Abstract
Autism spectrum disorders (ASD) is a complex neurodevelopmental disorder that may significantly impact on the affected individual’s life. Common variation (SNPs) could explain about 50% of ASD heritability. Despite this fact and the large size of the last GWAS meta-analysis, it is believed that hundreds of risk genes in ASD have yet to be discovered. New tools, such as TWAS (Transcriptome Wide Association Studies) which integrate tissue expression and genetic data, are a great approach to identify new ASD susceptibility genes. The main goal of this study is to use UTMOST with the publicly available summary statistics from the largest ASD GWAS meta-analysis as genetic input. In addition, an in silico biological characterization for the novel associated loci was performed. Our results have shown the association of 4 genes at the brain level (CIPC, PINX1, NKX2-2, and PTPRE) and have highlighted the association of NKX2-2, MANBA, ERI1, and MITF at the gastrointestinal level. The gastrointestinal associations are quite relevant given the well-established but unexplored relationship between ASD and gastrointestinal symptoms. Cross-tissue analysis has shown the association of NKX2-2 and BLK. UTMOST-associated genes together with their in silico biological characterization seems to point to different biological mechanisms underlying ASD etiology. Thus, it would not be restricted to brain tissue and it will involve the participation of other body tissues such as the gastrointestinal.
Subject terms: Genomics, Autism spectrum disorders
Introduction
Autism spectrum disorders (ASD) includes a range of neurodevelopmental disorders (NDDs) with onset in early development that are characterized by deficits in communication and social interactions, as well as by repetitive patterns of behavior and restrictive interests1. ASD is a complex genetic disorder, involving both environmental and genetic factors. Although an important part of the genetic architecture of ASD is unknown, it is considered that thousands of genes may be involved even most of them remain unidentified and functionally uncharacterized. Rare genetic variation only explains 3% of ASD genetic risk even if it confers a high individual risk2. However, common variation (SNPs; single nucleotide polymorphisms) could explain about 50% of ASD heritability. The most recent and the largest ASD GWAS meta-analysis done until now, including 18,381 ASD cases and 27,969 controls, has reported 93 genome-wide significant markers in three separate loci (top SNP: rs910805; p-value: 2.04 × 10−9)3. Another methodological approach for common variation are gene-based association analysis (GBA) methods that employ the p-values for each SNP within a gene to obtain a single statistic at this level. Thus, MAGMA has identified 15 genes, most of them located near the genome-wide significant SNPs identified in GWAS, but 7 genes have revealed association in four additional loci (KCNN2, MMP12, NTM, and a cluster of genes on chromosome 17)3. Additional GBA methods using other algorithms as PASCAL have helped to define the association of other genes located in the same LD region than those found by MAGMA (NKX2-4, NKX2-2, CRHR1-IT1, C8orf74, and LOC644172)4.
In addition to GBA, bioinformatic approaches that integrate functional data are increasingly used to highlight new genes underlying GWAS summary statistics. Transcriptome-wide association studies (TWAS) have emerged as useful tools to study the genetic architecture of complex traits. Among them, MetaXcan5 and FUSION6 are well-known TWAS methods.
UTMOST (unified test for molecular signatures) has been recently reported as a novel framework for single and cross-tissue expression imputation. UTMOST is able to consider the joint effect of SNPs (summary statistics) across LD regions (1000 Genomes) and to integrate tissue expression data (GTeX) creating single and cross-tissue covariance matrices that will help to define the gene-trait associations. UTMOST performance was demonstrated at several levels and its accuracy was also well proved as it was able to identify a greater number of associations in biologically relevant tissues for complex diseases7.
The main aim of this paper is to further mine the summary data from the largest ASD meta-analysis using UTMOST. In addition, an in silico biological characterization for the novel associated loci will be carried out using bioinformatic approaches (DEG, pathway, gene network, and an exploratory enhancer analysis).
Overall, our results have demonstrated the association of CIPC, PINX1, NKX2-2, and PTPRE at the brain level and have also revealed the relevance of gastrointestinal tissue in ASD etiology through the association of other genes (NKX2-2, MANBA, ERI1, and MITF).
Materials and methods
Datasets
Summary statistics from the latest ASD GWAS meta-analysis were obtained from the public repository available in the PGC website (http://www.med.unc.edu/pgc/results-and-downloads). The following data set was employed: iPSYCH_PGC_ASD_Nov2017.gz (Grove et al.3) which includes the meta-analysis of ASD by the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH) and the Psychiatric Genomics Consortium (PGC) released in November 2017. The data set comprises a total of 18,381 cases and 27,969 controls. Additional information about the genotyping, QC methods, and Ethics Commitees as well as informed consents employed in are available at the PGC website and in the previous Grove et al.3 study.
TWAS Analysis using UTMOST
UTMOST7 (https://github.com/Joker-Jerome/UTMOST) was run as a single tissue association test for 44 GTeX tissues (single_tissue_association_test.py) and a cross tissue association test combining gene-trait associations was run by the joint GBJ test (joint_GBJ_test.py). Both tests use the previous summary statistics of the ASD GWAS meta-analysis as an input3. UTMOST pre-calculated covariance matrices for single-tissue (covariance_tissue/) and joint test (covariance_joint/) were downloaded. Other necessary command parameters were used by default. Transcriptome-wide significance for single tissue analysis was established as p-value = 3.85 × 10−6 (0.05/12984(maximum number of genes tested) for brain tissues and p-value = 3.42 × 10−6 (0.05/14586 (maximum number of genes tested) for non-brain tissues after Bonferroni correction. Transcriptome-wide significance for joint test was established as p-value = 3.27 × 10−6 (0.05/15274) considering the number of effective test (Tables 1–3) (Supplementary.csv files for each tissue). Covariances matrices are only available for the 44 tissues of GteX v6: adipose subcutaneous, adipose visceral omentum, adrenal gland, artery aorta, artery coronary, artery tibial, brain anterior cingulate cortex BA24, brain caudate basal ganglia, brain cerebellar hemisphere, brain cerebellum, brain cortex, brain frontal cortex BA9, brain hippocampus, brain hypothalamus, brain nucleus accumbens basal ganglia, brain putamen basal ganglia, breast mammary tissue, cells EBV-transformed lymphocytes, cells transformed fibroblasts, colon sigmoid, colon transverse, esophagus gastroesophageal junction, esophagus mucosa, esophagus muscularis, heart atrial appendage, heart left ventricle, liver, lung, muscle skeletal, nerve tibial, ovary, pancreas, pituitary, prostate, skin not sun-exposed suprapubic, skin sun-exposed lower leg, small intestine, terminal ileum, spleen, stomach, testis, thyroid, uterus, vagina, whole blood.
Table 1.
UTMOST Single tissue analysis (Brain tissues).
| Loci | Location hg19 | MinP (UTMOST) | Tissue | Z score | SNPs in model | Other associations GWAS, GBA and/or TWAS |
|---|---|---|---|---|---|---|
| PTPRE | chr10:129705325-129884164 | 1.53 × 10−6 | Brain cortex | −4.8 | 6 | – |
| CIPC* | chr14:77564601-77583630 | 3.82 × 10−6 | Brain hipoccampus | −4.62 | 19 | – |
| NKX2-2 | chr20:21491660-21494664 | 9.38 × 10−9 | Brain nucleo accubens basal ganglia | −5.74 | 62 | Grove et al./Alonso-Gonzalez et al. |
| PINX1* | chr8:10622884-10697299 | 3.82 × 10−6 | Brain nucleo accubens basal ganglia | 4.62 | 14 | Grove et al./Alonso-Gonzalez et al. (neighbour gene C8orf74) |
List of independent significant loci.
Bonferroni threshold <3.85 × 10−6.
GWAS genome-wide association study, SNP single nucleotide polymorphism, TWAS transcriptome-wide association study.
*Marginally significant.
Table 3.
UTMOST Cross tissue analysis.
| Loci | Location hg19 | MinP (UTMOST) | test score | Other associations GWAS, GBA and/or TWAS |
|---|---|---|---|---|
| BLK | chr8:11351521-11422108 | 2.85 × 10−6 | 12.3 | Grove et al. |
| NKX2-2 | chr20:21491660-21494664 | 3.7 × 10−7 | 13.9 | Grove et al./Alonso-Gonzalez et al. |
List of independent significant loci.
Bonferroni threshold = 3.27 × 10−6.
GWAS genome-wide association study, SNP single nucleotide polymorphism, TWAS transcriptome-wide association study.
Morpheus software (https://software.broadinstitute.org/morpheus/) was used to display the Z scores of UTMOST significant genes across GTeX Brain tissues. UTMOST significance as a Z score in brain tissues is ∼4.6. Gray squares in the heatmap indicate that the gene weights were not available in the target tissue (Fig. 1).
Fig. 1. Heatmap representing UTMOST significant genes across weight sets and brain areas.
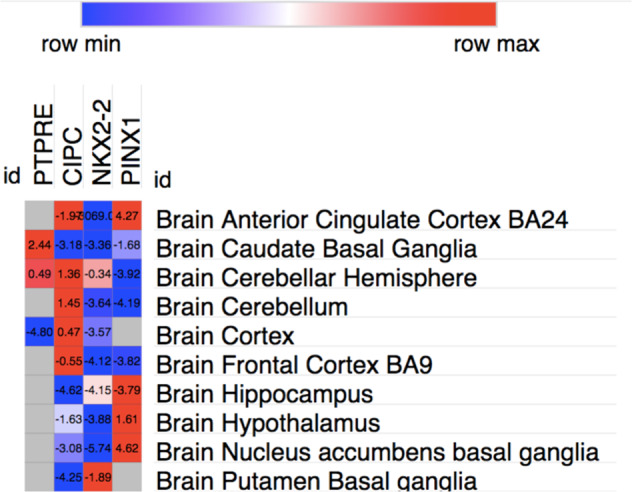
UTMOST significance as a Z score is ~4.6. Gray squares indicate that gene weights were not available in the target tissue.
DEG and gene expression analysis with FUMA
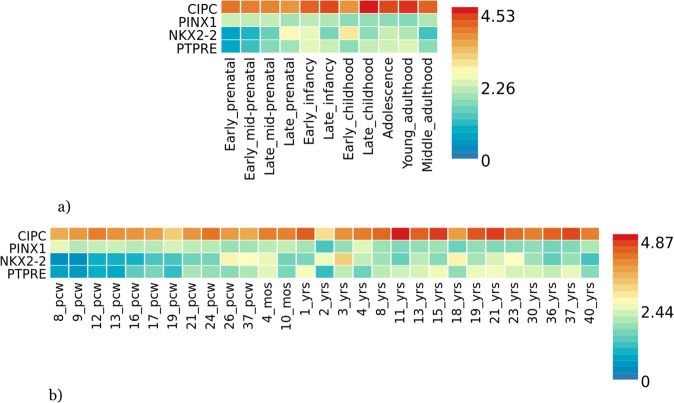
GENE2FUNC, a tool of FUMA8 (https://fuma.ctglab.nl/), was employed to carry out a gene expression heatmap and an enrichment analysis of differentially brain expressed genes (DEG) using BrainSpan RNA-seq data. Those genes represented in Table 4 (bold: PTPRE, CIPC, NKX2-2, PINX1) were used as an input. Expression values are TPM (Transcripts Per Million) for GTEx v6 and RPKM (Read Per Kilobase per Million). In order to define DEG sets, two-sided Student’s t-test were performed for these genes and per tissue against the different tissue types or developmental stages. Those genes with a p-value < 0.05 after Bonferroni correction and a log fold change ≥0.58 are defined as DEG. The direction of expression was considered. The −log10 (p-value) refers to the probability of the hypergeometric test DEG analysis was carried out creating differentially expressed genes for each expression data set (Fig. 2). Heatmaps display the normalized expression value (zero mean normalization of log2 transformed expression), and darker red means higher relative expression of that gene in each label, compared to a darker blue color in the same label (Fig. 3).
Table 4.
List of associated genes for each analysis (bold) and their predicted GeneMANIA interactors that were used as input for Metascape analysis.
| UTMOST Single brain | UTMOST Single Gastrointestinal | UTMOST Cross analysis |
|---|---|---|
| CIPC | NKX2-2 | NKX2-2 |
| PINX1 | MANBA | BLK |
| NKX2-2 | ERI1 | NKX6-1 |
| PTPRE | MITF | IAPP |
| NKX6-1 | NKX6-1 | MAFA |
| PTGES3 | TYRP1 | EGFR |
| DKC1 | TFE3 | PAX6 |
| IAPP | IAPP | OLIG2 |
| MAFA | TYR | PDX1 |
| TERT | MAFA | SYK |
| LMO4 | PAX6 | TRPV6 |
| GTPBP4 | TFEB | TDGF1 |
| SRC | LEF1 | CD79B |
| PDGFRB | HNRNPD | GAB1 |
| IL6R | GUSB | EPHA6 |
| PAX6 | PIAS3 | PLCG2 |
| IL6 | CREB3L4 | BLNK |
| OLIG2 | UBE2I | KIT |
| TERF1 | CREB3L3 | CD79A |
| PDX1 | CREB3L2 | NEUROG3 |
| HSP90AA1 | CREB3L1 | AR |
| SHC1 | TFEC | EPHA5 |
| DHX15 | SLBP | |
| CALM1 | OLIG2 |
Fig. 2. DEG analysis for PTPRE, CIPC, PINX1 and NKX2-2.
a DEG using BrainSpan 11 general developmental stages of brain samples and b DEG using BrainSpan 29 different ages of brain data. There are not significantly enriched DEG sets.
Fig. 3. Gene expression heatmaps for PTPRE, CIPC, PINX1 and NKX2-2.
a Gene expression heatmap using BrainSpan 11 general developmental stages of brain samples and b BrainSpan 29 differentes ages of brain data. Genes are ordered by expression clusteres and brain samples and ages by alphabetical order.
GeneMANIA and Metascape analysis
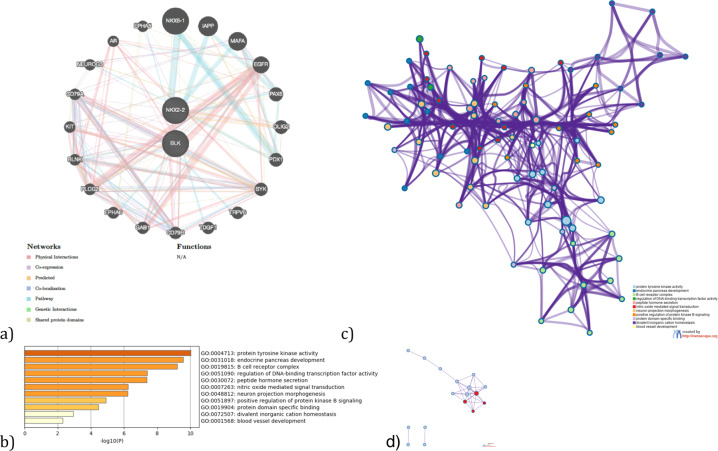
GeneMANIA9 (https://genemania.org/) was used to build a gene network for the UTMOST-associated genes by the single tissue analysis (brain and gastrointestinal tissues) and by the joint tissue analysis (Table 4). Each gene-network was subsequently analyzed with Metascape (https://metascape.org/)10 to carry out a pathway enrichment and a protein–protein interaction enrichment using the Evidence Counting (GPEC) prioritization tool. For each given gene list, pathway and process enrichment analysis has been carried out with the following ontology sources: GO biological processes, GO cellular components and GO molecular functions. The enrichment background includes all the genes in the genome. Terms with a p-value < 0.01, a minimum count of 3, and an enrichment factor >1.5 (the enrichment factor is the ratio between the observed counts and the counts expected by chance) are collected and grouped into clusters based on their membership similarities. More specifically, p-values are calculated based on the accumulative hypergeometric distribution, and q-values are calculated using the Benjamini–Hochberg procedure to account for multiple testings. Kappa scores are used as the similarity metric when performing hierarchical clustering on the enriched terms, and sub-trees with a similarity of >0.3 are considered a cluster. We select the terms with the best p-values from each of the 20 clusters, with the constraint that there are no more than 15 terms per cluster and no more than 250 terms in total (Figs. 4–6). The network is visualized using Cytoscape, where each node represents an enriched term and is colored first by its cluster ID (or each given gene list, protein–protein interaction enrichment analysis has been carried out with the following databases: BioGrid6, InWeb_IM7, OmniPath8. The resultant network contains the subset of proteins that form physical interactions with at least one other member in the list. If the network contains between 3 and 500 proteins, the Molecular Complex Detection (MCODE) algorithm has been applied to identify densely connected network components. The MCODE networks identified for individual gene lists are shown in Figs. 4–6.
Fig. 4. Gene networks, enriched ontology clusters, and PPI interaction analysis fot the brain-associated genes (CIPC, PINX1, NKX2-2, and PTPRE) by UTMOST (single tissue analysis).
a GeneMania gene network. b Bar graph of enriched terms across input gene lists, colored by p-values. c Network of enriched terms: colored by cluster ID, where nodes that share the same cluster ID are typically close to each other. d Protein–protein interaction network and MCODE components identified in the gene lists. (MCODE1:red).
Fig. 6. Gene network, enriched ontology clusters, and PPI interaction analysis for the associated genes by UTMOST cross. tissue analysis (BLK and NKX2-2) and their interactors.
a GeneMania gene network. b Bar graph of enriched terms across input gene lists, colored by p-values. c Network of enriched terms: colored by cluster ID, where nodes that share the same cluster ID are typically close to each other. d Protein–protein interaction network and MCODE components identified in the gene lists (MCODE1:red).
Enhancer analysis (dbSUPER)
dbSUPER (https://asntech.org/dbsuper/) was used to perform an exploratory enhancer analysis for the UTMOST-associated genes (single-tissue analysis) and for those tissues (brain and gastrointestinal) available at dbSUPER. The parameters selected were: SEs ranking method (H3K27ac), the peak calling was done with MACS (version 1.4.1) with parameters -p 1e-9,-keep-dup = auto, -w -S -space = 50, the stitching distance was established at 12,5 kb, the TSS exclusive zone was set at ±2 kb and the enhancer gene assignment was done within a 50 kb window.
Results and discussion
UTMOST analyses and comparison with previous results
UTMOST single tissue analysis (Brain tissues) showed association of two loci, NKX2-2 and PTPRE, while other two loci, CIPC and PINX1, showed a marginal association according to Bonferroni threshold (Table 1, Supplementary.csv files). NKX2-2 was previously identified as an associated gene by two different GBA algorithms, MAGMA, and PASCAL3,4. The results obtained by UTMOST also serve to indicate the nucleus accumbens basal ganglia as the brain area in which these genes may be acting. Although the association of PTPRE was not obtained as such in previous analyses, the association of its neighboring gene, C8orf74, was noted in a previous study and one of the index SNPs in the latest ASD GWAS was located near PINX1 (rs10099100) (Supplementary Fig. 1a, b)3,4.
NKX2-2 was also marginally associated by UTMOST within non-brain tissue together with other genes. It should be noted that some genes are tissue-specific for gastric and intestinal tissues such as stomach, esophagus and colon (MANBA, ERI1, MITF, and NKX2-2). The association of NKX2-2 in colon is noteworthy because NKX2-2 was previously reported as an ASD risk gene and now it is highlighted again by UTMOST in brain tissues. It seems that NKX2-2 together with BLK may play a role in ASD etiology but not only at the brain level since UTMOST cross-tissue analysis also found association for both genes (Tables 2, 3, Supplementary Figs. 1a, 2a, b, Supplementary.csv files)
Table 2.
UTMOST Single tissue analysis (Non Brain tisues).
| Loci | Location hg19 | MinP (UTMOST) | Tissue | Z score | SNPs in model | Other associations GWAS, GBA and/or TWAS |
|---|---|---|---|---|---|---|
| NKX2-2* | chr20:21491660-21494664 | 5.28 × 10−6 | Colon transverse | −4.55 | 2 | Grove et al./Alonso-Gonzalez et al. |
| MANBA | chr4:103552643-103682151 | 3.2 × 10−6 | Esophagus muscularis | 4.66 | 47 | – |
| ERI1* | chr8:8860314-8890849 | 3.84 × 10−6 | Esophagus muscularis | 4.62 | 124 | – |
| DOK5 | chr20:53092266-53267710 | 4.86 × 10−7 | Liver | −409.74 | 4 | – |
| ATXN1 | chr6:16299343-16761721 | 2.4 × 10−6 | Nerve tibial | −4.72 | 27 | – |
| FERMT2 | chr14:53323989-53417815 | 3.1 × 10−6 | Skin not sun exposed | −5.49 | 14 | – |
| MITF | chr3:69788586-70017488 | 2.34 × 10−6 | Stomach | 4.72 | 111 | – |
| CTSB | chr8:11700034-11725646 | 3.36 × 10−6 | Thyroid | 4.65 | 66 | – |
| ANGEL1 | chr14:77253586-77279283 | 1.89 × 10−6 | Uterus | 4.76 | 83 | – |
List of independent significant loci.
Bonferroni threshold = 3.42 × 10−6.
Genes in bold are associated within gastrointestinal tissues.
GWAS genome-wide association study, SNP single nucleotide polymorphism, TWAS transcriptome-wide association study.
*Marginally significant.
To evaluate the importance of each brain tissue in ASD etiology, a secondary analysis was performed using Z scores values for each ASD-associated gene across GTeX brain tissues. The heatmap showed that there is a wide specificity of association in terms of Z score for each gene and brain tissue indicating the importance of conducting tissue specific analyses such as UTMOST (Fig. 1).
DEG analysis with GENE2FUNC tool (FUMA)
DEG Analysis with BrainSpan data (11 general developmental stages of brain samples and 29 different ages of brain samples) for the brain associated set of genes have not shown any significant result (Fig. 2a, b). However, CIPC have shown overexpression across every single developmental stage in comparison with the remaining ASD-associated genes (Fig. 3a, b).
GeneMANIA and Metascape analysis
GeneMANIA was used to find out the possible interactors with the associated genes (Table 4). We have proposed three different analyses. One based on the genes associated in brain tissues; another one based on the associated genes at a gastrointestinal level, given the associations pointed out by UTMOST and the previous implications of gastrointestinal abnormalities and symptoms in ASD and the lack of biological knowledge about them. The final analysis is focused on the genes identified by the cross-tissue analysis and their interactors. The general goal is to delineate the biological pathways underlying each group of genes and the differences between them.
Gene network, enriched ontology clusters, and PPI interaction analysis for the brain-associated genes (CIPC, PINX1, NKX2-2, and PTPRE) by UTMOST Single tissue analysis and their interactors highlights different biological pathways mainly involved in telomere maintenance and transcription regulation (Tables 5, 6 and Fig. 4). However, associated genes in gastrointestinal tissue (NKX2-2, MANBA, ERI1, and MITF) and their interactors mainly regulate DNA transcription by RNA polymerase II and the fate of oligodendrocytes. This is an interesting finding given the possible involvement of these cells in the enteric nervous system in ASD (Tables 7, 8 and Fig. 5). Finally, NKX2-2 and BLK both associated in the cross-tissue analysis, seem to point to very diverse biological pathways such as protein tyrosine kinase activity, B cell receptor complex, regulation of DNA-binding transcription factor activity, and neuron projection morphogenesis, among others (Tables 8–10 and Fig. 6).
Table 9.
Top 11 clusters with their representative enriched terms (one per cluster) fot the associated genes by UTMOST Cross Tissue Analysis (BLK and NKX2-2) and their predicted interactors.
| GO | Category | Description | Count | % | Log10(P) | Log10(q) |
|---|---|---|---|---|---|---|
| GO:0004713 | GO Molecular functions | Protein tyrosine kinase activity | 6 | 42.86 | −10.03 | −5.68 |
| GO:0031018 | GO Biological processes | Endocrine pancreas development | 5 | 25.00 | −9.57 | −5.64 |
| GO:0019815 | GO Cellular components | B cell receptor complex | 3 | 21.43 | −9.21 | −5.56 |
| GO:0051090 | GO Biological processes | Regulation of DNA-binding transcription factor activity | 7 | 35.00 | −7.40 | −4.63 |
| GO:0030072 | GO Biological processes | Peptide hormone secretion | 6 | 30.00 | −7.37 | −4.63 |
| GO:0007263 | GO Biological processes | Nitric oxide mediated signal transduction | 3 | 21.43 | −6.25 | −3.89 |
| GO:0048812 | GO Biological processes | Neuron projection morphogenesis | 7 | 35.00 | −6.21 | −3.87 |
| GO:0051897 | GO Biological processes | Positive regulation of protein kinase B signaling | 4 | 20.00 | −4.91 | −2.92 |
| GO:0019904 | GO Molecular functions | Protein domain specific binding | 5 | 35.71 | −4.45 | −2.58 |
| GO:0072507 | GO Biological processes | Divalent inorganic cation homeostasis | 4 | 18.18 | −2.92 | −1.26 |
| GO:0001568 | GO Biological processes | Blood vessel development | 4 | 18.18 | −2.31 | −0.69 |
“Count” is the number of genes in the user-provided lists with membership in the given ontology term. “%“ is the percentage of all of the provided genes that are found in the given ontology term (only input genes with at least one ontology term annotation are included in the calculation). “Log10(P)” is the p-value in log base 10. “Log10(q)” is the multi-test adjusted p-value in log base 10.
Table 5.
Top 11 clusters with their representative enriched terms (one per cluster) for the associated genes (CIPC PINX1, NKX2-2, and PTPRE) (Single tissue analysis (Brain)) and their interactors.
| GO | Category | Description | Count | % | Log10(P) | Log10(q) |
|---|---|---|---|---|---|---|
| GO:0007004 | GO Biological processes | Telomere maintenance via telomerase | 7 | 43.75 | −13.85 | −9.50 |
| GO:0031018 | GO Biological processes | Endocrine pancreas development | 6 | 26.09 | −11.54 | −8.33 |
| GO:0051347 | GO Biological processes | Positive regulation of transferase activity | 8 | 50.00 | −8.34 | −5.76 |
| GO:0070851 | GO Molecular functions | Growth factor receptor binding | 5 | 31.25 | −7.69 | −5.24 |
| GO:0031647 | GO Biological processes | Regulation of protein stability | 6 | 26.09 | −6.61 | −4.30 |
| GO:0051098 | GO Biological processes | Regulation of binding | 6 | 26.09 | −6.00 | −3.83 |
| GO:0010035 | GO Biological processes | Response to inorganic substance | 6 | 37.50 | −5.98 | −3.83 |
| GO:0019904 | GO Molecular functions | Protein domain specific binding | 6 | 37.50 | −5.40 | −3.36 |
| GO:0050999 | GO Biological processes | Regulation of nitric−oxide synthase activity | 3 | 18.75 | −5.31 | −3.29 |
| GO:0009636 | GO Biological processes | Response to toxic substance | 6 | 26.09 | −5.08 | −3.10 |
| GO:0019932 | GO Biological processes | Second-messenger-mediated signaling | 4 | 17.39 | −3.12 | −1.51 |
“Count” is the number of genes in the user-provided lists with membership in the given ontology term. “%“ is the percentage of all of the provided genes that are found in the given ontology term (only input genes with at least one ontology term annotation are included in the calculation). “Log10(P)” is the p-value in log base 10. “Log10(q)” is the multi-test adjusted p-value in log base 10.
Table 6.
Protein–protein interaction network and MCODE components identified in the gene lists for the associated genes (CIPC PINX1, NKX2-2, and PTPRE) (Single tissue analysis (Brain)) and their interactors.
| GO | Description | Log10(P) | MCODE | GO | Description | Log10(P) |
|---|---|---|---|---|---|---|
| GO:0007004 | Telomere maintenance via telomerase | −13.0 | MCODE_1 | GO:0007169 | Transmembrane receptor protein tyrosine kinase signaling pathway | −4.5 |
| GO:0006278 | RNA-dependent DNA biosynthetic process | −12.7 | MCODE_2 | GO:0007004 | Telomere maintenance via telomerase | −7.6 |
| MCODE_2 | GO:0006278 | RNA-dependent DNA biosynthetic process | −7.5 | |||
| GO:0010833 | Telomere maintenance via telomere lengthening | −12.5 | MCODE_2 | GO:0010833 | Telomere maintenance via telomere lengthening | −7.4 |
Table 7.
Top 4 clusters with their representative enriched terms (one per cluster) fot the associated genes by UTMOST Single tissue analysis (GI tissues) (NKX2-2, MANBA, ERI1, and MITF) and their predicted interactors.
| GO | Category | Description | Count | % | Log10(P) | Log10(q) |
|---|---|---|---|---|---|---|
| GO:0000978 | GO Molecular functions | RNA polymerase II proximal promoter sequence-specific DNA binding | 12 | 57.14 | −13.89 | −9.63 |
| GO:0021778 | GO Biological processes | Oligodendrocyte cell fate specification | 3 | 25.00 | −9.03 | −5.82 |
| GO:0048066 | GO Biological processes | Developmental pigmentation | 4 | 33.33 | −8.21 | −5.12 |
| GO:0008134 | GO Molecular functions | Transcription factor binding | 6 | 28.57 | −4.81 | −2.47 |
“Count” is the number of genes in the user-provided lists with membership in the given ontology term. “%“ is the percentage of all of the provided genes that are found in the given ontology term (only input genes with at least one ontology term annotation are included in the calculation). “Log10(P)” is the p-value in log base 10. “Log10(q)” is the multi-test adjusted p-value in log base 10.
Table 8.
Protein–protein interaction network and MCODE components identified in the gene lists for the GI associated genes (and their interactors.
| GO | Description | Log10(P) | MCODE | GO | Description | Log10(P) |
|---|---|---|---|---|---|---|
| GO:0000978 | RNA polymerase II proximal promoter sequence-specific DNA binding | −11.2 | MCODE_1 | GO:0035497 | cAMP response element binding | −9.8 |
| GO:0000987 | proximal promoter sequence-specific DNA binding | −11.0 | MCODE_1 | GO:0030968 | Endoplasmic reticulum unfolded protein response | −6.9 |
| GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | −10.8 | MCODE_1 | GO:0034620 | Cellular response to unfolded protein | −6.7 |
Fig. 5. Gene networks, enriched ontology clusters for the associated genes by UTMOST single tissue analysis (gastrointestinal tissues) and their predicted interactors (NKX2-2, MANBA, ERI1, MITF).
a GeneMania gene network. b Bar graph of enriched terms across input gene lists, colored by p-values. c Network of enriched terms: colored by cluster ID, where nodes that share the same cluster ID are typically close to each other.
Table 10.
Protein–protein interaction network and MCODE components identified in the gene lists for the cross analysis associated genes and their interactors.
| GO | Description | Log10(P) | MCODE | GO | Description | Log10(P) |
|---|---|---|---|---|---|---|
| GO:0030183 | B cell differentiation | −9.6 | MCODE_1 | GO:0030183 | B cell differentiation | −9.1 |
| GO:0004713 | Protein tyrosine kinase activity | −9.4 | ||||
| GO:0007169 | Transmembrane receptor protein tyrosine kinase signaling pathway | −9.4 | MCODE_1 | GO:0042113 | B cell activation | −7.5 |
| MCODE_1 | GO:0030098 | Lymphocyte differentiation | −7.3 |
Enhancer analysis (dbSUPER)
Given the tissue specificity given by UTMOST associations, we found interesting to perform an exploratory analysis of enhancers. According to the dbSUPER dabase only CIPC could work as a superenhancer in the brain middle hippocampus (SE_06106; chr14: 77562660-77607123, size: 44463 pb) (Supplementary Fig. 3).
As far as we know, this is the first study that has employed the UTMOST framework combined with the summary statistics of the largest ASD meta-analysis. The main aim was to identify ASD tissue-specific genes in brain and/or other tissues. It should be noted at the outset that gene-level associations identified by UTMOST do not imply causality. However, looking at the regional plots for each loci associated by UTMOST, the results shown at brain and gastrointestinal level seems pretty consistent. Thus, UTMOST has served to identify new ASD-associated genes in brain tissues as PTPRE and CIPC. Furthermore, UTMOST has been useful to confirm and obtain information on the tissue in which other known ASD risk genes such as NKX2-2 and PINX1 may have functional relevance. Altogether, brain-associated genes seems to point to three brain areas: cortex, hippocampus and nucleo accumbens. Previous studies demonstrated the ASD-associated gene expression in brain cortex11. In addition, hippocampus underlie some of the featured social memory and cognitive behaviors both crucial aspects in ASD12. The nucleus accumbens is a key brain area also related with the social reward response in ASD13. It should be also noted some limitations of our study. UTMOST uses GTeX v6 data by default and it should be interesting to re-run the analysis once the tool will be updated. Thus, UTMOST could find novel associated genes when expression data from other relevant ASD brain tissues as the amygdala are included. Another limitation of our results is the small number of SNPs considered by UTMOST to create the statistic in some genes (PTPRE in brain tissues and NKX2-2 in non-brain tissues analysis). However, we have been very conservative in establishing Bonferroni correction for all tissues in an analysis group. Thus, for example, we always chose the maximum number of genes tested in one of the brain tissues and applied this to calcule Bonferroni for the whole group of brain tissues as a whole.
In relation to the biological pathways in which UTMOST-associated genes are involved, our results open possible avenues for future genetic and functional studies. Thus, the functional role in ASD of CIPC, PINX1, NKX2-2, and PTPRE has not yet been characterized in detail. Metascape analysis have provided an insight revealing their involvement in telomerase maintenance and transcription regulation. Thus, PINX1 (PIN2 (TERF1) Interacting Telomerase Inhibitor 1) enhances TRF1 binding to telomeres and inhibits telomerase activity. It was proved that its silencing compromises telomere length maintenance in cancer cells14. In addition, it was recently found that children with ASD and sensory symptoms have shorter telomeres, compared to those children exhibiting a typical development15. CIPC (CLOCK interacting pacemaker) is a mammalian circadian clock protein. Recently, circadian rithms were pointed out as involved in brain development and they could underly ASD etiology due to its implication in behavioral processes. Circadian rythms are regulated through several transcription factors in different cellular types, fact that might be related with the GO terms associated with transcription regulation in this study16. Moreover, the result of CIPC as a predicted superenhancer highlights its possible functional repercussion.
UTMOST found association of several genes in non-brain tissues. However, we found really interesting to study those genes related to gastrointestinal tissues (NKX2-2, MANBA, ERI1, and MITF). There is a well-known and established comorbidity among gastrointestinal symptoms and ASD17. These clinical associations suggest the implication of gastrointestinal populations of neuronal cells. A mutation in NLGN3 (R451C) has been recently identified in two ASD brothers with GI symptoms. Mice models have demonstrated that R451C alter the number of neuronal cells in the small intestine and impact fecal microbes18. These evidence suggest that the role of NKX2-2, MANBA, ERI1, and MITF in gastrointestinal tissue should be further studied.
BLK and NKX2-2 are both associated in the cross-tissue analysis. The importance of UTMOST approach is that is able to show the association of two previously known ASD risk genes but not restricted to brain tissue. These findings lead to a difficult question whether autism can be a multisystemic disorder, something that has been recently pointed out by some authors19.
In conclusion, UTMOST, a novel single and cross-tissue TWAS, has revealed new ASD-associated genes. These genes have been characterized at the pathway and gene network level using bioinformatic approaches. However, future tissue-specific functional studies will be key to properly determine their role in ASD etiology.
Supplementary information
Acknowledgements
We thank the Psychiatric Genomics Consortium Autism Spectrum Disorder Working Group for making the ASD genome-wide association study results publicly available. Instituto de Salud Carlos III (ISCIII)/PI1900809/Cofinanciado FEDER.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Cristina Rodriguez-Fontenla, Angel Carracedo
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-021-01378-8.
References
- 1.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (American Psychiatric Publishing) (2013).
- 2.Sanders SJ, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grove J, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso-Gonzalez A, Calaza M, Rodriguez-Fontenla C, Carracedo A. Novel gene-based analysis of ASD GWAS: insight into the biological role of associated genes. Front. Genet. 2019;10:733. doi: 10.3389/fgene.2019.00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbeira AN, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gusev A, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y, et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat. Genet. 2019;51:568–576. doi: 10.1038/s41588-019-0345-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hitti FL, Siegelbaum SA. The hippocampal CA2 region is essential for social memory. Nature. 2014;508:88–92. doi: 10.1038/nature13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park HR, et al. A short review on the current understanding of autism spectrum disorders. Exp. Neurobiol. 2016;25:1–13. doi: 10.5607/en.2016.25.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, et al. Silencing PinX1 compromises telomere length maintenance as well as tumorigenicity in telomerase-positive human cancer cells. Cancer Res. 2009;69:75–83. doi: 10.1158/0008-5472.CAN-08-1393. [DOI] [PubMed] [Google Scholar]
- 15.Lewis, C. R. et al. Telomere length and autism spectrum disorder within the family: relationships with cognition and sensory symptoms. Autism Res. 13, 1094–1101 (2020). [DOI] [PubMed]
- 16.Geoffray M-M, Nicolas A, Speranza M, Georgieff N. Are circadian rhythms new pathways to understand Autism Spectrum Disorder? J. Physiol. Paris. 2016;110:434–438. doi: 10.1016/j.jphysparis.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Buie T, et al. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics. 2010;125:S1–18. doi: 10.1542/peds.2009-1878C. [DOI] [PubMed] [Google Scholar]
- 18.Hosie S, et al. Gastrointestinal dysfunction in patients and mice expressing the autism‐associated R451C mutation in neuroligin‐3. Autism Res. 2019;12:1043–1056. doi: 10.1002/aur.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thom RP, et al. Beyond the brain: A multi-system inflammatory subtype of autism spectrum disorder. Psychopharmacol. (Berl.) 2019;236:3045–3061. doi: 10.1007/s00213-019-05280-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.