Abstract
Long non-coding RNAs (lncRNAs) are a large and diverse class of RNA molecules that are transcribed but not translated into proteins, with a length of more than 200 nucleotides. LncRNAs are involved in gene expression and regulation. The abnormal expression of lncRNAs is associated with disease pathogenesis. Small heterodimer partner (SHP, NR0B2) is a unique orphan nuclear receptor that plays a pivotal role in many biological processes by acting as a transcriptional repressor. In this review, we present the critical roles of SHP and summarize recent findings demonstrating the regulation between lncRNAs and SHP in liver disease.
Keywords: Non-coding RNAs, lncRNAs, Nuclear receptor, SHP
1. Introduction
Non-protein coding transcripts or non-coding RNAs (ncRNAs) account for 98% or so of the human transcriptome and play crucial roles in development, physiology, and disease (Esteller, 2011; Palazzo and Lee, 2015). Long non-coding RNAs (lncRNAs) are defined as those ncRNAs longer than 200 nucleotides, which are largely tissue-specifically expressed (Statello et al., 2021). LncRNAs participate in gene transcriptional, post-transcriptional, and epigenetic regulation and are essential in many physiological processes, such as X-chromosome inactivation in mammals (Cabili et al., 2011; Gomes et al., 2013; Zhao et al., 2017). Mechanistically, lncRNAs participate in histone modifications to remodel chromatin, direct the recruitment of RNA polymerase and cofactors, serve as scaffolds for the association of transcription factors (TFs) with other cofactors to mediate transactivation, act as decoys to prevent TFs from binding to their DNA elements, silence gene expression via modulation of translation and mRNA stability, or are involved in RNA alternative splicing (Fernandes et al., 2019; Wu et al., 2021; X. Zhang et al., 2019). About 20% of lncRNAs are derived from enhancer regions (termed eRNAs), participating in chromosomal enhancer-promoter looping (Bonasio and Shiekhattar, 2014). LncRNAs also cooperate with microRNAs (miRNAs) to regulate gene expression through competition of binding or acting as miRNA sponges (Lopez-Urrutia et al., 2019). LncRNAs can also be the precursors of miRNAs and serve as signals for the activation of specific biological events (P. Zhang et al., 2019).
Small heterodimer partner (SHP, NR0B2) belongs to the nuclear receptor (NR) superfamily. Generally, it acts as a transcriptional repressor through interaction with a variety of other NRs, including androgen receptor (AR), estrogen receptor alpha (ERα), hepatocyte nuclear factor 4 alpha (HNF4α), liver receptor homolog-1 (LRH-1), liver X receptor alpha (LXRα), peroxisome proliferator-activated receptor gamma (PPARγ), retinoic acid receptor alpha (RARα), and retinoid X receptor alpha (RXRα), to regulate diverse biological processes, such as bile acid synthesis, glucose/lipid metabolism, and drug metabolism (Zhang et al., 2011). As an orphan NR, SHP contains dimerization and ligand-binding domains but lacks a DNA-binding domain. SHP may also interact with other non-NR TFs to inhibit gene transcription and regulate diverse signaling pathways involved in metabolism, inflammation, and cell proliferation (Song et al., 2017b).
There is increasing recognition of the role of ncRNAs in diseases. The most well-studied ncRNAs are miRNAs, but lncRNAs also play critical roles in cellular homeostasis and are inherent to diseases. In addition to recapitulating the essential functions of SHP, this review summarizes the up-to-date findings on the crosstalk between lncRNAs and SHP and reveals their pathophysiological relevance to liver disease.
2. The primary function of SHP
2.1. SHP in cholesterol and bile acid homeostasis
Cholesterol homeostasis in mammals is maintained through biosynthesis, cellular uptake, and hepatic conversion to bile acids.
The well-established function of SHP is to suppress bile acid biosynthesis. Farnesoid X receptor (FXR) binds and activates the SHP promoter and SHP represses LRH-1-dependent activation of the cholesterol 7-alphahydroxylase (CYP7A1) promoter (Goodwin et al., 2000; Lu et al., 2000). CYP7A1 catalyzes the rate-limiting step in bile acid biosynthesis. FXR-mediated regulation of bile acid-related genes, including SHP and CYP7A1, depends on bromodomain-containing protein 4 (BRD4) that is required for the anti-inflammatory and anti-fibrotic actions of obeticholic acid (OCA), a potent and selective FXR agonist (Jung et al., 2020). SHP also interacts with HNF4α to repress the transcription of CYP8B1 that catalyzes the synthesis of cholic acid and determines the hydrophobicity of the bile acid pool (Zhang and Chiang, 2001). The role of SHP in hepatic bile acid biosynthesis is further elucidated in Shp knockout mice that exhibit mild defects in bile acid homeostasis, suggesting the existence of compensatory pathways of bile acid signaling (Wang et al., 2002). CYP8B1 is strongly induced in Shp knockout mice, which may increase the hydrophilicity of the bile acid pool and reduce the hepatotoxicity of bile acids (Wang et al., 2003). Besides, SHP maintains cholesterol homeostasis through repressing the expression of cholesterol biosynthesis enzyme, 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR), the rate-limiting enzyme of the mevalonate pathway in producing cholesterol and other isoprenoids; mechanistically, SHP inhibits LRH-1 and sterol regulatory element binding transcription factor 2 (SREBF2) from binding to the HMGCR promoter (Datta et al., 2006; Kim et al., 2015). The double knockout mice of SHP and FXR develop intrahepatic cholestasis, which recapitulates human progressive familial intrahepatic cholestasis (PFIC) and can be used to investigate the molecular pathogenesis of PFIC (K. H. Kim et al., 2018).
Hepatic miR-210 levels are elevated in cholestatic mouse models and patients with primary biliary cholangitis. MiR-210 promotes bile acid-induced liver injury in part by targeting the mixed-lineage leukemia-4 (MLL4) methyltransferase. SHP inhibits miR-210 expression by repressing a transcriptional activator, Kruppel-like factor-4 (KLF4), and nuclear levels of SHP are reduced in cholestatic livers (Kim et al., 2020a).
SHP is also highly expressed in the intestine. Postprandial fibroblast growth factor (FGF) 19 (human FGF19, mouse FGF15) induces SHP phosphorylation that inhibits the transcriptional activity of SREBF2, leading to the repression of intestinal NPC1-like intracellular cholesterol transporter 1 (NPC1L1) expression and cholesterol absorption (Kim et al., 2015; Y. C. Kim et al., 2019).
2.2. SHP in glucose and lipid metabolism
SHP has a major function in regulating glucose metabolism by inhibiting gluconeogenic gene expression. Glucose 6-phosphatase (G6Pase) and phosphoenolpyruvate carboxykinase (PEPCK) are the rate-limiting enzymes in hepatic gluconeogenesis. SHP represses G6Pase and PEPCK gene expression via inhibition of the forkhead transcription factors HNF3 and HNF6 (Kim et al., 2004; Lee et al., 2008). SHP antagonizes peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) coactivation of glucocorticoid receptor (GR), leading to the inhibition of PEPCK expression (Borgius et al., 2002). SHP represses CCAAT/enhancer-binding protein alpha (C/EBPα)-driven transcription of PEPCK and FOXO1-mediated transcription of G6Pase (Park et al., 2007; Yamagata et al., 2004). SHP also inhibits the transcriptional activity of LXRα and PPARγ by competing for binding to their common heterodimer partner RXRα to decrease hepatic glucokinase expression (Kim et al., 2009).
SHP is essential to maintain hepatic lipid homeostasis (Watanabe et al., 2004). SHP-deficient mice are protected against fatty liver in part by increasing very-low-density lipoprotein (VLDL) secretion. VLDL secretion is controlled by microsomal triglyceride transfer protein (MTTP), and SHP represses LRH-1-mediated transactivation of the MTTP promoter in hepatocytes (Huang et al., 2007; Lee et al., 2015; Wang et al., 2005). Mice with hepatocyte-specific deletion of SHP are protected against dyslipidemia induced by either a cholesterol/cholic acid diet or hypothyroidism (Hartman et al., 2009), and against fatty liver development by suppressing the expression of PPARγ and lipid-droplet protein fat-specific protein 27 (FSP27) (Akinrotimi et al., 2017). A recent study shows that hepatocyte-specific deletion of SHP reduces high-fat, -cholesterol, and -fructose (HFCF) diet-induced hepatic steatosis but aggravates the development of liver inflammation and fibrosis; interestingly, if the mice already present hepatic steatosis induced by HFCF diet, the adeno-associated virus-mediated hepatic depletion of SHP is no longer effective in reducing steatosis but still exacerbates liver inflammation and fibrosis (Magee et al., 2020). Indeed, SHP-deficient hepatocytes have an enhanced ability to recruit neutrophils to the injured liver, and SHP is a negative regulator of c-Jun-mediated transcription of chemokine (C-X-C motif) ligand 2 (CXCL2) and NF-κB p65-mediated induction of chemokine (C-C motif) ligand 2 (CCL2) (Noh et al., 2018; Zou et al., 2018).
As an integral component of the liver circadian network, SHP inhibits neuronal PAS domain-containing protein 2 (NPAS2) expression; they form a negative feedback loop to regulate the cyclic expression patterns of liver metabolic genes and maintain triglyceride and lipoprotein homeostasis (Lee et al., 2015). Constitutive SHP expression in transgenic mice can deplete the hepatic bile acid pool and induce triglyceride accumulation in the liver (Watanabe et al., 2004). This phenotype is attributable to SHP-mediated direct repression of downstream target genes including the bile acid sensor FXRα, and the indirect activation of the lipogenic PPARγ and SREBF1 gene expression (Boulias et al., 2005).
In contrast to the former findings that SHP contributes to or plays a promoting role in hepatic steatosis, a recent study focusing on the effect of FGF15/19 on hepatic lipogenesis reveals that FGF19 induces SHP phosphorylation to epigenetically silence the expression of lipogenic genes, such as SREBF1 and FASN, in a DNA methyltransferase-3a (DNMT3A)-dependent manner; further, the virus-mediated overexpression of SHP in obese mice substantially reduces liver triglyceride levels and inhibits lipogenesis in part by regulating phosphatidylcholine levels (Kim et al., 2020b; Y. C. Kim et al., 2018). In line with this, a decrease of FXR and SHP expression is found in beta-carotene oxygenase 1 and beta-carotene oxygenase 2 double knockout mice which develop hepatic steatosis (Lim et al., 2018). The phenotypic discrepancy between germline loss and adult manipulation of SHP expression in lipid metabolism is currently unclear. Possible compensatory changes in metabolic pathways in transgenic/knockout mice need to be taken into consideration.
Obesity-induced overexpression of miR-802 impairs hepatic insulin sensitivity and glucose metabolism (Kornfeld et al., 2013). SHP inhibits the transactivation of miR-802 by aromatic hydrocarbon receptor (AHR), which is attenuated in non-alcoholic fatty liver disease (NAFLD) patients and obese mice; activation of FXR by OCA reduces miR-802 expression and improves insulin resistance and hepatic steatosis (Seok et al., 2020). In accordance with this, as a repressor of AHR, SHP mitigates the AHR overexpression-induced hepatic increase of phosphatidylcholines and steatosis in obese mice (Y. C. Kim et al., 2018). These findings suggest that the FXR-SHP-miR-802 pathway may be targeted for the treatment of type 2 diabetes and NAFLD.
2.3. SHP in cell proliferation
SHP is identified as a tumor suppressor (He et al., 2008; Suresh et al., 2017). SHP expression is down-regulated in hepatocellular carcinoma (HCC). SHP inhibits hepatocyte proliferation and activates apoptosis to suppress tumor growth in HCC (He et al., 2008; Zhang et al., 2010, 2008). A small molecule activator of SHP, 5-(diethylsulfamoyl)-3-hydroxynaphthalene-2-carboxylic acid, has a strong inhibitory effect on HCC cell migration by suppressing CCL2 expression (Z. Yang et al., 2016). Interestingly, thymine DNA glycosylase (TDG) is essential for SHP expression in the liver in response to FXR agonists, and conditional deletion of TDG in adult mice results in a male-predominant onset of HCC (Hassan et al., 2020).
2.4. Other roles of SHP
2.4.1. SHP in mitosis
SHP displaces a major fraction of pregnane X receptor (PXR) and ERα from the mitotic chromatin via intermolecular interactions, resulting in attenuation of transcriptional activities during mitosis and implying the potential function of SHP in the regulation of “gene-bookmarking” events in cellular development (Kumar et al., 2021).
2.4.2. SHP in immunity
Specific roles of SHP in liver non-parenchymal cells have been described recently. FXR activation protects livers from ischemia/reperfusion injury (IRI) by up-regulating SHP in Kupffer cells (KCs) to inhibit the pro-inflammatory responses (Jin et al., 2020). SHP knockdown increases hepatic IRI in myeloid glycogen synthase kinase 3β (Gsk3β) knockout mice, suggesting a negative role of SHP in regulating innate immunity (Zhou et al., 2018). SHP overexpression can attenuate platelet-derived growth factor-BB (PDGF-BB)-stimulated activation of hepatic stellate cells (HSCs) in vitro (Ma et al., 2020), and a SHP agonist, ISO-COOH, attenuates HSCs trans-differentiation and ECM deposition in vitro and shows anti-fibrotic activity in carbon tetrachloride (CCl4)- or α-naphthyl-isothiocyanate (ANIT)-induced liver fibrosis in mice (Cipriani et al., 2017).
Other findings also support the emerging roles of SHP in immunity. For instance, the expression and activity of SHP within macrophages can alter T cell fate (Cipriani et al., 2017). SHP blocks the transcription of type I interferon (IFN) and serves as a potent negative regulator of the virus-mediated type I IFN signaling (J. H. Kim et al., 2019). SHP is a transcriptional target and repressor of LRH-1; the latter is the transcriptional regulator of intestinal glucocorticoid (GC) synthesis; the SHP/LRH-1 axis regulates virus-induced intestinal GC synthesis to maintain intestinal immune homeostasis (Huang et al., 2018).
2.4.3. SHP in autophagy
SHP regulates autophagy. FXR acts early, but SHP acts relatively late after feeding to epigenetically sustain postprandial inhibition of autophagy via a FGF19-SHP-LSD1 axis (Byun et al., 2017). Both global and hepatocyte-specific double knockout of FXR and SHP have a beneficial impact on glucose and fatty acid metabolism in aged mice, as shown by lower hepatic triglyceride accumulation, improved glucose/insulin tolerance, and accelerated fatty acid use, which are associated with enhanced expression of fatty acid metabolism and autophagy-machinery genes (Kim et al., 2017).
2.4.4. SHP in hepatotoxicity and endoplasmic reticulum (ER) stress
SHP participates in the circadian regulation of cytochrome P450 (CYP) enzymes, thereby impacting xenobiotic metabolism and drug-induced hepatotoxicity (T. Zhang et al., 2018). Indeed, hepatocyte SHP deficiency protects mice from acetaminophen APAP-induced liver injury (Y. H. Kim et al., 2018).
SHP interacts with and regulates the protein stability of the spliced form of X-box-binding protein 1 (XBP1s) to govern ER homeostasis (Sun et al., 2019). It has been demonstrated that FXR/SHP signaling activates XBP1s expression, and hepatic XBP1s expression is reduced in FXR- and SHP-null mice (X. Liu et al., 2018).
3. LncRNAs and SHP
It is well-established that SHP is a downstream target gene of FXR that activates SHP transcription to restrain bile acid synthesis and maintain the homeostasis of bile acid metabolism (Kim et al., 2017). SHP expression is rhythmically controlled by NPAS2, CLOCK-BMAL1, and LRH-1 (Lee et al., 2015; Oiwa et al., 2007). The reciprocal regulation between NRs and ncRNAs has emerged as essential mechanisms influencing diverse biological processes (Mahpour and Mullen, 2021; Wu et al., 2021). SHP inhibits the expression of several miRNAs, including miR-433, miR-127, miR-34a, and miR-200c, conforming to its general function of performing transcriptional repression (Song et al., 2017b). Intriguingly, it is notable that miR-433 plays an inhibitory role in liver cancer cell migration (Mansini et al., 2018; Yang et al., 2013), opposing SHP’s tumor suppression function. On the other hand, SHP is targeted by miR-142–3p in the regulation of cholestasis (Pan et al., 2017). Despite these findings, little is known about the crosstalk between lncRNAs and SHP. Several studies shed light on this in recent years.
3.1. MEG3 and SHP
Maternally expressed gene 3 (MEG3) is an imprinted gene encoding a lncRNA expressed in many normal tissues, and functions as a tumor suppressor (Al-Rugeebah et al., 2019; Zhou et al., 2012). MEG3 is required for embryonic development, as Meg3 knockout mice die prematurely. MEG3 expression is frequently lost in human cancers, possibly due to gene deletion and promoter methylation. Re-expression of MEG3 inhibits proliferation, induces apoptosis, and suppresses anchorage-independent growth of human tumor cells. MEG3 expression is frequently down-regulated in human HCC due to the methylation of DLK1-MEG3 locus on human chromosome 14q32 which encodes a cluster of metastasis-suppressive miRNAs predominantly regulated by DNA methylation (Anwar et al., 2012; Oshima et al., 2019). MEG3 also inhibits the growth of human liver cancer stem cells by reducing the activity of telomerase (Jiang et al., 2020).
MEG3 expression is down-regulated in murine and human fibrotic livers, which might be ascribed to the hypermethylation of gene promoter. Indeed, lncRNA HOX antisense intergenic RNA (HOTAIR) promotes the accumulation of polycomb repressive complex 2 (PRC2) and H3K27 trimethylation at the MEG3 promoter in LX-2 cells (a human HSC cell line) (Bian et al., 2017). MEG3 is a target gene of miR-212 and inhibits hedgehog-mediated epithelial-mesenchymal transition (EMT) in liver fibrosis (Yu et al., 2018). MEG3 overexpression inhibits HSC proliferation by activating the p53/caspase-3 signaling pathway (He et al., 2014). These findings suggest that MEG3 plays an inhibitory function in liver fibrosis.
Knockdown of MEG3 expression causes senescence in hepatic endothelial cells in diet-induced obese mice, potentiating obesity-induced insulin resistance and impairing glucose homeostasis (Cheng et al., 2021). However, hepatic expression of MEG3 is increased (about 2-fold) in patients with NFALD or nonalcoholic steatohepatitis (NASH), and obese mice, likely due to a compensatory regulation and suggesting a protective role of MEG3 in metabolic disorders (Cheng et al., 2021). Interestingly, another study shows that hepatic MEG3 is downregulated in murine NAFLD models and can bind to and antagonize the function of miR-21 to increase the expression of low-density lipoprotein receptor-related protein 6 (LRP6), a gene target of miR-21; this study also demonstrates a protective role of MEG3 in the pathogenesis of NAFLD (Huang et al., 2019). Nevertheless, MEG3 expression is up-regulated in the livers of ethanol-fed mice and induced by ethanol in AML-12 cells (a hepatocyte cell line) (Wang et al., 2018). The knockdown of MEG3 expression inhibits ethanol-induced steatosis and apoptosis and impairs the expression of NOD-like receptor family CARD domain containing 5 (NLRC5), a critical regulator of immune responses, in AML-12 cells; moreover, MEG3 is proposed as an endogenous competing lncRNA for miR-let-7c-5p that targets NLRC5. The MEG3/miR-let-7c-5p/NLRC5 axis might contribute to ethanol-induced liver injury but needs further investigation.
Forced overexpression of MEG3 in mouse livers causes rapid SHP mRNA decay, resulting in increased Cyp7a1 and Cyp8b1 expression, the disruption of bile acid homeostasis, and cholestatic liver injury (Zhang et al., 2017). There are multiple predicted RNA-binding protein polypyrimidine tract-binding protein 1 (PTBP1)-binding sites within the coding sequence and 3’-UTR of SHP mRNA; MEG3 interacts with PTBP1 and facilitates PTPB1 binding to SHP mRNA, which promotes SHP mRNA decay (Zhang et al., 2017). Despite the intramolecular interaction, how PTPB1 links the RNA-degradation machinery to SHP mRNA is still elusive. In fact, there are reports showing that PTBP1 protects transcripts from nonsense-mediated mRNA decay (Fritz et al., 2020; Ge et al., 2016). On the other hand, MEG3 RNA is dramatically elevated in the livers of Shp knockout mice and that SHP inhibits MEG3 expression by repressing cAMP response element-binding protein (CREB)-mediated transactivation of Meg3 gene promoter (Zhang et al., 2017). Thus, MEG3 and SHP constitute a feedback loop of reciprocal inhibition to maintain bile acid homeostasis, suggesting that MEG3 could be a therapeutic target to manage bile acid homeostasis and improve cholestatic liver injury.
3.2. H19 and SHP
H19 imprinted maternally expressed transcript (H19) is one of the earliest described lncRNAs (Mahpour and Mullen, 2021). The H19 locus is located on chromosome 11p15.5 in humans and on chromosome 7 in mice. It plays a pivotal role in embryonic development and growth control (Gabory et al., 2010; Monnier et al., 2013). The H19 gene cluster contains the insulin-like growth factor 2 (IGF2) gene located 90 kb upstream of H19. There is an intergenic differentially methylated region (DMR) upstream of IGF2, an imprinting control region (ICR) between IGF2 and H19, and an enhancer downstream of H19 (Thorvaldsen et al., 1998). The methylation status and the alternative binding of the enhancer to DMR or ICR determine the expression of these two imprinting genes, IGF2 and H19. H19 is expressed from the maternal allele, and IGF2 is expressed from the paternal allele (Kurukuti et al., 2006; Pope et al., 2017). H19 is highly expressed in embryonic tissues and the placenta. Its expression is drastically attenuated after birth in most tissues except for the skeletal muscle, cardiac muscle, and cartilage (Zeira et al., 2015).
The role of H19 is elucidated by its importance in diverse liver pathophysiology, including NAFLD (C. Liu et al., 2018; J. Liu et al., 2019; H. Wang et al., 2020; N. Zhang et al., 2018), cholestasis (Li et al., 2020, 2018, 2017; R. Liu et al., 2019; Song et al., 2017a; Xiao et al., 2019; L. Zhang et al., 2019; Zhang et al., 2016), fibrosis (Z. M. Wang et al., 2020; Xiao et al., 2019; J. J. Yang et al., 2016; Yang et al., 2018; Zhu et al., 2019), acute liver failure (Jin et al., 2018), hepatitis B viral (HBV) infection (Li et al., 2019; Y. Liu et al., 2019), and HCC (Matouk et al., 2007; Wei et al., 2019; Zhou et al., 2019). The level of specific lncRNAs in circulation can be useful biomarkers for the diagnosis and prognosis of liver disease. Indeed, high plasma H19 is associated with poor disease-free survival in patients with HCC and after curative hepatectomy (Yang et al., 2015).
H19 expression is reactivated and remarkably induced in adult human livers with cholestatic fibrosis and cirrhosis (Zhang et al., 2016). Bile duct ligation (BDL)-induced cholestasis activates hepatic H19 expression, which enhances intrahepatic inflammation, HSC activation, ductular reaction, and cholestatic liver fibrosis in mice; (Song et al., 2017a). BDL-induced cholestasis also reduces SHP mRNA expression, which can be blunted by H19 overexpression (Song et al., 2017a). Despite this, it seems that SHP and H19 antagonize the expression of each other (Li et al., 2018; Zhang et al., 2016). In mouse livers, forced overexpression of the anti-apoptotic protein BCL2 induces SHP protein degradation, leading to the re-expression of H19 due to the loss of SHP’s transcriptional repression function (Zhang et al., 2016). On the other hand, via exosomal transportation, cholangiocyte-derived H19 suppresses SHP expression in hepatocytes at both transcriptional and post-transcriptional levels (Li et al., 2018). Besides, cholangiocyte-derived H19 promotes the activation and proliferation of HSCs, which results in cholestatic liver injury in BDL and Mdr2−/− mice modeling biliary fibrosis (R. Liu et al., 2019). The inhibitory function of SHP in HSC activation has been shown in vitro (Cipriani et al., 2017; Ma et al., 2020). It is possible that cholangiocytes- or other types of liver cells-derived H19 also regulate SHP’s function in HSCs. It is obvious that intercellular communications are required for H19 and SHP to cooperatively control bile acid homeostasis and regulate the severity of cholestatic liver injury. It is noteworthy that a recent study using the RNAscope assay, a novel in situ RNA analysis platform, shows that H19 RNA is localized in HNF4α+ periportal hepatocytes, SOX9+ ductal progenitor cells, and F4/80+ KCs but not in CK19+ cholangiocytes and desmin+ HSCs in cholestatic livers (Jiang et al., 2018). In contrast, using immuno-purification or laser-capture microdissection, H19 RNA is shown to be predominantly expressed in cholangiocytes, about 100-fold higher than hepatocytes, HSCs, and KCs (R. Liu et al., 2019).
When expressed in the same cell type, how SHP and H19 mechanistically inhibit the expression of each other at the molecular level is still elusive. The upstream signaling pathways that activate H19 expression are still not fully understood (Chiang, 2017). It is known that H19 expression is regulated by multiple TFs, such as forkhead box A1 (FOXA1), hypoxia-inducible factor 1 subunit alpha (HIF1α), Paxillin (PXN), E2F transcription factor 1 (E2F1), SRY-sex determining region Y- box 2 (SOX2) (Yang et al., 2020). Defining the functional connections between these TFs and H19/SHP would help address the above question in various liver diseases. Interestingly, similar to MEG3, H19 likewise interacts with PTBP1 to modulate hepatic lipogenesis and glucose metabolism (C. Liu et al., 2018). However, H19 intriguingly decreases PTPB1 expression in cholestasis (L. Zhang et al., 2019). It is unknown whether this interaction can also accelerate SHP mRNA decay the same as MEG3 (Zhang et al., 2017).
4. Conclusions and Perspectives
The above critical roles and molecular connections of SHP in liver physiology and pathophysiology are summarized (Figure 1). Recent studies have demonstrated the association of lncRNAs with SHP expression in liver disorders (Chiang, 2017). Only a few studies reveal SHP regulation of lncRNAs, and the regulatory mechanism is confined to SHP’s canonical function as a transcriptional repressor (Zhang et al., 2017, 2016). Because SHP lacks a DNA-binding domain, it is reasonable to postulate that SHP interacts with other TFs to regulate lncRNA expression. Indeed, SHP physically interacts with CREB to inhibit CREB-dependent hepatic gluconeogenesis (Lee et al., 2010), which underlies the molecular basis of SHP inhibition of Meg3. Defining the genome landscape of DNA association regions for SHP and other TFs and distinguishing their colocalized genomic loci will be informative to reveal more SHP-regulated lncRNAs.
Figure 1.
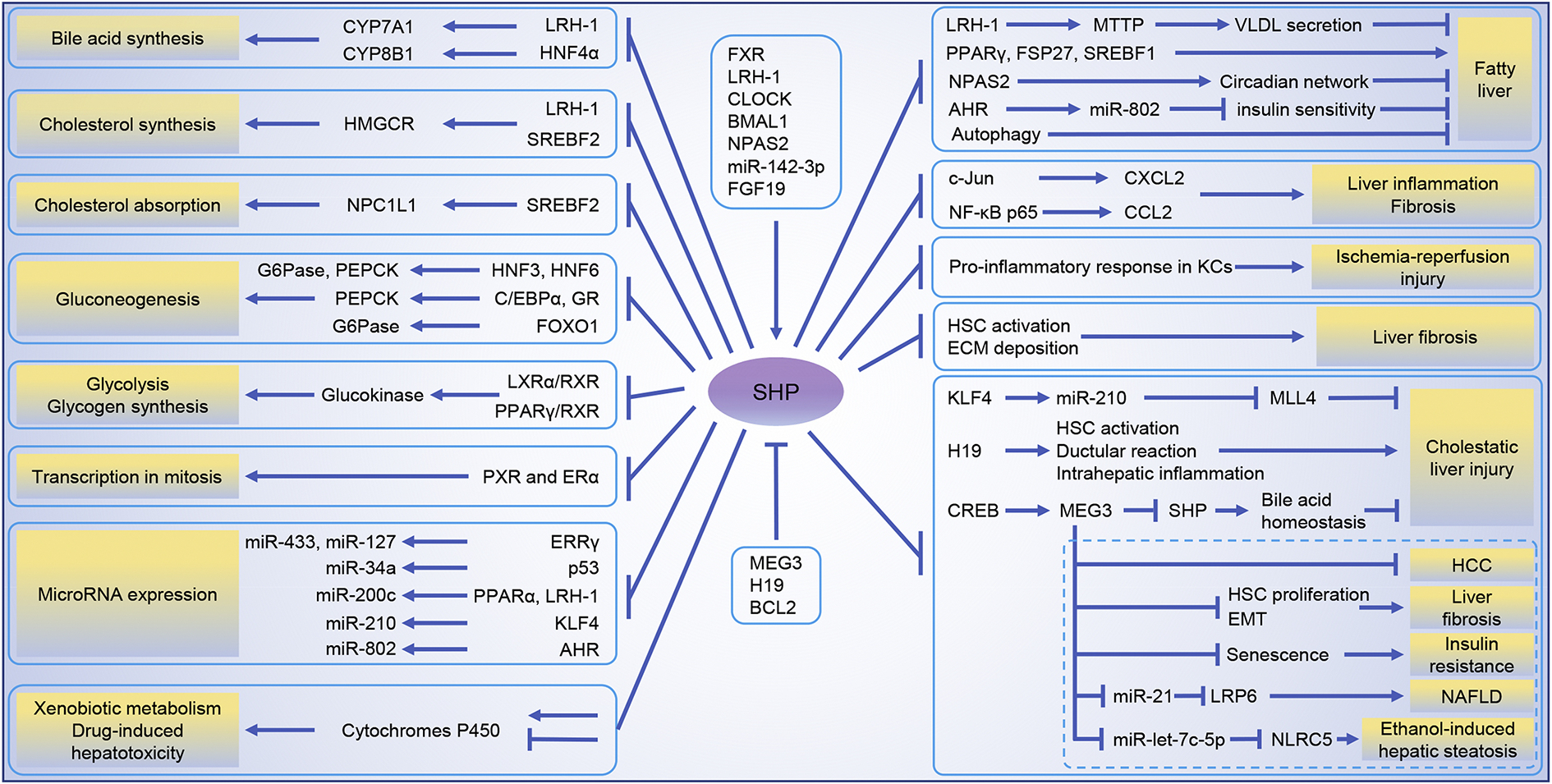
The critical roles and molecular connections of SHP in liver physiology and pathophysiology presented in this review.
SHP might indirectly regulate lncRNA expression through epigenetic mechanisms because it has been reported that SHP can impair estrogen-related receptor gamma (ERRγ)-mediated transcription of DNA (cytosine-5)-methyltransferase 1 (DNMT1) that silences gene expression through CpG island methylation, pointing to the potential role of SHP in gene transactivation (Zhang and Wang, 2011).
The pathophysiological significance of lncRNAs has been increasingly recognized (Pielok and Marycz, 2020). For instance, lncRNAs AK054921 and AK128652 are potential biomarkers to predict the progression of alcohol-associated liver disease (ALD) in individuals with excessive alcohol consumption; they are predictors of survival in patients with cirrhosis (Yang et al., 2017). The serum level of MEG3 is decreased in patients with chronic hepatitis B and negatively correlates with the severity of liver fibrosis (Chen et al., 2019). Multiple approaches have been proposed to regulate lncRNA expression to manage liver diseases, including RNA interference to target lncRNA, induction of lncRNA expression with agonistic or antagonistic compounds, manipulation of extracellular vesicles (EVs) (Sato et al., 2020). Studies to deorphanize SHP and further dissect the ncRNA-SHP network are warranted for developing lncRNA- or SHP-based novel diagnostics, therapeutics, and prevention strategies for liver disease.
Acknowledgements
This work is supported by NIH P50AA024333, R01AA027456, and U01AA026938 (to L.E.N.). J.W. is a recipient of the Pilot/Feasibility Award from the Cleveland Digestive Diseases Research Core Center (DDRCC) supported by NIH P30DK097948. We thank the Special Issue Guest Editor Dr. Warren B. Nothnick for handling the manuscript.
Footnotes
Declaration of competing interest
Authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akinrotimi O, Riessen R, VanDuyne P, Park JE, Lee YK, Wong LJ, Zavacki AM, Schoonjans K, Anakk S, 2017. Small heterodimer partner deletion prevents hepatic steatosis and when combined with farnesoid X receptor loss protects against type 2 diabetes in mice. Hepatology 66, 1854–1865. 10.1002/hep.29305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Rugeebah A, Alanazi M, Parine NR, 2019. MEG3: an Oncogenic Long Non-coding RNA in Different Cancers. Pathol Oncol Res 25, 859–874. 10.1007/s12253-019-00614-3 [DOI] [PubMed] [Google Scholar]
- Anwar SL, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, Kreipe H, Lehmann U, 2012. Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PLoS One 7, e49462. 10.1371/journal.pone.0049462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian EB, Wang YY, Yang Y, Wu BM, Xu T, Meng XM, Huang C, Zhang L, Lv XW, Xiong ZG, Li J, 2017. Hotair facilitates hepatic stellate cells activation and fibrogenesis in the liver. Biochim Biophys Acta Mol Basis Dis 1863, 674–686. 10.1016/j.bbadis.2016.12.009 [DOI] [PubMed] [Google Scholar]
- Bonasio R, Shiekhattar R, 2014. Regulation of transcription by long noncoding RNAs. Annu Rev Genet 48, 433–455. 10.1146/annurev-genet-120213-092323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgius LJ, Steffensen KR, Gustafsson JA, Treuter E, 2002. Glucocorticoid signaling is perturbed by the atypical orphan receptor and corepressor SHP. J Biol Chem 277, 49761–49766. 10.1074/jbc.M205641200 [DOI] [PubMed] [Google Scholar]
- Boulias K, Katrakili N, Bamberg K, Underhill P, Greenfield A, Talianidis I, 2005. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J 24, 2624–2633. 10.1038/sj.emboj.7600728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun S, Kim YC, Zhang Y, Kong B, Guo G, Sadoshima J, Ma J, Kemper B, Kemper JK, 2017. A postprandial FGF19-SHP-LSD1 regulatory axis mediates epigenetic repression of hepatic autophagy. EMBO J 36, 1755–1769. 10.15252/embj.201695500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL, 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927. 10.1101/gad.17446611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MJ, Wang XG, Sun ZX, Liu XC, 2019. Diagnostic value of LncRNA-MEG3 as a serum biomarker in patients with hepatitis B complicated with liver fibrosis. Eur Rev Med Pharmacol Sci 23, 4360–4367. 10.26355/eurrev_201905_17943 [DOI] [PubMed] [Google Scholar]
- Cheng X, Shihabudeen Haider Ali MS, Moran M, Viana MP, Schlichte SL, Zimmerman MC, Khalimonchuk O, Feinberg MW, Sun X, 2021. Long non-coding RNA Meg3 deficiency impairs glucose homeostasis and insulin signaling by inducing cellular senescence of hepatic endothelium in obesity. Redox Biol 40, 101863. 10.1016/j.redox.2021.101863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JYL, 2017. Linking long noncoding RNA to control bile acid signaling and cholestatic liver fibrosis. Hepatology 66, 1032–1035. 10.1002/hep.29289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriani S, Carino A, Masullo D, Zampella A, Distrutti E, Fiorucci S, 2017. Decoding the role of the nuclear receptor SHP in regulating hepatic stellate cells and liver fibrogenesis. Sci Rep 7, 41055. 10.1038/srep41055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Wang L, Moore DD, Osborne TF, 2006. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase promoter by nuclear receptors liver receptor homologue-1 and small heterodimer partner: a mechanism for differential regulation of cholesterol synthesis and uptake. J Biol Chem 281, 807–812. 10.1074/jbc.M511050200 [DOI] [PubMed] [Google Scholar]
- Esteller M, 2011. Non-coding RNAs in human disease. Nat Rev Genet 12, 861–874. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- Fernandes JCR, Acuna SM, Aoki JI, Floeter-Winter LM, Muxel SM, 2019. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA 5. 10.3390/ncrna5010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz SE, Ranganathan S, Wang CD, Hogg JR, 2020. The RNA-binding protein PTBP1 promotes ATPase-dependent dissociation of the RNA helicase UPF1 to protect transcripts from nonsense-mediated mRNA decay. J. Biol. Chem. 295, 11613–11625. 10.1074/jbc.RA120.013824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabory A, Jammes H, Dandolo L, 2010. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays 32, 473–480. 10.1002/bies.200900170 [DOI] [PubMed] [Google Scholar]
- Ge Z, Quek BL, Beemon KL, Hogg JR, 2016. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. Elife 5. 10.7554/eLife.11155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AQ, Nolasco S, Soares H, 2013. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci 14, 16010–16039. 10.3390/ijms140816010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA, 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 6, 517–526. 10.1016/s1097-2765(00)00051-4 [DOI] [PubMed] [Google Scholar]
- Hartman HB, Lai K, Evans MJ, 2009. Loss of small heterodimer partner expression in the liver protects against dyslipidemia. J Lipid Res 50, 193–203. 10.1194/jlr.M800323-JLR200 [DOI] [PubMed] [Google Scholar]
- Hassan HM, Isovic M, Kolendowski B, Bauer-Maison N, Onabote O, Cecchini M, Haig A, Maleki Vareki S, Underhill TM, Torchia J, 2020. Loss of Thymine DNA Glycosylase Causes Dysregulation of Bile Acid Homeostasis and Hepatocellular Carcinoma. Cell Rep 31, 107475. 10.1016/j.celrep.2020.03.039 [DOI] [PubMed] [Google Scholar]
- He N, Park K, Zhang Y, Huang J, Lu S, Wang L, 2008. Epigenetic inhibition of nuclear receptor small heterodimer partner is associated with and regulates hepatocellular carcinoma growth. Gastroenterology 134, 793–802. 10.1053/j.gastro.2008.01.006 [DOI] [PubMed] [Google Scholar]
- He Y, Wu YT, Huang C, Meng XM, Ma TT, Wu BM, Xu FY, Zhang L, Lv XW, Li J, 2014. Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis. Biochim Biophys Acta 1842, 2204–2215. 10.1016/j.bbadis.2014.08.015 [DOI] [PubMed] [Google Scholar]
- Huang J, Iqbal J, Saha PK, Liu J, Chan L, Hussain MM, Moore DD, Wang L, 2007. Molecular characterization of the role of orphan receptor small heterodimer partner in development of fatty liver. Hepatology 46, 147–157. 10.1002/hep.21632 [DOI] [PubMed] [Google Scholar]
- Huang J, Jia R, Brunner T, 2018. Local synthesis of immunosuppressive glucocorticoids in the intestinal epithelium regulates anti-viral immune responses. Cell Immunol 334, 1–10. 10.1016/j.cellimm.2018.08.009 [DOI] [PubMed] [Google Scholar]
- Huang P, Huang FZ, Liu HZ, Zhang TY, Yang MS, Sun CZ, 2019. LncRNA MEG3 functions as a ceRNA in regulating hepatic lipogenesis by competitively binding to miR-21 with LRP6. Metabolism 94, 1–8. 10.1016/j.metabol.2019.01.018 [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang L, Xie S, Chen Y, Song S, Lu Y, Lu D, 2020. Long noncoding RNA MEG3 blocks telomerase activity in human liver cancer stem cells epigenetically. Stem Cell Res Ther 11, 518. 10.1186/s13287-020-02036-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Huang Y, Cai S, Song Y, Boyer JL, Zhang K, Gao L, Zhao J, Huang W, Liang G, Liangpunsakul S, Wang L, 2018. H19 Is Expressed in Hybrid Hepatocyte Nuclear Factor 4alpha(+) Periportal Hepatocytes but Not Cytokeratin 19(+) Cholangiocytes in Cholestatic Livers. Hepatol Commun 2, 1356–1368. 10.1002/hep4.1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D, Lu T, Ni M, Wang H, Zhang J, Zhong C, Shen C, Hao J, Busuttil RW, Kupiec-Weglinski JW, Zhang J, Xu N, Zhai Y, 2020. Farnesoid X Receptor Activation Protects Liver From Ischemia/Reperfusion Injury by Up-Regulating Small Heterodimer Partner in Kupffer Cells. Hepatol Commun 4, 540–554. 10.1002/hep4.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wang J, Li H, Gao S, Shi R, Yang D, Wang X, Wang X, Zhu L, Wang X, Chen C, Ning K, Gao Z, Xu J, Fu Q, 2018. Extracellular Vesicles Secreted by Human Adipose-derived Stem Cells (hASCs) Improve Survival Rate of Rats with Acute Liver Failure by Releasing lncRNA H19. EBioMedicine 34, 231–242. 10.1016/j.ebiom.2018.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Chen J, Hu X, Sun H, Wu SY, Chiang CM, Kemper B, Chen LF, Kemper JK, 2020. BRD4 inhibition and FXR activation, individually beneficial in cholestasis, are antagonistic in combination. JCI Insight 6. 10.1172/jci.insight.141640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Yoon JE, Nikapitiya C, Kim TH, Uddin MB, Lee HC, Kim YH, Hwang JH, Chathuranga K, Chathuranga WAG, Choi HS, Kim CJ, Jung JU, Lee CH, Lee JS, 2019. Small Heterodimer Partner Controls the Virus-Mediated Antiviral Immune Response by Targeting CREB-Binding Protein in the Nucleus. Cell Rep 27, 2105–2118 e5. 10.1016/j.celrep.2019.04.071 [DOI] [PubMed] [Google Scholar]
- Kim JY, Kim HJ, Kim KT, Park YY, Seong HA, Park KC, Lee IK, Ha H, Shong M, Park SC, Choi HS, 2004. Orphan nuclear receptor small heterodimer partner represses hepatocyte nuclear factor 3/Foxa transactivation via inhibition of its DNA binding. Mol Endocrinol 18, 2880–2894. 10.1210/me.2004-0211 [DOI] [PubMed] [Google Scholar]
- Kim KH, Choi JM, Li F, Arizpe A, Wooton-Kee CR, Anakk S, Jung SY, Finegold MJ, Moore DD, 2018. Xenobiotic Nuclear Receptor Signaling Determines Molecular Pathogenesis of Progressive Familial Intrahepatic Cholestasis. Endocrinology 159, 2435–2446. 10.1210/en.2018-00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Choi S, Zhou Y, Kim EY, Lee JM, Saha PK, Anakk S, Moore DD, 2017. Hepatic FXR/SHP axis modulates systemic glucose and fatty acid homeostasis in aged mice. Hepatology 66, 498–509. 10.1002/hep.29199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Kim H, Park JM, Im SS, Bae JS, Kim MY, Yoon HG, Cha JY, Kim KS, Ahn YH, 2009. Interrelationship between liver X receptor alpha, sterol regulatory element-binding protein-1c, peroxisome proliferator-activated receptor gamma, and small heterodimer partner in the transcriptional regulation of glucokinase gene expression in liver. J Biol Chem 284, 15071–15083. 10.1074/jbc.M109.006742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Byun S, Seok S, Guo G, Xu HE, Kemper B, Kemper JK, 2019. Small Heterodimer Partner and Fibroblast Growth Factor 19 Inhibit Expression of NPC1L1 in Mouse Intestine and Cholesterol Absorption. Gastroenterology 156, 1052–1065. 10.1053/j.gastro.2018.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Byun S, Zhang Y, Seok S, Kemper B, Ma J, Kemper JK, 2015. Liver ChlP-seq analysis in FGF19-treated mice reveals SHP as a global transcriptional partner of SREBP-2. Genome Biol 16, 268. 10.1186/s13059-015-0835-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Jung H, Seok S, Zhang Y, Ma J, Li T, Kemper B, Kemper JK, 2020a. MicroRNA-210 Promotes Bile Acid-Induced Cholestatic Liver Injury by Targeting Mixed-Lineage Leukemia-4 Methyltransferase in Mice. Hepatology 71,2118–2134. 10.1002/hep.30966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Seok S, Byun S, Kong B, Zhang Y, Guo G, Xie W, Ma J, Kemper B, Kemper JK, 2018. AhR and SHP regulate phosphatidylcholine and S-adenosylmethionine levels in the one-carbon cycle. Nat Commun 9, 540. 10.1038/s41467-018-03060-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Seok S, Zhang Y, Ma J, Kong B, Guo G, Kemper B, Kemper JK, 2020b. Intestinal FGF15/19 physiologically repress hepatic lipogenesis in the late fed-state by activating SHP and DNMT3A. Nat Commun 11, 5969. 10.1038/s41467-020-19803-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Noh JR, Hwang JH, Kim KS, Choi DH, Kim JH, Moon SJ, Choi JH, Herault Y, Lee TG, Choi HS, Lee CH, 2018. Hepatocyte SHP deficiency protects mice from acetaminophen-evoked liver injury in a JNK-signaling regulation and GADD45beta-dependent manner. Arch Toxicol 92, 2563–2572. 10.1007/s00204-018-2247-3 [DOI] [PubMed] [Google Scholar]
- Kornfeld JW, Baitzel C, Konner AC, Nicholls HT, Vogt MC, Herrmanns K, Scheja L, Haumaitre C, Wolf AM, Knippschild U, Seibler J, Cereghini S, Heeren J, Stoffel M, Bruning JC, 2013. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature 494, 111–115. 10.1038/nature11793 [DOI] [PubMed] [Google Scholar]
- Kumar S, Vijayan R, Dash AK, Gourinath S, Tyagi RK, 2021. Nuclear receptor SHP dampens transcription function and abrogates mitotic chromatin association of PXR and ERalpha via intermolecular interactions. Biochim Biophys Acta Gene Regul Mech 1864, 194683. 10.1016/j.bbagrm.2020.194683 [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R, 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A 103, 10684–10689. 10.1073/pnas.0600326103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Seo WY, Song KH, Chanda D, Kim YD, Kim DK, Lee MW, Ryu D, Kim YH, Noh JR, Lee CH, Chiang JY, Koo SH, Choi HS, 2010. AMPK-dependent repression of hepatic gluconeogenesis via disruption of CREB.CRTC2 complex by orphan nuclear receptor small heterodimer partner. J Biol Chem 285, 32182–32191. 10.1074/jbc.M110.134890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Zhang Y, Tsuchiya H, Smalling R, Jetten AM, Wang L, 2015. Small heterodimer partner/neuronal PAS domain protein 2 axis regulates the oscillation of liver lipid metabolism. Hepatology 61, 497–505. 10.1002/hep.27437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Kim DK, Kim YD, Park KC, Shong M, Seong HA, Ha HJ, Choi HS, 2008. Orphan nuclear receptor SHP interacts with and represses hepatocyte nuclear factor-6 (HNF-6) transactivation. Biochem J 413, 559–569. 10.1042/BJ20071637 [DOI] [PubMed] [Google Scholar]
- Li L, Han T, Liu K, Lei CG, Wang ZC, Shi GJ, 2019. LncRNA H19 promotes the development of hepatitis B related hepatocellular carcinoma through regulating microRNA-22 via EMT pathway. Eur Rev Med Pharmacol Sci 23, 5392–5401. 10.26355/eurrev_201906_18208 [DOI] [PubMed] [Google Scholar]
- Li X, Liu R, Huang Z, Gurley EC, Wang X, Wang J, He H, Yang H, Lai G, Zhang L, Bajaj JS, White M, Pandak WM, Hylemon PB, Zhou H, 2018. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology 68, 599–615. 10.1002/hep.29838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu R, Wang Y, Zhu W, Zhao D, Wang X, Yang H, Gurley EC, Chen W, Hylemon PB, Zhou H, 2020. Cholangiocyte-Derived Exosomal lncRNA H19 Promotes Macrophage Activation and Hepatic Inflammation under Cholestatic Conditions. Cells 9. 10.3390/cells9010190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu R, Yang J, Sun L, Zhang L, Jiang Z, Puri P, Gurley EC, Lai G, Tang Y, Huang Z, Pandak WM, Hylemon PB, Zhou H, 2017. The role of long noncoding RNA H19 in gender disparity of cholestatic liver injury in multidrug resistance 2 gene knockout mice. Hepatology 66, 869–884. 10.1002/hep.29145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JY, Liu C, Hu KQ, Smith DE, Wang XD, 2018. Ablation of carotenoid cleavage enzymes (BCO1 and BCO2) induced hepatic steatosis by altering the farnesoid X receptor/miR-34a/sirtuin 1 pathway. Arch Biochem Biophys 654, 1–9. 10.1016/j.abb.2018.07.007 [DOI] [PubMed] [Google Scholar]
- Liu C, Yang Z, Wu J, Zhang L, Lee S, Shin DJ, Tran M, Wang L, 2018. Long noncoding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology 67, 1768–1783. 10.1002/hep.29654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Tang T, Wang GD, Liu B, 2019. LncRNA-H19 promotes hepatic lipogenesis by directly regulating miR-130a/PPARgamma axis in non-alcoholic fatty liver disease. Biosci Rep 39. 10.1042/BSR20181722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Li X, Zhu W, Wang Y, Zhao D, Wang X, Gurley EC, Liang G, Chen W, Lai G, Pandak WM, Robert Lippman H, Bajaj JS, Hylemon PB, Zhou H, 2019. Cholangiocyte-Derived Exosomal Long Noncoding RNA H19 Promotes Hepatic Stellate Cell Activation and Cholestatic Liver Fibrosis. Hepatology 70, 1317–1335. 10.1002/hep.30662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Guo GL, Kong B, Hilburn DB, Hubchak SC, Park S, LeCuyer B, Hsieh A, Wang L, Fang D, Green RM, 2018. Farnesoid X receptor signaling activates the hepatic X-box binding protein 1 pathway in vitro and in mice. Hepatology 68, 304–316. 10.1002/hep.29815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xu L, Lu B, Zhao M, Li L, Sun W, Qiu Z, Zhang B, 2019. LncRNA H19/microRNA-675/PPARalpha axis regulates liver cell injury and energy metabolism remodelling induced by hepatitis B X protein via Akt/mToR signalling. Mol Immunol 116, 18–28. 10.1016/j.molimm.2019.09.006 [DOI] [PubMed] [Google Scholar]
- Lopez-Urrutia E, Bustamante Montes LP, Ladron de Guevara Cervantes D, Perez-Plasencia C, Campos-Parra AD, 2019. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front Oncol 9, 669. 10.3389/fonc.2019.00669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ, 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell 6, 507–515. 10.1016/s1097-2765(00)00050-2 [DOI] [PubMed] [Google Scholar]
- Ma W, Cheng LS, Jiang W, Wu SD, 2020. The small heterodimer partner inhibits activation of hepatic stellate cells via autophagy. Adv Clin Exp Med 29, 683–693. 10.17219/acem/122175 [DOI] [PubMed] [Google Scholar]
- Magee N, Zou A, Ghosh P, Ahamed F, Delker D, Zhang Y, 2020. Disruption of hepatic small heterodimer partner induces dissociation of steatosis and inflammation in experimental nonalcoholic steatohepatitis. J Biol Chem 295, 994–1008. 10.1074/jbc.RA119.010233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahpour A, Mullen AC, 2021. Our emerging understanding of the roles of long non-coding RNAs in normal liver function, disease, and malignancy. JHEP Rep 3, 100177. 10.1016/j.jhepr.2020.100177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansini AP, Lorenzo Pisarello MJ, Thelen KM, Cruz-Reyes M, Peixoto E, Jin S, Howard BN, Trussoni CE, Gajdos GB, LaRusso NF, Perugorria MJ, Banales JM, Gradilone SA, 2018. MicroRNA (miR)-433 and miR-22 dysregulations induce histone-deacetylase-6 overexpression and ciliary loss in cholangiocarcinoma. Hepatology 68, 561–573. 10.1002/hep.29832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E, 2007. The H19 non-coding RNA is essential for human tumor growth. PLoS One 2, e845. 10.1371/journal.pone.0000845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L, 2013. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci U S A 110, 20693–20698. 10.1073/pnas.1310201110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh JR, Kim YH, Kim DK, Hwang JH, Kim KS, Choi DH, Lee SJ, Lee HG, Lee TG, Weng HL, Dooley S, Choi HS, Lee CH, 2018. Small heterodimer partner negatively regulates C-X-C motif chemokine ligand 2 in hepatocytes during liver inflammation. Sci Rep 8, 15222. 10.1038/s41598-018-33660-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oiwa A, Kakizawa T, Miyamoto T, Yamashita K, Jiang W, Takeda T, Suzuki S, Hashizume K, 2007. Synergistic regulation of the mouse orphan nuclear receptor SHP gene promoter by CLoCk-BMAL1 and LRH-1. Biochem Biophys Res Commun 353, 895–901. 10.1016/j.bbrc.2006.12.131 [DOI] [PubMed] [Google Scholar]
- Oshima G, Poli EC, Bolt MJ, Chlenski A, Forde M, Jutzy JMS, Biyani N, Posner MC, Pitroda SP, Weichselbaum RR, Khodarev NN, 2019. DNA Methylation Controls Metastasis-Suppressive 14q32-Encoded miRNAs. Cancer Res 79, 650–662. 10.1158/0008-5472.CAN-18-0692 [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES, 2015. Non-coding RNA: what is functional and what is junk? Front Genet 6, 2. 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Kent R, Won KJ, Jeong H, 2017. Cholic Acid Feeding Leads to Increased CYP2D6 Expression in CYP2D6-Humanized Mice. Drug Metab Dispos 45, 346–352. 10.1124/dmd.116.074013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MJ, Kong HJ, Kim HY, Kim HH, Kim JH, Cheong JH, 2007. Transcriptional repression of the gluconeogenic gene PEPCK by the orphan nuclear receptor SHP through inhibitory interaction with C/EBPalpha. Biochem J 402, 567–574. 10.1042/BJ20061549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielok A, Marycz K, 2020. Non-Coding RNAs as Potential Novel Biomarkers for Early Diagnosis of Hepatic Insulin Resistance. Int J Mol Sci 21. 10.3390/ijms21114182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope C, Mishra S, Russell J, Zhou Q, Zhong XB, 2017. Targeting H19, an Imprinted Long Non-Coding RNA, in Hepatic Functions and Liver Diseases. Diseases 5. 10.3390/diseases5010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Glaser S, Francis H, Alpini G, 2020. Concise Review: Functional Roles and Therapeutic Potentials of Long Non-coding RNAs in Cholangiopathies. Front Med 7, 48. 10.3389/fmed.2020.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok S, Sun H, Kim YC, Kemper B, Kemper JK, 2020. Defective FXR-SHP Regulation in Obesity Aberrantly Increases miR-802 Expression, Promoting Insulin Resistance and Fatty Liver. Diabetes 10.2337/db20-0856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Liu C, Liu X, Trottier J, Beaudoin M, Zhang L, Pope C, Peng G, Barbier O, Zhong X, Li L, Wang L, 2017a. H19 promotes cholestatic liver fibrosis by preventing ZEB1-mediated inhibition of epithelial cell adhesion molecule. Hepatology 66, 1183–1196. 10.1002/hep.29209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Lu S, Zhao J, Wang L, 2017b. Nuclear Receptor SHP: A Critical Regulator of miRNA and lncRNA Expression and Function. Nucl Recept. Res 4. 10.11131/2017/101312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statello L, Guo CJ, Chen LL, Huarte M, 2021. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22, 96–118. 10.1038/s41580-020-00315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Kelekar S, Kliewer SA, Mangelsdorf DJ, 2019. The orphan nuclear receptor SHP regulates ER stress response by inhibiting XBP1s degradation. Genes Dev 33, 1083–1094. 10.1101/gad.326868.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh S, Durakoglugil D, Zhou X, Zhu B, Comerford SA, Xing C, Xie XJ, York B, O’Donnell KA, 2017. SRC-2-mediated coactivation of anti-tumorigenic target genes suppresses MYC-induced liver cancer. PLoS Genet 13, e1006650. 10.1371/journal.pgen.1006650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS, 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 12, 3693–3702. 10.1101/gad.12.23.3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cao Y, Shu L, Zhu Y, Peng Q, Ran L, Wu J, Luo Y, Zuo G, Luo J, Zhou L, Shi Q, Weng Y, Huang A, He TC, Fan J, 2020. Long non-coding RNA (lncRNA) H19 induces hepatic steatosis through activating MLXIPL and mTORC1 networks in hepatocytes. J Cell Mol Med 24, 1399–1412. 10.1111/jcmm.14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Han Y, Kim CS, Lee YK, Moore DD, 2003. Resistance of SHP-null mice to bile acid-induced liver damage. J Biol Chem 278, 44475–44481. 10.1074/jbc.M305258200 [DOI] [PubMed] [Google Scholar]
- Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, Moore DD, 2002. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell 2, 721–731. 10.1016/s1534-5807(02)00187-9 [DOI] [PubMed] [Google Scholar]
- Wang L, Liu J, Saha P, Huang J, Chan L, Spiegelman B, Moore DD, 2005. The orphan nuclear receptor SHP regulates PGC-1alpha expression and energy production in brown adipocytes. Cell Metab 2, 227–238. 10.1016/j.cmet.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Wang Q, Li M, Shen Z, Bu F, Yu H, Pan X, Yang Y, Meng X, Huang C, Li J, 2018. The Long Non-coding RNA MEG3/miR-let-7c-5p Axis Regulates Ethanol-Induced Hepatic Steatosis and Apoptosis by Targeting NLRC5. Front Pharmacol 9, 302. 10.3389/fphar.2018.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZM, Xia SW, Zhang T, Wang ZY, Yang X, Kai J, Cheng XD, Shao JJ, Tan SZ, Chen AP, Wang SJ, Zhang F, Zhang ZL, Zheng SZ, 2020. LncRNA-H19 induces hepatic stellate cell activation via upregulating alcohol dehydrogenase III-mediated retinoic acid signals. Int Immunopharmacol 84, 106470. 10.1016/j.intimp.2020.106470 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, Auwerx J, 2004. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest 113, 1408–1418. 10.1172/JCI21025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LQ, Li L, Lu C, Liu J, Chen Y, Wu H, 2019. Involvement of H19/miR-326 axis in hepatocellular carcinoma development through modulating TWIST1. J Cell Physiol 234, 5153–5162. 10.1002/jcp.27319 [DOI] [PubMed] [Google Scholar]
- Wu J, Nagy LE, Liangpunsakul S, Wang L, 2021. Non-coding RNA Crosstalk with Nuclear Receptors in Liver Disease. Biochim Biophys Acta Mol Basis Dis 166083. 10.1016/j.bbadis.2021.166083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Liu R, Li X, Gurley EC, Hylemon PB, Lu Y, Zhou H, Cai W, 2019. Long Noncoding RNA H19 Contributes to Cholangiocyte Proliferation and Cholestatic Liver Fibrosis in Biliary Atresia. Hepatology 70, 1658–1673. 10.1002/hep.30698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Daitoku H, Shimamoto Y, Matsuzaki H, Hirota K, Ishida J, Fukamizu A, 2004. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem 279, 23158–23165. 10.1074/jbc.M314322200 [DOI] [PubMed] [Google Scholar]
- Yang JJ, Liu LP, Tao H, Hu W, Shi P, Deng ZY, Li J, 2016. MeCP2 silencing of LncRNA H19 controls hepatic stellate cell proliferation by targeting IGF1R. Toxicology 359–360, 39–46. 10.1016/j.tox.2016.06.016 [DOI] [PubMed] [Google Scholar]
- Yang JJ, She Q, Yang Y, Tao H, Li J, 2018. DNMT1 controls LncRNA H19/ERK signal pathway in hepatic stellate cell activation and fibrosis. Toxicol Lett 295, 325–334. 10.1016/j.toxlet.2018.07.013 [DOI] [PubMed] [Google Scholar]
- Yang Z, Koehler AN, Wang L, 2016. A Novel Small Molecule Activator of Nuclear Receptor SHP Inhibits HCC Cell Migration via Suppressing Ccl2. Mol Cancer Ther 15, 2294–2301. 10.1158/1535-7163.MCT-16-0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lu Y, Xu Q, Tang B, Park CK, Chen X, 2015. HULC and H19 Played Different Roles in Overall and Disease-Free Survival from Hepatocellular Carcinoma after Curative Hepatectomy: A Preliminary Analysis from Gene Expression Omnibus. Dis Markers 2015, 191029. 10.1155/2015/191029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ross RA, Zhao S, Tu W, Liangpunsakul S, Wang L, 2017. LncRNA AK054921 and AK128652 are potential serum biomarkers and predictors of patient survival with alcoholic cirrhosis. Hepatol Commun 1, 513–523. 10.1002/hep4.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L, 2013. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem 288, 28893–28899. 10.1074/jbc.M113.502682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhang T, Han S, Kusumanchi P, Huda N, Jiang Y, Liangpunsakul S, 2020. Long noncoding RNA H19 - a new player in the pathogenesis of liver diseases. Transl Res. 10.1016/j.trsl.2020.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Geng W, Dong P, Huang Z, Zheng J, 2018. LncRNA-MEG3 inhibits activation of hepatic stellate cells through SMO protein and miR-212. Cell Death Dis 9, 1014. 10.1038/s41419-018-1068-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeira E, Abramovitch R, Meir K, Even Ram S, Gil Y, Bulvik B, Bromberg Z, Levkovitch O, Nahmansson N, Adar R, Reubinoff B, Galun E, Gropp M, 2015. The knockdown of H19lncRNA reveals its regulatory role in pluripotency and tumorigenesis of human embryonic carcinoma cells. Oncotarget 6, 34691–34703. 10.18632/oncotarget.5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yang Z, Huang W, Wu J, 2019. H19 potentiates let-7 family expression through reducing PTBP1 binding to their precursors in cholestasis. Cell Death Dis 10, 168. 10.1038/s41419-019-1423-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yang Z, Trottier J, Barbier O, Wang L, 2017. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology 65, 604–615. 10.1002/hep.28882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Chiang JY, 2001. Transcriptional regulation of the human sterol alpha-hydroxylase gene (CYP8B1): roles of heaptocyte nuclear factor 4alpha in mediating bile acid repression. J Biol Chem 276, 41690–41699. 10.1074/jbc.M105117200 [DOI] [PubMed] [Google Scholar]
- Zhang N, Geng T, Wang Z, Zhang R, Cao T, Camporez JP, Cai SY, Liu Y, Dandolo L, Shulman GI, Carmichael GG, Taylor HS, Huang Y, 2018. Elevated hepatic expression of H19 long noncoding RNA contributes to diabetic hyperglycemia. JCI Insight 3. 10.1172/jci.insight.120304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wu W, Chen Q, Chen M, 2019. Non-Coding RNAs and their Integrated Networks. J Integr Bioinform 16. 10.1515/jib-2019-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Yu F, Guo L, Chen M, Yuan X, Wu B, 2018. Small Heterodimer Partner Regulates Circadian Cytochromes p450 and Drug-Induced Hepatotoxicity. Theranostics 8, 5246–5258. 10.7150/thno.28676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, Shen F, 2019. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int J Mol Sci 20. 10.3390/ijms20225573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hagedorn CH, Wang L, 2011. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta 1812, 893–908. 10.1016/j.bbadis.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu C, Barbier O, Smalling R, Tsuchiya H, Lee S, Delker D, Zou A, Hagedorn CH, Wang L, 2016. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Sci Rep 6, 20559. 10.1038/srep20559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Soto J, Park K, Viswanath G, Kuwada S, Abel ED, Wang L, 2010. Nuclear receptor SHP, a death receptor that targets mitochondria, induces apoptosis and inhibits tumor growth. Mol Cell Biol 30, 1341–1356. 10.1128/MCB.01076-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L, 2011. Nuclear receptor SHP inhibition of Dnmt1 expression via ERRgamma. FEBS Lett 585, 1269–1275. 10.1016/j.febslet.2011.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu P, Park K, Choi Y, Moore DD, Wang L, 2008. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology 48, 289–298. 10.1002/hep.22342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wu J, Liangpunsakul S, Wang L, 2017. Long Non-coding RNA in Liver Metabolism and Disease: Current Status. Liver Res 1, 163–167. 10.1016/j.livres.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wang H, Ni M, Yue S, Xia Y, Busuttil RW, Kupiec-Weglinski JW, Lu L, Wang X, Zhai Y, 2018. Glycogen synthase kinase 3beta promotes liver innate immune activation by restraining AMP-activated protein kinase activation. J Hepatol 69, 99–109. 10.1016/j.jhep.2018.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Fan RG, Qin CL, Jia J, Wu XD, Zha WZ, 2019. LncRNA-H19 activates CDC42/PAK1 pathway to promote cell proliferation, migration and invasion by targeting miR-15b in hepatocellular carcinoma. Genomics 111, 1862–1872. 10.1016/j.ygeno.2018.12.009 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Klibanski A, 2012. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol 48, R45–53. 10.1530/JME-12-0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Luo Z, Pan Y, Zheng W, Li W, Zhang Z, Xiong P, Xu D, Du M, Wang B, Yu J, Zhang J, Liu J, 2019. H19/miR-148a/USP4 axis facilitates liver fibrosis by enhancing TGF-beta signaling in both hepatic stellate cells and hepatocytes. J Cell Physiol 234, 9698–9710. 10.1002/jcp.27656 [DOI] [PubMed] [Google Scholar]
- Zou A, Magee N, Deng F, Lehn S, Zhong C, Zhang Y, 2018. Hepatocyte nuclear receptor SHP suppresses inflammation and fibrosis in a mouse model of nonalcoholic steatohepatitis. J Biol Chem 293, 8656–8671. 10.1074/jbc.RA117.001653 [DOI] [PMC free article] [PubMed] [Google Scholar]


