Abstract
Background
Imported malaria parasites with anti-malarial drug resistance (ADR) from Africa is a serious public health challenge in non-malarial regions, including Wuhan, China. It is crucial to assess the ADR status in African Plasmodium falciparum isolates from imported malaria cases, as this will provide valuable information for rational medication and malaria control.
Methods
During 2017–2019, a cross-sectional study was carried out in Wuhan, China. Peripheral blood 3 ml of returned migrant workers from Africa was collected. The target fragments from pfcrt, pfmdr1, and k13 propeller (pfk13) genes were amplified, sequenced, and analysed.
Results
In total, 106 samples were collected. Subsequently, 98.11% (104/106), 100% (106/106), and 86.79% (92/106) of these samples were successfully amplified and sequenced for the pfcrt (72–76), pfmdr1, and pfk13 genes, respectively. The prevalence of the pfcrt 76 T, pfmdr1 86Y, and pfmdr1 184F mutations was 9.62, 4.72, and 47.17%, respectively. At codons 72–76, the pfcrt locus displayed three haplotypes, CVMNK (wild-type), CVIET (mutation type), CV M/I N/E K/T (mixed type), with 87.50%, 9.62%, and 2.88% prevalence, respectively. For the pfmdr1 gene, NY (wild type), NF and YF (mutant type), N Y/F, Y Y/F, and N/Y Y/F (mixed type) accounted for 34.91, 43.40, 3.77, 15.09, 0.94, and 1.89% of the haplotypes, respectively. A total of 83 isolates with six unique haplotypes were found in pfcrt and pfmdr1 combined haplotypes, of which NY-CVMNK and NF-CVMNK accounted for 40.96% (34/83) and 43.37% (36/83), respectively. Furthermore, 90 cases were successfully sequenced (84.91%, 90/106) at loci 93, 97, 101, and 145, and 78 cases were successfully sequenced (73.58%, 78/106) at loci 343, 353, and 356 for pfcrt. However, the mutation was observed only in locus 356 with 6.41%. For pfk13, mutations reported in Southeast Asia (at loci 474, 476, 493, 508, 527, 533, 537, 539, 543, 553, 568, 574, 578, and 580) and Africa (at loci 550, 561, 575, 579, and 589) were not observed.
Conclusions
The present data from pfcrt and pfmdr1 demonstrate that anti-malarial drugs including chloroquine, amodiaquine, and mefloquine, remain effective against malaria treatment in Africa. The new mutations in pfcrt related to piperaquine resistance remain at relatively low levels. Another source of concern is the artemether-lumefantrine resistance-related profiles of N86 and 184F of pfmdr1. Although no mutation in pfk13 is detected, molecular surveillance must continue.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12936-021-03737-8.
Keywords: Imported malaria, Plasmodium falciparum, Anti-malarial resistance, Molecular surveillance, Wuhan
Background
Malaria is a mosquito-borne infectious disease that seriously threatens human health, among which falciparum malaria caused by Plasmodium falciparum is the most serious, mainly in tropical and subtropical regions in sub-Saharan Africa and Southeast Asia (SEA) [1]. In 2019, there was an estimated 229 million malaria cases from 87 malaria-endemic countries. Furthermore, approximately 94% of estimated cases were detected in Africa. The countries of Nigeria, Congo, Uganda, Mozambique, and Niger account for 51% of malaria cases (117 million). Additionally, it estimated approximately 409,000 deaths are estimated globally [1]. Although there have been no indigenous malaria cases reported in China for three consecutive years since 2017 [2], potential challenges remain in imported malaria cases. In recent years, with globalization, the number of migrant workers, tourists, and businesspeople in China has increased gradually, especially those returning from Africa and SEA [3], which has brought severe pressure for malaria eradication in China. Thus, it is necessary to strengthen surveillance for imported malaria.
Anti-malarial drugs are considered the major measure for malaria control [4]. However, with the continuous use of anti-malarial drugs, P. falciparum gradually achieves drug resistance and spreads rapidly [5]. Chloroquine (CQ) is a safe, inexpensive, and effective anti-malarial drug for malaria therapy. However, in the 1940s, P. falciparum parasites developed resistance to CQ. Since then, CQ-resistant (CQR) strains have begun to spread rapidly around the world [6]. After the discovery of artemisinin (ART) in the 1970s, malaria control was temporarily eased. To improve clinical efficacy and delay the emergence of parasite drug resistance, artemisinin-based combination therapy (ACT) have been recommended by the World Health Organization (WHO) since 2001 [7]. Unfortunately, ART resistance of P. falciparum isolates was reported in SEA [8–10]. Recently, dihydroartemisinin-piperaquine (DHA-PPQ) resistance has been detected in western Cambodia [11–14]. Although ACT remains effective in Africa and SEA, prolonged use of ART would lead to anti-malarial drug resistance. Anti-malarial drug resistance (ADR) would be disastrous for global malaria control. Therefore, in the absence of more choices, it is urgent to monitor the ADR status of P. falciparum parasites.
Mutations detected in P. falciparum essential genes including pfcrt, pfmdr1, pfdhfr, pfdhps, pfk13, and pfpm2 have been used as molecular markers of drug resistance. The pfk13 polymorphism has been considered to be related to ART resistance [9]. However, previous studies demonstrated that the distribution of alleles for pfk13 varies according to the mutations [15]. In SEA, the alleles of the 580Y mutation account for the vast majority [9]. In Africa, the mutation rate of pfk13 remained relatively low. In 2016, the newly discovered local ART resistance mutation 561H of pfk13 was reported from Rwanda, Africa [16]. The 72–76 amino acid mutation in pfcrt, especially the 76 T mutation, was the primary marker of CQR [17–19]. Several mutations in pfmdr1 are related to the resistance of P. falciparum to CQ, amodiaquine (AQ) and mefloquine (MQ) [20, 21]. At present, several newly detected mutations in pfcrt, including 93S, 97Y, 101F, 145I, 343L, 353 V and 356 T, have been identified to be associated to with PPQ with a decreasing trend for the susceptibility of P. falciparum strains in South America [13]. However, there was limited information on the effects of these alleles on PPQ in Africa, where malaria is endemic.
In the present study, polymorphisms of pfcrt, pfmdr1, and pfk13 for P. falciparum isolates imported from Africa in Wuhan, China were surveyed. This survey will provide valuable information for rational medication for malaria patients in clinical practice, preventing the spread of ADR P. falciparum in Africa and China.
Methods
Collection of samples
A cross-sectional study was performed in Wuhan, Hubei Province, China. Peripheral blood 3 ml of returned migrant workers from African countries was collected at major hospitals in the region from May 2017 to December 2019. These samples were collected from Wuhan Jinyintan Hospital, Wuhan Union Hospital and Zhongnan Hospital of Wuhan University, and Tongji Hospital. These samples were examined by an immuno-gold assay kit (ICT Diagnostics) for Plasmodium spp. antigen (HRPII). Then, blood smears were prepared and checked under a microscope. Finally, the species were identified by qPCR. Approximately 400 μl of blood was spotted on 3 MM Whatman filter paper and air-dried (identified and provided by the Center for Disease Control and Prevention of Wuhan City, Hubei Province). Then, these filter papers were numbered and stored at − 20 °C with in polyethylene bag. Consent of the owner and his legal guardian was obtained before sampling.
Determination of Plasmodium falciparum gene mutations
Genomic DNA (gDNA) was extracted from blood-spots by a TIANamp Blood Spots DNA Kit to yield approximately 50 μl of supernatant containing gDNA and stored at − 20 °C until further use. The target fragments of the pfcrt, pfmdr1, and pfk13 genes were amplified from the gDNA sample via nested PCR and traditional PCR. Following previously published primer information and a previous procedure [13, 16, 22, 23], the pfcrt, pfmdr1, and pfk13 genes were successfully amplified. The reaction system and procedure for PCR are listed in Additional file 2: Table S1. Loci 72–76 of the pfcrt gene and pfmdr1 gene were subjected to two rounds of PCR amplification (nested PCR). Loci 93–356 of the pfcrt gene were subjected to one round of PCR amplification (traditional PCR). After all the reactions finished, the 5.0 μl PCR products were analysed by 1.0% agarose gel electrophoresis. Then the remaining products were purified for Sanger sequencing (Genewiz, Soochow, China). The reference sequences of the P. falciparum 3D7 strain were downloaded from the database PlasmoDB (http://plasmodb.org/plasmo/) with the gene IDs: PF3D7_0709000 (pfcrt), PF3D7_0523000 (pfmdr1), and PF3D7_1343700 (pfk13). The sequencing data were analysed with Dnastar (DNASTAR Inc., Madison, WI, USA) and compared with the standard sequence. Synonymous mutations and nonsynonymous mutations were detected, and confirmed via traditional bidirectional Sanger sequencing. To avoid any kind of technical contamination, plasmids with known mutant alleles of pfcrt, pfmdr1, and pfk13 were used as the positive controls for these samples (Additional file 1: Fig. S1).
Data analysis
Excel software (Microsoft Excel; version 2016) was used to record the data and calculate the frequency of single nucleotide polymorphisms (SNPs) and haplotypes. The 95% confidence intervals of wild-type and mutant types for these genes were analysed with SPSS 25 (SPSS Inc., Chicago, IL, USA). Mixed-type genes, which include mixed infections of wild-type and mutant type, were excluded from the combined haplotype analysis [16].
Results
General information
A total of 106 of these returnees were diagnosed with P. falciparum from 2017 to 2019, all from Africa (Additional file 3: Table S2), including 48 cases in West Africa, 33 cases in Central Africa, 15 cases in South Africa, nine cases in East Africa, and one case in North Africa. These cases were mainly from 21 African countries, particularly concentrated in Congo (21.70%, 23/106), followed by Nigeria (16.98%, 18/106), Ivory Coast (9.43%, 10/106), and Mozambique (6.60%, 7/106).
Mutation prevalence of pfcrt and pfmdr1
The polymorphisms and haplotypes of pfcrt and pfmdr1 were analysed. For the pfcrt mutations C72S, M74I, N75E, and K76T, a total of 104 samples were successfully sequenced (98.11%, 104/106). Codon 72 in all samples had 100% wild type, and the mutation frequency of codons 74, 75, and 76 was 9.62% (10/104) (Table 1). These mutations were mainly concentrated in West Africa, followed by Central Africa. The results showed that the pfcrt genotype had polymorphisms at codons 72–76, including CVMNK (wild-type), CVIET (mutation type), and CV M/I N/E K/T (mixed type). Most isolates harboured parasites with the CVMNK (87.5%, 91/104). The proportion of CVIET among these isolates was 9.62% (10/104). The last case was CV M/I N/E K/T, accounting for 2.88% (3/104), and no haplotype of SVMNT was found (Additional file 4: Table S3).
Table 1.
Observed overall frequency of mutations in pfcrt and pfmdr1
| Gene | Mutations | Wild type(%, 95% CI) | Mutation(%, 95% CI) | Mixed type(%, 95% CI) | Total |
|---|---|---|---|---|---|
| pfcrt | C72S | 104(100.00, 1.00–1.00) | 0(0.00, 0.00–0.00) | 0(0.00, 0.00–0.00) | 104 |
| M74I | 91(87.5, 0.81–0.94) | 10(9.62, 0.039–0.15) | 3(2.88, − 0.004 to 0.062) | 104 | |
| N75E | 91(87.5, 0.81–0.94) | 10(9.62, 0.039–0.15) | 3(2.88, − 0.004 to 0.062) | 104 | |
| K76T | 91(87.5, 0.81–0.94) | 10(9.62, 0.039–0.15) | 3(2.88, − 0.004 to 0.062) | 104 | |
| I356T | 70(89.74, 0.39–0.55) | 5(6.41, 0.09–0.120) | 3(3.84, − 0.005 to 0.082) | 78 | |
| pfmdr1 | N86Y | 99(93.40, 0.89–0.98) | 5(4.72, 0.006–0.088) | 2(1.89, − 0.007 to 0.045) | 106 |
| Y184F | 37(34.91, 0.26–0.44) | 50(47.17, 0.38–0.57) | 19(17.92, 0.11–0.25) | 106 |
Mutations are shown in underline and bold
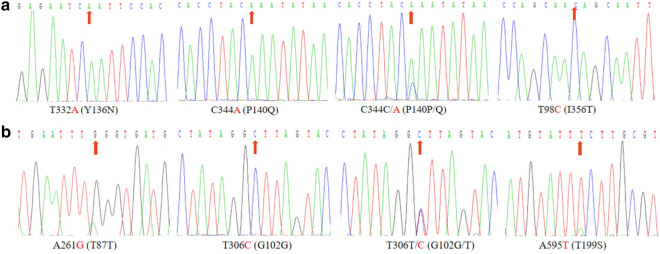
For several new alleles of pfcrt (Additional file 3: Table S2), 90 cases were successfully sequenced (84.91%, 90/106) at loci 93, 97, 101 and 145, and no mutation was found. Additionally, two isolates (2.22%, 2/90) were found to have mutations at locus 136 (Fig. 1a); these isolates come from Nigeria (1.11%, 1/90) of West Africa and Mozambique (1.11%, 1/90) in South Africa. At loci 140 (Fig. 1a), one isolate carried mutant allele and 1 isolate (1.28%) was mixed, from Gabon (1.11%, 1/90), Central Africa, Nigeria (1.11%, 1/90), and West Africa. In addition, 78 cases were successfully sequenced (73.58%, 78/106) at loci 343, 350, 353, and 356. No mutations were observed at loci 343, 350, and 353. For polymorphisms at locus 356, 70 isolates (89.74%) carried the wild-type allele, five isolates (6.41%) carried the mutant allele, and three isolates (3.84%) were mixed.
Fig. 1.
Sequence profile of PCR products with new mutation site in pfmdr1 and pfcrt genes. a The new mutation site detected in pfmdr1 gene; b The mutation site found in pfcrt gene
For the pfmdr1 gene, 100% of samples were obtained from the PCR products and sequenced successfully (Table 1). Sequencing data illustrated that the main epidemic mutation sites of pfmdr1 were concentrated at 86Y and 184F, with 4.72% and 47.17% mutations, respectively (Table 1). These mutations were also mainly focused in West Africa, followed by Central Africa and South Africa. Additionally, six haplotypes were detected in pfmdr1 gene (Additional file 4: Table S3), including NY (wild type), NF and YF (mutant type), NY/F, YY/F, and N/Y Y/F (mixed type), accounting for 34.91, 43.40, 3.77, 15.09, 0.94, and 1.89%, respectively. In addition, one nonsynonymous mutation at position 199 and several synonymous mutations at positions 87 and 102 were detected in these samples (Table 2; Fig. 1b).
Table 2.
Novel polymorphisms of pfmdr1 and pfcrt in Plasmodium falciparum isolates
| Gene | Referencea | Mutationb | Number of isolates | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Codon position | AAc | Codon | AAc | Codon | Base position | PCR positive | Sequencing | Mutation | Prevalence (%, 95% CI) | |
| pfmdr1 | 87 | L | tta | L | ttG | 261 | 106 | 106 | 1 | 0.94(− 0.009 to 0.028) |
| 102 | G | ggt | G | ggC | 306 | 2 | 1.89(− 0.007 to 0.045) | |||
| 102 | G | ggt | G | ggC/T | 306 | 1 | 0.94(− 0.009 to 0.028) | |||
| 199 | T | act | S | Tct | 595 | 1 | 0.94(− 0.009 to 0.028) | |||
| pfcrt | 136 | Y | tat | N | Aat | 332 | 90 | 90 | 2 | 2.22(− 0.009 to 0.053) |
| 140 | Q | caa | K | cCa | 344 | 1 | 1.11(− 0.011 to 0.033) | |||
| 140 | Q | caa | Q/K | cA/Ca | 344 | 1 | 1.11(− 0.011 to 0.033) | |||
aReference site is in bold type with lowercase
bMutation site is in bold type with capital letter
cAA amino acid residue
With these polymorphisms in the pfcrt and pfmdr1 genes, a total of six pfcrt/pfmdr1 combined haplotypes were assessed, namely, NF-CVIET, NF-CVMNK, NY-CVIET, NY-CVMNK, YF-CVIET, and YF-CVMNK, accounting for 7.23% (6/83), 43.37% (36/83), 3.61% (3/83), 40.96% (34/83), 1.20% (1/83), and 3.61% (3/83), respectively (Additional file 5: Table S4). These haplotypes were mainly concentrated in West Africa and Central Africa. Among them, Congo and Nigeria accounted for 21.69% (18/83) and 16.87% (14/83), respectively (Additional file 3: Table S2).
Analysis of mutation in pfk13 gene
For the pfk13, 93.40% (99/106) samples were amplified, and 92.93% (92/106) of the samples were sequenced successfully. Sequencing analysis showed that these isolates were all wild-type. The reported mutations in SEA at loci 474, 476, 493, 508, 527, 533, 537, 539, 543, 553, 568, 574, 578, and 580 were not detected in the current study. Furthermore, the previously detected mutations at positions 550, 561, 575, 579, and 589 in Africa were also not found.
Discussion
For the past several decades, the emergence and rapid transmission of P. falciparum ADR parasites has become a major cause of malaria burden globally [24]. In China, the continuous influx of imported malaria increases the possibility of malaria respreading [25]. The malaria-endemic area, including Africa and SEA, was the primary source of imported malaria in China including in Wuhan [16]. Thus, continuous surveillance of imported malaria and ADR profiles is essential for malaria eradication in the non-malarial regions, particularly Wuhan, China.
The mutation of 76 T in pfcrt was related to CQR [17]. For pfcrt, CVIET and SVMNT were the dominant mutant haplotypes. In Africa, mutant haplotype CVIET occurs more frequently [26]. CVIET (9.62%) was the most common mutant haplotype in the current study and was mainly distributed in West Africa (5.77%). SVMNT is mainly detected in South America and SEA and is rarely found in Africa [27]. The presence of SVMNT was not found in this survey. However, SVMNT was observed in Tanzania and Angola [28, 29]. In these regions, AQ was considered as the driving factor for haplotype selection of S VMNT [28, 29]. The CQ treatment resulted in high failure rates in southern Cameroon between 1999 and 2004 [30]. However, after an interval of 9 years, the frequency of CVMNK in southeastern Cameroon nearly doubled; Conversely, the CVIET decreased significantly [31]. Drug pressure caused by CQ declined during the period as a result of the cessation of drug imports to these countries. In the present study, haplotypes of CVMNK, CVIET, CV M/I N/E K/T with proportions of 87.50, 9.62, and 2.88% were observed during 2017–2019, respectively. Compared with the previous study [16], all current observed data indicate that the wild-type haplotype is increased and haplotypes of the mutation type and mixed type are decreased. In recent years, the frequency of CVMNK has increased in several regions of Africa [32, 33], which is consistent with this survey results. In the present data, CVMNK is mainly concentrated in West Africa (36.54%) and Central Africa (28.85%), especially in the Congo (21.15%) and Nigeria (15.38%). After CQ was discontinued in most countries in sub-Saharan Africa in the 1990s, the investigated isolates regained all or part of their sensitivity to anti-malarial drugs [34, 35]. It will offer the possibility for these areas to reintroduce CQ in the future for malaria control. Therefore, continuous monitoring of pfcrt to evaluate CQ resistance dynamics in a certain area is urgent.
DHA/PPQ is one of the ACT, effective against simple malaria. Thus, the effect of PPQ cannot be ignored. However, long-term use of anti-malarial drugs particularly PPQ induced ADR [14]. Previous studies indicated that several mutations of pfcrt (93S, 97Y, 101F, 145I, 343L, 353 V, and 356 T) were related to reducing parasite sensitivity to PPQ [13, 14, 36–39]. In this study, no mutations were detected at loci 93, 97, 101, 145, 343, 350, and 353. In an investigation of African isolates, consistent with the results of this study, no mutations at these sites were reported [13]. In this study, 5 isolates carried the mutant allele, and 3 isolates were mixed type at loci 356. In 2011–2012, the 356 T in Gambia and Congo were 78.7 and 36.5%, respectively [40]. The 356 T mutation was found in 54.7% of P. falciparum detected in Africa in 2017–2018. However, they also reported that the 356 T mutation was not associated with in vitro reduced susceptibility to PPQ [13]. Therefore, continuous observations of pfcrt mutations and susceptibility tests in vitro related to PPQ are necessary.
The pfmdr1 gene has been reported to be involved in regulating drug sensitivity or tolerance to several anti-malarial drugs, such as CQ, MQ, quinine (QN), artemether-lumefantrine (AL), and even ART [41]. The pfmdr1 gene 86Y mutation is a potential marker for CQR, while 184F may play a role in resistance to multiple anti-malarial drugs [41]. The previously reported 86Y and 184F mutations in pfmdr1 are most prevalent in Asia and Africa [42]. The frequencies of 86Y (4.72%) and 184F (47.17%) were monitored in this study, of which 184F was more prevalent. The results were similar to previous results in Nigeria and Senegal [43, 44]. In addition, compared with this previous survey in 2011–2016 [16], allele 86Y was significantly reduced. This is consistent with the results discussed above regarding the sensitivity of pfcrt gene recovery to CQ in recent years. Among the six observed haplotypes in this study, NF (43.40%) and NY (34.91%) were also the most frequent, mainly found in West Africa and Central Africa, especially in Congo and Nigeria, which could be a result of selective pressure by resistance to different drugs. In Nigeria, a previous study showed that NF was closely related to the sensitivity of AL [45]. It may be that the first-line drug CQ is replaced by AL, leading to an increased incidence of NF in these countries.
The pfk13 gene was crucial in the molecular surveillance of ADR for falciparum malaria parasites. To date, more than 200 nonsynonymous mutations of pfk13 have been reported [46]. In SEA and, more recently, South America, a number of these mutations have been associated with delayed parasite clearance following ACT, including mutations at loci 446, 458, 474, 476, 493, 508, 527, 533, 537 543, 553, 568, 574, 580 and so forth [46]. In Africa, a number of nonsynonymous mutations in pfk13 have been identified, including mutations at loci149, 189, 189, 561, 575, 579, 589, 578, 592, 637, 641, 656 and so forth [46, 47]. In this survey, no mutation was found in pfk13. Because these sample size was insufficient, it was not sufficient to say that the African plasmodium isolates were still highly sensitive to ART; it is necessary to carry out relevant tests with a larger sample size in the future. Although there is no mutation in the pfk13 gene to indicate ART resistance, it cannot be ignored that pfk13 is no longer the only biomarker of ART resistance, and there may be other genes as markers of ART resistance [48, 49]. Thus, genetic markers of ADR are urgently required. Previous studies have demonstrated that the new candidates pfubp-1 and pfap2mu are implicated in ART resistance in the P. falciparum [48, 49]. Alarming the high morbidity and mortality rates in Africa and the increased status of ADR in Africa could hamper malaria prevention, control, elimination, and even eradication. Therefore, it is critical to monitor mutations associated with ART resistance globally, especially in Africa, by delaying parasite clearance.
It is worth noting that several shortcomings of the current study cannot be neglected. First, affected by the epidemic of COVID-19, the sample size remains small. Thus, valuable information for the molecular surveillance of ADR is limited. Second, 17 samples failed SNPs analysis of pfk13 because of the failure of amplification and sequencing. In a further study, advanced gene-editing tools, particularly the CRISPR/Cas9 technique, should be considered using P. falciparum drug resistance genes [50, 51]. The CRISPR/Cas9 technique can rapidly locate the key sites related to ADR in targeted genes. Compared to natural mutation under long-term drug pressure, artificially introduced mutation by CRISPR/Cas9 can effectively shorten the process of discovering drug resistance sites. CRISPR/Cas9 will offer a useful measure for the discovery of novel mutations in drug resistance genes.
Conclusions
Overall, this study analysed the frequency and spatial distribution of mutations associated with ADR in the pfcrt, pfmdr1, and pfk13 genes from imported P. falciparum isolates in Wuhan, China. The wild-type pfcrt allele and pfmdr1 N86 were predominant in this study. These phenomena indicate that the cessation of CQ, AQ and MQ for a period of time may lead to the restoration of CQ, AQ and MQ sensitive parasites (at least partially). Moreover, it these drugs can continue to be effective for P. falciparum malaria case treatment in Africa. However, the increase in N86 and 184F mutations suggests a potential risk of drug pressure in AL. For new alleles with reduced sensitivity of pfcrt to PPQ, the expansion of these mutations further demonstrates their more significant survival advantage under strong and sustained PPQ pressure. If the development continues, this will lead to the failure of the first-line regimen DHA-PPQ. Thus, constant and careful worldwide surveillance for PPQ resistance is urgent. Although no mutation is detected in pfk13, caution should be made regarding ART therapy for P. falciparum in Africa, and continuous molecular surveillance is still urgently necessary.
Supplementary Information
Additional file 1: Fig. S1. Positive controls. A. pfcrt CVIET plasmid running in agarose gel electrophoresis and product sequence analysis; B. pfmdr1 NY plasmid with mutations at 86 and 184 sites running in agarose gel electrophoresis and product sequence analysis. C. Sequencing profile of pfk13 plasmid (JPG 3459 KB)
Additional file 2: Table S1. Reaction system and conditions for amplification of targeted fragments of pfmdr1 and pfcrt genes.
Additional file 3: Table S2. Haplotypes distribution of pfcrt and pfmdr1 during 2017-2019.
Additional file 4: Table S3. Haplotypes distribution of pfcrt and pfmdr1 from different areas.
Additional file 5: Table S4. Overall frequency of combined haplotypes in pfcrt and pfmdr1.
Acknowledgements
The authors would like to thank the Department of Schistosomiasis and Endemic Diseases, Wuhan Center for Disease Prevention and Control, and all participants who have contributed their blood samples.
Authors’ contributions
WJC and JL conceived and designed the study. KW coordinated the field collections of patient isolates. KW carried out a microscopic examination. WJC and XNS performed the experiments. WJC, HBT, and JL analysed the data. WJC and JL wrote the paper. All the authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Number 81802046) and Principle Investigator Program of Hubei University of Medicine (Grant Number HBMUPI202101), the Foundation for Graduate Science and Technology Innovation Project of Hubei University of Medicine (Grant Number YC2020003).
Availability of data and materials
The datasets analysed in this study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The current study was approved by the ethics committees of the Hubei University of Medicine and Wuhan City Center for Disease Prevention and Control Ethics Committee. Informed consent was obtained from all participated individuals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. World malaria report 2020. Geneva, World Health Organization, 2020.
- 2.Feng J, Zhang L, Huang F, Yin JH, Tu H, Xia ZG, et al. Ready for malaria elimination: zero indigenous case reported in the People's Republic of China. Malar J. 2018;17:315. doi: 10.1186/s12936-018-2444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng J, Xiao H, Zhang L, Yan H, Feng X, Fang W, et al. The Plasmodium vivax in China: decreased in local cases but increased imported cases from Southeast Asia and Africa. Sci Rep. 2015;5:8847. doi: 10.1038/srep08847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antony HA, Parija SC. Antimalarial drug resistance: an overview. Trop Parasitol. 2016;6:30–41. doi: 10.4103/2229-5070.175081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achan J, Mwesigwa J, Edwin CP, D'Alessandro U. Malaria medicines to address drug resistance and support malaria elimination efforts. Expert Rev Clin Pharmacol. 2018;11:61–70. doi: 10.1080/17512433.2018.1387773. [DOI] [PubMed] [Google Scholar]
- 6.Payne D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol Today. 1987;3:241–246. doi: 10.1016/0169-4758(87)90147-5. [DOI] [PubMed] [Google Scholar]
- 7.WHO. Antimalarial drug combination therapy. Geneva, World Health Organization, 2001.
- 8.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawkes M, Conroy AL, Kain KC. Spread of artemisinin resistance in malaria. N Engl J Med. 2014;371:1944–1945. doi: 10.1056/NEJMc1410735. [DOI] [PubMed] [Google Scholar]
- 11.Leang R, Taylor WR, Bouth DM, Song L, Tarning J, Char MC, et al. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in Western Cambodia: dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob Agents Chemother. 2015;59:4719–4726. doi: 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, et al. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis. 2016;16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foguim FT, Bogreau H, Gendrot M, Mosnier J, Fonta I, Benoit N, et al. Prevalence of mutations in the Plasmodium falciparum chloroquine resistance transporter, PfCRT, and association with ex vivo susceptibility to common anti-malarial drugs against African Plasmodium falciparum isolates. Malar J. 2020;19:201. doi: 10.1186/s12936-020-03281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton WL, Amato R, van der Pluijm RW, Jacob CG, Quang HH, Thuy-Nhien NT, et al. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect Dis. 2019;19:943–951. doi: 10.1016/S1473-3099(19)30392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnington CA, Phyo AP, Ashley EA, Imwong M, Sriprawat K, Parker DM, et al. Plasmodium falciparum Kelch 13 mutations and treatment response in patients in Hpa-Pun District, Northern Kayin State. Myanmar Malar J. 2017;16:480. doi: 10.1186/s12936-017-2128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao Y, Wu K, Xu M, Yang Y, Zhang Y, Yang W, et al. Surveillance of genetic variations associated with antimalarial resistance of Plasmodium falciparum isolates from returned migrant workers in Wuhan. Central China Antimicrob Agents Chemother. 2018;62:e02387–e2417. doi: 10.1128/AAC.02387-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 18.Djimde A, Doumbo OK, Steketee RW, Plowe CV. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet. 2001;358:890–891. doi: 10.1016/S0140-6736(01)06040-8. [DOI] [PubMed] [Google Scholar]
- 19.Cooper RA, Ferdig MT, Su XZ, Ursos LM, Mu J, Nomura T, et al. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol Pharmacol. 2002;61:35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- 20.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 21.Vinayak S, Alam MT, Sem R, Shah NK, Susanti AI, Lim P, et al. Multiple genetic backgrounds of the amplified Plasmodium falciparum multidrug resistance (pfmdr1) gene and selective sweep of 184F mutation in Cambodia. J Infect Dis. 2010;201:1551–1560. doi: 10.1086/651949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Chen J, Xie D, Eyi UM, Matesa RA, Obono MMO, et al. Molecular mutation profile of Pfcrt and Pfmdr1 in Plasmodium falciparum isolates from Bioko Island. Equatorial Guinea Infect Genet Evol. 2015;36:552–556. doi: 10.1016/j.meegid.2015.08.039. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Chen J, Xie D, Eyi UM, Matesa RA, Ondo Obono MM, et al. Limited artemisinin resistance-associated polymorphisms in Plasmodium falciparum K13-propeller and PfATPase6 gene isolated from Bioko Island, Equatorial Guinea. Int J Parasitol Drugs Drug Resist. 2016;6:54–59. doi: 10.1016/j.ijpddr.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis RE, Alonso P, Aponte J, Aregawi M, Barrette A, Bergeron L, et al. Malaria: global progress 2000–2015 and future challenges. Infect Dis Poverty. 2016;5:61. doi: 10.1186/s40249-016-0151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou S, Li Z, Cotter C, Zheng C, Zhang Q, Li H, et al. Trends of imported malaria in China 2010–2014: analysis of surveillance data. Malar J. 2016;15:39. doi: 10.1186/s12936-016-1093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Awasthi G, Satya Prasad GB, Das A. Pfcrt haplotypes and the evolutionary history of chloroquine-resistant Plasmodium falciparum. Mem Inst Oswaldo Cruz. 2012;107:129–134. doi: 10.1590/s0074-02762012000100018. [DOI] [PubMed] [Google Scholar]
- 27.Awasthi G, Das A. Genetics of chloroquine-resistant malaria: a haplotypic view. Mem Inst Oswaldo Cruz. 2013;108:947–961. doi: 10.1590/0074-0276130274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alifrangis M, Dalgaard MB, Lusingu JP, Vestergaard LS, Staalsoe T, Jensen AT, et al. Occurrence of the Southeast Asian/South American SVMNT haplotype of the chloroquine-resistance transporter gene in Plasmodium falciparum in Tanzania. J Infect Dis. 2006;193:1738–1741. doi: 10.1086/504269. [DOI] [PubMed] [Google Scholar]
- 29.Gama BE, Pereira-Carvalho GA, Lutucuta Kosi FJ, Almeida de Oliveira NK, Fortes F, et al. Plasmodium falciparum isolates from Angola show the StctVMNT haplotype in the pfcrt gene. Malar J. 2010;9:174. [DOI] [PMC free article] [PubMed]
- 30.Basco LK, Ngane VF, Ndounga M, Same-Ekobo A, Youmba JC, Abodo RT, et al. Molecular epidemiology of malaria in Cameroon. XXI. Baseline therapeutic efficacy of chloroquine, amodiaquine, and sulfadoxine-pyrimethamine monotherapies in children before national drug policy change. Am J Trop Med Hyg. 2006;75:388–95. [PubMed]
- 31.Ndam NT, Basco LK, Ngane VF, Ayouba A, Ngolle EM, Deloron P, et al. Reemergence of chloroquine-sensitive pfcrt K76 Plasmodium falciparum genotype in southeastern Cameroon. Malar J. 2017;16:130. doi: 10.1186/s12936-017-1783-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galatas B, Nhamussua L, Candrinho B, Mabote L, Cistero P, Gupta H, et al. In-Vivo Efficacy of chloroquine to clear asymptomatic infections in Mozambican adults: a randomized, placebo-controlled trial with implications for elimination strategies. Sci Rep. 2017;7:1356. doi: 10.1038/s41598-017-01365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Ruan W, Zhou S, Huang F, Lu Q, Feng X, et al. Molecular surveillance of pfcrt and k13 propeller polymorphisms of imported Plasmodium falciparum cases to Zhejiang Province, China between 2016 and 2018. Malar J. 2020;19:59. doi: 10.1186/s12936-020-3140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamya MR, Bakyaita NN, Talisuna AO, Were WM, Staedke SG. Increasing antimalarial drug resistance in Uganda and revision of the national drug policy. Trop Med Int Health. 2002;7:1031–1041. doi: 10.1046/j.1365-3156.2002.00974.x. [DOI] [PubMed] [Google Scholar]
- 35.Eriksen J, Nsimba SE, Minzi OM, Sanga AJ, Petzold M, Gustafsson LL, et al. Adoption of the new antimalarial drug policy in Tanzania–a cross-sectional study in the community. Trop Med Int Health. 2005;10:1038–1146. doi: 10.1111/j.1365-3156.2005.01486.x. [DOI] [PubMed] [Google Scholar]
- 36.Eastman RT, Dharia NV, Winzeler EA, Fidock DA. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob Agents Chemother. 2011;55:3908–3916. doi: 10.1128/AAC.01793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhingra SK, Redhi D, Combrinck JM, Yeo T, Okombo J, Henrich PP, et al. A variant PfCRT isoform can contribute to Plasmodium falciparum resistance to the first-line partner drug piperaquine. mBio. 2017;8:e00303–17. [DOI] [PMC free article] [PubMed]
- 38.Gupta H, Galatas B, Chidimatembue A, Huijben S, Cistero P, Matambisso G, et al. Effect of mass dihydroartemisinin-piperaquine administration in southern Mozambique on the carriage of molecular markers of antimalarial resistance. PLoS ONE. 2020;15:e0240174. doi: 10.1371/journal.pone.0240174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Pluijm RW, Imwong M, Chau NH, Hoa NT, Thuy-Nhien NT, Thanh NV, et al. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis. 2019;19:952–961. doi: 10.1016/S1473-3099(19)30391-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhingra SK, Gabryszewski SJ, Small-Saunders JL, Yeo T, Henrich PP, Mok S, et al. Global spread of mutant PfCRT and its pleiotropic impact on Plasmodium falciparum multidrug resistance and fitness. mBio. 2019;10:e02731–18. [DOI] [PMC free article] [PubMed]
- 41.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnadig N, Uhlemann AC, et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun. 2016;7:11553. doi: 10.1038/ncomms11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wurtz N, Fall B, Pascual A, Fall M, Baret E, Camara C, et al. Role of Pfmdr1 in in vitro Plasmodium falciparum susceptibility to chloroquine, quinine, monodesethylamodiaquine, mefloquine, lumefantrine, and dihydroartemisinin. Antimicrob Agents Chemother. 2014;58:7032–7040. doi: 10.1128/AAC.03494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Idowu AO, Oyibo WA, Bhattacharyya S, Khubbar M, Mendie UE, Bumah VV, et al. Rare mutations in pfmdr1 gene of Plasmodium falciparum detected in clinical isolates from patients treated with anti-malarial drug in Nigeria. Malar J. 2019;18:319. doi: 10.1186/s12936-019-2947-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Happi CT, Gbotosho GO, Folarin OA, Sowunmi A, Hudson T, O'Neil M, et al. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob Agents Chemother. 2009;53:888–895. doi: 10.1128/AAC.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.WHO. Artemisinin resistance and artemisinin-based combination therapy efficacy. Geneva, World Health Organization; 2018.
- 47.Gupta H, Macete E, Bulo H, Salvador C, Warsame M, Carvalho E, et al. Drug-resistant polymorphisms and copy numbers in Plasmodium falciparum, Mozambique, 2015. Emerg Infect Dis. 2018;24:40–48. doi: 10.3201/eid2401.170864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henriques G, Hallett RL, Beshir KB, Gadalla NB, Johnson RE, Burrow R, et al. Directional selection at the pfmdr1, pfcrt, pfubp1, and pfap2mu loci of Plasmodium falciparum in Kenyan children treated with ACT. J Infect Dis. 2014;210:2001–2008. doi: 10.1093/infdis/jiu358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Golassa L, Kamugisha E, Ishengoma DS, Baraka V, Shayo A, Baliraine FN, et al. Identification of large variation in pfcrt, pfmdr-1 and pfubp-1 markers in Plasmodium falciparum isolates from Ethiopia and Tanzania. Malar J. 2015;14:264. doi: 10.1186/s12936-015-0783-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee MCS, Lindner SE, Lopez-Rubio JJ, Llinas M. Cutting back malaria: CRISPR/Cas9 genome editing of Plasmodium. Brief Funct Genomics. 2019;18:281–289. doi: 10.1093/bfgp/elz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kudyba HM, Cobb DW, Florentin A, Krakowiak M, Muralidharan V. CRISPR/Cas9 gene editing to make conditional mutants of human malaria parasite P. falciparum. J Vis Exp. 2018;139:57747. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Positive controls. A. pfcrt CVIET plasmid running in agarose gel electrophoresis and product sequence analysis; B. pfmdr1 NY plasmid with mutations at 86 and 184 sites running in agarose gel electrophoresis and product sequence analysis. C. Sequencing profile of pfk13 plasmid (JPG 3459 KB)
Additional file 2: Table S1. Reaction system and conditions for amplification of targeted fragments of pfmdr1 and pfcrt genes.
Additional file 3: Table S2. Haplotypes distribution of pfcrt and pfmdr1 during 2017-2019.
Additional file 4: Table S3. Haplotypes distribution of pfcrt and pfmdr1 from different areas.
Additional file 5: Table S4. Overall frequency of combined haplotypes in pfcrt and pfmdr1.
Data Availability Statement
The datasets analysed in this study are available from the corresponding author on reasonable request.