Abstract
In order to define public health policies, simple, inexpensive and robust detection methods for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are vital for mass-testing in resource limited settings. The current choice of molecular methods for identification of SARS-CoV-2 infection includes nucleic acid-based testing (NAT) for viral genetic material and antigen-based testing for viral protein identification. Host exposure is detected using antibody detection assays. While NATs require sophisticated instrument and trained manpower, antigen tests are plagued by their low sensitivity and specificity. Thus, a test offering sensitive detection for presence of infection as a colorimetric readout holds promise to enable mass testing in resource constrained environments by minimally trained personnel. Here we present a novel HRPZyme Assisted Recognition of Infection by Optical Measurement (HARIOM) assay which combines specificity of NATs with sensitivity of enzymatic assays resulting in enhanced signal to noise ratios in an easily interpretable colorimetric readout. Using this assay, we could detect up to 102 copies of synthetic viral RNA spiked in saliva as a detection matrix. Validating our assay on suspected human subjects, we found concordance with PCR based readouts with visible colorimetric distinction between positive and negative samples in less than an hour. We believe that this assay holds the potential to aid in mass screening to detect SARS-CoV-2 infection by facilitating colorimetric detection with minimal resources and less trained personnel.
Keywords: DNAZyme, COVID-19, HRPZyme, Colorimetric detection, On-site diagnostics
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) led pandemic has severely impacted our civilization - cutting across nations, resulting in huge loss to world economy, and of precious lives. Early detection of infection via mass-screenings of societies is paramount in curtailing viral spread as mounting evidences suggest spread of infections by asymptomatic individuals spread of infections by asymptomatic individuals (Hogan et al., 2021; Yu et al., 2020). (Hogan et al., 2021). Such information is also pivotal for defining effective public health policies (Hinchman et al., 2020). Several methods are being developed for detection of SARS-CoV-2 infection. These methods rely on either detection of viral nucleic acids using RT-PCR, LAMP, SHERLOCK (Broughton et al., 2020; Esbin et al., 2020; Park et al., 2020; Shen et al., 2020), viral antigen (Mak et al., 2020; Porte et al., 2020) or host's immune response to infection (ELISA, antibody based rapid diagnostic tests) (Kontou et al., 2020; Petherick, 2020). Each of these tests has its own utility and utilization window. For example, nucleic acid and antigen-based tests are useful for detection of active infection while antibody-based tests can identify exposure to viruses and may indicate presence of protective antibodies (Manners et al., 2020).
Horseradish-peroxidase-mimicking DNAzyme (HRPzyme) is a short DNA sequence comprising of guanine (G)-rich sequence, which folds into a G-quadruplex structure which is stabilized upon forming a complex with hemin (Travascio et al., 1998). HRPzyme is able to catalyze the peroxidation of various HRP substrates such as 3,3′,5,5′-Tetramethylbenzidine (TMB) and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) in the presence of H2O2 giving a colorimetric readout. The short sequence of HRPZyme permits the possibility of primer integration in a manner such that the functional HRPZyme sequence forms only upon successful amplification of the target genomic region. Various HRPZyme and G-quadruplex based assays (Xi et al., 2020) have previously been developed for detection of pathogens such as Staphylococcus aureus (Mondal et al., 2018), Salmonella enterica Typhimurium (Kim et al., 2018), norovirus (Batule et al., 2018) and even coronavirus (Anantharaj et al., 2020) However their low sensitivity inhibited human applications. Here we report the use of a modified HRPZyme sequence (Li et al., 2016) with enhanced sensitivity for the detection of SARS-CoV-2 infection in human samples. HRPZyme Assisted Recognition of Infection by Optical Measurement (HARIOM) offers the dual advantages - specificity of PCR and enhanced sensitivity of an enzyme catalyzed reaction. Other notable features of the system include its robustness, requirement of minimal technical knowledge and usage of inexpensive reagents, making it ideal for use in low-resource settings and for mass screenings.
As HARIOM utilizes the power of oligo-embedded enzymatic activity, it offers the possibility of additive effect from simultaneous amplification of multiple transcripts in the same sample, hence enhancing sensitivity further. We demonstrate the same by using primers targeting ORF-1ab and N-gene in a single-pot reaction. The assay was able to detect 102 copies of viral RNA as determined by spiking copies of viral RNA in saliva samples. Furthermore, we validated our method on COVID-19 patient samples from nasopharyngeal swabs or oral swab. We found matching concordance with 2-gene RT-PCR and TrueNAT results and, thus we believe that HARIOM could be a valuable tool for the detection of SARS-CoV-2 infection.
2. Materials and methods
2.1. Chemicals
MES buffer (2-(N-morpholino) ethanesulfonic acid) (Cat no. M3671) and hemin (Cat no. 51280) were purchased from Sigma. TMB was purchased from ThermoFisher Scientific (Cat no. 34028). SARS-CoV-2 synthetic RNA was purchased from Twist Biosciences. Proteinase K was purchased from New England Biolabs (Cat no. P8107).
MES solution used in the reaction had the following composition: 25 mM MES salt, 200 mM NaCl, 10 mM KCl, 1% DMSO, 0.05% Triton X100.
2.2. Hemin stock
20 mM stock of hemin was prepared in DMSO. A sub-stock of 5 mM was freshly prepared for use which was added to MES buffer to make a final working stock solution of 50 μM.
2.3. Primers
Primers were ordered from Eurofins. Lyophilized pellets were dissolved in nuclease-free MilliQ water to make a stock of 100 μM. Final working concentration used was 0.5 μM per 20 μl reaction. Primer sequences used in the study are provided in Table 1 .
Table 1.
List of primers used in this study.
| Seq ID | Primer Name | Sequence |
|---|---|---|
| 1 | ACTB Fw | GCGAGAAGATGACCCAGAT |
| 2 | ACTB Rev | TCACCGGAGTCCATCACGAT |
| 3 | ACTB Rev-HRPZyme | TCCCAACCCGCCCTACCCTCACCGGAGTCCATCACGAT |
| 4 | 2019-nCoV_N2 Fw | TTACAAACATTGGCCGCAAA |
| 5 | 2019-nCOV_N2 Rev | CCAATTTGATGGCACCTGTG |
| 6 | 2019-nCOV_N2 Rev-HRPZyme | TCCCAACCCGCCCTACCCCCAATTTGATGGCACCTGTG |
| 7 | ORF-1ab Fw | ATGAGCTTAGTCCTGTTG |
| 8 | ORF-1ab Rev | CTCCCTTTGTTGTGTTGT |
| 9 | ORF-1ab Rev HRPZyme | TCCCAACCCGCCCTACCCCTCCCTTTGTTGTGTTGT |
2.4. PCR conditions for DNA amplification
PCR for HARIOM was performed with normal forward primer and HRPZyme complementary sequence containing reverse primer in a 20 μl reaction for Orf-1ab and/or N-gene, with annealing temperature of 55 °C. All the PCRs were carried out on a Veriti Thermal Cycler from Thermo Fisher Scientific.
2.5. Real-time PCR for detection of SARS-CoV-2 infection using commercial kits
Real-time PCR was performed with samples from symptomatic individuals who are suspected of having SARS-CoV-2 infection using FTD SARS-CoV-2 Assay from Siemens Healthineers using manufacturer's protocol. All the PCRs were performed using a LightCycler® 480 instrument from Roche Life Sciences.
2.6. Colorimetric readout
For colorimetric readout, 40 μl of 50 μM hemin -MES buffer was added to 18 μl of PCR amplified product. This was followed by addition of 25 μl of TMB spiked with 0.5 μl of 3% H2O2 and incubated in the dark for 30 min, or until color development took place for visual estimation.
2.7. Human samples
The work undertaken was done following ethical principles and guidelines, and was approved through Institute's Ethics Committee for MLP2005 whenever a human subjects or samples were utilized.
3. Results
3.1. HARIOM offers an easily interpretable detection of amplified SARS-CoV-2 genome from clinical samples
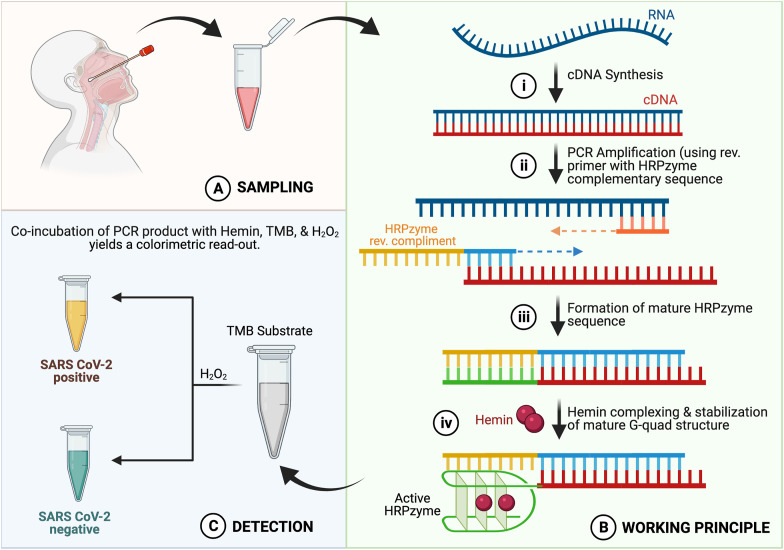
The assay principal relies upon detection of SARS-CoV-2 genomic fragments in a given sample using primers containing HRPZyme forming sequence upon amplification. The workflow consisted of collecting clinical specimen from suspected subjects in the form of nasopharyngeal swabs/oral swabs resuspended in viral transport medium (VTM) or saliva (Fig. 1 A). This is followed by extraction of viral RNA using extraction kit (Wyllie et al., 2020), and cDNA synthesis using random hexamers. cDNA thus synthesized is then amplified using SARS-CoV-2 genome specific primers, namely for ORF1ab and/or N-gene. The reverse primer consists of the HRPZyme complementary sequence (H-inv primer) in a manner that the functional HRPZyme sequence would only be formed upon amplification of the SARS-CoV-2 gene products (Fig. 1B). Upon incubation with hemin and TMB in the presence of H2O2, a color change is observed from colourless to yellow in samples containing SARS-CoV-2 genomic fragments (Fig. 1 C) and blue-green in samples negative for the presence of SARS-CoV-2.
Fig. 1.
Schematic illustration of the working principle of HARIOM assay. (A) Sampling: Nasophareangyl swab was taken. (B) Working principle: i. RNA isolated from nasopharegyl swab was subjected to cDNA synthesis, ii. PCR amplification using HRPZyme containing reverse primer, iii. Formation of mature HRPzyme sequence, iv. Hemin complexing and stabilization of mature G-quad structure. (C) Detection: Addition of TMB substrate and H2O2 provides a detetctable colorimetric output in samples positive for SARS-CoV-2 due to formation of mature HRPZyme sequence. Yellow color: SARS-CoV-2 positive; Blue color: SARS-CoV-2 negative. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. HRPZyme sequence enables colorimetric detection of amplified products
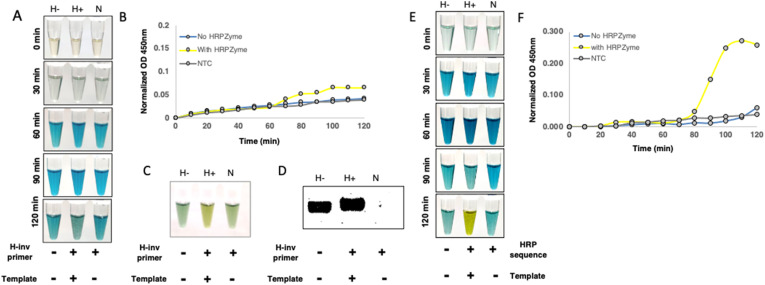
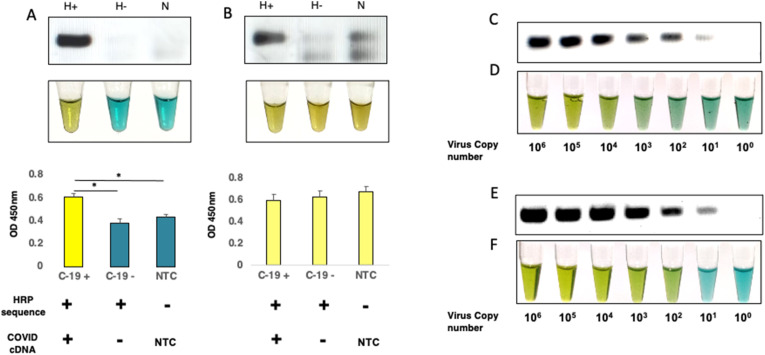
For biosafety considerations, sample availability and ease of handling, we decided to first test our reaction conditions for optimal peroxidase activity using primer targeting b-actin gene from human DNA. We PCR amplified a genomic region corresponding to beta-actin gene from human cDNA prepared from HEK 293T RNA as a template using normal forward primer and reverse primer with or without HRPZyme sequence. The PCR product was incubated first with hemin for 30 min at room temperature in accordance with various published protocols followed by incubation with HRP substrate, TMB and H2O2. Though in our hands, we couldn't find any significant colorimetric difference between various test groups until 120 min of addition of TMB and H2O2 (Fig. 2 A). Similar results were observed upon performing absorption kinetics of these samples at 450 nm, wherein the HRPZyme sequence containing primer product exhibited very minute difference in comparison to non-HRPZyme containing primer product and non-template control (Fig. 2B). However, after 180 min of incubation, we could see a difference in HRPZyme containing product, a distinguishing yellow color resulting from the two-electron loss oxidation state of TMB by peroxidase activity of HRPZyme (Fig. 2C). Despite having a significant amplification product on gel, the colorimetric profile of non-HRPZyme containing product was similar to non-template control of H-inv primer (Fig. 2D), suggesting that the colorimetric difference could be attributed to the presence of HRPZyme activity in the amplified product only. The HRPZyme containing product is slightly larger due to 18 bp HRPZyme sequence in the reverse primer. The HRPZyme sequence containing reverse primer was used in non-template control to account for primer dimer formation.
Fig. 2.
HRPZyme enables colorimetric detection of PCR amplicons. (A) Image displays colorimetric analysis of PCR amplicons with or without HRPZyme sequence incubated with hemin, TMB + H2O2. Samples were preincubated with hemin for 30 min followed by incubation with TMB and H2O2 at indicated time-points. (B) Graph shows absorption kinetics of H-, H+ and N at 450 nm after adding hemin followed by TMB and H2O2 at indicated time periods. (C) Image indicates colorimetric change in H-, H+ and N at 180 min. (D) Image displays agarose gel products corresponding to for H-, H+ and N PCR amplicons. (E) Colorimetric analysis of H-, H+ and N PCR amplicons co-incubated with hemin and TMB + H2O2. (F) Absorption kinetics of H-, H+ and N PCR amplicons co-incubated with hemin, TMB and H2O2 at 450 nm for different time intervals. H-: PCR amplicon without mature HRPZyme, H+: PCR amplicon with mature HRPZyme, N: NTC with H-inv primer, H-inv primer: Primer with HRPZyme complementary sequence.
Though we observe significantly higher peroxidase activity in amplicons containing HRPZyme sequence, the color development process was very slow, as it took around 3 h for clear differences to emerge, rendering it ineffective for rapid diagnostic purposes. The protocol for determination of peroxidase activity was adapted from existing literature (Li et al., 2016), where HRPZyme oligo was first incubated with hemin for 30 min at room temperature to form a stable HRPZyme complex followed by incubation with TMB substrate. We thought if we could expedite the process by coincubation of hemin and TMB substrate with the PCR product to take advantage of the peroxidase activity as soon as the HRPZyme starts maturing. Upon incorporation of these changes in the protocol we found that coincubation indeed accelerated the process significantly as we could now clearly see stark colorimetric difference between the groups in as early as 120 min (Fig. 2E). These differences were also reflected in absorption kinetics of these samples at 450 nm (Fig. 2F) wherein HRPZyme containing product started showing increased absorption after 80 min, and peaking around 100 min of incubation. This suggested to us that further optimization holds potential for improving this method further.
3.3. Optimizing hemin, TMB and H2O2 concentrations for most efficient HARIOM output
Encouraged by our coincubation results, we wondered if we could further enhance the contrast and speed up of the peroxidation reaction by optimizing other parameters, including concentrations of hemin, H2O2 and TMB in reaction mixture.
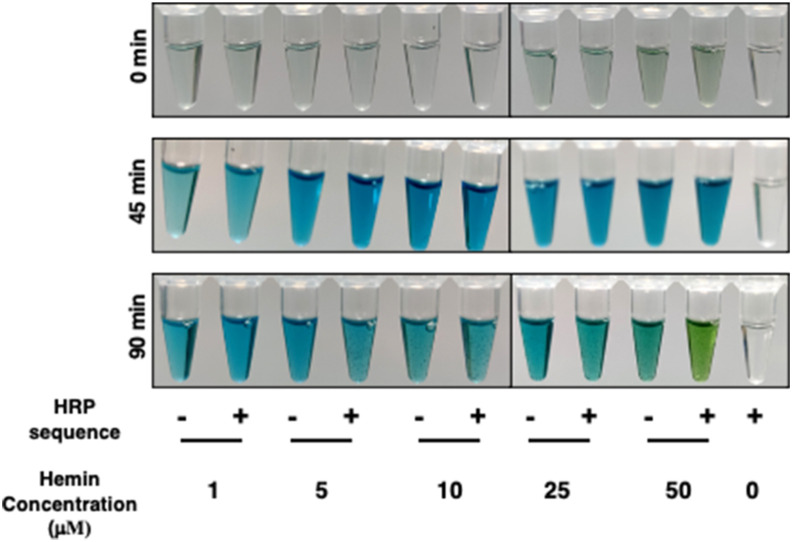
We begin our optimizations with varying hemin concentrations as past studies have used widely different range across applications. We used five concentrations of hemin starting from 1 μM to 50 μM, keeping TMB volume to 50 μl, and H2O2 concentration as 0.25 μl of 3% H2O2. We found that 50 μM of hemin gave the best resolution after 90 min of incubation (Fig. 3 ).
Fig. 3.
Optimization of conditions for best contrast. Hemin concentration ranging from 1 to 50 μM was used with 50 μl of TMB and 0.25 μl of 3% H2O2 per well.
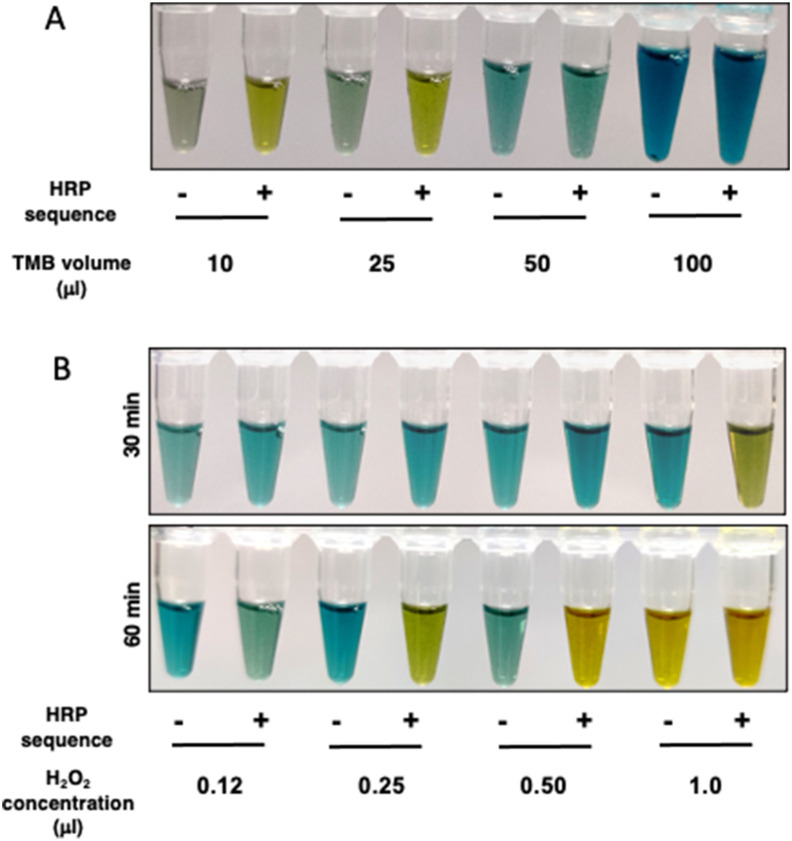
Next, we optimized the TMB volume per reaction, as different volumes of substrate may be utilized at differing rate accounting for the difference in reading output. We used TMB volume ranging from 10 μl to 100 μl per well, keeping hemin concentration at 50 μM, as optimized previously, PCR product volume as 20 μl, H2O2 concentration at 0.25 μl per reaction of 3% H2O2. This resulted in varying final volumes of reaction for same input ingredients, due to varying volume of TMB. We found that both, 10 and 25 μl TMB per reaction gave clearly distinguishable output after 60 min of incubation, with 25 μl providing better clarity between amplicons with or without HRPZyme (Fig. 4 A).
Fig. 4.
Optimization of conditions for best contrast. (A) Varying volume of TMB, from 10 to 100 μl was used with 50 μM of Hemin and 0.25 μl of 3% H2O2. (B) Indicated volume of H2O2 per well was used with 50 μM of hemin and 25 μl of TMB per well.
Finally, we sought to optimize the H2O2 volume per reaction. H2O2 acts as oxidizing substrate for HRP, that reduces colorless TMB to its blue-green (one electron loss oxidation state) or yellow (two electron loss oxidation state) form. We tested different volumes of 3% H2O2, ranging from 0.1 μl to 1 μl per well, keeping hemin concentration to 50 μM and TMB volume to 25 μl, in a final reaction volume of 75 μl. We found 0.25 μl of 3% H2O2 to give us the best results in as short as 60 min post reaction setup (Fig. 4B).
In summary, we identified that 20 μl PCR product upon coincubation with 40 μl of 50 μM hemin-MES buffer, 25 μl of TMB and 0.25 μl of 3% H2O2 provided us with the best contrast in a span of 60 min. At the same time, the modified HRPZyme sequence used provides much better sensitivity in comparison to the conventional sequence (Supplementary figure 1).
3.4. Determination of sensitivity of detection by spiking SARS-CoV-2 RNA in detection matrix
As the power of HARIOM lies in oligo-embedded enzymatic activity, we wondered if we could further enhance the sensitivity by amplifying two genes instead of just one. To test this, we used primers against ORF-1ab as well as N-gene. We tested two different PCR protocols for this using cDNA from nasopharyngeal swab preparation of SARS-CoV-2 positive patient, SARS-CoV-2 negative patient and non-template control (NTC). In one strategy, we used H-inv containing primers for both genes together in one-step PCR for 35 cycles, in another strategy, we used non-H-inv primers for first 10 cycles, followed by another PCR with this product using H-inv containing primers for next 30 cycles. We observed a clean single band corresponding to ORF-1ab as well as N gene in first strategy (both genes yielding almost similar product size 106 and 108 bps) with no amplification either in SARS-CoV-2 negative subject or NTC. These results were reflected in clean colorimetric distinction across samples and in absorption spectra at 450 nm (Fig. 5 A). However, in two-step PCR we observed a lot of background in SARS-CoV-2 negative and NTC samples, which was also reflected in colorimetric readouts and absorption spectra at 450 nm (Fig. 5B). Thus, we decided to follow the one-step PCR strategy moving forward.
Fig. 5.
Determination of limit of detection (LOD) with single primer and coupled primers. (A–B) Images represent analysis from (A) one-step PCR reaction, and (B) two-step PCR reaction for detection of SARSCoV-2 using HARIOM. Bands represent PCR amplicon. Tubes images represent colorimetric development corresponding to PCR amplicons. Graphs represent absorption at 450 nm for indicated samples. H+: PCR amplicon with mature HRPZyme, H-: PCR amplicon without mature HRPZyme, N: NTC with H-inv primer. (C) Bands represent PCR amplicons corresponding to indicated spiked copy number of synthetic viral genome using N-gene primer-set specific for SARS-CoV-2. (D) Colorimetric read-out corresponding to PCR products in panel C. (E) Bands indicate PCR amplicons obtained using both ORF-1ab and N-gene primer sets together. (F) Colorimetric detection of samples from panel E. Data shown are mean ± S.E.M. (n = 3). Statistical significance was determined using unpaired t-test. *p < 0.05.
During initial phases of testing, NP/oral swab suspended in VTM was standard matrix that was used for viral testing using RT-PCR. However, the sample collection method was inconvenient for the sample provider and at the same time posed exposure risk to health personnel. It has previously been reported that saliva can serve as a sensitive specimen for detection of respiratory viruses sensitive specimen for detection of respiratory viruses (Kim et al., 2018). , hence efforts across labs were made to explore saliva as an easily collectible biofluid for detection of SARS-CoV-2 viral RNA, and it was found to be a better and more sensitive alternative to NP swabs (Wyllie et al., 2020).
As salivary components have been shown to severely inhibit other colorimetric NAT detection assay, such as RT-LAMP, we sought to explore if we could use saliva as a specimen for our assay. We spiked serially diluted copies of viral synthetic RNA from 106 to 100 copies in inactivated saliva, performed cDNA synthesis and PCR, followed by detection using HARIOM. While using N-gene alone, we were able to detect 103 viral copies (Fig. 5C and D). Using the protocol optimized above of using both ORF-1ab and N-gene in a single-pot reaction, we could bring down our detection limit to 102 viral copies, which corresponds to 2 viral copies/μl of saliva, which is well within the clinical limit requirements for detection (Fig. 5E and F).
3.5. HARIOM can detect clinical infection in patient samples
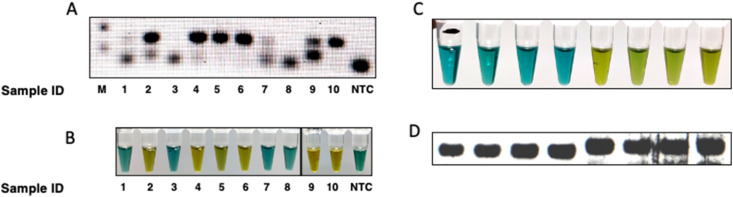
As our main objective was to assess the possibility of detection of SARS-CoV-2 genome in patient samples using HARIOM, we tested our optimized system on SARS-CoV-2 positive samples already validated by PCR. We took cDNA samples prepared from nasopharyngeal swabs of ten COVID-19 suspected subjects prepared using random hexamers. We then performed PCR, either using Real-Time PCR based 2-gene commercial kit from Siemens Healthineers, or with ORF-1ab and N-gene with H-inv reverse primer. These PCR products were then validated using HARIOM. We observed concordance in HARIOM results with PCR (Fig. 6 A and B; Supplementary Figure 2) and tested for the reproducibility of the method by performing HARIOM on a subset of samples after 3 and 7 days with consistent readout (Supplementary Figure 3).
Fig. 6.
Demonstration of HARIOM mediated detection of infection in clinical samples. (A) Bands represent PCR amplicons obtained from ten clinical using 2-gene commercial kit, (B) Colorimetric validation of clinical samples with HARIOM using ORF-1ab and N-gene with H-inv primers. (C) RNA extraction independent detection of beta-actin RNA from saliva of 4 healthy individuals using HARIOM. (D) PCR bands represent gel products corresponding to samples in C.
We previously established that HARIOM could detect purified viral RNA spiked in saliva matrix, but for clinical SARS-CoV-2 detection, viral isolation was still required which is technically challenging and time consuming. We sought to test a recently developed RNA extraction free protocol for detection of nucleic acids from saliva, that has been recently approved by FDA. We first tested the detection of beta-actin from saliva of four healthy individuals for presence of beta-actin RNA using previously used primer set. We used 5 μl of proteinase K treated saliva from healthy individuals for cDNA synthesis followed by amplification of beta-actin genomic region using reverse primer with or without HRPZyme sequence. We subjected the PCR product to HARIOM and could observe a clear signal in samples containing HRPZyme sequence, thus establishing that saliva can directly be used for RNA extraction free detection of nucleic acids (Fig. 6C and D).
4. Conclusion
In conclusion, we demonstrate that the modified HRPZyme sequence we used in conjunction with our optimized protocol presented here holds great promise for detection of nucleic-acids. HARIOM offers a powerful and generalized method for detection of nucleic-acids from a variety of sample matrices, ranging from nasopharyngeal swab, oral swab and direct saliva without the need for RNA isolation. We also demonstrated its capability in detection of SARS-CoV-2 infection from clinical specimen. We do acknowledge the limitation that the method presented here still is dependent on availability of PCR instrument, but we believe that in future iterations this could be taken to instrument-independent platforms, such as RPA, further enhancing the sensitivity of reaction while marinating the simplicity of colorimetric readout. One major limitation of the study that we feel is not being able to test on enough clinical samples so as to accurately gauge sensitivity and specificity as per clinical requirements, but as this is just a proof of principle demonstration of the technique, we hope to address this in future studies. We believe that the biosensor described here holds immense potential for the development of point-of-care diagnostics as well as offer the modularity to modify it as per need for detection of other pathogens.
CRediT authorship contribution statement
Mohd Ahmad: Data curation, Methodology. Pooja Sharma: Data curation, Methodology. Asangla Kamai: Data curation, Methodology. Anurag Agrawal: Supervision. Mohammed Faruq: Supervision. Ankur Kulshreshtha: Conceptualization, Data curation, Methodology, Writing-Reviewing and Editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
The authors have filed a patent based on the work described in this manuscript.
Acknowledgements
The authors gratefully acknowledge the financial support from Department of Science and Technology, Government of India (GAP-167) and Council of Scientific and Industrial Research (MLP2005). M.A was supported from GAP-167, P.S and AK acknowledges support from CSIR (MLP2005). Ankur K did not receive any financial support from any funding agency during the course of this project. We also acknowledge Dipika Nandi from University of Massachusetts Amherst and Dan O'Reilly from University of Massachusetts Medical School for critical review of the manuscript. Fig. 1 was created using Biorender. com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113280.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Anantharaj A., Das S.J., Sharanabasava P., Lodha R., Kabra S.K., Sharma T.K., Medigeshi G.R. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.586254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batule B.S., Kim S.U., Mun H., Choi C., Shim W.B., Kim M.G. J. Agric. Food Chem. 2018;66(11):3003–3008. doi: 10.1021/acs.jafc.7b05289. [DOI] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. Rna. 2020;26(7):771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchman A., Ali D., Goodwin B.W., Gillie M., Boudreaux J., Laborde Y. Ochsner J. 2020;20(2):123. doi: 10.31486/toj.20.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C.A. Department of pathology, S.U.S.o.M., stanford, California, sahoo, M.K., department of pathology, S.U.S.o.M., stanford, California, pinsky, B.A., department of pathology, S.U.S.o.M., stanford, California. J. Am. Med. Assoc. 2021;323(19):1967–1969. [Google Scholar]
- Kim Y.G., Yun S.G., Kim M.Y., et al. Comparison between saliva and nasopharyngeal swab specimens for detection of respiratory viruses by multiplex reverse transcription-PCR. J Clin Microbiol. 2017;55(1):226–233. doi: 10.1128/JCM.01704-16. PubMed PMID: 27807150; PubMed Central PMCID: PMCPMC5228234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.U., Batule B.S., Mun H., Shim W.-B., Kim M.-G. Food Contr. 2018;84:522–528. [Google Scholar]
- Kontou P.I., Braliou G.G., Dimou N.L., Nikolopoulos G., Bagos P.G. Basel) 2020;10(5) doi: 10.3390/diagnostics10050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li Y., Liu Z., Lin B., Yi H., Xu F., Nie Z., Yao S. Nucleic Acids Res. 2016;44(15):7373–7384. doi: 10.1093/nar/gkw634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C.S., Lam E.T., Chan R.C., Tsang D.N. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners C., Larios Bautista E., Sidoti H., Lopez O.J. Cureus. 2020;12(6):e8399. doi: 10.7759/cureus.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal B., N B., Ramlal S., Kingston J. J. Agric. Food Chem. 2018;66(6):1516–1522. doi: 10.1021/acs.jafc.7b04820. [DOI] [PubMed] [Google Scholar]
- Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T., Maeng J.S. J. Mol. Diagn. 2020;22(6):729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petherick A. Lancet. 2020;395:1101–1102. doi: 10.1016/S0140-6736(20)30788-1. 10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte L., Legarraga P., Vollrath V., Aguilera X., Munita J.M., Araos R., Pizarro G., Vial P., Iruretagoyena M., Dittrich S., Weitzel T. Int. J. Infect. Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Zhou Y., Ye J., Abdullah Al-Maskri A.A., Kang Y., Zeng S., Cai S. J Pharm Anal. 2020;10(2):97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travascio P., Li Y., Sen D. Chem. Biol. 1998;5(9):505–517. doi: 10.1016/s1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J.L., Geng B., Muenker M.C., Moore A.J., Vogels C.B.F., Petrone M.E., Ott I.M., Lu P., Venkataraman A., Lu-Culligan A., Klein J., Earnest R., Simonov M., Datta R., Handoko R., Naushad N., Sewanan L.R., Valdez J., White E.B., Lapidus S., Kalinich C.C., Jiang X., Kim D.J., Kudo E., Linehan M., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Weizman O.E., Wong P., Yang Y., Bermejo S., Odio C.D., Omer S.B., Dela Cruz C.S., Farhadian S., Martinello R.A., Iwasaki A., Grubaugh N.D., Ko A.I. N. Engl. J. Med. 2020:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H., Juhas M., Zhang Y. Biosens. Bioelectron. 2020;167 doi: 10.1016/j.bios.2020.112494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Xu S., Rong Z., Xu R., Liu X., Deng P., Liu H., Xu X. Delivery of infection from asymptomatic carriers of COVID-19 in a familial cluster. Int J Infect Dis. 2020 May;94:133–138. doi: 10.1016/j.ijid.2020.03.042. Epub 2020 Apr 2. PMID: 32247826; PMCID: PMC7129961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.