Abstract
Trillions of viruses inhabit the gastrointestinal tract. Some of them have been well-studied on their roles in infection and human health, but the majority remains unsurveyed. It has been established that the composition of the gut virome is highly variable based on the changes of diet, physical state, and environmental factors. However, the effect of host genetic factors, for example ethnic origin, on the gut virome is rarely investigated. Here, we characterized and compared the gut virome in a cohort of local Chinese residents and visiting Pakistani individuals, each group containing twenty-four healthy adults and six children. Using metagenomic shotgun sequencing and assembly of fecal samples, a huge number of viral operational taxonomic units (vOTUs) were identified for profiling the DNA and RNA viromes. National background contributed a primary variation to individuals’ gut virome. Compared with the Chinese adults, the Pakistan adults showed higher macrodiversity and different compositional and functional structures in their DNA virome and lower diversity and altered composition in their RNA virome. The virome variations of Pakistan children were not only inherited from that of the adults but also tended to share similar characteristics with the Chinese cohort. We also analyzed and compared the bacterial microbiome between two cohorts and further revealed numerous connections between viruses and bacterial host. Statistically, the gut DNA and RNA viromes were covariant to some extent (P < 0.001), and they both correlated the holistic bacterial composition and vice versa. This study provides an overview of the gut viral community in Chinese and visiting Pakistanis and proposes a considerable role of ethnic origin in shaping the virome.
Keywords: virus-like particle, gut virome, viral community, RNA virus, metagenomic sequencing, bacterial microbiome
1. Introduction
The human gut is a large reservoir of microorganisms, containing 1011–1012 bacterial cells (Sender, Fuchs, and Milo 2016; Vandeputte et al. 2017), 109–1012 viral particles (Castro-Mejia et al. 2015; Moreno-Gallego et al. 2019), and small quantities of archaea and eukaryotes in per gram of feces (Marchesi 2010). Benefiting from the development of high throughput sequencing techniques (e.g. amplicon or whole-metagenomic sequencing), the gut bacterial community has been well studied over the past years (Turnbaugh et al. 2006; Qin et al. 2010; Lloyd-Price et al. 2017). Gut bacteria was shown to exert profound effects on regulating host metabolism (Pedersen et al. 2016; Wang et al. 2020), and thereby had been linked to host health and diseases (Qin et al. 2012; Quigley 2013). However, as another part of the gut microbial ecosystem, the holistic viral community of enteric microbiome (or ‘gut virome’) was less well characterized (Handley 2016). Viruses have very flexible small genomes ranging from a few to several hundred kilobases (Shkoporov et al. 2019), which corresponds to approximately 1 per cent of the bacterial genome (in average, 2–4 Mbp) (Forster et al. 2019; Zou et al. 2019). The gut virome was predominantly composed of two bacteriophages, double-stranded DNA Caudovirales and single-stranded DNA Microviridae, which constituted over 80 per cent relative abundance of viral populations in the human intestine (Norman et al. 2015). The crAssphage and crAss-like phages, a type of Caudovirales members that characteristically infect Bacteroides spp., represented the highest abundance in healthy human gut (Shkoporov et al. 2018; Yutin et al. 2018). In addition to bacteriophages, eukaryotic viruses, archaeal viruses, and RNA viruses were also important components of gut virome (Minot et al. 2013; Lim et al. 2015).
Due to the low abundance of the virus in the human intestine, routine whole-metagenomic sequencing of feces is not adequate to fully depict the diversity and composition of gut virome (Qin Li et al. 2010). Recently, virus-like particle (VLP) enrichment and subsequently metagenomic sequencing provided a prospective application for fully delineating the gut virome (Hoyles et al. 2014; Kleiner et al. 2015). Based on the VLP technique, studies had shown that the normal gut virome was partly inherited from the mother (Pannaraj et al. 2018; Maqsood et al. 2019), potentially transferred between twins (Moreno-Gallego et al. 2019), and continuously expanded during the first years of life (Lim et al. 2015). In addition, longitudinal analysis revealed that the gut virome of healthy adults was highly diverse, temporally stable, and individually specific (Shkoporov et al. 2019). Disease-induced alterations of the gut virome had also been reported in multiple gastrointestinal and systemic disorders, including colorectal cancer (Hannigan et al. 2018; Nakatsu et al. 2018), inflammatory bowel disease (Norman et al. 2015; Zuo et al. 2019), type I diabetes (Zhao et al. 2017), and coronary heart disease (Guo et al. 2017). These studies suggest a significant role of gut virome in human disease and health. Besides of that, several recent studies had revealed the important role of host properties, including geography, lifestyle, and environment, in shaping the healthy gut virome (Hannigan et al. 2018; Zuo et al. 2020; Camarillo-Guerrero et al. 2021).
By studying the gut microbiome of migrated or short-term visiting peoples, previous studies had shown that their microbiota was markedly remodeled upon environmental change but yet accompanied with maintenance of numerous individual or ethnic microbial characteristics (Deschasaux et al. 2018; He et al. 2018; Vangay et al. 2018; Sun et al. 2020). To characterize the effect size of population heterogeneity on gut viral community, herein, we depicted the compositional differences of gut virome between Chinese residents (n = 24) and visiting Pakistani (n = 24) individuals living in the same city and also examined the repeatability of these differences in their child offsprings (respective n = 6). We quantified the DNA and RNA viromes from fecal VLPs, and parallelly measured the bacterial microbiome for virus–bacteria association analysis. This pilot study provided pieces of evidence for the effect of ethnic backgrounds on human gut virome.
2. Methods
2.1 Subject and sample collection
This study received approval from the ethics committee of Dalian Medical University, and written informed consent was obtained from each participant. The methods were carried out in accordance with the approved guidelines. Thirty healthy Pakistani from Dalian Medical University and thirty BMI-, dietary habit-, alcohol intake-, and frequency of smoking-matched Chinese healthy controls were recruited for this study. Only subjects with stable living and dietary habits were included. The exclusion criteria were 1, subjects with cancer, cardiovascular diseases, family hereditary diseases, communicable diseases, autoimmune diseases, liver cirrhosis, type 2 diabetes, and other systemic diseases; 2, immunosuppressor use within 2 months before enrollment; 3, history of gastrointestinal surgery; 4, tooth extraction or treatment for periodontal disease within 6 months; and 5, pregnancy. Each cohort was consisted of twenty-four healthy adults and six of their healthy child offsprings. Fresh fecal samples were collected from each subject and were immediately stored at an −80 °C freezer.
2.2 Experimental procedures for DNA and RNA viromes
2.2.1 Virus-like particles enrichment
Virus-like particles were enriched from the fecal samples according to the previously described protocol with minor modifications (edited by Andrés Moya 2018). The procedure of VLPs enrichment was performed on ice. One hundred milligrams of fecal sample was suspended in 1 ml HBSS buffer (137 mM NaCl, 5.4 mM KCl, 1.3 mM CaCl2, 0.3 mM Na2HPO4·2H2O, 0.5 mM MgCl2·7H2O, 0.4 mM KH2PO4, 0.6 mM MgSO4·7H2O, 4.2 mM NaHCO3, 5.6 mM D-glucose). Stool suspensions were then cleared by centrifuged twice at 10,000 g for 2 min in 8 °C to remove debris and cells. Supernatants were passed through 0.45-µm followed by 0.22-µm filters to further remove residual host and bacterial cells. The sterile filtrate was mixed with the equal volume of HBSS buffer and centrifugated at 750,000 g (Sorvall mTX150, Thermo Scientific) for 1 h. Any remaining nucleic acid in centrifugal precipitation that was not encapsulated was degraded by treating with a mixture of 2.4 μl TURBO DNase (4.8 U, Invitrogen), 8 μl RNase A/T1 Mix (16 μg RNase A, 40 U RNase T1, Thermo Scientific), and 1 μl Benzonase (5 U, EMD Millipore) followed by heat inactivation of nuclease at 65°C for 10 min. The enriched virus-like particles were used for DNA and RNA extraction immediately.
2.2.2 Viral DNA and RNA extraction
The DNA and RNA of virus were extracted by using TIANamp Virus DNA/RNA Kit (TIANGEN) according to the manufacturer’s protocols. The mixture contained extracted viral DNA, 4 μl viral genome, 1 μl 20 mM random primers D2-8N (5'-AAGCTAAGACGGCGGTTCGGNNNNNNNN-3'), 1 μl 10xRT mix, 1 μl 10 mM dNTP and 11.5 μl DEPC H2O was prepared. To synthesize the first strand of viral DNA, desaturated mixture at 95 °C for 5 min, add Klenow fragment solution (0.15 μl 10× Klenow Buffer, 0.5 μl Klenow fragment, 0.85 μl DEPC H2O) at 37 °C. The procedure should be performed twice to obtain two-strand viral DNA. The extracted RNA was reverse transcribed by using Vazyme HiScript II first-strand cDNA Synthesis Kit (+gDNA wiper) with the same random amplification primer. The two-strand of cDNA could be synthesized by the same approach.
2.2.3 cDNA preparation
Add the mixture contained rSAP and exonuclease-1 into viral two-strand DNA and cDNA at 37°C, respectively, to remove the remained dNTP and primer D2-8N. After 1 h, add 10 μl 5× Q5 Reaction Buffer, 3 μl 50 mM MgCl2, 1.5 μl 10 mM dNTP, 3 μl 20 mM primer D2 (5'-AAGCTAAGACGGCGGTTCGG-3′), 1.25 μl Q5 High-Fidelity DNA Polymerase and 23.25 μl DEPC H2O to amplify the viral DNA and cDNA by polymerase chain reaction. DNA and cDNA were stored at −20 °C freezer. The DNA and RNA concentration and purity were quantified with NanoDrop2000. DNA and cDNA quality were examined with a two per cent agarose gel electrophoresis system.
2.2.4 Shotgun sequencing of viromes
All the DNA and cDNA viral samples were subjected to shotgun metagenomic sequencing by using the Illumina HiSeq 3000 platform. Libraries were prepared with a fragment length of approximately 350 bp. Paired-end reads were generated using 150 bp in the forward and reverse directions.
2.3 Bioinformatic analysis of DNA and RNA viromes
2.3.1 DNA virome assembly, identification, clustering, and taxonomy
The quality control of DNA virome sequences was performed using fastp (Chen et al. 2018), and the high-quality reads were generated by 1, removing low-quality and low-complexity reads and 2, trimming the low-quality bases (Q < 20) at the end of reads and remove the trimmed reads with length <90 bp. The human contamination reads were removed based on Bowtie2 (Langmead and Salzberg 2012) alignment. Each sample was individually assembled using metaSPAdes (kmer = 21,33,55,77) (Nurk et al. 2017). After that, the assembled contigs (>1,000 bp) were identified as viruses when it satisfied one of the following criteria: 1, identified as virus in VIBRANT (Kieft et al. 2020) with default parameters (–virome mode); 2, the number of viral genes exceeded the microbial genes based on searching against the CheckV marker gene set (Nayfach et al. 2020); 3, score >0.9 and P <0.01 in the DeepVirFinder (Ren et al. 2020) (an upgraded version of VirFinder (Ren et al. 2017)). Viral contigs were pairwise blasted and the highly consistent viruses with ninety-five per cent nucleotide identity and eighty per cent coverage of the sequence were further clustered into vOTUs using inhouse scripts. The longest viral contig was defined as representative sequence for each vOTU. Taxonomic assignment of the DNA vOTUs was preformed based on the vConTACT2 pipeline (Bin Jang et al. 2019). Briefly, open reading frames were called from the vOTU sequences using Prodigal (Hyatt et al. 2010) and the resulting proteins were aligned against the vConTACT2 reference. Family-level taxonomy was assigned using a majority-rules approach, where if more than one-third of a vOTU’s proteins were assigned to the same viral family, it was considered part of that viral family.
2.3.2 Macrodiversity and microdiversity of DNA virome
The macrodiversity (Shannon diversity index) of virome was calculated using vegan package in R platform, with a uniformed number of reads (1 million) for each sample. The microdiversity (nucleotide diversity, π) for representative sequence in each vOTU was calculated based on the methodology developed by Schloissnig et al. (2013), and microdiversity of a sample was generated by averaging from the viruses that presented (depth >10×) in that sample.
2.3.3 Functional profiles of DNA virome
The viral proteins were aligned to KEGG (Kanehisa et al. 2017) database (blastp similarity >30%) for functional annotation. For functional profiling, the KEGG aligned proteins were dereplicated with CD-HIT (Li and Godzik 2006) (>95% identity and >90% sequence coverage) to construct the custom viral functional gene catalog, followed by mapping the reads to the catalog using the ‘very-sensitive-local’ setting in Bowtie2 (Langmead and Salzberg 2012). The relative abundance of each functional gene in sample was normalized by the total numbers of viral reads (the reads mapped to the viral sequence) in the sample and was transformed into centered log ratio coordinates using microbiome package in R platform. The carbohydrate-active enzymes and acquired antibiotic resistance genes (ARGs) for the viruses were predicted from the CAZy (Lombard et al. 2014) and CARD (Jia et al. 2017) databases, respectively, using the same manner as functional assignment.
2.3.4 RNA viromes assembly, identification, clustering, and taxonomy
The metatranscriptomic data of RNA virome reads were trimmed using fastp (Chen et al. 2018). The contamination of ribosomal RNA reads was identified and removed by mapping to the small subunit sequences (bacterial 16S and eukaryotic 18S) on the latest SILVA database (Quast et al. 2013). The rnaSPAdes was utilized in metatranscriptomic assembly for each sample (Bushmanova et al. 2019). To identify RNA viruses, the assembled contigs (>500 bp) was aligned to the reference RNA virus proteins downloaded from GenBank database using DIAMOND (blastx e-value <1e−5). We also identified the RNA viral contigs by searching the RNA-dependent RNA polymerase genes (RdRp genes, referred from Evan et al. (Starr et al. 2019)) using a Hidden Markov Model approach (Johnson et al. 2010). Then, the RNA viral sequences were clustered to generate vOTUs based on ninety-five per cent identity and ninety per cent coverage of the sequence. The family assignment of RNA vOTUs was based on the reference database (nucleotide similarity >70% to reference RNA viruses in a family), and the species assignment was based on nucleotide similarity more than ninety-five per cent.
2.4 Bacterial microbiome sequencing and analysis
All raw metagenomic data were trimmed and the human contamination sequences were removed using the same methods in virome. MetaPhlan2 (Truong et al. 2015) was employed to generate the taxonomic profile for each sample using default parameters. Enterotype analysis was performed at the bacterial genus level composition based on the methodology developed by Costea et al. (Costea et al. 2018). The high-quality microbiome data were assembled using metaSPAdes (Nurk et al. 2017), and the resulting contigs was searched against the NCBI-nt database to identity the bacteria sequence (>70% similarity and >70% coverage at the phylum level). To search the potential bacterial host of virus, the CRISPR spacers in bacteria sequence was predicted using MinCED (parameter ‘-minNR 2’) (Bland et al. 2007), and then the spacers were blasted to the viral sequences (‘blastn-short’ mode and bitscore >50) to identify the phage-bacterial host pairs. The matching bacterial host of viral sequence was summarized at the genus level. To avoid ambiguity, the bacterial genus producing the highest number of spacers hits was considered as the primary host. These procedures were used for host assignment of both DNA and RNA viruses.
2.5 Statistical analysis
Statistical analyses were implemented at the R platform (https://www.r-project.org/). Permutational multivariate analysis of variance (PERMANOVA) was performed with the adonis function of the vegan package, and the adonis P-value was generated based on 1,000 permutations. The method of effect size analysis was referred as Wang et al. (2020). The no-metric multidimensional scaling (NMDS) analysis was used as the ordination methods (metaMDS function in vegan package) for compositional data. The Procrustes coordinates analysis and significance were generated using the procuste and procuste.randtest functions in vegan package. The principal component analysis (PCA) was performed and visualized using the ade4 package. The Wilcoxon rank-sum test was used to measure statistical differences in diversity and taxonomic levels between two cohorts. P-values were corrected for multiple testing using the Benjamini–Hochberg procedure.
3. Results
This study included thirty Chinese residents and thirty visiting Pakistani individuals who were recruited at Dalian Medical University in March 2019. Both cohorts consisted of twenty-four healthy adults and six of their child offsprings (Table 1). All adults were students or young teachers of the Dalian Medical University, and the Pakistani adults and children had arrived in China for 0–18 months (average of 11 months) and 0–15 months (average of 9 months), respectively. Notably, the Chinese and Pakistani adults showed significant differences in their body mass index (BMI), dietary habit, and drinking and smoking rates (Table 1 and Supplementary Table S1), which seemed to be due to ethnic and lifestyle differences.
Table 1.
Characteristics of the subjects.
| Adults |
Children |
|||||
|---|---|---|---|---|---|---|
| Chinese | Pakistani | P-value | Chinese | Pakistani | P-value | |
| Number of subjects | 24 | 24 | 6 | 6 | ||
| Sex, F/M | 1/23 | 1/23 | 1.000 | 3/3 | 3/3 | 1.000 |
| Age, years | 26.0 ± 4.3 | 29.1 ± 3.7 | 0.011 | 2.8 ± 1.8 | 2.8 ± 1.7 | 1.000 |
| Weight, kg | 69.6 ± 11.0 | 76.7 ± 15.6 | 0.076 | 14 ± 5.4 | 13.3 ± 4.2 | 0.794 |
| BMI, kg/m2 | 22.8 ± 2.8 | 25.6 ± 4.5 | 0.011 | 16.0 ± 2.0 | 17.4 ± 3.1 | 0.396 |
| Drinking, % | 50% | 8.3% | 0.003 | 0% | 0% | 1.000 |
| Smoking, % | 16.7% | 33.3% | 0.030 | 0% | 0% | 1.000 |
| Antibiotics (≤2 mo), % | 8.3% | 8.3% | 1.000 | 0% | 0% | 1.000 |
| Prebiotics (≤2 mo), % | 58.3% | 41.7% | 0.387 | 66.7% | 50% | 1.000 |
| Living in China, mons | 11 ± 4 | 9 ± 6 | ||||
The data for age, weight, and BMI were presented as mean ± SD. P-values for age, weight, and BMI were calculated by Student’s t-test, and for sex, drinking, smoking, antibiotics, and prebiotics were calculated by Fisher’s exact test.
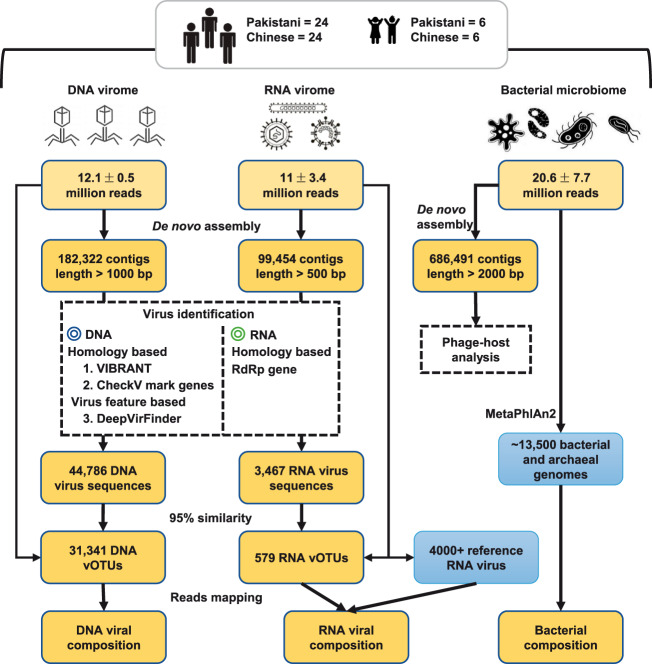
Fecal samples of all participants were collected and treated using a unified approach (see Section 2). To depict the gut viral characteristics of healthy individuals, we extracted DNA and RNA from fecal VLP factions and performed high throughput shotgun sequencing using the Illumina platform. To extend the content of total microbial community, the bacterial microbiome of feces was also profiled using whole-metagenomic sequencing. The analytical workflow of the DNA virome, RNA virome, and bacterial microbiome is shown in Fig. 1. Focusing on the comparison of gut viromes between Chinese and Pakistani individuals, overall, this study included six sections to elaborate the results:
Figure 1.
Overview of the workflow for analyzing of DNA virome, RNA virome, and bacterial microbiome.
1–2. DNA virome and its functional characteristics.
3–4. RNA virome and the concordance between DNA and RNA viromes.
5–6. Bacterial microbiome and the virus-bacteria associations.
3.1 Comparison of DNA viral community
We obtained 782 million high-quality non-human reads (12.1 ± 0.5 million per sample) through shotgun sequencing of the DNA viral community of sixty fecal samples. The reads were de novo assembled into 182,322 contigs with the minimum length threshold of 1 kbp, of which 24.6 per cent (44,786) were recognized as fragments with viral signals based on their sequence features and homology to known viral genomes (Fig. 1). Although the proportion of the viral contigs was low, on average, 72.1 per cent of sequencing reads in all samples were captured by the viral contigs (Supplementary Fig. S1a), revealing well representativeness of the high-abundance viral contents in human gut DNA virome. The viral contigs were further clustered into 31,341 ‘viral operational taxonomic units (vOTUs)’ (a phylogenetic definition of a discrete viral lineage that corresponds to ‘species’ in prokaryotes, also named ‘viral population’ (Gregory et al. 2019)) by removing the redundant contigs of ninety-five per cent nucleotide similarity. These vOTUs represented an average size of 3,836 ± 7,703 bp (Supplementary Fig. 1b), which was comparable with similar studies (Shkoporov et al. 2019) but remarkable lower than that of the available viral genomes (average 38.5 kbp for ∼6,500 complete virus isolates from the RefSeq database), suggesting that the vOTUs were mostly fragmented genomes. Only 43.3 per cent of vOTUs could be annotated into a specific family, highlighting a considerable novelty of gut virome.
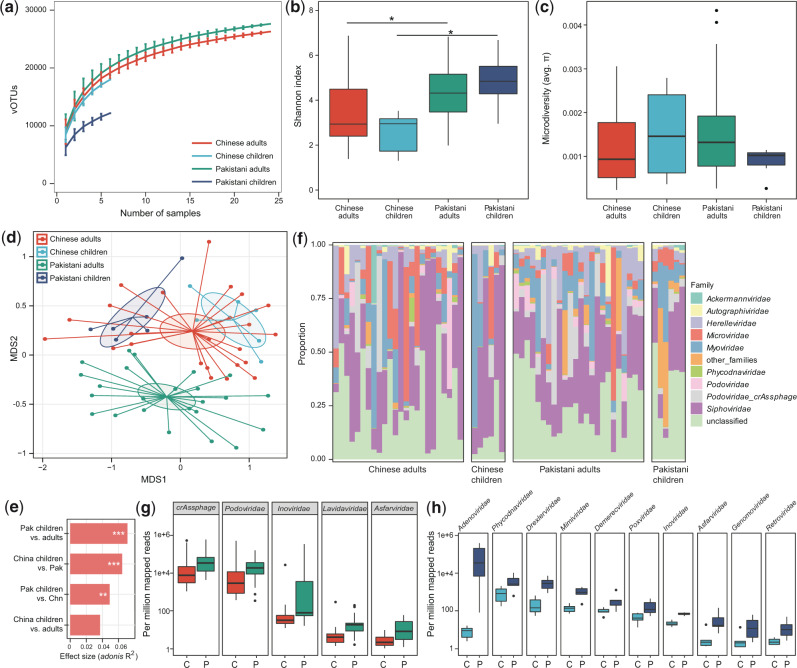
Rarefaction analysis showed that 1, the rarefaction curve was approximately saturated under the number of twenty samples in each group, and 2, the vOTU richness was significantly higher in Pakistani adults than in Chinese adults (P = 0.008, Fig. 2a). The within-sample diversity pattern of gut DNA viromes was assessed by macrodiversity (Shannon index) and microdiversity (nucleotide diversity or π (Gregory et al. 2019)) at the vOTU level. The Chinese adults showed a lower Shannon index than the Pakistani adults, similarly for the children (Fig. 2b), but no significant difference in microdiversity was detected between Chinese and Pakistanis (Fig. 2c).
Figure 2.
Differences in gut DNA virome between Chinese and Pakistanis. (a) Rarefaction curve analysis of number of vOTUs on each group of samples. The number of identified vOTUs in different groups is calculated based on a randomly selected specific number of samples with 30 replacements, and the median and quartiles numbers are plotted. Boxplot shows the macrodiversity (b) and microdiversity (c) that differ among four groups. The significance level in the Student’s t test is denoted as: *q < 0.05, **q < 0.01. (d) NMDS analysis based on the composition of virome, revealing the separations between different groups. The location of samples (represented by nodes) in the first two multidimensional scales are shown. Lines connect samples in the same group, and circles cover samples near the center of gravity for each group. (e) PERMANOVA analysis reveals that the virome of Pakistani children are more similar with the Chinese subjects. The effect sizes and P-values of the adonis analysis are shown. (f) Composition of gut virome at the family level. Boxplot shows the differential viral families of adults (g) and children (h) when compared between Chinese and Pakistanis. C, Chinese individuals; P, Pakistani individuals. For boxplot, boxes represent the interquartile range between the first and third quartiles and median (internal line); whiskers denote the lowest and highest values within 1.5 times the range of the first and third quartiles, respectively; and nodes represent outliers beyond the whiskers.
Next, we undertook an NMDS analysis to further understand the differences in fecal DNA viral communities between Chinese and Pakistanis. Clear separations were revealed in the viromes of both adults and children between Chinese and Pakistanis (q < 0.05 for both adults and children; Fig. 2d). Notably, we also found that 1, the viral communities of Chinese adults and children were similar, but those of Pakistani adults and children were different and 2, the viral communities of Pakistani children were closer to Chinese subjects when compared with those of Pakistani adults. PERMANOVA also revealed that nationality and age are the main reasons for shaping the gut DNA virome and validated the fashion of virome differences at Pakistani adults/children vs. Chinese adults/children (Fig. 2e).
We finally compared the DNA virome composition of Chinese and Pakistani at the family level, ignoring the family-level unclassified vOTUs (which represented only 26.1 per cent of total sequences). The most dominant viral families in all samples were Siphoviridae (average relative abundance, 31.0 ± 5.1%), Myoviridae (11.2 ± 2.4%), and Microviridae (8.1 ± 2.1%) (Fig. 2f). Compared with the Chinese adults, the viral communities of the Pakistani adults showed a significant increase in Podoviridae_crAssphage, Podoviridae, Inoviridae, Lavidaviridae, and Asfarviridae (Mann–Whitney U test, q < 0.05; Fig. 2g). Ten viral families, including Adenoviridae, Phycodnaviridae, Drexlerviridae, Inoviridae, and so on, were significantly higher in viral communities of Pakistani children (Fig. 2h), as compared with the Chinese children.
3.2 Functional analysis of DNA virome
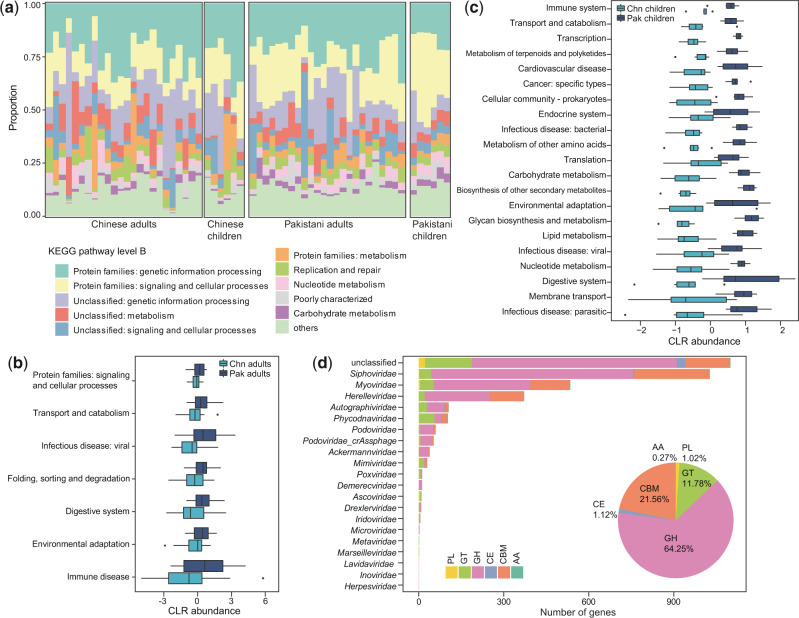
To better elucidate the functional capacity of the DNA viromes, we predicted a total of 166,098 protein-coding genes from the vOTUs (average of 5.3 genes per vOTU) and annotated functions of 11.7 per cent of these genes based on the KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa et al. 2017) database. Analysis on KEGG pathway level B showed that functions involved in genetic information procession and signal and cellar processes are dominant in all samples (Fig. 3a), suggesting that these are core functions of the gut DNA virome. Compared with the Chinese adults, viral-encoded functions in the Pakistani adults were significantly enriched in viral infectious and immune diseases, environmental adaptation, and several categories of signaling and cellular processes (transport and catabolism, folding, sorting, and degradation) (Mann–Whitney U test, q < 0.05; Fig. 3b). For example, a putative hemolysin enzyme (K03699) that encoded by several Myoviridae and Siphoviridae viruses showed over ten-fold enrichment in the virome of Chinese adults compared to that of Pakistani adults. When compared with the Chinese children, several important functions, including membrane transport, lipid metabolism, metabolism of terpenoids and polyketides, biosynthesis of other secondary metabolites, and transport and catabolism, were significantly higher in the viral communities of Pakistani children (Fig. 3c).
Figure 3.
Comparison of DNA viral functions between Chinese and Pakistanis. (a) Composition of viral functional categories at the KEGG pathway level B. b-c, Boxplot shows the KEGG pathways that differed in abundance between Chinese adults and Pakistani adults (b) and between Chinese children and Pakistani children (c). Boxes represent the interquartile range between the first and third quartiles and median (internal line); whiskers denote the lowest and highest values within 1.5 times the range of the first and third quartiles, respectively; and nodes represent outliers beyond the whiskers. (d) The taxonomic distribution of CAZymes. GH, glycoside hydrolase; GT glycosyltransferase; CBM, carbohydrate-binding module; CE, carbohydrate esterase; PL, polysaccharide lyase; AA auxiliary activity.
We identified a total of 3,484 CAZymes (Carbohydrate-active enzymes (Lombard et al. 2014)) from the viral genes, including 2,239 glycoside hydrolases, 751 carbohydrate-binding modules, 410 glycosyltransferases, 39 carbohydrate esterases, 35 polysaccharide lyases, and 10 auxiliary activities (Fig. 3d). The majority of CAZymes were encoded by unclassified vOTUs (31.6%) and Siphoviridae (29.5%), followed by Myoviridae (15.3%) and Herelleviridae (10.7%), suggesting their important roles in carbohydrate metabolism in the gut viral ecosystem. Moreover, we also identified 31 acquired ARGs from the DNA vOTUs (Supplementary Table S2). Most of these ARGs were related to rifampicin resistance, aminoglycoside resistance, macrolide-lincosamides-streptogramin resistance, and beta-lactamase. Taken together, these findings revealed that the DNA virus can widely express the carbohydrate metabolism-associated genes and are potentially involved in carrying and transmission of ARGs.
3.3 Comparison of RNA viral community
For RNA virome, we performed shotgun metatranscriptomic sequencing of sixty fecal samples described above and obtained 671 million reads (11 ± 3.4 million per sample) after removing the low-quality reads and bacterial ribosomal RNA contamination. A total of 99,454 contigs with minimum length threshold of 500 bp were assembled, 3,467 (3.5%) of which were identified as highly credible RNA viral fragments via blasting against the available RNA viral genomes and searching of the RNA-dependent RNA polymerase (RdRp) sequences (Fig. 1). 25.4% of these RNA viruses contained at least one RdRp gene, while twenty-eight viral RdRp genes had no homology with any known virus in the NCBI database. We obtained 579 RNA vOTUs based on clustering at ninety-five per cent nucleic acid level similarity. The average size of these vOTUs was 1,166 ± 915 bp, which was fragmented compared with the available RNA viral genomes (average 7.4 kbp from ∼4,000 isolates). Furthermore, considering that only average of 24.9 per cent reads of all samples were covered from the RNA vOTUs, we also used the available RNA viral genomes from the RefSeq database as a reference for analyzing the gut RNA virome. One hundred eighteen available RNA viruses were observed in our samples, which covered additional 1.3 per cent reads (on average) for further analysis.
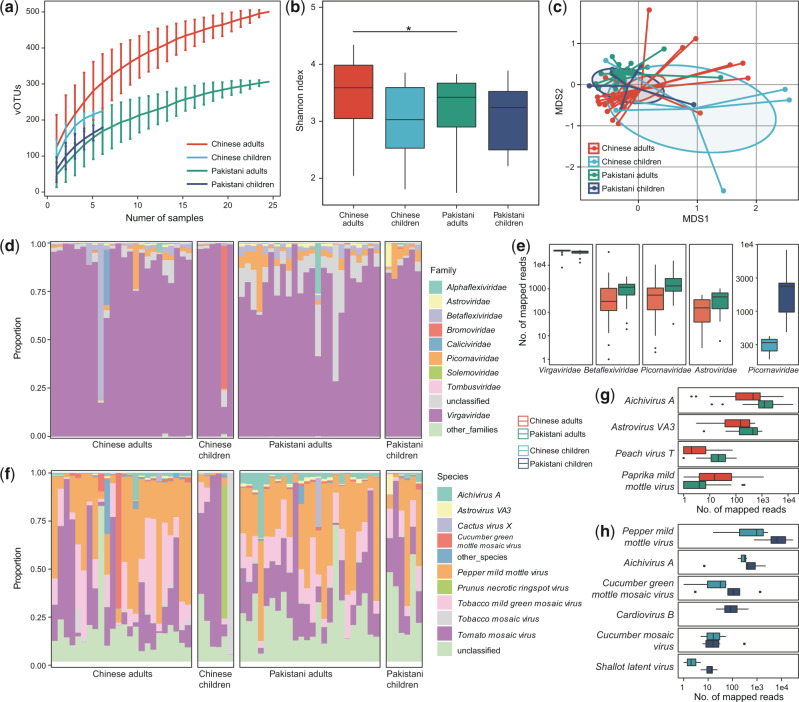
Rarefaction analysis showed that the detection of RNA virus was increased with the number of samples, and the accumulative curve was nearly saturated at nearly ten samples (Fig. 4a). This is due to our RNA virus pipeline mainly focused on the known species and the sequence containing an RdRp gene, but high proportions of viruses remain untagged and many of them are independent of the RdRp gene (Wolf et al. 2018). Compared with Pakistanis, the macrodiversity (Shannon index) was significantly higher in Chinese adults, but there was no statistical difference in that of children (Fig. 4b).
Figure 4.
Differences in gut RNA virome between Chinese and Pakistanis. (a) Rarefaction curve analysis of number of vOTUs on each group of samples. The number of identified vOTUs in different groups is calculated based on a randomly selected specific number of samples with 30 replacements, and the median and quartiles numbers are plotted. (b) Boxplot shows the Shannon diversity index among four groups. The significance level in the Student’s t test is denoted as: *q < 0.05; **q < 0.01. (c) NMDS analysis based on the composition of virome, revealing the separations between different groups. The location of samples (represented by nodes) in the first two multidimensional scales are shown. Lines connect samples in the same group, and circles cover samples near the center of gravity for each group. (d) Composition of gut virome at the family level. (e) Boxplot shows the differential viral families between Chinese and Pakistanis. (f) Composition of gut virome at the species level. Boxplot shows the differential viral families of adults (g) and children (h) when compared between Chinese and Pakistanis. For boxplot, boxes represent the interquartile range between the first and third quartiles and median (internal line); whiskers denote the lowest and highest values within 1.5 times the range of the first and third quartiles, respectively; and nodes represent outliers beyond the whiskers.
NMDS analysis on the overall RNA vOTUs composition captured significant separation of adults between Chinese and Pakistanis (adonis P < 0.001; Fig. 4c), but of children the separation was visible but not significant (adonis P = 0.2). Likewise, the viral communities of Chinese adults and children were closer, yet of Pakistani adults and children.
Finally, to investigate the gut RNA viral signatures between Chinese and Pakistanis, we compared two cohorts on viral composition. At the family level, the dominant family Virgaviridae consisted of an average 83.7 per cent relative abundance in all samples (Fig. 4d), which was slightly but significantly enriched in Chinese adults compared with that in Pakistani adults (Fig. 4e). Three other families, Betaflexiviridae, Picornaviridae, and Astroviridae, were reduced in Chinese adults than in Pakistani adults (Mann–Whitney U test, q < 0.05 for all), while Picornaviridae was also reduced in Chinese children than in Pakistani children. At the species level, the plant-associated virus, including Pepper mild mottle virus (average relative abundance, 29.1 ± 4.9%), Tomato mosaic virus (25.7 ± 6.3%), and Tobacco mild green mosaic virus (13.9 ± 1.5%), composed of the dominant species in all samples (Fig. 4f). Compared with the Chinese adults, the viral communities of the Pakistani adults showed a significant increase of Aichivirus A, Astrovirus VA3, and Peach virus T, and a remarkable depletion of Paprika mild mottle virus (Fig. 4g). When compared with the Chinese children, six species were significantly higher in viral communities of Pakistani children (Fig. 4h), with no species that was lower.
3.4 Concordance between DNA and RNA viromes
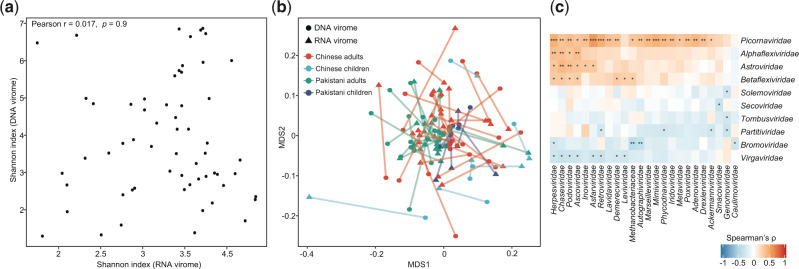
Having characterized the differences of DNA and RNA viromes between local Chinese residents and visiting Pakistanis, we wanted to examine the existence of concordance between DNA and RNA viromes. Although the DNA and RNA viromes were irrelevant in the Shannon diversity index (Pearson r = 0.02, P = 0.9; Fig. 5a), the overall compositions of the two types of viral community were strongly correlated (Procrustes correlation M2= 0.37, P < 0.001; Fig. 5b). And this correlation was reproducible across nationality and age. Moreover, we identified fifty-sixco-abundance correlations between twenty-four DNA and ten RNA viral families (Spearman correlation test q < 0.05; Fig. 5c), including some positive correlations between Adenoviridae and several RNA viruses and a negative correlation between Herpesviridae and Tombusviridae. The significance of these relationships required further studies.
Figure 5.
Correlations between DNA and RNA viromes. (a) Relationship of microdiversity between DNA and RNA virome. (b) Procrustes analysis of DNA virome versus RNA viromes. Samples for DNA and RNA viromes are shown as circles and blue triangles, respectively; and samples from the same individual are connected by lines. Colors represent samples belong to different groups. (c) Heatmap shows the co-abundance correlations between DNA and RNA viral families. The significance level in the Spearman correlation test is denoted as: *q < 0.05; **q < 0.01; ***q < 0.001.
3.5 Comparison of bacterial microbiome
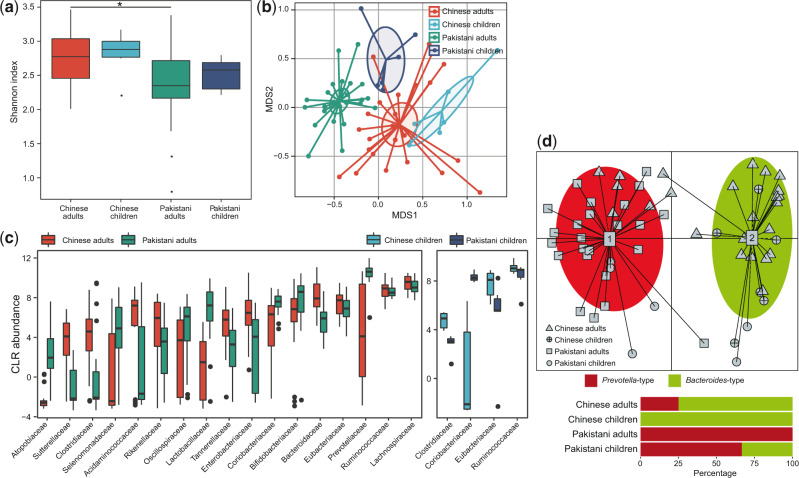
For the bacterial microbiome, we obtained a total of 1.2 billion reads (20.6 ± 7.7 million per sample) from the samples and quantified the relative abundances of a total of 833 taxa, including 12 phyla, 22 classes, 41 orders, 81 families, 179 genera, and 498 species, using MetaPhlAn2 (Truong et al. 2015). Comparison on Shannon index showed that the bacterial microbiome of Chinese adults exhibited significantly higher diversity than that of the Pakistanis (Fig. 6a), similarly but not significant trend was observed in that of children. NMDS analysis on the overall bacterial composition also revealed significant separation between Chinese and Pakistan adults (adonis P < 0.001; Fig. 6b), as well as between Chinese and Pakistan children (adonis P < 0.001). Consistent with the observations in DNA and RNA viromes, the bacterial microbiome of Pakistan children was also close to that of Chinese subjects in tendency.
Figure 6.
Differences in gut bacterial microbiome between Chinese and Pakistanis. (a) Boxplot shows the Shannon diversity index among four groups. The significance level in the Student’s t test is denoted as: *q < 0.05; **q < 0.01. (b) NMDS analysis based on the composition of bacterial microbiome, revealing the separations between different groups. The location of samples (represented by nodes) in the first two multidimensional scales are shown. Lines connect samples in the same group, and circles cover samples near the center of gravity for each group. (c) Boxplot shows the bacterial families that differed in abundance between two cohorts. Boxes represent the interquartile range between the first and third quartiles and median (internal line); whiskers denote the lowest and highest values within 1.5 times the range of the first and third quartiles, respectively; and nodes represent outliers beyond the whiskers. (d) Enterotype analysis of bacterial microbiome samples. The upper panel show the PCA of all samples, revealing the separation between two enterotypes. The lower panel show the composition of enterotypes in four groups.
Taxonomically, the bacterial microbiome of Chinese adults showed significant enrichment of Lachnospiraceae, Ruminococcaceae, Eubacteriaceae, Enterobacteriaceae, Tannerellaceae, Rikenellaceae, Acidaminococcaceae Clostridiaceae, and Sutterellaceae and depletion of Prevotellaceae, Bifidobacteriaceae, Coriobacteriaceae, Lactobacillaceae, Oscillospiraceae, Selenomonadaceae, and Atopobiaceae, compared with that of Pakistani adults (linear discriminant analysis [LDA] score >3; Fig. 6c). Similarly, Clostridiaceae, Eubacteriaceae, and Ruminococcaceae were enriched in Chinese children compared to Pakistani children, and Coriobacteriaceae was depleted. At the species level, the Chinese adults exhibited twenty-eight enriched bacterial species and nineteen decreased species when compared with the Pakistani adults, while the Chinese children showed eleven enriched species and twelve decreased species compared with the Pakistani children (Supplementary Table S3). The exhibition of enormous differential taxa led to a dramatic distinction of enterotype constitution between Chinese and Pakistanis. The Chinese subjects were characterized by a high proportion of Bacteroides -type (75% and 100% in adults and children, respectively), whereas almost of all Pakistani subjects were Prevotella-type (100% in adults and 66.7% in children) (Fig. 6d).
3.6 Virus–bacteria associations
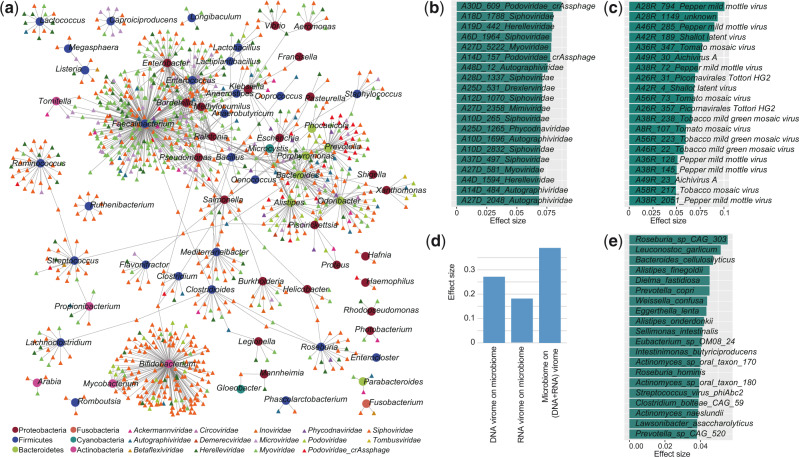
To study the virus-bacteria correlation, first, we predicted the bacterial hosts of the viruses by searching the potential viral CRISPR spacers from bacterial metagenomic assemblies (see Methods). This approach allowed host assignments for 4,143 DNA and 38 RNA vOTUs, representing 13.2 per cent and 6.6 per cent of all DNA and RNA viruses, respectively. A large connection network of viruses and its bacterial host was shown at Fig. 7a. Members of Faecalibacterium, Bifidobacterium, Prevotella, Ruminococcus, Enterobacter, and Alistipes were the most common host for human gut virome. The Siphoviridae and Myoviridae had infected the highest number of bacteria, while the crAss-like phages were more likely to infect Bacteroidetes members such as Prevotella and Bacteroides.
Figure 7.
Associations between virome and bacterial microbiome. (a) Host range of viruses predicted through CRISPR spacer matches. Circles and tringles represent the bacteria and viruses, respectively; and the colors represent their taxonomic assignment at the phylum (for bacteria) or family (for viruses) levels. The twenty DNA (b) and RNA families (c) for which the highest effect size that significant impact the bacterial microbiome communities. (d) The combined effect size of viruses on bacterial microbiome as well as bacteria on virome. To calculate the combined effect size, a set of non-redundant covariates (DNA vOTUs, RNA vOTUs, or bacterial species) is selected from the omic datasets, and then the accumulated effect size is calculated by adonis analysis using these selected covariates. (e) The twenty bacterial species with highest effect size for impacting the combined DNA and RNA viral communities.
Then, we performed the PERMANOVA-based effect size analysis between gut virome and microbiome. 226 DNA vOTUs (q < 0.05), including members of Siphoviridae, Phycodnaviridae, and Podoviridae-crAssphage, and twenty RNA vOTUs (q < 0.05) explained significant variance on the bacterial microbiome communities (Fig. 7b, c). More importantly, combination of these DNA and RNA vOTUs explained 27.2 per cent and 18.2 per cent of the microbiome variance, respectively (Fig. 7d), suggesting that the effect size of the gut virome on bacterial microbiome is considerable. Parallelly, fifty-two bacterial species were identified that significantly impact the holistic composition of DNA and RNA viromes, accounting for 39.0 per cent virome variance (Fig. 7d). These species included Roseburia sp. CAG-303, Leuconostoc garlicum, Bacteroides cellulosilyticus, Alistipes finegoldii, and Prevotella copri (Fig. 7e).
4. Discussion
Both ethnic origin and residential environment have unnegligible effects on individual’s gut microbiome (Gupta et al. 2017; Deschasaux et al. 2018; Gaulke and Sharpton 2018, Korpela et al. 2018). To extend this finding on gut virome, our study focused on the viral community of a cohort of Chinese and visiting Pakistanis. Despite sharing the residential environment, the viral diversity and composition of Chinese and Pakistanis were dramatically differed, suggesting that the nationality-specific characteristics of virome enable to maintain over an extended period (average 11 and 9 months for Pakistani adults and children, respectively). This result was in accordance with an earlier study showing that the individual characteristics of gut virome can be relatively stable for at least 1 year (Shkoporov et al. 2019).
Using de novo assembly and viral discovery approaches, we identified a huge number of viruses from the subjects’ fecal samples, including 31,341 non-redundant complete and partial DNA viral genomes and 579 non-redundant RNA viruses, particularly the number of DNA vOTUs increased over eightfold compared with the isolated viral sequences in RefSeq database. The majority of viruses were unclassified even at the family level, in agreement with previous observations of extensive novelty of viral world in multiple environments as well as in the human gut (Reyes et al. 2012; Paez-Espino et al. 2016; Rampelli et al. 2017; Camarillo-Guerrero et al. 2021).
The DNA viral macrodiversity of Chinese adults was lower than that of Pakistani adults, whereas an opposite phenomenon was observed in the diversity of bacterial community. This result conflicted with the observation in US adults, which exhibited a strong correlation between gut virome and microbiome diversities (Moreno-Gallego et al. 2019). As most of the DNA viruses were bacteriophages (in this study, the bacterial hosts of at least 7.2% DNA viruses were verified) (Moreno-Gallego et al. 2019), the bacterial microbiome may act as a key factor for virome diversity. The explanation for high DNA viral diversity in Pakistani adults was unknown, but the reason for the enrichment of some eukaryotic viruses in their gut was speculated (see the following discussion). In contrast to DNA virome, the RNA viral diversity was higher in Chinese adults than in Pakistani adults. This observation could be due to the difference of dietary habits between the two groups, as the gut RNA viruses were generally plant-associated in our cohort.
Significant compositional differences were observed in DNA and RNA viromes, so were bacterial microbiomes between Chinese residents and visiting Pakistanis. In DNA virome, the Pakistani adults showed remarkable enrichment of two representative viral families, Podoviridae_crAssphage and Podoviridae. crAss-like phages were the most abundant human-associated viruses, and recent studies had shown it is globally distributed with distinct distribution patterns which appear to be strongly influenced by human lifestyles (Guerin et al. 2018; Camarillo-Guerrero et al. 2021); while some members of Podoviridae were associated with gut disorders such as inflammatory bowel disease(Clooney, Sutton et al. 2019). Adenoviridae was highly abundant in the gut of Pakistani children but was rare in that of Chinese children, whereas some members of Adenoviridae were prevalent human-associated eukaryotic viruses that can cause respiratory infection, gastroenteritis, and multi-organ diseases (Wadell 1988; Centers for Disease and Prevention 2007; Jones et al. 2007). Inoviridae was enriched in both Pakistani adults and children when compared with Chinese subjects, suggesting potential transmission of such viruses from Pakistani parents to their offsprings. In RNA virome, some members of the plant-associated virus Virgaviridae were enriched in Pakistanis but some others were reduced. This finding was thought to be connected to the difference in dietary habits between two cohorts. For example, the abundance of Shallot latent virus was higher in Pakistani adults than in Chinese adults, as the shallot (e.g. onion, leek) is commonly used in halal foods in the school canteen but rarely appeared in Chinese foods (based on the authors' experience). In addition, some members of the Pakistani adult-enriched Picornaviridae, including Picornavirales Tottori-HG2, Enterovirus C, and Cosavirus A, and Astroviridae were well-known human enteroviruses that can cause diarrhea and enteric infections (Johnston et al. 1993; Monroe et al. 1993; Rotbart 2002). In the bacterial microbiome, the enterotype distribution of Chinese and Pakistanis was deviated, characterized by a high proportion of Bacteroides -type (associated with diets enriched animal carbohydrates (De Filippo et al. 2010; David et al. 2014)) and low proportion of Prevotella-type (associated with plant fiber-enriched diets (Costea et al. 2018)) in Chinese subjects. Combination of these findings suggested that dietary habits may be a key driver for shaping the gut RNA virome and bacterial microbiome. Of course, more proof-of-principle studies are needed in the future.
One striking observation was that the DNA virome of Pakistan children is closer to that of Chinese subjects when compared with the degree of deviation between Chinese and Pakistan adults. This phenomenon was also observed in RNA virome and bacterial microbiomes in tendency. These findings suggested that the virome and microbiome of children were more changeable than that of adults, even though the Pakistan adult participants seemed to live a bit longer in China. In accordance with the previous studies, the infant or child gut microbiome was less stable under the changes of environment, dietary pattern, and antibiotic usage (Koenig et al. 2011; Chu et al. 2016; Yassour et al. 2016). In addition, dynamic development of the infant gut virome towards a more stable adult-like gut virome was also confirmed by recent studies (Lim et al. 2015; Reyes et al. 2015; Beller and Matthijnssens 2019; Gregory, Zablocki et al. 2020).
We characterized the functional capacity of gut virome by identifying over 53,000 KEGG annotated protein-coding genes and found that the majority of viral-encoded functions were highly enriched genetic information processing signaling and cellular processes, in agreement with the findings in recent studies (Ma et al. 2018; Nayfach et al. 2020). Different from the observation in DNA viral composition, the Chinese adults revealed a more diver functional profile than that of the Pakistani adults, as revealed by more metabolism-associated genes in Chinese adults. In addition to general functions, we also identified over 11,000 CAZymes and 37 ARGs from all DNA viruses. To the best of our knowledge, the appearance of extensive CAZymes in gut virome was first found in this study. Potential viral contributions to complex carbon degradation were validated in ocean and soil ecosystems (Suttle 2007; Emerson et al. 2018). Thus, our findings further highlight the importance of viral carbohydrate metabolism capacity in the human gut. Moreover, the virus-encoded ARGs were also directly relevant to human health, consistent with previous studies (Ogilvie et al. 2013).
Not only bacteriophages but also free-living viruses in the human gut can influence bacterial microbiome structure and therefore indirectly affect health status (Foca et al. 2015; Scarpellini et al. 2015). We confirmed remarkable connections between viruses and bacterial hosts in our study cohort, including the previous-known parasitic relations (e.g. crAss-like phages and Bacteroidetes members (Guerin, Shkoporov et al. 2018, Shkoporov et al. 2018)) and many novel connections. Noticeably, the Pakistani-dominated genus Prevotella connected the largest number of viruses and was responsible for a large part of variance in the virome composition, in agreement with the previous studies showing that the high relative level of Prevotella lead to a higher prevalence of temperate bacteriophages and increased virome macrodiversity (Shkoporov et al. 2019). On the other hand, we also statistically revealed that the gut virome was also an important determinator of the bacterial microbiome.
As all participants shared the residential environment, we were only able to study the effect of nationality on their gut virome. Through collecting samples from the visiting Pakistani before they arrived in China or from other local Pakistani residents, future research is believed to confirm the effect of the environment on gut virome. Other limitations in this study included 1, the relatively small sample size; 2, the lack of longitudinal sampling for the individuals; and 3, the inadequacy of viral reference database. These limitations did not affect the robustness of results in the current cohort, but follow-up studies in wider populations will still complement some deficiencies of the current study and provide more new findings.
5. Conclusions
In summary, we systematically described the baseline gut DNA virome, RNA virome, and bacterial microbiome in a well-characterized cohort of Chinese and visiting Pakistanis and demonstrated that the national background contributed a primary variation to gut viral and bacterial communities. We also revealed numerous interconnections between DNA and RNA viruses, as well as between viruses and bacteria. The physiological mechanisms underlying the difference between the two cohorts remain unclear, but the ethnic factor must be proposed and considered in designing future studies of the virome.
Data availability
The raw sequencing dataset acquired in this study has been deposited to the NCBI SRA database under the accession code PRJNA641593. The sample metadata, vOTU and taxonomic composition data, and the statistical scripts are available from the corresponding authors on reasonable request.
Supplementary data
Supplementary data are available at Virus Evolution online.
Funding
This work was supported by grants from the Priority Academic Program Development of Jiangsu Higher Education Institutions (Integration of Chinese and Western Medicine), the National Natural Science Foundation of China (No. 81902037) and the Liaoning Provincial Natural Science Foundation (No. 20180530086).
Author Contributions
T.M., S.L., Y.M., and Q.Y. conceived and directed the study. Q.Y., Y.W., X.C., G.W., T.A., and X.L. developed and conducted the experiments. Q.Y., G.W., and T.A. performed sample collection and investigation. H.J., K.G., Y.Z., and P.Z. carried out data processing and analyses. S.L., Q.Y., and T.M. drafted the manuscript. Y.M., G.W., Y.L., J.W., G.C., A.Z., and P.L. participated in design and coordination, and helped draft the manuscript. P.Z., Y.S., and M.X. revised the manuscript. All authors read and approved the final manuscript.
Conflict of interest
None declared.
Supplementary Material
References
- Beller L., Matthijnssens J. (2019) ‘ What is (Not) Known about the Dynamics of the Human Gut Virome in Health and Disease’, Current Opinion in Virology, 37: 52–7. [DOI] [PubMed] [Google Scholar]
- Bin Jang H. et al. (2019) ‘ Taxonomic Assignment of Uncultivated Prokaryotic Virus Genomes is Enabled by Gene-Sharing Networks’, Nature Biotechnology, 37: 632–9. [DOI] [PubMed] [Google Scholar]
- Bland C. et al. (2007) ‘ CRISPR Recognition Tool (CRT): A Tool for Automatic Detection of Clustered Regularly Interspaced Palindromic Repeats’, BMC Bioinformatics, 8: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushmanova E. et al. (2019) ‘ rnaSPAdes: A De Novo Transcriptome Assembler and Its Application to RNA-Seq Data’, GigaScience, 8:giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarillo-Guerrero L. F. et al. (2021) ‘ Massive Expansion of Human Gut Bacteriophage Diversity’, Cell, 184: 1098–109.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Mejia J. L. et al. (2015) ‘ Optimizing Protocols for Extraction of Bacteriophages Prior to Metagenomic Analyses of Phage Communities in the Human Gut’, Microbiome, 3: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) (2007) ‘ Acute Respiratory Disease Associated with Adenovirus Serotype 14–Four States, 2006-2007’, MMWR Morb Mortal Wkly Rep, 56: 1181–4. [PubMed] [Google Scholar]
- Chen S. et al. (2018) ‘ Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor’, Bioinformatics (Oxford, England), 34: i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D. M. et al. (2016) ‘ The Early Infant Gut Microbiome Varies in Association with a Maternal High-Fat Diet’, Genome Medicine, 8: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clooney A. G. et al. (2019) ‘ Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflammatory Bowel Disease’, Cell Host & Microbe, 26: 764–78.e765. [DOI] [PubMed] [Google Scholar]
- Costea P. I. et al. (2018) ‘ Enterotypes in the Landscape of Gut Microbial Community Composition’, Nature Microbiology, 3: 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L. A. et al. (2014) ‘ Diet Rapidly and Reproducibly Alters the Human Gut Microbiome’, Nature, 505: 559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C. et al. (2010) ‘ Impact of Diet in Shaping Gut Microbiota Revealed by a Comparative Study in Children from Europe and Rural Africa’, Proceedings of the National Academy of Sciences of the United States of America, 107: 14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschasaux M. et al. (2018) ‘ Depicting the Composition of Gut Microbiota in a Population with Varied Ethnic Origins but Shared Geography’, Nature Medicine, 24: 1526–31. [DOI] [PubMed] [Google Scholar]
- Andrés Moya V. P. B. (ed.) (2018) The Human Virome: Methods and Protocols. New York, NY: Humana Press. [Google Scholar]
- Emerson J. B. et al. (2018) ‘ Host-Linked Soil Viral Ecology along a Permafrost Thaw Gradient’, Nature Microbiology, 3: 870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foca A. et al. (2015) ‘ Gut Inflammation and Immunity: What is the Role of the Human Gut Virome?’, Mediators Inflamm, 2015: 326032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster S. C. et al. (2019) ‘ A Human Gut Bacterial Genome and Culture Collection for Improved Metagenomic Analyses’, Nature Biotechnology, 37: 186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulke C. A., Sharpton T. J. (2018) ‘ The Influence of Ethnicity and Geography on Human Gut Microbiome Composition’, Nature Medicine, 24: 1495–6. [DOI] [PubMed] [Google Scholar]
- Gregory A. C. et al. (2020) ‘ The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut’, Cell Host & Microbe, 28: 724–40.e728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory A. C., Tara Oceans Coordinators. et al. (2019) ‘ Marine DNA Viral Macro- and Microdiversity from Pole to Pole’, Cell, 177: 1109–23.e1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin E. et al. (2018) ‘ Biology and Taxonomy of crAss-like Bacteriophages, the Most Abundant Virus in the Human Gut’, Cell Host & Microbe, 24: 653–64.e656. [DOI] [PubMed] [Google Scholar]
- Guo L. et al. (2017) ‘ Viral Metagenomics Analysis of Feces from Coronary Heart Disease Patients Reveals the Genetic Diversity of the Microviridae’, Virologica Sinica, 32: 130–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V. K., Paul S., Dutta C. (2017) ‘ Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity’, Frontiers in Microbiology, 8: 1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley S. A. (2016) ‘ The Virome: A Missing Component of Biological Interaction Networks in Health and Disease’, Genome Medicine, 8: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan G. D. et al. (2018) ‘ Biogeography and Environmental Conditions Shape Bacteriophage-Bacteria Networks Across the Human Microbiome’, PLoS Computational Biology, 14: e1006099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan G. D. et al. (2018) ‘ Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome’, mBio, 9:e02248–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. et al. (2018) ‘ Regional Variation Limits Applications of Healthy Gut Microbiome Reference Ranges and Disease Models’, Nature Medicine, 24: 1532–5. [DOI] [PubMed] [Google Scholar]
- Hoyles L. et al. (2014) ‘ Characterization of Virus-like Particles Associated with the Human Faecal and Caecal Microbiota’, Research in Microbiology, 165: 803–12. [DOI] [PubMed] [Google Scholar]
- Hyatt D. et al. (2010) ‘ Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification’, BMC Bioinformatics, 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B. et al. (2017) ‘ CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database’, Nucleic Acids Research, 45: D566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. S., Eddy S. R., Portugaly E. (2010) ‘ Hidden Markov Model Speed Heuristic and Iterative HMM Search Procedure’, Bmc Bioinformatics, 11: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. et al. (1993) ‘ Use of Polymerase Chain Reaction for Diagnosis of Picornavirus Infection in Subjects with and without Respiratory Symptoms’, Journal of Clinical Microbiology, 31: 111–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. S. et al. (2007) ‘ New Adenovirus Species Found in a Patient Presenting with Gastroenteritis’, Journal of Virology, 81: 5978–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. et al. (2017) ‘ KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs’, Nucleic Acids Research, 45: D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft K., Zhou Z., Anantharaman K. (2020) ‘ VIBRANT: automated Recovery, Annotation and Curation of Microbial Viruses, and Evaluation of Viral Community Function from Genomic Sequences’, Microbiome, 8: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner M., Hooper L. V., Duerkop B. A. (2015) ‘ Evaluation of Methods to Purify Virus-like Particles for Metagenomic Sequencing of Intestinal Viromes’, BMC Genomics, 16: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig J. E. et al. (2011) ‘ Succession of Microbial Consortia in the Developing Infant Gut Microbiome’, Proceedings of the National Academy of Sciences of the United States of America, 108 Suppl 1: 4578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpela K. et al. (2018) ‘ Selective Maternal Seeding and Environment Shape the Human Gut Microbiome’, Genome Research, 28: 561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012) ‘ Fast Gapped-Read Alignment with Bowtie 2’, Nature Methods, 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. (2006) ‘ Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences’, Bioinformatics (Oxford, England), 22: 1658–9. [DOI] [PubMed] [Google Scholar]
- Lim E. S. et al. (2015) ‘ Early Life Dynamics of the Human Gut Virome and Bacterial Microbiome in Infants’, Nature Medicine, 21: 1228–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J. et al. (2017) ‘ Strains, Functions and Dynamics in the Expanded Human Microbiome Project’, Nature, 550: 61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V. et al. (2014) ‘ The Carbohydrate-Active Enzymes Database (CAZy) in 2013’, Nucleic Acids Research, 42 (Database issue): D490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. et al. (2018) ‘ A Human Gut Phage Catalog Correlates the Gut Phageome with Type 2 Diabetes’, Microbiome, 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqsood R. et al. (2019) ‘ Discordant Transmission of Bacteria and Viruses from Mothers to Babies at Birth’, Microbiome, 7: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi J. R. (2010) ‘ Prokaryotic and Eukaryotic Diversity of the Human Gut’, Advances in Applied Microbiology, 72: 43–62. [DOI] [PubMed] [Google Scholar]
- Minot S. et al. (2013) ‘ Rapid Evolution of the Human Gut Virome’, Proceedings of the National Academy of Sciences of the United States of America, 110: 12450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe S. S. et al. (1993) ‘ Subgenomic RNA Sequence of Human Astrovirus Supports Classification of Astroviridae as a New Family of RNA Viruses’, Journal of Virology, 67: 3611–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Gallego J. L. et al. (2019) ‘ Virome Diversity Correlates with Intestinal Microbiome Diversity in Adult Monozygotic Twins’, Cell Host & Microbe, 25: 261–72.e265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu G. et al. (2018) ‘ Alterations in Enteric Virome Are Associated with Colorectal Cancer and Survival Outcomes’, Gastroenterology, 155: 529–41.e525. [DOI] [PubMed] [Google Scholar]
- Nayfach S. et al. (2020) ‘ CheckV Assesses the Quality and Completeness of Metagenome-Assembled Viral Genomes’, Nat Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman J. M. et al. (2015) ‘ Disease-Specific Alterations in the Enteric Virome in Inflammatory Bowel Disease’, Cell, 160: 447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S. et al. (2017) ‘ metaSPAdes: A New Versatile Metagenomic Assembler’, Genome Research, 27: 824–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie L. A. et al. (2013) ‘ Genome Signature-Based Dissection of Human Gut Metagenomes to Extract Subliminal Viral Sequences’, Nature Communications , 4: 2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D. et al. (2016) ‘ Uncovering Earth's Virome’, Nature, 536: 425–30. [DOI] [PubMed] [Google Scholar]
- Pannaraj P. S. et al. (2018) ‘ Shared and Distinct Features of Human Milk and Infant Stool Viromes’, Frontiers in Microbiology, 9: 1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen H. K., MetaHIT Consortium. et al. (2016) ‘ Human Gut Microbes Impact Host Serum Metabolome and Insulin Sensitivity’, Nature, 535: 376–81. [DOI] [PubMed] [Google Scholar]
- Qin J., MetaHIT Consortium. et al. (2010) ‘ A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing’, Nature, 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J. et al. (2012) ‘ A Metagenome-Wide Association Study of Gut Microbiota in Type 2 Diabetes’, Nature, 490: 55–60. [DOI] [PubMed] [Google Scholar]
- Quast C. et al. (2013) ‘ The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools’, Nucleic Acids Research, 41: D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley E. M. (2013) ‘ Gut Bacteria in Health and Disease’, Gastroenterol Hepatol (N Y), 9: 560–9. [PMC free article] [PubMed] [Google Scholar]
- Rampelli S. et al. (2017) ‘ Characterization of the Human DNA Gut Virome across Populations with Different Subsistence Strategies and Geographical Origin’, Environmental Microbiology, 19: 4728–35. [DOI] [PubMed] [Google Scholar]
- Ren J. et al. (2017) ‘ VirFinder: A Novel k-Mer Based Tool for Identifying Viral Sequences from Assembled Metagenomic Data’, Microbiome, 5: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J. et al. (2020) ‘ Identifying Viruses from Metagenomic Data Using Deep Learning’, Quantitative Biology, 8: 64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A. et al. (2015) ‘ Gut DNA Viromes of Malawian Twins Discordant for Severe Acute Malnutrition’, Proceedings of the National Academy of Sciences of the United States of America, 112: 11941–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A. et al. (2012) ‘ Going Viral: Next-Generation Sequencing Applied to Phage Populations in the Human Gut’, Nature Reviews. Microbiology, 10: 607–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotbart H. A. (2002) ‘ Treatment of Picornavirus Infections’, Antiviral Research, 53: 83–98. [DOI] [PubMed] [Google Scholar]
- Scarpellini E. et al. (2015) ‘ The Human Gut Microbiota and Virome: Potential Therapeutic Implications’, Digestive and Liver Disease, 47: 1007–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig S. et al. (2013) ‘ Genomic Variation Landscape of the Human Gut Microbiome’, Nature, 493: 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. (2016) ‘ Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans’, Cell, 164: 337–40. [DOI] [PubMed] [Google Scholar]
- Shkoporov A. N. et al. (2019) ‘ The Human Gut Virome is Highly Diverse, Stable, and Individual Specific’, Cell Host & Microbe, 26: 527–41.e525. [DOI] [PubMed] [Google Scholar]
- Shkoporov A. N. et al. (2018) ‘ PhiCrAss001 Represents the Most Abundant Bacteriophage Family in the Human Gut and Infects Bacteroides intestinalis’, Nature Communications, 9: 4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr E. P. et al. (2019) ‘ Metatranscriptomic Reconstruction Reveals RNA Viruses with the Potential to Shape Carbon Cycling in Soil’, Proceedings of the National Academy of Sciences of the United States of America, 116: 25900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. et al. (2020) ‘ Environmental Remodeling of Human Gut Microbiota and Antibiotic Resistome in Livestock Farms’, Nature Communications, 11: 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle C. A. (2007) ‘ Marine Viruses–Major Players in the Global Ecosystem’, Nature Reviews. Microbiology, 5: 801–12. [DOI] [PubMed] [Google Scholar]
- Truong D. T. et al. (2015) ‘ MetaPhlAn2 for Enhanced Metagenomic Taxonomic Profiling’, Nature Methods, 12: 902–3. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J. et al. (2006) ‘ An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest’, Nature, 444: 1027–31. [DOI] [PubMed] [Google Scholar]
- Vandeputte D. et al. (2017) ‘ Quantitative Microbiome Profiling Links Gut Community Variation to Microbial Load’, Nature, 551: 507–11. [DOI] [PubMed] [Google Scholar]
- Vangay P. et al. (2018) ‘ US Immigration Westernizes the Human Gut Microbiome’, Cell, 175: 962–72.e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadell G. (1988) Adenoviridae: The Adenoviruses. Laboratory Diagnosis of Infectious Diseases Principles and Practice. New York: Springer, pp. 284–300. [Google Scholar]
- Wang X. et al. (2020) ‘ An Aberrant Gut Microbiota Alters Host Metabolome and Impacts Renal Failure in Human and Rodents’, Gut, 69: 2131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Y. I. et al. (2018) ‘ Origins and Evolution of the Global RNA Virome’, mBio, 9:e02329–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M. et al. (2016) ‘ Natural History of the Infant Gut Microbiome and Impact of Antibiotic Treatment on Bacterial Strain Diversity and Stability’, Sci Transl Med, 8: 343ra381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N. et al. (2018) ‘ Discovery of an Expansive Bacteriophage Family That Includes the Most Abundant Viruses from the Human Gut’, Nature Microbiology, 3: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G. et al. (2017) ‘ Intestinal Virome Changes Precede Autoimmunity in Type I Diabetes-Susceptible Children’, Proceedings of the National Academy of Sciences of the United States of America, 114: E6166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y. et al. (2019) ‘ 1,520 Reference Genomes from Cultivated Human Gut Bacteria Enable Functional Microbiome Analyses’, Nature Biotechnology, 37: 179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T. et al. (2019) ‘ Gut Mucosal Virome Alterations in Ulcerative Colitis’, Gut, 68: 1169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T. et al. (2020) ‘ Human-Gut-DNA Virome Variations across Geography, Ethnicity, and Urbanization’, Cell Host & Microbe, 28: 741–51.e744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing dataset acquired in this study has been deposited to the NCBI SRA database under the accession code PRJNA641593. The sample metadata, vOTU and taxonomic composition data, and the statistical scripts are available from the corresponding authors on reasonable request.