Abstract
Genetics of traits related to fruit cuticle deposition and composition was studied in two red-fruited tomato species. Two mapping populations derived from the cross between the cultivated tomato (Solanum lycopersicum L.) and its closest relative wild species Solanum pimpinellifolium L. were employed to conduct a QTL analysis. A combination of fruit cuticle deposition, components and anatomical traits were investigated and the individual effect of each QTL evaluated. A total of 70 QTLs were identified, indicating that all the cuticle traits analyzed have a complex polygenic nature. A combination of additive and epistatic interactions was observed for all the traits, with positive contribution of both parental lines to most of them. Colocalization of QTLs for various traits uncovered novel genomic regions producing extensive changes in the cuticle. Cuticle density emerges as an important trait since it can modulate cuticle thickness and invagination thus providing a strategy for sustaining mechanical strength without compromising palatability. Two genomic regions, located in chromosomes 1 and 12, are responsible for the negative interaction between cuticle waxes and phenolics identified in tomato fruit. Several candidate genes, including transcription factors and structural genes, are postulated and their expression analyzed throughout development.
Subject terms: Plant breeding, Natural variation in plants
Introduction
The plant cuticle, as the interface between the plant and the environment, plays a role in plant performance, fruit quality and postharvest1,2. Among the functions of agronomic relevance are the protection from water loss and UV radiation, its function as a thermal regulator, and the mechanical protection against pathogens and abiotic stresses, either environmental or derived from internal tissues3. Thus, the cuticle participates in the protection from fungal penetration, fruit cracking, organ dehydration and fusion, and in controlling organ growth. The cuticle is composed of a lipid matrix of esterified polyhydroxy fatty acids named cutin, polysaccharides from the cell wall, phenolic compounds and waxes, a mixture of soluble lipids and triterpenoids that can be located within the cuticle matrix or deposited on the surface3. Waxes play a critical role in the control of water loss from internal tissues and can also confer some mechanical strength4,5. In tomato fruit cuticle, waxes play a small part in the mechanical resistance, being the phenolic fraction the main modulator of the cuticle’s stiffness, deformation, and strength6,7. This phenolic domain is composed of cinnamic acids derivatives present during fruit growth and increases significantly during ripening with the incorporation of the flavonoid chalconaringenin, responsible for the yellow-orange color of the cuticle in red ripe tomatoes6.
The cuticle is mainly located to the outer epidermal cell wall, and depending on the species, can cutinize the epidermal anticlinal walls and even collenchyma walls. In crops such as apple8, mango9, cucumber10, and tomato11, among others, various degrees of fruit cuticle invagination have been observed. A relation between cuticle thickness and invagination and the biomechanical properties of the cuticle has been established11. Moreover, a relation between cuticle thickness and invagination, skin strength and fruit cracking resistance has been indicated12,13. However, these two traits, cuticle thickness and invagination have a negative impact on the sensory properties of the skin, which are especially relevant in small fruits that are consumed unpeeled14. On the other hand, a thick cuticle that encapsulates the epidermal and some collenchyma cells would also have a negative impact on fruit peeling for processing and canning since it would difficult the removal of the skin and partially eliminate the collenchyma, a tissue that contributes to the mechanical properties of the fruit11,15,16.
Understanding the genetic basis of cuticle development and composition is needed for breeding in crop species. Although numerous genes related to cuticle biosynthesis have been identified, most of these have been uncovered after the analysis of mutagenized populations and/or gene silencing. However, their participation in the genetic variation present in natural populations, cultivars and wild species closely related to crops is uncertain. Natural variation is a valuable source of beneficial traits for breeding. Limited work based on variation in natural plant populations has been carried out to study the cuticle, and mainly focused on waxes17–21. Analysis of genes involved in anatomical and biophysical traits of the cuticle, many of great importance for plant performance and fruit quality, are scarce at most. In this sense it should be mentioned the relationship found between postharvest water loss in pepper and differences in wax composition and cutin amount22 and the identification of quantitative trait loci (QTL) for cuticle thickness and invagination in tomato and cucumber10,19.
Tomato genetics and breeding heavily relies on exploiting natural variation from related wild species. Wild tomato species have been reported to exhibit cuticle diversity for some anatomical and chemical traits18, however, its genetics has only been studied in lines derived from the green-fruited Solanum pennellii Dun.19. In this work, we have explored for the first time the cuticle variability of red-fruited species in two populations derived from the interspecific cross between the domesticated tomato (S. lycopersicum L.) and its closest wild relative S. pimpinellifolium L. A QTL analysis of the different cuticle components and anatomical traits was performed revealing a complex polygenic nature that combined additive and epistatic interactions among the different QTLs.
Results
Analysis of the parental lines
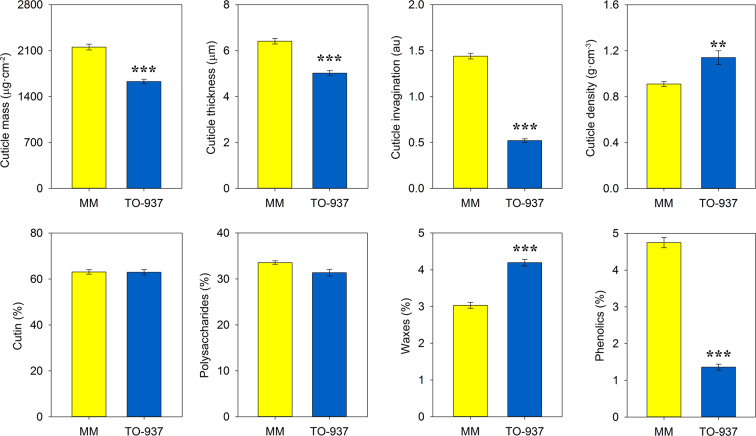
Cuticle analysis of the cultivated tomato “Moneymaker” (MM) and the wild accession TO-937 showed significant differences for most of the traits studied (Fig. 1). Thus, red ripe cuticles of TO-937 displayed a significantly lower cuticle mass, thickness and invagination. However, the decrease in thickness and invagination was not proportional to the reduction of cuticle mass, causing a significant increase in cuticle density compared to MM. The percentage of total waxes present in the cuticle of TO-937 was statistically higher compared to MM whereas phenolics were significantly lower. No significant differences in the percentages of the major cuticle components, cutin and polysaccharides, were detected.
Fig. 1. Comparison between the parental lines MM and TO-937 for the different cuticle traits studied.
Red ripe fruits were analyzed. Data are presented as means ± SE. Asterisks indicate significant differences between both lines. Arbitrary units (au)
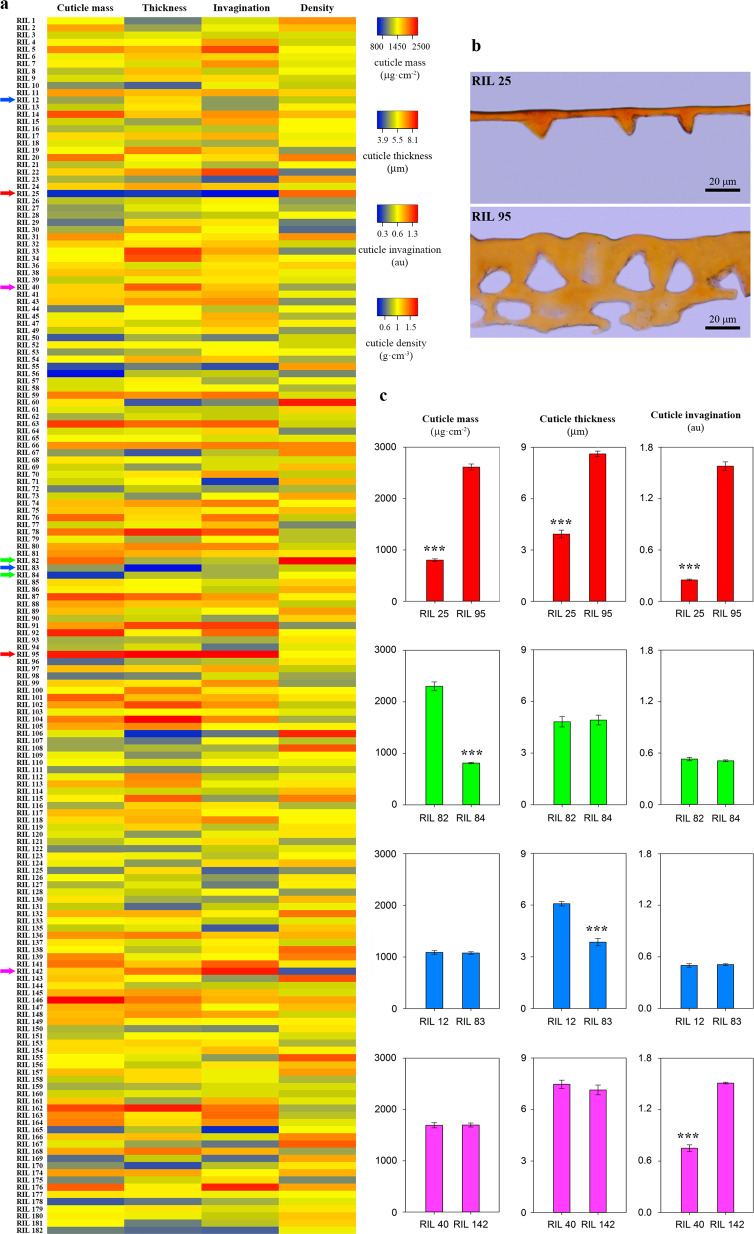
Cuticle characterization of the RIL (recombinant inbred line) population showed transgressive inheritance for most of the traits analyzed, with lines displaying values significantly higher and lower than the parental lines. This indicates that both parental lines have positive QTLs for the traits analyzed. For example, RILs with cuticle masses significantly lower than TO-937 (100 lines) and higher than MM (14 lines) were identified. Similarly, despite the very low percentage of cuticle phenolics detected in TO-937, 21 RILs showed values significantly higher than MM, indicating a positive contribution of TO-937 alleles. Cuticle mass was deconstructed into three parameters: thickness, degree of invagination and density. The combination of which provides a good approximation to the overall process of cuticle deposition. Results of the RIL population for these traits are shown in Fig. 2a. Although increasing cuticle masses could be expected to translate into thicker and more invaginated cuticles (Fig. 2b, c), this is not always the case (Fig. 2a). Comparison of RIL 82 and RIL 84 shows that significantly different cuticle masses can display similar thickness and invagination (Fig. 2c). At the same time, significant differences in cuticle thickness can be observed in lines with similar cuticle mass and invagination (RIL 12 vs. RIL 83), as well as significant changes in the degree of invagination without affecting cuticle mass or thickness (RIL 40 vs. RIL 142).
Fig. 2. Interaction among the different traits related to cuticle deposition.
a Heat map of the RIL population for cuticle mass, thickness, invagination and density. Arrows indicate the lines further shown in detail. b Cross-sections of the isolated cuticle of two RILs stained with Sudan IV. c Pair-wise comparisons of RILs with specific combinations of cuticle mass, thickness and invagination. Asterisks indicate significant differences with p < 0.001. Recombinant inbred line (RIL), arbitrary units (au)
Cuticle mass
Seventy QTLs were detected for different cuticle traits within both populations (Fig. 3). Statistically significant QTLs in the RIL population represent 34 of the total detected. The rest of the QTLs were identified as statistically significant in the IL population and most of them coincided with genomic regions that displayed non-statistically significant QTLs in the RIL population, that is, with probability below the 0.99 threshold. In these instances, the peak marker observed in the RIL population is given in the corresponding Tables. Out of the 15 QTLs identified for cuticle mass (cm) (Table 1), 13 had lyc effect that is, the MM allelic region increased cuticle mass whereas two other QTLs, cm5.2 and cm10.2, had pim effect, TO-937 alleles increased the cuticle mass. QTL epistatic analysis within the RIL population identified a significant interaction between cm1.1 and cm5.2 (LOD 5.18, 15% variance explained) with pim effect. This indicated that the lyc allelic cm1.1 region increases the pim effect of cm5.2.
Fig. 3. Map of tomato chromosomes showing QTL regions identified in the RIL and IL populations for the different cuticle traits.
Cuticle mass (cm), cuticle thickness (ct), cuticle invagination (ci), cuticle density (cd), percentage of cutin (cu), percentage of polysaccharides (ps), percentage of phenolics (ph), and percentage of waxes (wx). Bold lines represent QTL regions with p < 0.001. Symbols with the same shape and color located to the left of the chromosomes denote regions with epistatic interactions. Symbol color indicates the trait. Numbers to the left of chromosomes indicate genetic distances expressed in cM (Kosambi mapping function). Recombinant inbred line (RIL), inbred line (IL)
Table 1.
QTLs identified in the RIL and IL populations for cuticle mass
| QTL | Effect | LOD | PVE | P | Peak marker | Population |
|---|---|---|---|---|---|---|
| cm1.1 | lyc | – | – | – | solcap59975 | IL |
| cm1.2 | lyc | – | – | – | solcap48181 | IL |
| cm2.1 | lyc | 2.8 | 6.9 | 0.989 | solcap23851 | RIL/IL |
| cm2.2 | lyc | 3.67 | 9.8 | 0.997 | solcap42471 | RIL/IL |
| cm3.1 | lyc | 4.65 | 11.9 | 1.000 | solcap41353 | RIL/IL |
| cm3.2 | lyc | 6.85 | 17.0 | 1.000 | solcap62180 | RIL/IL |
| cm4.1 | lyc | 3.61 | 7.8 | 0.998 | solcap3108 | RIL/IL |
| cm5.1 | lyc | 3.04 | 6.8 | 0.992 | solcap23797 | RIL/IL |
| cm5.2 | pim | 3.66 | 8.9 | 0.998 | solcap37808 | RIL/IL |
| cm7.1 | lyc | – | – | – | – | IL |
| cm10.1 | lyc | 3.94 | 8.1 | 0.999 | solcap46305 | RIL/IL |
| cm10.2 | pim | 3.08 | 7.3 | 0.995 | solcap14879 | RIL/IL |
| cm11.1 | lyc | – | – | – | – | IL |
| cm11.2 | lyc | 6.02 | 13.8 | 1.000 | solcap100974 | RIL/IL |
| cm12.1 | lyc | 3.84 | 9.4 | 0.998 | solcap1497 | RIL/IL |
QTLs are named with a trait abbreviation followed by chromosome number and the number of the QTL within the chromosome. Effect refers to the allele (lyc/pim) that increases the trait. lyc, S. lycopersicum MM. pim, S. pimpinellifolium TO-937
PVE percentage of variance explained by the QTL in the RIL population, P probability. Marker closest to the maximum LOD (logarithm of the odds) score is also shown. cm cuticle mass, RIL recombinant inbred line, IL inbred line
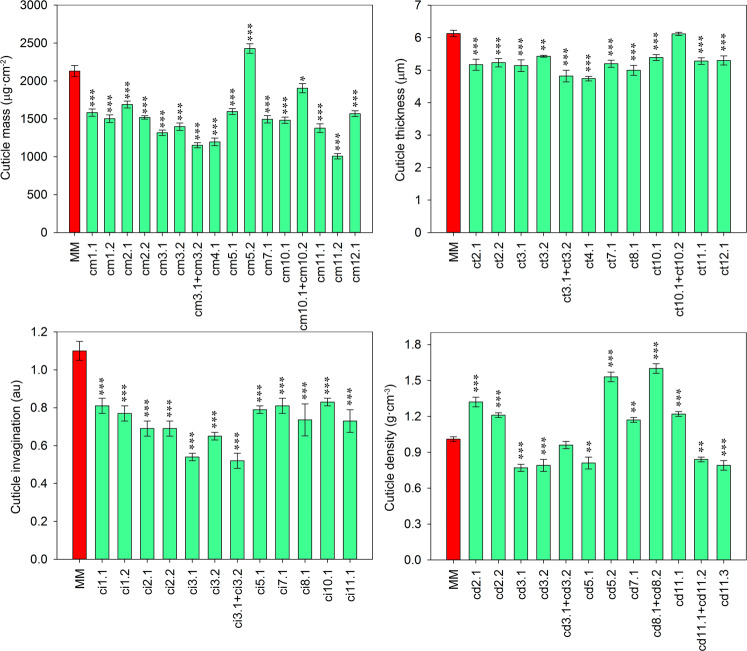
To study the individualized effect of cm3.1 and cm3.2, subILs were generated from the IL sp3-2 that separated and shortened these QTLs (Supplementary Fig. 1). The highest effect on cuticle mass was observed in the IL harboring cm11.2 with a 2-fold reduction compared to MM, followed by cm4.1 and the combination cm3.1 + cm3.2 (Fig. 4). Each of the QTLs of chromosome 3 showed a similar reduction in cuticle, ∼38%, whereas the combination showed a small additive effect, reducing 45% the cuticle mass. On the other hand, the pim effect of cm5.2 produced an increased 12–14% of cuticle mass. Although cm10.2 was not separated from cm10.1, comparison of cm10.1 + cm10.2 with cm10.1 showed an increase in cuticle mass to levels almost similar to those of MM, indicating that the pim effect of cm10.2 almost counteracted the lyc effect of cm10.1. In order to shorten cm11.1, overlapping subILs were generated from the sp11-1 line (Supplementary Fig. 2). Despite the significant decrease in cuticle mass observed in sp11-1 compared to MM, all the derived subILs were similar to MM, indicating that cm11.1 must be located to a lyc region present in all the subILs except sp11-1 (Supplementary Fig. 2).
Fig. 4. Comparison between the parental MM and the ILs harboring QTLs related to cuticle deposition.
Red ripe fruits were analyzed. Data are presented as means ± SE. Asterisks indicate significant differences with MM (*p < 0.05; **p < 0.01; ***p < 0.001). Inbred line (IL), cuticle mass (cm), cuticle thickness (ct), degree of cuticle invagination (ci), cuticle density (cd), and arbitrary units (au)
Of the 11 QTLs identified for cuticle thickness (ct) (Supplementary Table 1) the biggest decrease, ∼20%, was observed for ct4.1 and the combination of the two QTLs of chromosome 3 ct3.1 + ct3.2 (Fig. 4). Again, the combination ct3.1 + ct3.2 showed additive effect compared to the individual QTLs. With the exception of ct10.1 + ct10.2, all the lines harboring the individual or double QTLs showed a reduction in cuticle thickness compared to MM (Fig. 4). Comparison of the line harboring ct10.1 + ct10.2 with ct10.1 showed an increase in cuticle thickness to levels similar to MM, thus supporting the pim effect of ct10.2 observed in the RIL population. All the QTLs identified for cuticle invagination (ci) showed lyc effect (Supplementary Table 2) as it can be inferred from the significant decrease observed in comparison to MM (Fig. 4). The highest reduction in invagination, around 50%, was observed for ci3.1 and the combination ci3.1 + ci3.2, indicating that, contrary to what was observed for cuticle mass and thickness, ci3.1 had a dominant effect on the trait.
Regarding cuticle density (cd), QTLs with both lyc and pim effect were observed (Supplementary Table 3). The lowest decrease in density was observed in the ILs with cd3.1, cd3.2 and cd5.1 whereas those containing cd5.2 and cd8.2 showed the highest increase compared to MM (Fig. 4). Accumulation of cd3.1 + cd3.2 counteracted the effect on density of the individual QTLs and the combination was similar to MM. Despite cd8.1 was the only statistically significant QTL identified in the RIL population (Supplementary Table 3), its individualized effect could not be studied in the IL population since none of the lines carried the most proximal region of chromosome 8 as TO-937. However, since cd8.1 had lyc effect and cd8.2 pim, it is possible that the effect observed for cd8.2 (Fig. 4) is indeed the result of a lyc × pim interaction between both QTLs. Generation of new lines and further analyses will be necessary to understand the effect of this region. Cuticle density analysis of sp11-1 displayed a significant decrease in cuticle density whereas the derived subILs had a significantly higher density compared to MM (Supplementary Fig. 2). The decrease in density observed in sp11-1 could be the result of the lower cuticle mass associated with cm11.1, while the increase in cuticle density detected in the subILs could be explained as the result of a putative QTL (cd11.1) with pim effect located in the overlapping region present in all the subILs. Nevertheless, these two presumed QTLs need to be further individualized and their effects studied.
Phenolics
Nine QTL regions, in chromosomes 1, 4, 5, 7, 8, and 12, were identified in the RIL and IL populations (Table 2). Despite the very low percentage of phenolics (ph) present in TO-937 cuticle, four QTLs with pim effect were identified: ph4.1, ph5.1, ph5.2, and ph12.1. A significant epistatic interaction (LOD 5.57, 10.9% variance explained) with pim effect was identified between ph12.1 and a region in chromosome 11 (Fig. 3). This indicated that the lyc allelic region in chromosome 11 increases the pim effect of ph12.1 and could explain why ph12.1 was not statistically significant in the RIL population but showed marked differences in the IL population. This chromosome 11 region did not have an effect on phenolics by itself (data not shown).
Table 2.
QTLs identified in the RIL and IL populations for the percentage of cuticle components
| QTL | Effect | LOD | PVE | P | Peak marker | Population |
|---|---|---|---|---|---|---|
| ph1.1 | lyc | 11.37 | 26.7 | 1.000 | solcap457 | RIL/IL |
| wx1.1 | pim | 3.54 | 9.2 | 0.996 | solcap25914 | RIL |
| cu1.1 | lyc | 3.38 | 8.9 | 0.996 | solcap25922 | RIL |
| wx3.1 | pim | 3.13 | 7.4 | 0.992 | solcap62270 | RIL/IL |
| wx3.2 | pim | – | – | – | solcap61411 | IL |
| ph4.1 | pim | – | – | – | snp302a | IL |
| wx4.1 | pim | – | – | – | – | IL |
| ph5.1 | pim | 4.36 | 10.3 | 1.000 | solcap49234 | RIL |
| ph5.2 | pim | 2.90 | 7.2 | 0.991 | solcap37562 | RIL |
| ph7.1 | lyc | 3.39 | 9 | 0.998 | solcap68044 | RIL/IL |
| ph7.2 | lyc | 3.02 | 7.9 | 0.993 | solcap5853 | RIL/IL |
| ph8.1 | lyc | 5.64 | 14.3 | 1.000 | solcap48550 | RIL/IL |
| wx10.1 | lyc | 3.07 | 8 | 0.992 | snp112a | RIL/IL |
| cu10.1 | pim | – | – | – | snp112a | IL |
| ps10.1 | lyc | – | – | – | solcap59337 | IL |
| wx11.1 | lyc | – | – | – | – | IL |
| wx11.2 | pim | – | – | – | solcap100974 | IL |
| ph12.1 | pim | – | – | – | solcap1516 | IL |
| ph12.2 | lyc | 5.40 | 13.3 | 1.000 | solcap55706 | RIL/IL |
| wx12.1 | lyc | 5.84 | 12.6 | 1.000 | solcap45816 | RIL/IL |
| wx12.2 | pim | 3.48 | 7.2 | 0.997 | solcap55712 | RIL/IL |
QTLs are named with a trait abbreviation followed by chromosome number and the number of the QTL within the chromosome. Effect refers to the allele (lyc/pim) that increases the trait. lyc, S. lycopersicum MM. pim, S. pimpinellifolium TO-937
PVE percentage of variance explained by the QTL in the RIL population, P probability. Marker closest to the maximum LOD (logarithm of the odds) score is also shown. ph phenolics, wx waxes, cu cutin, ps polysaccharides, RIL recombinant inbred line, IL inbred line
In order to individually study ph7.1 and ph7.2, subILs derived from sp7-2 were generated (Supplementary Fig. 2). The effects of each phenolic QTL were analyzed in the IL population and compared with the recurrent parental line MM at different stages of development (Fig. 5). All the lines containing the QTLs showed significant differences in the percentage of phenolics at red ripe compared to MM, with the exception of ph5.1 and ph5.2 that only showed a slight but non-significant increase in phenolics. The other QTLs with pim effect, ph4.1 and ph12.1, did show a significant increase in the percentage of cuticle phenolics, 30 and 20% respectively, compared to MM. Regarding the QTLs with lyc effect, the biggest reduction of phenolics, ∼40%, was observed in ph12.2 and the combined effect of ph7.1 + ph7.2, whereas the remaining QTLs showed a similar reduction in the percentage of phenolics at red ripe. Surprisingly ph1.1, despite being the major QTL, showed an intermediate reduction in phenolics, comparable to ph7.1, ph7.2, and ph8.1. A noticeable additive effect was observed for the QTLs located in chromosome 7, with the combination ph7.1 + ph7.2 displaying a much lower percentage of phenolics than ph7.1 or ph7.2. Although the tomato fruit cuticle has a marked increase in phenolics during ripening, there is a small amount of phenolic acids that accumulate during development6. Hence, to extend the knowledge of the effect of these QTLs, the percentage of phenolics was also studied throughout development (Fig. 5). Interestingly, at the immature and mature green stages the ILs containing ph5.1 and ph5.2 showed significantly higher values than MM. However, these differences disappeared during ripening. It is precisely at the onset of ripening, when the differences between most of the other lines and MM became apparent. This suggests that, except for ph5.1 and ph5.2, the genes underneath these QTLs play an important role in the cuticle during ripening but not at earlier stages of development.
Fig. 5. Percentage of cuticle phenolics from fruits harvested at four stages of development.

The parental line MM and the ILs harboring QTL regions related to cuticle phenolics (ph) are shown. Data are presented as means ± SE. Asterisks indicate significant differences with MM (*p < 0.05; **p < 0.01; ***p < 0.001). Inbred line (IL), immature green (IG), mature green (MG), breaker (Br), and red ripe (RR)
Waxes, cutin, and polysaccharides
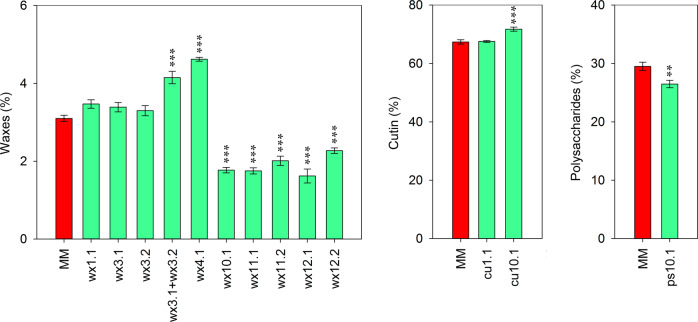
Nine QTLs were identified for the percentage of waxes (wx), six with pim effect, and 3 with lyc effect (Table 2). A percentage of waxes significantly higher than MM was observed for the ILs containing wx4.1 and the combination wx3.1 + wx3.2 (Fig. 6). However, the ILs that individualized the QTLs wx3.1 and wx3.2 did not show significant effects on waxes. Surprisingly, the line harboring wx1.1 did not show any difference with MM for waxes, and the two other QTLs with pim effect, wx11.2 and wx12.2 showed, contrary to what it was expected, lower percentage of waxes. Finally, wx10.1, wx11.1, and wx12.1 showed a significant decrease in waxes compared to MM. The QTL wx11.1 was identified and located after the analysis of the subILs derived from sp11-1 (Supplementary Fig. 2), similarly to what it has already been explained for cuticle mass and density. The significant epistatic interaction (LOD 5.28, 15.7% variance explained) identified between wx11.2 and a region in chromosome 7 (Fig. 3) could explain the decrease in waxes observed in the IL containing wx11.2 (Fig. 6). Whereas wx11.2 had pim effect and the chromosome 7 region lyc effect, the interaction showed an increased lyc effect. This region in chromosome 7 did not show an effect on waxes (data not shown). It is possible that a similar epistatic interaction, not detected in the QTL analysis were responsible for the change of effect detected in wx12.2 and the absence of effect of wx1.1.
Fig. 6. Comparison between the parental MM and the ILs with the QTLs related to the percentage of cutin, polysaccharides and waxes.
Red ripe fruits were analyzed. Data are presented as means ± SE. Asterisks indicate significant differences with MM (*p < 0.05; **p < 0.01; ***p < 0.001). Inbred line (IL), cutin (cu), polysaccharides (ps), and waxes (wx)
Despite no significant differences were detected for the percentage of cutin (cu) and polysaccharides (ps) between the parental lines, two QTLs for the percentage of cutin were identified in the RIL population, cu1.1 with lyc effect and cu10.1 with pim effect (Table 2). Additionally, one QTL for the percentage of polysaccharides was detected, ps10.1, with lyc effect that colocalized with cu10.1. Although a peak for percentage of polysaccharides was found in chromosome 1 in the QTL analysis, coinciding with the cu1.1 region, it was below the statistically significant threshold. Analysis of the ILs containing the different QTLs for cutin and polysaccharides and the parental line MM is shown in Fig. 6. The lines containing cu10.1 and ps10.1 displayed a statistically higher and lower percentage of cutin and polysaccharides, respectively. However, the line with cu1.1 did not show any significant difference in cutin compared with MM.
Analysis of the genomic regions
Genes located within each QTL region were investigated in order to identify candidate genes that were reported in the literature to affect cuticle synthesis (Table 3). Of the several known tomato genes involved in cuticle deposition, none of them located within any of the QTL regions. Indeed, only a few cuticle genes were identified in the QTLs. Despite the overlap between ph1.1 and wx1.1, each showed a different peak marker. MYB12, a transcription factor known to affect the phenolic metabolic pathway was closest to the ph1.1 and cu1.1 peak marker whereas LONG CHAIN ACYL SYNTHETASE 1 (LACS1), a gene involved in wax and cutin biosynthesis, was closer to the wx1.1 peak. Several β-KETOACYL-COENZYME A SYNTHASE (KCS) genes, involved in the elongation of very-long chain fatty acids, were identified in the QTLs. ECERIFERUM6 (CER6) was located in the peak of cm2.2, FIDDLEHEAD (FDH), and KCS11-like1, a gene very similar to Arabidopsis KCS11, were located in the ph8.1 peak marker or close to it, respectively; finally, KCS11-like2 was located within wx12.1. Additionally, putative tomato orthologs of Arabidopsis wax genes were also identified in QTLs related to cuticle mass, although far from the peak markers.
Table 3.
Cuticle related genes reported in the literature identified in the QTLs
| QTLs | Gene symbol | Gene locus | Species | Literature |
|---|---|---|---|---|
| ph1.1/wx1.1/cu1.1 | LACS1 | Solyc01g079240 | Arabidopsis | Lü et al.41 |
| MYB12 | Solyc01g079620 | Tomato | Adato et al.38 | |
| ph1.1 | DET1 | Solyc01g056340 | Tomato | Davuluri et al.40 |
| cm1.2/ci1.2 | CER9 | Solyc01g107880 | Arabidopsis | Lü et al.51 |
| LACS2 | Solyc01g109180 | Arabidopsis | Schnurr et al.52 | |
| cm2.2/ct2.2/ci2.2/cd2.2 | CER6 | Solyc02g085870 | Tomato | Leide et al.26 |
| CER13 | Solyc02g086500 | Arabidopsis | Rashotte et al.53 | |
| ph4.1/wx4.1 | PAS2 | Solyc04g014370 | Arabidopsis | Bach et al.54 |
| cm5.1/ph5.1 | GL1 | Solyc05g008250 | Arabidopsis | Xia et al.55 |
| cm5.2/ci5.1/cd5.2 | CER10 | Solyc05g054490 | Arabidopsis | Rashotte et al.53 |
| CYP77A6 | Solyc05g055400 | Arabidopsis | Li-Beisson et al.56 | |
| cm7.1/ct7.1/ci7.1/cd7.1 | TAGL1 | Solyc07g055920 | Tomato | Giménez et al.23 |
| ph8.1 | FDH | Solyc08g067260 | Arabidopsis | Voisin et al.27 |
| KCS11 like | Solyc08g067410 | Tomato | – | |
| wx12.1 | KCS11 like2 | Solyc12g006820 | Tomato | – |
Genes located on the peak marker, or <5 genes away from the peak marker, are shown in bold. Underlined genes are located 10–20 genes from the peak marker. Genes in regular case are located >20 genes away from the peak marker. When more than one QTL is shown, the gene/s indicated are located within the overlapping regions
cm cuticle mass, ct cuticle thickness, ci cuticle invagination, cd cuticle density, ph phenolics, cu cutin, wx waxes
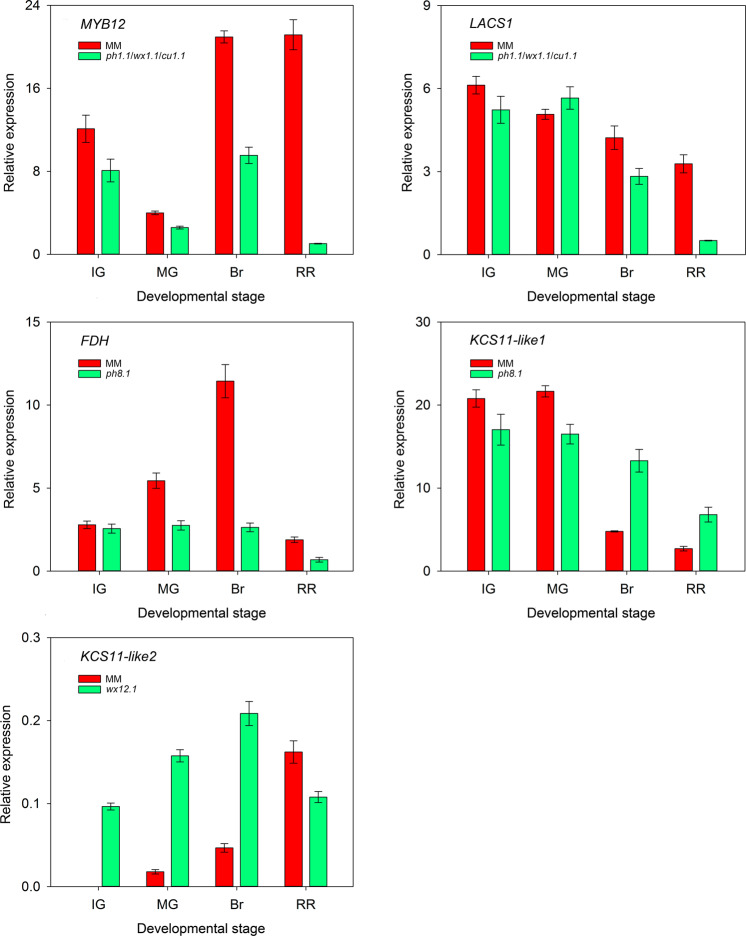
Comparative expression analyses of some putative candidate genes related to waxes and phenolics of MM and the corresponding ILs are shown in Fig. 7. The IL harboring ph1.1 showed a drastic decrease of MYB12 expression during ripening compared with MM, 2.8-fold at breaker and 32-fold at red ripe. These results are in accordance with the reduction in cuticle phenolics detected from the breaker stage until red ripe for ph1.1 (Fig. 5). LACS1 only showed a small decrease at breaker followed by a 8.7-fold reduction at red ripe (Fig. 7). FDH expression in MM increased during growth with a notable maximum at breaker whereas in the IL with ph8.1 the expression level did not change with growth and displayed a 5-fold decrease in expression at breaker compared to MM. Again, this profile coincided with the drastic decrease in cuticle phenolics already detected at breaker in the ph8.1 line (Fig. 5). On the other hand, the ph8.1 line showed a small increase of KCS11-like1 expression compared to MM during ripening. Finally, the IL containing wx12.1 showed an increased expression of KCS11-like2 compared to MM, especially during the whole fruit growth period until breaker. Inspection of changes in the mRNA sequences for these genes between the parental lines showed that the differences present produced conserved protein changes, with the only exception of LACS1 with a L183 (MM) to S183 (TO-937) change.
Fig. 7. Relative expression throughout epicarp development.
MM and the ILs containing the QTLs ph1.1/wx1.1/cu1.1, ph8.1, and wx12.1 were analyzed for the putative candidate genes MYB12, LACS1, FDH, KCS11-like1, and KCS11-like2. Data are presented as mean ± SE. Inbred line (IL), phenolics (ph), waxes (wx), cutin (cu), immature green (IG), mature green (MG), breaker (Br), and red ripe (RR)
Discussion
Natural variation of tomato fruit cuticle shows a combination of additive and epistatic traits
A comprehensive cuticle analysis has been carried out in two populations, RIL and IL, thus favoring the detection of additional QTLs that were not significant or present in the RIL population. Understanding the interaction of these epistatic QTLs with the genetic background is important in plant breeding for the introgression of specific traits. A combination of additive and epistatic interactions was detected for all the traits analyzed. Moreover, the transgressive inheritance found in the RIL population for most of the characters analyzed indicated the presence of QTLs with a positive contribution to the trait in both parents. Of the 25 genomic regions identified spanning 10 of the 12 chromosomes, some were specific and only affected one or two traits and others were more complex and produced global changes comprising many of the parameters studied. Uncovering the complex network of genetic interactions responsible for cuticle synthesis allows for a better understanding of its natural variability, provides an insight into the evolution of the traits and their potential for breeding purposes.
QTL analysis of a tomato IL population derived from the interspecific cross between cultivated tomato and the green-fruited species S. pennellii found three QTLs for the amount of specific wax and cutin components and another one for cuticle thickness19. With the exception of the thickness QTL, none of the other regions coincide with any of the QTLs herein identified. The thickness QTL coincided with the ct7.1 region found here. TOMATO AGAMOUS-LIKE1 (TAGL1) was postulated as one of the possible candidate genes for this trait19. Indeed, modification of TAGL1 expression in tomato has been shown to lead to significant changes in cuticle mass, thickness and invagination23. A QTL for cuticle thickness and invagination was reported in cucumber fruit and the transcription factor SHINE1 (SHN1) reported as the candidate gene10; however, SHN1 is neither located within any of the QTLs herein found nor expressed in tomato fruit epicarp24.
Of the numerous known genes involved in phenolic synthesis and regulation only MYB12 and DE-ETIOLATED1 (DET1) were located within one of the QTLs found for cuticle phenolics. Similarly, several genes have been found to participate in cuticle deposition25 however, none of these genes locate within the QTLs identified for any of the cuticle traits herein studied. Several genes involved in wax accumulation in Arabidopsis were identified in QTLs related to cuticle deposition (Table 3). Only two genes, LACS2 and the cytochrome P450 monooxigenase CYP77A6, reported to alter cutin deposition in Arabidopsis were located within cuticle QTLs. However, since none of these genes are located close to the QTL peak markers, their role as candidate genes needs to be further analyzed. Three KCSs were identified in the peak markers of QTLs unrelated to waxes, cm2.2 and ph8.1. In tomato, cer6 mutant has been shown to affect cuticle mass and reduce the synthesis of n-alkanes while increasing triterpenoids without altering the total amount of waxes26. This would be in agreement with its role as candidate gene for cm2.2. Of the two KCS located around ph8.1 peak marker, the notable reduction of FDH expression during ripening in the TO-937 allele indicates that this is the most likely candidate gene. Indeed, the maximum expression at breaker coincided with the massive incorporation of phenolics to the cuticle, although waxes also show a comparatively modest increase during ripening in MM (ref. 6). Mutation of FDH in Arabidopsis displayed an increase in cutin and waxes27 that was not observed in the ph8.1 line. This apparent unlikely role of a KCS in cuticle phenolics could be associated with transport. It is still unknown how phenolics are transported to the cuticle, although there are several reports of phenolics esterified to cutin and waxes28,29. It is plausible that they are transported already esterified to a fatty acid instead of the bonding occurring within the cuticle matrix. In this context, FDH could be involved in the synthesis of the fatty acid domain. Putative orthologs of KCS11 have been postulated as candidate genes for wax accumulation in eucalypt and banana leaves20,30. The notably increased expression throughout development of the TO-937 allele of KCS11-like2 and the reduction in waxes observed in the wx12.1 line suggests that this gene could be indirectly involved in wax synthesis through the shortage of substrate available for waxes. In this regard, the ubiquitous and relatively low expression of KCS11 in Arabidopsis has been suggested as an indication of a general role in the synthesis of very-long-fatty acids required for growth and development31.
Cuticle density as an emerging trait in fruit quality
Decomposition of cuticle mass into three anatomical and physical traits has shown that all the QTLs for cuticle mass colocalized with QTLs related to thickness, invagination and/or density (Fig. 3), indicating that changes in cuticle mass were always accompanied by changes in any of these other traits. However, changes in cuticle deposition did not always affect all the other three parameters, allowing the identification of groups of QTLs for cuticle mass with different effects. In one group of genomic regions, cm3.1, cm3.2, and cm11.2, cuticle mass had a small effect on thickness and invagination causing a decrease in cuticle density, whereas in another group, cm2.1, cm2.2, and cm7.1, the effect on thickness and invagination was more dramatic and increased the density. Also, some genomic regions for cuticle mass had only an effect on cuticle thickness, cm4.1, cm10.2, or cm12.1, and others such as cm1.1 and cm1.2 modified cuticle invagination without changing thickness. Overall, this indicates that alteration of cuticle mass does not always translate into parallel or even proportional changes in thickness, invagination and/or density. Similarly, in cucumber, a QTL for cuticle thickness and invagination and two additional ones for invagination only were reported, pointing that thickness and invagination do not always vary together10. The QTLs here identified provide genetic tools to independently modify some of these traits.
Differences in cuticle density among species have been reported in the literature32,33. However, its evolution throughout development has only been characterized in tomato, where variation between cultivars was detected as well as changes during ripening6. Density can modulate the mechanical properties of the cuticle, with a high density hindering the molecular motion of the polymer chains32. A comparative analysis of different tree species found that cuticle density only partially explained the mechanical strength of the cuticle33. However, a clear and significant correlation between these two parameters was reported in a study of numerous cultivars of Diospiros kaki34. Although a reduction in cuticle density was observed after removal of polysaccharides but not waxes32, the role of polysaccharides derived from the primary cell wall and their association and/or chemical connection to the cutin matrix still needs further investigation to properly understand their contribution to cuticle density. Additionally, several parameters such as cutin monomer composition, the relative concentration of functional groups able to crosslink, degree of polymerization and the number of secondary hydroxyl groups engaged in polymer chain interesterification could modify cutin 3D architecture and therefore affect cuticle density. However, a structural and compositional analysis of cuticle density is still missing. The material herein identified could be a potential source for these analyses.
Changes in cuticle mass through modification of cuticle density that do not translate into thicker and more invaginated cuticles, or modulation of cuticle density could provide a strategy to maintain mechanical strength while reducing undesirable cuticle traits for palatability that could be valuable for breeding purposes. In this sense, the QTL regions in chromosome 8 that provide a dense cuticle material with low thickness and invagination without affecting cuticle mass deserves further investigation.
Genetic interaction between cuticle phenolics and waxes
Cuticle phenolics and waxes play important biophysical roles that have great agronomic importance for fruit growth on the vine and also during postharvest2. However, a negative correlation between cuticle waxes and phenolics, based on the modification of the phenolic metabolic pathway, either silencing CHALCONE SYNTHASE (CHS), the first committed enzyme of the flavonoid pathway, or favoring the accumulation of anthocyanins in the fruit by expression of the transcription factors ROSEA and DELILA, has been reported35. The nature of this relation could have a critical impact in plant breeding since it would imply the necessity to choose between increasing mechanical resistance provided by phenolics and reduced water permeability driven by waxes. A small but significant and negative correlation was observed within the RIL population (r = −0.275, P < 0.01) between the percentages of phenolics and waxes. Analysis of the colocation of waxes and phenolics genetic determinants indicate that this relation is based on the QTLs present in chromosome 1 and 12 with overlapping regions for both compounds. However, identification of QTLs that modify either phenolics or waxes, such as ph7.1, ph7.2, ph8.1, and wx12.1, indicates that there are alternative genetic strategies to avoid a compromise between increased phenolics and waxes. Moreover, colocation of QTLs with similar effect for waxes and phenolics on chromosome 4 suggests a complex interaction between these pathways. In this sense, it has recently been reported a Kelch repeat F-box protein (SMALL AND GLOSSY LEAVES1) that negatively regulates both wax and phenylpropanoid biosynthesis in Arabidopsis36,37.
MYB12 was identified as the candidate gene behind ph1.1 since a notable downregulation of the pim allele was detected during ripening, coinciding with the phenotypic differences observed in the cuticle. MYB12 is the gene responsible for the colorless fruit epidermis (y) mutation in tomato and has been reported to dramatically decrease cuticle phenolics at ripe38. Cuticle analysis of Ailsa Craig y/y showed a significant decrease in phenolics and cutin and increase in waxes and polysaccharides compared to the wildtype (Supplementary Table 4), which would be in agreement with the overlap of the QTLs for phenolics, waxes and cutin and the almost significant QTL for polysaccharides present in chromosome 1. Transcriptome analyses of various y mutant alleles have shown an increase in various enzymes related to wax biosynthesis during ripening39 that would support a role of MYB12 in regulating the phenolic and fatty acid biosynthetic pathways. The reduced effect on phenolics (Fig. 5) observed for the line containing the pim allele of MYB12, despite ph1.1 being the major QTL with the highest additive effect, could be the result of an interaction between MYB12 and other region/s within this wide QTL that could be masking its individual effect. In this regard DET1, a regulator of light signal transduction whose downregulation increases flavonoid accumulation in tomato fruits, is the only gene with a known effect in the phenolic pathway located within ph1.1 region40. Also, this lesser effect on phenolics could be behind the absence of differences for waxes and cutin observed in the line harboring the overlapping QTLs ph1.1, wx1.1, and cu1.1 and would imply that a certain degree of phenolic reduction is needed for the effect on lipids to be significant. Additionally, an interaction between MYB12 and LACS1, the gene closest to wx1.1 peak marker, could also explain the results on waxes and cutin. Although LACS1 expression has not been reported to be modified in the transcriptomic analysis of y mutants39, the observed downregulation of the LACS1 pim allele during ripening indicates a potential role as candidate gene for waxes and cutin. Analysis of the Arabidopsis LACS family has uncovered a complex network of redundant activities, with overlapping functions of LACS1 and LACS2 on cutin and wax biosynthesis and LACS1 and LACS4 on wax biosynthesis41,42; hence the absence of effect on waxes and cutin could also be due to the involvement of other LACS genes. Individualized analysis of both genes and potential compensating roles of other LACS genes need to be addressed to clarify their effects.
Materials and methods
Plant material and subIL selection
A 169 RIL population was employed to carry out QTL analyses. This population is an F8 derived from the interspecific cross between Solanum lycopersicum L. “Moneymaker” (MM) and Solanum pimpinellifolium L. “TO-937” (TO-937) by single seed descent43. An inbred line (IL) population consisting of 52 lines derived from the recurrent backcross of TO-937 into MM background and selected for harboring specific introgressions,44 was also employed for QTL analysis and validation (Supplementary Fig. 3). SubILs with shorter introgressions were obtained for chromosomes 3, 7 and 11 from the F2 populations of the crosses of the corresponding ILs with MM. DNAzol (Life Technologies, USA) was used to isolate DNA from the F2 populations and Single Nucleotide Polymorphisms (SNP) SOLCAP markers (available at https://solgenomics.net/) were employed to select the plants. Markers were resolved using High Resolution Melting analysis. Ailsa Craig wildtype (wt) (LA2838A) and the nearly isogenic Ailsa Craig y/y (LA3189) harboring the colorless fruit epidermis (y) mutation were also studied. Five plants per RIL, 10 plants per IL and subIL, and 20 plants of MM, Ailsa Craig wt and Ailsa Craig y/y were grown in a polyethylene greenhouse at the Estación Experimental La Mayora, CSIC, Spain.
QTL analysis
The genotyping of the RIL population45 with the 8K SNP SOLCAP Infinium chip46 was employed to construct a linkage map containing 4885 SNP markers using JoinMap® 4.0 software (Kyazma BV, Wageningen, the Netherlands) with the maximum likelihood algorithm and the Kosambi mapping function. QTL analysis was carried out with the software MapQTL® 5.0 (Kyazma BV, Wageningen, the Netherlands) using interval mapping and subsequent QTL modeling by multiple QTL mapping after automatic cofactor selection. Permutation tests with 10,000 resamplings were employed to determine the probability associated to each QTL. A reduction of the map to 1302 informative SNP markers was performed for computational purposes to carry out a composite interval mapping47 and epistatic analysis using IciMapping48. QTL map was drawn with Mapchart 2.2 (ref. 49).
Any change in the amount of cuticle deposited (cuticle mass) also modifies the amount of cuticle components, unless their percentage distribution is modified. For this reason, the QTL analysis of the different cuticle components was carried out with the percentage and not the amount. Unless indicated otherwise, all the QTLs identified for the amount of cuticle also affected the amount of phenolics, waxes, cutin and polysaccharides.
Gene expression analyses
Trizol® Reagent (Life Technologies, USA) was used to isolate RNA from tomato fruit peels at different stages of development (immature green, mature green, breaker, and red ripe). Genomic DNA was removed with RNase-free DNase and RNA was cleaned with the Nucleospin RNA clean-up kit (Macherey-Nagel, Germany). First-strand cDNA synthesis was carried out with the Super Script III First-Strand Synthesis Super Mix for qRT-PCR (Invitrogen, USA). Relative transcript amount of the different genes was measured by RT-qPCR using SsoAdvanced™ SYBR® Green Supermix (Bio-Rad, USA) following the ΔCt method with three control genes, CAC, EXPRESSED, and SAND previously employed for tomato peel analyses24. Three biological replicates corresponding to pools of epicarp pieces from different tomatoes and plants were analysed. For each biological replicate three technical replicates were performed. Primers used for amplification are shown in Supplementary Table 5. Comparison of the mRNA sequences of MM and TO-937 genes was carried out to identify changes that would result in non-conserved amino acid substitutions.
Tissue sectioning and staining
Small pericarp pieces of three fruits per genotype, each fruit from a different plant, were fixed in a formaldehyde, acetic acid and ethanol solution (1:1:18), dehydrated in an ethanol dilution series (70–95%) and embedded in a commercial resin. Samples were cross-sectioned into slices 4 µm thick using a microtome (Leica, Germany) and stained with Sudan IV to visualize the cuticle. For the comparison of IL and subILs with MM, six fruits of MM, each from a different plant, were studied. Cuticle thickness was estimated from 30 measurements of cross-sectioned samples using the image analysis program Fiji. Ten non-seriated sections per biological replicate were analysed and 1–2 measures taken in each section. To avoid the effect of cutinized anticlinal cell walls or ridges, cuticle thickness was measured in the central region of cells, since this area remains constant throughout fruits. Cuticle area of 10 non-seriated cross-sections per biological replicate were calculated with the software Fiji and referred to the weight of the cuticle surface area to estimate cuticle density, following the protocol already established in6. Cuticle invagination was estimated from a minimum of 15 non-seriated cross-sections per biological replicate using an arbitrary index, as reported in7. This index accounts for how much the anticlinal and inner periclinal walls of epidermal and subepidermal cells are cutinized. 0 means no cutinization of the epidermal anticlinal cell walls; 0.25: the upper half of the epidermal anticlinal cell walls is cutinized; 0.5: epidermal anticlinal cell walls are completely cutinized; 0.75: additionally, half of the inner periclinal epidermal cell wall is also cutinized; 1: all the epidermal cell walls are cutinized. Values >1 indicate additional cell wall cutinization of the first hypodermal cell layer following the same criteria as for the epidermal cell layer.
Cuticle isolation and cuticle component analyses
Cuticles were enzymatically isolated from tomato fruits following the protocol already stablished6 using an aqueous solution of sodium citrate with mixture of cellulase and pectinase and NaN3 to prevent microbial growth. Isolated cuticles were air dried and stored under dry conditions. Cuticle surface area was calculated using Fiji and 20 measures per genotype, 40 in the case of MM. Samples were then weighed in order to determine the mass of cuticle per unit surface area. Cuticular waxes were removed by heating at 60 °C 0.1 g of isolated cuticles in 100 mL chloroform: methanol (2:1 v:v) for 2 h. Cutin isolates were obtained after refluxing the dewaxed cuticles in a 6 M HCl aqueous solution for 12 h. Phenolic components were estimated after cutin depolymerization in a UV-VIS spectrophotometer (Pharmacia Biotech, NJ, USA) following a protocol previously described50. Percentages of cuticle components were estimated from five samples per genotype, except for MM where 10 samples were determined.
Statistics
Data are expressed as means ± standard error (SE). T tests were used to compare the parental lines MM and TO-937, and Ailsa Craig wt and y/y. One-way ANOVA and Dunnett’s tests were used to compare the ILs or subILs with MM. Percentages were arcsin-transformed for statistical analyses, although they are presented untransformed. Pearson’s coefficient was employed to correlate the different variables studied. Asterisks indicate differences at *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary information
Acknowledgements
This work was supported by grant RTI2018-094277-B/AEI/10.13039/501100011033 from Agencia Estatal de Investigación, Ministerio de Ciencia e Innovación, Spain co-financed by the European Regional Development Fund (ERDF).
Author contributions
R.B. obtained the subILs, performed the cuticle mass and phenolic analyses in the IL and subIL populations, participated in the QTL analyses and analyzed data; P.S. carried out the invagination and density analysis of the RIL population and all the microscopy analyses in the IL and subIL populations, R.M.G. carried out the sectioning and cuticle thickness analyses of the RIL population; A.H. participated in the conceptualization and funding acquisition; R.F.M. constructed the linkage maps and participated in the conceptualization, funding acquisition, and the QTL analyses; E.D. wrote the paper draft and coordinated its preparation, performed the gene expression analyses, and participated in the conceptualization, funding acquisition, and QTL analyses. All authors contributed in the discussion of results and participated in the revision of the paper.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflict of interest
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00548-5.
References
- 1.Petit J, Bres C, Mauxion JP, Bakan B, Rothan C. Breeding for cuticle-associated traits in crop species: traits, targets, and strategies. J. Exp. Bot. 2017;68:5369–5387. doi: 10.1093/jxb/erx341. [DOI] [PubMed] [Google Scholar]
- 2.Lara I, Heredia A, Domínguez E. Shelf life potential and the fruit cuticle: the unexpected player. Front. Plant Sci. 2019;10:770. doi: 10.3389/fpls.2019.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domínguez E, Heredia-Guerrero JA, Heredia A. The biophysical design of plant cuticles: an overview. N. Phytol. 2011;189:938–949. doi: 10.1111/j.1469-8137.2010.03553.x. [DOI] [PubMed] [Google Scholar]
- 4.Khanal BP, Grimm E, Finger S, Blume A, Knoche M. Intracuticular wax fixes and restricts strain in leaf and fruit cuticles. N. Phytol. 2013;200:134–143. doi: 10.1111/nph.12355. [DOI] [PubMed] [Google Scholar]
- 5.Jetter R, Riederer M. Localization of the transpiration barrier in the epi- and intracuticular waxes of eight plant species: water transport resistances are associated with fatty acyl rather than alicyclic components. Plant Physiol. 2016;170:921–934. doi: 10.1104/pp.15.01699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.España L, et al. Biomechanical properties of tomato fruit cuticle during development are modulated by changes in the relative amount of their components. N. Phytol. 2014;202:790–802. doi: 10.1111/nph.12727. [DOI] [PubMed] [Google Scholar]
- 7.España L, et al. Transient silencing of CHALCONE SYNTHASE during fruit ripening modifies tomato epidermal cells and cuticle. Plant Physiol. 2014;166:1371–1386. doi: 10.1104/pp.114.246405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konarska A. The structure of the fruit peel in two varieties of Malus domestica Borkh. (Rosaceae) before and after storage. Protoplasma. 2013;250:701–714. doi: 10.1007/s00709-012-0454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camacho-Vázquez C, et al. Filling gaps in our knowledge on the cuticle of mangoes (Mangifera indica) by analyzing six fruit cultivars: architecture/structure, postharvest physiology and possible resistance to fruit fly (Tephritidae) attack. Postharvest Biol. Technol. 2019;148:83–96. doi: 10.1016/j.postharvbio.2018.10.006. [DOI] [Google Scholar]
- 10.Rett-Cadman S, et al. QTL and transcriptomic analyses implicate cuticle transcription factor SHINE as a source of natural variation for epidermal traits in cucumber fruit. Front. Plant Sci. 2019;10:1536. doi: 10.3389/fpls.2019.01536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matas AJ, Cobb ED, Bartsch JA, Paolillo DJ, Niklas KJ. Biomechanics And anatomy of Lycopersicon esculentum fruit peels and enzyme-treated samples. Am. J. Bot. 2004;91:352–360. doi: 10.3732/ajb.91.3.352. [DOI] [PubMed] [Google Scholar]
- 12.Voisey PW, Lyall LH, Kloek M. Tomato skin strength: Its measurement and relation to cracking. J. Am. Soc. Hortic. Sci. 1970;95:485–488. [Google Scholar]
- 13.Matas AJ, Cobb ED, Paolillo DJ, Niklas KJ. Crack resistance in cherry tomato fruit correlates with cuticular membrane thickness. HortSci. 2004;39:1354–1358. doi: 10.21273/HORTSCI.39.6.1354. [DOI] [Google Scholar]
- 14.Amos R. Sensory properties of fruit skins. Postharvest Biol. Technol. 2007;44:307–311. doi: 10.1016/j.postharvbio.2006.12.007. [DOI] [Google Scholar]
- 15.Barrett DM, Garcia E, Wayne JE. Textural modification of processing tomatoes. Crit. Rev. Food Sci. Nutr. 1998;38:173–258. doi: 10.1080/10408699891274192. [DOI] [PubMed] [Google Scholar]
- 16.Ayvaz H, Santos AM, Rodriguez-Saona LE. Understanding tomato peelability. Compr. Rev. Food Sci. Food Saf. 2016;15:619–632. doi: 10.1111/1541-4337.12195. [DOI] [PubMed] [Google Scholar]
- 17.Riedel M, et al. Cuticular wax composition in Cocos nucifera L.: physicochemical analysis of wax components and mapping of their QTLs onto the coconut molecular linkage map. Tree Genet. Genomes. 2009;5:53–69. doi: 10.1007/s11295-008-0168-7. [DOI] [Google Scholar]
- 18.Yeats TH, et al. The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. Plant J. 2012;69:655–666. doi: 10.1111/j.1365-313X.2011.04820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernández-Moreno JP, et al. Uncovering tomato quantitative trait loci and candidate genes for fruit cuticular lipid composition using the Solanum pennellii introgression line population. J. Exp. Bot. 2017;68:2703–2716. doi: 10.1093/jxb/erx134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosney BJ, et al. Genetic control of cuticular wax compounds in Eucalyptus globulus. N. Phytol. 2016;209:202–215. doi: 10.1111/nph.13600. [DOI] [PubMed] [Google Scholar]
- 21.Würschum T, Langer SM, Longin CFH, Tucker MR, Leiser WL. Refining the genetic architecture of flag leaf glaucousness in wheat. Theor. Appl. Genet. 2020;133:981–991. doi: 10.1007/s00122-019-03522-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons EP, et al. Fruit cuticle lipid composition and fruit post-harvest water loss in an advanced backcross generation of pepper (Capsicum sp.) Physiol. Plant. 2012;146:15–25. doi: 10.1111/j.1399-3054.2012.01592.x. [DOI] [PubMed] [Google Scholar]
- 23.Giménez E, et al. Transcriptional activity of the MADS box ARLEQUIN/TOMATO AGAMOUS-LIKE1 gene is required for cuticle development of tomato fruit. Plant Physiol. 2015;168:1036–1048. doi: 10.1104/pp.15.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segado P, Heredia-Guerrero JA, Heredia A, Domínguez E. Sequential involvement of cutinsomes and CUS1 in tomato fruit cutin deposition. Plant Physiol. 2020;183:1622–1637. doi: 10.1104/pp.20.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hen-Avivi S, Lashbrooke J, Costa F, Aharoni A. Scratching the surface: genetic regulation of cuticle assembly in fleshy fruit. J. Exp. Bot. 2014;65:4653–4664. doi: 10.1093/jxb/eru225. [DOI] [PubMed] [Google Scholar]
- 26.Leide J, Hildebrandt U, Reussing K, Riederer M, Vogg G. The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: effects of a deficiency in a β-ketoacyl-coenzyme A synthase (LeCER6) Plant Physiol. 2007;144:1667–1679. doi: 10.1104/pp.107.099481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voisin D, et al. Dissection of the complex phenotype in cuticular mutants of Arabidopsis reveals a role of SERRATE as a mediator. PLOS Genet. 2009;5:e1000703. doi: 10.1371/journal.pgen.1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karabourniotis, G. & Liakopoulos, G. In Advances in Plant Physiology, 8 (ed Hemantaranjan, A.) Ch. 3 (Scientific Publishers, 2005).
- 29.Leide J, de Souza AX, Papp I, Riederer M. Specific characteristics of the apple fruit cuticle: investigation of early and late season cultivars ‘Prima’ and ‘Florina’ (Malus domestica Borkh.) Sci. Hortic. 2018;229:137–147. doi: 10.1016/j.scienta.2017.10.042. [DOI] [Google Scholar]
- 30.Sampangi-Ramaiah MH, et al. Barrier against water loss: relationship between epicuticular wax composition, gene expression and leaf water retention capacity in banana. Funct. Plant Biol. 2016;43:492–501. doi: 10.1071/FP15296. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, et al. Arabidopsis 3-ketoacyl-Coenzyme A synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 2013;162:567–580. doi: 10.1104/pp.112.210450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreiber L, Schönherr J. Phase transitions and thermal expansion coefficients of plant cuticles: The effects of temperature on structure and function. Planta. 1990;182:186–193. doi: 10.1007/BF00197109. [DOI] [PubMed] [Google Scholar]
- 33.Onoda Y, Richards L, Westoby M. The importance of leaf cuticle for carbon economy and mechanical strength. N. Phytol. 2012;196:441–447. doi: 10.1111/j.1469-8137.2012.04263.x. [DOI] [PubMed] [Google Scholar]
- 34.Tsubaki S, Ozaki Y, Yonemori K, Azuma J. Mechanical properties of fruit-cuticular membranes isolated from 27 cultivars of Diospyros kaki Thunb. Food Chem. 2012;132:2135–2139. doi: 10.1016/j.foodchem.2011.12.039. [DOI] [Google Scholar]
- 35.Heredia A, Heredia-Guerrero JA, Domínguez E. CHS silencing suggests a negative cross-talk between wax and flavonoid pathways in tomato fruit cuticle. Plant Signal. Behav. 2015;10:e1019979. doi: 10.1080/15592324.2015.1019979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H, Yu S, Jung SH, Lee B, Suh MC. The F-Box protein SAGL1 and ECERIFERUM3 regulate cuticular wax biosynthesis in response to changes in humidity in Arabidopsis. Plant Cell. 2019;31:2223–2240. doi: 10.1105/tpc.19.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu S, Kim H, Yun DJ, Suh MC, Lee B. Post-translational and transcriptional regulation of phenylpropanoid biosynthesis pathway by Kelch repeat F-box protein SAGL1. Plant Mol. Biol. 2019;99:135–148. doi: 10.1007/s11103-018-0808-8. [DOI] [PubMed] [Google Scholar]
- 38.Adato A, et al. Fruit surface flavonoid accumulation in tomato is controlled by a SlMYB12 regulated transcriptional network. PLOS Genet. 2009;5:e1000777. doi: 10.1371/journal.pgen.1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernández-Moreno JP, et al. Characterization of a new pink-fruited tomato mutant results in the identification of a null allele of the SlMYB12 transcription factor. Plant Physiol. 2016;171:1821–1836. doi: 10.1104/pp.16.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davuluri GR, et al. Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 2005;23:890–895. doi: 10.1038/nbt1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lü S, et al. Arabidopsis CER8 encodes LONGCHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2009;59:553–564. doi: 10.1111/j.1365-313X.2009.03892.x. [DOI] [PubMed] [Google Scholar]
- 42.Jessen D, et al. Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. Plant J. 2011;68:715–726. doi: 10.1111/j.1365-313X.2011.04722.x. [DOI] [PubMed] [Google Scholar]
- 43.Alba JM, Montserrat M, Fernández-Muñoz R. Resistance to the two-spotted spider mite (Tetranychus urticae) by acylsucroses of wild tomato (Solanum pimpinellifolium) trichomes studied in a recombinant inbred line population. Exp. Appl. Acarol. 2009;47:35–47. doi: 10.1007/s10493-008-9192-4. [DOI] [PubMed] [Google Scholar]
- 44.Barrantes W, et al. Highly efficient genomics-assisted development of a library of introgression lines of Solanum pimpinellifolium. Mol. Breed. 2014;34:1817–1831. doi: 10.1007/s11032-014-0141-0. [DOI] [Google Scholar]
- 45.Rambla JL, et al. Identification, introgression, and validation of fruit volatile QTLs from a red-fruited wild tomato species. J. Exp. Bot. 2017;68:429–442. doi: 10.1093/jxb/erw455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sim SC, et al. Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS ONE. 2012;7:e0040563. doi: 10.1371/journal.pone.0040563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng L, Li H, Zhang L, Wang J. QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in bi-parental populations. Crop J. 2015;3:269–283. doi: 10.1016/j.cj.2015.01.001. [DOI] [Google Scholar]
- 49.Voorrips REMapChart. Software for the graphical presentation of linkage maps and QTLs. J. Heredity. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 50.Domínguez E, España L, López-Casado G, Cuartero J, Heredia A. Biomechanics of isolated tomato (Solanum lycopersicum) fruit cuticles during ripening: the role of flavonoids. Funct. Plant Biol. 2009;36:613–620. doi: 10.1071/FP09039. [DOI] [PubMed] [Google Scholar]
- 51.Lü S, et al. Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 2012;159:930–944. doi: 10.1104/pp.112.198697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schnurr J, Shockey J, Browse J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell. 2004;16:629–642. doi: 10.1105/tpc.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rashotte A, Jenks MA, Feldmann KA. Cuticular waxes on eceriferum mutants of Arabidopsis thaliana. Phytochemistry. 2001;57:115–123. doi: 10.1016/S0031-9422(00)00513-6. [DOI] [PubMed] [Google Scholar]
- 54.Bach L, et al. The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl Acad. Sci. U.S.A. 2008;105:14727–14731. doi: 10.1073/pnas.0805089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia Y, et al. The glabra1 mutation affects cuticle formation and plant responses to microbes. Plant Physiol. 2010;154:833–846. doi: 10.1104/pp.110.161646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li-Beisson Y, et al. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc. Natl Acad. Sci. U.S.A. 2009;106:22008–22013. doi: 10.1073/pnas.0909090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.