Abstract
Introduction
We evaluated for two novel automated biomarker assays how cerebrospinal fluid (CSF) amyloid beta (Aβ)1– 42‐ratios improved the concordance with amyloid positron emission tomography (PET) positivity compared to Aβ1– 42 alone.
Methods
We selected 288 individuals from the Amsterdam Dementia Cohort across the Alzheimer's disease clinical spectrum when they had both CSF and amyloid PET visual read available, regardless of diagnosis. CSF Aβ1– 42, phosphorylated tau (p‐tau), and total tau (t‐tau) were measured with Elecsys and Lumipulse assays, and Aβ1–40 with Lumipulse. CSF cut‐points were defined using receiver operating characteristic (ROC) for amyloid PET positivity.
Results
For both Elecsys and Lumipulse the p‐tau/Aβ1– 42, Aβ1– 42/Aβ1– 40, and t‐tau/Aβ1– 42 ratios showed similarly good concordance with amyloid PET (Elecsys: 93,90,90%; Lumipulse: 94,92,90%) and were higher than Aβ1– 42 alone (Elecsys 85%; Lumipulse 84%).
Discussion
Biomarker ratios p‐tau/Aβ1– 42, Aβ1– 42/Aβ1– 40, t‐tau/Aβ1– 42 on two automated platforms show similar optimal concordance with amyloid PET in a memory clinic cohort.
Keywords: Alzheimer's disease, amyloid‐beta, amyloid positron emission tomography, biomarkers, cerebrospinal fluid, concordance cut‐points, Elecsys, Lumipulse
1. INTRODUCTION
Cerebrospinal fluid (CSF) biomarkers for amyloid beta(1‐42) (Aβ1‐42), phosphorylated tau (p‐tau), and total tau (t‐tau) are part of recent research criteria to support a diagnosis of Alzheimer's disease (AD). 1 CSF Aβ1‐42 concentrations decrease in the disease process when Aβ aggregates into plaques, while CSF p‐tau concentrations increase along the formation of AD‐specific tangle pathology and increases in CSF t‐tau concentrations may in addition reflect other aspects of neurodegeneration. 1 , 2 These biomarkers are altered in very early, pre‐clinical stages of AD, when cognition is still normal. 1 Therefore, CSF biomarkers have been proven to be useful tools for AD diagnostics. Still, their implementation in clinical practice has been a difficult trajectory marked by obstacles such as inter‐laboratory and intra‐laboratory variation. 3 , 4 Efforts of collaborative initiatives such as BIOMARK‐ADP, 5 together with technological innovations, have led to the development of standardized operating procedures for CSF collection and storage, 6 , 7 a certified reference measurement procedure for Aβ1‐42, 8 and fully automated assays for the CSF biomarkers that have been calibrated against this reference method. 9 , 10 , 11 , 12 These achievements greatly reduced the variation in CSF biomarker results across and within laboratories. 13 The final goal of successful biomarker implementation is to establish global cut‐off values that are independent of assay platform, cohort, or laboratory.
Because next generation automated assays seem to detect Aβ1‐42 more accurately than the older manual immunoassays, that is, with less interference of Aβ1‐40, and show different concentration ranges, 9 , 14 re‐establishment of biomarker cut‐offs is essential. The current gold standard for cut‐off determination of CSF biomarkers is amyloid positron emission tomography (PET), which is approved by the Food and Drug Administration (FDA) for in vivo amyloid pathology, or clinical diagnosis, because a definite diagnosis of AD can only be made at autopsy. Previous studies using either Elecsys or Lumipulse assays showed that CSF biomarker ratios improved the agreement with amyloid PET compared to single biomarker cut‐points. 15 , 16 , 17 , 18 , 19 , 20 For example, using the Aβ1‐42/Aβ1‐40 ratio compared to Aβ1‐42 alone improved the concordance, which is hypothesized to be due to Aβ1‐40 correcting for inter‐individual biological variation in amyloid production and/or clearance, and/or Aβ1‐40 correcting for artificial decrease of Aβ1‐42 concentrations during the pre‐analytical phase. 21 , 22 , 23 Ratios of Aβ1‐42 with p‐tau or t‐tau also improved the concordance with amyloid PET in research cohorts using either Elecsys or Lumipulse biomarkers. 15 , 16 , 17 , 18 , 19 , 20 These findings call for a head‐to‐head validation of the performance of the different biomarker interpretation modalities on the different platforms in the clinical setting, such as the memory clinic. Also, it remains to be addressed whether similar cut‐offs for the automated assays can be applied. Introduction of these automated biomarker assays in diagnostic practice calls for a re‐evaluation of the use of application and optimal interpretation of biomarker results in a clinical setting.
RESEARCH IN CONTEXT
Systematic review: Literature was reviewed using PubMed and meeting abstracts or presentations. The cerebrospinal fluid (CSF) Elecsys and Lumipulse biomarkers have separately been studied in a few recent publications, which are cited, but were not previously compared in a head‐to‐head comparison.
Interpretation: Our findings show that for both Elecsys and Lumipulse assays phosphorylated tau/amyloid beta (Aβ)1– 42 ratios outperformed CSF Aβ1‐42 alone in detecting positive amyloid positron emission tomography (PET) in a clinical diagnostic setting. Of note, concordance improved similarly for both the Aβ1‐42/Aβ1‐40 and total tau/Aβ1‐42 ratios. Cut‐offs were platform specific, but biomarker concordance with amyloid PET positivity did not depend on the platform used in this head‐to‐head comparison.
Future directions: For clinical implementation, future studies should perform multicenter comparisons to further address the feasibility of determining universal cut‐points for these ratios, independent of assay platform.
We aimed to determine how the ratios of biomarkers, Aβ1– 42/Aβ1– 40, p‐tau/Aβ1– 42, and t‐tau/Aβ1– 42 improved discrimination of amyloid PET positivity compared to Aβ1‐42 alone in a retrospective memory clinic cohort including AD and other types of dementia, to evaluate whether the improved performance was dependent on the automated platform used, and to define cut‐off values for all biomarker combinations. Last, we compared our biomarker cut‐offs to previously published cut‐offs defined on the same platforms to evaluate the feasibility of a future universal cut‐off.
2. METHODS
2.1. Study population
We selected CSF samples from patients from the Amsterdam Dementia Cohort 24 that visited the memory clinic between 2006 and 2016 when they had an amyloid PET scan within one year of CSF collection available. All subjects underwent extensive neurological examination, neuropsychological testing, neuroimaging, and CSF biomarker testing as part of the diagnostic work‐up. Clinical diagnoses were established by consensus during a multidisciplinary meeting according to consensus criteria. 25 , 26 , 27 , 28 Diagnostic groups included in the current study were subjects presenting with subjective cognitive decline (SCD, n = 58), mild cognitive impairment (MCI, n = 42), possible/probable AD (n = 145), frontotemporal dementia (FTD, n = 23), dementia with Lewy bodies (DLB, n = 6), vascular dementia (VaD, n = 5), or other dementia (n = 9). All patients signed written informed consent to use medical data and biomaterials for research purposes and the study was approved by the local ethical committee in accordance with the Declaration of Helsinki.
2.2. CSF biomarker measurements
2.2.1. CSF collection and processing
CSF samples were obtained by lumbar puncture using a 25‐gauge needle and syringe between the L3/L4, L4/L5, or L5/S1 intervertebral space, collected in polypropylene tubes and processed as previously described. 29
2.2.2. Elecsys assays
Aβ1– 42, p‐tau (181P), and t‐tau (Roche Diagnostics GmbH) were analyzed in CSF samples by board‐certified technicians using the fully automated Elecsys biomarker assays. 14 CSF of 17 samples (6%) needed transfer to a 0.5 mL Sarstedt tube as the original 2.0 mL Sarstedt tubes evoked an error on the Cobas e601 analyzer due to low sample volume (< 0.5 mL). No systematic effect in Aβ1– 42 results was observed between the transferred and the non‐transferred samples (data not shown). The Elecsys Aβ1‐42 concentration exceeded the upper limit of detection of the assay at 1700 pg/mL in 42 cases (15%); these concentrations were included in all further analyses and graphs as 1700 pg/mL.
2.2.3. Lumipulse assays
Next, pristine aliquots of the same samples were measured for Aβ1– 42, Aβ1– 40, p‐tau (181P), and t‐tau on the Lumipulse G 1200 system (Fujirebio Diagnostics, Inc.) 19 , 20 , 30 , 31 by board‐certified technicians according to manufacturer's instructions. CSF of 16 samples (6%) needed transfer to a 1.7 mL polystyrene Hitachi tube as the original 2.0 mL Sarstedt tubes evoked an error on the Lumipulse analyzer. No systematic effect in Aβ1– 42 results was observed between the transferred and the non‐transferred samples (data not shown).
2.3. PET amyloid imaging
Amyloid PET imaging was conducted using 11C‐PiB (n = 86), 18F‐florbetaben (n = 133), 18F‐flutemetamol (n = 64), and 18F‐florbetapir (n = 5) tracers. 32 , 33 , 34 , 35 PET scans were evaluated based on visual reading according to the manufacturers’ guidelines by an experienced nuclear medicine physician (BvB) and included as dichotomized scores (i.e., positive and negative).
2.4. Statistical analyses
Groups were dichotomized for amyloid PET status, and pair‐wise comparisons of demographic characteristics and biomarker concentrations were performed with chi‐square (for categorical variables), Student's t (for continuous variables with normal distribution), and Mann Whitney U (for continuous variables with non‐normal distribution) tests. Biomarker cut‐points were calculated based on optimal Youden's index in receiver operating curve (ROC) analyses with amyloid PET result as gold standard. Areas under the curve (AUCs) were compared pair‐wise across Aβ1– 42, Aβ1– 42/Aβ1– 40, p‐tau/Aβ1– 42, and t‐tau/Aβ1– 42 and were statistically compared per platform using 2000 bootstrapping iterations in the “roc.test” function of the “pROC” package (version 1.16.2) with D‐statistic indicating the difference between the two AUCs. 36 As the Elecsys assays did not include Aβ1– 40, the Elecsys Aβ1– 42/Aβ1– 40 ratio was calculated with the Lumipulse Aβ1– 40 result. The AUC comparisons were corrected for multiple testing using Bonferroni correction: per assay six ratios were pair‐wise compared, P‐value threshold was 0.00833 (= 0.05/6); between assays (Elecsys versus Lumipulse) four ratios were pair‐wise compared, P‐value threshold was 0.0125 (= 0.05/4). Sensitivity, specificity, and overall percentage agreements (OPAs) were calculated for all biomarkers and biomarker ratios to detect positive amyloid PET status. Spearman correlations and Passing‐Bablok regression analyses for direct comparison between Elecsys and Lumipulse biomarker concentrations were performed using the “mcr” package in R (version 1.2.1). 37 Data analysis was performed using R statistical programming (version 3.6.1) 38 and if not mentioned otherwise, P‐values below 0.05 were considered statistically significant.
2.5. Comparison of cut‐points across global cohorts
We searched the literature for publications of CSF Aβ cut‐points to determine amyloid PET positivity in other cohorts using Elecsys or Lumipulse assays to evaluate the comparability of cut‐points across these different settings and cohorts. We excluded the Elecsys Aβ1– 42/Aβ1– 40 cut‐point from our cohort in the comparison, because this ratio was calculated using the Lumipulse Aβ1– 40 result and no previous literature was available. Literature was selected by searching the PubMed database with combinations of terms “Elecsys” OR “Lumipulse,” AND “amyloid imaging” AND “concordance.” Papers that established cut‐off values for Elecsys or Lumipulse assays to determine amyloid PET positivity, assessed by title and abstract screening, were included. We chose to report only one cut‐point per cohort with preference for cut‐points based on Youden's index and preference for amyloid PET outcomes based on visual reads to align with the approach of the current study.
3. RESULTS
3.1. Cohort characteristics
We included 288 individuals in the present study, who were on average 63 ± 7 years old; 131 (45%) were female and 179 (62%) had a positive amyloid PET read (Table 1). Compared to negative, amyloid PET–positive subjects had lower Mini‐Mental State Examination (MMSE) scores, more often carried one or two apolipoprotein E (APOE) ε4 allele(s), and most often had AD‐type dementia. Median time delay between CSF collection and PET imaging was 29 days and did not differ between the amyloid PET–positive and ‐negative groups. Compared to those with normal amyloid PET, patients with abnormal amyloid PET showed decreased CSF Aβ1‐42, increased CSF t‐tau and p‐tau concentrations (Table 1; Figure 1), but no significant difference in CSF Aβ1‐40 concentrations. In 42 cases (15%), the Elecsys Aβ1‐42 concentration exceeded the upper limit of detection of the assay at 1700 pg/mL, resulting in artificially skewed distributions. CSF Aβ1‐42 concentrations and the ratios of Aβ1‐42/Aβ1‐40, p‐tau/Aβ1‐42, and t‐tau/Aβ1‐42 for both Elecsys and Lumipulse assays all showed different values for amyloid PET–positive cases compared to amyloid PET–negative cases (P < 0.001; Figure 1).
TABLE 1.
Cohort characteristics, stratified for amyloid PET visual read status
| Total | Amyloid PET – | Amyloid PET + | P‐value of pair‐wise comparison | |
|---|---|---|---|---|
| N | 288 | 109 | 179 | |
| Sex = f (%) | 131 (45%) | 43 (39) | 88 (49) | 0.114 |
| Age (mean [SD]) | 63 (7) | 62 (8) | 63 (6) | 0.086 |
| MMSE (mean [SD)) | 24 (4) | 26 (3) | 23 (4) | <0.001 |
| APOE ε4 carrier (%) | <0.001 | |||
| Unknown | 8 (3) | 0 (0) | 8 (5) | |
| Non‐carrier | 122 (42) | 67 (62) | 55 (31) | |
| Carrier | 158 (55) | 42 (39) | 116 (65) | |
| Days between CSF collection and PET imaging (median [IQR]) | 29 [15, 57] | 24 [14, 62] | 30 [16, 52] | 0.435 |
| Diagnosis (%) | 2.044 | |||
| SCD | 58 (20) | 44 (40) | 14 (8) | |
| MCI | 42 (15) | 17 (16) | 25 (14) | |
| AD | 145 (50) | 10 (9) | 135 (75) | |
| FTD | 23 (8) | 22 (20) | 1 (1) | |
| DLB | 6 (2) | 4 (4) | 2 (1) | |
| VaD | 5 (2) | 5 (5) | 0 (0) | |
| Dementia other | 9 (3) | 7 (6) | 2 (1) | |
| Elecsys CSF Aβ1‐42 (pg/mL, median [IQR]) | 852 [681, 1230] | 1522 [1097, 1700] | 742 [608, 872] | <0.001 |
| Elecsys CSF p‐tau (pg/mL, median [IQR]) | 27 [17, 39] | 15 [12, 20] | 35 [26, 44] | <0.001 |
| Elecsys CSF t‐tau (pg/mL, median [IQR]) | 282 [196, 368] | 195 [145, 262] | 336 [268, 405] | <0.001 |
| Lumipulse CSF Aβ1‐42 (pg/mL, median [IQR]) | 606 [478, 838] | 983 [692, 1312] | 529 [438, 616] | <0.001 |
| Lumipulse CSF Aβ1‐40 (pg/mL, median [IQR]) | 11770 [9874, 14064] | 11853 [8739, 14106] | 11744 [10090, 14048] | 0.247 |
| Lumipulse CSF p‐tau (pg/mL, median [IQR]) | 70 [38, 115] | 33 [25, 46] | 101 [74, 129] | <0.001 |
| Lumipulse CSF t‐tau (pg/mL, median [IQR]) | 520 [355, 755] | 355 [290, 442] | 656 [502, 852] | <0.001 |
Pair‐wise comparisons were performed using chi‐square tests for categorical variables, T‐tests for continuous normally distributed variables and Mann‐Whitney U test for non‐normally distributed variables.
Abbreviations: Aβ, amyloid beta; AD, Alzheimer's disease; APOE, apolipoprotein E; CSF, cerebrospinal fluid; DLB, dementia with Lew bodies; FTLD, frontotemporal lobar dementia; IQR, interquartile ratio; MCI, mild cognitive impairment; MMSE, Mini‐Mental State Examination; PET, positron emission tomography; p‐tau, phosphorylated tau; SCD, subjective cognitive decline; SD, standard deviation; t‐tau, total tau; VaD, vascular dementia.
FIGURE 1.
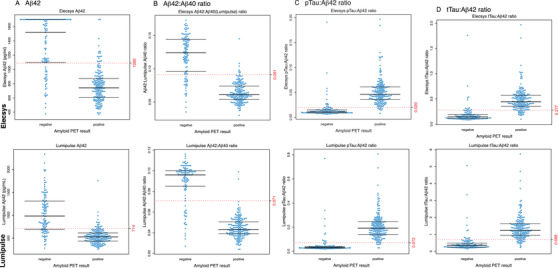
Distribution of biomarkers and biomarker ratios between the amyloid positron emission tomography (PET)‐positive and ‐negative groups. Boxplot with beeswarm 52 for Elecsys (upper row) and Lumipulse (bottom row) biomarkers amyloid beta (Aβ)1‐42 (A), Aβ1‐42/Aβ1‐42 ratio (B), phosphorylated tau/Aβ1‐42 ratio (C), and total tau/Aβ1‐42 ratio (D) in relation to an amyloid PET‐negative or ‐positive result. Dotted lines represent the cut‐point obtained through receiver operating characteristic analysis (Table 2)
3.2. Biomarker ratios are better predictors of PET amyloid positivity than Aβ1‐42 alone
We performed ROC analyses per assay with amyloid PET as reference and compared AUCs of single CSF biomarker Aβ1‐42, with ratios of Aβ1‐42 with Aβ1‐40, p‐tau, or t‐tau (Figure 2, Tables 2 and 3). For both platforms, the p‐tau/Aβ1‐42 ratio resulted in the highest AUCs (95% confidence interval [CI]) and overall percentage agreements (95% CI): 0.95 (0.89–0.96) and 93 (90–96)% for Elecsys, 0.96 (0.93–0.99) and 94 (92–97)% for Lumipulse. AUCs and overall percentage agreements were also high for both Aβ1‐42/Aβ1‐40 (0.93 [0.89–0.96] and 90 [86–93]% for Elecsys; 0.94 [0.91–0.98] and 92 [89–96]% for Lumipulse) and t‐tau/Aβ1‐42 (0.94 [0.91–0.98] and 90 [86–94]% for Elecsys; 0.94 [0.90–0.97] and 90 [87–94]% for Lumipulse). For both Elecsys and Lumipulse assays, ratios with p‐tau, t‐tau, or Aβ1‐40 performed better than Aβ1‐42 alone (P < 0.01). Sensitivity, specificity, and OPA percentages were largely overlapping across Aβ1‐42 and the Aβ1‐42 ratios, except for p‐tau/Aβ1‐42 versus Aβ1‐42, for which the 95% CI of the OPA was higher and not overlapping with that of Aβ1‐42 for either Elecsys or Lumipulse. For Lumipulse, additionally, the 95% CI of the OPA for the Aβ1‐42/Aβ1‐40 ratio (upper limit at 89%) did not overlap with that of Aβ1‐42 (lower limit at 89%).
FIGURE 2.
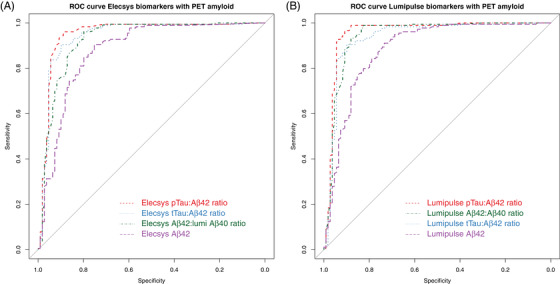
Receiver operating characteristic curves of amyloid beta (Aβ)1‐42 alone and as ratio of Aβ1‐40, phosphorylated tau, or total tau to predict positron emission positron emission tomography (PET) amyloid positivity for the Elecsys (A) and Lumipulse (B) assays. See Table 2 for areas under the curves and concordance percentages. The gray line represents the identity line
TABLE 2.
Concordance of Elecsys and Lumipulse biomarker concentrations and ratios with amyloid PET result
| Biomarker | Method | AUC [95% CI] | Cut‐point [95% CI] | Sensitivity [95% CI] | Specificity [95% CI] | OPA [95% CI] |
|---|---|---|---|---|---|---|
| Aβ1‐42 | Elecsys | 0.88 [0.83–0.92] | 1089 [864–1120] pg/mL | 91 [77–95] % | 75 [69–89] % | 85 [80–89] % |
| Lumipulse | 0.88 [0.84–0.93] | 714 [606–798] pg/mL | 91 [75–98] % | 73 [65–91] % | 84 [79–89] % | |
| Aβ1‐42/Aβ1‐40 | Elecsys Aβ1‐42; Lumipulse Aβ1‐40 | 0.93 [0.89–0.96] | 0.091 [0.080–0.10] | 96 [86–99] % | 80 [73–91] % | 90 [86–93] % |
| Lumipulse | 0.94 [0.91–0.98] | 0.071 [0.056–0.073] | 99 [89–100] % | 83 [79–94] % | 92 [89–96] % | |
| p‐tau/Aβ1‐42 | Elecsys | 0.95 [0.92–0.98] | 0.020 [0.020–0.027] | 96 [90–98] % | 89 [84–96] % | 93 [90–96] % |
| Lumipulse | 0.96 [0.93–0.99] | 0.072 [0.052–0.095] | 97 [91–100] % | 91 [85–97] % | 94 [92–97] % | |
| t‐tau/Aβ1‐42 | Elecsys | 0.94 [0.91–0.98] | 0.277 [0.194–0.313] | 89 [83–98] % | 90 [81–97] % | 90 [86–94] % |
| Lumipulse | 0.94 [0.90–0.97] | 0.688 [0.54–0.83] | 91 [83–96] % | 90 [84–97] % | 90 [87–94] % |
Abbreviations: Aβ, amyloid beta; AUC, area under the curve; CI, confidence interval; OPA, overall percentage agreement; PET, positron emission tomography; p‐tau, phosphorylated tau; t‐tau, total tau.
TABLE 3.
Pair‐wise statistical comparisons of AUCs from ROC analyses across biomarker concentrations and ratios
| Biomarker (ratio) comparison | Biomarker platform | D statistic | P‐value |
|---|---|---|---|
| Aβ1‐42 vs. Aβ1‐42/Aβ1‐40 | Elecsys | 3 | 0.005 |
| Aβ1‐42 vs. p‐tau/Aβ1‐42 | Elecsys | 4.27 | 0.00002 |
| Aβ1‐42 vs. t‐tau/Aβ1‐42 | Elecsys | 4 | 0.00006 |
| Aβ1‐42/Aβ1‐40 vs. p‐tau/Aβ1‐42 | Elecsys | 2.42 | 0.02 |
| Aβ1‐42/Aβ1‐40 vs. t‐tau/Aβ1‐42 | Elecsys | 2 | 0.1 |
| p‐tau/Aβ1‐42 vs. t‐tau/Aβ1‐42 | Elecsys | 2.25 | 0.02 |
| Aβ1‐42 vs. Aβ1‐42/Aβ1‐40 | Lumipulse | 3 | 0.001 |
| Aβ1‐42 vs. p‐tau/Aβ1‐42 | Lumipulse | 4 | 0.00002 |
| Aβ1‐42 vs. t‐tau/Aβ1‐42 | Lumipulse | 3 | 0.002 |
| Aβ1‐42/Aβ1‐40 vs. p‐tau/Aβ1‐42 | Lumipulse | 1 | 0.2 |
| Aβ1‐42/Aβ1‐40 vs. t‐tau/Aβ1‐42 | Lumipulse | –0.6 | 0.6 |
| p‐tau/Aβ1‐42 vs. t‐tau/Aβ1‐42 | Lumipulse | 2 | 0.02 |
| Aβ1‐42 | Elecsys vs. Lumipulse | –0.9 | 0.4 |
| Aβ1‐42/Aβ1‐40 | Elecsys vs. Lumipulse | –2.0 | 0.02 |
| p‐tau/Aβ1‐42 | Elecsys vs. Lumipulse | –1.4 | 0.2 |
| t‐tau/Aβ1‐42 | Elecsys vs. Lumipulse | 0.9 | 0.4 |
Note: AUC distributions were compared using 2000 bootstrapping iterations. Bonferroni correction for multiple testing was applied; p‐values that were below threshold are indicated in bold.
Abbreviations: Aβ, amyloid beta; AUC, area under the curve; p‐tau, phosphorylated tau; ROC, receiver operating characteristic; t‐tau, total tau.
Sensitivity analyses including only patients with SCD, MCI, or AD‐type dementia showed essentially similar results (Table S1 in supporting information). Biomarker cut‐points and overall percentage agreements, and their 95% CIs, for Aβ1‐42 and ratios were nearly identical. Again, for both Elecsys and Lumipulse, the p‐tau:Aβ1‐42 ratio had the highest overall percentage agreement with 95% CI not overlapping with that of Aβ1‐42.
3.3. Direct comparison Aβ1‐42, p‐tau, and t‐tau between Elecsys and Lumipulse
Biomarkers Aβ1‐42, p‐tau, and t‐tau correlated well between Elecsys and Lumipulse assays, with Spearman correlations of 0.97, 0.96, and 0.89, respectively (all P < 0.001). Conversion formulas to translate Elecsys to Lumipulse biomarker results obtained by Passing‐Bablok regression analyses are presented in Figure S1 in supporting information.
3.4. Comparison of cut‐points with literature
Finally, Table 4 shows our cut‐points listed together with those of previous studies. Five cohorts other than the current study reported cut‐points for Elecsys biomarkers and three cohorts did so for Lumipulse biomarkers (Table 4). The majority of studies (four out of six studies; including in total 1392 patients from five independent cohorts) have used Elecsys, and only two other studies used Lumipulse (two out of six studies, including in total 411 patients from three independent cohorts). Previous determined cut‐points used the optimized Youden's index, except for the BioFinder and Alzheimer's Disease Neuroimaging Initiative cohorts, which were calculated based on optimized performance (positive predictive agreement [PPA] and negative predictive agreement [NPA]) and stability of PPA and NPA when varying cut‐offs slightly. For Elecsys, cut‐offs showed comparable values for different markers, except for Aβ1‐42 in the Expedition cohorts that had a much higher cut‐point. For Lumipulse, cut‐offs for biomarker ratios were very comparable, but that of Aβ1‐42 was similar to Knight's Alzheimer's Disease Research Center cohort, but lower than the Sant Pau Initiative on Neurodegeneration (SPIN) and Eisai cohorts, probably due to differences in cohort composition.
TABLE 4.
Cut‐points for Aβ1‐42 and Aβ1‐42 ratios using Elecsys and Lumipulse assays in comparison to amyloid PET imaging across global cohorts
| Cohort | Cohort composition | N (%) PET + | Biomarker method | Cut‐point method | Cut‐point Aβ1‐42 (pg/mL) | Cut‐point Aβ1‐42/Aβ1‐40 | Cut‐point p‐tau/Aβ1‐42 | Cut‐point t‐tau/Aβ1‐4 | Amyloid PET method |
|---|---|---|---|---|---|---|---|---|---|
| ADC | n = 58 SCD; n = 42 MCI; n = 145 AD; n = 23 FTD; n = 6 DLB; n = 5 VaD; n = 9 other dementia | 179 (62%) | Elecsys | Youden's |
1089 [95% CI: 864–1120] |
0.02 [95% CI: 0.020–0.028] |
0.277 [95% CI: 0.194–0.313] |
Visual reads | |
| AIBL 15 | n = 140 CN; n = 33 MCI; n = 27 AD; n = 2 FTD | 84 (42%) | Elecsys | Youden's | 1054 | 0.064 | 0.0183 | 0.258 | Quantitative SUVR, dichotomized on tracer‐specific threshold |
| BioFINDER 16 | n = 120 SCD; n = 153 MCI | 110 (40%) | Elecsys | Optimized for (1) performance (PPA and NPA) and (2) stability of PPA and NPA when varying cut‐offs slightly | 1100 | n/a | 0.022 | 0.26 | Visual reads |
| ADNI 16 | n = 94 SMC (= significant memory concern); n = 272 EMCI; n = 152 LMCI; n = 128 AD | 347 (54%) | Elecsys | Optimized for (1) performance (PPA and NPA) and (2) stability of PPA and NPA when varying cut‐offs slightly | 977 | n/a | 0.025 | 0.27 | Visual reads |
| Knight's ADRC 17 | Community‐dwelling volunteers involved in normal aging and dementia studies; CDR 0/0.5/1/2/3: n = 176/18/3/1/0 | 50 (25%) | Elecsys | Youden's | 1098 | 0.075 | 0.0198 | 0.211 | Quantitative SUVR, dichotomized on tracer‐specific threshold |
| EXPEDITION and EXPEDITION2 18 | n = 55 mild AD; n = 20 moderate AD, participating in phase 3, double‐blind, placebo‐controlled international trials of solanezumab | Not reported for visual reads | Elecsys | Youden's | 1198 | 0.0233 | 0.289 | Visual reads | |
| ADC | n = 58 SCD; n = 42 MCI; n = 145 AD; n = 23 FTD; n = 6 DLB; n = 5 VaD; n = 9 other dementia | 179 (62%) | Lumipulse | Youden's | 714 [95% CI: 606–798] | 0.071 [95% CI: 0.056–0.073] | 0.072 [95% CI: 0.052–0.095] | 0.688 [95% CI: 0.54–0.83] | Visual reads |
| SPIN 19 | n = 6 CN; n = 35 MCI; n = 12 AD; n = 30 DLB; n = 9 FTD; n = 2 other diagnoses | 59 (63%) | Lumipulse | Youden's | 916 | 0.062 | 0.068 | 0.62 | Visual reads |
| Eisai 20 | Subjects with early AD included in the BAN2401‐201 and MISSION AD E2609‐301/302 clinical trials; CDR 0/0.5/1/2: n = 0/120/10/0 | 81 (62%) | Lumipulse | Youden's | 818 | n/a | n/a | 0.53 | Visual reads |
| Knight's ADRC 20 | Community‐dwelling volunteers involved in normal aging and dementia studies; CDR 0/0.5/1/2: n = 165/18/3/1 | 49 (26%) | Lumipulse | Youden's | 732 | n/a | n/a | 0.54 | Quantitative SUVR, dichotomized on tracer‐specific threshold |
Note: The ADC cohort (current study) is taken as the reference; cut‐points of other cohorts that exceed the 95% CI range of the ADC cohort are underlined.
Abbreviation: Aβ, amyloid beta; AD, Alzheimer's disease; ADC, Amsterdam Dementia Cohort; ADNI, Alzheimer's Disease Neuroimaging Initiative; ADRC, Alzheimer's Disease Research Center; CDR, Clinical Dementia Rating; CI, confidence interval; CN, cognitively normal; DLB, dementia with Lewy bodies; FTD, frontotemporal dementia; MCI, mild cognitive impairment; NPA, negative predictive agreement; PET, positron emission tomography; PPA, positive predictive agreement; p‐tau, phosphorylated tau; SCD, subjective cognitive decline; SPIN, Sant Pau Initiative on Neurodegeneration; SUVR, standardized uptake value ratio; t‐tau, total tau; VaD, vascular dementia.
4. DISCUSSION
In this large clinical sample set with CSF and PET measures for amyloid, we found that next generation fully automated Elecsys and Lumipulse assays showed similar high concordance with amyloid PET (OPA: 90%–94%) when using biomarker ratios with either Aβ1– 40 or t‐tau or p‐tau, and improved concordance compared to CSF Aβ1– 42 alone (OPA: 84%–85%). Cut‐points for Elecsys and Lumipulse biomarkers were largely comparable within each assay, but not across the two assays in a cross‐cohort literature comparison.
Our finding that CSF biomarker ratios of Aβ1– 42 show better concordance with the amyloid PET outcome than Aβ1– 42 alone for Elecsys and Lumipulse assays is in line with previous findings. 9 , 15 , 16 , 17 , 18 , 19 , 39 , 40 This suggests that method calibration of the next‐generation assays has indeed increased the consistency in biomarker results reported across studies. It is supposed that decreases in CSF Aβ1– 42 reflect aggregation of soluble Aβ1– 42 into plaques. Our, and other, results suggest that apparently the improved measurements of the soluble part of Aβ1– 42 makes it more difficult to measure aggregation. The concordance of CSF Aβ1– 42 with amyloid PET results was ±93% with the older generation Innotest assay, but decreased to ±85% using the next‐generation assays (current study and Janelidze et al. 9 and Doecke et al. 41 ). Direct comparison studies of soluble Aβ1– 42 measured with older versus newer generation assays showed an r2 of 0.8 to 0.9, which was lower than Aβ1– 42 correlations between next‐generation assays only, 9 , 14 , 42 suggesting that older and newer assays may not identically reflect Aβ1– 42. We and others collectively show that ratios with Aβ1– 42 for the next‐generation assays strongly improve concordance with amyloid PET to 90% to 95%. 15 , 16 , 17 , 18 , 19 , 20 For Aβ1– 42/Aβ1– 40 an explanation might be that this ratio better reflects the rate of amyloid precursor protein metabolism and as such correct for physiological Aβ1–42 effects. Aβ1– 42 as ratio of p‐tau or t‐tau might give a better reflection of aggregation likely due to the correlation of high p‐tau and t‐tau levels with amyloid plaques. 43 It might seem less intuitive to combine CSF p‐tau with Aβ1‐42 for prediction of amyloid PET, although for clinical use combining two hallmark pathologies instead of only the amyloid pathology contributes to a more accurate risk prediction of developing AD in preclinical stages. 44
We achieved a concordance of 90% to 94% for CSF Aβ1‐42 biomarkers and ratios compared to PET. The small number of cases with discordant CSF and PET results could be explained by either changes in CSF Aβ1‐42 preceding those in amyloid PET 45 or amyloid PET changes preceding those in CSF. 46 Longitudinal studies showed that patients with CSF+/PET– amyloid status seem to be in the earliest stages of AD development, as they turned amyloid positive on PET within the next years. 46 , 48 , 49 Patients with CSF–/PET+ discordant amyloid status did not develop amyloid or tau accumulation on PET in the next five years, 48 but did deteriorate on cognition, 47 suggesting that CSF and PET amyloid reflect different aspects of amyloid pathology.
Comparison of biomarker cut‐points across assays and cohorts (Table 4) suggests similar performance of Elecsys and Lumipulse assays; these assays can thus be used interchangeably to detect amyloid positivity, provided that assay‐specific cut‐offs are used. For multicenter studies, we recommend using one type of assay or using dichotomized biomarker results based on assay‐specific cut‐points. It is important to mention that cut‐points and corresponding sensitivity and specificity percentages when based on Youden's index will naturally show variation across cohorts that is inherent to differences in cohort compositions (i.e., diagnoses, disease severity, and age). For Lumipulse in particular, larger cohorts are required to assess the across‐cohort stability of biomarker cut‐points. Also, cut‐points will depend on pre‐analytical conditions. Pre‐analytics were the same for the analyses within the current study, but not completely similar compared to the other studies presented in Table 4 nor to the situation deemed ideal in routine diagnostics, which is direct biomarker analysis without sample freezing. The latter would, however, be hard to implement in view of analyses of samples that are shipped, for example, from smaller memory clinics without biomarker lab facilities or for centralized biomarker analyses performed in clinical trials. Lumipulse assay standards were recalibrated against the certified reference material at time of biomarker analysis in this study, 49 but the Elecsys assays were not. Recent recalibration of the Elecsys assay standards compared to Lumipulse and another assay showed the promising result of < 9% between‐assay bias in Aβ1‐42 concentrations measured in the certified reference materials. 50 Assay comparison studies in clinical cohorts should further examine the feasibility of using global cut‐points for CSF biomarker interpretation with these recalibrated assays.
Our study was performed in a real‐world memory clinic setting as we did not only include patients in the AD dementia spectrum, but also other dementias such as FTD, DLB, and VaD, which do not typically show amyloid pathology. The agreement of the CSF amyloid and amyloid PET results was however not different in our cohort when we excluded diagnoses other than AD, MCI, or SCD (15% of the original cohort), suggesting that CSF biomarkers perform well for amyloid PET positivity regardless of clinical diagnosis. This supports the use of CSF biomarkers in clinical diagnostic settings.
Because the Aβ1‐40 assay is not commercially available for Elecsys for use in clinical practice, we here combined the Lumipulse Aβ1‐40 result with the Aβ1‐42 result from Elecsys. This resulted in an AUC of 0.93, similar to AUCs reported in studies that measured the amyloid peptides on the same platform. 15 , 17 Potential noise due to differences in reagents and protocols between platforms was thus not reflected in the performance of this combined ratio. To enable use of amyloid ratios in clinical practice, we therefore suggest that the Elecsys biomarkers can be combined with the Aβ1‐40 result from another platform, such as Lumipulse, to obtain a ratio of Aβ1‐42/Aβ1‐40.
The major strength of this study is that we compared CSF biomarker results between two next‐generation (Elecsys and Lumipulse) assays in the same dataset. As both assays are applied in a clinical setting and for clinical trial analyses, such head‐to‐head comparison is important for future alignment of biomarker results interpretation. Our results show a strong agreement between biomarker ratios and amyloid PET for both platforms, meaning that biomarker outcomes from either platform reliably reflect the presence of amyloid pathology, as long as the platform‐specific cut‐points are applied.
A limitation of the current study is that the Elecsys Aβ1‐42 assay has its upper limit of detection at 1700 pg/mL. Although for diagnostic purposes (when biomarker status is determined for dichotomized values) this is not an issue, it hampers research on better understanding continuous CSF concentrations. 51 The performance of the Elecsys ratios might be slightly underestimated by including these values as 1700 pg/mL, instead of their actual, higher concentration, because for two to five cases the resulting biomarker ratio was classified as pathological (which was not the case when entering a hypothetical higher value, e.g., 2500 pg/mL). Another limitation could be that quantitative analyses for the PET scans were not available in this study. Visual reads, however, are the FDA‐approved method of identifying amyloid positivity; moreover, scans were read by one experienced nuclear medicine physician (BvB) and according to standardized procedures, which increases robustness of the reading results. Furthermore, different tracers were used for amyloid PET scoring, but any potential variation was minimized by using visual reads of PET results and the intra‐rater reliability of different tracers applied within one subject was 100% (BvB).
Altogether, based on the data here presented we recommend using the p‐tau/Aβ1‐42, Aβ1‐42/Aβ1‐40, or t‐tau/Aβ1‐42 ratio for AD pathology when using the automated assays Elecsys or Lumipulse, as these most accurately reflect the amyloid PET result. These ratios can be used for CSF biomarker interpretation in routine clinical settings or for clinical trial evaluation.
CONFLICTS OF INTEREST
Eline A.J. Willemse, Betty M. Tijms, Bart N.M. van Berckel, Wiesje M. van der Flier, Philip Scheltens, and Charlotte E. Teunissen have no competing interests to declare. Nathalie Le Bastard is a full‐time employee of Fujirebio Europe NV, Gent, Belgium.
AUTHOR CONTRIBUTIONS
Study supervision: Philip Scheltens and Charlotte E. Teunissen. Study conception and design: Eline A.J. Willemse, Betty M. Tijms, Nathalie Le Bastard, Wiesje M. van der Flier, Philip Scheltens, and Charlotte E. Teunissen. Data acquisition: Eline A.J. Willemse, Bart N.M. van Berckel, Wiesje M. van der Flier, Philip Scheltens. Analysis and interpretation of data: Eline A.J. Willemse, Betty M. Tijms, Wiesje M. van der Flier, Philip Scheltens, and Charlotte E. Teunissen. Drafting of the manuscript: Eline A.J. Willemse. Obtaining funding: Bart N.M. van Berckel, Wiesje M. van der Flier, Philip Scheltens, Nathalie Le Bastard, Charlotte E. Teunissen. All authors critically revised the manuscript for its intellectual content.
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
The authors thank Joop Nijhof, Kees van Uffelen, and Lynn Boonkamp for their excellent technical assistance. We are thankful for receiving in‐kind contributions in the form of assay kits from Roche Diagnostics and Fujirebio Europe N.V. These contributions had no influence on the results or conclusions from this study. Part of this study was funded by a ZonMW Memorabel grant (project number 733050206). Research of Alzheimer Center Amsterdam is part of the neurodegeneration research program of Amsterdam Neuroscience. Alzheimer Center Amsterdam is supported by Stichting Alzheimer Nederland and Stichting VUmc fonds. The chair of Wiesje van der Flier is supported by the Pasman stichting. PET scan costs were funded by grants of ZonMW, Piramal Imaging (now Life‐MI), GE Healthcare, Avid Radiopharmaceuticals, and the Center for Translational Molecular Medicine.
Willemse EAJ, Tijms BM, van Berckel BNM, et al. Comparing CSF amyloid‐beta biomarker ratios for two automated immunoassays, Elecsys and Lumipulse, with amyloid PET status . Alzheimer's Dement. 2021;13:e12182. 10.1002/dad2.12182
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Jack CR, Bennett DA, Blennow K, et al. NIA‐AA Research Framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14:535‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scheltens P, Blennow K, Breteler MMB, et al. Alzheimer's disease. Lancet. 2016;388:505‐517. [DOI] [PubMed] [Google Scholar]
- 3. Mattsson N, Andreasson U, Persson S, et al. CSF biomarker variability in the Alzheimer's Association quality control program. Alzheimers Dement. 2013;9:251‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuhlmann J, Andreasson U, Pannee J, et al. CSF Aβ1–42 – an excellent but complicated Alzheimer's biomarker – a route to standardisation. Clin Chim Acta. 2016;467:27‐33. [DOI] [PubMed] [Google Scholar]
- 5. Winblad B. BIOMARKAPD | JPND 2012. https://www.neurodegenerationresearch.eu/initiatives/annual‐calls‐for‐proposals/closed‐calls/biomarkers‐transnational‐call/results‐of‐biomarker‐call/biomarkapd/ Accessed April 30, 2019.
- 6. del Campo M, Mollenhauer B, Bertolotto A, et al. Recommendations to standardize preanalytical confounding factors in Alzheimer's and Parkinson's disease cerebrospinal fluid biomarkers: an update. Biomark Med. 2012;6:419‐430. [DOI] [PubMed] [Google Scholar]
- 7. Janelidze S, Stomrud E, Brix B, Hansson O. Towards a unified protocol for handling of CSF before β‐amyloid measurements. Alzheimers Res Ther. 2019;11:63. 10.1186/s13195-019-0517-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pannee J, Blennow K, Zetterberg H, Portelius E. Absolute quantification of Aβ1‐42 in CSF using a mass spectrometric reference measurement procedure. J Vis Exp. 2017;. 121:55386. 10.3791/55386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Janelidze S, Pannee J, Mikulskis A, et al. Concordance between different amyloid immunoassays and visual amyloid positron emission tomographic assessment. JAMA Neurol. 2017;74:1492‐1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bjerke M, Andreasson U, Kuhlmann J, et al. Assessing the commutability of reference material formats for the harmonization of amyloid beta measurements. Clin Chem Lab Med. 2015;54:1177‐1191. [DOI] [PubMed] [Google Scholar]
- 11. Pannee J, Gobom J, Shaw LM, et al. Round robin test on quantification of amyloid‐β 1‐42 in cerebrospinal fluid by mass spectrometry. Alzheimers Dement. 2016;12:55‐59. [DOI] [PubMed] [Google Scholar]
- 12. Bittner T, Zetterberg H, Teunissen CE, et al. Technical performance of a novel, fully automated electrochemiluminescence immunoassay for the quantitation of β‐amyloid (1‐42) in human cerebrospinal fluid. Alzheimers Dement. 2016;12:517‐526. 10.1016/j.jalz.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 13. The Alzheimer's Association QC program for CSF biomarkers n.d. https://neurophys.gu.se/english/departments/psychiatry_and_neurochemistry/Neurochemical_pathophysiology_and_diagnostics/TheAlzAssQCProgram Accessed April 30, 2019.
- 14. Willemse EAJ, van Maurik IS, Tijms BM, et al. Diagnostic performance of Elecsys immunoassays for cerebrospinal fluid Alzheimer's disease biomarkers in a nonacademic, multicenter memory clinic cohort: the ABIDE project. Alzheimers Dement (Amst). 2018;10:563‐572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doecke JD, Ward L, Burnham SC, et al. Elecsys CSF biomarker immunoassays demonstrate concordance with amyloid‐PET imaging. Alzheimers Res Ther. 2020;12:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hansson O, Seibyl J, Stomrud E, et al. CSF biomarkers of Alzheimer's disease concord with amyloid‐β PET and predict clinical progression: a study of fully automated immunoassays in BioFINDER and ADNI cohorts. Alzheimers Dement. 2018;14:1470‐1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schindler SE, Gray JD, Gordon BA, et al. Cerebrospinal fluid biomarkers measured by Elecsys assays compared to amyloid imaging. Alzheimers Dement. 2018;14:1460‐1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shaw LM, Waligorska T, Fields L, et al. Derivation of cutoffs for the Elecsys® amyloid β (1–42) assay in Alzheimer's disease. Alzheimer's dement diagnosis. Assess Dis Monit. 2018;10:698‐705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alcolea D, Pegueroles J, Muñoz L, et al. Agreement of amyloid PET and CSF biomarkers for Alzheimer's disease on Lumipulse. Ann Clin Transl Neurol. 2019;6:1815‐1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaplow J, Vandijck M, Gray J, et al. Concordance of Lumipulse cerebrospinal fluid t‐tau/Aβ42 ratio with amyloid PET status. Alzheimer's. Dement. 2020;16:144‐152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vanderstichele HMJ, Janelidze S, Demeyer L, et al. Optimized standard operating procedures for the analysis of cerebrospinal fluid Aβ42 and the ratios of Aβ isoforms using low protein binding tubes. J Alzheimers Dis. 2016;53:1121‐1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Willemse E, van Uffelen K, Brix B, Engelborghs S, Vanderstichele H, Teunissen C. How to handle adsorption of cerebrospinal fluid amyloid β (1–42) in laboratory practice? Identifying problematic handlings and resolving the issue by use of the Aβ42/Aβ40 ratio. Alzheimers Dement. 2017;13. 10.1016/j.jalz.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 23. Delaby C, Muñoz L, Torres S, et al. Impact of CSF storage volume on the analysis of Alzheimer's disease biomarkers on an automated platform. Clin Chim Acta. 2019;490:98‐101. [DOI] [PubMed] [Google Scholar]
- 24. van der Flier WM, Scheltens P. Amsterdam dementia cohort: performing research to optimize care. J Alzheimers Dis. 2018;62(3):1091‐1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Neary D, Snowden JS, Gustafson L, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546‐1554. [DOI] [PubMed] [Google Scholar]
- 27. McKeith IG, Dickson DW, Lowe J, et al. Diagnosis and management of dementia with Lewy bodies: third report of the DLB Consortium. Neurology. 2005;65:1863‐1872. [DOI] [PubMed] [Google Scholar]
- 28. Román GC, Tatemichi TK, Erkinjuntti T, et al. Vascular dementia: diagnostic criteria for research studies. Report of the NINDS‐AIREN International Workshop. Neurology. 1993;43:250‐260. [DOI] [PubMed] [Google Scholar]
- 29. van der Flier WM, Pijnenburg YAL, Prins N, et al. Optimizing patient care and research: the Amsterdam Dementia Cohort. J Alzheimers Dis. 2014;41:313‐327. [DOI] [PubMed] [Google Scholar]
- 30. Paciotti S, Sepe FN, Eusebi P, et al. Diagnostic performance of a fully automated chemiluminescent enzyme immunoassay for Alzheimer's disease diagnosis. Clin Chim Acta. 2019;494:74‐78. [DOI] [PubMed] [Google Scholar]
- 31. Leitão MJ, Silva‐Spínola A, Santana I, et al. Clinical validation of the Lumipulse G cerebrospinal fluid assays for routine diagnosis of Alzheimer's disease. Alzheimers Res Ther. 2019;11:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ossenkoppele R, Prins ND, Pijnenburg YAL, et al. Impact of molecular imaging on the diagnostic process in a memory clinic. Alzheimers Dement. 2013;9:414‐421. [DOI] [PubMed] [Google Scholar]
- 33. de Wilde A, van Maurik IS, Kunneman M, et al. Alzheimer's biomarkers in daily practice (ABIDE) project: rationale and design. Alzheimers Dement. 2017;6:143‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zwan MD, Bouwman FH, Konijnenberg E, et al. Diagnostic impact of [18F]flutemetamol PET in early‐onset dementia. Alzheimers Res Ther. 2017;9:2. 10.1186/s13195-016-0228-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ten Kate M, Visser PJ, Bakardjian H, et al. Gray matter network disruptions and regional amyloid beta in cognitively normal adults. Front Aging Neurosci. 2018;10:67. 10.3389/fnagi.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robin X, Turck N, Hainard A, et al. pROC: an open‐source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Manuilova E, Schuetzenmeister A, Model F. mcr: Method Comparison Regression. R package version 1.2.1. 2014. https://CRAN.R-project.org/package=mcr
- 38. R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 39. Janelidze S, Zetterberg H, Mattsson N, et al. CSF Aβ42/Aβ40 and Aβ42/Aβ38 ratios: better diagnostic markers of Alzheimer disease. Ann Clin Transl Neurol. 2016;3:154‐165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lewczuk P, Matzen A, Blennow K, et al. Cerebrospinal fluid Aβ42/40 corresponds better than Aβ42 to amyloid PET in Alzheimer's disease. J Alzheimers Dis. 2017;55:813‐822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Doecke JD, Rembach A, Villemagne VL, et al. Concordance between cerebrospinal fluid biomarkers with Alzheimer's disease pathology between three independent assay platforms. J Alzheimers Dis. 2017;61:169‐183. [DOI] [PubMed] [Google Scholar]
- 42. Bjerke M, Andreasson U, Kuhlmann J, et al. Assessing the commutability of reference material formats for the harmonization of amyloid‐β measurements. Clin Chem Lab Med. 2016;54:1177‐1191. [DOI] [PubMed] [Google Scholar]
- 43. van der Kant R, Goldstein LSB, Ossenkoppele R. Amyloid‐β‐independent regulators of tau pathology in Alzheimer disease. Nat Rev Neurosci. 2020;21:21‐35. [DOI] [PubMed] [Google Scholar]
- 44. Duits FH, Teunissen CE, Bouwman FH, et al. The cerebrospinal fluid "Alzheimer profile": easily said, but what does it mean?. Alzheimers Dement. 2014;10:713‐723. [DOI] [PubMed] [Google Scholar]
- 45. Tijms BM, Vermunt L, Zwan MD, et al. Pre‐amyloid stage of Alzheimer's disease in cognitively normal individuals. Ann Clin Transl Neurol. 2018;5(9):1037‐1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Palmqvist S, Mattsson N, Hansson O. Cerebrospinal fluid analysis detects cerebral amyloid‐β accumulation earlier than positron emission tomography. Brain. 2016;139:1226‐1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Landau SM, Lu M, Joshi AD, et al. Comparing positron emission tomography imaging and cerebrospinal fluid measurements of β‐amyloid. Ann Neurol. 2013;74:826‐836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guo T, Shaw LM, Trojanowski JQ, Jagust WJ, Landau SM, Alzheimer's Disease Neuroimaging Initiative . Association of CSF Aβ, amyloid PET and cognition in cognitively unimpaired elderly adults. Neurology. 2020;95. 10.1212/WNL.0000000000010596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reimand J, Collij L, Scheltens P, Bouwman F, Ossenkoppele R, for the Alzheimer's Disease Neuroimaging Initiative . Association of amyloid‐beta CSF/PET discordance and tau load five years later. Neurology. 2020;. 95(19):e2648‐e2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Degrieck R, Dauwe M, Dekeyser F, et al. Implementation of IFCC‐endorsed CRMs in the LUMIPULSE® G β‐Amyloid 1‐42 assay, Lissabon: 14th International Conference on Alzheimer's & Parkison's Diseases; 2019.
- 51. Boulo S, Kuhlmann J, Andreasson U, et al. First amyloid β1‐42 certified reference material for re‐calibrating commercial immunoassays. Alzheimers Dement. 2020;. 16(11):1493‐1503. 10.1002/alz.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eklund A (2021). beeswarm: The Bee Swarm Plot, an Alternative to Stripchart. R package version 0.3.1. https://CRAN.R-project.org/package=beeswarm
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


