Abstract
Despite their economic importance and well-characterized domestication syndrome, the genomic impact of domestication and the identification of variants underlying the domestication traits in Cucurbita species (pumpkins and squashes) is currently lacking. Cucurbita argyrosperma, also known as cushaw pumpkin or silver-seed gourd, is a Mexican crop consumed primarily for its seeds rather than fruit flesh. This makes it a good model to study Cucurbita domestication, as seeds were an essential component of early Mesoamerican diet and likely the first targets of human-guided selection in pumpkins and squashes. We obtained population-level data using tunable Genotype by Sequencing libraries for 192 individuals of the wild and domesticated subspecies of C. argyrosperma across Mexico. We also assembled the first high-quality wild Cucurbita genome. Comparative genomic analyses revealed several structural variants and presence/absence of genes related to domestication. Our results indicate a monophyletic origin of this domesticated crop in the lowlands of Jalisco. We found evidence of gene flow between the domesticated and wild subspecies, which likely alleviated the effects of the domestication bottleneck. We uncovered candidate domestication genes that are involved in the regulation of growth hormones, plant defense mechanisms, seed development, and germination. The presence of shared selected alleles with the closely related species Cucurbita moschata suggests domestication-related introgression between both taxa.
Subject terms: Population genetics, Genomics, Plant domestication
Introduction
Domestication is an evolutionary process where human societies select, modify and eventually assume control over the reproduction of useful organisms. A mutualistic relationship emerges from this interaction, where humans exploit a particular resource of interest, while the domesticated organism benefits from increased fitness and extended geographical range1,2. This is well illustrated in Cucurbita L. (pumpkins, squashes, and some gourds), where human-guided domestication and breeding have considerably extended their distribution despite the extinction of their natural dispersers (e.g., mastodons and similar megafauna)3. Today, Cucurbita stand as successful crops grown and consumed worldwide, with a global annual production of ~24 million tons4.
With ca. 21 taxa, the Cucurbita genus has experienced independent domestication events in five species5,6. Each Cucurbita crop experienced a unique selection for specific traits, predominantly defined by the nutritional and cultural needs of early human populations in America7. However, many domestication traits are common to domesticated Cucurbita, including the loss of bitter compounds (cucurbitacins), the loss of physical defense mechanisms (e.g., urticating trichomes), the loss of seed dormancy, the enlargement of fruits and seeds, and the diversification of fruit morphology4,8.
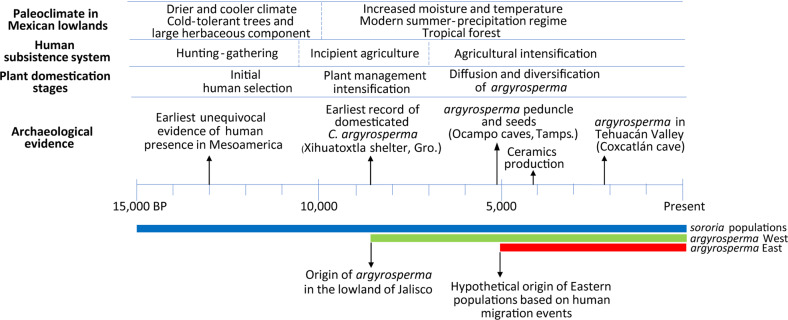
The initial steps of Cucurbita domestication were most likely directed towards seed rather than flesh consumption9. Seeds are rich in both carbohydrates and fatty acids, and cucurbitacins can be removed through boiling and washing; processes that are still employed for the consumption of wild Cucurbita seeds in Western Mexico7. Because the cultivation of C. argyrosperma (pipiana squash, cushaw pumpkin, or silver-seed gourd) is directed towards seed production rather than fruit flesh, it stands as an excellent model to investigate the early steps of Cucurbita domestication. Cucurbita argyrosperma subsp. argyrosperma (argyrosperma hereafter) was domesticated in Mesoamerica from its wild relative Cucurbita argyrosperma subsp. sororia (sororia hereafter), according to archaeological and genetic evidence6,10,11. Argyrosperma exhibits morphological differences from sororia, including larger fruits, larger seeds, and lack of urticating trichomes (Fig. 1). The earliest archaeological record of argyrosperma is presumed to be from 8700-year-old phytoliths in the Central Balsas Valley (Guerrero), although its taxonomic identity remains uncertain10. C. argyrosperma is a monoecious outcrossing species and gene flow has been previously described between the domesticated and wild subspecies12. Both subspecies are sympatric throughout the Pacific Coast of Mexico and Central America, with a few populations scattered in the coast of the Gulf of Mexico11,13. The domesticated taxon is also distributed in the Yucatan Peninsula, where its wild counterpart is absent11.
Fig. 1. Some morphological differences between C. argyrosperma subsp. sororia (left) and C. argyrosperma subsp. argyrosperma (right).

Differences in a fruit size, b seed size and shape, c and in the presence of urticating trichomes
Despite the economic importance and the growing genomic resources in Cucurbita species8,13, studies aimed at understanding their domestication are still lacking. To start filling this gap, we report here the first genome assembly of the wild relative sororia, which complements the existing assembly of argyrosperma14. The comparison between the genomes allowed us to find genomic structural variants between both subspecies. We characterized a large sample of argyrosperma landraces (117 individuals from 19 locations) and sororia accessions (50 individuals from 4 locations) using genome-wide data to investigate their demographic history and propose a domestication scenario. We also performed selection scans throughout the genome of C. argyrosperma to detect candidate regions associated with the domestication of this species.
Results
Genome assembly of C. argyrosperma subsp. sororia
We sequenced the genome of a wild individual of subspecies sororia using Illumina HiSeq4000 (213x coverage) and PacBio Sequel (75.4x coverage). The genome was assembled in 828 contigs with an N50 contig size of 1.3 Mbp and an L50 of 58 contigs (Table S1). A BUSCO analysis15 against the embryophyte odb9 database detected 92.8% of complete BUSCOs, 1.2% fragmented BUSCOs, and 6.0% missing BUSCOs within the genome assembly, similarly to other Cucurbita genome assemblies14,16. We predicted 30,592 protein-coding genes within the genome assembly using BRAKER217. The BUSCO completeness of the gene predictions (92.5% complete BUSCOs, 3.1% fragmented BUSCOs) is comparable to that of the genome assembly and that of other genome annotations14,16, despite using RNA-seq data of a different individual from which the genome was assembled (see “Methods”). Around 35.8% of our sororia genome assembly is composed of transposable elements (TEs), slightly higher than the 34.1% of TEs found in a previous argyrosperma assembly14.
The genome of argyrosperma was previously assembled in 920 scaffolds14, so we aimed at improving the assemblies for both the sororia and the argyrosperma genomes using a reference-guided scaffolding step against the genome assembly of C. moschata16. We anchored 99.97% of the argyrosperma genome assembly and 98.8% of the sororia genome assembly into 20 pseudomolecules using RaGOO18, which corresponds to the haploid chromosome number in Cucurbita19. Both assemblies show high synteny conservation across the genus (Fig. S1) and confirm a previously reported inversion in chromosome four that is shared with C. moschata16.
Structural variants between the wild and domesticated genomes
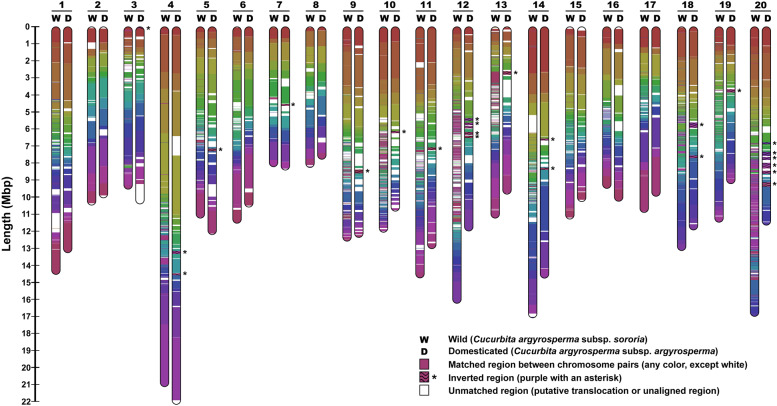
We compared the genome of argyrosperma against the genome of sororia (Fig. 2). Some of the centromeres in the genome of sororia were larger than in argyrosperma, possibly due to a better assembly of the repetitive regions. We found 443 high-confidence structural variants (SVs) such as copy-number variants (CNVs), inversions, and translocations between the wild and the domesticated genomes (Table S2). We also found several regions that could not be aligned between both genomes (Table S2). The size of the sororia genome assembly is ~254 Mbp, 9.23% larger than the genome assembly of argyrosperma14, which could be partially explained by these SVs. We found that two copies of thaumatin-like protein 1a (TL1) and auxin-responsive protein SAUR32 were disrupted by inversions in the argyrosperma genome. We also found a CNV loss spanning the transcription factor PIF1 and a translocation containing the gene LONG AFTER FAR-RED 3 (LAF3). The genomes of argyrosperma and sororia share some common genes within their unaligned regions, such as microtubule-associated proteins and genes related to tryptophan biosynthesis (Table S2), suggesting that those regions contain highly divergent sequences and are not limited to presence/absence variants. However, other unaligned regions contain more genes in sororia than in argyrosperma, including the major pollen allergen Ole e 6 (OLE6), some proteolytic enzymes and sucrose biosynthetic genes that are absent in the argyrosperma genome (Table S2), suggesting that presence/absence variants are also included within the unaligned regions.
Fig. 2. Chromosome map representing the matching regions and putative structural variants between the genome assemblies of C. argyrosperma subsp. sororia (wild) and C. argyrosperma subsp. argyrosperma (domesticated).
Matching colors represent the aligned homologous regions between both genomes, while white segments represent regions that could not be aligned to the other genome. Inverted regions are highlighted with an asterisk
Population data and SNP genotyping
We used samples previously collected throughout Mexico11 corresponding to 117 individuals of argyrosperma, 50 individuals of sororia, 19 feral individuals of argyrosperma previously reported to have a semi-wild phenotype and a cultivated genotype based on microsatellite data11 and 6 individuals of C. moschata (the domesticated sister species of C. argyrosperma), that were used as outgroups (Table S3). The samples were sequenced using the tunable Genotype by Sequencing (tGBS) method20 to obtain genome-wide genetic information of the C. argyrosperma populations. The reads were quality-filtered and mapped against the genome assemblies of argyrosperma and sororia to predict single nucleotide polymorphisms (SNPs) and assess possible reference biases in the SNP prediction. Using the reference genome of argyrosperma, we obtained an initial dataset consisting of 12,813 biallelic SNPs with a mean read depth of 50 reads per SNP and a minor allele frequency (MAF) of at least 1% (13k dataset). We also mapped the whole-genome Illumina reads of argyrosperma and sororia, as well as the whole-genome sequencing of a C. moschata individual and a C. okeechobeensis subsp. martinezii individual (a closely related wild Cucurbita species), against the reference genome of argyrosperma to obtain a dataset of 11,498,421 oriented biallelic variants (SNPs and indels) across the genome that was used to assess introgression and incomplete lineage sorting.
Demographic history of C. argyrosperma during its domestication
We eliminated the SNPs that deviated from Hardy–Weinberg equilibrium (exact test with p < 0.01) and pruned nearby SNPs under linkage disequilibrium (LD with an r2 > 0.25 in 100 kbp sliding windows) from the 13k dataset to retrieve a set of 2861 independent SNPs that could be used for demographic analyses.
We found similar genetic variation in sororia and argyrosperma, regardless of the reference genome used (average nucleotide diversity π range 0.095–0.098 for both taxa, see Table S4). At a population scale, the wild population in Jalisco had the highest genetic diversity within sororia, while the highest diversity in argyrosperma was found in the Pacific Coast of Mexico (Table S5). The domesticated and wild populations of C. argyrosperma displayed low genetic differentiation (FST = 0.0646; 95% confidence interval from 0.0565 to 0.0751), while feral populations were more closely related to argyrosperma (FST = 0.0479) than with sororia (FST = 0.1006).
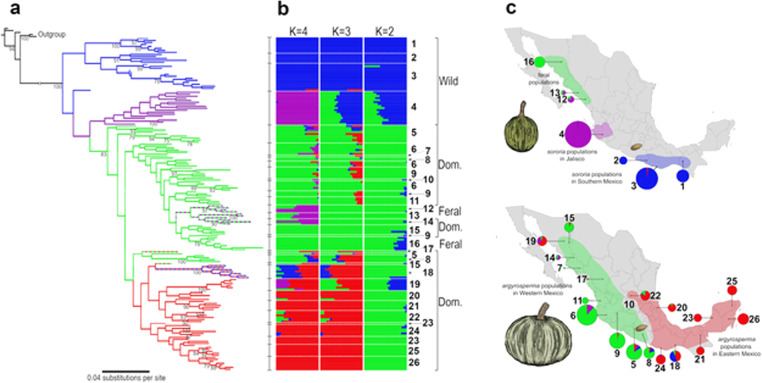
We used SNPhylo21 and ADMIXTURE22 to evaluate the genealogical relationships and genetic structure among the wild and domesticated populations of C. argyrosperma. We confirmed the genetic differentiation between sororia and argyrosperma, as detected by the FST analyses. Our Maximum-Likelihood (ML) tree groups all the argyrosperma populations in a single monophyletic clade (Fig. 3a). We found additional genetic differentiation between the sororia populations in Southern Mexico (populations 1–3) and the sororia populations in Jalisco (population 4), in both the ADMIXTURE assignations (Fig. 3b) and their positions in the ML tree (Fig. 3a). The sororia populations of Jalisco are genetically closer to argyrosperma, as shown by their paraphyletic position in the ML tree (Fig. 3a). Consistent with a domestication in the lowlands of Western Mexico, the argyrosperma populations of Guerrero and Jalisco represent the basal branches of the argyrosperma clade (Fig. 3a), all showing instances of genetic similarity to the sororia populations in Jalisco in the four genetic groups (K) of ADMIXTURE (Fig. 3b). The argyrosperma populations in Western Mexico (populations 5–17) are genetically differentiated from the Eastern populations (populations 18–26), with a possible recent anthropogenic dispersion event of Eastern populations into Onavas, Sonora (population 19; Fig. 3b, c). These four genetic groups are also retrieved in a principal component analysis (PCA; Fig. S2). The ADMIXTURE results (Fig. 3b) uncovered admixture events between some sororia and argyrosperma populations. This pattern is evident in the populations of Oaxaca and Sinaloa (populations 14 and 18; Fig. 3c). The feral populations are consistently grouped alongside their sympatric domesticated populations within argyrosperma (Fig. 3a, b), indicating that these populations diverged recently from nearby domesticated populations. These demographic patterns are robust to different SNP filters, such as different thresholds for missing data, or filtering for significant deviations from Hardy–Weinberg equilibrium (Fig. S3).
Fig. 3. Genetic structure and phylogenetic relationships between the wild and domesticated populations of Cucurbita argyrosperma based on 2861 SNPs.
a Maximum-Likelihood tree with 100 bootstraps (only bootstrap values > 70 shown). b ADMIXTURE analysis using K values ranging from 2 to 4. c Geographic distribution of the wild (top) and domesticated (bottom) populations, with pie chart colors representing the ADMIXTURE assignation of the individuals in K = 4 genetic groups (size of pie charts proportional to sample size). The seed in the maps represents the earliest archaeological record of argyrosperma from Xihuatoxtla, Guerrero (dated 8700 years BP)10
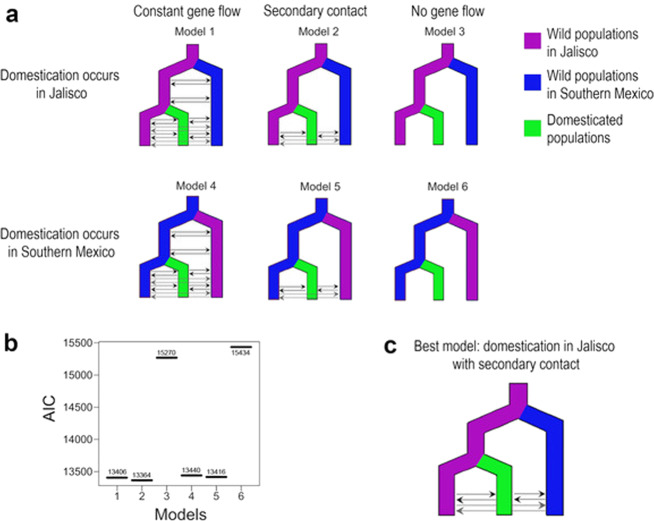
We used Fastsimcoal 223 to explicitly test whether argyrosperma was domesticated from a sororia population in Southern Mexico or from a sororia population in Jalisco. Given that gene flow has been previously observed between argyrosperma and sororia12, we compared three different gene flow models (continuous gene flow, secondary contact, or no gene flow) for each scenario (Fig. 4a). A comparison between models using the Akaike Information Criterion indicates that the Jalisco domestication model with secondary contact (i.e., extant gene flow after initial genetic isolation between subspecies) is the most likely of the domestication scenarios assayed. We were able to discard the other unrealistic domestication scenarios (i.e., domestication in southern Mexico and lack of gene flow between subspecies), further supporting a domestication event in Jalisco.
Fig. 4. Coalescent simulations and most likely domestication scenario of Cucurbita argyrosperma.
a The six models assessed against the unfolded multidimensional Site Frequency Spectrum of our data. b Comparison of the Akaike Information Criterion (AIC) of all the models. c The domestication model that best fits the data
Selection scans in C. argyrosperma
In order to perform the tests to detect selective signals associated with the domestication of C. argyrosperma, we removed from the 13k dataset the C. moschata individuals as well as the feral individuals of C. argyrosperma. We used the 1% MAF threshold for this subset, obtaining a 10,617 SNP dataset suitable to detect selective signals, with a marker density of 44 SNPs per Mb. LD was limited within the dataset, with a mean pairwise r2 of 0.1, and with argyrosperma showing a faster LD decay than sororia (Fig. S4). We performed three outlier tests as implemented in BayeScEnv24, PCAdapt25, and LFMM 226,27 to detect selective signals between the domesticated and the wild populations of C. argyrosperma (Fig. 5). BayeScEnv is an FST-based method that tests correlation with other variables, in our case the wild or domesticated nature of each population (coded as 0 and 1, respectively). PCAdapt does not require an a priori grouping of individuals into wild/domesticated, as we used the two principal components of a PCA to control for the underlying genetic structure between subspecies (Fig. S2). LFMM 2 identifies the number of latent factors in the populations through least square estimates to find correlations between genetic variants and the domesticated or wild phenotypes in the dataset26. In order to reduce the number of false positives, we only analyzed the SNPs that were detected as outliers by at least two of the tests.
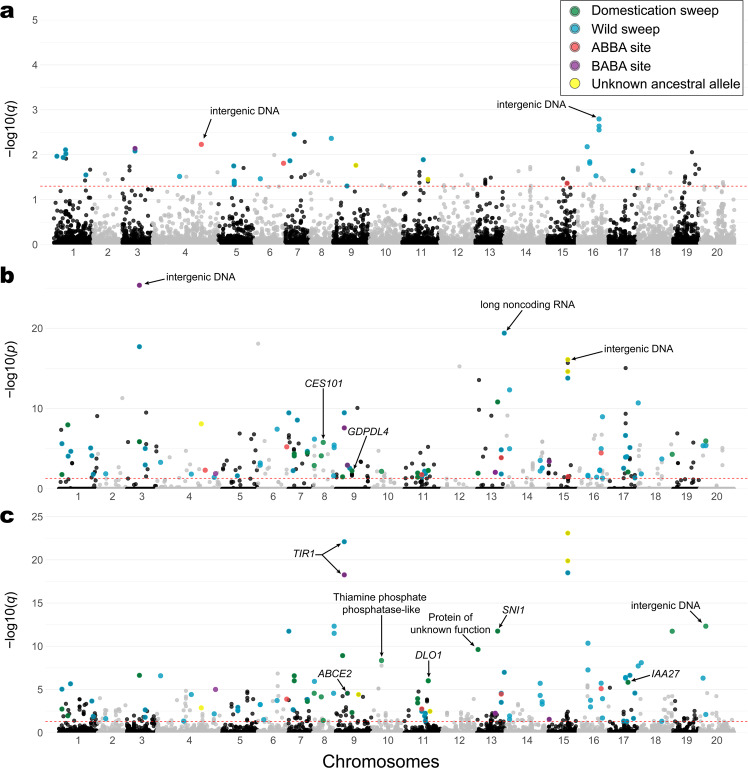
Fig. 5. Putative footprints of selection associated with the domestication of Cucurbita argyrosperma.
Manhattan plots representing the a BayeScEnv, b PCAdapt, and c LFMM 2 tests in each chromosome of the genome. The red dotted line indicates the cutoff value (q-value or Bonferroni-corrected p-value < 0.05) to determine a candidate locus. Only 110 loci were retrieved as candidate SNPs by more than one test, which are highlighted depending on whether the putative signal of positive selection corresponds to argyrosperma (green), sororia (blue), an ABBA site (red) a BABA site (purple) or an unknown selective direction (yellow). The arrows indicate the position of some of the discovered candidate genes
We discovered 110 SNPs that converged as candidates in at least two tests (Fig. 5). We used C. moschata and C. okeechobeensis subsp. martinezii as outgroups to determine the direction of the putative selective pressures for each candidate SNP, as well as determining possible introgression or incomplete lineage sorting between sororia, argyrosperma and C. moschata. We found that 22 of the candidate SNPs corresponded to selective signals in argyrosperma, while 70 candidate SNPs corresponded to selective signals in sororia (Fig. 5). We could not determine the direction of selection for 5 candidate SNPs, and 13 showed signals of introgression or incomplete lineage sorting.
We identified several instances of either genetic introgression or incomplete lineage sorting between C. moschata and both argyrosperma (ABBA sites) or sororia (BABA sites) (Fig. S5). We performed a genome-wide D-statistic analysis and found significantly more instances of shared derived variants between argyrosperma and C. moschata than between sororia and C. moschata (block-jackknife p-value = 0.0014), obtaining an overall admixture fraction fG of 0.01 (Table S6). From the 13 candidate SNPs with signals of introgression, 6 correspond to ABBA sites and 7 correspond to BABA sites (Fig. 5).
We found 45 protein-coding genes and one long-noncoding RNA including candidate SNPs within their structure (i.e., introns, exons, UTRs), which were assigned according to the observed direction of the putative selective signals (Table S7). Among the genes under putative selection in sororia were four serine/threonine-protein kinases, including PBL10 and PBL23 (Table S7). Among the genes under putative selection in argyrosperma, we found glycerophosphodiester phosphodiesterase GDPDL4, auxin-responsive protein IAA27, ABC transporter E family member 2 (ABCE2), DMR6-like oxygenase 1 (DLO1), and serine/threonine-protein kinase CES101 (Table S7). We also found several genes under putative selection overlapping ABBA and BABA sites (Table S7). We found a transport inhibitor response 1 (TIR1) homolog under putative selection in both sororia and as a BABA site. Curiously, the gene MKP1 was found under selection in both argyrosperma and in sororia.
Discussion
The genome assembly of sororia represents the first high-quality sequenced genome of a wild Cucurbita, which allowed us to detect structural and functional differences with the argyrosperma genome. The genome assembly of argyrosperma was smaller than the sororia assembly, which is possibly caused by the loss of structural variants during its domestication, as has been reported in pan-genome studies28. Many of these unaligned regions contain entire genes in sororia, making this wild taxon a reservoir of potentially adaptive presence/absence variants. However, the extant genetic diversity of argyrosperma is similar to that of sororia, which suggests that the effects of the domestication bottleneck were alleviated by the current gene flow between both subspecies as suggested by our coalescent simulations and by the results of the previous studies12. The fast LD decay in argyrosperma further supports the limited effect of the domestication bottleneck. This gene flow may be related to the sympatric distribution of the wild and domesticated populations of C. argyrosperma throughout the Pacific Coast of Mexico11, where their coevolved pollinator bees Peponapsis spp. and Xenoglossa spp. are found29. Traditional agricultural practices are another fundamental force that maintains the diversity of crop species30. Since argyrosperma is a traditional crop cultivated for both self-supply and local markets where it has a specialized gastronomic niche13, the genetic diversity in argyrosperma is also maintained by the conservation of local landrace varieties at local scales31,32.
Our demographic analyses suggest that the extant populations from Jalisco are the closest modern relatives of the initial population of sororia from which argyrosperma originated. The genetic relatedness between the sororia populations from Western Mexico and argyrosperma was also observed with mitochondrial markers6. The domestication of C. argyrosperma likely started around 8,700 years ago, as suggested by the earliest, albeit taxonomically ambiguous, archaeological record of argyrosperma10,33. Since crop domestication in Mesoamerica is linked to migration patterns and cultural development of early human populations in America7,34, we expected argyrosperma to share historical demography patterns with human history. Our data shows that the argyrosperma populations found in Guerrero and Jalisco are the most closely related to the sororia populations from Jalisco. This means that even if the closest wild relatives of argyrosperma are currently found in Jalisco, the domestication process may have occurred throughout the lowlands of Jalisco and the Balsas basin10. Ancient human migration events have been proposed to occur along the river basins in Southwestern Mexico, which may explain the genetic cohesiveness among the argyrosperma populations of that area that represent the first fully domesticated lineage of the species34. Previous studies based on 8,700 years old phytoliths found in Xihuatoxtla, Guerrero, suggest the co-occurrence of Zea mays and C. argyrosperma in the Balsas region within this time period10,33. Overall, the patterns of genetic structure for C. argyrosperma are coherent with the archaeological evidence of early human migration throughout Mesoamerica35,36 (Fig. 6).
Fig. 6. Model of Cucurbita argyrosperma domestication based on archaeological6,33–35 and genetic data.
The colored lines represent the presence of each C. argyrosperma genetic cluster in Mesoamerica (blue = sororia populations, green = argyrosperma populations in Western Mexico, red = argyrosperma populations in Eastern Mexico)
We found several signals of putative selection between argyrosperma and sororia, even though tGBS sequencing has a limited capacity to detect selective signals across the genome37. The SNP density for our selection tests was of 44 SNPs per Mb, which is one order of magnitude denser than other studies using reduced-representation genome sequencing to detect selective signals37. This is a consequence of the relatively small genome size of C. argyrosperma14. Nonetheless, the LD in C. argyrosperma decays at a shorter length than our SNP density, so several signals of selection might be missing from our scans, and some signals might not correspond to the actual gene under selection, but rather to a neighboring region in the genome37. Therefore, our genome scans should be interpreted as a partial representation of the selective signals associated with the domestication process37.
Most of the putative selective signals were attributed to sororia, probably because wild taxa are subject to many natural selective pressures. Many of the SNPs that were retrieved as outliers were found on genes involved in biotic and abiotic plant defense responses. For example, PBL10 and PBL23 have been suggested to be involved in plant defense pathways due to their similarity to other serine/threonine-protein kinases38.
However, we also found some genes under putative selection in argyrosperma that could be attributed to defense mechanisms such as DLO1, which is involved in pathogenic defense responses39. We found GDPDL4 under putative selection, which is involved in trichome differentiation40, a morphological characteristic that differentiates argyrosperma from sororia (Fig. 1). We also found two disrupted copies of TL1 and the absence of OLE6 within the argyrosperma genome, both of which cause allergic reactions in humans41,42. This is concordant with previous studies showing that selective pressures during domestication actively purge these defense mechanisms, as the products of these responses are usually unpleasant or harmful to humans when the plant is consumed43. This is particularly important for breeding programs, since wild Cucurbita such as sororia harbor loci associated with disease resistance that their domesticated counterparts have lost4. The selective pressures found in MKP1 suggest a disruptive selection regime between sororia and argyrosperma. Since MKP1 modulates defense responses44, it is possible that both subspecies adapted to differential environmental pressures as domestication took place.
We found several candidate genes involved in the regulation of ABA. The alteration of growth hormones may play an important role in C. argyrosperma domestication. ABA is involved in a myriad of functions, such as the regulation of plant growth, plant development, seed dormancy, and response to biotic/abiotic stress45. In this sense, the lack of dormancy in seeds and gigantism are both common domestication changes that are present in domesticated cucurbits that may be caused by changes in the regulation of ABA and brassinosteroids8,46. We found SAUR32, PIF1, and LAF3 within the high-confidence SVs in argyrosperma, all of which act as inhibitors of seed germination under dark conditions47–49 and may explain the lack of seed dormancy in argyrosperma. Likewise, we found IAA27 under selection in argyrosperma, which is involved in plant growth and development50.
Selection over seed size has been particularly important in the domestication of C. argyrosperma9. We found ABCE2 under selection in argyrosperma. Some ABC transporters are involved in the transmembrane transport of ABA-GE, an ABA conjugate that is usually attributed to the plant response against water stress51. However, previous studies in Hordeum vulgare suggest that the transport of ABA-GE may play a role in seed development alongside de novo ABA synthesis within the developing seed52, suggesting a role of ABC transporters in the seed development of C. argyrosperma. Previous studies have also identified an association between variants in ABC transporter proteins and seed size in Cucurbita maxima53 and Linum usitatissimum54, further suggesting that ABA may be deregulated via selective pressures on ABC transporters to enhance seed size in C. argyrosperma during its domestication. Variants in a serine/threonine-protein kinase, as the one we found in our selective scans, have also been associated with seed size in C. maxima53.
We found shared derived variants between C. moschata and C. argyrosperma under putative selection. This suggests that, given the close relationship between C. argyrosperma and C. moschata, both species may share domestication loci involved in common domestication traits. However, we also found TIR1 under selection in sororia and as a BABA site, which is an auxin receptor involved in ethylene signaling and antibacterial resistance in roots55. The introgression of wild alleles into domesticated Cucurbita has been previously reported as an effective method to improve the resistance of domesticated crops56. Along this line, our results suggest that sororia is an important source of adaptive alleles for C. moschata. ABBA and BABA sites under selection may be shared with C. moschata either due to incomplete lineage sorting or by adaptive introgression with the wild and domesticated populations of C. argyrosperma. The incorporation of domesticated loci between C. argyrosperma and C. moschata through introgression may have been an effective way for Mesoamerican cultures to domesticate multiple Cucurbita taxa. This hypothesis is supported by the significant amount of ABBA sites shared between the genomes of C. argyrosperma and C. moschata. However, this hypothesis needs to be further addressed using population-level data of C. moschata with other wild and domesticated Cucurbita species.
Materials and methods
Genome assembly and annotation of Cucurbita argyrosperma subsp. sororia
We sequenced and assembled de novo the genome of a sororia individual collected in Puerto Escondido (Oaxaca, Mexico). Its DNA was extracted from leaf tissue and sequenced using PacBio Sequel at the University of Washington PacBio Sequencing Services and using Illumina HiSeq4000 at the Vincent J. Coates Genomics Sequencing Laboratory in UC Berkeley (NIH S10 Instrumentation Grants S10RR029668 and S10RR027303). We filtered the Illumina sequences using the qualityControl.py script (https://github.com/Czh3/NGSTools) to retain the reads with a PHRED quality ≥ 30 in 85% of the sequence and an average PHRED quality ≥ 25. The Illumina adapters were removed using SeqPrep (https://github.com/jstjohn/SeqPrep) and the paired reads that showed overlap were merged. The chloroplast genome was assembled using NOVOplasty57 and the organellar reads were filtered using Hisat258 against the chloroplast genome of argyrosperma14 and the mitochondrial genome of C. pepo59. We assembled the nuclear genome into small contigs using the Illumina reads and the Platanus assembler60. The Platanus contigs were assembled into larger contigs using the PacBio Sequel reads and DBG2OLC61. We performed two iterations of minimap2 and racon62 to obtain a consensus genome assembly by mapping the PacBio reads and the Platanus contigs against the DBG2OLC backbone. We performed three additional polishing steps using PILON63 by mapping the Illumina reads against the consensus genome assembly with BWA mem64.
The genome annotation processes were performed using the GenSAS v6.0 online platform65. The transposable elements within the genome were predicted and masked using RepeatModeler (http://www.repeatmasker.org/RepeatModeler/). We downloaded five RNA-seq libraries of C. argyrosperma available on the Sequence Read Archive (accessions SRR7685400, SRR7685404–SRR7685407) to use them as RNA-seq evidence for the gene prediction. We performed the same quality filters described above for the RNA-seq data and aligned the high-quality reads against the masked genome of C. argyrosperma subsp. sororia using STAR v2.766. We used filterBAM from the Augustus repository67 to filter low-quality alignments and used the remaining alignments as RNA-seq evidence to predict the gene models using BRAKER217. The gene predictions were functionally annotated using InterProScan68 and by aligning the gene models against the SwissProt database69 using BLASTp70 with an e-value < 1e−6. We performed a manual assessment of the predicted gene models to eliminate annotation artifacts.
Anchoring the reference genomes into pseudomolecules
We aimed to improve the genome assemblies of argyrosperma and sororia, which were assembled in 920 scaffolds14 and 817 contigs, respectively. Thus, we generated PacBio corrected reads from the published PacBio RSII reads of argyrosperma (NCBI SRA accession SRR7685401) and the PacBio Sequel reads of sororia (sequenced for this study at the University of Washington PacBio Sequencing Services) using CANU71. The macrosynteny of Cucurbita genomes is largely conserved between species72, so we performed a reference-guided scaffolding step using RaGOO18 to anchor the genome assemblies of argyrosperma14 and sororia into pseudomolecules. We used the genome assembly of C. moschata16 as the reference genome for RaGOO18 and we used the PacBio corrected reads of each taxon to detect and correct misassemblies, using a gap size of 2600 bp for chromosome padding (i.e., we filled the gaps between contigs with 2600 stretches of Ns, corresponding to the average gap length of the argyrosperma genome assembly). The chromosome numbers in both assemblies were assigned in correspondence to the genome assembly of C. moschata16.
Structural variant analysis
We evaluated the synteny between Cucurbita genomes using Synmap273. We predicted the SVs between argyrosperma and sororia using SyRI74 alongside nucmer75 with a minimum cluster length of 500 bp, an alignment extension length of 500 bp, a minimum match length of 100 bp and a minimum alignment identity of 90%. We also used Sniffles alongside NGMLR76 as an additional SV predictor, by aligning the argyrosperma PacBio reads against the genome assembly of sororia (only the SVs with a minimum support of 6 reads were retained). We only analyzed the SVs that were independently predicted by SyRI and Sniffles. Only the SVs that overlapped within a range of ±100 bp at the start and end positions of each prediction were retained as high-confidence SVs. Due to the limitations of read-mapping techniques to predict presence–absence variants, we only analyzed the unaligned regions predicted by SyRI for this type of variants.
The gene content associated with each type of structural variant was considered either as the overlap between genes and variants (inversions and translocations) or as the genes contained entirely within the structural variants (CNVs and unaligned regions). We performed a Gene Ontology enrichment analysis using topGO and the weight01 algorithm77 to find enriched biological functions associated with each type of structural variant. We determined the significantly enriched biological functions by performing Fisher’s exact test (p-value < 0.05). We plotted the genome rearrangements and SVs between sororia and argyrosperma using Smash78 with a minimum block size of 100,000 bp, a threshold of 1.9, and a context of 28.
Data filtering and SNP genotyping
We used previously collected seeds from 19 populations of argyrosperma landraces, four populations of sororia, and three feral populations11, covering most of the reported distribution of this species throughout Mexico5 (Table S3). Each of the collected seeds came from a different maternal plant, in order to avoid signals of inbreeding. The seeds were germinated in a greenhouse and total DNA was extracted from fresh leaves using a DNeasy Plant MiniKit (Qiagen) of 192 individuals across the collected populations (Table S3), including five individuals of C. moschata to be used as outgroup. All 192 individuals were sequenced by Data2Bio LLC using the tunable Genotyping by Sequencing (tGBS) method20 with an Ion Proton instrument and two restriction enzymes (Sau3AI/BfuCI and NspI). The wild and domesticated populations were randomly assigned to the plate wells before library preparation to avoid sequencing biases.
The raw reads of the tGBS sequencing were trimmed using LUCY279, removing bases with PHRED quality scores < 15 using overlapping sliding windows of 10 bp. Trimmed reads shorter than 30 bp were discarded. The trimmed reads were mapped against the genome assembly of argyrosperma using segemehl80, since empirical studies suggest this read-mapping software outperforms others for Ion Torrent reads81. We only retained the reads that mapped uniquely to one site of the reference genome for subsequent analyses.
We used BCFtools82 for an initial variant calling step, retaining variants with at least 6 mapped reads per individual per site where the reads had a minimum PHRED quality score of 20 in the called base and a minimum mapping quality score of 2083. We used plink84 to perform additional filters, such as retaining only biallelic SNPs, retaining SNPs with no more than 50% of missing data, individuals with no more than 50% of missing data and sites with a minor allele frequency (MAF) of at least 1% (13k dataset). After eliminating individuals with missing data, 109 individuals of argyrosperma, 44 individuals of sororia, 14 feral individuals and 5 individuals of C. moschata remained for the subsequent analyses. We repeated the SNP prediction using the reference genome of sororia to evaluate potential reference biases. We found a similar number of SNPs (10,990) and comparable estimates of genetic diversity (see Table S4), suggesting that reference bias does not have a meaningful impact on our results. Thus, we employed the domesticated genome as the reference for the rest of the population analyses. We repeated our analyses using a separate filtering step of missing data for the domesticated and the wild populations, retrieving 84.18% of the SNPs from the 13k dataset and obtaining the same results (Fig. S3).
In order to obtain an adequate SNP dataset to infer the demographic history of C. argyrosperma, we performed additional filters to the 13k dataset with plink84, including (i) the elimination of all the SNPs that diverged significantly (p < 0.01) from the Hardy–Weinberg equilibrium exact test85 to remove potential allelic dropouts, (ii) the elimination of adjacent SNPs with a squared correlation coefficient (r2) larger than 0.25 within 100 kbp sliding windows with a step size of 100 bp. We repeated the demographic analyses without filtering the SNPs with significant deviations from Hardy–Weinberg equilibrium, obtaining the same results (Fig. S3).
We also generated a SNP dataset to detect selective signals associated with the domestication of C. argyrosperma by eliminating all the feral individuals of C. argyrosperma, which could not be assigned to either a wild or a domesticated population, as well as the five individuals of C. moschata. We also eliminated the SNP sites with more than 50% missing data and performed a MAF filter of 1% after reducing the number of individuals in the 13k dataset. The SNP density was calculated with VCFtools86 and the LD decay was calculated using plink84 with a minimum r2 threshold of 0.001.
We also sequenced the genome of a C. moschata individual from Chiapas (Mexico) and the genome of a C. okeechobeensis subsp. martinezii individual from Coatepec (Veracruz, Mexico) using the Illumina HiSeq4000 platform in UC Berkeley, to evaluate possible introgression and incomplete lineage sorting with C. argyrosperma. We downloaded the genome sequences of argyrosperma14 from the Sequence Read Archive (accessions SRR7685402 and SRR7685403). The Illumina whole-genome sequences were filtered using the same quality parameters as the ones used in the genome assembly of sororia (see above) and were aligned against the genome assembly of argyrosperma using BWA mem64. We only retained the reads that mapped uniquely to one site of the reference genome and retained only the biallelic sites with a sequencing depth > =10 reads per genome.
Population structure
We used diveRsity87 to calculate the pairwise FST statistics, using 100 bootstraps to calculate the 95% confidence intervals. We estimated the genetic variation in the wild, domesticated and feral populations with STACKS88. Using ADMIXTURE22, we evaluated the genetic structure among the sororia and argyrosperma populations, evaluating their individual assignment into one (CV error = 0.26205), two (CV error = 0.25587), three (CV error = 0.25806) and four (CV error = 0.26658) genetic groups (K). We reconstructed a maximum-likelihood tree with SNPhylo21, based on substitutions per site between all the individuals with 100 bootstraps to assess the reliability of the tree topology. We used plink84 to perform a principal component analysis (PCA) using 10 principal components.
Coalescent simulations
We used Fastsimcoal 223 to determine the parameters that maximize the composite likelihood of each model given the unfolded multidimensional SFS. The unfolded multidimensional SFS was obtained with DADI89, using 17 sororia individuals of Jalisco, 27 sororia individuals of Southern Mexico, 109 argyrosperma individuals and 5 C. moschata individuals as an outgroup to unfold the SFS. We ran 100,000 simulations with 20 replicates for each model (two divergence scenarios and three gene flow scenarios) using the following settings: a parameter estimation by Maximum Likelihood with a stopping criterion of 0.001 difference between runs, a minimum SFS count of 1, a maximum of 40 loops to estimate the SFS parameters and a maximum of 200,000 simulations to estimate the SFS parameters. We also selected log-uniform priors for parameter estimations, setting times of divergence between 1000 and 200,000 generations (domestication times are expected to fall within this interval, given that the presumed most ancient evidence of the human presence in America is 33,000 years old90; while the split between the wild populations is expected to coincide with the extinction of the megafauna in America, which acted as the natural dispersers of wild Cucurbita during the Pleistocene3), effective population sizes (Ne) between 100 and 60,000 individuals and migration rates (m) between 0.0001 and 0.5. The constant gene flow scenarios calculated a migration rate matrix throughout the simulation, from the present back to the common ancestral population of all lineages, with independent migration rates for each possible direction of gene flow. The secondary contact scenarios simulated a migration matrix only at the start of the simulation, before the coalescence event between the wild and domesticated lineages (see Data S1–S6 for detailed model parameters). We also constrained the times of divergence in all scenarios by forcing the domesticated taxa to diverge after the wild one. Each generation in the model corresponds to one calendar year, as this species displays an annual life cycle13. The 20 replicates of each model converged to similar likelihoods, indicating that the simulations performed well. After corroborating that all replicates converged to similar likelihoods, we combined all replicates and retained all outputs that were above 95% of the likelihood distribution. We found that the Jalisco model of divergence with secondary contact had the lowest Akaike Information Criterion values for all the tested models.
Tests to detect selective signals and introgression
We used BayeScEnv24 to detect putative regions under selection that were differentiated between sororia and argyrosperma. For the “environmental” values used by BayeScEnv, we assigned each population as either wild (0) or domesticated (1). We ran two independent MCMC analyses with 20 initial pilot runs with a length of 10,000 generations and a main run with an initial burn-in of 100,000 generations and a subsequent sampling step for 100,000 generations sampling every 20 generations. We confirmed the convergence between both chains using the Gelman and Rubin statistic91. The SNPs with q-values < 0.05 were regarded as candidate loci under selection.
The Mahalanobis distances implemented in PCAdapt25 were used to detect candidate SNPs after controlling for the first two principal components in our dataset, which correspond to the subspecies and geographical differentiation observed among the populations (see Fig. S2). We performed Bonferroni corrections to adjust the p-values obtained from PCAdapt and the SNPs with Bonferroni-corrected p-values < 0.05 were regarded as candidate loci under selection.
We used LFMM 226 to identify candidate SNPs differentiating the wild and domesticated phenotypes of C. argyrosperma. We tested K number of latent factors from 1 to 10 using sNMF27 and determined an optimal K = 6. We used 6 latent factors and a ridge penalty to identify significant associations between the response (wild or domesticated phenotypes) and the genetic variants. Finally, we performed FDR corrections to obtain q-values, with q-values < 0.05 being regarded as candidate loci under selection.
We performed an ABBA-BABA test using Dsuit92 against the 11,498,421 whole-genome variants to evaluate signals of introgression or incomplete lineage sorting between argyrosperma, sororia and C. moschata, while using C. okeechobeensis subsp. martinezii as an outgroup. We calculated a global D-statistic by performing a SNP-by-SNP analysis to determine the amount of ABBA and BABA sites throughout the entire genome. We also calculated local D-statistics within 500 SNP windows with a step size of 250 SNPs. We used the five tGBS data of C. moschata, as well as the whole-genome sequences of C. moschata and C. okeechobeensis subsp. martinezii, to define the ancestral state of each candidate locus from the selection scans and determine the direction of the selective signals or whether they corresponded to ABBA or BABA sites. We used snpEff93 to associate the candidate loci found in at least two tests with the genome annotation of argyrosperma14.
Supplementary information
Acknowledgements
This work was funded by Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO) KE004 “Diversidad genética de las especies de Cucurbita en México e hibridación entre plantas genéticamente modificadas y especies silvestres de Cucurbita” and CONABIO PE001 “Diversidad genética de las especies de Cucurbita en México. Fase II. Genómica evolutiva y de poblaciones, recursos genéticos y domesticación” (both awarded to R.L-S. and L.E.E.). J.B-R. and G.S-V. are doctoral students from Programa de Doctorado en Ciencias Biomédicas and Programa de Doctorado en Ciencias Biológicas, Universidad Nacional Autónoma de México, and received fellowships 583146 and 292164 from CONACyT. We acknowledge the Doctorado en Ciencias Biológicas and the Doctorado en Ciencias Biomédicas for the support provided during the development of this project. We thank Jodi Lynn Humann for her technical support while using the GenSAS web server. Special thanks to Laura Espinosa-Asuar and Silvia Barrientos for their technical support. We thank Rodrigo García Herrera, head of the Scientific Computing Department at LANCIS, Instituto de Ecología UNAM, for running the HTC infrastructure we used for the genome assembly and chromosome anchoring. The population genomic analyses were carried out using CONABIO’s computing cluster, which was partially funded by SEMARNAT through the grant “Contribución de la Biodiversidad para el Cambio Climático” to CONABIO. We acknowledge the technical support of MSc Emanuel Villafán and the resources for high-performance computing that the Institute of Ecology (INECOL) made available for conducting the functional gene annotations.
Author contributions
L.E.E., M.I.T., G.S-V., and J.B-R. designed the research; G.S-V. and G.C-M. collected the biological material; G.S-V. extracted DNA for the tGBS data; G.C-M. and G.S-V. coordinated the sequencing procedures of the tGBS data; J.B-R. and X.A-D. extracted DNA for the reference genome and coordinated its sequencing procedures; E.A-P. coordinated the administrative and lab work; J-B-R., J.A.A-L. and Y.T.G-G. performed the bioinformatic analyses; G.S-V., J.B-R., J.A.A-L., M.I.T., and L.E.E. analyzed the results; J.B-R., G.S-V., and L.E.E. drafted the manuscript; R.L-S. and L.E.E. obtained the funding. All authors revised the final version of the manuscript.
Data availability
All the raw sequencing data and genome assemblies are available in the National Center of Biotechnology Information under the BioProject accession PRJNA485527 (genome accesions SDJN00000000 and JAGKQH010000000; SRA accesions available in Table S3). The genome assemblies are also available in Figshare (https://doi.org/10.6084/m9.figshare.14370584.v1) and at the Cucurbit Genomics Database94.
Conflict of interest
The authors declare no competing interest.
Footnotes
These authors contributed equally: Josué Barrera-Redondo, Guillermo Sánchez-de la Vega
Contributor Information
Josué Barrera-Redondo, Email: josue_barrera@comunidad.unam.mx.
Rafael Lira-Saade, Email: rlira@unam.mx.
Luis E. Eguiarte, Email: fruns@unam.mx
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00544-9.
References
- 1.Meyer RS, Purugganan MD. Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 2013;14:840–852. doi: 10.1038/nrg3605. [DOI] [PubMed] [Google Scholar]
- 2.Zeder MA. Core questions in domestication research. Proc. Natl Acad. Sci. USA. 2015;112:3191–3198. doi: 10.1073/pnas.1501711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kistler L, et al. Gourds and squashes (Cucurbita spp.) adapted to megafaunal extinction and ecological anachronism through domestication. Proc. Natl Acad. Sci. USA. 2015;112:15107–15112. doi: 10.1073/pnas.1516109112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paris, H. S. in Genetics and Genomics of Cucurbitaceae 111–154 (Springer International Publishing, 2016). 10.1007/7397_2016_3.
- 5.Castellanos-Morales G, et al. Historical biogeography and phylogeny of Cucurbita: Insights from ancestral area reconstruction and niche evolution. Mol. Phylogenet Evol. 2018;128:38–54. doi: 10.1016/j.ympev.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 6.Sanjur OI, Piperno DR, Andres TC, Wessel-Beaver L. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: implications for crop plant evolution and areas of origin. Proc. Natl Acad. Sci. USA. 2002;99:535–540. doi: 10.1073/pnas.012577299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zizumbo-Villarreal D, Flores-Silva A, Marín PC-G. The Archaic Diet in mesoamerica: incentive for milpa development and species domestication. Economic Bot. 2012;66:328–343. doi: 10.1007/s12231-012-9212-5. [DOI] [Google Scholar]
- 8.Chomicki, G., Schaefer, H. & Renner, S. S. Origin and domestication of Cucurbitaceae crops: insights from phylogenies, genomics and archaeology. New Phytol.226, 1240–1255 (2019). [DOI] [PubMed]
- 9.Whitaker TW, Cutler HC. Cucurbits and cultures in the Americas. Economic Bot. 1965;19:344–349. doi: 10.1007/BF02904804. [DOI] [Google Scholar]
- 10.Piperno DR, Ranere AJ, Holst I, Iriarte J, Dickau R. Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc. Natl Acad. Sci. USA. 2009;106:5019–5024. doi: 10.1073/pnas.0812525106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sánchez-de la Vega G, et al. Genetic resources in the Calabaza Pipiana Squash (Cucurbita argyrosperma) in Mexico: genetic diversity, genetic differentiation and distribution models. Front Plant Sci. 2018;9:400. doi: 10.3389/fpls.2018.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montes-Hernandez S, Eguiarte LE. Genetic structure and indirect estimates of gene flow in three taxa of Cucurbita (Cucurbitaceae) in western Mexico. Am. J. Bot. 2002;89:1156–1163. doi: 10.3732/ajb.89.7.1156. [DOI] [PubMed] [Google Scholar]
- 13.Lira, R. et al. in Ethnobotany of Mexico 389–401 (Springer New York, 2016). 10.1007/978-1-4614-6669-7_15.
- 14.Barrera-Redondo J, et al. The genome of Cucurbita argyrosperma (Silver-Seed Gourd) reveals faster rates of protein-coding gene and long noncoding RNA turnover and neofunctionalization within Cucurbita. Mol. Plant. 2019;12:506–520. doi: 10.1016/j.molp.2018.12.023. [DOI] [PubMed] [Google Scholar]
- 15.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 16.Sun H, et al. Karyotype stability and unbiased fractionation in the Paleo-Allotetraploid Cucurbita genomes. Mol. Plant. 2017;10:1293–1306. doi: 10.1016/j.molp.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 2016;32:767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alonge M, et al. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 2019;20:224. doi: 10.1186/s13059-019-1829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitaker, T. W. & Bemis, W. P. Origin and evolution of the cultivated Cucurbita. Bull. Torrey Bot. Club102, 362–368 (1975).
- 20.Ott A, et al. tGBS® genotyping-by-sequencing enables reliable genotyping of heterozygous loci. Nucleic Acids Res. 2017;45:e178. doi: 10.1093/nar/gkx853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee TH, Guo H, Wang X, Kim C, Paterson AH. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics. 2014;15:162. doi: 10.1186/1471-2164-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Excoffier L, Foll M. Fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics. 2011;27:1332–1334. doi: 10.1093/bioinformatics/btr124. [DOI] [PubMed] [Google Scholar]
- 24.de Villemereuil P, Gaggiotti OE. A new FST-based method to uncover local adaptation using environmental variables. Methods Ecol. Evol. 2015;6:1248–1258. doi: 10.1111/2041-210X.12418. [DOI] [Google Scholar]
- 25.Luu K, Bazin E, Blum MG. PCAdapt: an R package to perform genome scans for selection based on principal component analysis. Mol. Ecol. Resour. 2017;17:67–77. doi: 10.1111/1755-0998.12592. [DOI] [PubMed] [Google Scholar]
- 26.Caye K, Jumentier B, Lepeule J, François O. LFMM 2: fast and accurate inference of gene-environment associations in genome-wide studies. Mol. Biol. Evol. 2019;36:852–860. doi: 10.1093/molbev/msz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frichot E, Mathieu F, Trouillon T, Bouchard G, François O. Fast and efficient estimation of individual ancestry coefficients. Genetics. 2014;196:973–983. doi: 10.1534/genetics.113.160572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan AW, et al. Super-Pangenome by integrating the wild side of a species for accelerated crop improvement. Trends Plant Sci. 2020;25:148–158. doi: 10.1016/j.tplants.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson HD. Gene flow in squash species. BioScience. 1990;40:449–455. doi: 10.2307/1311392. [DOI] [Google Scholar]
- 30.Jarvis DI, et al. A global perspective of the richness and evenness of traditional crop-variety diversity maintained by farming communities. Proc. Natl Acad. Sci. USA. 2008;105:5326–5331. doi: 10.1073/pnas.0800607105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montes-Hernández S, Merrick LC, Eguiarte LE. Maintenance of squash (Cucurbita spp.) landrace diversity by farmers activities in Mexico. Genet. Resour. Crop Evol. 2005;52:697–707. doi: 10.1007/s10722-003-6018-4. [DOI] [Google Scholar]
- 32.Barrera-Redondo J, et al. Landrace diversity and local selection criteria of domesticated squashes and gourds (Cucurbita) in the central Andean mountain range of Peru: Tomayquichua, Huánuco. Bot. Sci. 2020;98:101–116. doi: 10.17129/botsci.2239. [DOI] [Google Scholar]
- 33.Ranere AJ, Piperno DR, Holst I, Dickau R, Iriarte J. The cultural and chronological context of early Holocene maize and squash domestication in the Central Balsas River Valley, Mexico. Proc. Natl Acad. Sci. USA. 2009;106:5014–5018. doi: 10.1073/pnas.0812590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zizumbo-Villarreal D, Colunga-GarcíaMarín P. Origin of agriculture and plant domestication in West Mesoamerica. Genet. Resour. Crop Evol. 2010;57:813–825. doi: 10.1007/s10722-009-9521-4. [DOI] [Google Scholar]
- 35.Stinnesbeck W, et al. The earliest settlers of Mesoamerica date back to the late Pleistocene. PLoS ONE. 2017;12:e0183345. doi: 10.1371/journal.pone.0183345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piperno DR. The origins of plant cultivation and domestication in the new world tropics. Curr. Anthropol. 2011;52:S453–S470. doi: 10.1086/659998. [DOI] [Google Scholar]
- 37.Lowry DB, et al. Responsible RAD: striving for best practices in population genomic studies of adaptation. Mol. Ecol. Resour. 2017;17:366–369. doi: 10.1111/1755-0998.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Zeilmaker T, et al. DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2015;81:210–222. doi: 10.1111/tpj.12719. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi S, et al. The glycerophosphoryl diester phosphodiesterase-like proteins SHV3 and its homologs play important roles in cell wall organization. Plant Cell Physiol. 2008;49:1522–1535. doi: 10.1093/pcp/pcn120. [DOI] [PubMed] [Google Scholar]
- 41.de Jesús-Pires C, et al. Plant thaumatin-like proteins: function, evolution and biotechnological applications. Curr. Protein Pept. Sci. 2020;21:36–51. doi: 10.2174/1389203720666190318164905. [DOI] [PubMed] [Google Scholar]
- 42.Batanero E, Ledesma A, Villalba M, Rodríguez R. Purification, amino acid sequence and immunological characterization of Ole e 6, a cysteine-enriched allergen from olive tree pollen. FEBS Lett. 1997;410:293–296. doi: 10.1016/S0014-5793(97)00582-6. [DOI] [PubMed] [Google Scholar]
- 43.Moreira X, Abdala-Roberts L, Gols R, Francisco M. Plant domestication decreases both constitutive and induced chemical defenses by direct selection against defensive traits. Sci. Rep. 2018;8:12678. doi: 10.1038/s41598-018-31041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulm R, et al. Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 2002;21:6483–6493. doi: 10.1093/emboj/cdf646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen K, et al. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020;62:25–54. doi: 10.1111/jipb.12899. [DOI] [PubMed] [Google Scholar]
- 46.Martínez AB, et al. Differences in seed dormancy associated with the domestication of Cucurbita maxima: elucidation of some mechanisms behind this response. Seed Sci. Res. 2017;28:1–7. doi: 10.1017/S0960258517000320. [DOI] [Google Scholar]
- 47.Park JE, Kim YS, Yoon HK, Park CM. Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci. 2007;172:150–157. doi: 10.1016/j.plantsci.2006.08.005. [DOI] [Google Scholar]
- 48.Yang L, Jiang Z, Jing Y, Lin R. PIF1 and RVE1 form a transcriptional feedback loop to control light-mediated seed germination in Arabidopsis. J. Integr. plant Biol. 2020;62:1372–1384. doi: 10.1111/jipb.12938. [DOI] [PubMed] [Google Scholar]
- 49.Hare PD, Møller SG, Huang LF, Chua NH. LAF3, a novel factor required for normal phytochrome A signaling. Plant Physiol. 2003;133:1592–1604. doi: 10.1104/pp.103.028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002;49:387–400. doi: 10.1023/A:1015255030047. [DOI] [PubMed] [Google Scholar]
- 51.Burla B, et al. Vacuolar transport of abscisic acid glucosyl ester is mediated by ATP-binding cassette and proton-antiport mechanisms in Arabidopsis. Plant Physiol. 2013;163:1446–1458. doi: 10.1104/pp.113.222547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seiler C, et al. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J. Exp. Bot. 2011;62:2615–2632. doi: 10.1093/jxb/erq446. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, et al. Construction of a high-density genetic map and analysis of seed-related traits using specific length amplified fragment sequencing for Cucurbita maxima. Front Plant Sci. 2019;10:1782. doi: 10.3389/fpls.2019.01782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo D, et al. Resequencing 200 flax cultivated accessions identifies candidate genes related to seed size and weight and reveals signatures of artificial selection. Front Plant Sci. 2019;10:1682. doi: 10.3389/fpls.2019.01682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Navarro L, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 56.Holdsworth WL, LaPlant KE, Bell DC, Jahn MM, Mazourek M. Cultivar-based introgression mapping reveals wild species-derived Pm-0, the major powdery mildew resistance locus in squash. PloS ONE. 2016;11:e0167715. doi: 10.1371/journal.pone.0167715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45:e18. doi: 10.1093/nar/gkw1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alverson AJ, et al. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae) Mol. Biol. Evol. 2010;27:1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kajitani R, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye C, Hill CM, Wu S, Ruan J, Ma ZS. DBG2OLC: efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci. Rep. 2016;6:31900. doi: 10.1038/srep31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Humann JL, Lee T, Ficklin S, Main D. Structural and functional annotation of eukaryotic genomes with GenSAS. Methods Mol. Biol. 2019;1962:29–51. doi: 10.1007/978-1-4939-9173-0_3. [DOI] [PubMed] [Google Scholar]
- 66.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider M, et al. The UniProtKB/Swiss-Prot knowledgebase and its Plant Proteome Annotation Program. J. Proteom. 2009;72:567–573. doi: 10.1016/j.jprot.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinforma. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koren S, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gong L, Pachner M, Kalai K, Lelley T. SSR-based genetic linkage map of Cucurbita moschata and its synteny with Cucurbita pepo. Genome. 2008;51:878–887. doi: 10.1139/G08-072. [DOI] [PubMed] [Google Scholar]
- 73.Haug-Baltzell A, Stephens SA, Davey S, Scheidegger CE, Lyons E. SynMap2 and SynMap3D: web-based whole-genome synteny browsers. Bioinformatics. 2017;33:2197–2198. doi: 10.1093/bioinformatics/btx144. [DOI] [PubMed] [Google Scholar]
- 74.Goel M, Sun H, Jiao WB, Schneeberger K. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 2019;20:277. doi: 10.1186/s13059-019-1911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sedlazeck FJ, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 78.Pratas D, Silva RM, Pinho AJ, Ferreira PJ. An alignment-free method to find and visualize rearrangements between pairs of DNA sequences. Sci. Rep. 2015;5:10203. doi: 10.1038/srep10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li S, Chou HH. LUCY2: an interactive DNA sequence quality trimming and vector removal tool. Bioinformatics. 2004;20:2865–2866. doi: 10.1093/bioinformatics/bth302. [DOI] [PubMed] [Google Scholar]
- 80.Hoffmann S, et al. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caboche S, Audebert C, Lemoine Y, Hot D. Comparison of mapping algorithms used in high-throughput sequencing: application to Ion Torrent data. BMC Genomics. 2014;15:264. doi: 10.1186/1471-2164-15-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy–Weinberg equilibrium. Am. J. Hum. Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl P. A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evolution. 2013;4:782–788. doi: 10.1111/2041-210X.12067. [DOI] [Google Scholar]
- 88.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Mol. Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ardelean, C. F. et al. Evidence of human occupation in Mexico around the Last Glacial Maximum. Nature548, 87–92 (2020). [DOI] [PubMed]
- 91.Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Stat. Sci. 1992;7:457–472. [Google Scholar]
- 92.Malinsky, M., Matschiner, M. & Svardal, H. Dsuite—fast D-statistics and related admixture evidence from VCF files. Mol. Ecol. Resour. (2020). [DOI] [PMC free article] [PubMed]
- 93.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng Y, et al. Cucurbit Genomics Database (CuGenDB): a central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 2019;47:D1128–D1136. doi: 10.1093/nar/gky944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the raw sequencing data and genome assemblies are available in the National Center of Biotechnology Information under the BioProject accession PRJNA485527 (genome accesions SDJN00000000 and JAGKQH010000000; SRA accesions available in Table S3). The genome assemblies are also available in Figshare (https://doi.org/10.6084/m9.figshare.14370584.v1) and at the Cucurbit Genomics Database94.