Abstract
Aging is a critical factor affecting physical health and disease in mammals. Emerging evidence indicates that aging may affect the gut bacteriome in cynomolgus macaques, but little is known about whether or how the gut virome changes with age. Here, we compared the DNA gut viral composition of 16 female cynomolgus monkeys (Macaca fascicularis) at three life stages (young, adult, and old) using the shotgun metagenome sequencing method. We found that the DNA gut virome from these monkeys differed substantially among the three groups. The gut viruses were dominated by bacteriophages, the most abundant of which was the Caudovirales order (i.e., Siphoviridae, Myoviridae, and Podoviridae families). Additionally, the co‐occurrence analysis revealed that the age‐related bacteriophages were correlated in an extensive and complex manner with the main intestinal bacteria (i.e., Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria phyla). Furthermore, the age‐related DNA gut viral functions were enriched for genetic information processing, nucleotide, and folate metabolism. Our gut virome analysis provides new insight into how aging influences the gut virome of non‐human primates.
Keywords: aging, bacteriophages, cynomolgus macaques, DNA gut virome, shotgun metagenome sequencing
We characterized changes of the DNA gut virome associated with age in female crab‐eating macaques. The structural and functional profiles of the gut DNA virome differed significantly among the three age groups. The gut virome was dominated by dsDNA phages, especially the Caudovirales order, which showed strong correlations with the highly prevalent gut bacteria. Our findings suggest that modulating the intestinal virome may be beneficial for the treatment of aging.

1. INTRODUCTION
Aging often negatively impacts the vitality and health of human beings. For example, previous studies have shown that aging is implicated in the development of atherosclerosis (Wang & Bennett, 2012), Alzheimer's disease (Qiu et al., 2009), and innate immune dysregulation (Shaw et al., 2013). Accompanied by a broad influence on the general physiology of the gastrointestinal tract, the aging process also inevitably affects gut microbes in human beings (Biagi et al., 2010) or non‐human primates (Duan et al., 2019). The gut microbiome is composed of bacteria, eukaryotic viruses, bacterial viruses (bacteriophages), fungi, and archaea. Numerous studies have reported that gut microbial homeostasis plays an essential part in maintaining human health (Gentile & Weir, 2018; Lynch & Pedersen, 2016). Disturbance of gut microbiota is also implicated in a wide range of human diseases such as cancer (Flemer et al., 2017; Matson et al., 2018), obesity (Sharma et al., 2018), and neuropsychiatric conditions (Sampson et al., 2016; Sharon et al., 2019). Some studies have focused on how aging modulates the gut bacteriome. Our previous research, for example, found that the gut bacteriome changed significantly with age, as demonstrated by enriched Veillonellaceae, Coriobacteriaceae, and Succinivibrionaceae, and depleted Ruminococcaceae and Rikenellaceae in old monkeys relative to their young counterparts (Duan et al., 2019). However, it remains unknown whether and how aging shapes the gut virome.
Recently, some studies have shown that the gut virome plays a crucial role in maintaining human health and that chronic viral infections can confer symbiotic protection from bacterial infection (Barton et al., 2007), suggesting that not all viruses are harmful to their hosts (Virgin, 2014). However, investigations on the human gut virome are still in their infancy, and a great host of human viruses await identification. To date, two studies have characterized the human intestinal virome and bacteriome in infant and adult monozygotic twins. Lim et al. (Lim et al., 2015) investigated the dynamic changes occurring in the gut virome and bacteriome at six time points from birth to 2 years of age in infant monozygotic twins. Moreno‐Gallego et al. (2019) explored the interactions occurring between the gut virome and bacteriome in adult monozygotic twins. Both studies highlight the dynamic nature of the human gut virome and the association between gut viruses and microbes in infant and adult monozygotic twins during a relatively short period, but information on the human gut virome from youth to old age is eagerly awaited. The human gut virome is complex, and its composition is relatively unstable because of the numerous confounding factors that can affect it (e.g., diet, antibiotics use, and environmental and geographical factors; Górska et al., 2018; Minot et al., 2011; Rampelli et al., 2017). Instead, as non‐human primates, cynomolgus macaques are not only closely related to human beings in genetics but unlike humans, they are more likely to keep their diet, nutritional status, health condition, and geographical location consistent.
To elucidate the impact of aging on the gut virome in non‐human primates, we investigated the composition and function of the gut virome from five young (2–4 years), six adult (5–15 years), and five old (17–20 years) female cynomolgus macaques using shotgun metagenome sequencing. We found that the DNA gut virome in the monkeys was dominated by phages, most of which belonged to the Caudovirales order, including Myoviridae, Siphoviridae, and Podoviridae families. Moreover, a complex and diverse relationship was identified between age‐related bacteriophages and bacteria. Collectively, our findings augment the current understanding of the relationship between the gut virome and aging in non‐human primates.
2. MATERIALS AND METHODS
2.1. Study subject selection and fecal sample collections
All the cynomolgus monkeys (crab‐eating macaques, Macaca fascicularis) used in the experiments were provided by Zhongke Experimental Animal Co., Ltd. The animals lived under the following standard conditions: 22 ± 1°C temperature, 50 ± 5% relative humidity, and 12‐h light/12‐h dark cycle with lights on at 07:00 (Zheng et al., 2020). Stool samples were obtained from 5 young (2–4 years), 6 adult (5–15 years), and 5 old (17–20 years) female cynomolgus macaques (Appt et al., 2010; Drevon‐Gaillot et al., 2006; Wang et al., 2020), all of which had the same living conditions, daily diet and health qualities (Xu et al., 2012). All the stool samples from the selected monkeys were collected with a sterile device without any medium and stored at −80°C until subsequent processing.
2.2. Shotgun metagenome sequencing and taxonomic assignments
The shotgun metagenome sequencing protocol was based on our previous published work (Yang et al., 2020; Zheng et al., 2020). Briefly, microbial DNA was extracted from the fecal samples using the E.Z.N.A® DNA kit (Omega Bio‐Tek) according to the manufacturer's instructions. The concentration and purity of the DNA extracted from the stool samples were qualified and assessed by a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) for the construction of the metagenome shotgun sequencing libraries. Each library was sequenced on the Illumina HiSeq 4000 platform (Illumina Inc.) at Majorbio Bio‐Pharm Technology Co., Ltd. using the HiSeq 3000/4000 PE Cluster Kit and the HiSeq 3000/4000 SBS Kit according to the manufacturer's instructions (www.illumina.com). Host reads were determined and removed according to the sequence alignments with the Bayesian model averaging (Zheng et al., 2020). Low‐quality sequences were discarded using Sickle (https://github.com/najoshi/sickle). The remaining high‐quality sequences were assembled using SOAPaligner (Li et al., 2008) to evaluate the gene abundance in each sample.
To cluster the viral sequences, the representative sequences of a nonredundant gene catalog were aligned against the National Center for Biotechnology Information (NCBI) NR database using an e‐value cutoff of 1e‐5 with BLASTP (Version 2.2.28+) for taxonomic annotations. Kyoto encyclopedia of genes and genomes (KEGG) annotation was conducted using BLASTP (Version 2.2.28+) against the KEGG database (Xie et al., 2011) using an e‐value cutoff of 1e˗5. The unit of gene abundance was unified using reads per kilobase million.
2.3. Metagenomic analysis of fecal samples
Based on the taxonomy annotation for viruses, α‐diversity, a measure of viral community richness (Chao and Ace index) and diversity (Shannon and Invsimpson index) at the species level, was assessed and visualized using the vegan and fossil packages in R, respectively. β‐diversity was assessed based on Abund–Jaccard distance and visualized by principal coordinate analysis (PCoA). ANOSIM (analysis of similarities) was performed to identify differences in β‐diversity among the three age groups. The key viral and bacterial taxa and the KEGG categories responsible for discrimination among the three age groups were identified using linear discriminant analysis effective size, LEfSe (Segata et al., 2011). Only linear discriminant analysis (LDA) values >2.0 and p values <0.05 were considered to represent significantly enriched taxa.
2.4. Statistical analysis
Statistical analyses were carried out using SPSS version 21.0 (SPSS), and plots were generated from R packages (pheatmap, ggplot2), GraphPad Prism (version 8.0), and Cytoscape (version 3.7.2). Continuous variables such as age were analyzed using ANOVA followed by LSD multiple comparison tests. We applied the non‐parametric factorial Kruskal–Wallis test followed by Dunn's multiple comparison tests to compare the three groups in cases of heteroscedasticity or non‐normally distributed variables. Correlations between age‐related bacteriophages and bacteria were tested by Spearman's correlation analysis. Statistical significance was set at <0.05.
3. RESULTS
3.1. Shotgun metagenomes of cynomolgus stool samples in DNA virus domain
We obtained fecal samples from 16 female cynomolgus macaques between the ages of 2 and 20. The detailed characteristics of all the study subjects were listed in our previously published report (Duan et al., 2019). On average, we obtained 101,290,630 ± 7,898,542 (mean ±s.d.) paired reads per sample. After quality control, 100,140,304 ± 7,837,876 paired reads per sample were used to de novo assemble the microbial contigs. We identified 18,773,882 predicted genes from the filtered sequence data, from which a nonredundant gene set containing 9317 genes was assigned to the viral microbiome which comprised only the DNA gut virome and used for subsequent analyses.
3.2. Similar α‐diversity of the gut DNA virome among the three age groups
Initially, a downward trend in α‐diversity was detected for the viral richness (Chao and Ace index) at increased age, but no significant difference was observed (one‐way ANOVA, all p values >0.05; Figure 1a). Furthermore, our pan analysis illustrated that the rate at which viruses accumulated declined with age, suggesting that the downward trend in viral richness was not likely attributable to different sample sizes (Figure 1b). Similarly, we found that the gut viral richness was highest in the young and gradually decreased with age, although no statistical difference was detected at the quantified levels (one‐way ANOVA, p = 0.206; Figure 1c). Intriguingly, viral diversity (Invsimpson and Shannon index) decreased in the adult age‐group versus the young, a finding reversed in the oldest age‐group (one‐way ANOVA, all p values >0.05; Figure 1a).
FIGURE 1.
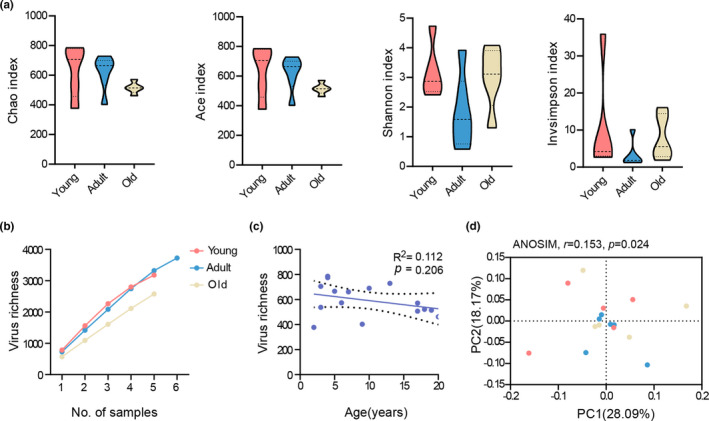
The DNA virome biodiversity within the gut metagenome of cynomolgus macaques in the different age‐groups. (a) α‐diversity analysis showing the lack of statistically significant differences in viral richness (Chao and Ace index) and diversity (Invsimpson and Shannon index) among the three age groups. (b) Rarefaction curves showing the acquisition of viral species richness. (c) Viral species richness at the indicated ages. Linear regression, R 2 value, and 95% confidence intervals were shown. (d) Principal coordinate analysis (PCoA) based on the Abund–Jaccard distances showed a clear separation among the three age groups at the species level (ANOSIM, r = 0.153, p = 0.024). Young, n = 5; adult, n = 6; and old, n = 5
3.3. The global composition of gut DNA virome shifted with age
We compared β‐diversity at the species level using the Abund–Jaccard distance metric. PCoA of the gut DNA virome revealed clear segregation among the young, adult, and old female cynomolgus macaques (ANOSIM, r = 0.153, p = 0.024; Figure 1d). This suggested that aging contributed to the variation in the global phenotypes of gut DNA virome in the macaques.
To determine the shared and distinct intestinal viruses among the three groups, the gut viral composition at different levels was then compared. At the species level, the Venn diagram showed that 647 of 1048 viral species were shared among the three age groups, while 73, 71, and 40 were exclusive to young, adult, and old cohorts, respectively (Figure 2a). At the family level, the gut DNA virome from the macaques mainly comprised seven families, which was dominated by Podoviridae, Siphoviridae, Myoviridae, and unclassified_d_virus families (Figure 2b). Among them, Podoviridae, Siphoviridae, and Myoviridae were assigned to the Caudovirales order, the main phage members. To further quantify age‐related changes in the gut virome, we compared the relative abundance of the aforementioned phages among the three age groups. Consequently, only one family, Myoviridae, was statistically enriched in the young age‐group relative to the adult group (one‐way ANOVA, p = 0.027), but no significant difference was observed between the young or adult group and the old age‐group. The remaining families were not found to differ statistically among the three age groups (one‐way ANOVA, all p values >0.05; Figure 2c; Figure A1).
FIGURE 2.
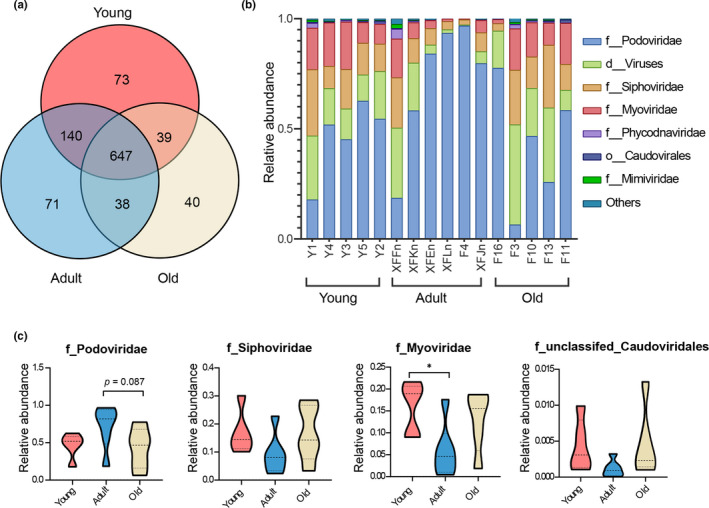
The DNA virome at different levels in the gut microbiome of young, adult, and old female cynomolgus. (a) Venn diagram depicting the viral richness and the overlap in viral communities among young (red), adult (blue), and old (yellow) monkeys at the species level. (b) The community bar plot illustrated that the gut DNA virome from cynomolgus macaques mainly contained seven families. (c) Quantification of the relative abundance of the four families assigned to the Caudovirales order. Only the Myoviridae family was significantly enriched in the young age‐group versus the adult group (p = 0.027), not the old group. p values were determined by one‐way ANOVA followed by LSD’s multiple comparison tests. The following n values represented the number of independent animals used for statistical evaluation: young, n = 5; adult, n = 6; and old, n = 5
3.4. Bacteriophage dominance and age‐related alterations in the gut DNA virome
The LEfSe analysis was conducted to confirm the profiles of the gut DNA virome in the different age‐groups. Altogether, we identified 45 differentially represented viral species responsible for this discrimination (Figure 3a; Table A1). These different species were similarly dominated by phages, most of which were assigned to the Caudovirales order, which included Myoviridae (19 species, 42.22%), Siphoviridae (14 species, 31.11%), and Podoviridae (6 species, 13.33%) families (Figure 3b). Furthermore, the relative abundance of the families (Myoviridae r = −0.662, p = 0.005; Siphoviridae r = −0.562, p = 0.024; and Podoviridae r = −0.535, p = 0.033) decreased significantly with age (Figure 3c).
FIGURE 3.
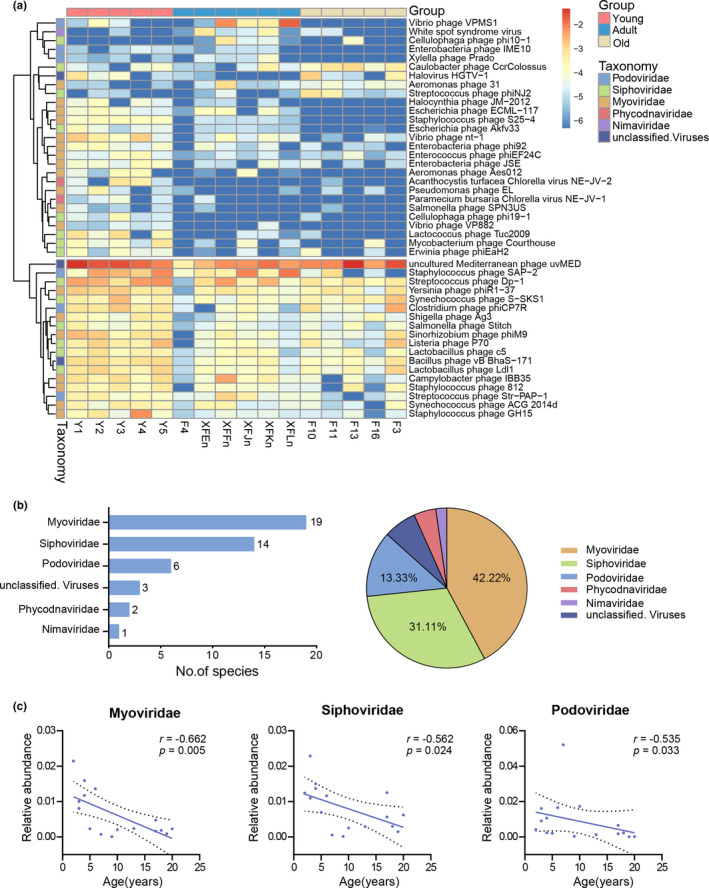
The differentially represented viral species among the three groups. (a) Heatmap of the 45 discriminative species’ abundances among the young, adult, and old age‐groups (LDA > 2.0). Species (raw) were sorted at the family level, and samples (column) were sorted by age. Color intensity (blue to yellow) indicated the score‐normalized abundance for each species. (b) The absolute and relative proportion of discriminative species in different families. The intestinal viral species from the monkeys were dominated by phages, the overwhelming majority of which were from the Caudovirales order, which comprised Myoviridae (42.22%), Siphoviridae (31.11%), and Podoviridae (13.33%) families. (c) Scatter diagrams showing the relative abundances of the age‐related Myoviridae, Siphoviridae, and Podoviridae families, as determined by Spearman's correlation analysis
3.5. Functional profiles of the age‐related gut DNA virome based on KEGG pathway analysis
To outline the functional profiles of the gut DNA virome, we annotated the nonredundant gene set from the gut virome using the KEGG database. In the first‐level KEGG pathways, the intestinal viral functions were mainly enriched in genetic information processing and metabolism (Figure 4a). To further explore the functional profiles of the age‐related gut viruses, we performed a PCoA on the third‐level KEGG pathways using the Abund–Jaccard distance metric. The ANOSIM results indicated that the KEGG pathways were separated among the three age groups (r = 0.369, p = 0.001; Figure 4b). Using LEfSe analysis, we identified five differential pathways that were mainly involved in pyrimidine metabolism, DNA replication, and folate metabolism among the three age groups (LDA >2.0; Figure 4c; Table A2). Importantly, the broader conclusions were not driven by outliers, which was supported even following the removal of outliers.
FIGURE 4.
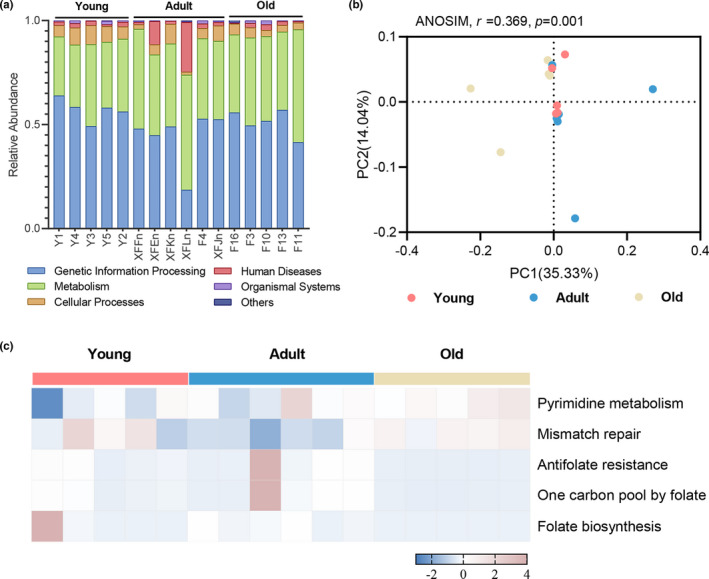
The functional profiles of age‐related gut DNA virome in the KEGG pathway. (a) Relative abundances of the first‐level KEGG pathways in the gut DNA virome. (b) PCoA based on the Abund–Jaccard distance showed clear discrimination among the three groups in the third‐level KEGG pathways. (c) Heatmap of the relative abundances of the five differential pathways among the young, adult, and old age‐groups (LDA > 2.0). Functional profiles in the DNA virome were enriched in pyrimidine metabolism, DNA replication, and folate metabolism
3.6. The relationship between age‐related bacteriophages and bacteria
We sought to determine whether the changes we detected in the bacteriophage sequences from the monkey feces at different ages were related to disturbances in the gut microbes. Interestingly, bacteriophage richness was positively correlated with bacterial richness (Spearman's correlation, r = 0.779, p < 0.001; Figure A2a). In contrast, previous research has shown that phage richness is negatively correlated with bacterial diversity in infant monozygotic twins (Moreno‐Gallego et al., 2019). Likewise, we found that bacteriophage and bacterial diversity were positively related in an age‐dependent manner, although no statistical difference was found (Spearman's correlation, r = 0.147, p = 0.587; Figure A2b). Thus, the intestinal viruses and microbes from cynomolgus monkeys synchronously changed with age.
To further investigate possible correlations between age‐related gut phages and bacteria in the healthy monkeys, a co‐occurrence network analysis was conducted. We observed that age‐related bacteriophages were broadly related to symbiotic bacteria (Figure 5a), with 64.10% (25/39 viral species) of the altered phages showing strong correlations with multiple gut bacteria (related to more than one bacterium r > ±0.7, p < 0.001). Likewise, these phages were mainly assigned to Myoviridae, Siphoviridae, and Podoviridae families. The age‐related intestinal bacteriophage members also showed clear correlations with multiple symbiotic bacteria, which were mainly assigned as highly prevalent gut bacteria phyla, including Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria phyla (Figure 5b; Table A3). We next identified the hosts of these phages in the NCBI database and, interestingly, most of the hosts were Firmicutes or Proteobacteria members, but these bacterial hosts and the age‐related gut bacteria were inconsistent at the species level (Table A4).
FIGURE 5.
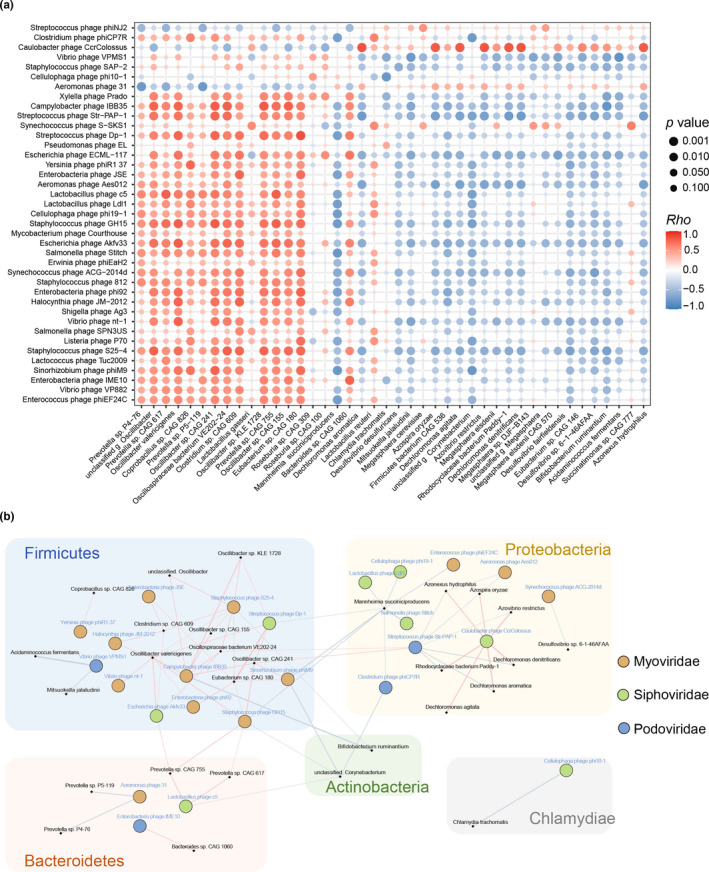
Correlations between gut bacteriophages and bacteria. (a) Association between 39 different bacteriophages and 42 discriminative bacteria identified by LEfSe analysis of the metagenomic sequences (LDA > 2.0). The size and color of each scatter plot point showed the p values of Spearman's correlation (ranging from 9.87e–06 to 0.99) and correlation coefficient values (ranging from −0.867 to 0.874) between the different bacteriophages and bacteria, respectively. (b) Co‐occurrence network deduced from the relationship between the age‐related bacteriophages and bacteria. Circles and dots represented the different bacteriophages and bacteria, respectively. Circle colors varied according to the viral family. Bacterial species that were annotated at the phylum level were marked. Lines between nodes indicated Spearman's negative (light blue) or positive (light red) correlation, and line thickness indicated the p‐value (p < 0.001)
4. DISCUSSION
We have previously reported that the gut bacteriome in cynomolgus macaques changes with age (Duan et al., 2019). Recently, some studies have reported that a variety of gut viruses, especially tailed bacteriophages, are present in the intestines of healthy mammals (Lawrence et al., 2019). Furthermore, the gut virome is believed to affect mammalian health, but whether dynamic changes occur in them during aging remains unknown. Here, we compared the structure and function of the gut DNA virome in 16 female monkeys at three representative life stages (young, adult, and old), and found that the gut DNA virome dramatically changed with age. The most abundant gut viruses, which were bacteriophages within the Caudovirales order, included Siphoviridae, Myoviridae, and Podoviridae families, a finding consistent with previous reports (Norman et al., 2015; Shkoporov & Hill, 2019). Moreover, there was a broad and strong interaction between the age‐related gut bacteriophages and bacteria, the latter of which were highly prevalent gut microbes such as Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria phyla. Overall, our findings augment the current understanding of the effect of aging on the structure and function of the gut virome in non‐human primates.
The gut virome in humans is personalized and stable and it is dominated by phages (Shkoporov et al., 2019). Previous studies have reported that bacteriophages from the human intestinal mucosa or stools mainly contain double‐stranded (ds) DNA and single‐stranded (ss) DNA phages. dsDNA phages generally comprise Siphoviridae, Podoviridae, and Myoviridae families, whereas ssDNA phages are mostly Microviridae family members (Shkoporov & Hill, 2019 ). Here, we found that the gut DNA virome in healthy female monkeys was dominated by Siphoviridae, Myoviridae, and Podoviridae families, which was partly consistent with previous studies, underscoring the notion that virus–host interactions cannot be solely considered as pathogenic. Nonetheless, the fact that we were unable to identify ssDNA bacteriophages in the gut DNA virome was probably accounted for low viral loads and inadequate nucleic acid extraction procedures. For instance, it is known that the ssDNA genome is unstable and easily degraded (Minot et al., 2013), and Microviridae virions form circular DNA packaged in icosahedral capsids.
To date, two major bacteriome–phageome dynamics have been detected by researchers that elucidate some aspects of the dynamic changes that occur in the phage community and phage–bacteria coevolution. Among them, one is the “piggyback‐the‐winner” model (Shkoporov & Hill, 2019; Silveira & Rohwer, 2016), whereby intestinal phages are dominated by lysogenic phages, resulting in a stable virus to microbe ratio; the other is the “kill‐the‐winner” model (Beller & Matthijnssens, 2019; Rodriguez‐Valera et al., 2009), in which virulent phages replicate and proliferate within host bacteria, leading to decreased bacterial loads by lysing their hosts. Our results showed that bacteriophage richness was positively correlated with bacterial richness, which was consistent with the former model, suggesting that most intestinal bacteriophages in our study might retain lysogenic or lysogeny‐like interactions with their hosts (Mills et al., 2013). Despite their high metabolic consumption requirements, lysogenic phages can beneficially alter the physiological characteristics of host bacteria (Brussow et al., 2004). Therefore, intestinal phages do more than just act as predators; in some cases, they help to maintain gut microbial homeostasis by integrating genes that carry competitive advantages into the symbiotic genome of the host (Mills et al., 2013; Modi et al., 2013).
Herein, age‐related bacteriophages formed a broad and strong co‐occurring relationship with highly prevalent gut microbes from the Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria phyla. Consistent with this, previous studies have found that members of the Siphoviridae family are capable of infecting some intestinal microbes such as Bacteroides and Clostridium in humans (Gilbert et al., 2017; Gómez‐Doñate et al., 2011; Ramírez‐Vargas et al., 2018). A recent study also reported that Myoviridae viral particle types can be induced and identified from human intestinal Bifidobacterium (Mavrich et al., 2018). This inferred that the abundance of phages in a given environment may reflect the abundance of their symbiotic host bacteria (Breitbart & Rohwer, 2005).
Unlike the gut bacteriome, which was enriched in genes associated with arginine biosynthesis, purine metabolism, microbial polysaccharide metabolism, and gut viral functions were enriched for genetic information processing and nucleotide and folate metabolism. Intestinal bacteriophages are capable of modulating the functions of prokaryotic communities by the integration into prokaryotic genomes. For example, gene integration can directly affect bacterial hosts, resulting in cascading effects on other intestinal bacterial species, with consequential alteration of the gut microbiome (Hsu et al., 2019). Thus, further efforts will be required to clarify the complex interacting effect of the gut virome on the gut bacteriomes.
Interestingly, we found that the richness of gut DNA virome in female cynomolgus generally tended to decline with age, especially the Caudovirales order, resulting in lower bacterial biodiversity in old monkeys. Generally speaking, high biodiversity is equal to good health status (Huttenhower et al., 2012). The downregulated Caudovirales order was related to the altered intestinal folate metabolism. A multicenter study has shown that functional folate deficiency increases with age in inpatients (Mézière et al., 2014). Our findings suggest that modulating the intestinal virome may be beneficial for the treatment of aging.
There were some limitations in our study. First, we did not sequence the RNA virome, which is increasingly recognized as an important component of the gut microbiome that is involved in health and disease (Virgin, 2014). Therefore, future studies should investigate changes in the gut RNA virome with age. Second, due to the low reproductive rate and morbidity, as well as ethical considerations, the sample sizes were relatively small; thus, the reliability of the association reported may be impacted. Finally, the dynamic changes we observed in the gut virome with age require in‐depth validation.
5. CONCLUSIONS
In summary, we profiled structural and functional alterations of the gut DNA virome associated with age in non‐human primates. We found that the gut DNA virome differed significantly among the three age groups, as observed in alterations in the viral functional pathways related to nucleotide and folate metabolism. The gut virome was dominated by dsDNA phages, especially the Caudovirales order, which showed strong correlations with the highly prevalent gut bacteria. Herein, age‐related gut bacteriophages and bacteria were in a complementary relationship, and both of them decreased in richness during aging. We thus speculated that certain bacteriophage supplementation may be beneficial for the treatment of aging through modulating the structure and function of the intestinal flora in mammals.
ETHICS STATEMENT
This study was conducted following the Guide for the Care and Use of Laboratory Animals of the Institute of Neuroscience at Chongqing Medical University (#20100031). The living environments and animal care procedures were in line with Chinese regulatory requirements and were accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Xunmin Tan: Formal analysis (equal); Visualization (equal); Writing‐original draft (equal). Tingjia Chai: Formal analysis (equal); Visualization (equal); Writing‐original draft (equal). Jiajia Duan: Writing‐review & editing (equal). Jing Wu: Data curation (equal); Software (equal). Hanping Zhang: Data curation (equal); Software (equal). Yifan Li: Data curation (equal); Software (equal). Yu Huang: Data curation (equal); Software (equal). Xi Hu: Data curation (equal); Software (equal). Peng Zheng: Writing‐review & editing (equal). Jinlin Song: Validation (equal). Ping Ji: Validation (equal). Xin Jin: Validation (equal). Hongmei Zhang: Validation (equal). Peng Xie: Conceptualization (lead); Funding acquisition (lead); Supervision (lead); Writing‐review & editing (equal).
ACKNOWLEDGEMENTS
This research was funded by the National Key R&D Program of China (2017YFA0505700), Non‐profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2019PT320002), Projects of International Cooperation and Exchanges NSFC (81820108015), and the Natural Science Foundation Project of China (81971296, 81771490, 81371310, and 81200899). We thank the Zhongke Experimental Animal Co., Ltd. for providing the stool samples of cynomolgus macaques.
APPENDIX 1.
TABLE A1.
The discriminative species of the gut DNA virome among the young, adult, and old groups
| Species | Family | Relative abundance | p‐value | LDA | Enrichment | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Young (mean) | Young (SD) | Adult (mean) | Adult (SD) | Old (mean) | Old (SD) | |||||
| Enterococcus phage phiEF24C | Myoviridae | 1.8E‐04 | 1.7E‐04 | 2.0E‐05 | 1.8E‐05 | 3.9E‐05 | 4.7E‐05 | 0.015 | 2.124 | Young |
| Vibrio phage VP882 | Myoviridae | 3.3E‐05 | 2.1E‐05 | 9.3E‐06 | 2.1E‐05 | 0.0E+00 | 0.0E+00 | 0.008 | 2.383 | Young |
| Enterobacteria phage IME10 | Podoviridae | 2.2E‐05 | 1.8E‐05 | 1.6E‐05 | 1.1E‐05 | 0.0E+00 | 0.0E+00 | 0.029 | 2.888 | Young |
| Sinorhizobium phage phiM9 | Myoviridae | 2.2E‐03 | 1.4E‐03 | 2.1E‐04 | 2.4E‐04 | 5.0E‐04 | 6.8E‐04 | 0.028 | 2.964 | Young |
| Lactococcus phage Tuc2009 | Siphoviridae | 8.3E‐04 | 8.2E‐04 | 1.4E‐05 | 1.9E‐05 | 4.1E‐06 | 8.2E‐06 | 0.041 | 2.594 | Young |
| Staphylococcus phage S25‐4 | Myoviridae | 3.3E‐04 | 5.8E‐05 | 2.1E‐05 | 1.2E‐05 | 0.0E+00 | 0.0E+00 | 0.002 | 2.806 | Young |
| Listeria phage P70 | Siphoviridae | 1.9E‐03 | 1.5E‐03 | 1.4E‐04 | 1.2E‐04 | 9.7E‐04 | 1.4E‐03 | 0.046 | 2.949 | Young |
| Salmonella phage SPN3US | Myoviridae | 2.2E‐05 | 2.3E‐05 | 1.7E‐06 | 3.7E‐06 | 1.6E‐06 | 3.1E‐06 | 0.047 | 2.644 | Young |
| Paramecium bursaria Chlorella virus NE‐JV‐1 | Phycodnaviridae | 4.5E‐05 | 4.5E‐05 | 6.7E‐06 | 1.5E‐05 | 0.0E+00 | 0.0E+00 | 0.023 | 2.563 | Young |
| Vibrio phage nt‐1 | Myoviridae | 1.4E‐03 | 1.3E‐03 | 1.1E‐04 | 1.1E‐04 | 2.0E‐05 | 2.6E‐05 | 0.016 | 2.866 | Young |
| Shigella phage Ag3 | Myoviridae | 5.4E‐04 | 3.2E‐04 | 8.3E‐05 | 8.8E‐05 | 1.8E‐04 | 1.8E‐04 | 0.034 | 2.505 | Young |
| Halocynthia phage JM‐2012 | Myoviridae | 1.2E‐04 | 1.0E‐04 | 1.1E‐05 | 1.2E‐05 | 0.0E+00 | 0.0E+00 | 0.020 | 2.097 | Young |
| Enterobacteria phage phi92 | Myoviridae | 2.1E‐04 | 1.2E‐04 | 4.6E‐05 | 5.2E‐05 | 1.5E‐05 | 1.5E‐05 | 0.016 | 2.312 | Young |
| Acanthocystis turfacea Chlorella virus NE‐JV‐2 | Phycodnaviridae | 4.4E‐04 | 5.5E‐04 | 0.0E+00 | 0.0E+00 | 1.6E‐06 | 3.1E‐06 | 0.011 | 2.387 | Young |
| Staphylococcus phage 812 | Myoviridae | 6.6E‐04 | 2.9E‐04 | 1.4E‐04 | 8.1E‐05 | 2.9E‐04 | 3.5E‐04 | 0.037 | 2.454 | Young |
| Synechococcus phage ACG 2014d | Myoviridae | 6.4E‐04 | 3.4E‐04 | 2.2E‐04 | 2.5E‐04 | 1.1E‐04 | 1.4E‐04 | 0.031 | 2.483 | Young |
| Erwinia phage phiEaH2 | Siphoviridae | 1.4E‐04 | 1.0E‐04 | 1.1E‐05 | 2.0E‐05 | 4.8E‐05 | 8.0E‐05 | 0.035 | 2.345 | Young |
| Salmonella phage Stitch | Siphoviridae | 4.6E‐04 | 2.9E‐04 | 7.8E‐05 | 7.7E‐05 | 1.3E‐04 | 1.1E‐04 | 0.032 | 2.336 | Young |
| Escherichia phage Akfv33 | Siphoviridae | 1.4E‐04 | 1.4E‐04 | 1.6E‐05 | 8.9E‐06 | 3.6E‐07 | 7.2E‐07 | 0.004 | 2.529 | Young |
| Mycobacterium phage Courthouse | Siphoviridae | 4.1E‐04 | 4.8E‐04 | 3.8E‐05 | 6.4E‐05 | 3.8E‐05 | 7.6E‐05 | 0.026 | 2.459 | Young |
| Staphylococcus phage GH15 | Myoviridae | 2.8E‐03 | 4.6E‐03 | 6.0E‐05 | 3.5E‐05 | 4.1E‐05 | 2.8E‐05 | 0.010 | 3.224 | Young |
| Cellulophaga phage phi19‐1 | Siphoviridae | 1.2E‐04 | 1.7E‐04 | 0.0E+00 | 0.0E+00 | 4.1E‐06 | 8.2E‐06 | 0.001 | 2.038 | Young |
| Lactobacillus phage Ldl1 | Siphoviridae | 1.5E‐03 | 8.3E‐04 | 2.0E‐04 | 1.3E‐04 | 7.6E‐04 | 9.4E‐04 | 0.022 | 2.803 | Young |
| Lactobacillus phage c5 | Siphoviridae | 8.0E‐04 | 5.3E‐04 | 2.3E‐04 | 2.0E‐04 | 2.3E‐04 | 1.8E‐04 | 0.020 | 2.518 | Young |
| Aeromonas phage Aes012 | Myoviridae | 1.9E‐04 | 2.0E‐04 | 3.7E‐05 | 8.1E‐05 | 0.0E+00 | 0.0E+00 | 0.031 | 2.346 | Young |
| Enterobacteria phage JSE | Myoviridae | 2.1E‐04 | 1.1E‐04 | 3.6E‐05 | 4.3E‐05 | 2.5E‐05 | 3.0E‐05 | 0.013 | 2.355 | Young |
| uncultured Mediterranean phage uvMED | unclassified Viruses | 4.2E‐02 | 2.1E‐02 | 1.0E‐02 | 6.7E‐03 | 3.5E‐02 | 3.2E‐02 | 0.029 | 4.202 | Young |
| Bacillus phage vB BhaS‐171 | unclassified Viruses | 1.8E‐03 | 4.7E‐04 | 2.5E‐04 | 1.9E‐04 | 4.7E‐04 | 4.4E‐04 | 0.009 | 2.906 | Young |
| Yersinia phage phiR1‐37 | Myoviridae | 2.2E‐03 | 1.0E‐03 | 5.3E‐04 | 5.0E‐04 | 8.6E‐04 | 6.0E‐04 | 0.041 | 2.898 | Young |
| Escherichia phage ECML‐117 | Myoviridae | 1.6E‐04 | 1.1E‐04 | 1.5E‐04 | 1.5E‐04 | 0.0E+00 | 0.0E+00 | 0.013 | 2.210 | Young |
| Pseudomonas phage EL | Myoviridae | 1.5E‐04 | 2.3E‐04 | 2.3E‐06 | 3.3E‐06 | 9.7E‐06 | 1.2E‐05 | 0.049 | 2.364 | Young |
| Streptococcus phage Dp‐1 | Siphoviridae | 7.3E‐03 | 3.5E‐03 | 2.8E‐03 | 3.3E‐03 | 8.8E‐04 | 4.2E‐04 | 0.033 | 3.526 | Young |
| Synechococcus phage S‐SKS1 | Siphoviridae | 1.3E‐03 | 1.5E‐03 | 2.0E‐04 | 1.3E‐04 | 1.2E‐03 | 1.3E‐03 | 0.025 | 2.749 | Young |
| Streptococcus phage Str‐PAP‐1 | Podoviridae | 4.5E‐04 | 2.4E‐04 | 6.0E‐04 | 8.7E‐04 | 5.4E‐05 | 5.3E‐05 | 0.028 | 2.504 | Adult |
| Campylobacter phage IBB35 | Myoviridae | 1.3E‐03 | 7.5E‐04 | 1.4E‐03 | 2.7E‐03 | 7.9E‐05 | 7.0E‐05 | 0.020 | 2.865 | Adult |
| Xylella phage Prado | Podoviridae | 9.4E‐06 | 5.9E‐06 | 5.0E‐05 | 6.8E‐05 | 0.0E+00 | 0.0E+00 | 0.042 | 3.018 | Adult |
| Aeromonas phage 31 | Myoviridae | 8.4E‐06 | 6.9E‐06 | 3.7E‐04 | 7.7E‐04 | 1.1E‐04 | 4.3E‐05 | 0.014 | 2.558 | Adult |
| Cellulophaga phage phi10‐1 | Siphoviridae | 0.0E+00 | 0.0E+00 | 2.1E‐04 | 4.1E‐04 | 3.5E‐05 | 7.0E‐05 | 0.029 | 2.248 | Adult |
| White spot syndrome virus | Nimaviridae | 3.2E‐06 | 6.4E‐06 | 3.0E‐04 | 4.5E‐04 | 1.6E‐05 | 2.0E‐05 | 0.042 | 2.430 | Adult |
| Staphylococcus phage SAP‐2 | Podoviridae | 6.3E‐03 | 5.0E‐03 | 6.4E‐03 | 8.3E‐03 | 4.4E‐04 | 8.1E‐04 | 0.035 | 3.472 | Adult |
| Vibrio phage VPMS1 | Podoviridae | 7.4E‐05 | 1.2E‐04 | 7.7E‐03 | 1.2E‐02 | 0.0E+00 | 0.0E+00 | 0.023 | 3.522 | Adult |
| Caulobacter phage CcrColossus | Siphoviridae | 8.5E‐05 | 7.5E‐05 | 1.3E‐04 | 2.4E‐04 | 9.5E‐04 | 7.9E‐04 | 0.020 | 2.728 | Old |
| Halovirus HGTV‐1 | unclassified Viruses | 3.6E‐04 | 5.5E‐04 | 4.3E‐06 | 7.1E‐06 | 7.2E‐04 | 1.1E‐03 | 0.050 | 2.381 | Old |
| Clostridium phage phiCP7R | Podoviridae | 1.6E‐03 | 1.0E‐03 | 1.5E‐04 | 1.5E‐04 | 1.7E‐03 | 2.4E‐03 | 0.042 | 2.884 | Old |
| Streptococcus phage phiNJ2 | Siphoviridae | 7.5E‐06 | 9.3E‐06 | 1.5E‐05 | 2.6E‐05 | 4.6E‐04 | 6.1E‐04 | 0.034 | 2.449 | Old |
TABLE A2.
The differential KEGG pathways among the three age groups
| KEGG pathway | Relative abundance | p‐value | LDA | Enrichment | |||||
|---|---|---|---|---|---|---|---|---|---|
| Young (mean) | Young (SD) | Adult (mean) | Adult (SD) | Old (mean) | Old (SD) | ||||
| Mismatch repair | 1.4E‐01 | 3.3E‐02 | 9.9E‐02 | 1.7E‐02 | 1.4E‐01 | 1.1E‐02 | 0.034 | 4.366 | Young |
| Folate biosynthesis | 1.6E‐02 | 3.1E‐02 | 3.3E‐03 | 2.3E‐03 | 0.0E+00 | 0.0E+00 | 0.014 | 4.293 | Young |
| Antifolate resistance | 4.5E‐03 | 3.8E‐03 | 1.7E‐02 | 3.0E‐02 | 3.0E‐04 | 2.7E‐04 | 0.018 | 4.511 | Adult |
| One carbon pool by folate | 4.5E‐03 | 3.7E‐03 | 1.8E‐02 | 3.1E‐02 | 3.0E‐04 | 2.7E‐04 | 0.009 | 4.516 | Adult |
| Pyrimidine metabolism | 1.8E‐01 | 4.6E‐02 | 2.2E‐01 | 4.5E‐02 | 2.3E‐01 | 2.1E‐02 | 0.017 | 4.801 | Old |
TABLE A3.
The discriminative bacterial species among the three groups
| Species | Phylum | Relative abundance | p‐value | LDA | Enrichment | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Young (mean) | Young (SD) | Adult (mean) | Adult (SD) | Old (mean) | Old (SD) | |||||
| Bifidobacterium ruminantium | Actinobacteria | 6.59E‐06 | 3.09E‐06 | 2.52E‐05 | 4.15E‐05 | 2.40E‐04 | 4.13E‐04 | 0.024 | 2.103 | Old |
| unclassified g Corynebacterium | Actinobacteria | 5.86E‐08 | 5.07E‐08 | 1.60E‐06 | 3.13E‐06 | 6.25E‐04 | 1.25E‐03 | 0.049 | 2.451 | Old |
| Bacteroides sp. CAG 1060 | Bacteroidetes | 5.08E‐03 | 2.12E‐03 | 5.21E‐03 | 2.60E‐03 | 1.12E‐03 | 6.64E‐04 | 0.015 | 3.317 | Adult |
| Prevotella sp. CAG 617 | Bacteroidetes | 7.26E‐04 | 2.18E‐04 | 4.69E‐04 | 1.81E‐04 | 3.17E‐04 | 1.65E‐04 | 0.039 | 2.298 | Young |
| Prevotella sp. CAG 755 | Bacteroidetes | 1.03E‐03 | 3.96E‐04 | 6.87E‐04 | 3.07E‐04 | 4.71E‐04 | 2.31E‐04 | 0.035 | 2.439 | Young |
| Prevotella sp. P4‐76 | Bacteroidetes | 1.79E‐03 | 4.11E‐04 | 1.10E‐03 | 4.68E‐04 | 8.80E‐04 | 4.25E‐04 | 0.028 | 2.647 | Young |
| Prevotella sp. P5‐119 | Bacteroidetes | 1.82E‐03 | 4.25E‐04 | 1.25E‐03 | 4.52E‐04 | 9.92E‐04 | 4.97E‐04 | 0.039 | 2.602 | Young |
| Chlamydia trachomatis | Chlamydiae | 6.01E‐03 | 1.23E‐03 | 3.48E‐03 | 1.17E‐03 | 7.23E‐03 | 2.04E‐03 | 0.013 | 3.252 | Old |
| Acidaminococcus fermentans | Firmicutes | 1.12E‐04 | 6.96E‐05 | 2.29E‐04 | 3.51E‐04 | 5.68E‐03 | 9.68E‐03 | 0.038 | 3.418 | Old |
| Clostridium sp. CAG 609 | Firmicutes | 3.71E‐04 | 2.08E‐04 | 1.15E‐04 | 4.87E‐05 | 5.44E‐05 | 2.99E‐05 | 0.011 | 2.225 | Young |
| Coprobacillus sp. CAG 826 | Firmicutes | 5.98E‐04 | 1.67E‐04 | 2.81E‐04 | 1.05E‐04 | 3.13E‐04 | 2.74E‐04 | 0.038 | 2.194 | Young |
| Eubacterium sp. CAG 146 | Firmicutes | 1.78E‐04 | 4.84E‐05 | 1.93E‐04 | 4.27E‐05 | 4.93E‐04 | 3.52E‐04 | 0.027 | 2.169 | Old |
| Eubacterium sp. CAG 180 | Firmicutes | 2.44E‐03 | 1.04E‐03 | 8.47E‐04 | 5.44E‐04 | 6.32E‐04 | 4.63E‐04 | 0.024 | 2.972 | Young |
| Firmicutes bacterium CAG 536 | Firmicutes | 1.14E‐04 | 3.48E‐05 | 1.64E‐04 | 4.38E‐05 | 6.81E‐04 | 4.71E‐04 | 0.034 | 2.453 | Old |
| Lactobacillus gasseri | Firmicutes | 3.27E‐04 | 3.31E‐04 | 2.20E‐05 | 2.05E‐05 | 2.53E‐04 | 3.60E‐04 | 0.041 | 2.211 | Young |
| Lactobacillus reuteri | Firmicutes | 1.42E‐02 | 9.52E‐03 | 1.26E‐03 | 1.09E‐03 | 2.28E‐02 | 2.81E‐02 | 0.034 | 3.998 | Old |
| Megasphaera cerevisiae | Firmicutes | 5.33E‐04 | 4.16E‐04 | 2.97E‐04 | 1.68E‐04 | 1.21E‐03 | 8.64E‐04 | 0.046 | 2.633 | Old |
| Megasphaera elsdenii | Firmicutes | 1.82E‐03 | 1.99E‐03 | 1.99E‐03 | 2.02E‐03 | 1.16E‐02 | 1.20E‐02 | 0.029 | 3.664 | Old |
| Megasphaera elsdenii CAG 570 | Firmicutes | 2.00E‐04 | 1.99E‐04 | 2.24E‐04 | 2.19E‐04 | 1.23E‐03 | 1.26E‐03 | 0.038 | 2.686 | Old |
| Megasphaera sp. DJF‐B143 | Firmicutes | 1.64E‐04 | 6.49E‐05 | 1.10E‐04 | 3.26E‐05 | 5.04E‐04 | 4.56E‐04 | 0.031 | 2.271 | Old |
| Mitsuokella jalaludinii | Firmicutes | 1.27E‐04 | 4.85E‐05 | 2.14E‐04 | 2.06E‐04 | 1.66E‐03 | 1.57E‐03 | 0.020 | 2.854 | Old |
| Oscillibacter sp. CAG 155 | Firmicutes | 2.35E‐03 | 4.04E‐04 | 1.82E‐03 | 6.16E‐04 | 1.17E‐03 | 1.78E‐04 | 0.048 | 2.763 | Young |
| Oscillibacter sp. CAG 241 | Firmicutes | 6.86E‐03 | 1.65E‐03 | 5.43E‐03 | 3.69E‐03 | 3.33E‐03 | 9.12E‐04 | 0.041 | 3.234 | Young |
| Oscillibacter sp. KLE 1728 | Firmicutes | 5.26E‐04 | 9.06E‐05 | 4.01E‐04 | 1.38E‐04 | 2.89E‐04 | 5.85E‐05 | 0.034 | 2.071 | Young |
| Oscillibacter valericigenes | Firmicutes | 1.32E‐03 | 2.94E‐04 | 9.74E‐04 | 4.08E‐04 | 7.99E‐04 | 1.70E‐04 | 0.034 | 2.396 | Young |
| Oscillospiraceae bacterium VE202‐24 | Firmicutes | 9.14E‐04 | 1.20E‐04 | 6.76E‐04 | 2.36E‐04 | 4.59E‐04 | 9.12E‐05 | 0.024 | 2.348 | Young |
| Roseburia sp. CAG 100 | Firmicutes | 9.12E‐04 | 3.51E‐04 | 1.48E‐03 | 7.93E‐04 | 5.80E‐04 | 2.39E‐04 | 0.034 | 2.672 | Adult |
| Roseburia sp. CAG 309 | Firmicutes | 4.90E‐04 | 2.39E‐04 | 9.72E‐04 | 3.34E‐04 | 3.22E‐04 | 2.96E‐04 | 0.018 | 2.506 | Adult |
| unclassified g Megasphaera | Firmicutes | 2.04E‐04 | 1.33E‐04 | 1.45E‐04 | 7.83E‐05 | 8.10E‐04 | 6.76E‐04 | 0.027 | 2.499 | Old |
| unclassified g Oscillibacter | Firmicutes | 2.53E‐03 | 4.62E‐04 | 1.91E‐03 | 6.65E‐04 | 1.31E‐03 | 2.70E‐04 | 0.035 | 2.776 | Young |
| Mannheimia succiniciproducens | Proteobacteria | 2.48E‐06 | 1.08E‐06 | 2.98E‐04 | 5.79E‐04 | 7.25E‐05 | 9.56E‐05 | 0.019 | 2.126 | Adult |
| Azonexus hydrophilus | Proteobacteria | 8.67E‐05 | 1.05E‐04 | 1.44E‐04 | 1.73E‐04 | 1.53E‐03 | 1.47E‐03 | 0.024 | 2.839 | Old |
| Azospira oryzae | Proteobacteria | 1.90E‐05 | 2.26E‐05 | 3.02E‐05 | 3.55E‐05 | 3.34E‐04 | 3.30E‐04 | 0.024 | 2.179 | Old |
| Azovibrio restrictus | Proteobacteria | 4.35E‐05 | 4.18E‐05 | 6.33E‐05 | 7.60E‐05 | 6.64E‐04 | 6.44E‐04 | 0.028 | 2.472 | Old |
| Dechloromonas agitata | Proteobacteria | 1.22E‐04 | 1.41E‐04 | 2.01E‐04 | 2.37E‐04 | 2.12E‐03 | 2.09E‐03 | 0.024 | 2.979 | Old |
| Dechloromonas aromatica | Proteobacteria | 6.68E‐05 | 7.96E‐05 | 1.14E‐04 | 1.37E‐04 | 1.21E‐03 | 1.18E‐03 | 0.018 | 2.738 | Old |
| Dechloromonas denitrificans | Proteobacteria | 9.56E‐05 | 1.15E‐04 | 1.55E‐04 | 1.88E‐04 | 1.67E‐03 | 1.66E‐03 | 0.018 | 2.875 | Old |
| Desulfovibrio desulfuricans | Proteobacteria | 4.74E‐05 | 2.22E‐05 | 1.03E‐04 | 5.25E‐05 | 8.36E‐04 | 1.41E‐03 | 0.049 | 2.568 | Old |
| Desulfovibrio fairfieldensis | Proteobacteria | 4.56E‐05 | 2.25E‐05 | 1.22E‐04 | 5.55E‐05 | 9.50E‐04 | 1.57E‐03 | 0.018 | 2.631 | Old |
| Desulfovibrio sp. 6–1‐46AFAA | Proteobacteria | 3.20E‐05 | 1.47E‐05 | 7.36E‐05 | 3.55E‐05 | 5.15E‐04 | 8.26E‐04 | 0.020 | 2.358 | Old |
| Rhodocyclaceae bacterium Paddy−1 | Proteobacteria | 1.59E‐05 | 1.85E‐05 | 2.69E‐05 | 3.25E‐05 | 2.82E‐04 | 2.70E‐04 | 0.024 | 2.107 | Old |
| Succinatimonas sp. CAG 777 | Proteobacteria | 1.18E‐04 | 8.64E‐05 | 5.14E‐05 | 2.27E‐05 | 7.80E‐04 | 7.57E‐04 | 0.043 | 2.561 | Old |
TABLE A4.
The hosts of 39 differential bacteriophages based on the NCBI database
| Bacteriophage | Bacterial phylum | Bacterial species | Reference |
|---|---|---|---|
| Mycobacterium phage Courthouse | Actinobacteria | Mycobacterium smegmatis | https://www.ncbi.nlm.nih.gov/nuccore/NC_023690.1 |
| Cellulophaga phage phi19‐1 | Bacteroidetes | Cellulophaga baltica | https://www.ncbi.nlm.nih.gov/nuccore/NC_021799.1 |
| Cellulophaga phage phi10‐1 | Bacteroidetes | Cellulophaga baltica | https://www.ncbi.nlm.nih.gov/nuccore/NC_021802.1 |
| Synechococcus phage S‐SKS1 | Cyanobacteria | Synechococcus sp. WH7803 | https://www.ncbi.nlm.nih.gov/nuccore/NC_020851.1 |
| Synechococcus phage ACG‐2014d | NA | NA | https://www.ncbi.nlm.nih.gov/nuccore/NC_026923.1 |
| Clostridium phage phiCP7R | Firmicutes | Clostridium perfringens | https://www.ncbi.nlm.nih.gov/nuccore/NC_017980.1 |
| Enterococcus phage phiEF24C | Firmicutes | Enterococcus fecalis | https://www.ncbi.nlm.nih.gov/nuccore/NC_009904.1 |
| Listeria phage P70 | Firmicutes | Listeria sp. | https://www.ncbi.nlm.nih.gov/nuccore/NC_018831.1 |
| Lactobacillus phage Ldl1 | Firmicutes | Lactobacillus delbrueckii subsp. Lactis | https://www.ncbi.nlm.nih.gov/nuccore/NC_026609.1 |
| Lactobacillus phage c5 | Firmicutes | Lactobacillus delbrueckii subsp. bulgaricus | https://www.ncbi.nlm.nih.gov/nuccore/NC_019449.1 |
| Staphylococcus phage S25‐4 | NA | NA | https://www.ncbi.nlm.nih.gov/nuccore/NC_022918.1 |
| Staphylococcus phage 812 | Firmicutes | Staphylococcus aureus | https://www.ncbi.nlm.nih.gov/nuccore/NC_029080.1 |
| Staphylococcus phage GH15 | Firmicutes | Staphylococcus aureus | https://www.ncbi.nlm.nih.gov/nuccore/NC_019448.1 |
| Staphylococcus phage SAP‐2 | Firmicutes | Staphylococcus aureus | https://www.ncbi.nlm.nih.gov/nuccore/NC_009875.1 |
| Lactococcus phage Tuc2009 | Firmicutes | Lactococcus lactis | https://www.ncbi.nlm.nih.gov/nuccore/NC_002703.1 |
| Streptococcus phage Dp‐1 | Firmicutes | Streptococcus pneumoniae | https://www.ncbi.nlm.nih.gov/nuccore/NC_015274.1 |
| Streptococcus phage Str‐PAP‐1 | Firmicutes | Streptococcus parauberis | https://www.ncbi.nlm.nih.gov/nuccore/NC_028666.1 |
| Streptococcus phage phiNJ2 | Firmicutes | Streptococcus suis NJ2 serotype 9 | https://www.ncbi.nlm.nih.gov/nuccore/NC_019418.1 |
| Aeromonas phage Aes012 | Proteobacteria | Aeromonas sp. | https://www.ncbi.nlm.nih.gov/nuccore/NC_020879.1 |
| Aeromonas phage 31 | Proteobacteria | Aeromonas salmonicida | https://www.ncbi.nlm.nih.gov/nuccore/NC_007022.1 |
| Campylobacter phage IBB35 | NA | NA | https://www.ncbi.nlm.nih.gov/nuccore/NC_041833.1 |
| Caulobacter phage CcrColossus | Proteobacteria | Caulobacter crescentus | https://www.ncbi.nlm.nih.gov/nuccore/NC_019406.1 |
| Enterobacteria phage IME10 | Proteobacteria | Escherichia coli | https://www.ncbi.nlm.nih.gov/nuccore/NC_019501.1 |
| Salmonella phage SPN3US | NA | NA | https://www.ncbi.nlm.nih.gov/nuccore/NC_027402.1 |
| Shigella phage Ag3 | Proteobacteria | Shigella boydii | https://www.ncbi.nlm.nih.gov/nuccore/NC_013693.1 |
| Enterobacteria phage phi92 | Proteobacteria | Escherichia coli K92 | https://www.ncbi.nlm.nih.gov/nuccore/NC_023693.1 |
| Erwinia phage phiEaH2 | Proteobacteria | Erwinia amylovora | https://www.ncbi.nlm.nih.gov/nuccore/NC_019929.1 |
| Salmonella phage Stitch | Proteobacteria | Salmonella typhimurium | https://www.ncbi.nlm.nih.gov/nuccore/NC_027297.1 |
| Escherichia phage Akfv33 | Proteobacteria | Escherichia coli | https://www.ncbi.nlm.nih.gov/nuccore/NC_017969.1 |
| Enterobacteria phage JSE | Proteobacteria | Escherichia coli K12 | https://www.ncbi.nlm.nih.gov/nuccore/NC_012740.1 |
| Yersinia phage phiR1 37 | Proteobacteria | Yersinia enterocolitica. | https://www.ncbi.nlm.nih.gov/nuccore/NC_016163.1 |
| Escherichia phage ECML‐117 | Proteobacteria | Escherichia coli | https://www.ncbi.nlm.nih.gov/nuccore/NC_025441.1 |
| Pseudomonas phage EL | Proteobacteria | Pseudomonas aeruginosa | https://www.ncbi.nlm.nih.gov/nuccore/NC_007623.1 |
| Sinorhizobium phage phiM9 | Proteobacteria | Sinorhizobium meliloti | https://www.ncbi.nlm.nih.gov/nuccore/NC_028676.1 |
| Vibrio phage VP882 | Proteobacteria | Vibrio parahaemolyticus | https://www.ncbi.nlm.nih.gov/nuccore/NC_009016.1 |
| Vibrio phage nt‐1 | Proteobacteria | Vibrio natriegens HER1138 | https://www.ncbi.nlm.nih.gov/nuccore/NC_021529.2 |
| Vibrio phage VPMS1 | Proteobacteria | Vibrio parahaemolyticus Ex‐1 | https://www.ncbi.nlm.nih.gov/nuccore/NC_021776.1 |
| Xylella phage Prado | Proteobacteria | Xylella fastidiosa | https://www.ncbi.nlm.nih.gov/nuccore/NC_022987.1 |
| Halocynthia phage JM‐2012 | NA | NA | https://www.ncbi.nlm.nih.gov/nuccore/NC_017975.1 |
FIGURE A1.
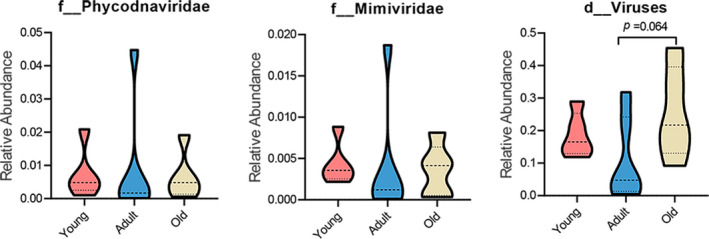
Quantification of the relative abundance of the three unassigned families. No significant difference was found among the three agegroups. p values were determined by one‐way ANOVA followed by LSD’s multiple comparison tests. The following n values represented the number of independent animals for statistical evaluation: young, n = 5; adult, n = 6; and old, n = 5
FIGURE A2.
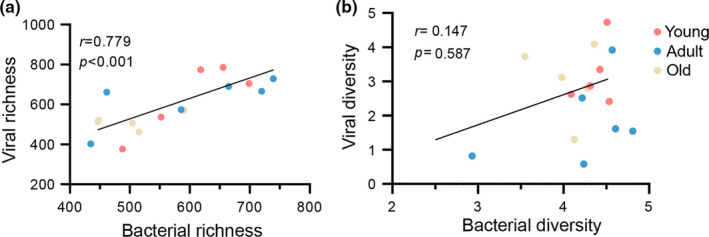
Positive relationships between the gut DNA viruses and bacteria. (a) Correlation plot between viral richness and bacterial richness. (b) Correlation between viral diversity and bacterial diversity. The lines indicate linear regression, and Spearman's correlation coefficient is shown. The color spectrum indicates age progression from young to old
Xunmin Tan and Tingjia Chai contributed equally.
DATA AVAILABILITY STATEMENT
The raw data were uploaded in the NCBI Sequence Read Archive (SRA) database under the accession number PRJNA560277, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA560277.
REFERENCES
- Appt, S. E., Chen, H., Goode, A. K., Hoyer, P. B., Clarkson, T. B., Adams, M. R., Wilson, M. E., Franke, A. A., & Kaplan, J. R. (2010). The effect of diet and cardiovascular risk on ovarian aging in cynomolgus monkeys (Macaca fascicularis). Menopause, 17, 741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, E. S., White, D. W., Cathelyn, J. S., Brett‐Mcclellan, K. A., Engle, M., Diamond, M. S., Miller, V. L., & Virgin, H. W. T. (2007). Herpesvirus latency confers symbiotic protection from bacterial infection. Nature, 447, 326–329. [DOI] [PubMed] [Google Scholar]
- Beller, L., & Matthijnssens, J. (2019). What is (not) known about the dynamics of the human gut virome in health and disease. Current Opinion in Virology, 37, 52–57. [DOI] [PubMed] [Google Scholar]
- Biagi, E., Nylund, L., Candela, M., Ostan, R., Bucci, L., Pini, E., Nikkila, J., Monti, D., Satokari, R., Franceschi, C., Brigidi, P., & de Vos, W. (2010). Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One, 5, e10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart, M., & Rohwer, F. (2005). Here a virus, there a virus, everywhere the same virus? Trends in Microbiology, 13, 278–284. [DOI] [PubMed] [Google Scholar]
- Brussow, H., Canchaya, C., & Hardt, W. D. (2004). Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiology and Molecular Biology Reviews, 68(3), 560–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevon‐Gaillot, E., Perron‐Lepage, M. F., Clement, C., & Burnett, R. (2006). A review of background findings in cynomolgus monkeys (Macaca fascicularis) from three different geographical origins. Experimental and Toxicologic Pathology, 58, 77–88. [DOI] [PubMed] [Google Scholar]
- Duan, J., Yin, B., Li, W., Chai, T., Liang, W., Huang, Y., Tan, X., Zheng, P., Wu, J., Li, Y., Li, Y., Zhou, W., & Xie, P. (2019). Age‐related changes in microbial composition and function in cynomolgus macaques. Aging, 11, 12080–12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemer, B., Lynch, D. B., Brown, J. M., Jeffery, I. B., Ryan, F. J., Claesson, M. J., O'Riordain, M., Shanahan, F., & O'Toole, P. W. (2017). Tumour‐associated and non‐tumour‐associated microbiota in colorectal cancer. Gut, 66, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile, C. L., & Weir, T. L. (2018). The gut microbiota at the intersection of diet and human health. Science, 362, 776–780. [DOI] [PubMed] [Google Scholar]
- Gilbert, R. A., Kelly, W. J., Altermann, E., Leahy, S. C., Minchin, C., Ouwerkerk, D., & Klieve, A. V. (2017). Toward understanding phage: Host interactions in the rumen; complete genome sequences of lytic phages infecting rumen bacteria. Frontiers in Microbiology, 8, 2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Doñate, M., Payán, A., Cortés, I., Blanch, A. R., Lucena, F., Jofre, J., & Muniesa, M. (2011). Isolation of bacteriophage host strains of Bacteroides species suitable for tracking sources of animal fecal pollution in water. Environmental Microbiology, 13, 1622–1631. [DOI] [PubMed] [Google Scholar]
- Górska, A., Peter, S., Willmann, M., Autenrieth, I., Schlaberg, R., & Huson, D. H. (2018). Dynamics of the human gut phageome during antibiotic treatment. Computational Biology and Chemistry, 74, 420–427. [DOI] [PubMed] [Google Scholar]
- Hsu, B. B., Gibson, T. E., Yeliseyev, V., Liu, Q., Lyon, L., Bry, L., Silver, P. A., & Gerber, G. K. (2019). Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host & Microbe, 25, 803–814.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower, C., Gevers, D., Knight, R., Abubucker, S., Badger, J. H., & Chinwalla, A. T. (2012). Structure, function and diversity of the healthy human microbiome. Nature, 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, D., Baldridge, M. T., & Handley, S. A. (2019). Phages and Human Health: More Than Idle Hitchhikers. Viruses, 11(7), 587. 10.3390/v11070587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., Li, Y., Kristiansen, K., & Wang, J. (2008). SOAP: Short oligonucleotide alignment program. Bioinformatics, 24, 713–714. [DOI] [PubMed] [Google Scholar]
- Lim, E. S., Zhou, Y., Zhao, G., Bauer, I. K., Droit, L., Ndao, I. M., Warner, B. B., Tarr, P. I., Wang, D., & Holtz, L. R. (2015). Early life dynamics of the human gut virome and bacterial microbiome in infants. Nature Medicine, 21, 1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, S. V., & Pedersen, O. (2016). The human intestinal microbiome in health and disease. New England Journal of Medicine, 375, 2369–2379. [DOI] [PubMed] [Google Scholar]
- Matson, V., Fessler, J., Bao, R., Chongsuwat, T., Zha, Y., Alegre, M. L., Luke, J. J., & Gajewski, T. F. (2018). The commensal microbiome is associated with anti‐PD‐1 efficacy in metastatic melanoma patients. Science, 359, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich, T. N., Casey, E., Oliveira, J., Bottacini, F., James, K., Franz, C., Lugli, G. A., Neve, H., Ventura, M., Hatfull, G. F., Mahony, J., & van Sinderen, D. (2018). Characterization and induction of prophages in human gut‐associated Bifidobacterium hosts. Scientific Reports, 8, 12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mézière, A., Audureau, E., Vairelles, S., Krypciak, S., Dicko, M., Monié, M., & Giraudier, S. (2014). B12 deficiency increases with age in hospitalized patients: a study on 14,904 samples. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences, 69, 1576–1585. [DOI] [PubMed] [Google Scholar]
- Mills, S., Shanahan, F., Stanton, C., Hill, C., Coffey, A., & Ross, R. P. (2013). Movers and shakers: Influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes, 4, 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot, S., Bryson, A., Chehoud, C., Wu, G. D., Lewis, J. D., & Bushman, F. D. (2013). Rapid evolution of the human gut virome. Proceedings of the National Academy of Sciences of the United States of America, 110, 12450–12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot, S., Sinha, R., Chen, J., Li, H., Keilbaugh, S. A., Wu, G. D., Lewis, J. D., & Bushman, F. D. (2011). The human gut virome: Inter‐individual variation and dynamic response to diet. Genome Research, 21, 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi, S. R., Lee, H. H., Spina, C. S., & Collins, J. J. (2013). Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature, 499, 219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno‐Gallego, J. L., Chou, S. P., di Rienzi, S. C., Goodrich, J. K., Spector, T. D., Bell, J. T., Youngblut, N. D., Hewson, I., Reyes, A., & Ley, R. E. (2019). Virome diversity correlates with intestinal microbiome diversity in adult monozygotic twins. Cell Host & Microbe, 25, 261–272.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman, J. M., Handley, S. A., Baldridge, M. T., Droit, L., Liu, C. Y., Keller, B. C., Kambal, A., Monaco, C. L., Zhao, G., Fleshner, P., Stappenbeck, T. S., McGovern, D. P., Keshavarzian, A., Mutlu, E. A., Sauk, J., Gevers, D., Xavier, R. J., Wang, D., Parkes, M., & Virgin, H. W. (2015). Disease‐specific alterations in the enteric virome in inflammatory bowel disease. Cell, 160, 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, C., Kivipelto, M., & von Strauss, E. (2009). Epidemiology of Alzheimer's disease: Occurrence, determinants, and strategies toward intervention. Dialogues in Clinical Neuroscience, 11, 111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez‐Vargas, G., Goh, S., & Rodríguez, C. (2018). The novel phages phiCD5763 and phiCD2955 represent two groups of big plasmidial siphoviridae phages of Clostridium difficile . Frontiers in Microbiology, 9, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelli, S., Turroni, S., Schnorr, S. L., Soverini, M., Quercia, S., Barone, M., Castagnetti, A., Biagi, E., Gallinella, G., Brigidi, P., & Candela, M. (2017). Characterization of the human DNA gut virome across populations with different subsistence strategies and geographical origin. Environmental Microbiology, 19, 4728–4735. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Valera, F., Martin‐Cuadrado, A. B., Rodriguez‐Brito, B., Pasić, L., Thingstad, T. F., Rohwer, F., & Mira, A. (2009). Explaining microbial population genomics through phage predation. Nature Reviews Microbiology, 7, 828–836. [DOI] [PubMed] [Google Scholar]
- Sampson, T. R., Debelius, J. W., Thron, T., Janssen, S., Shastri, G. G., Ilhan, Z. E., Challis, C., Schretter, C. E., Rocha, S., Gradinaru, V., Chesselet, M. F., Keshavarzian, A., Shannon, K. M., Krajmalnik‐Brown, R., Wittung‐Stafshede, P., Knight, R., & Mazmanian, S. K. (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell, 167, 1469–1480.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., & Huttenhower, C. (2011). Metagenomic biomarker discovery and explanation. Genome Biology, 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, V., Smolin, J., Nayak, J., Ayala, J. E., Scott, D. A., Peterson, S. N., & Freeze, H. H. (2018). Mannose alters gut microbiome, prevents diet‐induced obesity, and improves host metabolism. Cell Reports, 24, 3087–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon, G., Cruz, N. J., Kang, D. W., Gandal, M. J., Wang, B., Kim, Y. M., Zink, E. M., Casey, C. P., Taylor, B. C., Lane, C. J., Bramer, L. M., Isern, N. G., Hoyt, D. W., Noecker, C., Sweredoski, M. J., Moradian, A., Borenstein, E., Jansson, J. K., Knight, R., … Mazmanian, S. K. (2019). Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell, 177(1600–1618), e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, A. C., Goldstein, D. R., & Montgomery, R. R. (2013). Age‐dependent dysregulation of innate immunity. Nature Reviews Immunology, 13, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkoporov, A. N., Clooney, A. G., Sutton, T. D. S., Ryan, F. J., Daly, K. M., Nolan, J. A., McDonnell, S. A., Khokhlova, E. V., Draper, L. A., Forde, A., Guerin, E., Velayudhan, V., Ross, R. P., & Hill, C. (2019). The human gut virome is highly diverse, stable, and individual specific. Cell Host & Microbe, 26, 527–541.e5. [DOI] [PubMed] [Google Scholar]
- Shkoporov, A. N., & Hill, C. (2019). Bacteriophages of the human gut: The "known unknown" of the microbiome. Cell Host & Microbe, 25, 195–209. [DOI] [PubMed] [Google Scholar]
- Silveira, C. B., & Rohwer, F. L. (2016). Piggyback‐the‐Winner in host‐associated microbial communities. NPJ Biofilms Microbiomes, 2, 16010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin, H. W. (2014). The virome in mammalian physiology and disease. Cell, 157, 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. C., & Bennett, M. (2012). Aging and atherosclerosis: mechanisms, functional consequences, and potential therapeutics for cellular senescence. Circulation Research, 111, 245–259. [DOI] [PubMed] [Google Scholar]
- Wang, S., Zheng, Y., Li, J., Yu, Y., Zhang, W., Song, M., Liu, Z., Min, Z., Hu, H., Jing, Y., He, X., Sun, L., Ma, L., Esteban, C. R., Chan, P., Qiao, J., Zhou, Q., Izpisua Belmonte, J. C., Qu, J., Tang, F., & Liu, G. H. (2020). Single‐cell transcriptomic atlas of primate ovarian aging. Cell, 180(585–600), e19. [DOI] [PubMed] [Google Scholar]
- Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., Kong, L., Gao, G., Li, C. Y., & Wei, L. (2011). KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research, 39, W316–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F., Xie, L., Li, X., Li, Q., Wang, T., Ji, Y., Kong, F., Zhan, Q., Cheng, K., Fang, L., & Xie, P. (2012). Construction and validation of a systematic ethogram of Macaca fascicularis in a free enclosure. PLoS One, 7, e37486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Zheng, P., Li, Y., Wu, J., Tan, X., Zhou, J., Sun, Z., Chen, X., Zhang, G., Zhang, H., Huang, Y., Chai, T., Duan, J., Liang, W., Yin, B., Lai, J., Huang, T., Du, Y., Zhang, P., … Xie, P. (2020). Landscapes of bacterial and metabolic signatures and their interaction in major depressive disorders. Science Advances, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, P., Wu, J., Zhang, H., Perry, S. W., Yin, B., Tan, X., Chai, T., Liang, W., Huang, Y., Li, Y., Duan, J., Wong, M. L., Licinio, J., & Xie, P. (2020). The gut microbiome modulates gut‐brain axis glycerophospholipid metabolism in a region‐specific manner in a nonhuman primate model of depression. Molecular Psychiatry. 10.1038/s41380-020-0744-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data were uploaded in the NCBI Sequence Read Archive (SRA) database under the accession number PRJNA560277, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA560277.


