Abstract
Tea plants (Camellia sinensis) are commercially cultivated in >60 countries, and their fresh leaves are processed into tea, which is the most widely consumed beverage in the world. Although several chromosome-level tea plant genomes have been published, they collapsed the two haplotypes and ignored a large number of allelic variations that may underlie important biological functions in this species. Here, we present a phased chromosome-scale assembly for an elite oolong tea cultivar, “Huangdan”, that is well known for its high levels of aroma. Based on the two sets of haplotype genome data, we identified numerous genetic variations and a substantial proportion of allelic imbalance related to important traits, including aroma- and stress-related alleles. Comparative genomics revealed extensive structural variations as well as expansion of some gene families, such as terpene synthases (TPSs), that likely contribute to the high-aroma characteristics of the backbone parent, underlying the molecular basis for the biosynthesis of aroma-related chemicals in oolong tea. Our results uncovered the genetic basis of special features of this oolong tea cultivar, providing fundamental genomic resources to study evolution and domestication for the economically important tea crop.
Subject terms: Genomics, Plant genetics
Introduction
The development of high-throughput sequencing has vigorously promoted research progress in plant genomics and genetics, especially single-molecule long-read sequencing and high-throughput chromatin conformation capture (Hi-C) technology, which have allowed complete plant genome sequencing and assembly at a low cost1. Diploid or polyploid genomes consist of two or more homologous chromosome sets. Notably, most plant reference genome assemblies are collapsed homologous copies of each chromosome. Although such assembled genomes are convenient for comparison and inference, they ignore allelic variants that may have potential biological functions and affect the quality of heterozygous plant genomes2. Unzipping haplotypes into two or more sequence sets has become a new trend in genome assembly that is conducive to research on plant heterosis and evolution and provides a solid foundation for accurate and reliable genome editing. ALLHiC is a new algorithm for constructing allele-aware, chromosome-scale assemblies based on Hi-C data3, and it has been successfully applied to the genome assembly of several plant species, including an autotetraploid4 and an autooctoploid3 sugarcane genome, an autotetraploid cultivated alfalfa genome5, an allotetraploid rapeseed genome6, and two Ficus genomes7. ALLHiC is sensitive to the quality of input assembly and becomes powerless when the proportion of chimeric contigs and collapsed regions increases. However, high-precision sequencing technology and advanced assembly algorithms will help ALLHiC cross this barrier. Therefore, ALLHiC is a powerful tool for genome research on heterozygous diploid and polyploid plants.
The tea plant Camellia sinensis L. O. Kuntze is a perennial evergreen woody plant that is commercially cultivated in >60 countries and accounts for >5 million tons of leaves picked each year8. At present, >2 billion cups of tea are consumed worldwide every day. These teas are mainly manufactured by processing fresh tea leaves and are rich in characteristic secondary compounds, including theanine, caffeine, catechins, and volatiles, which are beneficial to human health9. The United Nations General Assembly (UNGA) designated May 21 as “International Tea Day” to praise the high value of tea to the global society, economy, and culture, especially its significant role in rural poverty reduction and development in developing countries. However, compared with the tremendous contribution of tea to the global economic industry, fundamental biological research on tea plants and breeding efficiency are still lagging10.
Tea plants are an ideal model for investigating the molecular regulation mechanism of secondary metabolites11. The rich collection of secondary metabolites endow tea with a distinctive taste, aroma and color and are involved in the responses to various environmental stresses12,13; thus, these metabolites are attracting increasing research attention. Great advances in tea genomics have effectively facilitated researchers’ understanding of tea quality and adaptability. The genomic sequences of two major tea plant varieties, namely, Camellia sinensis vars. sinensis (CSS)9,14–17 and assamica (CSA)18 have been resolved, especially those of three CSS cultivars (cultivars Shuchazao9,15, Biyun16, and Longjin 4317) whose high-precision chromosome-level genomic sequences have been obtained. In addition, the high-quality genome of an ancient tea tree has been published19. However, these genomic assemblies have collapsed haplotypes and ignore the large number of allelic variations that exist in the highly heterozygous tea genome.
In general, tea plants exhibit self-incompatibility20, which leads to high levels of heterozygosity and allelic variation in their genomes. The allelic variations between two haplotypes within a single genome may play a crucial role in phenotypic variation, heterosis, and evolution2,3. Several studies support the key point that hybrid rice exhibits heterosis owing to alleles with high transcriptional activity and dominant expression21,22. In this study, we generated a chromosome-scale reference genome of CSS Huangdan (HD, 2n = 2x = 30) by PacBio HiFi (high-fidelity) sequencing and Hi-C technology. A phased genome with two fully represented haplotypes was also generated to facilitate the mining of allelic variation and allele-specific expression (ASE). HD is an elite tea cultivar originating from Anxi County, Fujian Province, China. Commercially, HD is suitable for processing into oolong, green, and black teas and was approved by the Chinese Crop Variety Approval Committee in 1985 (No. GS13008-1985). HD is not only a standard reference species for breeding oolong tea cultivars but also a high-aroma backbone parent that has been used to breed a series of excellent hybrid offspring23,24, including CSS Jinguanyin (JGY, No. GS2002017), CSS Huangguanyin (HGY, No. GS2002015), and CSS Jinmudan (JMD, No. GS2010024). Our research will enhance the understanding of genetic variation and heterosis of tea plants and lay the foundation for precise gene editing and molecular breeding in the future.
Results
Genome assembly and annotation
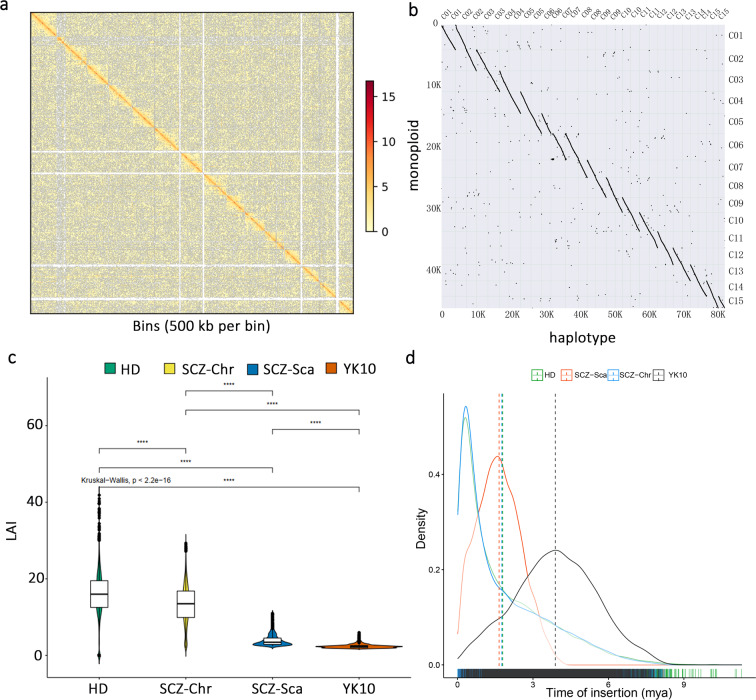
A total of 54.90 Gb (~18.67-fold) of high-fidelity (HiFi) PacBio reads were sequenced on the PacBio Sequel II platform. In addition, chromosomal-level genome assembly was achieved using 327.80 Gb (~111.50-fold) of Illumina short reads generated from two high-throughput conformation capture (Hi-C) libraries (Supplementary Table 1). To overcome the assembly problem caused by the high level of heterozygosity (2.79–3.40%) of the tea plant genome, we generated nonredundant genome sequences using Hifiasm25 and Purge_Dups26, yielding a contig-level assembly with a total size of 2.94 Gb. The assembled genome size was consistent with the genome size estimated using flow cytometry (Supplementary Table 2). Compared with early published tea plant genomes, the contig N50 of the HD genome was 2.61 Mb, which is ~4.3-fold, ~38.1-fold, and ~127.7-fold that of chromosome-level CSS-Shuchazao (SCZ-Chr)15, scaffold-level CSS-Shuchazao (SCZ-Sca)14, and CSA-Yunkang 10 (YK 10)18, respectively (Table 1). BUSCO27 analysis showed that the HD contig set contained 95.0% complete core orthologous genes in plants (Supplementary Table 3). Subsequently, we anchored the nonredundant contig set onto 15 pseudochromosomes based on Hi-C contacts, representing a high-quality chromosome-scale monoploid genome assembly (Fig. 1). The length of the pseudochromosomes ranges from 140 Mb (Chr11) to 247 Mb (Chr1) (Supplementary Table 4). The interaction signals were enriched in chromosomes, and the intensity of interaction along the diagonal was relatively smooth, showing well-organized contig orderings (Fig. 2a). Assembly assessment through long-terminal repeat (LTR) annotation showed that the LTR Assembly Index (LAI) score28 of the HD monoploid assembly was 16.6 (Table 1 and Fig. 2c), indicating that more LTRs were recovered in our assembly than in other published tea genomes (Supplementary Table 5).
Table 1.
Comparison of contig assemblies among tea plant genomes
| HD | SCZ-Chr | SCZ-Sca | YK 10 | |
|---|---|---|---|---|
| Contig assembly size (Gb) | 2.94 | 2.94 | 2.89 | 2.57 |
| Max length (Mb) | 20.56 | 2.89 | 0.54 | 0.26 |
| Contig N50 (kb) | 2610.56 | 600.46 | 67.07 | 19.96 |
| Complete BUSCO ratio (%) | 95.0% | 90.6% | 93.1% | 90.2% |
| Raw LAI | 13.75 | 12.35 | 1.93 | 2.32 |
| LAI | 16.16 | 14.19 | 3.89 | 5.17 |
| GC content (%) | 38.82 | 38.25 | 37.84 | 42.31 |
| No. of genes | 43,779 | 50,525 | 33,932 | 36,951 |
| Average gene length (bp) | 5452 | 4906 | 6821 | 3290 |
| Average CDS length (bp) | 1092 | 1086 | 1345 | 990 |
| Average exon length (bp) | 342 | 245 | 238 | 237 |
| Average exon number per gene | 4.4 | 5.1 | 5.7 | 4.8 |
| Exon GC content (%) | 43.26 | 43.83 | 44.50 | 44.55 |
| Average intron length (bp) | 1223 | 973 | 1298 | 640 |
| Intron GC content (%) | 34.57 | 35.38 | 33.94 | 33.50 |
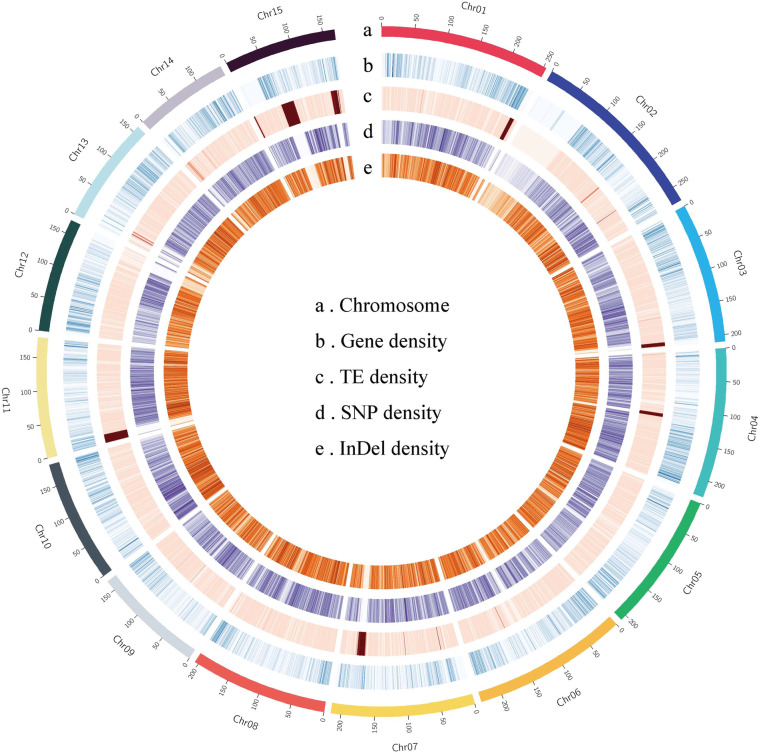
Fig. 1. Genomic features of the HD genome along 15 pseudochromosomes.
The tracks from outermost to innermost indicate a pseudochromosomes of the monoploid genome, b gene density, c transposable element (TE) density, d single-nucleotide polymorphism (SNP) density, and e insertion-deletion (InDel) density
Fig. 2. Assembly and LTR features of HD genome.
a Chromatin interaction at 500-Kb resolution in the HD genome. b Syntenic analysis of HD monoploid genome assembly and HD-phased haplotypes. c Assessment of assembly quality with LAI value, where the y-axis represents LAI. d Distribution of LTR insertion time in HD and three published tea plant genomes. Dashed lines represent the mean insertion time in each genome
Although high heterozygosity presents difficulties in resolving the tea plant genome, it stores abundant genetic resources that the monoploid genome cannot decipher. The recent emergence of HiFi reads and the Hifiasm algorithm help us to overcome the difficulties of phasing highly heterozygous genomes. On this basis, the ALLHiC algorithm3 aids in constructing a haplotype-resolved chromosome-level assembly by separating homologous chromosomes according to the strength of interaction signals. We finally divided the HD genome into two haplotypes (A and B) with the assistance of the monoploid reference assembly. The two haplotypes comprised a total of 30 pseudochromosomes with a total length of 5.88 GB (2.92 Gb for HA and 2.95 Gb for HB). To assess the quality of the two haplotypes, we first compared them with the aforementioned HD monoploid reference assembly, showing good collinearity (Fig. 2b). BUSCO analysis showed that 89.9% and 92.5% conserved single-copy genes were recalled from haplotype A and haplotype B, respectively, and a total of 95.1% were complete BUSCO genes (Table 2).
Table 2.
Phased chromosome-scale assembly and annotation
| Haplotype A | Haplotype B | |
|---|---|---|
| Length of chromosomes (Gb) | 2.92 | 2.95 |
| BUSCO completeness of assembly (%) | 89.9 | 92.5 |
| Number of genes | 44,105 | 44,409 |
| Number of genes with >2 alleles | 7211 | |
| Number of genes with 2 alleles | 19,723 | |
| Number of genes with 1 allele | 1066 | |
| Total number of genes with alleles | 26,000 |
Combining information such as orthologous proteins, de novo prediction and expression evidence, we annotated 43,779 protein-coding genes through the MAKER pipeline29, of which 98.20% (42,981/43,779) of the genes were supported by transcript evidence, and 49.85% (21,826/43,779) and 41.44% (22,139/43,779) genes could be functionally annotated by the GO and KEGG databases, respectively. The average length of the protein-encoding genes was 5452 bp. In addition, the average length of introns was 1223 bp with 35.85% GC content, and the average length of exons was 342 bp with 43.26% GC content (Table 1). We further used TEclass30, a transposable element (TE) automatic classification tool, to enhance the repeatMasker annotation result. Finally, 70.75% of the HD genome was annotated as repetitive sequences (2.08/2.94 Gb) (Supplementary Table 6), and LTR retrotransposons (LTR-RTs) belonged to the largest class of TEs, accounting for 48.64% of the genomes (1.43/2.94 Gb). Meanwhile, 24,741 intact LTR-RTs were identified in the HD genome using LTR_retriever31 (Supplementary Table 5). Similar to the SCZ-Chr genome15, the HD genome showed more abundant LTR-RTs than two previously published draft genomes assembled based on Illumina short reads14,18, indicating that long-read sequencing technology provided a better solution for TE annotation in the highly repetitive genome. We also observed a significant enrichment of LTR-RT insertion events in the HD and SCZ-Chr genomes (Fig. 2d), dating a recent LTR burst back to 0.6 mya in the tea plant.
Alleles associated with stress tolerance and aroma-related components
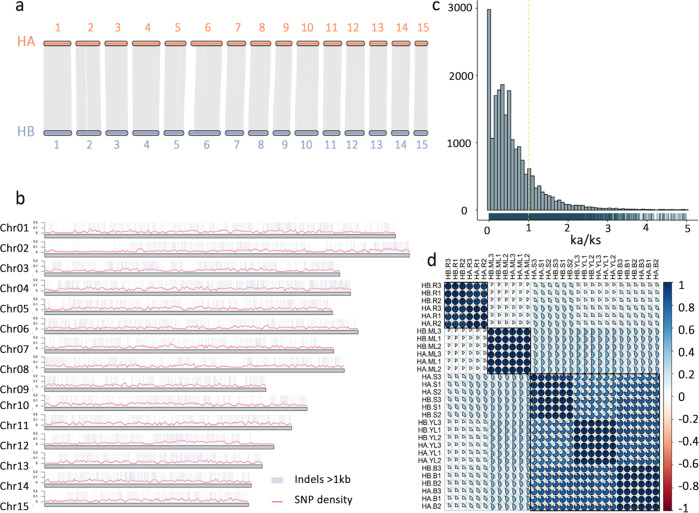
Comparison between the two haplotypes revealed a high level of synteny (Fig. 3a). However, a large number of haplotypic variations were detected, including 23.57 million SNPs, 1.14 million insertions, and 1.13 million deletions (Supplementary Table 7). The genetic variants were widespread and evenly distributed across the 15 pseudochromosomes (Fig. 3b). The phased chromosome-scale genome assembly allows us to separate alleles from 59.38% (26,000/43,779) protein-coding genes with at least one base substitution. These alleles showed an average similarity of 92.6% and Ka/Ks ratio of 0.61 (Fig. 3c), suggesting relatively rapid evolution. Functional annotation revealed that the genes under positive selection (Ka/Ks>1) were enriched in multiple biological processes, such as the response to reactive oxygen species (ROS) and the metabolism of multiple terpenoids (Supplementary Fig. 1), indicating their important roles in responses to stress and likely association with aroma-related volatile organic compounds (VOCs). RNA-seq reads from five different tissues, including buds, roots, stems, young leaves, and mature leaves, were also mapped against the allelic genes from HA and HB. We evaluated and clustered the correlation between the expression of 26,000 allelic genes in five tissues and found that the expression of these alleles in the same tissue was highly correlated and clustered together (Fig. 3d).
Fig. 3. Allelic variations and expression between two HD haplotypes.
a Synteny block between allele gene pairs. b Genetic variations between the two haplotypes. c Distribution of ka/ks between allele gene pairs. d Correlation analysis and clustering of the expression of alleles in five tissues, including roots (R), stems (S), buds (B), young leaves (YL), and mature leaves (ML)
Extensive structural variations affected aroma-related genes
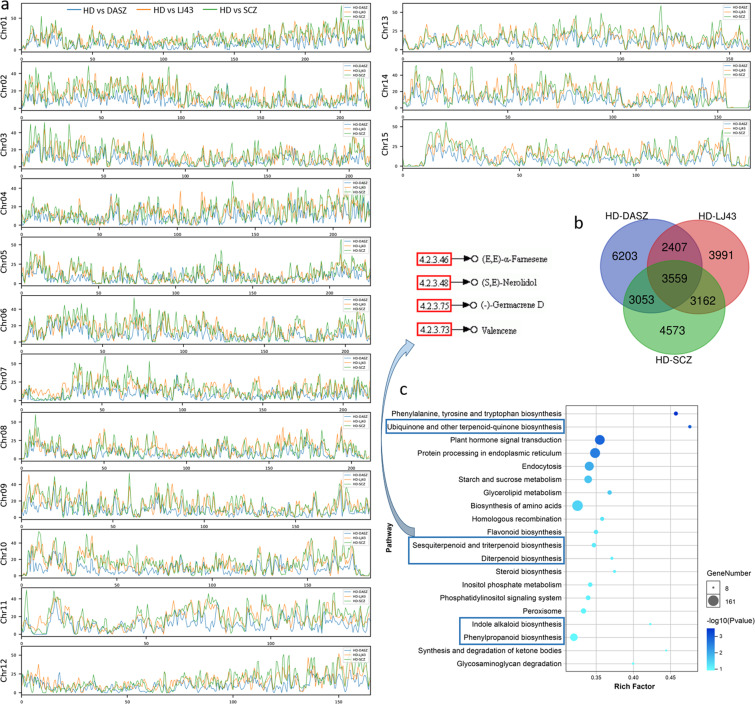
Structural variants (SVs) represent genomic rearrangements greater than 50 bp, which are an important basis for crop improvement and domestication traits32. However, SVs have not yet been investigated in tea plants by comparing high-quality genomes. Here, we identified 45,304, 44,358, and 26,754 SVs by comparing HD to the three published SCZ-Chr15, LJ4317 and DASZ19 genomes (Table 3 and Fig. 4a). A similar level of SV was observed in the three genomes, ranging from 294.08 (9.77%) to 380.32 (12.63%) Mb. The SVs were not evenly distributed across the 15 chromosomes. For instance, chromosome 1 has a high SV frequency and the longest total length, and chromosome 12 has the lowest SV frequency (Fig. 4a). These SVs affected 14,347, 13,119, and 15,222 protein-coding genes in SCZ-Chr, LJ43 and DASZ, respectively, suggesting that the SVs had a large effect on gene function. Among these genes, 3559 were shared (Fig. 4b). These genes were involved in a variety of metabolic pathways in our KEGG analysis (Supplementary Fig. 2), including zeatin biosynthesis, starch and sucrose metabolism, amino acid biosynthesis, carotenoid biosynthesis, glycosphingolipid biosynthesis, and diterpenoid biosynthesis, indicating the metabolic differentiation of tea germplasm resources. While HD is suitable for high-aroma oolong tea, SCZ and LJ43 are famous for processing into green tea. We analyzed the SV annotation genes of HD vs. SCZ-chr and HD vs. LJ43 and found that these genes were annotated in many aroma-related pathways (Fig. 4c and Supplementary Fig. 3), which indicated that SVs may affect the high-aroma characteristics of HD. We also observed that many terpenoids, which are important components of the aroma of oolong tea, exhibited structural variations in their synthetase genes (Fig. 4c), including (E)-nerolidol synthase (NES) and α-farnesene synthase (AFS), which can be used as SV resources for high-aroma tea breeding.
Table 3.
SV statistics of the HD genome vs. three tea plant genomes
| HD vs. SCZ | HD vs. LJ43 | HD vs. DASZ | |
|---|---|---|---|
| No. of InDels (50–500 bp) | 8174 | 8351 | 3718 |
| No. of InDels (500–10,000 bp) | 4539 | 5132 | 2023 |
| No. of InDels (10,000–50,000 bp) | 456 | 400 | 246 |
| No. of InDels (50,000–100,000 bp) | 21 | 19 | 9 |
| No. of tandem expansion/contraction | 5209 | 4998 | 3653 |
| No. of repeat expansion/contraction | 26,905 | 25,458 | 17,105 |
| Size of InDels (Mb) | 24.21 | 24.17 | 11.64 |
| Size of tandem expansion/contraction (Mb) | 75.76 | 41.46 | 46.79 |
| Size of repeat expansion/contraction (Mb) | 280.36 | 254.62 | 235.65 |
| Total SV number | 45,304 | 44,358 | 26,754 |
| Total SV size (Mb) | 380.32 | 320.25 | 294.08 |
| Gene count | 14,347 | 13,119 | 15,222 |
Fig. 4. Analysis of SVs and related genes.
a SV frequency plot of HD and three other tea plant genomes on 15 chromosomes. HD: Huangdan, SCZ: Suchazao15, LJ43: Longjing 4317, DASZ: An ancient tea tree19. b Venn plot of three sets of SV-annotated genes. c Top 20 KEGG pathways of SV-annotated genes between the HD and SCZ genomes. Pathways related to aroma are marked with blue boxes, and SV-related terpene synthases (TPSs) and their corresponding terpenoids are highlighted
Expansion of the terpene synthase (TPS) family contributes to the high-aroma characteristics of HD
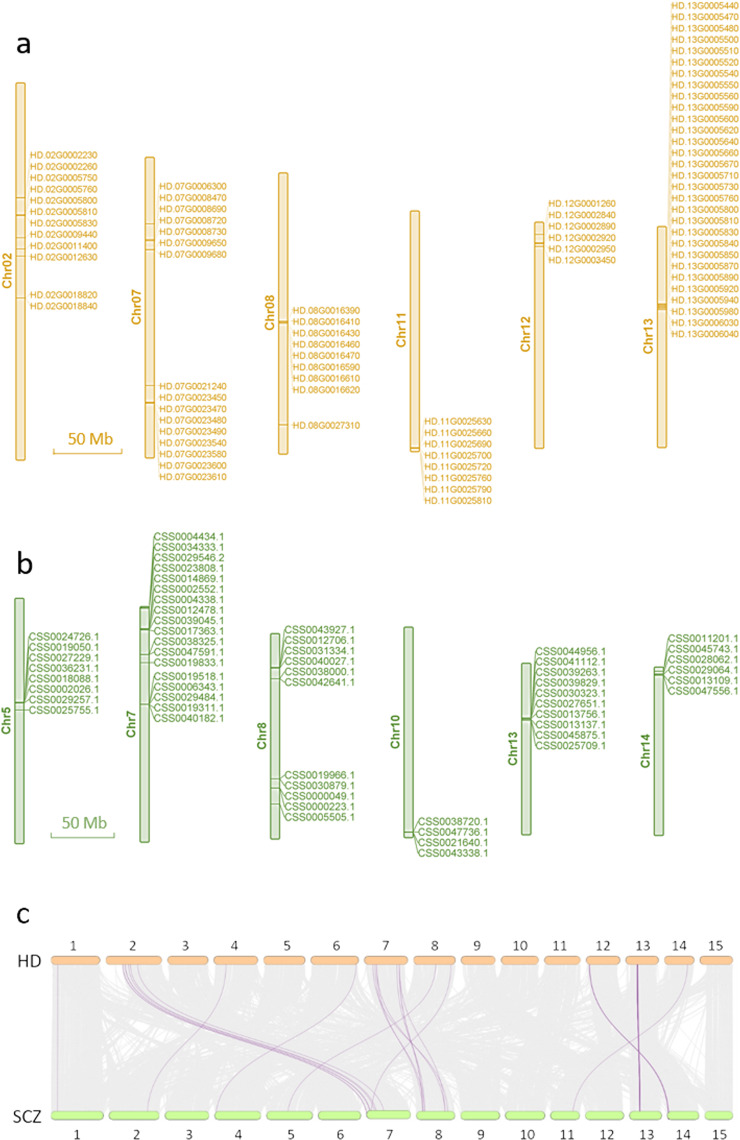
As a famous high-aroma backbone parent, HD has been used to breed a series of high-quality offspring. Compared with ordinary tea cultivars, the molecular mechanism of high-aroma formation is worthy of in-depth exploration. Our previous research24 showed that the most important aroma components of HD leaves are monoterpenoids and sesquiterpenoids, such as linalool and its oxides, geraniol, nerolidol, and α-farnesene, which are the quality basis for HD that is suitable for processing oolong tea. TPS is an important enzyme that can catalyze the production of a variety of terpenoids, which are essential for the tea aroma from the substrate. Compared with coffee and cacao, the newly released CSS-Suchazao (SCZ) genome showed that the amplification of TPS genes was driven by recent tandem replication events15. Here, we identified 93 HDTPS genes in the HD genome (Supplementary Table 8). This number is greater than the number identified in the SCZ genome (72 SCZTPSs)15, suggesting that TPS genes have expanded considerably in the HD genome. Obviously, compared to Arabidopsis, kiwifruit, and coffee (Supplementary Fig. 4), the TPSs expanded with tandem duplication were specific to tea plants. HDTPSs were widely distributed on 14 chromosomes (Supplementary Table 8); however, they showed a similar clustered pattern and high collinearity across six chromosomes compared with SCZTPSs (Fig. 5). Notably, 30 HDTPSs were enriched on chromosome 13, and this number was substantially higher than that in the SCZ genome.
Fig. 5. Comparison of TPS genes between HD and SCZ.
a The TPS genes of HD were mainly distributed on six chromosomes. b The TPS genes of SCZ were mainly distributed on six chromosomes. c Collinearity analysis of HD and SCZ, in which the purple line indicates the collinearity of TPS genes
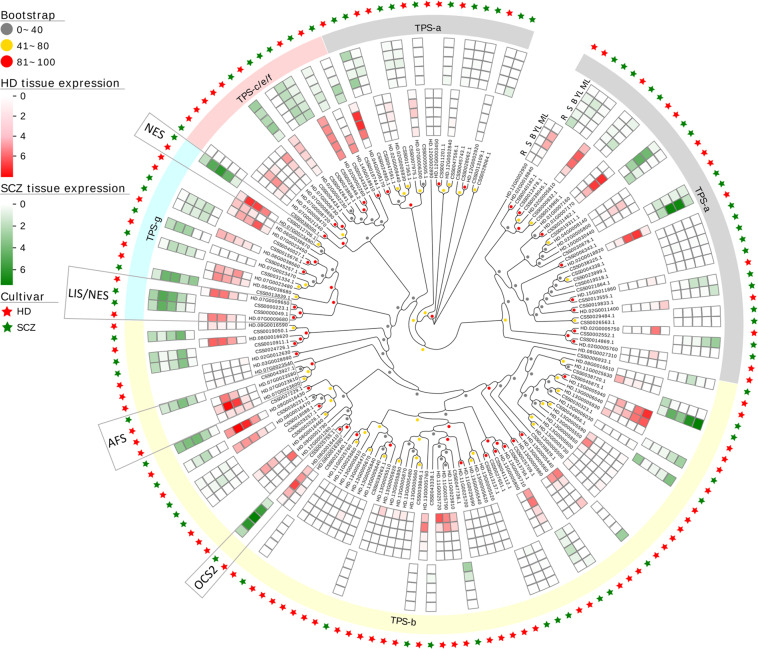
To compare the transcription abundance of TPS genes in HD and SCZ tissues, we mapped the transcriptome data of the five tissues (root, stem, apical bud, young leaf, and mature leaf) to their respective genomes using the same methods aforementioned. Phylogenetic analysis divided HDTPSs into four groups (Fig. 6). TPS-b contained the most genes (55, 59.14%), followed by TPS-a (22, 23.66%). Classification of other plants showed that TPS-a is mainly composed of sesquiterpene synthases, TPS-b and TPS-g are mainly monoterpene synthases, and TPS-c/e/f include diterpene synthases33. Our results showed that 27 HDTPSs were highly expressed in at least one tissue (TPM > 10), whereas 12 were expressed in SCZ (Supplementary Table 8). The apical buds and young leaves of tea plants are the major economically important tissues used for processing tea beverages. We further analyzed the expression of TPS genes in buds and young leaves and discovered that 18 TPSs were highly expressed in HD (TPM > 10) and 10 in SCZ. We also compared four homologous TPS genes, including linalool and (E)-nerolidol synthases (LIS/NES, HD.07G0009650, HD.07G0009680, CSS0000049.1, and CSS0000223.1)34, (E)-NES (HD.07G0023490 and CSS0012706.1)35, α-farnesene synthase (AFS, HD.07G0023580, HD.07G0023600, HD.07G0023610, and CSS0043927.1)36, and ocimene synthase 2 (OCS2, HD.08G0016410 and CSS0025755.1)37, that have been identified in vitro or in vivo (Fig. 6). Among them, the expression levels of the LIS/NES homologous transcripts of HD and SCZ were similar, whereas the expression level of SCZOCS2 in buds and leaves was higher than that of HDOCS2. However, the TPM values of the apical buds and young leaves of one HDAFS were 6-fold and 80-fold greater than in SCZ, respectively. The expression of NES of HD in young leaves was nearly 3-fold that of SCZ. In addition, we tested the aroma components of the leaves of HD and SCZ in October. The changes in the marked terpenoids were consistent at the genetic level, except for (E)-nerolidol, which may be due to the season and the unprocessed state of the tissues (Supplementary Fig. 5). These results indicate that the widespread and specifically high expression of TPS genes in HD tissues may be an important factor affecting their high-aroma characteristics.
Fig. 6. Phylogenetic and tissue expression analysis of TPS genes from HD and SCZ.
The legends are shown on the left. The TPM values of five tissues (root, stem, apical bud, young leaf, and mature leaf) from HD and SCZ were normalized. Four homologous TPS genes, including linalool and (E)-nerolidol synthase (LIS/NES)34, (E)-nerolidol synthase (NES)35, α-farnesene synthase (AFS)36, and ocimene synthase 2 (OCS2)37, that have been identified in vitro or in vivo are marked
Identification and comparison of C-repeat binding factors (CBFs)
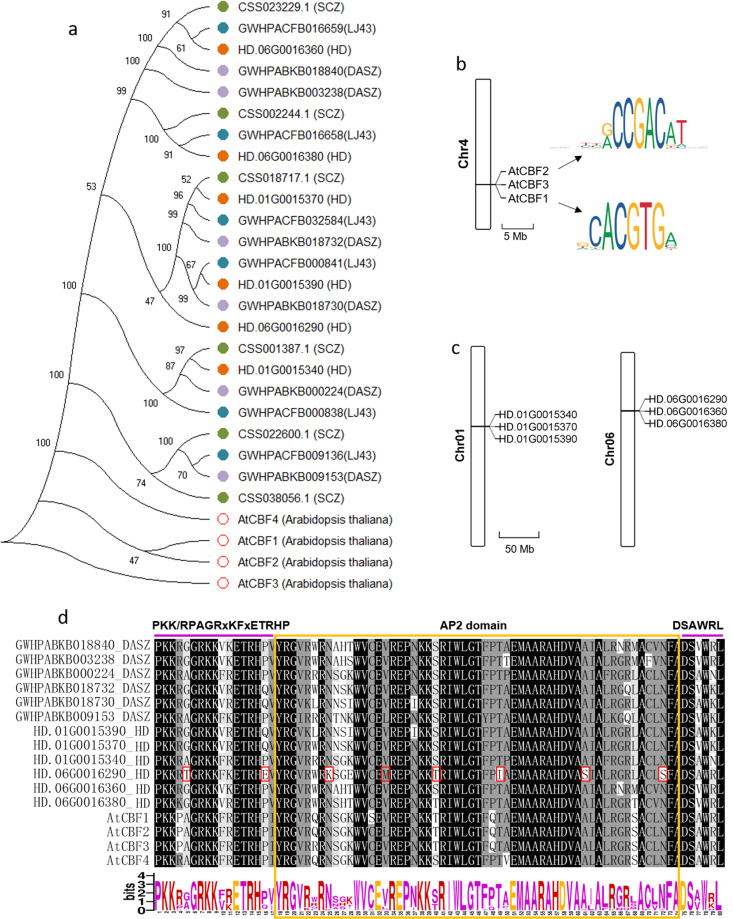
C-repeat binding factors (CBFs) are the core regulators in response to cold stress in plants; they can be rapidly induced by cold and accompany the C-repeat/dehydration response motif (CRT/DRE) in the promoter region of the target cold-response (COR) genes38–40. The CBFs of tea plant were initially identified in the previous draft genome and were found to be rapidly induced by cold stress and to regulate target genes in multiple pathways41,42. We are interested in the number of CBF regulatory factors in tea plants, and the recently published chromosome-level genomes of tea plants provide convenience for our identification and comparison. We identified six CBFs in the four tea plant genomes, two more than in Arabidopsis, which was consistent with previous experimental verification42. The CBFs of tea plant and Arabidopsis clustered into different clades, indicating species conservation (Fig. 7a). However, the CBF clades from the four tea germplasms were different, which may be related to the different degrees of cold resistance of various tea plant resources. Arabidopsis AtCBF1-3 is recognized as an important cascade core that regulates cold acclimation. They are arranged in tandem on chromosome 4 to regulate target genes in response to cold stress through binding G/ACCGAC motifs (Fig. 7b)40,43. Interestingly, we observed that the six CBFs in the HD genome were arranged tandemly on chromosomes 1 and 6 (Fig. 7c), which may play an important role in the regulation of cold response in tea plants. Tea plants are believed to originate from southwestern China, and the recently sequenced ancient tea plant (DASZ) genome19 from Yunnan Province allows us to compare the evolution and variation of the conserved domain of CBFs. By comparing the AP2 DNA-binding domain and two flanking signature sequences of CBFs among HD, DASZ, and Arabidopsis, we observed that the overall CBF protein sequences were highly conserved, and the similarity reached 89.77% (Fig. 7d). Among them, CBFs from tea plants and Arabidopsis showed some amino acid differences at multiple positions. Compared with the ancient tea tree, one of the CBF genes (HD.06G0016290) from the elite tea cultivar HD showed amino acid mutations at 8 positions, which may affect its cold response and DNA-binding function. In summary, these results provide genomic evidence for CBF-mediated resistance to cold stress in tea plants.
Fig. 7. Comparison of C-repeat binding factors (CBFs).
a Phylogenetic tree analysis of CBFs from four tea resources and Arabidopsis thaliana. HD: Huangdan, SCZ: Suchazao15, LJ43: Longjing 4317, DASZ: An ancient tea tree19. b Three AtCBFs arranged in tandem on chromosome 4 in Arabidopsis. The binding sites of AtCBF1 and AtCBF2 verified by DNA affinity purification sequencing (DAP-seq) are presented in the form of sequence logos43. c Six CBFs arranged in tandem on chromosomes 1 and 6 in the HD genome. d Multisequence alignment of the conserved binding domains of CBFs from HD, DASZ, and Arabidopsis. The sequence logo reflects the conservation of the amino acid at each position. The yellow box represents the AP2 DNA-binding domain; the purple overlines indicate two signature sequences; the small red box represents the amino acid mutation of CBFs of HD compared to DASZ
Discussion
Oolong tea is one of six major types of tea production and is known for its high levels of aroma23. Our study decoded the fully phased genome of an elite oolong tea cultivar, HD. Analysis of the genome revealed extensive genetic variations compared with other published tea genomes and frequent allelic imbalances associated with or likely contributing to high-aroma characteristics and stress tolerance. In addition, we observed a tandem evolution pattern of CBF genes, indicating their potential roles in the stress tolerance.
Aroma plays a vital role in the quality of tea, and in addition to the tea manufacturing process, tea cultivars are the basis for the formation of tea aroma. HD is always well known for its high-aroma characteristics. Our team has participated in the breeding project with HD and CSS Tieguanyin as the backbone parents and has cultivated a series of high-aroma and high-quality offspring, including CSS Jinguanyin (JGY, No. GS2002017), CSS Huangguanyin (HGY, No. GS2002015), and CSS Jinmudan (JMD, No. GS2010024), which are certified by the Chinese Crop Variety Approval Committee as one of the most important pedigrees in Chinese tea breeding19. Our previous study showed that sesquiterpenes and monoterpenes are the core aroma components of HD and its progeny24. Terpenoids are an important component of tea aroma due to their pleasant aroma and low odor threshold44. Comparative genomics of HD and two green tea cultivars revealed extensive structural variations in TPS genes, which may affect the high-aroma characteristics of oolong tea cultivars. The number of TPS genes in SCZ (72) is notably higher than that in coffee (53), cacao (36), and kiwifruit (34)15. Our results showed that HD has 21 more TPS genes than SCZ, and they are mainly clustered on six chromosomes, especially chromosome 13, which may enhance the synthesis of terpenoids in HD. Remarkably, compared to SCZ, the TPS genes in HD were more widely and highly expressed in buds and leaves, which are economically important tissues. As a recent study has revealed, the increase in TPS members, the formation of clusters, the differentiation of catalytic functions, and differences in in gene expression regulation may all affect the production and concentration of terpenes in plants45. Although the number of TPS genes in tea plants has expanded to varying degrees, the function of most TPS genes is still unknown46, except for several TPSs that have been shown to be involved in the formation of linalool34, (E)-nerolidol34,35, (E)-β-ocimene37,47, and α-farnesene36,47. Oolong tea has more pleasant or distinctive aroma qualities than green tea48, possibly due to higher levels of (E)-nerolidol and α-farnesene35,49. This is likely attributable to the significantly upregulated expression of two TPS genes (NES and AFS). Overall, our results suggest that the expansion and specifically high expression of TPS family genes are the molecular basis of HD’s high-aroma characteristics.
Materials and methods
Tea plant materials and sequencing
Fresh leaves of an individual CSS cultivar HD planted in the tea plantation of Fujian Agriculture and Forestry University (Fuzhou, China; 26°08′19″ N, 119°24′06″ E) were collected for genome sequencing. The high-quality genomic DNA of HD samples was extracted using a DNeasy Plant Mini Kit (Qiagen, USA) and sent to Annoroad (Ningbo, China) to construct 10–25 kb SMRTbellTM libraries and sequenced using the PacBio Sequel II platform. The tender leaves of HD were also plucked and sent to Annoroad for Hi-C library construction and sequencing through an Illumina HiSeq Nova-seq system. In addition, five HD tissues, including tender roots, young stems, apical buds, tender leaves, and mature leaves, were collected, and the total RNA was extracted by an RNAprep Pure Plant kit (TIANGEN, Beijing, China). A total of 3 μg RNA was used for each sample to generate a transcriptome library and sequenced using the Illumina platform.
Genome assembly
We analyzed the genome size of the sequenced individuals by flow cytometry (BD FACSCalibur, BD Bioscience, USA) using tomato and maize as internal controls50. The CSS HD genome was assembled by incorporating sequencing data from PacBio circular consensus sequencing technology51 and the Hi-C method. First, HiFi reads were assembled with Hifiasm with the default parameters. We assembled two levels of chromosome-scale genomes, including a monoploid genome and an allele-defined haplotype-resolved assembly. Briefly, for monoploid assembly, although Purge_dups was contained in Hifiasm, we checked the read depth and filtered the primary contigs of the initial Hifiasm assembly by Purge_dups (v1.25) with the default parameters and evaluated the results by the Benchmarking Universal Single-Copy Orthologs (BUSCO) completeness27 and duplication score. Hi-C reads were assessed by the HiC-Pro (v2.11.4) program52 and were uniquely mapped to contig assemblies. Meanwhile, Juicer tools (v1.11.08)53 and 3D-DNA pipelines (v180114)54 were used to detect and correct misassembled contigs. To distribute contigs into the appropriate groups, we aligned the set of Hi-C corrected contigs against the “Shuchazao” chromosome-scale assembly using RaGOO (v1.1). Finally, the ALLHiC optimize algorithm (v0.9.13) was used to adjust the order and orientation of contigs in each group. We merged the haplotig sequence and alternative assembly to redo Purge_dups (v1.25) with the cutoff parameter “2 6 10 12 20 70”. The purged primary contigs and alternative contigs were merged and regarded as a draft contig assembly for the monoploid genome. The resulting contigs were subjected to haplotype phasing using the ALLHiC algorithm with default parameters, and the monoploid genome sequences were selected as a reference to identify allelic contigs. Chromosome-level haplotype A and haplotype B of CSS HD were fully resolved and released. In addition, we calculated the heterozygosity through GenomeScope255 with 33 k-mers. Chromosome localization and collinearity analysis of genes were visualized in TBtools56.
Repetitive sequences and gene annotation
We first ran RepeatModeler (v1.08), which can automatically execute RECON57 and RepeatScout58, to construct a de novo repeat library of the HD genome. RepeatMasker (v4.07) was utilized to identify and cluster repetitive sequences, and Teclass (v2.1.3)30 was used to further classify unknown repetitive sequences. The TRF package (v4.09)59 with modified parameters (“trf 1 1 2 80 5 200 2000”) from previous research4 was used to find tandem repeats.
We ran two rounds of the MAKER pipeline29, which can integrate predictions based on orthologous proteins, ab initio calculations and transcriptome data to better annotate protein-coding genes. We first assembled transcriptome data through de novo assembly and genome-guided assembly by Trinity (v2.20)60. Then, the assembled transcripts were subjected to calculation and proper filtering of FPKM expression values with the same standards as in previous research4. The filtered data were imported into the PASA program (v2.1.0)61 and compared with the UniProt plant protein database for evaluation. Finally, the remaining transcripts were further trained4. The evidence was integrated by the MAKER pipeline and used to generate a preliminary set of HD-encoding genes. The predicted gene models with low AED scores were selected for retraining using GENEMARK62, SNAP (https://github.com/KorfLab/SNAP)63, and AUGUSTUS (v3.3.1)64 in the second round of MAKER running. Additionally, the transcriptome data were mapped to the genome via HISAT2 (v2.10) and reassembled via StringTie (v1.3.4)65. The assembled data were imported into the MAKER pipeline together with the homologous proteins of Arabidopsis thaliana, rice, grape, tomato, Morus notablis, and papaya. After filtering putative transposon-derived gene models, a set of HD protein-coding genes was generated. Then, the selected protein-coding gene set was functionally annotated with high-confidence orthologs in the eggNOG database66.
Allele gene and genetic variation identification
Initially, we annotated the HD A/B subgenome with the MAKER pipeline described above. The identification of alleles was based on the methods of sugarcane genome research. We first produced a table of alleles with two columns (including HA and HB). Two main approaches, including the synteny-based approach and the GMAP-based approach67, were used to generate the backbone of the allelic table. Initially, we used JCVI (https://github.com/tanghaibao/jcvi)68, the Python version of MCSCAN, to scan synteny blocks between two sets of genes. The cscore = 0.99 parameter enabled us to obtain 1 vs. 1 comparison genes between two sets of haplotypes, while genes from the same synteny block were considered alleles to be classified. Subsequently, the gene models excluded from the synteny block were aligned to the monoploid assembly with GMAP (version 2013-10-28). If gene pairs share more than half of the reference genome coordinates, they can exist as alleles. The unfiltered alleles were compared in pairs using the mafft (v7.471) L-Ins-I model to enable accurate alignment between gene pairs. By comparing the alignment results, we were able to calculate the similarity between alleles. Only gene pairs with high similarity (>0.7) were considered to form a pair of alleles, while a pair of identical genes was considered to be the same allele. In the other case, there were multiple genes (>2) aligned to the same location using GMAP. We sorted these genes according to their similarity, removed the gene pairs with low similarity, and added the filtered genes as paralogs of alleles to the allele table.
To explore the genetic difference and evolutionary relationship between two sets of haplotypes, we identified SNPs and InDels between them. SNP identification was performed by the Nucmer utility69 with parameter settings consistent with the generation of pseudohaplotypes. The InDels between two subgenomes were found by Assemblytics (http://assemblytics.com/)70 with the default parameters. The analysis of structural variants (SVs) among tea plant genomes was also carried out by Nucmer69 and Assemblytics70.
Analysis of allele expression
A pipeline built into Trinity software60, including bowtie, RNA-Seq by Expectation Maximization (RSEM) and edgeR, was used to analyze the difference in the expression of each pair of alleles in the allele table. In detail, for RNA-seq sequencing reads in five tissues, bud, root, stem, young leaf and mature leaf, we used trim-galore to remove their adaptations before mapping. Subsequently, reads were mapped to allelic transcripts, and the parameters “--all --best --strata -- M 300 --k 1” were used to obtain the optimal results. Then, RSEM was used to calculate the allele transcripts per kilobase of exon model per million mapped reads (TPM) values, and edegR was used to quantitatively evaluate the allelic differences of expression level.
Analysis of volatile compounds (VOCs)
We collected the leaves of HD and SCZ in October, and the samples were ground to a powder in liquid nitrogen. One gram of the powder was transferred to a 20 ml headspace vial. The vials were sealed using crimp-top caps with TFE-silicone headspace septa. At the time of SPME analysis, each vial was placed at 60 °C for 10 min, and then a 65 µm divinylbenzene/carboxene/polydimethylsilioxane fiber (Supelco, Bellefonte, PA, USA) was exposed to the headspace of the sample for 20 min at 60 °C. VOCs were detected by MetWare (http://www.metware.cn/) based on the Agilent 7890B-7000D platform. Desorption of the VOCs from the fiber coating was carried out in the injection port of the GC apparatus at 250 °C for 5 min in splitless mode. The identification and quantification of VOCs was carried out with a 30 m x 0.25 mm × 1.0 µm DB-5MS (5% phenyl-polymethylsiloxane) capillary column. Helium was used as the carrier gas at a linear velocity of 1.0 ml/min. The oven temperature was programmed from 40 °C (5 min), increased at 6 °C/min to 280 °C, and held for 5 min. Mass spectra were recorded in electron impact (EI) ionization mode at 70 eV. The quadrupole mass detector, ion source and transfer line temperatures were set at 150, 230, and 280 °C, respectively. Mass spectra were scanned in the range m/z 30–350 amu at 1 s intervals. Volatile compounds were identified by comparing the mass spectra with the data system library (MWGC) and linear retention index.
Supplementary information
Acknowledgements
This research was funded by the Fujian Province “2011 Collaborative Innovation Center”, the Chinese Oolong Tea Industry Innovation Center special project (J2015-75), the National Natural Science Foundation of China (31701874), the Major Special Project of Scientific and Technological Innovation on Anxi Tea (AX2021001), the Earmarked Fund for the China Agriculture Research System (CARS-19), the Scientific Research Foundation of the Graduate School of Fujian Agriculture and Forestry University (324-1122yb060), and the Scientific Research Foundation of Horticulture College of Fujian Agriculture and Forestry University (2018B02).
Author contributions
N.Y., X.Z., and P.W. conceived and designed the research. P.W. and X.Z. collected the samples. P.W., J.Y., X.Z., S.C., S.J., C.Y., W.W., S.G., H.C., Y.Z., M.G., and X.C. performed the genome assembly and data analysis. P.W. and J.Y. wrote the manuscript. N.Y., X.Z., S.J., Y.S., Y.G., J.Y., and C.Y. revised the manuscript.
Data availability
All data are publicly available in the BIG Data Center (https://bigd.big.ac.cn/) under project number PRJCA003382. The raw genome and transcriptome sequencing data are publicly available under sub-number CRA003208. The assembly and annotation of the HD diploid genome are available under sub-number GWHAZTZ00000000. The assembly and annotation of the HD haplotype genome are available under sub-number GWHBAUV00000000.
Conflict of interest
The authors declare no competing interests.
Footnotes
These authors contributed equally: Pengjie Wang, Jiaxin Yu
Contributor Information
Xingtan Zhang, Email: zhangxingtan@caas.cn.
Naixing Ye, Email: ynxtea@126.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00542-x.
References
- 1.Chen F, et al. The sequenced angiosperm genomes and genome databases. Front. Plant Sci. 2018;9:418. doi: 10.3389/fpls.2018.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang XT, Wu RX, Wang YB, Yu JX, Tang HB. Unzipping haplotypes in diploid and polyploid genomes. Comput. Struct. Biotechnol. J. 2019;18:66–72. doi: 10.1016/j.csbj.2019.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang XT, Zhang SC, Zhao Q, Ming R, Tang HB. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants. 2019;5:833–845. doi: 10.1038/s41477-019-0487-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhang JS, et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 2018;50:1565–1573. doi: 10.1038/s41588-018-0237-2. [DOI] [PubMed] [Google Scholar]
- 5.Chen HT, et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020;11:2494. doi: 10.1038/s41467-020-16338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, X. Q. et al. A high-quality Brassica napus genome reveals expansion of transposable elements, subgenome evolution and disease resistance. Plant Biotechnol. J. 10.1111/pbi.13493 (2020). [DOI] [PMC free article] [PubMed]
- 7.Zhang XT, et al. Genomes of the banyan tree and pollinator wasp provide insights into fig-wasp coevolution. Cell. 2020;183:875–889. doi: 10.1016/j.cell.2020.09.043. [DOI] [PubMed] [Google Scholar]
- 8.Drew L. The growth of tea. Nature. 2019;566:S2–S4. doi: 10.1038/d41586-019-00395-4. [DOI] [PubMed] [Google Scholar]
- 9.Chen JD, et al. The chromosome-scale genome reveals the evolution and diversification after the recent tetraploidization event in tea plant. Hortic. Res. 2020;7:63. doi: 10.1038/s41438-020-0288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia EH, Tong W, Wu Q, Wei S, Wan XC. Tea plant genomics: achievements, challenges and perspectives. Hortic. Res. 2020;7:7. doi: 10.1038/s41438-019-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y, Zeng LT, Hou XL, Liao YY, Yang ZY. Low temperature synergistically promotes wounding-induced indole accumulation by INDUCER OF CBF EXPRESSION-mediated alterations of jasmonic acid signaling in Camellia sinensis. J. Exp. Bot. 2020;71:2172–2185. doi: 10.1093/jxb/erz570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 13.Zeng LT, Watanabe N, Yang ZY. Understanding the biosyntheses and stress response mechanisms of aroma compounds in tea (Camellia sinensis) to safely and effectively improve tea aroma. Crit. Rev. Food Sci. Nutr. 2018;59:2321–2334. doi: 10.1080/10408398.2018.1506907. [DOI] [PubMed] [Google Scholar]
- 14.Wei CL, et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl Acad. Sci. USA. 2018;115:E4151–E4158. doi: 10.1073/pnas.1719622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia EH, et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into genome evolution and adaptation of tea plants. Mol. Plant. 2020;13:1013–1026. doi: 10.1016/j.molp.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Zhang QJ, et al. The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons to drive genome size evolution. Mol. Plant. 2020;13:935–938. doi: 10.1016/j.molp.2020.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Feng H, Chang Y, Ma C, Yang Y. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020;11:4447. doi: 10.1038/s41467-020-18228-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia EH, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant. 2017;10:866–877. doi: 10.1016/j.molp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Zhang WY, et al. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat. Commun. 2020;11:3719. doi: 10.1038/s41467-020-17498-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchinoue Y. Analysis of self-incompatibility alleles of major varieties of tea. Jpn. Agr. Res. Q. 1979;13:43–48. [Google Scholar]
- 21.Huang XH, et al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 2015;6:6258. doi: 10.1038/ncomms7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao L, et al. Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc. Natl Acad. Sci. USA. 2019;116:5653–5658. doi: 10.1073/pnas.1820513116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng L, Zhou X, Su X, Yang Z. Chinese oolong tea: An aromatic beverage produced under multiple stresses. Trends Food Sci. Technol. 2020;106:242–253. doi: 10.1016/j.tifs.2020.10.001. [DOI] [Google Scholar]
- 24.Zheng YC, et al. Transcriptome and metabolite profiling reveal novel insights into volatile heterosis in the tea plant (Camellia Sinensis) Molecules. 2019;24:3380. doi: 10.3390/molecules24183380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods18, 170–175 (2021). [DOI] [PMC free article] [PubMed]
- 26.Guan D, et al. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020;36:2896–2898. doi: 10.1093/bioinformatics/btaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simão FA, Waterhouse RM, Panagiotis I, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 28.Ou S, Chen J, Jiang N. Assessing genome assembly quality using the LTR Assembly Index (LAI) Nucleic Acids Res. 2018;46:e126. doi: 10.1093/nar/gky730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantarel BL, et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abrusán G, Grundmann N, DeMester L, Makalowski W. TEclass–a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics. 2009;25:1329–1330. doi: 10.1093/bioinformatics/btp084. [DOI] [PubMed] [Google Scholar]
- 31.Ou SJ, Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal-repeat retrotransposons. Plant Physiol. 2017;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alonge M, Wang X, Benoit M, Soyk S, Pereira L. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell. 2020;182:145–161. doi: 10.1016/j.cell.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen F, Tholl D, Bohlmann JR, Pichersky E. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66:212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu G, et al. Implementation of CsLIS/NES in linalool biosynthesis involves transcript splicing regulation in Camellia sinensis. Plant Cell Environ. 2017;41:176–186. doi: 10.1111/pce.13080. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Y, et al. Formation of (E)-nerolidol in tea (Camellia sinensis) leaves exposed to multiple stresses during tea manufacturing. Food Chem. 2017;231:78–86. doi: 10.1016/j.foodchem.2017.03.122. [DOI] [PubMed] [Google Scholar]
- 36.Wang XW, et al. Formation of α-Farnesene in tea (Camellia sinensis) leaves induced by herbivore-derived wounding and its effect on neighboring tea plants. Int. J. Mol. Sci. 2019;20:4151. doi: 10.3390/ijms20174151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu QS, et al. Unraveling a crosstalk regulatory network of temporal aroma accumulation in tea plant (Camellia sinensis) leaves by integration of metabolomics and transcriptomics. Environ. Exp. Bot. 2018;149:81–94. doi: 10.1016/j.envexpbot.2018.02.005. [DOI] [Google Scholar]
- 38.Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl Acad. Sci. USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilmour SJ, et al. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang PJ, et al. Identification of CBF transcription factors in tea plants and a survey of potential CBF target genes under low temperature. Int. J. Mol. Sci. 2019;20:5137. doi: 10.3390/ijms20205137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Z, Ban Q, Hao J, Zhu X, Li Y. Genome-wide characterization of the c-repeat binding factor (CBF) gene family involved in the response to abiotic stresses in tea plant (Camellia sinensis) Front. Plant Sci. 2020;11:921. doi: 10.3389/fpls.2020.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Malley RC, et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;166:1598. doi: 10.1016/j.cell.2016.08.063. [DOI] [PubMed] [Google Scholar]
- 44.Schuh C, Schieberle P. Characterization of the key aroma compounds in the beverage prepared from Darjeeling black tea: quantitative differences between tea leaves and infusion. J. Agric. Food Chem. 2006;54:916–924. doi: 10.1021/jf052495n. [DOI] [PubMed] [Google Scholar]
- 45.Chen H, et al. Combinatorial evolution of a terpene synthase gene cluster explains terpene variations in Oryza. Plant Physiol. 2020;182:480–492. doi: 10.1104/pp.19.00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou HC, Shamala LF, Yi XK, Yan Z, Wei S. Analysis of terpene synthase family genes in Camellia sinensis with an emphasis on abiotic stress conditions. Sci. Rep. 2020;10:933. doi: 10.1038/s41598-020-57805-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin J, et al. Scenarios of genes-to-terpenoids network led to the identification of a novel α/β-Farnesene/β-ocimene synthase in Camellia sinensis. Int. J. Mol. Sci. 2020;21:655. doi: 10.3390/ijms21020655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang ZY, Baldermann S, Watanabe N. Recent studies of the volatile compounds in tea. Food Res. Int. 2013;53:585–599. doi: 10.1016/j.foodres.2013.02.011. [DOI] [Google Scholar]
- 49.Lin J, Shi YX, Dong CW, Wang XC. Headspace volatiles influenced by infusion matrix and their release persistence: a case study of oolong tea. Food Sci. Biotechnol. 2019;28:1349–1358. doi: 10.1007/s10068-019-00587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dolezel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- 51.Wenger AM, Peluso P, Rowell WJ, Chang PC, Hunkapiller MW. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019;37:1155–1162. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Servant N, et al. HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durand NC, et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dudchenko O, et al. De novo assembly of the aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ranallo-Benavidez TR, Jaron KS, Schatz MC. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 2020;11:1432. doi: 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen CJ, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 57.Bao ZR, Eddy SR. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:351–358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 59.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haas BJ, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33:6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korf I. Gene finding in novel genomes. BMC Bioinforma. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinforma. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huerta Cepas J, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2018;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 68.Tang HB, et al. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 69.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maria N, Schatz MC. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics. 2016;32:3021–3023. doi: 10.1093/bioinformatics/btw369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are publicly available in the BIG Data Center (https://bigd.big.ac.cn/) under project number PRJCA003382. The raw genome and transcriptome sequencing data are publicly available under sub-number CRA003208. The assembly and annotation of the HD diploid genome are available under sub-number GWHAZTZ00000000. The assembly and annotation of the HD haplotype genome are available under sub-number GWHBAUV00000000.