Abstract
Serine proteases catalyze a multi-step covalent catalytic mechanism of peptide bond cleavage. It has long been assumed that serine proteases including thrombin carry-out catalysis without significant conformational rearrangement of their stable two-β-barrel structure. We present nuclear magnetic resonance (NMR) and hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments on the thrombin-thrombomodulin (TM) complex. Thrombin promotes procoagulative fibrinogen cleavage when fibrinogen engages both the anion binding exosite 1 (ABE1) and the active site. It is thought that TM promotes cleavage of protein C by engaging ABE1 in a similar manner as fibrinogen. Thus, the thrombin-TM complex may represent the catalytically active, ABE1-engaged thrombin. Compared to apo- and active site inhibited-thrombin, we show that thrombin-TM has reduced μs-ms dynamics in the substrate binding (S1) pocket consistent with its known acceleration of protein C binding. Thrombin-TM has increased μs-ms dynamics in a β-strand connecting the TM binding site to the catalytic aspartate. Finally, thrombin-TM had doublet peaks indicative of dynamics that are slow on the NMR timescale in residues along the interface between the two β-barrels. Such dynamics may be responsible for facilitating the N-terminal product release and water molecule entry that are required for hydrolysis of the acyl-enzyme intermediate.
Subject terms: Biochemistry, Biophysics
Introduction
Serine proteases have provided textbook examples of enzyme catalysis since the first crystal structure of chymotrypsin was solved in 19671. The well-known catalytic triad, the S1 substrate-binding pocket, and the oxy-anion hole provide nucleophilic and acid–base catalysis. The myriad of serine protease crystal structures show that the arrangement of these critical structural elements is highly similar leading to the assumption that serine proteases transition between their catalytic steps without significant structural rearrangements. We have been studying the serine protease, thrombin, which catalyzes the last step of the blood-clotting protease cascade. The crystal structure of α-thrombin reveals the canonical chymotrypsin (CT) fold, comprised of an N-terminal and a C-terminal β-barrel, as well as an aligned catalytic triad containing His 57CT (79seq), Asp 102CT (135seq), and Ser 195CT (241seq)2. It should be noted that the thrombin residues numbered according to the chymotrypsin system will be referenced with a “CT” subscript, while those numbered using the sequential system will include “seq”. The proteolytic cleavage of the zymogen, prothrombin, generates the new N-terminus of the thrombin heavy chain, which is observed to be inserted into the Ile cleft forming a key interaction with Asp 194 and positioning Ser 195 for catalysis3–5. Using hydrogen–deuterium exchange mass spectrometry (HDX-MS), we previously showed, however, that the new N-terminal peptide is highly exchanging in the absence of the covalent active site inhibitor D-Phe-Pro-Arg-chloromethylketone (PPACK)6. In fact, binding of PPACK reduced exchange throughout the thrombin molecule. Similarly, NMR experiments comparing apo- and PPACK-thrombin showed that the presence of PPACK leads to the appearance of many, strong resonances that are either weak or absent in the apo-thrombin spectrum7,8. A number of thrombin resonances remained absent from heteronuclear single quantum coherence (HSQC) spectra of PPACK thrombin in multiple NMR experiments9,10. These missing resonances were largely localized to the loop regions of thrombin, which are the main structural elements that distinguish thrombin from trypsin and chymotrypsin, suggesting that the thrombin loops remain dynamic even when an inhibitor is bound at the active site. HDX-MS and accelerated molecular dynamics (AMD) simulations also reported the loops of thrombin to be conformationally dynamic6–8,11. In addition, Carr-Purcell-Meiboom-Gill (CPMG) experiments revealed a large number of backbone NH groups undergo μs-ms dynamics throughout apo thrombin and a pathway of residues connecting the allosteric site (ABE1) to the active site remain mobile in PPACK thrombin8.
The loops of thrombin have been shown to play important roles in thrombin activity. Residues from the 170sCT, 180sCT, and 220sCT loops have been implicated in the Na+-mediated allosteric activation of thrombin through the structuring of the S1 binding site that results from the presence of a Na+ ion bound in this region12,13. Residues within the 30sCT and 70sCT loops interact with the thrombin cofactor thrombomodulin (TM). TM binding toggles the substrate specificity of thrombin away from pro-coagulative substrates that encourage clotting once proteolytically cleaved by thrombin, and towards the anticoagulative thrombin substrate protein C, which acts to shut down the clotting cascade once activated by thrombin14. Though TM contains multiple domains, the TM456 fragment containing only the 4th, 5th, and 6th EGF-like domains of TM is able to invoke the full anticoagulative activity of thrombin15,16. The active site inhibitor, D-Glu-Gly-Arg-chloromethylketone (GGACK), was required to solve the crystal structure of thrombin bound to TM45617. Surface plasmon resonance reported rapid association and dissociation rates (6.7 × 106 M-1 s-1 and 0.033 s-1 respectively) for the interaction of TM456 with thrombin18, and Isothermal titration calorimetry (ITC) showed that TM456-binding is primarily driven by a change in entropy rather than enthalpy19,20. HDX-MS experiments on thrombin in the presence and absence of TM have reported reductions in amide exchange in regions of thrombin –such as the active site- that are distant from the TM-binding site, consistent with the fact that TM is an allosteric modulator of thrombin function6,11.
Although multiple studies have investigated the impact of TM binding on thrombin dynamics, there is still no clear explanation as to how TM binding modulates the catalytic activity of the protease. Here we report NMR CPMG experiments to characterize the dynamic consequences of TM456 binding to thrombin on the µs-ms timescale21,22. Complementary use of HDX-MS reveals changes in the dynamics of some of the loops for which NMR resonances were missing23,24. Through the complementary use of these two biophysical methods, we report the identities of the thrombin residues that facilitate allosteric communication between the TM binding site, the catalytic residues, and the S1 binding pocket. TM binding remodels thrombin dynamics on multiple timescales; ordering the substrate binding pocket while also promoting the conformational flexibility required for the mechanistic steps of the serine protease reaction.
Results
Resonance assignments and chemical shift perturbations
We previously reported resonance assignments, backbone dynamics and CPMG analysis of the μs-ms dynamics for apo thrombin8 and PPACK thrombin8,10. In this previous work, we were able to assign 82% of the thrombin sequence in PPACK-thrombin and 73% in apo-thrombin. Here we have analyzed the complex between apo thrombin and the fragment of thrombomodulin, TM456, which is fully active in promoting protein C cleavage/activation6. The sample used for NMR experimentation contained a 1.5 molar excess of unlabeled TM to labeled thrombin. The thrombin-TM456 spectra more closely resembled the apo-thrombin and a similar percentage of amino acids could be assigned (Supplementary Fig. 1). Every single cross peak in the spectra was assigned suggesting that missing cross peaks are likely due to conformational heterogeneity. Nine residues- clustered around the C-terminal -barrel and the S1 pocket- which were missing from the apo-thrombin spectra could be assigned in thrombin-TM456. These nine residues were also observed in PPACK-thrombin HSQC spectra. Interestingly, four assignments were present in the apo-thrombin spectrum that were missing from the thrombin-TM456 spectra, and these were all located within the N-terminal -barrel.
There were 23 residues in the thrombin-TM456 spectra that showed multiple peaks, while the resonances for these same assignments in the apo and PPACK spectra were singlets. Often, one peak of the multiplet corresponded to the equivalent resonance in apo-thrombin (Supplementary Fig. 2). All of these assignments were verified by backbone walks using HNCA, HNCO, and HNCOCA spectra. The appearance of these multiplet resonances suggested that the presence of TM456 induces dynamics that are slow on the NMR timescale (< 80 s-1) in these residues.
The TM binding site remains dynamic when TM is present
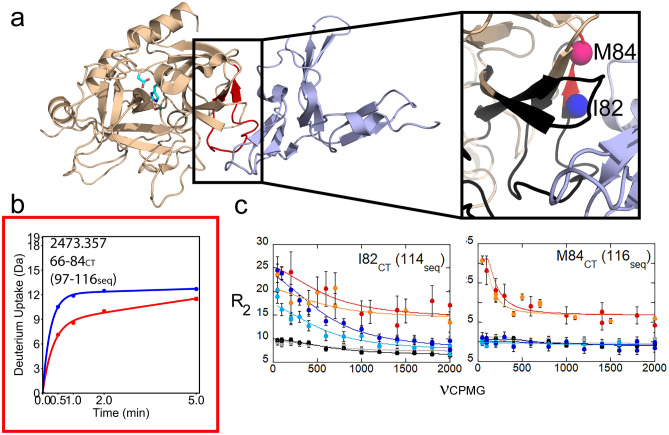
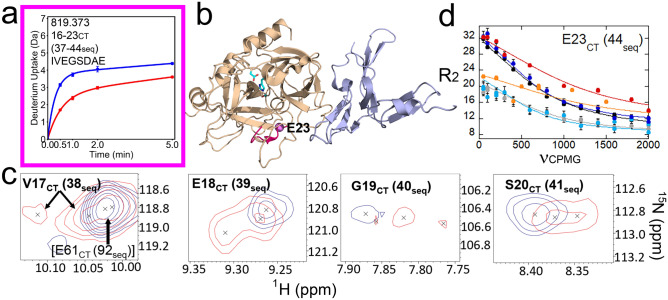
Despite the stabilization of thrombin afforded by PPACK binding20, resonances corresponding to the N-terminus of the heavy chain Ile 16CT (37seq) and nearly all of the residues in the 30sCT (residues 54–61seq) and 70sCT (residues 97–113seq) loops as well as residues 151-156CT (193-197seq), and the 220sCT loop 221-221ACT (268-269seq) were not observed in previous NMR experiments on PPACK-thrombin or apo-thrombin8,10. Cross peaks for these residues were also not observed in the thrombin-TM456 spectra. We were, however, able to learn about the solvent accessibility/dynamics of these loops from HDX-MS, which reported on 99% of the thrombin sequence for experiments on WT-thrombin with TM456 either present or absent (Supplementary Fig. 3). The HDX-MS experiments showed a ~ 3 deuteron decrease in exchange of residues 66-84CT (97-116seq) in the 70sCT loop within 1 min when TM456 was present, consistent with the exclusion of solvent due to TM456 binding (Fig. 1a,b). Interestingly, this binding site “protection” was transient, becoming less over the 5 min time course of the HDX experiment suggesting that this region remains dynamic, which is consistent with our inability to observe resonances for the 70sCT loop in the NMR. We did observe resonances for Ile 82CT (114seq) and Met 84CT (116seq) in the β-sheet following the 70sCT loop and CPMG curves for these two residues showed that their chemical environments are changing on the μs-ms timescale (Fig. 1a,c).
Figure 1.
(a) Structure of thrombin (wheat) bound to TM456 (light blue) [PDB ID: 1DX5]. The side chains of the catalytic triad are shown as cyan sticks. Thrombin residues 66-84CT (97-116seq) of the 70sCT loop are colored red (left). A close-up of the 30sCT and 70sCT loops (right) has residues missing from the thrombin-TM456 HSQC colored black, and the amides in this strand for which CPMG data was obtained as spheres. For all figures, pink spheres indicate residues with NH resonances experiencing increased μs-ms dynamics in thrombin-TM456 and blue spheres indicate residues with NH resonances experiencing reduced μs-ms dynamics as compared to apo-thrombin. (b) Deuterium uptake plots for the peptide spanning residues 66-84CT (97-116seq; MH + 2473.357) in apo-thrombin (blue) compared to thrombin bound to TM456 (red). Error bars (standard deviation of three replicates) are shown. (c) CPMG plots for resonances corresponding to Ile 82CT (112seq) and Met 84CT (116seq). For all figures, the red and orange curves are from spectra collected on thrombin-TM456 at 800 MHz and 600 MHz respectively, the blue and cyan curves are from apo-thrombin at 800 MHz and 600 MHz respectively, and the black and grey curves are from PPACK-thrombin at 800 MHz and 600 MHz respectively. For all figures, molecular structures were produced in PyMol, deuterium uptake plots were produced in DECAv112 (
available at https://github.com/komiveslab/DECA), NMR spectra were plotted in SPARKY (available at https://nmrfam.wisc.edu/nmrfam-sparky-distribution/), and NMR CPMG curves were plotted in Kaleidagraph v4.5.3.
The thrombin N-terminal β-barrel becomes more dynamic when TM is bound
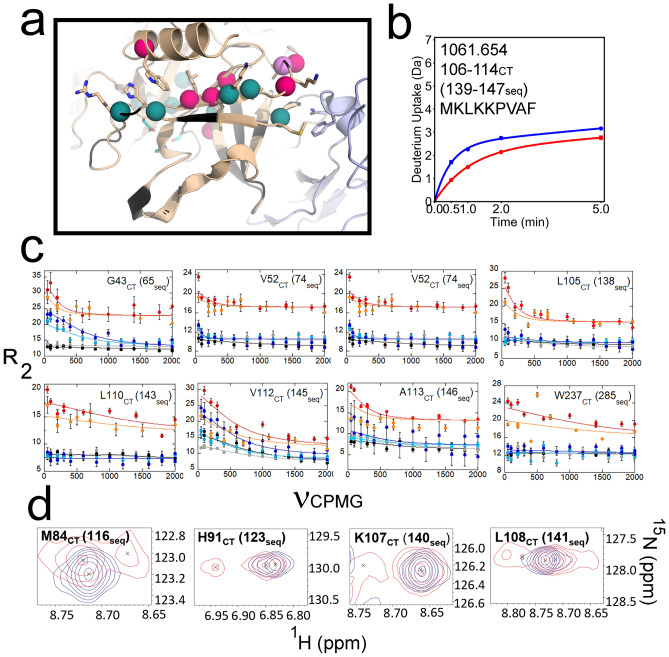
Ten residues within the thrombin N-terminal β-barrel experienced more significant dynamics on the μs-ms timescale when TM456 was present. A subgroup of these residues was distributed along two antiparallel -strands that connect the thrombin 70sCT and 90sCT loops (Fig. 2a). HDX-MS showed residues 106-114CT (139-147seq) were more protected from deuterium exchange (by ~ 1 deuteron) when TM456 was present (Fig. 2b). Interestingly, CPMG experiments showed that TM456 induces μs-ms dynamics in four residues within this segment (Fig. 2c). This subgroup of residues also included Leu 105CT (138seq), suggesting that the dynamic modulation induced by TM456 may be transferred into the catalytic Asp 102CT (189seq) through these antiparallel β-strands. Resonances corresponding to Ile 88 and Tyr 89CT (120-121seq) located directly across from Leu 105CT and Met 106CT in the neighboring β-strand could be assigned in the apo-thrombin spectrum, but they were absent in the thrombin-TM456 spectrum supporting the claim that the residues in this region are dynamic. Although we could assign Asp 102CT (189seq), CPMG analysis could not be performed due to the presence of overlapped resonances. In addition, a few residues within these same antiparallel β-strands showed peak multiplets (Fig. 2d). Thus, TM456 appeared to stabilize the end of the β-strand leading to the catalytic Asp 102CT (189seq), yet also induced μs-ms dynamics in this strand and the neighboring strand. These results provide strong evidence that TM binding at the 70sCT loop induces dynamics along the antiparallel β-strands that connect the TM binding site to the active site catalytic residues.
Figure 2.
(a) Structure of thrombin (wheat) bound to TM456 (light blue) [PDB ID: 1DX5]. The residues corresponding to resonances missing from the thrombin-TM456 HSQC are colored black. The residues corresponding to multiplet resonances (teal spheres), resonances experiencing increased μs-ms dynamics in thrombin-TM456 compared to apo-thrombin (pink spheres), and resonances experiencing similar μs-ms dynamics in thrombin-TM456 and apo-thrombin (violet spheres) and the catalytic triad (cyan sticks) are shown. (b) Deuterium uptake plot for the peptide spanning residues 106-114CT (139-147seq; MH + 1061.654). (c) CPMG plots for resonances in the thrombin N-terminal β-barrel (symbols as in Fig. 1). (d) Examples of thrombin-TM456 HSQC (red) doublet resonances compared to apo-thrombin (blue) resonances for residues in the N-terminal β-barrel.
TM binding allosterically reduces the dynamics of the S1 pocket, and also induces a slow time scale conformational change in the C-terminal -barrel
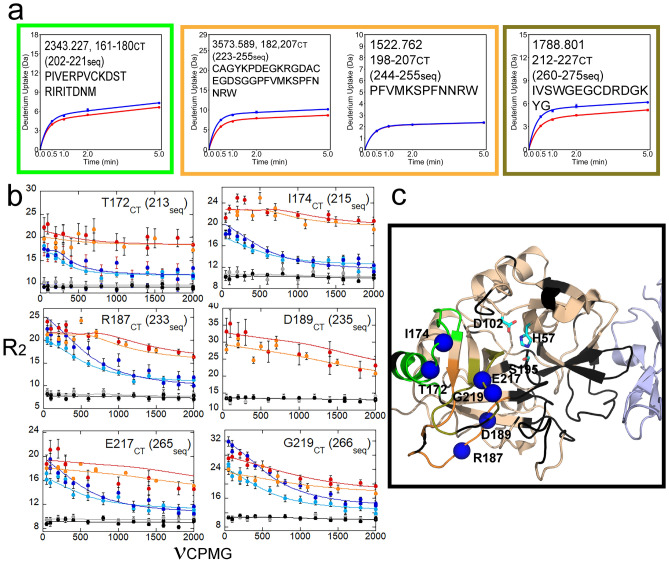
HDX-MS revealed that TM456 binding reduced exchange in the 170sCT, 180sCT, and 220sCT loops of thrombin despite the > 20 Å distance between the TM binding site and these loops. Upon TM456 binding, decreased exchange of ~ 1 deuteron was observed for the 170sCT loop residues 161-180CT (202-221seq) and of ~ 2 deuterons for the 180sCT loop residues 182-198CT based on subtraction of deuterium uptake into residues 198-207CT (244-255seq) from uptake into residues 182-207CT (223-255seq). Finally, a decreased exchange of ~ 1.5 deuterons was observed for the 220sCT loop residues 212-227CT (260-275seq) (Fig. 3a,c). CPMG analysis identified seven residues within these loops that showed significantly decreased μs-ms dynamics when TM456 was present as compared to apo-thrombin (Fig. 3b,c).
Figure 3.
(a) Deuterium uptake plots for the peptides spanning residues 161-180CT (202-221seq; MH + 2343.227), 182-207CT (223-255seq; MH + 3575.589), 198-207CT (244-255seq; MH + 1522.762), and 212-227CT (260-275seq; MH + 1788.801). (b) CPMG plots for resonances experiencing reduced μs-ms dynamics around the S1 pocket when TM456 is bound compared to apo-thrombin (symbols as in Fig. 1). (c) Structure of thrombin (wheat) bound to TM456 (light blue) [PDB ID: 1DX5]. The residues within the peptides identified in panel (A) are colored accordingly. Residues with missing HSQC resonances (black) and residues corresponding to resonances experiencing reduced μs-ms dynamics in thrombin-TM456 compared to apo-thrombin (blue spheres) are shown.
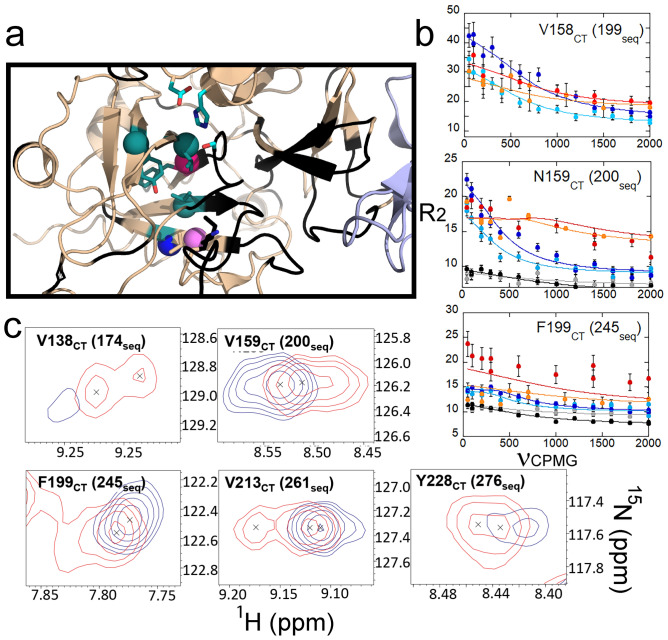
Val 158CT and Asn 159CT are in the center of the β-strand following the 140sCT loop and leading to the 170sCT loop (Fig. 4a). A stack of three valines (residues 158CT (199seq), 138CT (174seq) and 213CT (261seq)) forms one side of the hydrophobic core of the C-terminal β-barrel (Fig. 4a). Val 158CT (199seq) experiences different chemical environments on the μs-ms timescale in all three thrombin states (Fig. 4b). Val 138CT (174seq) also showed μs-ms timescale dynamics in apo thrombin, but this resonance became a doublet in the thrombin-TM456 HSQC spectrum (Fig. 4c). Finally, V213CT (261seq) was also a doublet only in the thrombin-TM456 HSQC spectrum (Fig. 4c). Adjacent to Val 158CT, Asn 159CT was a doublet only in the thrombin-TM456 complex, and it was experiencing different chemical environments on the μs-ms timescale in apo-thrombin but not in the thrombin-TM456 complex (Fig. 4b,c). The core of the C-terminal β-barrel is flanked by Phe 199CT (245seq) which also was a doublet in the thrombin-TM456 HSQC spectrum and showed μs-ms timescale dynamics in all three thrombin states (Fig. 4b,c). Tyr 228CT (276seq) could be assigned to both a doublet resonance and a singlet resonance with notably different chemical shifts (Supplementary Fig. 4). Thus, the addition of TM456 causes decreased H/D exchange in the loops around the thrombin S1 pocket, reduces μs-ms timescale dynamics, and also induces dynamics that are slow on the NMR timescale in the residues that make up the hydrophobic core of the C-terminal β-barrel.
Figure 4.
(a) Structure of thrombin (wheat) bound to TM456 (light blue) [PDB ID: 1DX5] showing the C-terminal β-barrel. Residues with missing resonances (black), doublet resonances (teal spheres), resonances experiencing increased μs-ms dynamics (pink spheres), resonances experiencing similar μs-ms dynamics (violet spheres) and resonances experiencing decreased μs-ms dynamics (blue spheres) in thrombin-TM456 compared to apo-thrombin are shown. (b) CPMG plots for resonances in the thrombin C-terminal β-barrel (symbols as in Fig. 1). (c) Examples of thrombin-TM456 HSQC (red) doublet resonances compared to apo-thrombin (blue) resonances.
TM induces slow dynamics in the N-terminus of the heavy chain and the 140sCT loop
HDX-MS showed that residues 16-23CT (36-44seq) at the N-terminus of the thrombin heavy chain showed decreased exchange when TM456 was bound to thrombin (Fig. 5a). The x-ray crystal structure of thrombin-TM456 (PDB ID: 1DX517), shows the hydrophobic side chains of Val 158CT (199seq) and Val 138CT (174seq) interacting with the side chain of Ile 16CT (36seq) at the N-terminus of the thrombin heavy chain, which inserts into the Ile cleft to form the catalytically active conformation. Adjacent to the new N-terminus belonging to Ile 16CT, cross peaks for Val 17CT- Ser 20CT (38-41seq) also were doublets in the thrombin-TM456- spectrum (Fig. 5b). The cross peak for Val 17CT (38seq) could not be assigned in apo-thrombin, but it could be assigned in the thrombin-TM456 spectrum suggesting that it adopts a more defined conformation(s) in the thrombin-TM456 complex. Glu 23CT (44seq) showed dynamics on the μs-ms timescale in all three thrombin states, providing more evidence that the N-terminus of the heavy chain remains somewhat mobile in the thrombin-TM456 complex (Fig. 5c). Thus, TM456 modulates the dynamics of the N-terminus of the heavy chain decreasing amide exchange and inducing dynamics that are slow on the NMR time scale.
Figure 5.
(a) Deuterium uptake plot for the peptide spanning residues 16-23CT (37-44seq; MH + 819.373). (b) Location of residues 16-23CT(37-44seq) in the structure of thrombin (wheat) bound to TM456 (light blue) [PDB ID: 1DX5]. (hot pink). The resonance corresponding to Glu 23CT (44seq) experienced similar μs-ms dynamics in thrombin-TM456 and apo-thrombin (purple sphere). (c) Examples of thrombin-TM456 HSQC (red) doublet resonances compared to apo-thrombin (blue) resonances for residues at the N-terminus of the heavy chain. (d) CPMG plot for the resonance corresponding to residue Glu 23CT (44seq) (symbols as in Fig. 1).
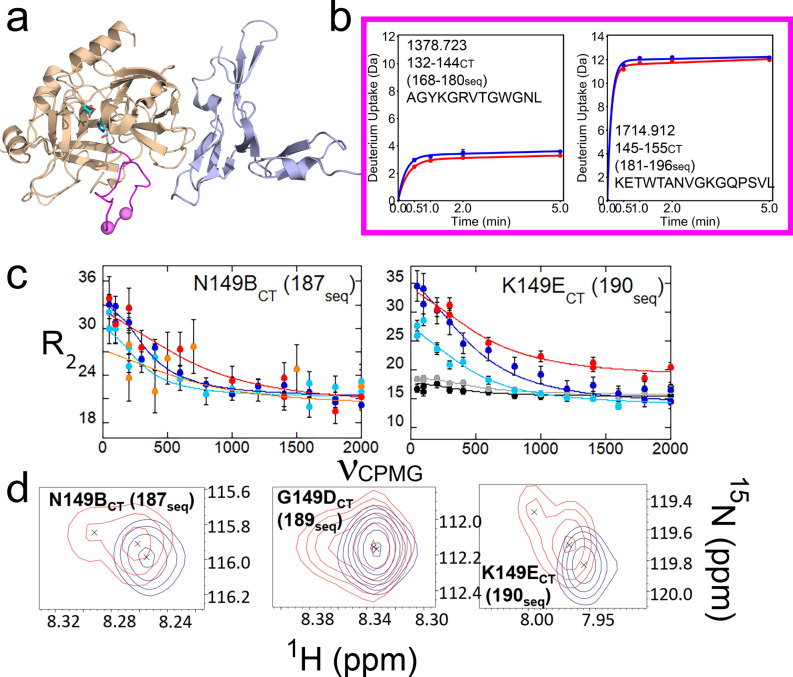
Residues 139-149ACT (175-186seq), the N-terminal half of the 140sCT loop could not be assigned in any thrombin samples (Fig. 6a). The dynamics of this part of the 140sCT loop were determined instead from HDX-MS experiments. Residues 145-155CT (181-196seq) showed nearly complete exchange, consistent with this loop being very dynamic and solvent-exposed whereas residues 132-144CT (168-180seq) exchanged much less (Fig. 6b). The deuterium exchange of the 140sCT loop did not change with the addition of TM456. Residues N149BCT and K149ECT at the tip of the 140sCT loop could be assigned and CPMG data revealed they were undergoing similar μs-ms timescale dynamics in all thrombin states (Fig. 6c). Residues 149B-150CT (187-191seq) all became doublets in the thrombin-TM456 HSQC (Fig. 6d) but were singlets in the other thrombin forms. Thus, TM456 induces slow timescale dynamics in the C-terminal half of the 140sCT loop.
Figure 6.
(a) Structure of thrombin (wheat) bound to TM456 (light blue) [PDB ID: 1DX5]. Residues 140-155CT (176-196seq) of the 140sCT loop are colored magenta. The amides of residues corresponding to resonances experiencing μs-ms dynamics in both thrombin-TM456 and apo-thrombin are shown as violet spheres. Sidechains are shown for the catalytic triad (cyan). (b) Deuterium uptake plots for the peptides spanning residues 132-144CT (168-180seq; MH + 1378.723) and 145-155CT (181-196seq; MH + 1714.912). The colors are the same as in previous figures. (c) CPMG plots for thrombin resonances corresponding to residues Asn 149BCT (187seq) and Lys 149ECT (190seq). The colors of the curves are the same as in previous figures. CPMG data could not be obtained from the 600 MHz spectra for Thr 149CT (187seq). (d) Examples of thrombin-TM456 HSQC (red) doublet resonances compared to apo-thrombin (blue) resonances for residues in the 140sCT loop.
Discussion
For thrombin to catalyze fibrinogen cleavage, fibrinogen must engage both the anion binding exosite 1 (where TM binds) and the active site25. It is thought that TM promotes cleavage of protein C by engaging anion binding exosite 1 in a manner similar to fibrinogen. Thus, one can think of the thrombin-TM complex as revealing the catalytically active form of thrombin via engagement of anion binding exosite 1. Previous studies have highlighted the dynamics of thrombin loops7,9,11, 26, and suggested that loop dynamics may allow it to bind different substrates. This has led some to speculate that TM may take advantage of modulating loop dynamics to direct catalysis toward protein C, its anticoagulant substrate26–29. Here we present a combination of HDX-MS and NMR CPMG data yielding a much more complete understanding of dynamics throughout the thrombin-TM complex. The rich dynamic behavior of the thrombin-TM complex appears to be related to the complicated serine protease multi-step catalytic mechanism.
A remarkable finding is that entropy compensation appears to occur between the N-terminal and C-terminal β-barrels of thrombin when TM binds. Ten residues showed increased dynamics in the N-terminal β-barrel when compared to apo- and PPACK thrombin. Conversely, nine residues showed decreased dynamics in the C-terminal β-barrel. Thus, TM appears to order the C-terminal β-barrel and disorder the N-terminal β-barrel to which it is bound. This result was completely unexpected considering that TM binding reduces amide exchange throughout thrombin.
HDX-MS experiments showed reduced exchange at the TM binding site, but this could be due either to reduced dynamics or to interface protection6. The H/D exchange protection did not persist after 5 min of incubation, which is likely due to the very rapid association and dissociation kinetics observed for thrombin-TM binding18. NMR signals were not observed for much of the 70sCT loop in apo and PPACK-thrombin8, and here we show that these cross peaks are also missing even when TM is bound to thrombin. The thrombin-TM interaction is entirely entropically-driven20, which is also consistent with the binding site remaining disordered. Apo thrombin has a large number of residues that experience different chemical environments on the μs-ms timescale throughout the molecule8.
Interestingly, the reduction in μs-ms timescale dynamics observed in the C-terminal β-barrel involves residues in the loops implicated in the formation of the S1 pocket12,13. Six residues within these loops were experiencing different chemical environments on the μs-ms timescale in apo-thrombin but their dynamics were quenched by binding PPACK. Remarkably, the dynamics of all of these residues were also quenched in the thrombin-TM complex, strongly indicating that TM binding over 25 Å away allosterically organizes the S1 pocket. These residues include Asp 189 at the bottom of the S1 substrate binding pocket and the sodium-binding site. The fact that this part of thrombin is significantly rigidified by TM was also observed through crystallographic studies on thrombin-TM in the absence of Na+29, and explains the insensitivity of the thrombin-TM complex to Na+ concentration 30, as well as the increased binding rate of thrombin active-site inhibitors observed when TM is bound 31–35. Such a pre-formation of this critical substrate-binding pocket may account for the marked improvement of the association rate of protein C into the thrombin active site from 27 M-1 s-1 in the absence of TM to 105 M-1 s-1 in the presence of TM (Fig. 7a)36.
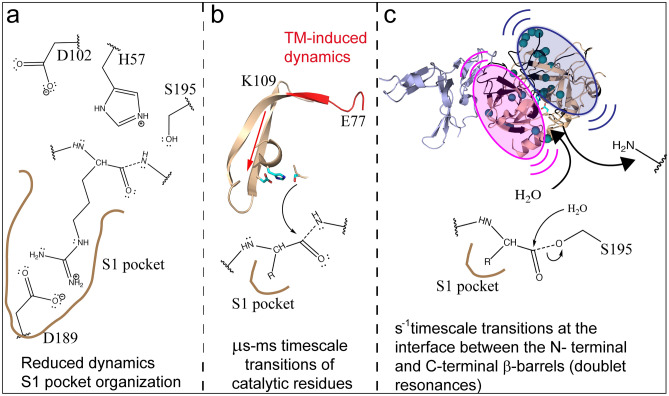
Figure 7.
A model that describes the steps of the serine protease mechanism. (a) TM decreases dynamics in the substrate binding pocket to promote protein C binding. (b) TM-induces μs-ms dynamics that may promote the optimal activity of the catalytic triad. The backbone of residues 77-84CT (108-116seq) is colored red, and the backbone of residues 56-58CT (78-80seq), 85-113CT (117–146), and 195CT (241seq) are colored wheat. The side chains of the catalytic triad are shown as cyan sticks. (c) Doublet resonances indicate TM induced dynamics on the seconds time scale of the N-terminal (pink oval) and C-terminal (blue oval) β-barrels that may be required for the release of the first product from Ser 195CT and entry of H2O into the active site for acyl enzyme hydrolysis.
The next step in the proteolytic cleavage of protein C by thrombin is formation of the first covalently-bound intermediate. This intermediate is probably mimicked by the PPACK-thrombin complex6–9,11, which was observed to have the lowest degree of conformational heterogeneity7,8. A similar conformationally rigid, covalently-bound intermediate species was recently reported for the UMP/CMP kinase from D. discoideum37, suggesting that rigidity may be a common feature of covalently-bound reaction intermediates.
TM improves the kcat for thrombin cleavage of protein C by over 200 fold38. We found that TM induces dynamics in a pair of β-strands that link the TM binding site to the catalytic residues through the N-terminal β-barrel. This pair of β-strands, which connect the 70sCT and 90sCT loops, was only dynamic in the thrombin-TM complex, but not in apo-thrombin or PPACK-thrombin. Community network analysis of accelerated MD simulations had indicated that several small communities of residues were moving independently of one another in this region of apo-thrombin, whereas these communities coalesce into a single community when TM binds11. We now have evidence that these predicted correlated motions involve the pair of β-strands, one of which leads to the catalytic Asp 102CT (189seq). It is interesting to speculate that these motions may be important for catalysis since Asp 102CT must closely approach His 57CT in order to raise its pKa to promote formation of both tetrahedral intermediates in the catalytic mechanism (Fig. 7b).
Remarkably, many cross peaks in the HSQC were doublets only in thrombin-TM456 but not in apo-thrombin or PPACK thrombin. When we mapped all of the residues for which doublets are observed onto the structure of thrombin, we saw that they fall all along the interface between the N-terminal and C-terminal β-barrels (Fig. 7c). Doublets represent dynamics that are slow on the NMR time scale, < 80 s-1 yet this is still fairly fast compared to the kcat for protein C cleavage of 5 s-116. It is tempting to speculate that the doublets reveal thrombin dynamics that are important for catalysis of the slow steps of peptide bond cleavage. For serine proteases, the slow steps are thought to be release of the first product, the N-terminal portion after cleavage of the scissile bond, and subsequent binding of a water molecule that is required for hydrolysis of the acyl-enzyme intermediate which remains bound in the S1 pocket (Fig. 7c).
These data support a model of the serine protease reaction coordinate that explains how the conformational heterogeneity induced by TM facilitates effective catalysis of protein C activation. TM binding allosterically remodels apo-thrombin dynamics to accomplish three catalytic functions: (1) TM decreases dynamics in the C-terminal β-barrel forming the S1 pocket to accelerate protein C binding, (2) TM enhances μs-ms timescale dynamics in a pair of β-strands connecting the TM binding site to the 90sCT loop in the N-terminal β-barrel. The dynamics of these strands likely alter Asp 102CT to promote catalysis, and (3) TM causes dynamics that are slow on the NMR timescale along the interface between the N-terminal and C-terminal β-barrels. These slower time scale dynamics are likely required for release of the first product and subsequent hydrolysis of the covalently bound acyl-enzyme intermediate.
Methods
Thrombin expression and purification
The S195M mutant and WT human thrombin were expressed and refolded from E. coli as previously described8,10. After initial refolding and initial purification, the S195M thrombin was mixed with WT thrombin at a ratio of 1:30 WT:S195M and was left to rock for 12–16 h at 30 °C to convert the meizothrombin species formed to α-thrombin. After activation, the WT α-thrombin was removed by addition of biotinyl-PPACK (Haematologic Technologies) then captured on streptavidin resin (Thermo Scientific) and removed. The α-form of the S195M thrombin was isolated from other forms by chromatography on a 10/100 GL MonoS cation exchange column (GE Healthcare Life Sciences) using a gradient of 100 mM–500 mM NaCl in 25 mM phosphate pH 6.5. This method of thrombin purification has been shown to result in > 95% α-thrombin, and previous NMR analysis of isotopically labeled thrombin prepared this way demonstrated that the species present was α-thrombin10,18.
Production of TM456
The TM456 species used for all thrombin-TM456 experiments was “TM456m” which ends at Gly449 and has Cys448 changed to Ser as previously described16. Protein C activation assays were performed to assess the specific activity of the expressed TM456 as previously described16.
Hydrogen–deuterium exchange mass spectrometry
The α-thrombin-TM456 complex was prepared at a molar ratio of 1:10 thrombin:TM456, to ensure 99% of thrombin would be bound to TM456 at the protein concentrations used in the experiment. HDX-MS was performed using a Waters Synapt G2Si system with HDX technology (Waters Corporation)39. Deuterium exchange reactions were measured in triplicate and experiments were performed and analyzed as described previously40. HDX-MS was performed using a Waters Synapt G2Si system with HDX technology (Waters Corporation)39. Deuterium exchange reaction were prepared using a Leap HDX PAL autosampler (Leap technologies, Carrboro, NC). D2O buffer was prepared by lyophilizing 1 mL of 250 mM phosphate pH 6.5 along with 850 mM NaCl for the apo-thrombin experiments and 1000 mM NaCl for the thrombin-TM456 experiments, before being resuspended in 10 mL 99.96% D2O immediately before use. For each deuteration time point (performed in triplicate), 5 µL of protein was held at 25 °C for 5 min before being mixed with 55 µL of D2O buffer, then quenched for 1 min at 1 °C by mixing 1:1 with ice cold 250 mM TCEP pH 2.5. The quenched sample was then injected into a 50 µL sample loop, followed by digestion on an in-line pepsin column (immobilized pepsin, Pierce, Inc.) at 15 °C. The resulting peptides were captured on a BEH C18 Vanguard pre-column, separated by analytical chromatography (Acquity UPLC BEH C18, 1.7 µM, 1.0 × 50 mm, Waters Corporation) using a 7–50% acetonitrile in 0.1% formic acid over 7.5 min, and electrosprayed into the Waters Synapt G2Si quadrupole time-of-flight mass spectrometer. The mass spectrometer was set to collect data in the Mobility, ESI+ mode; mass acquisition range 200–2,000 (m/z); scan time 0.4 s with continuous lock mass correction. For peptide identification, the mass spectrometer was set to collect data in MSE, mobility ESI + mode and 10 μM α-thrombin. Peptides masses were identified using PLGS 2.5 (Waters Corporation) requiring a minimum number of 250 ion counts for low energy peptides and 50 ion counts for their fragment ions; the peptides also had to be larger than 1500 Da.
DynamX 3.0 (Waters Corporation) was used to determine deuterium uptake based on the peptides identified by PLGS, but incorporating additional filters including a cut-off score of 7, minimum products per amino acid of 0.2, maximum MH + error tolerance of 5 ppm, retention time standard deviation of 5%, and requiring that the peptide be present in at least 2 of the 3 peptide identification runs. The deuterium uptake for each peptide was calculated by comparing the centroids of the mass envelopes of the deuterated samples vs. the undeuterated controls. For all HDX-MS data, at least 2 biological replicates were analyzed, each with 3 technical replicates. Data are represented as mean values + /- SEM of 3 technical replicates due to processing software limitations, however biological replicates were highly reproducible due to use of the LEAP robot for all experiments. The deuterium uptake was corrected for back-exchange using a global back exchange correction factor (typically 25%) determined from the average percent exchange measured in disordered termini of various proteins. ANOVA analyses and t tests with a p value cutoff of 0.05 implemented in the program, DECA, were used to determine the significance of differences between HDX data points40. The peptides reported on the coverage maps are actually those from which deuterium uptake data were obtained. Deuterium uptake plots were generated in DECA (github.com/komiveslab/DECA).
NMR sample preparation
Purified 2H-15 N-S195M or 2H-15 N-13C-S195M thrombin was added to unlabeled TM456 at a molar ratio of 1:1.5 thrombin:TM456 to ensure 99% of thrombin would be bound to TM456 at the protein concentrations used in the experiment. The proteins were suspended in buffer containing 25 mM phosphate pH 6.5,150 mM NaCl, 0.05% sodium azide and 10% D2O at a final concentration of 0.12 mM thrombin.
NMR data collection
For all experiments, Shigemi salt-tolerant susceptibility matched NMR tubes were used. The sample volume was 170 μL and the tube was properly oriented inside the magnet to optimize S/N and minimize sample heating 41. All NMR experiments were acquired on Varian VNMRS (Agilent Technologies, Santa Clara, CA) and Bruker Avance III spectrometers operating at 600, 800 and 900 MHz (1H), and equipped with cryogenic triple-resonance probes at 298 K.
For the sequence specific assignment of backbone resonances, the TROSY version of 2D 1H,15 N-HSQC and 3D HNCA, 3D HN(CO)CA and 3D HNCO spectra were recorded42,43. The acquisition parameters for these spectra are reported in Table S1. The 3D spectra were recorded with Non-Uniform Sampling (NUS) using optimized Poisson-gap distribution schedules44 with sampling rates ranging between 36 and 38%. Spectra were processed using NMRPipe. The 3D spectra recorded with NUS were reconstructed using the SMILE package available in NMRPipe45, and analyzed with NMRFAM-SPARKY.
To identify conformational exchange, spectra were recorded at 600 and 800 MHz (1H) using a CPMG relaxation dispersion experiment with TROSY selection. The total relaxation delay for CPMG was fixed to 40 ms and multiple 2D spectra were recorded in an interleaved manner by changing the number of CPMG pulses within the constant relaxation delay. A total of 10 ʋCPMG values ranging from 50 to 2000 Hz were recorded both at 600 and 800 MHz, with 2 additional ʋCPMG values recorded in duplicate for error estimation. All relaxation dispersion spectra were processed using NMRPipe and peak intensities were extracted using NMRFAM-SPARKY46.
NMR resonance assignments
Assignments were transferred from the previously assigned PPACK-thrombin10 and apo-thrombin8. All transferred and new assignments were confirmed with the 3D experimental data. Twenty-seven residues for which N–H peaks were visible in PPACK-bound thrombin did not have visible peaks in the HSQC-TROSY spectrum of thombin-TM456. These included residues: Gln 30CT (51seq), Val 66CT (97seq), Lys 81CT (113seq), Arg 97A – Leu 99CT (130-132seq), Thr 139 – Ala 149ACT (175-186seq), Gln 151 – Gln 156CT (192-197seq), Glu 192CT (238seq), Gly 196CT (242seq), Asp 221CT (268seq), and Arg 221ACT (269 seq). Four resonances were visible in the HSQC-TROSY spectrum of apo-thrombin, but these were not observed in the thrombin-TM456m spectrum. These included resonances for residues: Ser 45CT (67seq), Val 66CT (97seq), Ile 88CT (120seq), and Tyr 89CT (121seq). Nine residues were visible in the HSQC-TROSY thrombin-TM456m spectrum that were not observed in the apo-thrombin spectrum. These included residues: Glu 18CT (38seq), Gly 188 – Cys 191CT (234-237seq), Trp 215CT (263seq), Cys 220CT (267seq), Tyr 225CT (273seq), and Gly 226CT (274seq).
CPMG Experiments
The effective relative relaxation rates (R2,eff) due to contributions from conformational exchange on μs-ms time scale were evaluated with CPMG experiments collected at NMRFAM using a TROSY-CPMG pulse sequence47. The 600 MHz and 800 MHz R2,eff relaxation dispersion data for all thrombin-TM456 residues with Rex > 6 Hz (24 residues) were individually fit to the Richard-Carver Eq. 48 by using the software package CPMGFit (http://www.palmer.hs.columbia.edu/software/cpmgfit.html). We attempted to globally fit the thrombin-TM456 CPMG data using GLOVE, which minimizes global and local parameters alternately, and incorporates a Monte-Carlo minimization method to allow fitting parameters to pass through local minima49. However, none of the residues with Rex > 6 s−1 could be globally fit within an acceptable χ2 value.
Supplementary Information
Acknowledgements
This work was supported by NIH R01HL127041. RP was supported by the Molecular Biophysics Training Grant, NIH Grant T32 GM008326. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant P41GM103399 (NIGMS), old number: P41RR002301. Equipment was purchased with funds from the University of Wisconsin-Madison, the NIH P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA.
Author contributions
R.B.P. designed and performed experiments, analyzed data, and wrote manuscript. T.M. helped with protein purification, M.T. collected all the NMR data, E.A.K. designed experiments and wrote manuscript.
Data availability
The Chemical Shift Assignments for thrombin in complex with TM456 have been deposited with BMRB ID 50,678.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88432-z.
References
- 1.Matthews BW, Sigler PB, Henderson R, Blow DM. Three-dimensional structure of tosyl-alpha-chymotrypsin. Nature. 1967;214:652–656. doi: 10.1038/214652a0. [DOI] [PubMed] [Google Scholar]
- 2.Bode W, et al. The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. Embo. J. 1989;8:3467–3475. doi: 10.1002/j.1460-2075.1989.tb08511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bode W, Schwager P, Huber R. The transition of bovine trypsinogen to a trypsin-like state upon strong ligand binding The refined crystal structures of the bovine trypsinogen-pancreatic trypsin inhibitor complex and of its ternary complex with Ile-Val at 19 A resolution. J. Mol. Biol. 1978;118:99–112. doi: 10.1016/0022-2836(78)90246-2. [DOI] [PubMed] [Google Scholar]
- 4.Huber R, Bode W. Structural basis of the activation and action of trypsin. Acc Chem Res. 1978;11:114–122. doi: 10.1021/ar50123a006. [DOI] [Google Scholar]
- 5.Stojanovski BM, Chen Z, Koester SK, Pelc LA, Di Cera E. Role of the I16–D194 ionic interaction in the trypsin fold. Sci. Rep. 2019;9:18035. doi: 10.1038/s41598-019-54564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handley LD, Treuheit NA, Venkatesh VJ, Komives EA. Thrombomodulin binding selects the catalytically active form of thrombin. Biochemistry. 2015;54:6650–6658. doi: 10.1021/acs.biochem.5b00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuglestad B, Gasper PM, McCammon JA, Markwick PR, Komives EA. Correlated motions and residual frustration in thrombin. J. Phys. Chem. B. 2013;117:12857–12863. doi: 10.1021/jp402107u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Handley, L. D. et al. NMR reveals a dynamic allosteric pathway in thrombin. Sci Rep, (2017). [DOI] [PMC free article] [PubMed]
- 9.Lechtenberg BC, Johnson DJ, Freund SM, Huntington JA. NMR resonance assignments of thrombin reveal the conformational and dynamic effects of ligation. Proc. Natl. Acad. Sci. USA. 2010;107:14087–14092. doi: 10.1073/pnas.1005255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuglestad B, et al. The dynamic structure of thrombin in solution. Biophys. J. 2012;103:79–88. doi: 10.1016/j.bpj.2012.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasper PM, Fuglestad B, Komives EA, Markwick PRL, McCammon JA. Allosteric networks in thrombin distinguish procoagulant vs. anticoagulant activities. Proc. Natl. Acad. Sci. USA. 2012;109:21216–21222. doi: 10.1073/pnas.1218414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pineda AO, et al. Molecular dissection of Na+ binding to thrombin. J. Biol. Chem. 2004;279:31842–31853. doi: 10.1074/jbc.M401756200. [DOI] [PubMed] [Google Scholar]
- 13.Dang QD, Guinto ER, Cera ED. Rational engineering of activity and specificity in a serine protease. Nat. Biotech. 1997;15:146–149. doi: 10.1038/nbt0297-146. [DOI] [PubMed] [Google Scholar]
- 14.Esmon NL, Owen WG, Esmon CT. Isolation of a membrane-bound cofactor for thrombin-catalyzed activation of protein C. J. Biol. Chem. 1982;257:859–864. doi: 10.1016/S0021-9258(19)68276-1. [DOI] [PubMed] [Google Scholar]
- 15.Kurosawa S, Galvin JB, Esmon NL, Esmon CT. Proteolytic formation and properties of functional domains of thrombomodulin. J. Biol. Chem. 1987;262:2206–2212. doi: 10.1016/S0021-9258(18)61640-0. [DOI] [PubMed] [Google Scholar]
- 16.White CE, Hunter MJ, Meininger DP, White LR, Komives EA. Large-scale expression, purification and characterization of small fragments of thrombomodulin: the roles of the sixth domain and of methionine 388. Protein. Eng. 1995;8:1177–1187. doi: 10.1093/protein/8.11.1177. [DOI] [PubMed] [Google Scholar]
- 17.Fuentes-Prior P, et al. Structural basis for the anticoagulant activity of the thrombin–thrombomodulin complex. Nature. 2000;404:518–525. doi: 10.1038/35006683. [DOI] [PubMed] [Google Scholar]
- 18.Baerga-Ortiz A, Rezaie AR, Komives EA. Electrostatic dependence of the thrombin-thrombomodulin interaction. J. Mol. Biol. 2000;296:651–658. doi: 10.1006/jmbi.1999.3447. [DOI] [PubMed] [Google Scholar]
- 19.Baerga-Ortiz A, Bergqvist S, Mandell JG, Komives EA. Two different proteins that compete for binding to thrombin have opposite kinetic and thermodynamic profiles. Protein. Sci. 2004;13:166–176. doi: 10.1110/ps.03120604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Treuheit NA, Beach MA, Komives EA. Thermodynamic compensation upon binding to exosite 1 and the active site of thrombin. Biochemistry. 2011;50:4590–4596. doi: 10.1021/bi2004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleckner IR, Foster MP. An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta. 2011;1814:942–968. doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farber PJ, Mittermaier A. Relaxation dispersion NMR spectroscopy for the study of protein allostery. Biophys. Rev. 2015;7:191–200. doi: 10.1007/s12551-015-0166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giladi M, Khananshvili D. Hydrogen-deuterium exchange mass-spectrometry of secondary active transporters: from structural dynamics to molecular mechanisms. Front Pharmacol. 2020;11:70. doi: 10.3389/fphar.2020.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 2011;40:1224–1234. doi: 10.1039/C0CS00113A. [DOI] [PubMed] [Google Scholar]
- 25.Berliner LJ, Sugawara Y, Fenton JW. Human.alpha.-thrombin binding to nonpolymerized fibrin-sepharose: evidence for an anionic binding region. Biochemistry. 1985;24:7005–7009. doi: 10.1021/bi00345a038. [DOI] [PubMed] [Google Scholar]
- 26.Lechtenberg BC, Freund SM, Huntington JA. An ensemble view of thrombin allostery. Biol. Chem. 2012;393:889–898. doi: 10.1515/hsz-2012-0178. [DOI] [PubMed] [Google Scholar]
- 27.Huntington JA. How Na+ activates thrombin–a review of the functional and structural data. Biol. Chem. 2008;389:1025–1035. doi: 10.1515/BC.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai MT, Di Cera E, Shafer JA. Kinetic pathway for the slow to fast transition of thrombin. Evidence of linked ligand binding at structurally distinct domains. J. Biol. Chem. 1997;272:30275–30282. doi: 10.1074/jbc.272.48.30275. [DOI] [PubMed] [Google Scholar]
- 29.Adams TE, Li W, Huntington JA. Molecular basis of thrombomodulin activation of slow thrombin. J. Thromb. Haemost. 2009;7:1688–1695. doi: 10.1111/j.1538-7836.2009.03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vindigni A, White CE, Komives EA, Di Cera E. Energetics of thrombin-thrombomodulin interaction. Biochemistry. 1997;36:6674–6681. doi: 10.1021/bi962766a. [DOI] [PubMed] [Google Scholar]
- 31.Myles T, Church FC, Whinna HC, Monard D, Stone SR. Role of thrombin anion-binding exosite-I in the formation of thrombin-serpin complexes. J. Biol. Chem. 1998;273:31203–31208. doi: 10.1074/jbc.273.47.31203. [DOI] [PubMed] [Google Scholar]
- 32.Rezaie AR, Cooper ST, Church FC, Esmon CT. Protein C inhibitor is a potent inhibitor of the thrombin-thrombomodulin complex. J. Biol. Chem. 1995;270:25336–25339. doi: 10.1074/jbc.270.43.25336. [DOI] [PubMed] [Google Scholar]
- 33.Rezaie AR, He X, Esmon CT. Thrombomodulin increases the rate of thrombin inhibition by BPTI. Biochemistry. 1998;37:693–699. doi: 10.1021/bi971271y. [DOI] [PubMed] [Google Scholar]
- 34.De Cristofaro R, Landolfi R. Allosteric modulation of BPTI interaction with human alpha- and zeta-thrombin. Eur. J. Biochem. 1999;260:97–102. doi: 10.1046/j.1432-1327.1999.00118.x. [DOI] [PubMed] [Google Scholar]
- 35.van de Locht A, et al. The thrombin E192Q-BPTI complex reveals gross structural rearrangements: implications for the interaction with antithrombin and thrombomodulin. Embo. J. 1997;16:2977–2984. doi: 10.1093/emboj/16.11.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu H, Bush LA, Pineda AO, Caccia S, Di Cera E. Thrombomodulin changes the molecular surface of interaction and the rate of complex formation between thrombin and protein C. J. Biol. Chem. 2005;280:7956–7961. doi: 10.1074/jbc.M412869200. [DOI] [PubMed] [Google Scholar]
- 37.Zeymer C, Werbeck ND, Zimmermann S, Reinstein J, Hansen DF. Characterizing active site conformational heterogeneity along the trajectory of an enzymatic phosphoryl transfer reaction. Angew Chem. Int. Ed. Engl. 2016;55:11533–11537. doi: 10.1002/anie.201606238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esmon NL, DeBault LE, Esmon CT. Proteolytic formation and properties of gamma-carboxyglutamic acid-domainless protein C. J. Biol. Chem. 1983;258:5548–5553. doi: 10.1016/S0021-9258(20)81925-5. [DOI] [PubMed] [Google Scholar]
- 39.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal. Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lumpkin R, Komives EA. DECA, a comprehensive, automatic post-processing program for HDX-MS data. Mol. Cell Proteom. 2019;18:2516–2523. doi: 10.1074/mcp.TIR119.001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda M, et al. Construction and performance of an NMR tube with a sample cavity formed within magnetic susceptibility-matched glass. J. Magn. Reson. 2011;209:167–173. doi: 10.1016/j.jmr.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kay LE, Ikura M, Tschudin R, Bax A. Three-dimensional triple-resonance NMR Spectroscopy of isotopically enriched proteins. J. Magn.. Reson. 2011;213:423–441. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Yamazaki T, Lee W, Arrowsmith CH, Muhandiram D, Kay LE. A suite of triple resonance NMR experiments for the backbone assignment of 15N, 13C, 2H labeled proteins with high sensitivity. J. Am. Chem. Soc. 1994;116:11655–11666. doi: 10.1021/ja00105a005. [DOI] [Google Scholar]
- 44.Hyberts SG, Takeuchi K, Wagner G. Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J. Am. Chem. Soc. 2010;132:2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ying J, Delaglio F, Torchia DA, Bax A. Sparse multidimensional iterative lineshape-enhanced (SMILE) reconstruction of both non-uniformly sampled and conventional NMR data. J. Biomol. NMR. 2017;68:101–118. doi: 10.1007/s10858-016-0072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loria JP, Rance M, Palmer AG., 3rd Transverse-relaxation-optimized (TROSY) gradient-enhanced triple-resonance NMR spectroscopy. J. Magn. Reson. 1999;141:180–184. doi: 10.1006/jmre.1999.1891. [DOI] [PubMed] [Google Scholar]
- 48.Carver J, Richards R. A general two-site solution for the chemical exchange produced dependence of T2 upon the Carr-Purcell pulse separation. J. Magn. Reson. 1972;1969(6):89–105. [Google Scholar]
- 49.Sugase K, Konuma T, Lansing JC, Wright PE. Fast and accurate fitting of relaxation dispersion data using the flexible software package GLOVE. J. Biomol. NMR. 2013;56:275–283. doi: 10.1007/s10858-013-9747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Chemical Shift Assignments for thrombin in complex with TM456 have been deposited with BMRB ID 50,678.