Abstract
H9N2 subtype avian influenza virus (AIV) is an influenza A virus that is widely spread throughout Asia, where it jeopardizes the poultry industry and provides genetic material for emerging human pathogens. To better understand the epidemicity and genetics of H9 subtype AIVs, we conducted active surveillance in live poultry markets (LPMs) in Hubei Province from 2013 to 2017. A total of 4798 samples were collected from apparent healthy poultry and environment. Real-time RT-PCR revealed that the positivity rate of influenza A was 26.6% (1275/4798), of which the H9 subtype accounted for 50.3% (641/1275) of the positive samples. Of the 132 H9N2 viral strains isolated, 48 representative strains were subjected to evolutionary analysis and genotyping. Phylogenetic analysis revealed that all H9N2 viral genes had 91.1%–100% nucleotide homology, clustered with genotype 57, and had high homology with human H9N2 viruses isolated from 2013 to 2017 in China. Using a nucleotide divergence cutoff of 95%, we identified ten distinct H9N2 genotypes that continued to change over time. Molecular analysis demonstrated that six H9N2 isolates had additional potential glycosylation sites at position 218 in the hemagglutinin protein, and all isolates had I155T and Q226L mutations. Moreover, 44 strains had A558V mutations in the PB2 protein and four had E627V mutations, along with H9N2 human infection strains A/Beijing/1/2016 and A/Beijing/1/2017. These results emphasize that the H9N2 influenza virus in Hubei continues to mutate and undergo mammalian adaptation changes, indicating the necessity of strengthening the surveillance of the AIV H9N2 subtype in LPMs.
Electronic supplementary material
The online version of this article (10.1007/s12250-020-00260-z) contains supplementary material, which is available to authorized users.
Keywords: Avian influenza, H9N2, Genetic variation, Phylogenetic analysis, Live poultry markets (LPMs)
Introduction
The influenza A virus has eight distinct gene segments that can be divided into 18 hemagglutinin (HA) and 11 neuraminidase (NA) subtypes, of which H1–H16 are found in birds and H17 and H18 are found in bats (Webster et al. 1992; Tong et al. 2013). H9N2, H5Nx, and H7N9 subtype influenza A viruses cause the most harm to the poultry industry; however, as H9N2 subtype avian influenza viruses (AIVs) have low pathogenicity in poultry, their control has not been prioritized (Li et al. 2014). The first H9N2 influenza virus was isolated in the United States in 1966 (Homme and Easterday 1970) and appeared in China’s Guangdong Province in 1994, where it has been widely prevalent since 1998 (Chen et al. 1994; Liu et al. 2003). The H9N2 influenza virus not only causes substantial damage to the poultry industry by direct infection or stimulating infection, but also has the potential to infect mammals, and occasionally humans, and cause a pandemic (Peiris et al. 1999; Fusaro et al. 2011).
Influenza viruses can produce new viruses via mutation, recombination, and reassortment. H9N2 influenza viruses can provide some or all of their internal genes to produce new lethal reassortants that could infect humans, such as H5N1, H7N9, H10N8, and H5N6 (Guan et al. 1999; Chen et al. 2014; Yang et al. 2015; Feng et al. 2016; Pu et al. 2017). Based on genetic and phylogenetic analyses, the HA gene of H9N2 influenza viruses can be divided into Eurasian and American avian lineages. The Eurasian avian lineage mainly consists of three distinct lineages: A/chicken/Beijing/1/94-like (BJ/94-like); A/quail/Hong Kong/G1/97-like (G1-like); and A/duck/Hong Kong/Y439/97 (Y439-like) (Sun et al. 2010). A recent study identified 117 different genotypes from 730 H9N2 influenza viruses in Chinese birds, mammals, and human hosts from 1994 to 2013 (Li et al. 2017). Continuous research on the genetic evolution of H9N2 avian influenza virus is of great significance.
Live poultry markets (LPMs) provide a convenient location for the reorganization of influenza viruses from different species of poultry (Liu et al. 2014). As human-infecting AIVs have a history of exposure in LPMs, influenza surveillance in LPMs plays a key role in public health security. Although the H9N2 virus has been studied in LPMs in other regions of China and short-term local studies have been carried out in Hubei Province (Zhou et al. 2012; Huang et al. 2015; Wu et al. 2015; Chen et al. 2016), the presence and genetic structure of H9N2 viruses in LPMs in Hubei Province is still poorly understood. This is the first study to perform a comprehensive genetic evolution analysis of H9N2 influenza viruses in LPMs in Hubei Province, central China. Our results revealed the prevalence and genetic characteristics of H9N2 influenza viruses. In addition, our findings can help assess the potential pandemic risks of H9N2 influenza viruses.
Materials and Methods
Sample Collection
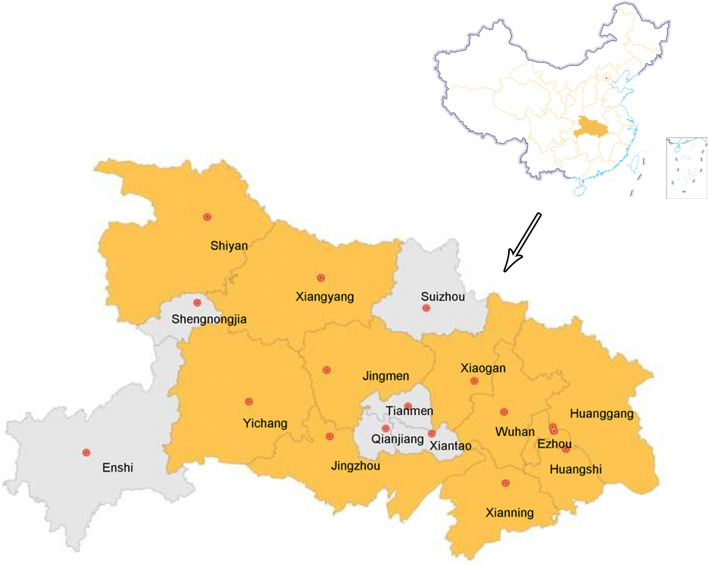
From October 2013 to December 2017, we carried out active surveillance in LPMs in 11 districts of Hubei Province, including: Wuhan, Huanggang, Huangshi, Ezhou, Jingzhou, Yichang, Xiangyang, Jingmen, Xianning, Xiaogan, and Shiyan (Fig. 1). A total of 4798 samples were collected from apparent healthy poultry, including 4342 swab samples (3036 chickens, 1261 ducks, 35 pigeons, 10 geese) and 456 environmental samples (Table 1). Throat and cloaca swabs from the same bird were placed in one preservation tube according to previously described sampling methods (Deng et al. 2013). Poultry samples were collected in duplicate: one to detect AIV RNA and the other to inoculate chicken embryos. Each sample was placed in phosphate-buffered saline (PBS, pH 7.4) containing 10% glycerol, ampicillin (2000 U/mL), gentamycin (2000 μg/mL), and streptomycin (2000 μg/mL). The samples were immediately cryopreserved in the laboratory and stored at −70 °C.
Fig. 1.
Map of sampling sites in Hubei Province, China. The eleven districts where samples were collected are shown in yellow.
Table 1.
Detection of influenza A viruses by real-time RT-PCR in samples from live poultry markets in Hubei Province.
| Samples (n) | Influenza A (%) | H5 (%) | H7 (%) | H9 (%) | H5 + H9 (%) | |
|---|---|---|---|---|---|---|
| Chicken | 3036 | 763 (25.1) | 54 (1.9) | 10 (0.3) | 575 (18.9) | 63 (2.1) |
| Duck | 1261 | 399 (31.6) | 79 (6.2) | 0 | 43 (3.4) | 61 (4.8) |
| Pigeon | 35 | 2 (5.7) | 0 | 0 | 2 (5.7) | 0 |
| Goose | 10 | 0 | 0 | 0 | 0 | 0 |
| Environment | 456 | 111 (24.3) | 16 (3.5) | 7 (1.5) | 21 (4.6) | 34 (7.4) |
| Total | 4798 | 1275 (26.6) | 149 (3.1) | 17 (0.4) | 641 (13.4) | 158 (3.3) |
Detection of AIV RNA
RNA was extracted from the collected samples using a Thermo KingFisher Flex Purification System (ThermoFisher, Vantaa, Finland) with a Mag Pure Viral Nucleic Acid KF Kit (Mag, Guangzhou, China). Influenza A, H5, H7, and H9 subtypes were detected using TransScriptII Probe One-Step qRT-PCR SuperMix (Transgene, Beijing, China). Primers and probes were selected based on the Chinese National Influenza Center (CNIC) national influenza monitoring technical guide and the World Health Organization (WHO) information for the molecular diagnosis of influenza virus (CNIC 2011; WHO 2013, 2014). Samples with a cycle threshold (Ct) value of < 35 were considered positive.
Virus Isolation and Sequencing
To isolate the viruses, H9-subtype positive samples were inoculated into 9-day-old specific pathogen-free chicken embryos, and allantoic fluid was harvested as described previously (Deng et al. 2013) and tested for HA activity. HA-positive samples were characterized by real-time RT-PCR (CNIC 2011; WHO 2013, 2014). Briefly, viral RNA was extracted from allantoic fluid using a High Pure Viral RNA kit (Roche, Mannheim, Germany), and the surface genes and internal genes coding sequences were amplified by RT-PCR using PrimeScript™ One Step RT-PCR Kit Ver. 2 (TaKaRa, Dalian, China) with specific primers (Hoffmann et al. 2001). RT-PCR was performed in a 50 μL reaction mix containing 5 μL of RNA, 25 μL of PrimeScript One step Enzyme Mix, 14 μL of RNase-Free dH2O, 2 μL of 10 pmol forward primer, and 2 μL of 10 pmol reverse primer. Amplification parameters were as follows: initial step of 30 min at 50 °C, then 5 min at 94 °C, 40 cycles of 30 s at 94 °C, 30 s at 50 °C, and 2 min at 72 °C, followed by 10 min extension at 72 °C. The amplified products were sent to Shanghai Sangon Biotech for sequencing. AIV isolation and identification were conducted in Physical Containment Level 2 Plus laboratory.
Genetic and Phylogenetic Analyses
Nucleotide sequences were edited using the SeqMan module in DNAstar (7.0), and phylogenetic analyses were performed using MAGE 6.0 with the neighbor-joining algorithm and bootstrap analysis with 1000 replicates. Multiple sequence alignments were compiled using Clustal W (Tamura et al. 2013). Phylogenetic analyses were based on the following protein coding nucleotide sequences: PB2, 49-2289; PB1, 25-2233; polymerase acidic (PA), 25–2151; HA, 43-1572; nucleoprotein (NP), 46-1527; NA, 20–1408; matrix (M), 26-1001; and nonstructural (NS), 29-859. Potential HA glycosylation sites were predicted using NetNGlyc1.0 (https://www.cbs.dtu.dk/services/NetNGlyc/). The “group” of each segment of H9N2 influenza viruses in the study was categorized using 95% sequence identity cutoffs refers to previously described (Deng et al. 2013). Genotyping was determined by the combination of group assignments of each of eight segments (Li et al. 2014). Reference strain sequences were obtained from the Influenza Virus Database (https://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi). Nucleotide sequences were deposited in GenBank under accession numbers MN647131 to MN647514 (Supplementary Table S1).
Results
Prevalence of Influenza A Virus in Hubei Province
From October 2013 to December 2017, we collected 4798 swabs and environmental samples from LPMs throughout Hubei Province (Central China) and screened the samples for influenza A H5, H7, and H9 subtypes using real-time RT-PCR. A total of 1275 samples were positive for influenza A (26.6%), of which 641 were positive for the H9 subtype (13.4%), 149 were positive for the H5 subtype (3.1%), 158 were positive for the H5 + H9 subtype (3.3%), and 17 were positive for the H7 subtype (Table 1).
Phylogenetic Analysis and Genotyping of H9N2 Subtype
After molecular identification and virus isolation, 132 H9 subtype viruses were isolated and their HA and NA genes were sequenced. H9N2 subtype influenza viruses were the main subtype, and there were mixed infections. Highly homologous strains were removed based on sampling time, location, and poultry breed, leaving 48 H9N2 influenza viruses that underwent full-segment genetic analysis. The reference sequences included classic H9N2 strains, three genotype 57 strains, all full-length human H9N2 influenza virus strains from 1999 to 2017, and other reference strains.
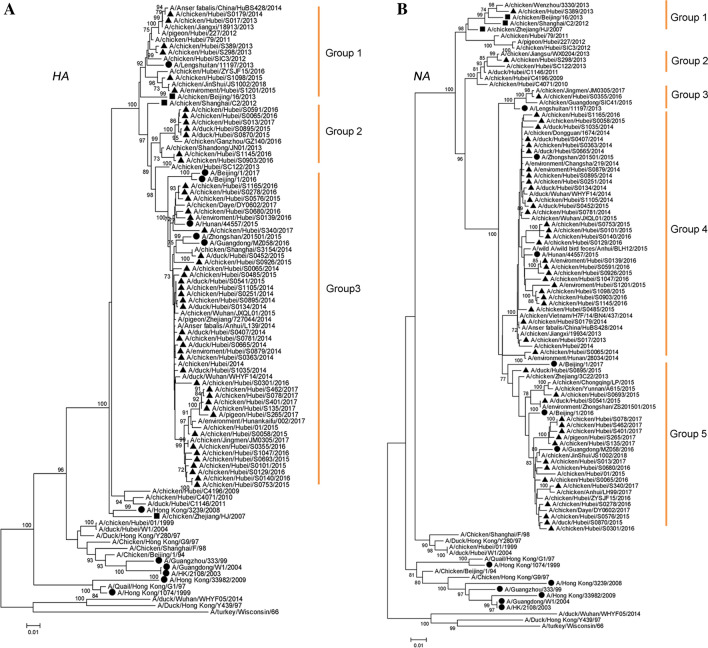
To better understand the genetic evolution of the H9N2 AIVs isolated from LPMs, we performed phylogenetic analysis on all eight genes of the 48 H9N2 isolates. Genetic evolution analysis revealed that all gene segments of the 48 H9N2 viruses belonged to the Eurasian lineage and genotype 57. The surface genes of all H9N2 viruses analyzed in the study were highly homologous with the human strains after 2013, while the HA gene shared 93.4%–99.9% identity at the nucleotide level and formed three phylogenetic groups (Groups 1–3). Group 1 contained six isolates and human origin strain A/Lengshuitan/11197/2013 (H9N2). Group 2 contained seven isolates. Group 3 contained 35 isolates and five H9N2 human strains (Fig. 2A). The NA genes shared 91.1%–100% identity at the nucleotide level and formed five phylogenetic groups (Groups 1–5). Groups 1, 2, and 3 each contained one isolate and group 3 contained the human strain A/Lengshuitan/11197/2013 (H9N2), while groups 4 and 5 contained the other five H9N2 human strains (Fig. 2B).
Fig. 2.
Phylogenetic analysis of the surface genes of H9N2 avian influenza viruses collected from live poultry markets in Hubei Province between 2013 to 2017. A HA gene. B NA gene. Solid triangles indicate strains isolated in this study. Solid circles indicate H9N2 strains that caused human infection. Solid squares indicate genotype 57 strains. For each node, bootstrap values ≥ 70% are shown. The tree was created using the neighbor-joining method and bootstrapped with 1000 replicates using MEGA6.0.
The internal genes displayed respective nucleotide homologies of 94.4%–100% for the PB2 gene, 94.8%–99.9% for the PB1 gene, 93.2%–100% for the PA gene, 93.6%–100% for the NP gene, 95.3%–100% for the M gene, and 94.3%–100% for the NS gene. Based on their nucleotide identity, the PA and NP genes were divided into two phylogenetic groups, while the PB2, PB1, M, and NS genes were categorized into one phylogenetic group. Group 2 of the PA gene contained only one A/chicken/Hubei/S389/2013 isolate (H9N2). The internal genes of all H9N2 viruses were clustered into genotype 57, whose representative strain is A/chicken/Zhejiang/HJ/2007 (H9N2). These results indicated that the internal genes of the H9N2 viruses were highly homologous with the human H9N2 strains from 2013 to 2017 and AIVs such as H7N9, H10N8, H5N2, H5N6, and H10N6 (Supplementary Figure S1 and S2).
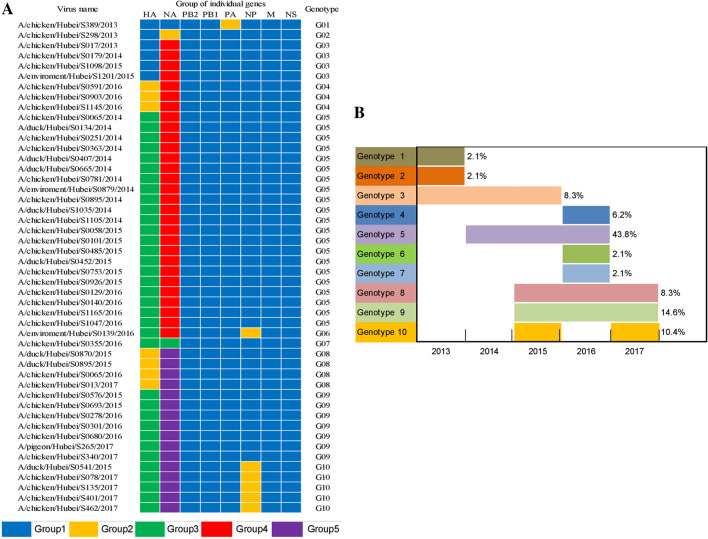
Based on the 95% genetic diversity of each segment, the 48 H9N2 viruses in this study were divided into ten genotypes (Fig. 3A). Genotypes 1 (2013), 2 (2013), 6 (2016), and 7 (2016) each contained one isolate and were thus deemed to be transient genotypes. Genotype 4 only appeared in 2016. Genotype 5 (2014–2016) was the main genotype, accounting for 43.8% of the isolated strains. Genotypes 8 and 9 existed in 2015–2017, whereas genotype 10 was detected in 2015 and 2017, with a significant change in its NP gene (Fig. 3B). These findings indicated that the genotypes of H9N2 influenza viruses continued to change over time.
Fig. 3.
The genotypes and timeline of 48 H9N2 viruses collected in Hubei Province from 2013 to 2017. A The genotypes of 48 H9N2 viruses. B Timeline.
Molecular Characterization
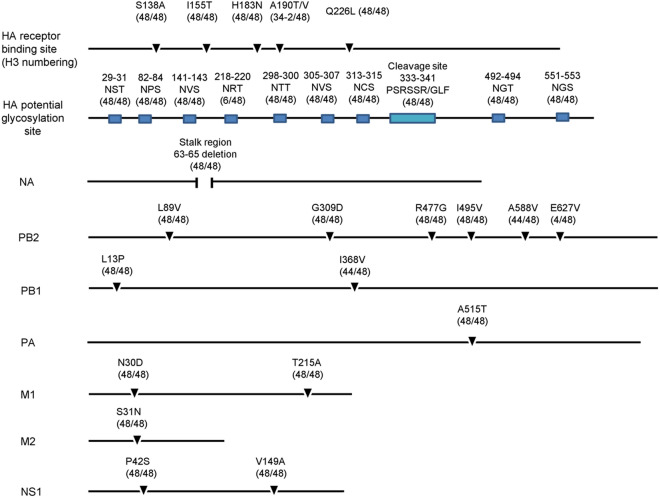
All H9N2 isolates possessed a “PSRSSR/GLF” motif at the HA cleavage site (Fig. 4), which is a typical characteristic of low pathogenicity in chickens (Guo et al. 2000; Ge et al. 2009). The HA amino acid receptor binding sites were S138A, I155T, H183N, A190V/T, Q226L, 227M, and 228G (H3 numbering, as used throughout the manuscript). I155T and Q226L mutations were detected in all strains, which favor the affinity of influenza viruses for human-type receptors (Matrosovich et al. 2001; Wan et al. 2008). In all viruses, HA has eight known potential glycosylation sites (Asn-X-Ser/Thr, where X =/=P) at positions 29, 82, 141, 298, 305, 313, 492, and 551 (Wu et al. 2015; Zhu et al. 2018). Six of the H9N2 viruses in this study had an additional potential glycosylation site at position 218, which was detected in the human H9N2 influenza viruses A/Hong Kong/1074/1999 and A/Hong Kong/3239/2008 (Supplementary Table S2A).
Fig. 4.
Summary of molecular features of H9N2 isolates. Key amino acids and their change frequencies are indicated.
In NA, three amino acids (sites 63–65) were deleted at the stalk region in all virus strains in this study, as described previously (Huang et al. 2015; Wu et al. 2015; Ge et al. 2018; Wang et al. 2018). However, no H274Y or R292K substitutions were observed, indicating that isolated viruses would be sensitive to NA inhibitors such as oseltamivir (Hurt et al. 2009) (Supplementary Table S2B).
In PB2, the A588V mutation, which is a new marker of mammalian AIV adaptation, was detected in 44 of the 48 H9N2 virus strains (Xiao et al. 2016). L89V, G309D, R477G, and I495V mutations were also detected in PB2, which have been shown to increase AIV virulence in mice (Li et al. 2009). No E627K or D701N mutations were detected in PB2; however, four viruses possessed an E627V mutant, which was identified in human H9N2 influenza virus A/Beijing/1/2016 and A/Beijing/1/2017.
In PB1, L13P and 368V mutation were found. A515T was found in PA. The mutations N30D and T215A in the M1 and the P42S and V149A substitutions in NS1 were detected in all strains, which associated with increased virulence in mice (Fan et al. 2009; Li et al. 2006; Jiao et al. 2008). In addition, S31N was identified in M2 in all isolates, suggesting that these viruses were resistant to antiviral drugs (Belshe et al. 1988). (Supplementary Table S2B). The results indicated that the H9N2 avian influenza virus in this study had undergone mutations adapted to mammals (Fig. 4).
Discussion
The H9N2 virus poses a threat to public health as it can act as a genetic donor and directly infect humans as an emerging human influenza virus (Gu et al. 2017). In this study, we found that the rate of avian influenza A virus positivity in LPMs was 26.57% and H9N2 was the main subtype, consistent with a previous report from Wuhan in Hubei Province over the same period (Chen et al. 2016). In addition, the positivity rate in chickens was higher than that in waterfowl duck, and mixed infections of different influenza viruses were observed, as reported previously (Wu et al. 2015). As such, LPMs likely played a key role in the emergence of new influenza viruses, and disinfection should be used to stop the spread of the disease.
H9N2 influenza virus has low pathogenicity in birds and humans; however, its gene segments can reassort with other influenza viruses with the potential to produce new virus epidemics that can cause greater harm. The H5N6, H7N9, and H10N8 subtypes that infect humans possess all the H9N2 influenza virus internal genes (Gao et al. 2013; Qi et al. 2014; Shen et al. 2016), while the PB2 and PB1 genes of early human H5N1 virus infections are highly homologous to those in H9N2 (Guan et al. 1999). In this study, we found that the internal genes of the H9N2 isolates were highly homologous with many other influenza viruses, posing the risk of a new influenza virus epidemic. Genotype 57 appeared in China in 2007 and dominated from 2010 to 2013 (Pu et al. 2015; Li et al. 2017), providing a genetic source for more than ten influenza reassortants, including H7N9 and H10N8, and thus posing a potential influenza pandemic concern (Pu et al. 2015). The H9N2 viral genes in this study belonged to genotype 57 and were divided into ten genotypes based on 95% nucleotide homology, indicating a high degree of genetic diversity. Studies carried out over the same period in Hunan and Jiangxi Provinces found that the isolated H9N2 viruses also belonged to genotype 57 (Huang et al. 2015; Han et al. 2018). Recently, new H9N2 genotypes were identified from environmental samples in China (Zou et al. 2019). In this study, the surface genes of the H9N2 isolates displayed great genetic diversity; however, there were only a few changes in the internal genes and continuous changes in genotypes. Together, these results indicate that the H9N2 AIV in Hubei continues to evolve and should be given high priority.
HA is an AIV receptor binding and membrane fusion glycoprotein that is the main inducer of neutralizing antibodies against viral infection (Skehel and Wiley 2000). HA receptor binding preferences play an important role in the replication and spread of influenza viruses. For instance, the characteristics of H9N2 viruses in southern China from 2009 to 2013 indicated that natural chicken-derived H9N2 isolates gradually gained preference for human α-2,6 sialic acid receptors, with several variants displaying airborne transmission in ferrets (Li et al. 2014). Previous studies have shown that the HA I155T mutation is important for the binding of H9N2 to mammalian receptors (Li et al. 2014). In this study, all H9N2 strains had I155T mutations, indicating that all these viruses had changes that enable mammalian infection. The HA Q226L mutation, which has been shown to contribute to the binding between human receptor and the H9N2 virus (Vines et al. 1998), was detected in all H9N2 isolates in our study. Thus, our findings indicate that H9N2 has the potential for cross-species transmission.
The E627K and D701N mutations in influenza virus PB2 proteins are critical for enhancing mammalian pathogenicity (Chen et al. 2013; Nieto et al. 2017). Previous studies have shown that H7N9 and H10N8 viruses readily acquire E627K or D701N mutations in PB2 during human replication (Qi et al. 2014; Nieto et al. 2017; Qi et al. 2018), and PB2 E627K mutations have recently been discovered in wild birds for the first time (Ge et al. 2018). Although no E627K or D701N mutations were identified in PB2 in this study, four of the isolates had E627V mutations similar to those in the H9N2 influenza virus that infected humans in 2016 and 2017. A recent study found that PB2 E627V mutations could increase H9N2 virus replication and enhance its pathogenicity (Arai et al. 2019). Similarly, we found that most isolates in this study had recently acquired the mammalian adaptation mutation A588V in PB2 protein, which can enhance polymerase activity, virus replication, and virulence of influenza H7N9, H10N8, and H9N2 viruses since 2013.
In conclusion, we conducted an epidemiological survey of AIVs in LPMs in Hubei Province and performed genetic analysis on representative H9N2 isolated strains. We revealed that the H9N2 subtype virus is the dominant influenza virus in LPMs and mixed infections are prevalent, posing a greater risk of genetic recombination to generate new influenza viruses. Phylogenetic analysis indicated that all H9N2 influenza viruses in this study were divided into multiple genotypes that change over time. Their internal genes were highly homologous with H7N9, H10N8, H5N6, and other influenza viruses that infect humans, thus may act as a gene donor to promote the emergence of new influenza viruses. Multiple specific amino acid mutations were observed in HA and PB2 proteins, among which the E627V mutation in PB2 was also identified in the H9N2 influenza strains that infected humans in 2016 and 2017, indicating that H9N2 has the potential for cross-species transmission. Together, our findings demonstrate that the H9N2 subtype influenza virus in central China continues to mutate and displays the potential for human infection. Subsequent studies on the biological characteristics of the isolates are required to further assess the risk of infection. In addition, the surveillance of H9N2 avian influenza virus should be strengthened to elucidate the genetic characteristics.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Dr. Yushan Chen from Wuhan University of Science and Technology for her help with data analysis. This study was supported by the National Key Research and Development Program of China (Grant No. 2016YFD0500800 to ZL).
Author Contributions
DRB and NS designed and coordinated the study. ZH, XJ, JX, JH, and HX collected samples. ZH, FP, and ZX carried out most of the experiments. ZH, TL, YS, and ZL analyzed the data. ZH, WZ, and ZL wrote and finalized the manuscript. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
The study was approved by the Hubei Center for Animal Disease Control and Prevention. All institutional and national guidelines for the care and use of animals were followed.
Contributor Information
Nianhua Song, Email: snh082@163.com.
Zili Li, Email: lizili@mail.hzau.edu.cn.
References
- Arai Y, Kawashita N, Ibrahim MS, Elgendy EM, Daidoji T, Ono T, Takagi T, Nakaya T, Matsumoto K, Watanabe Y. PB2 mutations arising during H9N2 influenza evolution in the middle east confer enhanced replication and growth in mammals. PLoS Pathog. 2019;15:e1007919. doi: 10.1371/journal.ppat.1007919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshe RB, Smith MH, Hall CB, Betts R, Hay AJ. Genetic basis of resistance to rimantadine emerging during treatment of influenza virus infection. J Virol. 1988;62:1508–1512. doi: 10.1128/jvi.62.5.1508-1512.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Zhang Z, Chen W. The study of avian influenza: I. The isolation and preliminary serological identification of avian influenza virus in chicken. Chin J Vet Med. 1994;20:3–5. [Google Scholar]
- Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, Yao H, Wo J, Fang Q, Cui D, Li Y, Yao X, Zhang Y, Wu H, Zheng S, Diao H, Xia S, Zhang Y, Chan KH, Tsoi HW, Teng JL, Song W, Wang P, Lau SY, Zheng M, Chan JF, To KK, Chen H, Li L, Yuen KY. Human infections with the emerging avian influenza a H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381:1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, Zou S, Yang L, Chen T, Dong L, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang S, Zhang Y, Li H, Gong T, Shi Y, Ni X, Li J, Zhou J, Fan J, Wu J, Zhou X, Hu M, Wan J, Yang W, Li D, Wu G, Feng Z, Gao GF, Wang Y, Jin Q, Liu M, Shu Y. Clinical and epidemiological characteristics of a fatal case of avian influenza a H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- Chen LJ, Lin XD, Guo WP, Tian JH, Wang W, Ying XH, Wang MR, Yu B, Yang ZQ, Shi M, Holmes EC, Zhang YZ. Diversity and evolution of avian influenza viruses in live poultry markets, free-range poultry and wild wetland birds in China. J Gen Virol. 2016;97:844–854. doi: 10.1099/jgv.0.000399. [DOI] [PubMed] [Google Scholar]
- Chinese National Influenza Center (CNIC) (2011) national influenza monitoring technical guide. Available: http://www.chinaivdc.cn/cnic/zyzx/jcfa/201605/t20160520_129702.htm. Accessed 9 Aug 2011
- Deng G, Tan D, Shi J, Cui P, Jiang Y, Liu L, Tian G, Kawaoka Y, Li C, Chen H. Complex reassortment of multiple subtypes of avian influenza viruses in domestic ducks at the Dongting Lake Region of China. J Virol. 2013;87:9452–9462. doi: 10.1128/JVI.00776-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Deng G, Song J, Tian G, Suo Y, Jiang Y, Guan Y, Bu Z, Kawaoka Y, Chen H. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology. 2009;384:28–32. doi: 10.1016/j.virol.2008.11.044. [DOI] [PubMed] [Google Scholar]
- Feng Y, Guan W, Yuan B, Wang Y, Li Z, Song Y, Li S, Yang Z, Zhong N, Zhang Y, Xia X. Emergence of triple-subtype reassortants of fatal human H5N6 avian influenza virus in Yunnan, China. J Infect. 2016;72:753–756. doi: 10.1016/j.jinf.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Fusaro A, Monne I, Salviato A, Valastro V, Schivo A, Amarin NM, Gonzalez C, Ismail MM, Al-Ankari AR, Al-Blowi MH, Khan OA, Maken Ali AS, Hedayati A, Garcia Garcia J, Ziay GM, Shoushtari A, Al Qahtani KN, Capua I, Holmes EC, Cattoli G. Phylogeography and evolutionary history of reassortant H9N2 viruses with potential human health implications. J Virol. 2011;85:8413–8421. doi: 10.1128/JVI.00219-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. Human infection with a novel Avian-origin Influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Ge FF, Zhou JP, Liu J, Wang J, Zhang WY, Sheng LP, Xu F, Ju HB, Sun QY, Liu PH. Genetic evolution of H9 subtype influenza viruses from live poultry markets in Shanghai, China. J Clin Microbiol. 2009;47:3294–3300. doi: 10.1128/JCM.00355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Yao Q, Chai H, Hua Y, Deng G, Chen H. A 627 K variant in the PB2 protein of H9 subtype influenza virus in wild birds. Influenza Other Respir Viruses. 2018;12:728–741. doi: 10.1111/irv.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Xu L, Wang X, Liu X. Current situation of H9N2 subtype avian influenza in China. Vet Res. 2017;48:49. doi: 10.1186/s13567-017-0453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YJ, Krauss S, Senne DA, Mo IP, Lo KS, Xiong XP, Norwood M, Shortridge KF, Webster RG, Guan Y. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- Han L, He W, Yan H, Li X, Wang C, Shi Q, Zhou T, Dong G. The evolution and molecular characteristics of H9N2 avian influenza viruses in Jiangxi of China. J Med Virol. 2018;91(4):711–716. doi: 10.1002/jmv.25363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- Homme PJ, Easterday BC. Avian influenza virus infections. I. Characteristics of influenza A/Turkey/Wisconsin/1996 virus. Avian Dis. 1970;14:66–74. [PubMed] [Google Scholar]
- Huang Y, Zhang H, Li X, Hu S, Cai L, Sun Q, Li W, Deng Z, Xiang X, Zhang H, Li F, Gao L. Detection and genetic characteristics of H9N2 avian influenza viruses from live poultry markets in Hunan Province, China. PLoS One. 2015;10:e0142584. doi: 10.1371/journal.pone.0142584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt AC, Holien JK, Parker MW, Barr IG. Oseltamivir resistance and the H274Y neuraminidase mutation in seasonal, pandemic and highly pathogenic influenza viruses. Drugs. 2009;69:2523–2531. doi: 10.2165/11531450-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Jiao P, Tian G, Li Y, Deng G, Jiang Y, Liu C, Liu W, Bu Z, Kawaoka Y, Chen H. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol. 2008;82(3):1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jiang Y, Jiao P, Wang A, Zhao F, Tian G, Wang X, Yu K, Bu Z, Chen H. The nS1 gene contributes to the virulence of H5N1 avian influenza viruses. J Virol. 2006;80(22):11115–11123. doi: 10.1128/JVI.00993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ishaq M, Prudence M, Xi X, Hu T, Liu Q, Guo D. Single mutation at the amino acid position 627 of PB2 that leads to increased virulence of an H5N1 avian influenza virus during adaptation in mice can be compensated by multiple mutations at other sites of PB2. Virus Res. 2009;144:123–129. doi: 10.1016/j.virusres.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Li X, Shi J, Guo J, Deng G, Zhang Q, Wang J, He X, Wang K, Chen J, Li Y, Fan J, Kong H, Gu C, Guan Y, Suzuki Y, Kawaoka Y, Liu L, Jiang Y, Tian G, Li Y, Bu Z, Chen H. Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 avian influenza viruses. PLoS Pathog. 2014;10:e1004508. doi: 10.1371/journal.ppat.1004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang S, Bing G, Carter RA, Wang Z, Wang J, Wang C, Wang L, Wu G, Webster RG, Wang Y, Sun H, Sun Y, Liu J, Pu J. Genetic evolution of influenza H9N2 viruses isolated from various hosts in China from 1994 to 2013. Emerg Microbes Infect. 2017;6:e106. doi: 10.1038/emi.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu X, Cheng J, Peng D, Jia L, Huang Y. Phylogenetic analysis of the hemagglutinin genes of twenty-six avian influenza viruses of subtype H9N2 isolated from chickens in China during 1996–2001. Avian Dis. 2003;47:116–127. doi: 10.1637/0005-2086(2003)047[0116:PAOTHG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Liu D, Shi W, Gao GF. Poultry carrying H9N2 act as incubators for novel human avian influenza viruses. Lancet. 2014;383:869. doi: 10.1016/S0140-6736(14)60386-X. [DOI] [PubMed] [Google Scholar]
- Matrosovich MN, Krauss S, Webster RG. H9N2 influenza a viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281:156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- Nieto A, Pozo F, Vidal-Garcia M, Omenaca M, Casas I, Falcon A. Identification of rare PB2-D701NMutation from a patient with severe influenza: contribution of the PB2-D701NMutation to the pathogenicity of human influenza. Front Microbiol. 2017;8:575. doi: 10.3389/fmicb.2017.00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, Xu G, Sun H, Wang M, Wen C, Wei Y, Wang D, Zhu B, Lemmon G, Jiao Y, Duan S, Wang Q, Du Q, Sun M, Bao J, Sun Y, Zhao J, Zhang H, Wu G, Liu J, Webster RG. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci USA. 2015;112:548–553. doi: 10.1073/pnas.1422456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Sun H, Qu Y, Wang C, Gao W, Zhu J, Sun Y, Bi Y, Huang Y, Chang KC, Cui J, Liu J. M gene reassortment in H9N2 influenza virus promotes early infection and replication: contribution to rising virus prevalence in chickens in China. J Virol. 2017 doi: 10.1128/JVI.02055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Zhou X, Shi W, Huang L, Xia W, Liu D, Li H, Chen S, Lei F, Cao L, Wu J, He F, Song W, Li Q, Li H, Liao M, Liu M. Genesis of the novel human-infecting influenza A(H10N8) virus and potential genetic diversity of the virus in poultry, China. Euro Surveill. 2014;19:20841. doi: 10.2807/1560-7917.es2014.19.25.20841. [DOI] [PubMed] [Google Scholar]
- Qi W, Jia W, Liu D, Li J, Bi Y, Xie S, Li B, Hu T, Du Y, Xing L, Zhang J, Zhang F, Wei X, Eden JS, Li H, Tian H, Li W, Su G, Lao G, Xu C, Xu B, Liu W, Zhang G, Ren T, Holmes EC, Cui J, Shi W, Gao GF, Liao M. Emergence and adaptation of a novel highly pathogenic H7N9 influenza virus in birds and humans from a 2013 human-infecting low-pathogenic ancestor. J Virol. 2018 doi: 10.1128/JVI.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YY, Ke CW, Li Q, Yuan RY, Xiang D, Jia WX, Yu YD, Liu L, Huang C, Qi WB, Sikkema R, Wu J, Koopmans M, Liao M. Novel reassortant avian influenza A(H5N6) viruses in humans, Guangdong, China, 2015. Emerg Infect Dis. 2016;22:1507–1509. doi: 10.3201/eid2208.160146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Sun Y, Pu J, Jiang Z, Guan T, Xia Y, Xu Q, Liu L, Ma B, Tian F, Brown EG, Liu J. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Vet Microbiol. 2010;146:215–225. doi: 10.1016/j.vetmic.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. New World Bats Harbor Diverse Iinfluenza AViruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vines A, Wells K, Matrosovich M, Castrucci MR, Ito T, Kawaoka Y. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J Virol. 1998;72:7626–7631. doi: 10.1128/jvi.72.9.7626-7631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, Stevens J, Cattoli G, Capua I, Chen LM, Donis RO, Busch J, Paulson JC, Brockwell C, Webby R, Blanco J, Al-Natour MQ, Perez DR. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS ONE. 2008;3:e2923. doi: 10.1371/journal.pone.0002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Wang J, Bi Y, Fan D, Liu H, Luo N, Yang Z, Wang S, Chen W, Wang J, Xu S, Chen J, Zhang Y, Yin Y. Characterization of avian influenza H9N2 viruses isolated from ostriches (Struthio camelus) Sci Rep. 2018;8:2273. doi: 10.1038/s41598-018-20645-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) (2013) Real-time RT-PCR Protocol for the detection of Avian Influenza A (H7N9) virus. Available: https://www.who.int/influenza/gisrs_laboratory/cnic_realtime_rt_pcr_protocol_a_h7n9.pdf?ua=1. Accessed Apr 2013
- World Health Organization (WHO) (2014) WHO information for molecular diagnosis of influenza virus—update. Available: https://www.who.int/influenza/gisrs_laboratory/molecular_diagnosis/en/. Accessed Mar 2014
- Wu H, Peng X, Peng X, Cheng L, Lu X, Jin C, Xie T, Yao H, Wu N. Genetic and molecular characterization of H9N2 and H5 avian influenza viruses from live poultry markets in Zhejiang Province, Eastern China. Sci Rep. 2015;5:17508. doi: 10.1038/srep17508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Ma W, Sun N, Huang L, Li Y, Zeng Z, Wen Y, Zhang Z, Li H, Li Q, Yu Y, Zheng Y, Liu S, Hu P, Zhang X, Ning Z, Qi W, Liao M. PB2-588 V promotes the mammalian adaptation of H10N8, H7N9 and H9N2 avian influenza viruses. Sci Rep. 2016;6:19474. doi: 10.1038/srep19474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZF, Mok CK, Peiris JS, Zhong NS. Human infection with a Novel Avian Influenza A (H5N6) virus. N Engl J Med. 2015;373:487–489. doi: 10.1056/NEJMc1502983. [DOI] [PubMed] [Google Scholar]
- Zhou JP, Ge FF, Liu J, Ju HB, Yang DQ, Wang J, Zhang WY, Liu PH. Epidemiological survey and genetic evolution of H9 subtype influenza viruses in Shanghai, China, from 2006 to 2010. Arch Virol. 2012;157:1193–1198. doi: 10.1007/s00705-012-1266-2. [DOI] [PubMed] [Google Scholar]
- Zhu R, Yang X, Zhang J, Xu D, Fan J, Shi H, Wang S, Liu X. Identification, sequence analysis, and infectivity of H9N2 avian influenza viruses isolated from geese. J Vet Sci. 2018;19:406–415. doi: 10.4142/jvs.2018.19.3.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Zhang Y, Li X, Bo H, Wei H, Dong L, Yang L, Dong J, Liu J, Shu Y, Wang D. Molecular characterization and receptor binding specificity of H9N2 avian influenza viruses based on poultry-related environmental surveillance in China between 2013 and 2016. Virology. 2019;529:135–143. doi: 10.1016/j.virol.2019.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.