1% agarose gels. Signature PCR amplicons were then gel-extracted with the PureLink PCR Purification Kit (Shanghai, China; Invitrogen, CAT K310001) and cloned into the pEASY-T1 Cloning Vector (Beijing, China; TRANS, Cat: CT101). At least five clones for each sample were subjected to Sanger sequencing, and the resulting sequences were aligned to known AAV serotypes available using Vector NTI software package (Bethesda, MD, USA; Informax, Inc) to validate amplicons. PCR reactions were accompanied with negative template controls performed in triplicate to exclude the possibility of “environmental” AAV DNA contamination.
Genome copy numbers of tissues were assessed by quantitative TaqMan PCR analysis using TaqMan® Universal PCR Master Mix ABI #4326614 on a CFX96 instrument (Hercules, CA, Bio-Rad). The primers and probe set was designed to target the AAV Rep sequence:
Forward5’-GTGCCCTTCTACGGST-3’
Reverse5’-CCAGATCACCATCTTGTCGA-3’
Probe5’-6-FAM-AACTGGACCAATGAGAACT-MGB-3’ (Invitrogen)
The pAAVrep2/cap2 plasmid (8 011 bp) was linearized by HindIII digestion and serially diluted from 1x108 copies/5 μl to 10 copies/5 μl and used as the standard curve for qPCR analysis.
High-throughput sequencing of signature PCR amplicons
Signature amplicons were generated as described above. Since the signature PCR amplicons contained mostly conserved nucleotides, staggered sequences at both ends of amplicons were introduced by PCR to maximize the diversity of the amplicon-seq libraries library prep for HiSeq X Ten paired-end, 150-bp sequencing.
Primers for generating signature amplicon libraries:
1st round PCR
Forward primers:
| Primer name | Sequence (5’ → 3’) |
|---|---|
| I5-AAV-1 (52 nt) | CACTCTTTCCCTACACGACGCTCTTCCGATCTTGCCTCAGGAAATTGGCATT |
| I5-AAV-2 (53 nt) | CACTCTTTCCCTACACGACGCTCTTCCGATCTaTGCCTCAGGAAATTGGCATT |
| I5-AAV-3 (54 nt) | CACTCTTTCCCTACACGACGCTCTTCCGATCTgaTGCCTCAGGAAATTGGCATT |
| I5-AAV-4 (55 nt) | CACTCTTTCCCTACACGACGCTCTTCCGATCTagaTGCCTCAGGAAATTGGCATT |
| I5-AAV-5 (56 nt) | CACTCTTTCCCTACACGACGCTCTTCCGATCTtagaTGCCTCAGGAAATTGGCATT |
| I5-AAV-6 (57 nt) | CACTCTTTCCCTACACGACGCTCTTCCGATCTctagaTGCCTCAGGAAATTGGCATT |
| I5-AAV-7 (58 nt) | CACTCTTTCCCTACACGACGCTCTTCCGATCTcctagaTGCCTCAGGAAATTGGCATT |
| I5-AAV-8 (59 nt) | CACTCTTTCCCTACACGACGCTCTTCCGATCTgcctagaTGCCTCAGGAAATTGGCATT |
Reverse primer: equal molar mix of the following 8 primers
| Primer name | Sequence (5’ → 3’) |
|---|---|
| I7-AAV-1 (52 nt) | GACTGGAGTTCAGACGTGTGCTCTTCCGATCTCCAGTTGTTGTTGATGAGTC |
| I7-AAV-2 (53 nt) | GACTGGAGTTCAGACGTGTGCTCTTCCGATCTaCCAGTTGTTGTTGATGAGTC |
| I7-AAV-3 (54 nt) | GACTGGAGTTCAGACGTGTGCTCTTCCGATCTgaCCAGTTGTTGTTGATGAGTC |
| I7-AAV-4 (55 nt) | GACTGGAGTTCAGACGTGTGCTCTTCCGATCTtgaCCAGTTGTTGTTGATGAGTC |
| I7-AAV-5 (56 nt) | GACTGGAGTTCAGACGTGTGCTCTTCCGATCTttgaCCAGTTGTTGTTGATGAGTC |
| I7-AAV-6 (57 nt) | GACTGGAGTTCAGACGTGTGCTCTTCCGATCTgttgaCCAGTTGTTGTTGATGAGTC |
| I7-AAV-7 (58 nt) | GACTGGAGTTCAGACGTGTGCTCTTCCGATCTagttgaCCAGTTGTTGTTGATGAGTC |
| I7-AAV-8 (59 nt) | GACTGGAGTTCAGACGTGTGCTCTTCCGATCTcagttgaCCAGTTGTTGTTGATGAGTC |
An equimolar mix of the eight primer pairs were used for reactions.
(Illumina adapter sequence(uppercase)/stagger region (lowercase)/AAV primer binding sequence (underlined))
2nd round PCR
| Primer name | Sequence (5’ → 3’) |
|---|---|
| TruSeq HT P5 primer (70 nt) | AATGATACGGCGACCACCGAGATCTACACxrefXXacactctttccctacacgacgctcttccgatct |
| TruSeq HT P7 primer (66 nt) | CAAGCAGAAGACGGCATACGAGATxrefXXgtgactggagttcagacgtgtgctcttccgatct |
(P5/P7 flow-cell attachment sequence (uppercase)/barcode (X)/Illumina adapter sequence (lowercase))
Purification with one volume of AMPure XP beads (Brea, CA, Beckman Coulter, A63880) following manufacturer’s recommendations was used before and after the 1st round of PCR. PCR amplicons were performed using Q5 High-Fidelity 2X Master Mix (NEB, M0492L) and amplicons were visually validated by standard gel electrophoresis. Hiseq reads were aligned to the signature regions of 14 serotypes: AAV1, AAV2, AAV2/3-hybrid, AAV3, AAV4, AAV5, AAV6, AAV7, AAV8, AAV9, AAVrh.10, AAVrh.39, and AAVrh·43. The abundance of reads mapping to each serotype’s signature region were tabulated using a custom workflow on the Galaxy web platform at http://usegalaxy.org (30). Unique amino acid sequences were defined and tabulated by USEARCH (usearch11.0.667) with zero-radius operational-taxonomic units (ZOTUs) (31). Data were displayed using GraphPad Prism (v8.4).
Statistics analysis.
Quantification of AAV genome copy numbers by qPCR was performed in technical triplicates for each sample. Data was converted to copy number/cell and shown as mean ±SD. Prism software was used to calculate statistical significance by paired two-tailed Student’s t-test.
Results:
Tumor and adjacent non-tumor tissues show no significant differences in the molecular prevalence of AAV
Our aim was to profile the epidemiology of AAV in cancer patients. To do so, we opted to quantify AAV positivity by molecular signature via PCR detection (Figure 1). We obtained 728 tumor and adjacent non-lesion resection samples from 413 cancer patients receiving care at West China Hospital (Chengdu, China) (Table 1). Specifically, specimens from brain tumors (n=54, four cases with unknown tumor grade), and pancreatic carcinomas (n=34) were collected. Tumor mass (TM) tissues and matched adjacent non-tumor (NT) tissues were collected from rectal carcinomas (TM=50; NT=50), gastric carcinomas (TM=21; NT=21, four are gastrointestinal stromal tumors), lung adenocarcinomas (TM=50; NT=50), lung squamous carcinomas (TM=50; NT=50), hepatocellular carcinomas (HCC) (TM=49; NT=49), hepatoid adenocarcinomas (HAC) (TM=45; NT=43, eight cases with unknown tumor grade), breast carcinomas (TM=50; NT=50, one case with unknown age). A select sample of hematoxylin and eosin stained sections used to verify and index tumor grades are provided in Figure 2. For our investigation, a qPCR primer set spanning a highly variable sequence of the AAV cap gene, hence referred to as the “signature” PCR region (29), was used to assess AAV-positivity among tumor and tumor adjacent samples. We found that among the cohort of patients, approximately 80% were positive for AAV (Figure 3A). Interestingly, different carcinoma/tissue types varied in AAV positivity. For example, lung squamous carcinomas were 98% positive for AAV (96% in adjacent non-lesion tissues), while lung adenocarcinomas were 58% positive (56% in adjacent non-lesion tissues). We next aimed to compare AAV genome copy number variation between tumor masses and adjacent non-tumor tissues in patient samples that were positive for AAV, using a set of primers spanning a conserved region of rep gene (copy number PCR region, Figure 1). If AAV positivity and presumed integration is correlated with tumorigenesis, we expected that tumor samples would display AAV genome copies per host cell genome closer to a theoretical ratio of 1:1, resulting from integration and clonal expansion within the tumor mass. In contrast, adjacent non-tumor tissues should display drastically less AAV genome copies per cell. Tissue pairs that did not exhibit AAV positivity in both tumor mass and adjacent tissues were not included in this comparison. We found that among all tissues quantified, we did not find any samples with more than 0.003 AAV genome copies per host cell genome (Figure 3B). We also did not observe copy number differences between tumor mass and adjacent non-tumor tissues among all tissue/tumor types, on average.
Table 1.
Molecular detection of AAV in tumor and tumor adjacent tissues from cancer patients
| Organ | Number positive/total | Breast | Rectum | Pancreas | Brain | Lung ad | Lung sq | HCC | HAC | Stomach | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TM | NT | TM | NT | TM | NT | TM | NT | TM | NT | TM | NT | TM | NT | TM | NT | TM | NT | ||
| Total | 591/728 | 46/50 | 45/50 | 36/50 | 33/50 | 40/45 | - | 52/55 | - | 29/50 | 28/50 | 49/50 | 48/50 | 37/49 | 35/49 | 37/45 | 36/43 | 19/21 | 21/21 |
| Gender | |||||||||||||||||||
| Male | 350/429 | 18/25 | 15/25 | 23/27 | - | 21/22 | - | 13/22 | 13/22 | 47/48 | 46/48 | 31/41 | 29/41 | 34/40 | 32/38 | 13/15 | 15/15 | ||
| Female | 240/298 | 46/50 | 45/50 | 18/25 | 18/25 | 17/18 | - | 30/32 | - | 16/28 | 15/28 | 2/2 | 2/2 | 6/8 | 6/8 | 3/5 | 4/5 | 6/6 | 6/6 |
| Age† | |||||||||||||||||||
| <20 years | 1/1 | - | - | - | - | - | - | 1/1 | - | - | - | - | - | - | - | - | - | - | - |
| 20-40 years | 61/71 | 7/7 | 7/7 | 1/2 | 2/2 | - | - | 8/9 | - | 1/1 | 0/1 | 1/1 | 1/1 | 12/14 | 11/14 | 3/5 | 5/5 | 1/1 | 1/1 |
| 40-60 years | 298/372 | 23/27 | 25/27 | 20/28 | 16/28 | 15/19 | - | 33/34 | - | 13/24 | 14/24 | 29/29 | 29/29 | 19/23 | 15/23 | 20/25 | 20/24 | 3/4 | 4/4 |
| >60 years | 230/283 | 15/16 | 14/16 | 15/20 | 15/20 | 25/26 | - | 9/10 | - | 15/25 | 14/25 | 19/20 | 18/20 | 6/12 | 9/12 | 14/15 | 11/14 | 15/16 | 16/16 |
One unknown age
Abbreviations: TM, tumor mass; NT, adjacent non-tumor tissue. Lung ad, Lung adenocarcinoma; Lung sq, Lung squamous carcinoma. HCC, Hepatocellular carcinoma; HAC, hepatocellular adenocarcinoma.
Figure 2. Representative H&E-stained sections of tissue resections from cancer patients.
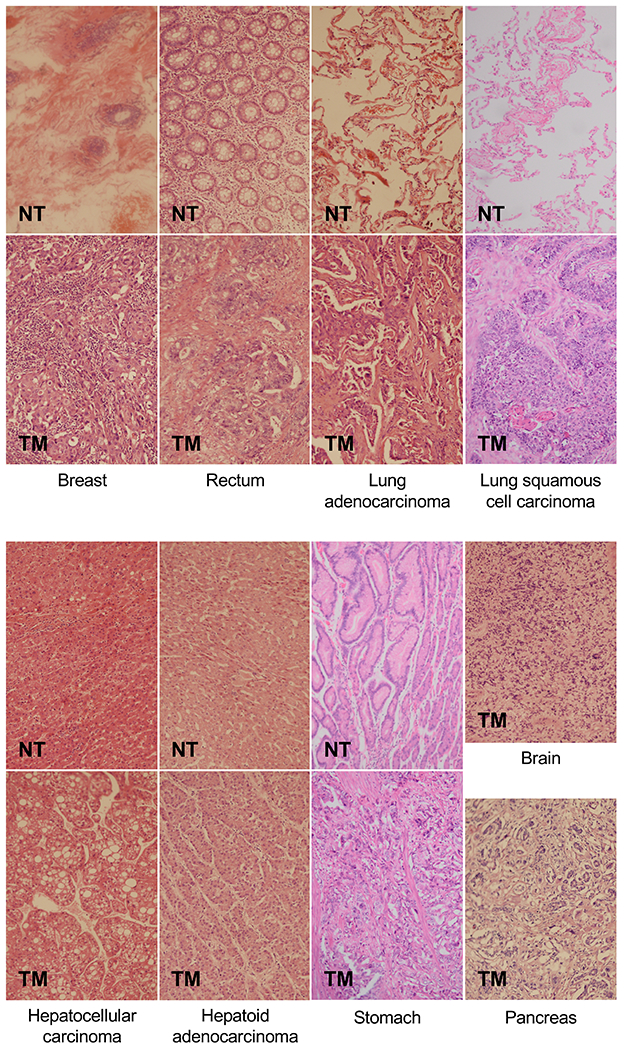
Tumor mass (TM) samples from patients with breast cancer, rectal cancer, lung adenocarcinoma, lung squamous cell carcinoma, hepatocellular carcinoma hepatoid adenocarcinoma, gastric (stomach) cancer, glioma (brain), and pancreatic cancer tissues were obtained, sectioned, and stained to determine tumor grades. Adjacent non-tumor tissues (NT) for all tumor types, except for glioma and pancreas tissues were also obtained and examined to verify normal morphology of samples.
Figure 3. Detection of AAV abundance across tumor and adjacent non-tumor tissues.
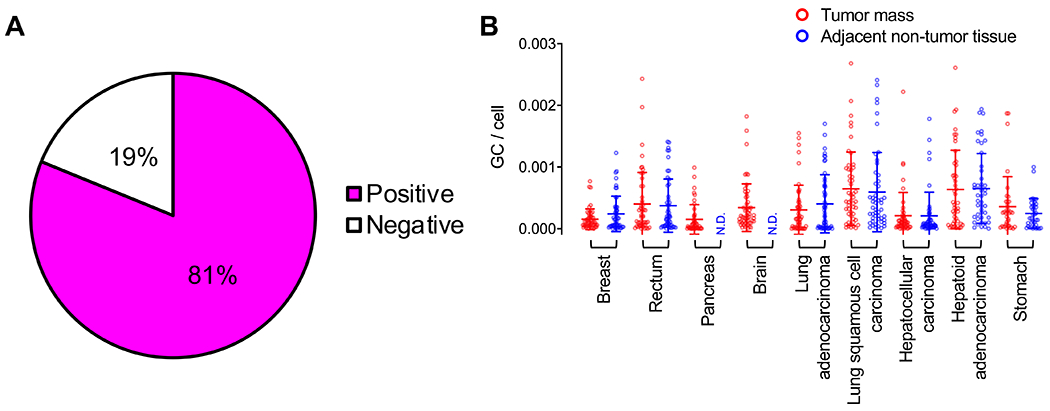
(A) Pie chart of 728 tumorectomy samples (tumor masses and adjacent non-lesion tissues) that are positive or negative for AAV, as assessed by signature PCR. (B) Quantification of AAV genome copy per host cell genome of positive samples in both tumor mass and adjacent non-tumor tissues. Each dot represents one patient tissue sample. Tumor mass (red), adjacent non-tumor tissues (blue). Means ±SD are displayed.
We also partitioned the analysis to determine whether gender- or age-specific differences in AAV copy number was present between tumor and non-tumor tissues. When stratified by gender, no specific differences were observed (Figure 4). However, when stratified by age, we only observed significant difference between three groups (Figure 5): in the 41-60 age group with HCC, where there is a significantly higher copy number in adjacent non-tumor tissues than in tumor masses (p = 0.043); and in the over 60-year-old patient group, where adjacent non-tumor breast tissues had a higher copy number than the tumor mass (p = 0.005), and lung squamous cell carcinoma tissues displayed a higher copy number than adjacent non-tumor tissues (p = 0.047). Although notable, the genome copy numbers detected in lung cancer samples were still under 0.003 genomes per host cell genome. These data again indicate that tumor tissues, aside from squamous lung carcinoma, did not exhibit higher AAV copy numbers compared with non-tumor tissues.
Figure 4. Detection of AAV abundance across tissues distributed by gender.
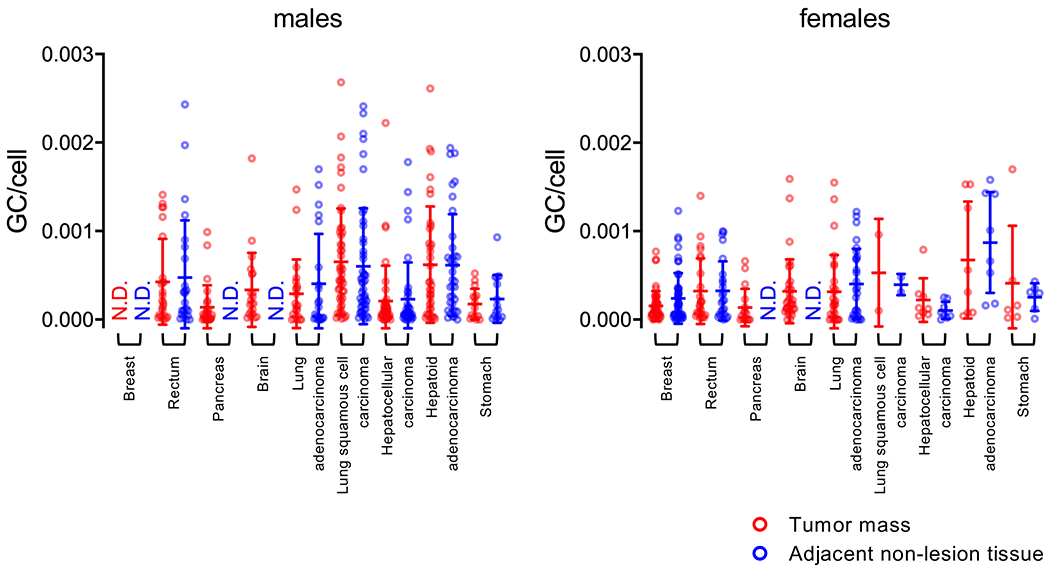
Quantification of AAV genome copy per host cell genome among positive samples in both tumor mass and adjacent non-tumor tissues. The data is partitioned by gender (males, left graph; females right graph). Each dot represents one patient tissue sample. Tumor mass (red), adjacent non-tumor tissues (blue). Means ±SD are displayed.
Figure 5. Detection of AAV abundance across tissues distributed by age.
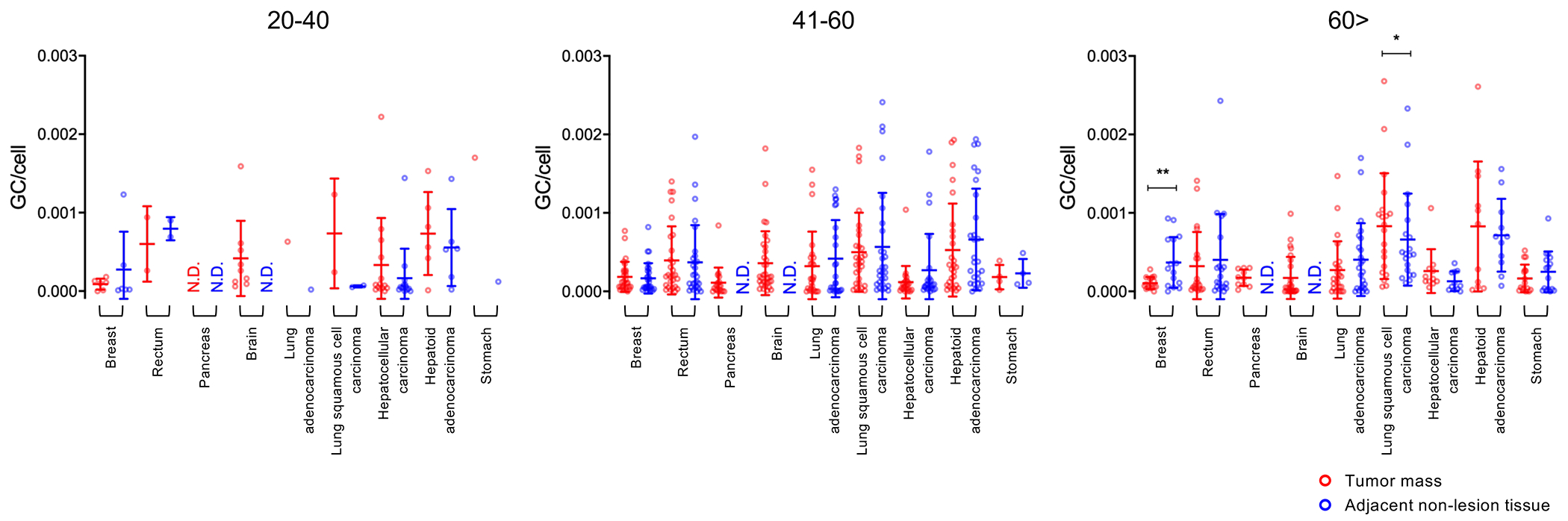
Quantification of AAV genome copy per host cell genome among positive samples in both tumor mass and adjacent non-tumor tissues split. The data is partitioned by age (between 20 and 40, left graph; between 41 and 60, middle graph; over 60, right graph). Each dot represents one patient tissue sample. Tumor mass (red), adjacent non-tumor tissues (blue). Means ±SD are displayed. *, p<0.05; **, p<0.01
To further explore the relationship between AAV copy number and tumor progression, we stratified samples by tumor grade as diagnosed by frozen sections (Figures 2 and 6). Once again, pairwise comparison between tumor mass and adjacent non-tumor tissues did not reveal any significant differences. Notably, both brain and pancreas samples were not accompanied by adjacent non-tumor tissues (Figure 6C, D). Nonetheless, there were no copy number trends when tissues were grouped by cancer grade in either glioma or pancreas samples.
Figure 6. Detection of AAV abundance across tissues distributed by tumor grade.
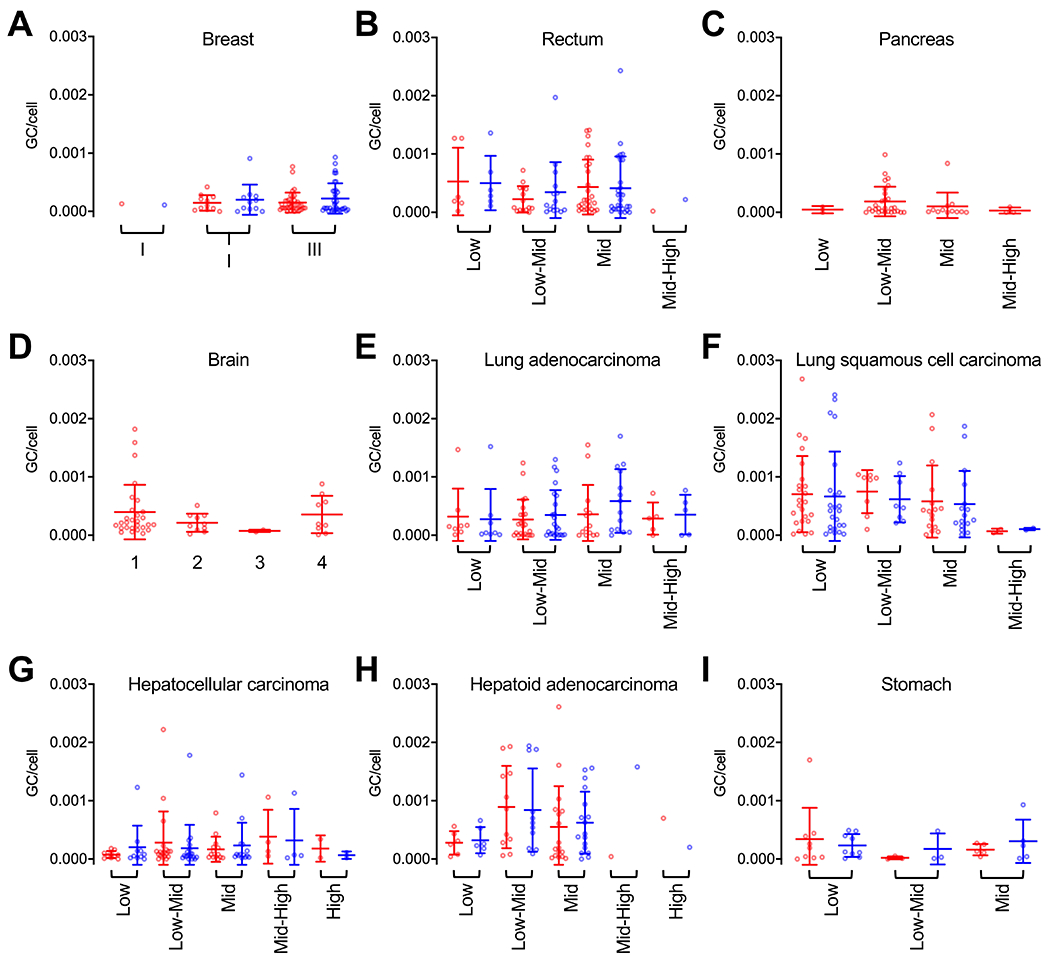
Quantification of AAV genome copy per host cell genome among positive samples in both tumor mass and adjacent non-tumor tissues. Data is grouped by tumor grade for each cancer type. Sample tissues: breast cancer (A), rectal cancer (B), pancreatic cancer (C), glioma (brain, D), adenocarcinoma of the lung (E), lung squamous cell carcinoma (F), hepatocellular carcinoma (G), hepatoid adenocarcinoma (H), gastric (stomach) cancer (I). Each dot represents one patient tissue sample. Tumor mass (red), adjacent non-tumor tissues (blue). Means ±SD are displayed.
High-throughput sequencing of signature regions show a high degree of viral variation
We next aimed to address whether the low copy numbers we detected in tissues were due to infiltrating cell types or cells of the desmoplastic stroma that lack integrated AAV genomes of the tumor mass, which could dilute AAV-positive cells. These can be endothelial cells, immune cells, or cancer-associated fibroblasts. To address this in an unbiased fashion, we aimed to determine the diversity of the AAV genomes found within the tissues. In these experiments, we opted to specifically investigate sample pairs that successfully yielded amplicons from both tissues. This approach rules out the few samples in which AAV is detected in tumors but absent in non-tumor samples (Table 1), which may miss some interesting molecular representation. Nonetheless, this strategy overcomes false-negative PCR amplification that may lead to misinterpretation of data. The unique approach indirectly shows whether integrants in the tumor mass are present by querying whether AAV sequences exhibit clonality. We generated libraries for next-gen sequencing with signature region PCR amplicons. All reads were mapped to a DNA reference containing the signature regions of 14 known serotypes (Figure 7). What we observed was quite surprising. Many of the tissues analyzed exhibited signature sequences mapping to several serotypes. For example, sample C364 (breast cancer, Figure 7A) contains reads mapping to AAV2, AAV2/3-hybrid, AAV8, and AAVrh.43. This finding was unexpected; we predicted that some tissues could contain perhaps two serotypes, but the diversity observed here was remarkable. Despite the serotype diversity found within individual tissues, the majority were positive for AAV2/3-hybrid variants. The next most abundant serotype present in tissues was AAV2, followed by AAV8 and finally AAVrh.43. Of note, a single HCC sample (C447) and a gastric cancer sample (C128) and were comprised of 40.4% and 59.2% AAV1, respectively (Figure 7E, F). We did not observe any trends in serotype profile among the different tissues of origin or type. Signature reads were also analyzed by variant diversity. We translated the DNA reads to amino acid sequences to infer the diversity of capsids in tissues by probing amino acid sequence variant across over the signature region as a proxy. Again, we predicted if AAV integration is correlated with tumorigenesis, we would expect that diversity in signature reads to be significantly less in tumor samples than in non-adjacent tumor tissues. On average, we observed that tissues had about 20 unique signature sequences in tissue libraries (Figure 7). The highest amount of diversity was observed among adenocarcinoma of the lung samples, where more than 80 unique signature sequences were observed (Figure 7C). Above all, we did not see a statistically significant difference in the diversity of signature sequences between tumor and adjacent non-tumor tissues.
Figure 7. Serotype diversity and sequence variation of AAV signature region obtained from patient tissues.
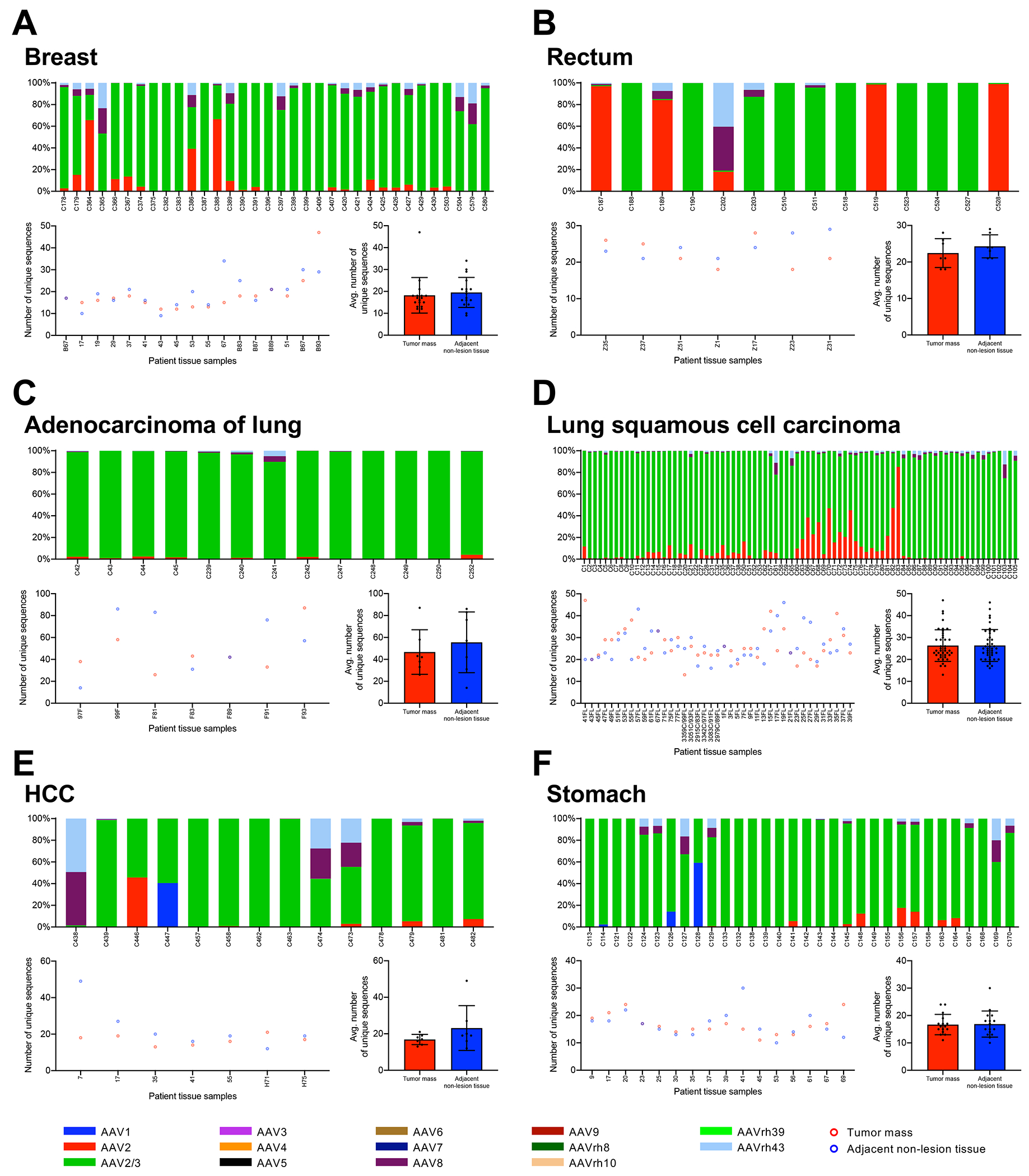
Signature PCR amplicons were subjected to high-throughput sequencing and aligned to 14 known serotype signature regions. Sample tissues: breast cancer (A), rectal cancer (B), adenocarcinoma of the lung (C), lung squamous cell carcinoma (D), hepatocellular carcinoma (E), gastric cancer (F). Data for each tissue of origin is displayed in three ways, stacked histogram of the percentage of reads mapping to known serotypes (top); number of unique amino acid sequences tabulated for tumor (red) and adjacent non-tumor (blue) samples (lower left); and mean ±SD of unique amino acid sequences (lower right).
Discussion:
Gene therapy has given hope for treating genetic diseases. However, one of the prominent concerns for AAV vectors has been their low integration profiles, which may lead to genotoxicity in the form of cancer. These concerns were attributed to early work, which showed in mouse models that integration into the Rian locus leads to HCC (9, 13, 32). Thus far, such outcomes have yet to emerge in human clinical trials. Reports, including the recent study by La Belle et al., which queried 109 AAV-positive HCC tumors, indicated that wild-type AAV infection is indirectly linked with liver cancer in humans has re-raised the concern (16, 17). Furthermore, demonstration that the wild-type sequence element proximal to the 3’-ITR carries liver-related enhancer function, lends further mechanistic support that spurious integration events into cancer driver genes can distinctively result in HCC (33). It is notable that this element is removed in many vector designs, and these findings once again do not directly inform on the potential for rAAVs to induce tumors in patient tissues. Although these examples were not cases of patients receiving gene therapy vectors, critics of AAV gene therapy have used these lines of evidence as proof that AAV-based vectors are cancer risk factors by transitive inference. However, recent demonstration that canine models receiving gene therapy have vector genome integration into cancer driver genes in the liver have reignited this conversation (23), and have forced re-visitation of investigations regarding whether wild-type AAVs can integrate into cancer driver genes as a natural phenomenon.
The current controversy that cancer can be attributed to AAV-mediated genotoxicity still stands within the field. The studies suggesting that AAV is a potential risk factor for cancer conflicts with the high rate of AAV sero-positivity in the healthy population, which can range from 40-80% (24). In addition, these studies are heavily biased by the methodology employed in the investigation. Integration events were queried by high-throughput sequencing that specifically identified and interrogated integrants in HCC samples and does not explore whether these events are causative or concurrent with tumor incidents (16). In other words, it is possible that the promoters of cancer drivers are more accessible to AAV genome integration, therefore events observed are a result of opportunistic integration following tumorigenesis, and not a cause of it. It is therefore not clear whether such observations fall in line with the natural distribution of integration events seen in the high degree of natural infection occurring in the human population, and whether this necessarily implicates AAV as a significant risk factor for HCC. Despite the uncertainty with pre-existing reports, concerns of AAV causing HCC still affect how these biotherapies are perceived. We therefore sought to take a critical look at AAV positivity within a large population of cancer patients. We compared viral genome copy number between tumor tissues and corresponding adjacent non-tumor tissues and in this way, clonal expansion can be assessed as a hallmark of AAV integration-mediated cancer. As expected, we found that a large percentage of cancer patients were positive for AAV. In total, we detected proviral sequences for serotypes AAV2, AAV2/3-hybrid, AAV1, AAV8, and AAVrh.43 among all tissues. The most frequently detected serotype was AAV2/3, which was found in all tissue samples. We note that this distribution only represents a small portion of the Chinese population within the Sichuan province. This is the first molecular seroprevalence for AAV among this population group; thus, we could not validate our findings with previous studies. There have been reports from 2012 to 2019 of AAV seroprevalence within China’s Anhui province, Beijing, and Shanghai, which reported >90% AAV2, >80% AAV3, >60% AAV8, and ~70% AAV1, depending on the region (34–36). Interestingly, AAV5 was reported to persist in 35% and 47% of the population in Beijing and the Anhui province, respectively (35). However, AAV5 was not identified in our molecular analysis of tissues. These differences may be due to the unique method of how serotype prevalence was determined in our study. Nonetheless, our work adds to the wealth of knowledge pertaining to AAV epidemiology. Importantly, comparing tumor and adjacent-non-tumor tissues revealed very low copy numbers, demonstrating a lack of clonality in resections. Recognizing that tumor tissue includes both malignant and tumor stromal cells, these data still are not consistent with clonal integration within the malignant population of tumor cells, which is most commonly 31% to 50% in common cancers, such as colon and breast cancer (37, 38). Thus, copy numbers within the range observed in this study are not consistent with clonal integration, even within the malignant subpopulation of tumors. We therefore emphasize that if AAV was a causative factor for tumorigenesis, the virus copy number, when normalized to cell number, should be significantly higher than non-tumor tissues. This outcome would be the case, since the expansion of primary tumors conforms to clonal expansion despite cellular heterogeneity, which may occur during progressive stages. If AAV integration resulted in tumorigenesis in these patients, the number of detected AAV genome copies would be significantly higher. Stratifying the data in different ways also indicated a lack of significant difference in the types and abundances of AAV between tumor and adjacent normal tissues. Finally, high-throughput sequencing of the AAV signature region also showed a high diversity of reads, further demonstrating a lack of clonality.
We note that our means of validating the presence of AAV genomes in tissues is limited to what can be detected by the signature region and a segment of the rep ORF. It has been shown that AAV integration events may lack sequences encompassing the ORFs, but ITR sequences are maintained and are thus a more reliable marker for integration. Unfortunately, the sequences interrogated in our study do not span subgenomic fragments, opening up to the possibility that integration events might have been missed in our study. In the recent La Bella et al. report, among 233 AAV-positive non-tumor liver tissues, only 27.5% of tissues demonstrated amplification of other genomic regions, hinting at the presence of full-length genomes, while 26% of tissues harbored detectable episomal forms as determined by PCR amplification spanning across the recombined ITR. In addition, among 109 positive tumor tissues, they reported that 76% of tissues only amplified one or two viral regions, with observational enrichment of the 3’-ITR. Therefore, our querying of specific ORF regions could potentially confound our interpretations and overall conclusions that the presence of AAV did not reveal any clonal events. Notwithstanding, further exploration into “ITR-only” integrants of AAV genomes is needed to complement our findings.
In conclusion, we show via unbiased profiling of AAV genomes between tumor masses and adjacent non-tumor tissues that there were no statistically significant differences among the tissues evaluated at the levels of both genome abundance and variability. These new findings, in part, substantiates the lack of causative correlation between AAV and cancer including, HCC, and provides further insights into the ongoing controversy surrounding whether or not AAV is a cancer risk factor.
Acknowledgements:
This work was supported by grants from the University of Massachusetts Medical School (an internal grant) and by the NIH (R01NS076991-01, P01AI100263-01, P01HL131471-02, R01AI121135, UG3HL147367-01, and R01HL097088).
Footnotes
Competing interests
G.G. work has been funded by the NIH, and is a scientific co-founder of Voyager Therapeutics and Aspa Therapeutics, and holds equity in these companies. G.G. is an inventor on patents with potential royalties licensed to Voyager Therapeutics, Aspa Therapeutics, and other biopharmaceutical companies. The remaining authors declare no competing interests.
References:
- 1.Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogden PJ, Kelsic ED, Sinai S, Church GM. Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science. 2019;366(6469):1139–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duan D, Sharma P, Yang J, Yue Y, Dudus L, Zhang Y, et al. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J Virol. 1998;72(11):8568–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnepp BC, Jensen RL, Chen CL, Johnson PR, Clark KR. Characterization of adeno-associated virus genomes isolated from human tissues. J Virol. 2005;79(23):14793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakai H, Montini E, Fuess S, Storm TA, Meuse L, Finegold M, et al. Helper-independent and AAV-ITR-independent chromosomal integration of double-stranded linear DNA vectors in mice. Mol Ther. 2003;7(1):101–11. [DOI] [PubMed] [Google Scholar]
- 6.Berns KI. The Unusual Properties of the AAV Inverted Terminal Repeat. Hum Gene Ther. 2020;31(9-10):518–23. [DOI] [PubMed] [Google Scholar]
- 7.Kotin RM, Menninger JC, Ward DC, Berns KI. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics. 1991;10(3):831–4. [DOI] [PubMed] [Google Scholar]
- 8.Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317(5837):477. [DOI] [PubMed] [Google Scholar]
- 9.Wang PR, Xu M, Toffanin S, Li Y, Llovet JM, Russell DW. Induction of hepatocellular carcinoma by in vivo gene targeting. Proc Natl Acad Sci U S A. 2012;109(28):11264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pachori AS, Melo LG, Zhang L, Loda M, Pratt RE, Dzau VJ. Potential for germ line transmission after intramyocardial gene delivery by adeno-associated virus. Biochem Biophys Res Commun. 2004;313(3):528–33. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Malani N, Hamilton SR, Schlachterman A, Bussadori G, Edmonson SE, et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood. 2011;117(12):3311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong L, Malani N, Li M, Brady T, Xie J, Bell P, et al. Recombinant adeno-associated virus integration sites in murine liver after ornithine transcarbamylase gene correction. Hum Gene Ther. 2013;24(5):520–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandler RJ, LaFave MC, Varshney GK, Trivedi NS, Carrillo-Carrasco N, Senac JS, et al. Vector design influences hepatic genotoxicity after adeno-associated virus gene therapy. J Clin Invest. 2015;125(2):870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosas LE, Grieves JL, Zaraspe K, La Perle KM, Fu H, McCarty DM. Patterns of scAAV vector insertion associated with oncogenic events in a mouse model for genotoxicity. Mol Ther. 2012;20(11):2098–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gil-Farina I, Schmidt M. Interaction of vectors and parental viruses with the host genome. Curr Opin Virol. 2016;21:35–40. [DOI] [PubMed] [Google Scholar]
- 16.Nault JC, Datta S, Imbeaud S, Franconi A, Mallet M, Couchy G, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47(10):1187–93. [DOI] [PubMed] [Google Scholar]
- 17.La Bella T, Imbeaud S, Peneau C, Mami I, Datta S, Bayard Q, et al. Adeno-associated virus in the liver: natural history and consequences in tumour development. Gut. 2020;69(4):737–47. [DOI] [PubMed] [Google Scholar]
- 18.Berns KI, Byrne BJ, Flotte TR, Gao G, Hauswirth WW, Herzog RW, et al. Adeno-Associated Virus Type 2 and Hepatocellular Carcinoma? Hum Gene Ther. 2015;26(12):779–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buning H, Schmidt M. Adeno-associated Vector Toxicity-To Be or Not to Be? Mol Ther. 2015;23(11):1673–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nault JC, Mami I, La Bella T, Datta S, Imbeaud S, Franconi A, et al. Wild-type AAV Insertions in Hepatocellular Carcinoma Do Not Inform Debate Over Genotoxicity Risk of Vectorized AAV. Mol Ther. 2016;24(4):660–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanlon KS, Kleinstiver BP, Garcia SP, Zaborowski MP, Volak A, Spirig SE, et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat Commun. 2019;10(1):4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller DG, Petek LM, Russell DW. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat Genet. 2004;36(7):767–73. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen GN, Everett JK, Kafle S, Roche AM, Raymond HE, Leiby J, et al. A long-term study of AAV gene therapy in dogs with hemophilia A identifies clonal expansions of transduced liver cells. Nat Biotechnol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calcedo R, Vandenberghe LH, Gao G, Lin J, Wilson JM. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis. 2009;199(3):381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huser D, Khalid D, Lutter T, Hammer EM, Weger S, Hessler M, et al. High Prevalence of Infectious Adeno-associated Virus (AAV) in Human Peripheral Blood Mononuclear Cells Indicative of T Lymphocytes as Sites of AAV Persistence. J Virol. 2017;91(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CL, Jensen RL, Schnepp BC, Connell MJ, Shell R, Sferra TJ, et al. Molecular characterization of adeno-associated viruses infecting children. J Virol. 2005;79(23):14781–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boutin S, Monteilhet V, Veron P, Leborgne C, Benveniste O, Montus MF, et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther. 2010;21(6):704–12. [DOI] [PubMed] [Google Scholar]
- 28.Halbert CL, Miller AD, McNamara S, Emerson J, Gibson RL, Ramsey B, et al. Prevalence of neutralizing antibodies against adeno-associated virus (AAV) types 2, 5, and 6 in cystic fibrosis and normal populations: Implications for gene therapy using AAV vectors. Hum Gene Ther. 2006;17(4):440–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao G, Alvira MR, Somanathan S, Lu Y, Vandenberghe LH, Rux JJ, et al. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc Natl Acad Sci U S A. 2003;100(10):6081–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–W44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. [DOI] [PubMed] [Google Scholar]
- 32.Chandler RJ, LaFave MC, Varshney GK, Burgess SM, Venditti CP. Genotoxicity in Mice Following AAV Gene Delivery: A Safety Concern for Human Gene Therapy? Mol Ther. 2016;24(2):198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Logan GJ, Dane AP, Hallwirth CV, Smyth CM, Wilkie EE, Amaya AK, et al. Identification of liver-specific enhancer-promoter activity in the 3’ untranslated region of the wild-type AAV2 genome. Nat Genet. 2017;49(8):1267–73. [DOI] [PubMed] [Google Scholar]
- 34.Ling C, Wang Y, Feng YL, Zhang YN, Li J, Hu XR, et al. Prevalence of neutralizing antibodies against liver-tropic adeno-associated virus serotype vectors in 100 healthy Chinese and its potential relation to body constitutions. J Integr Med. 2015;13(5):341–6. [DOI] [PubMed] [Google Scholar]
- 35.Liu Q, Huang W, Zhang H, Wang Y, Zhao J, Song A, et al. Neutralizing antibodies against AAV2, AAV5 and AAV8 in healthy and HIV-1-infected subjects in China: implications for gene therapy using AAV vectors. Gene Ther. 2014;21(8):732–8. [DOI] [PubMed] [Google Scholar]
- 36.Liu Q, Huang W, Zhao C, Zhang L, Meng S, Gao D, et al. The prevalence of neutralizing antibodies against AAV serotype 1 in healthy subjects in China: implications for gene therapy and vaccines using AAV1 vector. J Med Virol. 2013;85(9):1550–6. [DOI] [PubMed] [Google Scholar]
- 37.Kramer CJH, Vangangelt KMH, van Pelt GW, Dekker TJA, Tollenaar R, Mesker WE. The prognostic value of tumour-stroma ratio in primary breast cancer with special attention to triple-negative tumours: a review. Breast Cancer Res Treat. 2019;173(1):55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mesker WE, Junggeburt JM, Szuhai K, de Heer P, Morreau H, Tanke HJ, et al. The carcinoma-stromal ratio of colon carcinoma is an independent factor for survival compared to lymph node status and tumor stage. Cell Oncol. 2007;29(5):387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]


