Abstract
Severe fever with thrombocytopenia syndrome virus (SFTSV), the causative agent of a febrile human disease, was first identified from central and eastern provinces in China, and later in Japan and South Korea. Hubei Province is one of the major SFTS epidemic areas in the central part of China. This study reported the isolation of 11 new SFTSV strains from patients in Hubei Province collected in 2017. Extensive phylogenetic analyses were conducted based on the complete coding sequences of SFTSV segments including the new strains. It was suggested that five different SFTSV genotypes were circulating in Hubei, and 15 reassortment patterns and migration pathways correlated with each genotype were identified, which was more than previously recognized. Hubei Province was more involved in the evolutionary events of SFTSV than that previously thought in which the evolutionary events of SFTSV were reported to be independent from those in other epidemic regions. Further divergence of SFTSV strains was suggested by pairwise comparison of SFTSV sequences from each genotype and sequence identity normalized to representative strain in genotype C1. Subsequently, amino acid variations specific for genotype(s), strain(s), or cluster(s) were inspected, which may be related to differential biological activity of SFTSV strains/genotypes. In conclusion, we analyzed the current status of SFTSV phylogeny in Hubei Province and discussed the possible events correlated to SFTSV evolution. It provided an in-depth insight into SFTSV evolution, raising concerns for the use of proper SFTSV strains in future studies.
Electronic supplementary material
The online version of this article (10.1007/s12250-020-00289-0) contains supplementary material, which is available to authorized users.
Keywords: Severe fever with thrombocytopenia syndrome virus (SFTSV), Genotypes, Genomic reassortment, Virus migration, Amino acid variations
Introduction
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging severe human disease with presenting symptoms such as high fever and thrombocytopenia, accompanied by leucopenia, central nervous system symptoms, gastrointestinal symptoms, and even multiple organ failure (Yu et al. 2011). SFTS is characterized by a mortality rate ranging from 6% to 30%, depending on the geographic location (McMullan et al. 2012; Zhan et al. 2017). SFTS was initially reported in central and eastern regions in China and later discovered in South Korea and Japan (Reece et al. 2018). Occurrence of SFTS was recently suspected in Vietnam and Pakistan (Mahmood et al. 2019; Tran et al. 2019), indicating the potential of SFTSV wider distribution in the Asia. In China, the high numbers of SFTS cases occurred in Henan, Shandong, Anhui, and Hubei Provinces, accounting for over 90% of the total cases (Zhan et al. 2017). SFTS virus (SFTSV) is the causative agent of SFTS. SFTSV is an emerging virus belonging to the Dabie bandavirus species of Bandavirus genus, family Phenuiviridae according to the recent International Committee on Taxonomy of Viruses (ICTV) taxonomy (https://talk.ictvonline.org/taxonomy/). The SFTSV genome contains three segments: L segment encoding RNA-dependent RNA polymerase (RdRp), M segment encoding glycoprotein (GP), and S segment encoding non-structural protein (NSs) and nucleoprotein (NP) in reverse direction. SFTSV evolution might be associated with geographic distributions (Yoshikawa et al. 2015). Known strains can be divided into the Chinese lineage, containing most strains from China and a few from South Korea, and the Japanese lineage, containing strains isolated in South Korea and Japan, and a few strains isolated in China. The Chinese lineage could be divided into 4 genotypes (C1 to C4), while the Japanese lineage is divided into 3 genotypes (J1 to J3) (Shi et al. 2017; Yoshikawa et al. 2015). Genomic recombination and reassortment events are believed to play an important role in SFTSV evolution. Moreover, SFTSV evolution may also be affected by virus migration between epidemic areas in China, South Korea, and Japan (Lv et al. 2017; Shi et al. 2017).
In 2010, the first SFTS case was reported in Hubei Province in central China (Yu et al. 2011). All the strains initially identified in Hubei Province belonged to the C3 genotype (Shi et al. 2017). Later, in 2016, strains belonging to the C2 genotype were found in Hubei, suggesting the presence of multiple genotypes in the Province (Zhang et al. 2017). This finding also suggests that SFTSV strains are evolving in Hubei Province and likely also in other epidemic areas.
In this study, 11 new SFTSV strains isolated from patients in Hubei Province in 2017 were analyzed. We conducted extensive phylogenetic and genotypic analyses to characterize the new strains and investigate their genetic evolution and diversity.
Materials and Methods
Virus Isolation from the Blood Serum of SFTS Patients
Blood serum samples of 54 SFTS patients were collected in 2017 during the acute phase of the disease by the Department of Infectious Disease in Union Hospital (Wuhan City, Hubei Province). Patients were selected according to their clinical manifestations, routine blood tests, and detection of SFTSV by quantitative Reverse Transcription-PCR (RT-PCR) using the SFTSV probe qRT-PCR detection kit (Daan Gene, Guangzhou, China) according to the manufacturer’s instructions. Demographic features about gender, age, occupation, location, and the disease outcome of all patients enrolled in the study are summarized in Supplementary Table S1. Vero cells (ATCC number: CCL-81) were incubated with each sample to isolate the virus by blind passage, as previously described (Zhang et al. 2017). After three culture passages, indirect immunofluorescence assay (IFA) was performed to detect SFTSV antigen expression in cells and RT-PCR was carried out to detect SFTSV RNA in the cell culture supernatants, as previously described (Zhang et al. 2017). The complete genome of the isolated strains was sequenced using previously described primers and methods (Zhang et al. 2017). Personal information, clinical manifestations, and laboratory test results of each patient from whom SFTSV was isolated are listed in Table 1 and Supplementary Table S2.
Table 1.
Personal information, and laboratory tests for SFTS patients.
| Case1 | Case2 | Case3 | Case4 | Case5 | Case6 | Case7 | Case8 | Case9 | Case10 | Case11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Personal information | |||||||||||
| Gender | Female | Female | Female | Male | Male | Male | Male | Female | Male | Female | Male |
| Age | 54 | 43 | 34 | 72 | 69 | 69 | 76 | 47 | 72 | 66 | 45 |
| Location | XG | SZ | HG | HG | SZ | HG | HG | HG | HG | SZ | HG |
| Occupation | Farmer | Others | Civil servant | Farmer | Farmer | Farmer | Farmer | Others | Others | Farmer | Others |
| Outcome | Survived | Survived | Survived | Fatal | Lost at follow-up | Survived | Fatal | Survived | Fatal | Survived | Survived |
| Lab tests | |||||||||||
| Virus Load in serum sample (RNA copies/mL) | 1.41 × 104 | 1.14 × 103 | 3.69 × 105 | < 100 | 4.83 × 104 | < 100 | 6.94 × 104 | 1.4 × 103 | 5.57 × 106 | < 100 | 5.62 × 104 |
| Strains | HBXG51 | HBSZ52 | HBHG54 | HBHG8 | HBSZ11 | HBHG29 | HBHG30 | HBHG35 | HBHG36 | HBSZ55 | HBHG38 |
| Genotypes | |||||||||||
| L segment | C2 | C2 | C2 | C3 | C3 | C3 | C3 | C3 | C4 | C2 | C4 |
| M segment | C2 | C2 | C2 | C3 | C3 | C3 | C3 | C3 | C4 | C2 | C4 |
| S segment | C2 | C2 | C2 | C3 | C3 | C3 | C3 | C3 | C4 | C3 | C3 |
XG Xiaogan City, SZ Suizhou City, HG Huanggang City.
SFTSV Phylogeny, Reassortment, Migration, and Amino Acid Variations Analyses
To analyze SFTSV phylogeny, a dataset was established, including the complete SFTSV coding sequence (CDS) (L, M, and S segments) deposited in GenBank until December 2018 and the new strains reported in this study. In total, 287 L segment sequences, 309 M segment sequences, and 512 S segment sequences were analyzed. The GenBank accession numbers and locations of strains used for the analysis are shown in Supplementary Table S3. Phylogenetic trees were constructed using Mega 6.0, as previously described (Shi et al. 2017). Reassortment events were detected using the RDP software package as previously described (Shi et al. 2017). To analyze SFTSV migration, viral sequences were aligned by using Clustal Omega (https://www.ebi.ac.uk/), and phylogeography trees based on alignment of L, M, and S segments, respectively, were constructed by BEAST 2.3.x (Ancestral Reconstruction/Discrete Phylogeography method).
Pairwise comparison of nucleotide sequences of the RdRp, GP, NSs, and NP ORFs was performed using the Megalin software (Version 7.1) in the DNAStar package. Sequence identity was normalized using the C1 genotype SD4/China/2010 strain (GenBank number: HM802202-HM802204) as the reference. This strain was among the first batch of strains isolated from patients in 2010 when the SFTS disease was first reported (Yu et al. 2011). Sequence identity within each genotype and normalized against the SD4/China/2010 strain are shown using box-plot and violin-plot, respectively. Amino acid (aa) variations of the four SFTSV proteins (RdRp, GP, NP, and NSs) were analyzed by aligning all the protein sequences in the dataset using Clustal W. Variations associated with each SFTSV genotypes were plotted using ggtree and ggplot packages in R Studio (version 3.5.2). The relevant aa positions in the GP 3D structure (PDB code: Gn, 5Y10; Gc, 5G47) were visualized using Pymol (version 4.40).
Results
Isolation of 11 New SFTSV Strains from SFTS Patients
In 2017, 54 patients (34–77 years old) were diagnosed with SFTS in Union hospital of Huazhong University of Science and Technology in Wuhan, China. Twenty-six of them were men (48.1%), and about half of them were farmers (26, 48.1%). Most patients came from cities located in Hubei Province (44/54): 26 patients (48.1%) came from Huanggang City, 10 (18.5%) from Suizhou City, 6 (11.1%) from Xiaogan City, 1 (1.9%) from Jingmen City, and 1 (1.9%) from Wuhan City. The remaining 10 patients (18.5%) came from Xinyang City, located in Henan Province. Among the 54 patients, 3 died, 4 patients were lost at follow-up, and 47 patients recovered (Supplementary Table S1).
After three blind passages in Vero cells, SFTSV was isolated from the blood serum of 11 patients living in 3 cities of Hubei Province (Table 1 and Supplementary Figure S1). These patients had typical SFTS manifestations and the presence of the virus was confirmed by RNA detection (Table 1). Sequencing results showed that the 11 isolates (GenBank numbers: MT320787 to MT320819) were new strains, sharing a very high nucleotide identity (99.94%–95.53%) with previously isolated SFTSV strains. They were designated as strains HBHG8, HBSZ11, HBHG29, HBHG30, HBHG35, HBHG36, HBHG38, HBXG51, HBSZ52, HBHG54, and HBSZ55, according to the locations of the patients (Table 1) and were sent to the National Virus Resource Center for preservation (NVRC preservation numbers: IVCAS 6.7514 to IVCAS 6.7524).
Phylogenetic Analysis Suggests Genomic Reassortment Events Occurred in the New SFTSV Strains
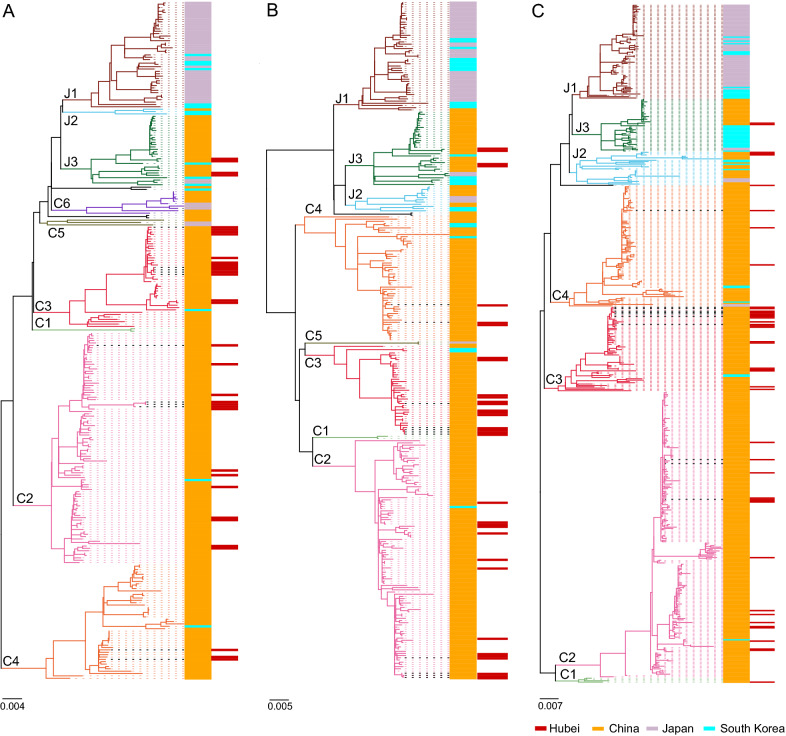
Extensive phylogenetic analyses of the L, M, and S segments of SFTSV strains were performed (Fig. 1). Of the 512 S segment sequences, 406 were derived from different provinces and areas in China. As revealed by the S segment tree in Fig. 1C, 368 (90.6%) of the 406 China-derived SFTSV strains belonged to the Chinese lineage, which is generally composed of the C1–C4 genotypes, while the other 38 strains (9.4%) from China belonged to the J2 and J3 genotypes of the Japanese lineage. We also found two additional genotypes (C5 and C6) in the L tree and one additional genotype (C5) in the M tree, as suggested in the previous study (Lv et al. 2017). Sixty-four of the 66 strains (96.9%) derived from Japan belonged mainly to the J1 genotype, with 2 strains (3.1%) belonging to the Chinese lineage. 42 of the 47 strains (89.4%) from South Korea mainly belonged to the Japanese lineage, with 5 strains (10.6%) belonging to different genotypes of the Chinese lineage. There have been 45 strains derived from Hubei Province including the 11 new strains in this study. Strains from Hubei were assigned to genotypes C2, C3, C4, and J3, both in the L and M trees (Fig. 1A and 1B) and to genotypes C1, C2, C3, C4, J2, and J3 in the S tree (Fig. 1C). This result suggests that multiple genotypes have been circulating in Hubei. Moreover, of the 11 new strains reported in this study, 3 strains (HBXG51, HBSZ52, HBHG54) belonged to genotype C2, 5 strains (HBHG8, HBSZ11, HBHG29, HBHG30, HBHG35) belonged to genotype C3, and 1 strain (HBHG36) belonged to genotype C4 (Fig. 1). According to the S tree, the remaining two strains, HBHG38 and HBSZ55, were assigned to the genotype C3. However, the M and L segments of HBHG38 belonged to genotype C4, while those of HBSZ55 belonged to genotype C2 (Table 1). This suggests the occurrence of genetic reassortment events among SFTSV strains isolated in the Hubei Province.
Fig. 1.
Phylogenetic analysis of SFTSV strains. The ML trees were constructed based on the sequence alignment of L, M, and S segments (A), (B) and (C), respectively. The 11 strains isolated in this study are indicated by bold black dotted lines. Genotypes (C1 to C6 and J1 to J3) are distinguished in branches of different colors. SFTSV strains from China, Japan, and South Korea are labeled in orange, light purple, and cyan, respectively. Strains from the Hubei Province are indicated in red in a different panel.
An Increased Rate of Genetic Reassortment and Migration Events May Affect SFTSV Evolution
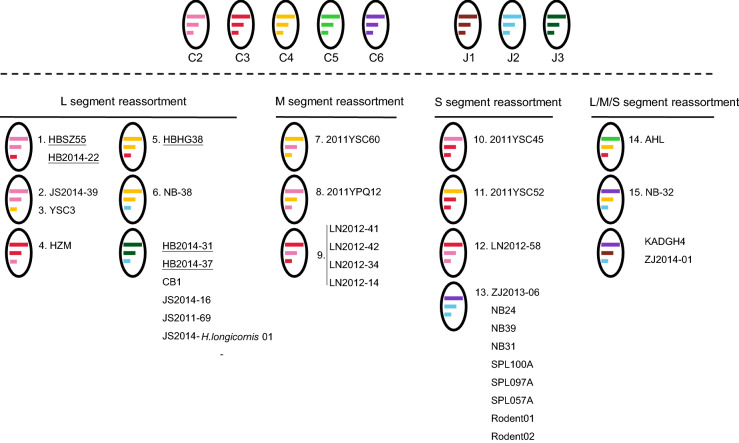
The L, M, and S trees had similar topological structures. However, 34 strains showed shifted evolutionary positions among the trees, which was consistent with potential reassortment events. Thirty strains had one segment belonging to one genotype and the other two segments belonging to a distinct genotype: 6 reassortment events involved in the S segment, 3 events in the M segment, and 4 events in the L segment (Fig. 2). In addition, 4 strains (AHL, NB-32, KADGH4, and ZJ2014-01) had all the three segments belonging to a different genotype (Supplementary Table S4 and Fig. 2). All genotypes, except for C1, were involved in reassortment events. Furthermore, 15 reassortment events of 18 strains were identified by using RDP packages, including 6 events involving the S segment, 3 events involving the M segment, 4 events involving the L segment, and 2 events involving more than one segment (Supplementary Table S5 and Fig. 2). Five strains from Hubei, including the HBHG38 and HBSZ55 strains, reported for the first time in this study, and the HB2014-22, HB2014-31, and HB2014-37 strains, which were reported in 2018 (Hu et al. 2018) were identified as potential reassortants, due to the discrepancy in their positions among the three phylogenetic trees (Supplementary Table S4 and Fig. 2). Moreover, the HBHG38 and HBSZ55 strains were identified as potential reassortants by using RDP packages (Supplementary Table S5 and Fig. 2). Our phylogenetic analysis of the available SFTSV strains and 11 new strains is to date the most comprehensive analysis of the evolutionary status of SFTSV.
Fig. 2.
Graphical representation of SFTSV reassortment events listed with the potential reassortants. Segments of each genotype are indicated by different colors. The L, M, and S segments are indicated by different line lengths. Reassortants from the Hubei Province are underlined. Reassortants identified by RDP packages are numbered in accordance with the events listed in Supplementary Table S5.
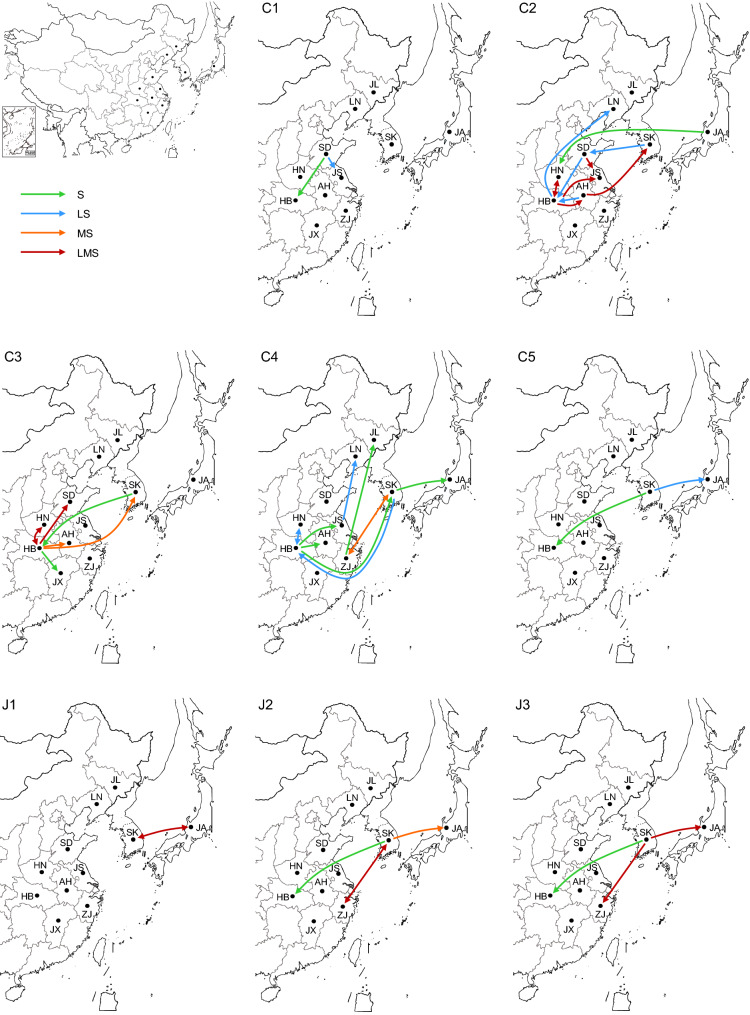
The analysis of the parental strain of the reassortants showed that the reassortment events could have occurred between strains isolated from the same province (event 1, 2, 3, 4, 6, 7, 10, and 15) as well as between strains isolated from two different areas (event 5, 9, 11, 12, 13, and 14) (Supplementary Table S5), suggesting that the occurrence of genetic exchange among different areas. As virus migration may play a role in the observed exchange (Shi et al. 2017), the migration pathways of each genotype were further characterized. Multiple migration pathways were found for C2 and C4 strains, whereas J1 strains migrated only between South Korea and Japan (Fig. 3 and Supplementary Table S6). Hubei and Shandong Provinces in China and South Korea were critical locations for SFTSV spreading. Viruses belonging to the C1, C2, and C3 genotypes were exported and imported in Shandong Province. Chinese genotypes (C1 to C5) are shown to spread from Hubei Province to adjacent provinces and other countries such as South Korea and Japan. Moreover, Hubei Province received all the known genotypes from other epidemic regions. South Korea was a critical location for the export and import of viruses mainly from the Japanese lineage (J1–J3 genotypes) and genotype C5, and was also involved in migration related to viruses of C2, C3, and C4 genotypes (Fig. 3 and Supplementary Table S6). These results suggest an increase in the migration of the virus in China and South Korea, which may have influenced the SFTSV evolution.
Fig. 3.
Migration pathways of the different SFTSV genotypes in China, Japan, and South Korea. Migration pathways of each SFTSV genotype were shown in an enlarged map showing areas including provinces in China, South Korea, and Japan. One-way arrows indicate export or import-only events, and double-sided arrows indicate a virus export and import pathway. Migrations involving the S segment only are shown in green, involving the L and S segments in blue, involving the M and S segments in orange, and involving all three segments in red.
Analysis of Sequence Identity and Amino Acid Variations of SFTSV Genotypes
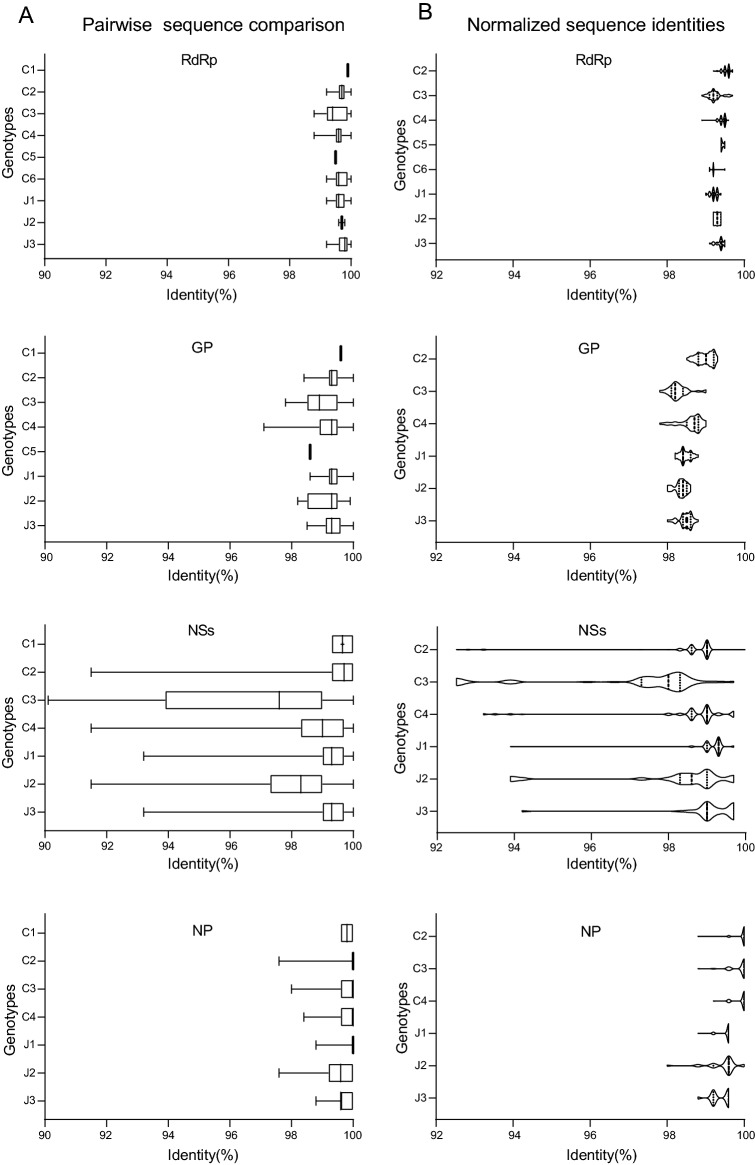
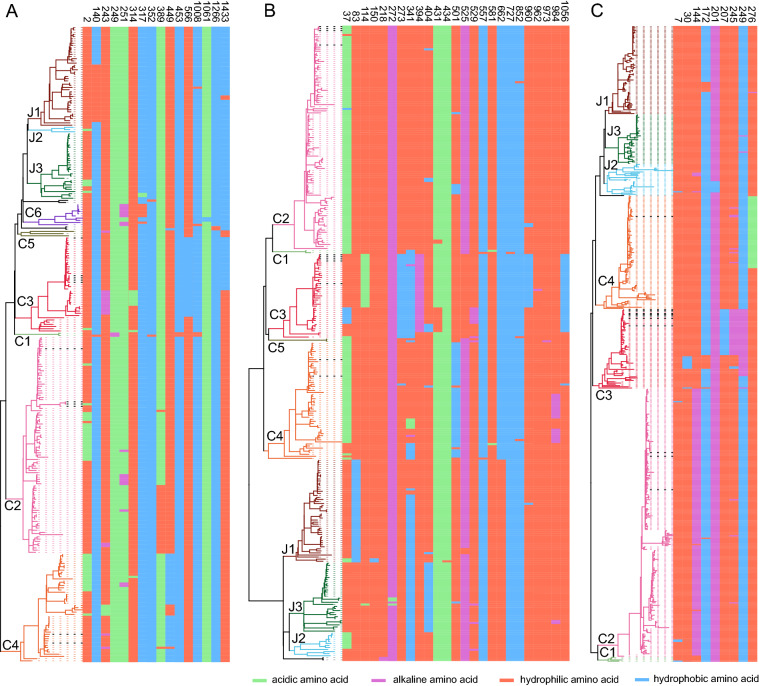
We further analyzed the genomic identity among the SFTSV genotypes. Pairwise comparison of RdRp, GP, NSs, and NP ORFs within each genotype showed that NSs is less conserved compared to other ORFs, probably because it is a non-structural protein with multiple functions. The C1 genotype comprises 2–4 strains according to the sequences involved in S, M, and L trees with a narrow range of sequence identity (99.3%–100%). Genotype C2 comprised the majority of SFTSV strains (98, 108, and 215 sequences involved in RdRp, GP, and both NSs and NP ORFs, respectively). However, in this genotype, the sequence identity is distributed in a narrow range from 91.5% to 100% differing for RdRp, GP, NSs, and NP ORFs. Despite a moderate number of strains (43, 41, and 66 sequences for analysis of the RdRp, GP, and both NSs and NP ORFs, respectively) belonged to the C3 genotype, it was characterized by a wider sequence identity range, particularly for RdRp (98.8%–100%), GP (97.8%–100%), NSs (90.1%–100%), and NP (98.0%–100%), suggesting a higher genetic divergence of this genotype. SFTSV strains in Genotype J2 is (3, 11, and 26 sequences for the analyses of RdRp, GP, and both NSs and NP ORFs, respectively) less than those in the J1 and J3 genotypes. However, it also presented a broader range of sequence identity for the GP (98.2%–99.9%), NSs (91.5%–100%), and NP (97.6%–100%) ORFs (Fig. 4A). These results suggested the presence of different genetic variations among SFTSV strains belonging to the same genotype. Sequence identities were normalized by comparing all strains to the C1 genotype SD4/China/2010 strain, which formed several clusters for most genotypes (Fig. 4B). It suggests the presence of genetic divergence in SFTSV strains belonging to the same genotype. Further, we identified hotspot mutation sites in RdRp, GP, NSs, and NP proteins involving aa substitutions changing the characteristic of the aa residue (hydrophilic, hydrophobic, acidic, and alkaline). We found 16 hotspot sites in RdRp, 25 sites in GP, and 10 sites in NSs (Fig. 5, Supplementary Table S7). If sites of the strain belonging to a certain genotype had amino acid substitutions of different characteristics, it is shown with colored patterns in Fig. 5. For example, in most strains belonging to C4 genotype, the aa in position 449 in the RdRp protein is hydrophobic, while strains belonging to other genotypes have a hydrophilic aa. Moreover, in the RdRp protein, most of the strains belonging to genotype C6 have an alkaline aa in position 251 and a hydrophilic aa in position 317, while other strains mostly have an acidic aa and a hydrophobic aa, respectively, at the same two sites (Fig. 5A). The GP protein had the highest numbers of hotspots: 16 sites were identified in the N-terminus fragment Gn and 9 in the C-terminus fragment Gc (Table S7, Supplementary Figure S2A). Interestingly, the C3 genotype presented different aa patterns in 5 different positions (273, 341, 394, 525, and 1056), distinct from any other genotype. Differential patterns within the C3 genotype were also observed at positions 31, 114, 404, 431, 529, and 960. This is consistent with the higher genetic divergence found for the C3 genotype (Fig. 4A). Specific patterns in the GP were also identified in other genotypes, albeit for a smaller number of positions. The C4 genotype showed specific patterns for most strains at positions 37 and 501 and, the J3 genotype has a hydrophobic aa in position 404 while other genotypes have a hydrophilic aa. Interestingly, we also found that viruses belonging to the Japanese lineage have a hydrophilic aa in position 662, while viruses belonging to the Chinese lineage have a hydrophobic aa, suggesting that SFTSV divergence could be affected by geographic distribution. Although the C2 genotype had the most number of strains, it also showed the most conserved pattern than the other genotypes (Fig. 5B). Genotype-specific patterns were also observed in the NSs protein (Fig. 5C). For example, in the C2 genotype, an alkaline aa is in position 144, while others have a hydrophilic aa. Variable patterns were also observed within the C3 genotype in positions 30, 172, 207, 245, and 249, more than any other genotype. This further supports the higher divergence of SFTSV strains belonging to the C3 genotype. NP was highly conserved among all the SFTSV strains. Besides the 51 hotspots of shifted aa characteristics identified in RdRp, GP, and NSs, other sites having aa mutations of same characteristic were also found (Supplementary Table S7). It is worth noting that, although aa substitution with shifted characteristics was not found in NP, positions 52 and 95 of almost all viruses of the Chinese lineage have a Lysine, while viruses of the Japanese lineage use Arginine (Supplementary Table S7). These results further suggest that SFTSV evolution appears to be correlated to geographic differentiation.
Fig. 4.
Comparison of genomic sequence identity of SFTSV strains. A Pairwise sequence comparison of SFTSV strains within the same genotype. The identity range is plotted as a box plot for each genotype. The upper and lower bars represent the maximum and minimum values, respectively. The box shows identities ranging from 25% to 75% of the total values, and the solid line in the middle indicates the median. B Normalized sequence identities of SFTSV strains to the C1 genotype SD4/China/2010 strain, plotted as violin plots. The median and quartile identities are indicated by “(solid dotted lines)” and “(dotted lines)”, respectively.
Fig. 5.
Analysis of hotspot sites for amino acid variations in (A) RdRp, (B) GP, and (C) NSs proteins among the indicated genotype. Substitutions are shown in different colors: green for acidic amino acids, purple for alkaline amino acids, red for hydrophilic amino acids, and blue for hydrophobic amino acids.
Discussion
The SFTS infectious disease was first reported in Hubei and Henan Provinces in 2010 and SFTSV was identified as the causative agent (Yu et al. 2011). Since then, an increasing number of SFTSV strains were detected and/or isolated from ticks, animal hosts, and human patients (Hu et al. 2018; Lee et al. 2019; Park et al. 2019; Yang et al. 2019; Zhang et al. 2011). Based on the comparison of SFTSV sequences, SFTSV evolution was found to be associated with geographic distribution (Yoshikawa et al. 2015). Further analyses of SFTSV phylogeny suggested the occurrence of reassortment and recombination events (Hu et al. 2018; Zhang et al. 2017). In 2017, Lv and his colleagues found two potential novel SFTSV genotypes (C6 and J4) (Lv et al. 2017). These findings suggested that SFTSV has always been evolving and resulted in diverging phylogeny.
Hubei Province is one of the major SFTS epidemic regions, accounting for more than 10% of the Chinese total cases (Sun et al. 2017). Initially, only one of the genotypes (C3) was found in Hubei Province, and import and export events were not reported there (Shi et al. 2017). However, in 2016, SFTSV strains belonging to the C2 genotype were identified in Hubei (Zhang et al. 2017). Later on, a retrospective study reported the presence of 4 different genotypes in Hubei, as early as in 2014 (Hu et al. 2018). In this study, we found an increased genetic diversity, with the co-prevalence of 6 genotypes in the region, according to the S tree analysis (Fig. 1C). As expected, genetic reassortment played an important role in SFTSV evolution. We found 35 potential reassortants, classified into 16 events (Fig. 2, Supplementary Table S4 and Table S5), of which 18 were further supported by RDP packages. Compared to a previous phylogenetic study (Lv et al. 2017), we found novel reassortants and more genotypes were involved in reassortment events. Moreover, we found that 31 reassortants had a single segment substitution, while 4 reassortants had all the three segments belonging to a different genotype. These results suggest an increasing genetic exchange among different SFTSV genotypes, resulting in an increased viral genetic diversity. This increase is likely caused by virus migration among different geographic locations. Genotype C2, C3, and C4 are characterized by more migrating pathways, suggesting that these genotypes might be more involved in genetic exchange events than other genotypes. Consistently with a previous study (Shi et al. 2017), we found that Shandong Province in China and South Korea are still critical regions for the import and export of SFTSV. Moreover, in this study, we found that most of the identified migration pathways are related to Hubei Province (Fig. 3 and Supplementary Table S6), in contrast to a previous 2016 study, which reported that the Hubei Province was isolated from other provinces (Shi et al. 2017). Collectively, our findings indicate that the SFTSV phylogeny has changed in recent years. It is unclear which factors might have contributed to the role of Hubei Province as a hub for SFTSV spread. Nevertheless, further investigation of the potential factors, such as human trafficking and transport of animals, and meteorological factors in the epidemic region, are needed.
Analyses of sequence identities and aa variations specific for genotype(s) and strain(s) revealed further genetic divergence of SFTSV. Most of the substitutions were found on GP, the structural glycoprotein critical for the virus entry into host cells, assembly of progeny viruses, and antibody recognition and neutralization by the host. These sites were mapped in the linear diagram and the 3D structures of Gn and Gc fragments (Supplementary Figure S2). Of the 16 sites located on Gn, four sites (37, 83, 114, and 150) are on the surface of Domain I (D I), one (218) on D II, and two (272 and 273) on D III. Of the 9 sites located on Gc, two sites (581 and 727) are on D I, two (662 and 852) on D II, and four (960, 962, 973, and 984) on D III (Supplementary Figure S2). Gn is critical for antibodies recognition and neutralization by the host (Halldorssona et al. 2016; Kim et al. 2019; Wu et al. 2017). So far, two neutralizing antibody epitopes (positions 247 to 272, and 284 to 290) were identified on Gn D III (Kim et al. 2019; Wu et al. 2017) and three other epitopes recognized by patient serum samples were recently identified (unpublished data). All the epitopes are in conserved regions, which do not contain any variation sites. This may indicate that SFTSV antigenicity could be consistent, although further dedicated studies are needed. Amino acid variations may affect protein function and virus properties. Of the variable sites identified in Gc, position 962 is known to be important for glycoprotein-mediated fusion ability. Replacement of Arginine (R) with Serine (S) at this site could induce a more efficient syncytium formation (Tani et al. 2019). In this position, all the analyzed strains use S, except the HB29 strain, which uses R (Fig. 5B, Supplementary Table S7). HB29 belongs to the C3 genotype and is often being used for in vitro studies, despite it exhibits no or a weak syncytium formation only (Tani et al. 2019). Therefore, the use of HB29 strain as an SFTSV model to investigate the entry, fusion, and other SFTSV-related capabilities need careful consideration. The impact of other amino acid variations on the virus function is still unclear. Likely, the reported amino acid variations in the RdRp, GP, NP, and NSs proteins could affect the protein function and thus have an impact on the regulation of SFTSV infection and proliferation. Therefore, further studies are needed to characterize the impact of these mutations on the efficiency of entry, replication, and viral production. These amino acid substitutions may determine a different biological activity of the different SFTSV genotypes. For this reason, the choice of SFTSV strain for future studies should be taken carefully. In conclusion, our results provide insights into SFTSV evolution and improve our knowledge of SFTSV epidemiology. The rapid evolution of SFTSV needs further studies to characterize the impact of genetic diversity on virus pathogenicity and immunogenicity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Mr. Zhong Zhang and Ms. Min Zhou for the help of cell culture. This work was supported by the National Key R&D Programme of China (2018ZX10734404-010), the Nature Science Foundation of Hubei Province (2019CFB790 and 2018CFB471), the Innovation Team Project of Hubei Provincial Health Commission (WJ2019C003), the Open Research Fund of State Key Laboratory of Virology (2018IOV004), and the Special Project of Technical Conditions (2060503).
Author Contributions
XW, FD, and SS conceived and designed the experiments; ML, BL, ML, LX, WZ, and CP collected samples and information, and performed clinical tests; XW, YZ, JS, YF, ZS, JW, QW, ST, and SS performed the experiments including data collection, virus isolation and identification, and phylogeny analyzing; WXL and SS wrote the manuscript; XZ, FD, and SS revised the manuscript. HLW, TZ, XZ, FD, and SS finalized the manuscript. All authors approved the final version of the manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they no conflict of interest.
Human and Animal Rights
The collection and tests with human serum samples were reviewed and approved both by the Ethics Committees of Wuhan Institute of Virology, Chinese Academy of Sciences (Approval Number: WIVH33201801) and the Ethics Committees of Tongji Medical College, HuaZhong University of Science and Technology (Approval Number: 2017-S326). Written informed content was provided by adult participants.
Contributor Information
Xin Zheng, Email: xinsunshine1011@aliyun.com.
Fei Deng, Email: df@wh.iov.cn.
Shu Shen, Email: shenshu@wh.iov.cn.
References
- Halldorssona S, Behrensb A, Harlosa K, Huiskonena J, Elliottc R, Crispinb M, Brennanc B, Bowdena T. Structure of a phleboviral envelope glycoprotein reveals a consolidated model of membrane fusion. PNAS. 2016;113:7154–7159. doi: 10.1073/pnas.1603827113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Cai K, Liu M, Li W, Xu J, Qiu F, Zhan J. Laboratory detection and molecular phylogenetic analysis of severe fever with thrombocytopenia syndrome virus in Hubei Province, central China. Arch Virol. 2018;163:3243–3254. doi: 10.1007/s00705-018-3993-5. [DOI] [PubMed] [Google Scholar]
- Kim K, Kim J, Ko M, Chun J, Kim H, Kim S, Min J, Park W, Oh M, Chung J. An anti-Gn glycoprotein antibody from a convalescent patient potently inhibits the infection of severe fever with thrombocytopenia syndrome virus. PLoS Pathog. 2019;15:e1007375. doi: 10.1371/journal.ppat.1007375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cho Y, Jeong H, Kim W, Son K, Kim J, Oh S, Jheong W, Chaeb J. Complete genome sequences of two severe fever with thrombocytopenia syndrome virus strains isolated from a human and a dog in the republic of Korea. Microbiol Resour Announc. 2019;8:e01695-18. doi: 10.1128/MRA.01695-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Zhang H, Tian L, Zhang R, Zhang Z, Li J, Tong Y, Fan H, Carr M, Shi W. Novel sub-lineages, recombinants and reassortants of severe fever with thrombocytopenia syndrome virus. Ticks Tick Borne Dis. 2017;8:385–390. doi: 10.1016/j.ttbdis.2016.12.015. [DOI] [PubMed] [Google Scholar]
- Mahmood N, Khurram M, Khan M, Umar M, Jalil A, Sharif S. Characteristics of probable severe fever with thrombocytopenia syndrome patients: a perspective study from Pakistan. Int J Med Devel Ctries. 2019;3:8. [Google Scholar]
- McMullan L, Folk S, Kelly A, MacNeil A, Goldsmith C, Metcalfe M, Batten B, Albariño C, Zaki S, Rollin P, Nicholson W, Nichol S. A new phlebovirus associated with severe febrile illness in Missouri. N Engl J Med. 2012;367:834–841. doi: 10.1056/NEJMoa1203378. [DOI] [PubMed] [Google Scholar]
- Park E, Shimojima M, Nagata N, Ami Y, Yoshikawa T, Iwata-Yoshikawa N, Fukushi S, Watanabe S, Kurosu T, Kataoka M, Okutani A, Kimura M, Imaoka K, Hanaki K, Suzuki T, Hasegawa H, Saijo M, Maeda K, Morikawa S. Severe fever with thrombocytopenia syndrome phlebovirus causes lethal viral hemorrhagic fever in cats. Sci Rep. 2019;9:11990. doi: 10.1038/s41598-019-48317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece L, Beasley D, Milligan G, Sarathy V, Barrett A. Current status of severe fever with thrombocytopenia syndrome vaccine development. Curr Opin Virol. 2018;29:72–78. doi: 10.1016/j.coviro.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Shi J, Hu S, Liu X, Yang J, Liu D, Wu L, Wang H, Hu Z, Deng F, Shen S. Migration, recombination, and reassortment are involved in the evolution of severe fever with thrombocytopenia syndrome bunyavirus. Infect Genet Evol. 2017;47:109–117. doi: 10.1016/j.meegid.2016.11.015. [DOI] [PubMed] [Google Scholar]
- Sun J, Lu L, Wu H, Yang J, Ren J, Liu Q. The changing epidemiological characteristics of severe fever with thrombocytopenia syndrome in China, 2011–2016. Sci Rep. 2017;7:9236. doi: 10.1038/s41598-017-08042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Kawachi K, Kimura M, Taniguchi S, Shimojima M, Fukushi S, Igarashi M, Morikawa S, Saijo M. Identification of the amino acid residue important for fusion of severe fever with thrombocytopenia syndrome virus glycoprotein. Virology. 2019;535:102–110. doi: 10.1016/j.virol.2019.06.014. [DOI] [PubMed] [Google Scholar]
- Tran X, Yun Y, Le Van An SHK, Kim S, Thao N, Man P, Yoo J, Heo S, Cho N, Lee K. Endemic severe fever with thrombocytopenia syndrome, Vietnam. Emerg Infect Dis. 2019;25:1029–1031. doi: 10.3201/eid2505.181463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhu Y, Gao F, Jiao Y, Oladejo BO, Chai Y, Bi Y, Lu S, Dong M, Zhang C, Huang G, Wong G, Li N, Zhang Y, Li Y, Feng WH, Shi Y, Liang M, Zhang R, Qi J, Gao GF. Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope. Proc Natl Acad Sci USA. 2017;114:E7564–E7573. doi: 10.1073/pnas.1705176114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhao Z, Hou G, Zhang C, Liu J, Xu L, Li W, Tan Z, Tu C, He B. Genomes and seroprevalence of severe fever with thrombocytopenia syndrome virus and Nairobi sheep disease virus in Haemaphysalis longicornis ticks and goats in Hubei, China. Virology. 2019;529:234–245. doi: 10.1016/j.virol.2019.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Shimojima M, Fukushi S, Tani H, Fukuma A, Taniguchi S, Singh H, Suda Y, Shirabe K, Toda S, Shimazu Y, Nomachi T, Gokuden M, Morimitsu T, Ando K, Yoshikawa A, Kan M, Uramoto M, Osako H, Kida K, Takimoto H, Kitamoto H, Terasoma F, Honda A, Maeda K, Takahashi T, Yamagishi T, Oishi K, Morikawa S, Saijo M. Phylogenetic and geographic relationships of severe fever with thrombocytopenia syndrome virus in China, South Korea, and Japan. J Infect Dis. 2015;212:889–898. doi: 10.1093/infdis/jiv144. [DOI] [PubMed] [Google Scholar]
- Yu X, Liang M, Zhang S, Liu Y, Li J, Sun Y, Zhang L, Zhang Q, Popov V, Li C, Qu J, Li Q, Zhang Y, Hai R, Wu W, Wang Q, Zhan F, Wang X, Kan B, Wang S, Wan K, Jing H, Lu J, Yin W, Zhou H, Guan X, Liu J, Bi Z, Liu G, Ren J, Wang H, Zhao Z, Song J, He J, Wan T, Zhang J, Fu X, Sun L, Dong X, Feng Z, Yang W, Hong T, Zhang Y, Walker D, Wang Y, Li DX. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med. 2011;364:1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J, Wang Q, Cheng J, Hu B, Li J, Zhan F, Song Y, Guo D. Current status of severe fever with thrombocytopenia syndrome in China. Virol Sin. 2017;32:51–62. doi: 10.1007/s12250-016-3931-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhou D, Xiong Y, Chen X, He Y, Sun Q, Yu B, Li J, Dai Y, Tian J, Qin X, Jin D, Cui Z, Luo X, Li W, Lu S, Wang W, Peng J, Guo W, Li M, Li Z, Zhang S, Chen C, Wang Y, Xu J. Hemorrhagic fever caused by a novel tick—borne Bunyavirus in Huaiyangshan, China. Chin J Epidemiol. 2011;32:209–220. [PubMed] [Google Scholar]
- Zhang Y, Shen S, Shi J, Su Z, Li M, Zhang W, Li M, Hu Z, Peng C, Zheng X, Deng F. Isolation, characterization, and phylogenic analysis of three new severe fever with thrombocytopenia syndrome bunyavirus strains derived from Hubei Province, China. Virol Sin. 2017;32:89–96. doi: 10.1007/s12250-017-3953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.