Abstract
Little is known about the functions of the crustacean gut microbiome, but environmental parameters and habitat are known to affect the composition of the intestinal microbiome, which may in turn affect the physiological status of the host. The mud crab Scylla serrata is an economically important species, and is wild‐caught, and farmed across the Indo‐Pacific region. In this study, we compared the composition of the gut microbiome (in terms of gut microbial species richness and abundance) of S. serrata collected from wild sites, and farms, from the east and west coast of India, and also tested the effects of the environment on the composition. The water temperature had a statistically significant effect on gut microbiome composition, with microbial biodiversity decreasing with increasing water temperature. This could have negative effects on both wild and farmed mud crabs under future climate change conditions, although further research into the effects of temperature on gut microbiomes is required. By comparison, salinity, crab mass and carapace width, geographical location as well as whether they were farmed or wild‐caught crabs did not have a significant impact on gut microbiome composition. The results indicate that farming does not significantly alter the composition of the gut microbiome when compared to wild‐caught crabs.
Keywords: 16S rRNA, aquaculture, bacterial diversity, gut microbiome, mud crab, nanopore sequencing
The effects of habitat (wild vs farmed), location, and environment on the composition of the gut microbiome of the mud crab Scylla serrata in India were tested. Location and habitat are not associated with differences in intestinal microbial composition. However, the water temperature had a significant effect and microbial diversity decreases with increasing temperature. This could have negative effects on both wild and farmed mud crabs under future climate change conditions.

1. INTRODUCTION
It is now well established that the gut microbiome of humans and other vertebrates is involved in various physiological processes such as development, nutrition, and the immune response, including the production of vitamins and exogenous enzymes (e.g. Belkaid & Hand, 2014; Brestoff & Artis, 2013; Rowland et al., 2018), all of which play an important role in maintaining the internal environment of the host. Whilst it has been hypothesized that the crustacean gut microbiome positively contributes to crustacean physiological and metabolic status (Cornejo‐Granados et al., 2018), and any disturbance in the delicate balance of the gut microbial composition can affect their susceptibility to pathogens (Shi et al., 2019), relatively little is known about the structure and function of the intestinal microbiota in this group. The composition of the crustacean gut microbiome depends on several internal and external factors such as the developmental stage of the host (e.g. Rungrassamee et al., 2013), host anatomy (e.g. Apprill, 2017) environmental conditions that are either seasonal or sudden and extreme events such as prevalent rainfalls, increased temperature, as well as their habitat, availability of feed (e.g. Sullam et al., 2012; Xia et al., 2014) and stress related to, for instance, territorialism (Moloney et al., 2014).
Crabs from the genus Scylla are currently the only farmed commercial crab species in India and the mud crab Scylla serrata is a particularly economically important species due to its large size and high meat content (Le Vay, 2001). Crab farming is a growing sector, especially in the state of Andhra Pradesh on the east coast that is considered the “cradle of Indian aquaculture” (Belton et al., 2017). On the other hand, local communities along the state of Karnataka on the west coast are involved in sporadic marine and inland fishing, rather than the farming of crabs on a large scale (Government of Karnataka, 2016). Studies on fish have shown that hatchery‐reared and/or captive fish have microbiomes that differ from their wild counterparts with reduced biodiversity or significantly different composition that potentially can lead to disadvantages to the host, such as altered metabolic pathways, and reduced immunity (e.g. Lavoie et al., 2018; Ramirez & Romero, 2017).
In this study, we characterized the gut microbiome of the mud crab S. serrata, and compared the microbial composition in animals from wild sites and crab farms, from the east and west coast of India. To quantify any differences in the microbiome of crabs, we used long read 16S rRNA nanopore sequencing. Further, we aimed to examine the role of geographical location, habitat (estuaries or aquaculture pond), and environmental conditions (salinity and temperature) on their impact on gut microbial diversity and quantity and how it relates to the physiological status of the animal.
2. MATERIALS AND METHODS
2.1. Sample collection
Twenty four male S. serrata crabs (with no signs of disease) were collected from the west and east coasts of South India (Figure 1). This included animals from the wild catch and also from crab farms. Crabs (n = 3, C1‐3) from each sampling coast were collected from two sites (estuaries) representing wild samples (WW1‐2, west coast, and EW1‐2, east coast) and two culture farms (WF 1–2, west coast, and EF1‐2, east coast). Water temperature and salinity were recorded at each site (Table 1). Animals in both farms on the west coast from where samples were collected were fed with fresh bycatch, mainly sardines. Crabs on the east coast were fed with fresh tilapia in the farm EF1 and dried sardines in the farm EF2. Apart from the site EF2 where animals were fed a mix of probiotics, yeast, and jaggery (unrefined cane sugar) once a month, no additives were given at the other farms. Crabs in the sites EF2, WF1, and WF2 were kept in earthen ponds, while site EF1 was connected to the estuary. The crabs were transported to the laboratory as soon as possible and subjected to cryoanesthesia. After the measurement of weight and carapace width, the animals were thoroughly washed with sterile water and disinfected with 75% ethanol for 2–3 minutes. The animals were dissected using sterile lancets and the gut (midgut and hindgut) was separated using sterile forceps and immediately placed in sterile 1.5 mL microcentrifuge tubes. All dissecting tools were alcohol flame sterilized between dissecting each sample. Samples were stored at −80°C until further analysis.
FIGURE 1.
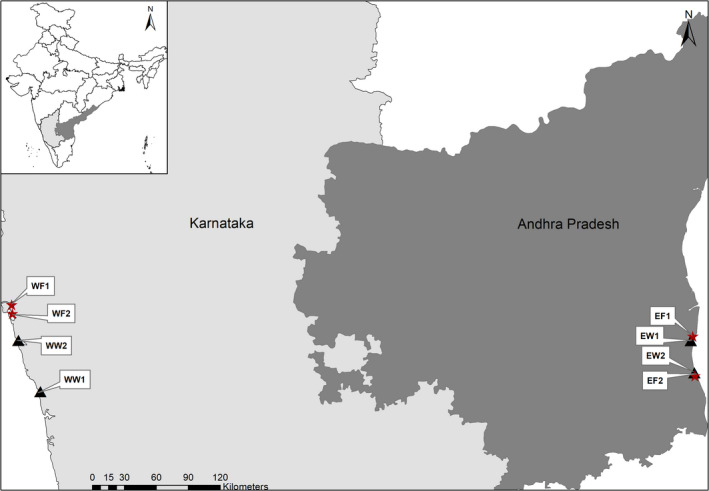
Sampling sites: triangle—wild sites, star—farms. WF—west coast farm, WW—west coast wild site, EF—east coast farm, EW—east coast wild site. Three crabs (C1‐C3) were collected from each sampling site.
TABLE 1.
Characteristics of sampling sites and crabs.
| Sample ID | Coast | Site type | Latitude | Longitude | Temperature (oC) |
Salinity (ppt) |
Crab mass (g) | Carapace width (mm) |
|---|---|---|---|---|---|---|---|---|
| WW1 | West | Wild | 13o50’53.52”N | 74o37’52.089” E | 30 | 27 | 450.88 ± 98.55 | 140.00 ± 14.79 |
| WW2 | West | Wild | 14o16’47.496” N | 74o26’37.679” E | 29 | 33 | 699.56 ± 215.63 | 160 ± 17.32 |
| WF1 | West | Farm | 14o34’26.364” N | 74o22’28.938” E | 28 | 35 | 148.93 ± 30.54 | 91.33 ± 4.93 |
| WF2 | West | Farm | 14o30’19.296” N | 74o23’38.151” E | 27 | 10 | 815.26 ± 33.15 | 158.00 ± 2.00 |
| EW1 | East | Wild | 14o16’43.86” N | 80o7’19.436” E | 31 | 21 | 200.00 ± 164.62 | 109.00 ± 29.51 |
| EW2 | East | Wild | 14o0’24.948” N | 80o9’10.411” E | 30 | 33 | 103.33 ± 40.41 | 87.33 ± 10.11 |
| EF1 | East | Farm | 14o18’48.168” N | 80o8’20.893” E | 27 | 27 | 366.66 ± 81.44 | 130.00 ± 6.55 |
| EF2 | East | Farm | 13o58’46.272” N | 80o9’27.586” E | 35 | 36 | 190.00 ± 52.91 | 101.33 ± 4.16 |
2.2. DNA extraction, PCR amplification and sequencing of 16S rRNA amplicon
Total DNA from gut samples was extracted using the QIAamp DNA Stool Mini Kit (QIAGEN, Germany) and DNeasy PowerSoil Kit (QIAGEN, Germany) following the manufacturer's instructions. Intestines were firstly lysed in InhibitEX Buffer and then purified on QIAamp spin columns. Purification includes digesting proteins with Proteinase K, binding DNA to the QIAamp silica membrane, washing away impurities and eluting pure DNA from the spin column with water. The quality and quantity of extracted DNA were determined in a NanoPhotometer N60 (Implen, Germany). Samples were stored at −20°C until amplification.
The 16S rRNA gene was then amplified using forward primer 16F‐ 5’ AGAGTTTGATCMTGGCTCAG 3’ and the reverse primer 16R‐ 5’ TACGGYTACCTTGTTACGACTT 3’. The PCR mixture contained high‐fidelity DNA polymerase, 0.5 mM dNTPs, 3.2 mM MgCl2 and PCR enzyme buffer, 40 ng of extracted DNA and 10 pM of each primer. The reaction conditions included an initial denaturation at 95°C for 3 minutes followed by 25 cycles each of denaturation at 95°C for 15 seconds, annealing at 60°C for 15 seconds and elongation at 72°C for 2 minutes followed by a final extension at 72°C for 10 minutes. The PCR products were purified using the QIAGEN Gel Purification Kit (QIAGEN, Germany). The amplified products were outsourced for the library preparation and Oxford Nanopore Technology (ONT) 1‐D sequencing using GridION device to the Biokart India Pvt. Ltd., Bangalore, India according to the manufacturer's instruction. Briefly, amplicons were purified using the QIAGEN Gel Purification Kit (QIAGEN, Germany). DNA concentration was estimated by using a Qubit dsDNA HS assay kit and Qubit 4.0 Fluorometer (Thermo Fisher Scientific, USA). Purified PCR products from each sample were end‐repaired and dA tailing using NEBNext Ultra II End Repair/dA‐Tailing Module (New England Biolabs, USA) was performed according to the protocol described by the manufacturer. The dA tailed PCR products were ligated with barcode adaptors using the Oxford Nanopore Native Barcode kit (EXP‐PBC096) and the Oxford Nanopore 1D Ligation Sequencing kit (SQK‐LSK109). The DNA library was loaded into a flow cell for 24–48 h run on the GridION portable sequencer for sequencing (Oxford Nanopore Technologies, UK).
2.3. Data analysis
After base‐calling raw FAST5 files, trimming and alignment of the reads along with the operational taxonomic unit (OTU) picking was performed using GAIA 2.0 workflow (Paytuví et al., 2019). The length of the sequences varied mainly between 100 and 1600 base pairs. Sample WF2C1 was excluded from further analyses as it was a statistically significant outlier due to low quality reads according to Grubb's test (p < 0.05). Alpha diversity and beta diversity at the genus level were calculated in PAST (Hammer et al., 2001). METAGENassist (Arndt et al., 2012) was used to map OTUs to phenotype. Statistical analyses and plotting were carried out in PRIMER‐E (Clarke and Gorley, 2006) and the R Studio using Bray‐Curtis similarity of square root transformed data. The genera abundant less than 1% were combined in the group designated as “Other”. Values of p < 0.05 were considered significant (95% confidence interval). SIMPER test was used to calculate (dis)similarity between groups using the average of Bray–Curtis dissimilarity. An unconstrained hierarchical divisive clustering routine UNCTREE was used to cluster samples based on alpha diversity. As for the multivariate analysis, we chose distance‐based linear model (DistLM) in PRIMER‐E and permutational multivariate analysis of variance (PERMANOVA) using community ecology package “vegan” in the R Studio (Oksanen et al., 2017) to evaluate the significance of environmental parameters, crab mass and carapace width, geographical location and type. Chi‐square test was used to assess associations between alpha biodiversity indices and variable factors.
3. RESULTS
3.1. The composition of the gut microbiome
The 16S rRNA amplicon sequencing on Nanopore GridION generated a total of 530,355 OTUs, from which 32% could not be assigned to any taxonomic unit. Acquired OTUs were assigned to 19 phyla, 45 classes, 88 orders, 160 families, 317 genera, and 430 species. The OTUs were assigned to five main phyla: Proteobacteria (51.8% ±9.7%), Actinobacteria (10.9% ±8.3%), Cyanobacteria, (7.3% ±4.2%) Firmicutes (4.6% ±1.1%) and Bacteriodetes (3.2% ±0.8%); five classes: Betaproteobacteria (43% ±12%), Alphaproteobacteria (5.7% ± 1.6%), Actinobacteria (5.1% ±3.9%), Bacilli (4.1% ±1.4%), and Rubrobacteria (3.3% ±2.0%); five major genera: Massilia (25% ±11.5%), Pseudoduganella (8.1% ±3.5%), Microcoleus (4.3% ±2.3%), Bacillus (3.1% ±1.0%), and Gaiella (2.9% ±1.4%) (Figure 2). At the species level, OTUs were assigned to five main species: Massilia albidiflava (25.2%±7.3%), Massilia sp. NCCP 1146 (2.6% ±0.4%), Microcoleus sp. HTT‐U‐KK5 (2.6% ±1.6%), Pseudoduganella violaceinigra (9.3% ±2.1%), and Aciditerrimonas ferrireducens (1.4%± 0.9%).
FIGURE 2.
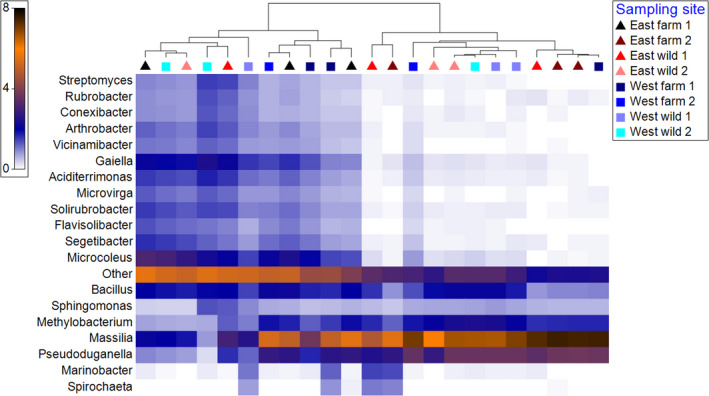
Shade plots of relative abundance of operational taxonomic units OTUs (%) assigned to 20 most abundant genera of individual crab gut microbiomes from 8 different sampling sites. Triangles represent east coast samples and squares represent west coast samples. The samples are clustered with unconstrained hierarchical divisive clustering routine UNCTREE. The relative abundance is square root transformed. The taxa present less than 1% are combined under “Other”.
Geographical location or habitat (wild or pond cultivated) did not have a significant impact on gut microbial biodiversity. On the other hand, a distance‐based linear model (DistLM) showed that temperature had a statistically significant effect on the OTU abundance (%) at the genus level (p = 0.018). There was a trend of decreased OTU richness with increasing temperature (Figure 3). This was also confirmed by PERMANOVA (p = 0.032). However, salinity, crab mass, and carapace width were not statistically significant (p > 0.05). Calculated alpha diversity analysis showed that the number of bacterial genera found in mud crab guts varied from 92 (EF2C1) to 289 (WW1C3). While the temperature was the only statistically significant factor that affected Shannon's diversity index (H), the number of taxa alone was also significantly affected by the coast (p = 0.0117) and the interaction between crab body mass and carapace width (p = 0.0231).
FIGURE 3.
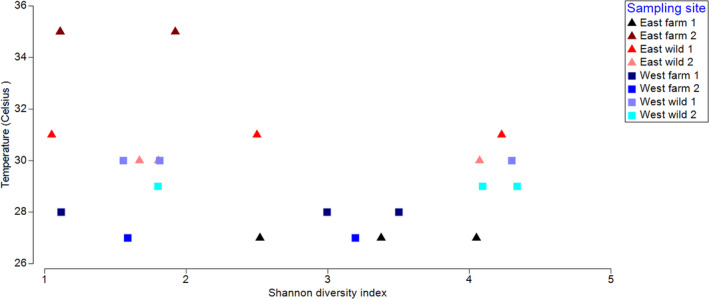
Shannon's diversity index (H’) at the genus level of individual crab gut microbiomes from 8 different sampling sites plotted against the temperature of their sampling sites. Triangles represent east coast samples and squares represent west coast samples. The samples EF2C1 and EF2C2 have similar Shannon diversity index, thus have overlapped and appear as one triangle. A higher number indicates higher biodiversity based on the OTU abundance and richness. The results of the distance‐based linear model showed that temperature had a statistically significant effect on the OTU abundance (%) at the genus level (p = 0.018).
Although microbial composition varied between individuals, all animals from the site EF1 presented consistently high OTU richness and evenness. Yet, in the case of the second farm on the east coast EF2, the OTU richness and evenness were the lowest (Table 2). We clustered samples based on the alpha diversity indices using unconstrained hierarchical divisive clustering routine UNCTREE and obtained two main clusters (Figure 4). The SIMPER analysis showed that the greatest dissimilarity of OTUs present in the eight sites sampled was between farms on the east coast EF1 and EF2 (62.53%) and the farm on the east coast and the wild site on the west coast EF2 and WF2 (64.36%). Examining similarity between wild and farmed animals, it was seen that OTUs varied more in wild animals (66.20% similarity within the group) than in the pond cultivated animals (71.39% similarity within the group).
TABLE 2.
Alpha diversity indices for individual animals at the genus level.
| Number of taxa | Individuals | Simpson 1‐D | Shannon H | Evenness eH/S | Chao−1 | |
|---|---|---|---|---|---|---|
| EF1C1 | 215 | 13299 | 0.9589 | 4.05 | 0.2669 | 218 |
| EF1C2 | 245 | 15040 | 0.7343 | 2.521 | 0.05076 | 251.3 |
| EF1C3 | 244 | 19057 | 0.879 | 3.377 | 0.1201 | 248.2 |
| EF2C1 | 92 | 15504 | 0.4555 | 1.109 | 0.03294 | 99.5 |
| EF2C2 | 95 | 15635 | 0.452 | 1.111 | 0.03198 | 107.7 |
| EF2C3 | 125 | 11575 | 0.6277 | 1.923 | 0.05474 | 137.7 |
| EW1C1 | 158 | 11594 | 0.7624 | 2.5 | 0.07707 | 182.5 |
| EW1C2 | 281 | 14622 | 0.965 | 4.228 | 0.2441 | 291 |
| EW1C3 | 57 | 3144 | 0.4295 | 1.05 | 0.05014 | 72.4 |
| EW2C1 | 112 | 6508 | 0.6009 | 1.67 | 0.04744 | 149.1 |
| EW2C2 | 143 | 12590 | 0.6397 | 1.804 | 0.04249 | 174.7 |
| EW2C3 | 252 | 18948 | 0.9579 | 4.072 | 0.2329 | 258.1 |
| WF1C1 | 83 | 9338 | 0.4551 | 1.117 | 0.03682 | 99.5 |
| WF1C2 | 246 | 12184 | 0.8933 | 3.502 | 0.1349 | 252.7 |
| WF1C3 | 262 | 14370 | 0.802 | 2.995 | 0.07626 | 263.8 |
| WF2C2 | 251 | 15716 | 0.8471 | 3.195 | 0.09721 | 259.7 |
| WF2C3 | 185 | 13056 | 0.5497 | 1.587 | 0.02642 | 222.6 |
| WW1C1 | 133 | 15533 | 0.5723 | 1.556 | 0.03563 | 177 |
| WW1C2 | 145 | 15569 | 0.6444 | 1.813 | 0.04227 | 178.2 |
| WW1C3 | 289 | 13056 | 0.9627 | 4.3 | 0.2549 | 292.7 |
| WW2C1 | 141 | 18257 | 0.6429 | 1.801 | 0.04295 | 153.2 |
| WW2C2 | 253 | 13369 | 0.9578 | 4.094 | 0.2371 | 263.9 |
| WW2C3 | 256 | 19567 | 0.973 | 4.337 | 0.2988 | 265.8 |
Simpson's index (1‐D) indicates evenness, Shannon's diversity index (H’) accounts for both species richness and abundance, Buzas and Gibson's evenness index (eH/S) implies evenness, Chao1 estimates based on the abundance of less present taxa.
FIGURE 4.
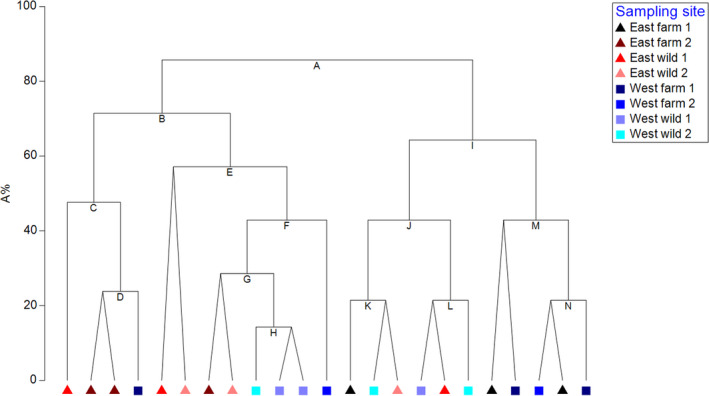
Unconstrained hierarchical divisive UNCTREE clusters based on alpha diversity indices of individual crab gut microbiomes at the genus level. Triangles represent east coast samples and squares represent west coast samples. The dendrogram is plotted against an arbitrary equi‐stepped scale (A%) in which the divisions sum up to 100.
3.2. Phenotypic characterization of the gut microbiome
The results of the mapping of obtained OTUs to phenotypic categories showed about 7% of bacteria found in crabs from sites EF1, EW2, and WW2 were potential human pathogens. However, enteric bacteria derived from the gut of warm‐blooded animals and the pathogenic genus like Salmonella was less than 0.1% and genera Staphylococcus and Streptococcus were less than 0.8%. In addition, no crab pathogens such as Aeromonas, Rickettsia, and Spiroplasma were found in any of the samples. Less than 0.1% of OTUs were identified as Vibrio parahaemolyticus.
Only between 8 to 22% of OTUs on an individual level could be mapped to a specific metabolic pathway. By mapping OTUs to phenotypic characteristics, the main five metabolic processes the mud crab gut microbiome is involved were ammonia oxidation, dehalogenation, sulfate reduction, nitrite reduction, and sulfide oxidation (Figure 5). A very low percentage of lignin degraders were mapped only to wild crab gut samples. Other metabolic processes identified included iron oxidation, lignin degradation, selenate reduction, sulfur reduction, and storage of polyhydroxybutyrate. PERMANOVA showed that temperature (p = 0.029) and habitat (p = 0.038) significantly affect differences between animals. The coast and salinity did not show any statistically significant difference.
FIGURE 5.
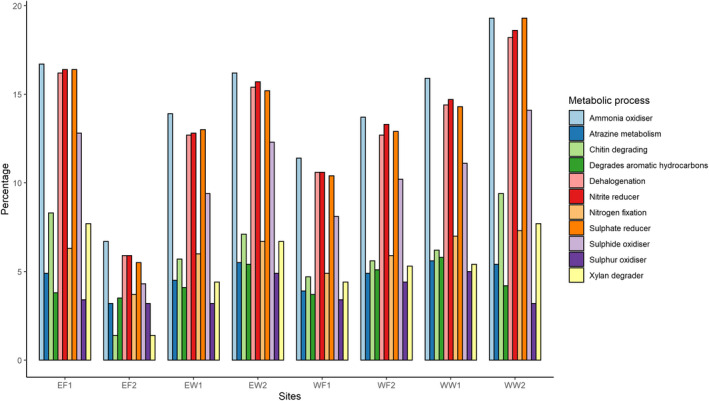
This figure indicates eleven main metabolic processes in which gut bacteria of mud crabs were involved in. Operational taxonomic units OTUs were mapped to phenotypic characteristics with the help of METAGENassist. The results shown, are the average for the sampling site, no individual data were given. To be recognized as one of the eleven main metabolic processes, 5% of OTUs of at least one sample had to be assigned to the process.
4. DISCUSSION
Due to the recognized contribution of the intestinal microbiome to host health, it is essential to assess the bacterial composition of aquaculture species as it plays a significant role in determining their physiological status. Studies on the gut microbiome of aquatic animals and especially fish show that trophic level, season, development, sex, habitat, and life stage are among the factors affecting the composition of the gut microbiome at the interspecies level (Butt & Volkoff, 2019). However, some studies report high individual variability of the crustacean gut microbiome within groups (Ding et al., 2017; Li et al., 2007; Wei et al., 2019). This was observed in this study too and could be explained by the fact that S. serrata is an omnivorous and opportunistic scavenger. We did not find any significant differences in the gut microbiome between wild and pond‐cultivated crabs and these results corroborated with the observation in Eriocheir sinensis (Li et al., 2007) and black tiger shrimp Penaeus monodon (Rungrassamee et al., 2014). On the other hand, higher diversity and higher bacterial load were observed in wild S. paramamoisam crabs than in the healthy and diseased pond‐raised crabs (Li et al., 2012). The similarity between groups suggests that environmental conditions might play an essential role in forming the gut microbiome (Fraune & Bosch, 2007). Furthermore, there is no formulated feed for S. serrata and the use of probiotics is not common; therefore, wild and pond raised crabs more likely have an identical kind of diet.
The most common phyla in the S. serrata gut microbiome were Proteobacteria, Actinobacteria, Cyanobacteria, Firmicutes, and Bacteriodetes, while the studies on S. paramamoisam from China found Fusobacteria and Tenericutes to be among the core gut microbiome phyla (Deng et al., 2019; Li et al., 2012; Wei et al., 2019). Yet, in this study, the gut microbiome of S. serrata comprised <0.08% Tenericutes, and no Fusobacteria were identified in any of the samples. Fusobacteria, Gram‐negative obligate anaerobic bacilli have been associated with colorectal adenoma and colorectal carcinoma (e.g. Kostic et al., 2012; Saito et al., 2019). Tenericutes, a Gram‐negative obligate cell‐associated bacteria have been recorded in all vertebrate guts examined. Although it is one of the least abundant phyla in mammalian gut microbiota, it has been found in dolphins in a relatively high proportion (Bik et al., 2016). Tenericutes is also one of the most abundant phyla in the gut of the Chinese mitten crab, Eriocheir sinensis (Ding et al., 2017; Dong et al., 2018; Zhang et al., 2016). In a meta‐analysis study of marine and freshwater shrimp microbiota, Tenericutes and Fusobacteria were twenty five and five times, respectively, more abundant in marine shrimps compared to freshwater shrimps (Cornejo‐Granados et al., 2018). Estuaries in south India are subject to highly variable salinity due to the heavy monsoon, which can vary from 0 to 35 ppt (Ramachandra et al., 2013; Shruthi et al., 2011), and this could explain the absence of these two phyla in the S. serrata gut microbiome. Although variations in the gut microbial composition in different geographical locations are often explained by the differences in the diet and behavior, and not by the location per se (Ye et al., 2014), it is not clear how these differences would affect animal health if crab seed (juveniles for farm rearing) were imported into India, in this instance, from China. Further research is required to determine differences in gut microbial composition at different developmental stages and whether changes in diet and environmental factors induce any alterations. Additionally, it would be interesting to analyze the implications of the above factors on host physiology.
Proteobacteria, Firmicutes, Bacteriodetes, and Actinobacteria comprise core components of the gut microbiome in humans (Hugon et al., 2015; Lawley & Walker, 2013), fish (e.gSandve et al., 2017; Sullam et al., 2012) and crustaceans. However, the crustaceans have less of Actinobacteria (e.g. Ding et al., 2017; Dong et al., 2018; Shi et al., 2019; Zhang et al., 2014, 2016) when compared to the other three groups. The abundance of Cyanobacteria in the gut could be linked with the host trophic level. A study on fish with different diets showed Cyanobacteria to be abundant in filter‐feeding fish, less in herbivorous and omnivorous fish and very little in carnivorous fish (Liu et al., 2016). Scylla serrata juveniles and small adult crabs (up to 99 mm CW) are omnivorous, whereas middle‐ and large‐sized crabs are top benthic predators, opportunistic scavengers and exhibit cannibalistic behavior (Alberts‐Hubatsch et al., 2016).
From informal enquiries with crab farmers in India, we are aware that rising temperatures that have been observed in recent years are perceived as one of the reasons for high crab mortality, and ultimately a threat to their livelihoods. Elevated water temperature has been shown to significantly reduce the bacterial diversity in the gut of mussels Mytilus coruscus, yet simultaneously increase the abundance of opportunistic bacteria, such as Bacteroides and Arcobacter, which could result in higher host susceptibility to disease (Li et al., 2018). Furthermore, the diversity of planktonic bacteria has been found to decrease in the Atlantic Ocean toward the equator (Milici et al., 2016). Thus, as the sea surface temperature (SST) is projected to increase (IPCC, 2014) as a result of global climate change, changes in the crab gut microbiome could be expected, and as a consequence, could negatively affect the physiological and immune status of crabs. This could be detrimental to crabs facing the twin threats of increasing SSTs and increasing pathogen levels such as Vibrio spp. due to warm temperatures (Semenza et al., 2017). Yet, the temperature is only one of many environmental factors that could determine microbial richness and abundance, thus more detailed studies considering various physiochemical data are required to understand the role of water temperature in altering the gut microbiome (Thompson et al., 2017). Further investigation is also required to assess the effects of probiotics and other additives such as yeast and jaggery applied in sampling site EF2 in interaction with physiochemical factors.
By mapping OTUs to phenotypic characteristics, almost none of the OTUs were assigned to ammonia‐oxidizing bacteria (AOB) such as Nitrosphira, Nitrosomonas, and Nitrosococcus (Burrel et al., 2001). Thus, we hypothesize that most nitrogen fixation, ammonia oxidation and nitrite reduction in the guts of S. serrata is performed by Cyanobacteria as reported in some studies (Andriesse et al., 1990; Herrero et al., 2001) evidence to which is indicated by their significant presence in the gut microbiome. The heterotrophic bacteria, B. subtilis, found in soil has also been reported to be involved in nitrogen fixation (Beneduzi et al., 2008; Hashem et al., 2019) and Bacillus was one of the main genera found in the crab gut. Ammonia, nitrite, and nitrate are common and essential components in the aquatic environment, yet elevated levels can be toxic to aquatic animals (Romano & Zeng, 2013). Therefore, the results indicate that gut bacteria are strongly involved in mineralization by processing these compounds to avoid toxic effects. Microbial oxidation of sulfur is carried out to produce energy that is further used for synthesizing their structural components and it is possible that Bacillus (Friedrich et al., 2001) and Microcoleus (Fike et al., 2016) could be responsible for these functions in the crab samples analyzed.
5. CONCLUSIONS
This study, to our knowledge, is the first to identify the composition of the gut microbiome of the mud crab, Scylla serrata using long read 16S rRNA Oxford Nanopore Sequencing Technology, and assess the impact of geographical location, habitat, and environmental conditions on bacterial diversity and abundance. By comparing the relative abundance and bacterial diversity of crab guts from wild and pond cultivated crabs, from both the east and west coasts of South India, it was observed that the geographical location, habitat, crab body mass and carapace width, and water salinity do not induce changes in the gut microbiome. However, the water temperature was shown to influence gut bacterial diversity, which tended to decrease with increasing water temperature. Human and animal pathogens made up less than 0.1% of the gut samples studied. Thus, the findings suggest that current practices of crab farming result in healthy crabs and that geographical location does not impact farm success. Yet, in the context of climate change, further research is required to assess the effects of temperature on gut microbiomes, and their functions, and whether and how controlling temperature in aquaculture settings might help offset changes associated with variability in climate. In addition to overexploitation, we perceive increased temperature as a result of climate change, to be another potential threat to wild S. serrata populations. Furthermore, India has developed a central hatchery for S. serrata seed production to promote mud crab aquaculture. The results obtained do not indicate that farmed crabs will be disadvantaged compared to their wild counterparts in terms of their gut microbiome composition.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Elina Apine: Conceptualization (supporting); Formal analysis (lead); Investigation (lead); Methodology (equal); Writing‐original draft (lead). Praveen Rai: Resources (lead); Supervision (supporting); Writing‐review & editing (equal). Madhu K Mani: Investigation (supporting); Project administration (supporting); Resources (equal); Writing‐review & editing (equal). Vikram Subramanian: Data curation (lead); Formal analysis (equal); Writing‐review & editing (equal). Indrani Karunasagar: Conceptualization (equal); Funding acquisition (supporting); Methodology (equal); Project administration (lead); Supervision (lead); Writing‐review & editing (equal). Anna Godhe: Conceptualization (lead); Funding acquisition (supporting); Methodology (lead). Lucy Turner: Conceptualization (equal); Funding acquisition (lead); Methodology (equal); Supervision (equal); Writing‐review & editing (lead).
ETHICS STATEMENT
None required.
ACKNOWLEDGEMENTS
This work was supported by a PhD scholarship granted by the University of Plymouth to EA (PI LMT). We would like to thank fishers and crab farmers for their help with animal collection. We also would like to thank laboratory technicians at NUCSER, Nitte (Deemed to be University), research scholar, Aloysius, for his assistance in crab collection on the west coast and Dr T. Neeraja at the College of Fishery Science, Muthukur for her help in Andhra Pradesh. The services of sequencing by Biokart Ltd. Laboratory, Bengaluru, is gratefully acknowledged.
DATA AVAILABILITY STATEMENT
The sequence datasets generated during the current study are available at NCBI Sequence Read Archive (SRA) under BioProject PRJNA691201: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA691201. BioSample accession numbers are SAMN17283444 ‐ SAMN17283464.
REFERENCES
- Alberts‐Hubatsch, H., Lee, S. Y., Meynecke, J. O., Diele, K., Nordhaus, I., & Wolff, M. (2016). Life‐history, movement, and habitat use of Scylla serrata (Decapoda, Portunidae): current knowledge and future challenges. Hydrobiologia, 763(1), 5–21. 10.1007/s10750-015-2393-z [DOI] [Google Scholar]
- Andriesse, X., Bakker, H., & Weisbeek, P. (1990). Analysis of Nitrate Reduction Genes in Cyanobacteria. In Ullrich W. R., Rigano C., Fuggi A., & Aparicio P. J. (Eds.), Inorganic Nitrogen in Plants and Microorganisms. Springer, Berlin. [Google Scholar]
- Apprill, A. (2017). Marine animal microbiomes: toward understanding host‐microbiome interactions in a changing ocean. Frontiers in Marine Science, 4, 1–9. 10.3389/fmars.2017.00222 [DOI] [Google Scholar]
- Arndt D., Xia J., Liu Y., Zhou Y., Guo A. C., Cruz J. A., Sinelnikov I., Budwill K., Nesbo C. L., & Wishart D. S. (2012). METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Research, 40,(W1), W88–W95. 10.1093/nar/gks497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid, Y., & Hand, T. W. (2014). Role of the microbiota in immunity and inflammation. Cell, 157(1), 121–141. 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton, B., Padiyar, A., Ravibabu, G., & Gopal Rao, K. (2017). Boom and bust in Andhra Pradesh: Development and transformation in India’s domestic aquaculture value chain. Aquaculture, 470, 196–206. 10.1016/j.aquaculture.2016.12.019 [DOI] [Google Scholar]
- Beneduzi, A., Peres, D., Vargas, L. K., Bodanese‐Zanettini, M. H., & Passaglia, L. M. P. (2008). Evaluation of genetic diversity and plant growth promoting activities of nitrogen‐fixing bacilli isolated from rice fields in South Brazil. Applied Soil Ecology, 39(3), 311–320. 10.1016/j.apsoil.2008.01.006 [DOI] [Google Scholar]
- Bik, E. M., Costello, E. K., Switzer, A. D., Callahan, B. J., Holmes, S. P., Wells, R. S., Carlin, K. P., Jensen, E. D., Venn‐Watson, S., & Relman, D. A. (2016). Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nature Communications, 7, 10156, 10.1038/ncomms10516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff, J. R., & Artis, D. (2013). Commensal bacteria at the interface of host metabolism and the immune system. Nature Immunology, 14(7), 676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrel, P. C., Phalen, C. M., & Hovanec, T. A. (2001). Identification of Bacteria Responsible for Ammonia Oxidation in Freshwater Aquaria. Applied and Environmental Microbiology, 67(12), 5791–5800. 10.1128/AEM.67.12.5791-5800.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt, R. L., & Volkoff, H. (2019). Gut microbiota and energy homeostasis in fish. Frontiers in Endocrinology, 10, 6–8. 10.3389/fendo.2019.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo‐Granados, F., Gallardo‐Becerra, L., Leonardo‐Reza, M., Ochoa‐Romo, J. P., & Ochoa‐Leyva, A. (2018). A meta‐analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota. PeerJ, 6, e5382. 10.7717/peerj.5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y., Cheng, C., Xie, J., Liu, S., Ma, H., Feng, J., Su, Y., & Guo, Z. (2019). Coupled changes of bacterial community and function in the gut of mud crab (Scylla Paramamosain) in response to Baimang disease. AMB Express, 9(1), 1–9. 10.1186/s13568-019-0745-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Z. F., Cao, M. J., Zhu, X. S., Xu, G. H., & Wang, R. L. (2017). Changes in the gut microbiome of the Chinese mitten crab (Eriocheir sinensis) in response to White spot syndrome virus (WSSV) infection. Journal of Fish Diseases, 40(11), 1561–1571. 10.1111/jfd.12624 [DOI] [PubMed] [Google Scholar]
- Dong, J., Li, X., Zhang, R., Zhao, Y., Wu, G., Liu, J., Zhu, X., & Li, L. (2018). Comparative analysis of the intestinal bacterial community and expression of gut immunity genes in the Chinese Mitten Crab (Eriocheir sinensis). AMB Express, 8(1), 10.1186/s13568-018-0722-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fike, D. A., Bradley, A. S., & Leavitt, W. D. (2016). Geomicrobiology of sulfur. In Ehrlich H. L., Newman D. K., & Kappler A. (Eds.), Ehrlich's Geomicrobiology, (6th ed.). Taylor and Francis Group. [Google Scholar]
- Fraune, S., & Bosch, T. C. G. (2007). Long‐term maintenance of species‐specific bacterial microbiota in the basal metazoan Hydra. Proceedings of the National Academy of Sciences of the United States of America, 104(32), 13146–13151. 10.1073/pnas.0703375104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, C. G., Rother, D., Bardischewsky, F., Quentmeier, A., & Fischer, J. (2001). Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Applied and Environmental Microbiology, 67(7), 2873–2882. 10.1128/AEM.67.7.2873-2882.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Government of Karnataka (2016). Fisheries Department. Introduction. Available at. http://www.karnataka.gov.in/fisheries/english/Pages/Introduction.aspx (Accessed 10 December 2019). [Google Scholar]
- Hammer, Ø., Harper, D. A. T., & Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica, 4(1), 9. [Google Scholar]
- Hashem, A., Tabassum, B., & Fathi Abd_Allah, E. (2019). Bacillus subtilis: A plant‐growth promoting rhizobacterium that also impacts biotic stress. Saudi Journal of Biological Sciences, 26(6), 1291–1297. 10.1016/j.sjbs.2019.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero, A., Muro‐Pastor, A. M., & Flores, E. (2001). Nitrogen control in cyanobacteria. Journal of Bacteriology, 183(2), 411–425. 10.1128/JB.183.2.411-425.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugon, P., Dufour, J. C., Colson, P., Fournier, P. E., Sallah, K., & Raoult, D. (2015). A comprehensive repertoire of prokaryotic species identified in human beings. The Lancet Infectious Diseases, 15(10), 1211–1219. 10.1016/S1473-3099(15)00293-5 [DOI] [PubMed] [Google Scholar]
- IPCC (2014). Intergovernmental Panel on Climate Change 5th Assessment Report. Cambridge and New York. [Google Scholar]
- Kostic, A. D., Gevers, D., Pedamallu, C. S., Michaud, M., Duke, F., Earl, A. M., Ojesina, A. I., Jung, J., Bass, A. J., Tabernero, J., Baselga, J., Liu, C., Shivdasani, R. A., Ogino, S., Birren, B. W., Huttenhower, C., Garrett, W. S., & Meyerson, M. (2012). Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research, 22(2), 292–298. 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie, C., Courcelle, M., Redivo, B., & Derome, N. (2018). Structural and compositional mismatch between captive and wild Atlantic salmon (Salmo salar) parrs’ gut microbiota highlights the relevance of integrating molecular ecology for management and conservation methods. Evolutionary Applications, 11(9), 1671–1685. 10.1111/eva.12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley, T. D., & Walker, A. W. (2013). Intestinal colonization resistance. Immunology, 138(1), 1–11. 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Vay, L. (2001). Ecology and management of mud crab scylla spp. Asian Fisheries Science, 14, 101–111. [Google Scholar]
- Li, K., Guan, W., Wei, G., Liu, B., Xu, J., Zhao, L., & Zhang, Y. (2007). Phylogenetic analysis of intestinal bacteria in the Chinese mitten crab (Eriocheir sinensis). Journal of Applied Microbiology, 103(3), 675–682. 10.1111/j.1365-2672.2007.03295.x [DOI] [PubMed] [Google Scholar]
- Li, S., Sun, L., Wu, H., Hu, Z., Liu, W., Li, Y., & Wen, X. (2012). The intestinal microbial diversity in mud crab (Scylla paramamosain) as determined by PCR‐DGGE and clone library analysis. Journal of Applied Microbiology, 113(6), 1341–1351. 10.1111/jam.12008 [DOI] [PubMed] [Google Scholar]
- Li, Y. F., Yang, N., Liang, X., Yoshida, A., Osatomi, K., Power, D., Batista, F. M., & Yang, J. L. (2018). Elevated seawater temperatures decrease microbial diversity in the gut of Mytilus coruscus. Frontiers in Physiology, 9, 1–9. 10.3389/fphys.2018.00839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Guo, X., Gooneratne, R., Lai, R., Zeng, C., Zhan, F., & Wang, W. (2016). The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Scientific Reports, 6, 1–12. 10.1038/srep24340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milici, M., Tomasch, J., Wos‐Oxley, M. L., Wang, H., Jáuregui, R., Camarinha‐Silva, A., Deng, Z.‐L., Plumeier, I., Giebel, H.‐A., Wurst, M., Pieper, D. H., Simon, M., & Wagner‐Döbler, I. (2016). Low diversity of planktonic bacteria in the tropical ocean. Scientific Reports, 6(1), 1–9. 10.1038/srep19054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney, R. D., Desbonnet, L., Clarke, G., Dinan, T. G., & Cryan, J. F. (2014). The microbiome: stress, health and disease. Mammalian Genome, 25, 49–74. 10.1007/s00335-013-9488-5 [DOI] [PubMed] [Google Scholar]
- Oksanen, J., Blanchet, G. F., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., Minchin, P. R., O'Hara, R. B., Simpson, G. L., Solymos, P., Stevens, M. H. H., Szoecs, E., & Wagner, H. (2017). vegan: Community Ecology Package. R package version 2.4‐3. https://CRAN.R‐project.org/package=vegan
- Paytuví, A., Battista, E., Scippacercola, F., Aiese Cigliano, R., & Sanseverino, W. (2019). GAIA: an integrated metagenomics suite. bioRxiv. 10.1101/804690 [DOI] [Google Scholar]
- Ramachandra, T. V., Subash Chandran, M. D., Joshi, N. V., Bhat, M., Mesta, P. N., & Naik, S. (2013). Estuarine Fish Diversity and Livelihoods in Uttara Kannada District, Karnataka State. Sahyadri Conservation Series 34, ENVIS Technical Report 64, CES, Indian Institute of Science. Bangalore, India. [Google Scholar]
- Ramirez, C., & Romero, J. (2017). Fine Flounder (Paralichthys adspersus) Microbiome Showed Important Differences between Wild and Reared Specimens. Frontiers in Microbiology, 8, 271. 10.3389/fmicb.2017.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, N., & Zeng, C. (2013). Toxic Effects of Ammonia, Nitrite, and Nitrate to Decapod Crustaceans: A Review on Factors Influencing their Toxicity, Physiological Consequences, and Coping Mechanisms. Reviews in Fisheries Science, 21(1), 1–21. 10.1080/10641262.2012.753404 [DOI] [Google Scholar]
- Rowland, I., Gibson, G., Heinken, A., Scott, K., Swann, J., Thiele, I., & Tuohy, K. (2018). Gut microbiota functions: metabolism of nutrients and other food components. European Journal of Nutrition, 57(1), 1–24. 10.1007/s00394-017-1445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungrassamee, W., Klanchui, A., Chaiyapechara, S., Maibunkaew, S., Tangphatsornruang, S., Jiravanichpaisal, P., & Karoonuthaisiri, N. (2013). Bacterial Population in Intestines of the Black Tiger Shrimp (Penaeus monodon) under Different Growth Stages. PLoS One, 8(4), e60802. 10.1371/journal.pone.0060802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungrassamee, W., Klanchui, A., Maibunkaew, S., Chaiyapechara, S., Jiravanichpaisal, P., & Karoonuthaisiri, N. (2014). Characterization of intestinal bacteria in wild and domesticated adult black tiger shrimp (Penaeus monodon). PLoS One, 9(3), e91853. 10.1371/journal.pone.0091853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., Koido, S., Odamaki, T., Kajihara, M., Kato, K., Horiuchi, S., Adachi, S., Arakawa, H., Yoshida, S., Akasu, T., Ito, Z., Uchiyama, K., Saruta, M., Xiao, J.‐Z., Sato, N., & Ohkusa, T. (2019). Metagenomic analyses of the gut microbiota associated with colorectal adenoma. PLoS One, 14(2), 1–18. 10.1371/journal.pone.0212406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandve, S. R., Angell, I. L., Vik, J. O., Pope, P. B., Snipen, L.‐G., & Rudi, K. (2017). Stable core gut microbiota across the freshwater‐to‐saltwater transition for farmed atlantic salmon. Applied and Environmental Microbiology, 84(2), 1–9. 10.1128/aem.01974-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza, J. C., Trinanes, J., Lohr, W., Sudre, B., Löfdahl, M., Martinez‐Urtaza, J., Nichols, G. L., & Rocklöv, J. (2017). Environmental suitability of vibrio infections in a warming climate: An early warning system. Environmental Health Perspectives, 125(10), 1–12. 10.1289/EHP2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, C. E., Xia, M., Li, R., Mu, C., Zhang, L., Liu, L., Ye, Y., & Wang, C. (2019). Vibrio alginolyticus infection induces coupled changes of bacterial community and metabolic phenotype in the gut of swimming crab. Aquaculture, 499, 251–259. 10.1016/j.aquaculture.2018.09.031 [DOI] [Google Scholar]
- Shruthi, M. S., Sushanth, V. R., & Rajashekhar, M. (2011). Diatoms as indicators of water quality deterioration in the estuaries of Dakshina Kannada and Udupi Districts of Karnataka. International Journal of Environmental Sciences, 2(2), 996–1006. 10.6088/ijes.00202020056 [DOI] [Google Scholar]
- Sullam, K. E., Essinger, S. D., Lozupone, C. A., O’Connor, M. P., Rosen, G. L., Knight, R., Kilham, S. S., & Russell, J. A. (2012). Environmental and ecological factors that shape the gut bacterial communities of fish: A meta‐analysis. Molecular Ecology, 21(13), 3363–3378. 10.1111/j.1365-294X.2012.05552.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J., Prill, R. J., Tripathi, A., Gibbons, S. M., Ackermann, G., Navas‐Molina, J. A., Janssen, S., Kopylova, E., Vázquez‐Baeza, Y., González, A., Morton, J. T., Mirarab, S., Zech Xu, Z., Jiang, L., … Knight, R. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature, 551(7681), 457–463. 10.1038/nature24621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, H., Wang, H., Tang, L., Mu, C., Ye, C., Chen, L., & Wang, C. (2019). High‐throughput sequencing reveals the core gut microbiota of the mud crab (Scylla paramamosain) in different coastal regions of southern China. BMC Genomics, 20(1), 1–12. 10.1186/s12864-019-6219-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, J. H., Lin, G., Fu, G. H., Wan, Z. Y., Lee, M., Wang, L., Liu, X. J., & Yue, G. H. (2014). The intestinal microbiome of fish under starvation. BMC Genomics, 15(1), 1–11. 10.1186/1471-2164-15-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, L., Amberg, J., Chapman, D., Gaikowski, M., & Liu, W. T. (2014). Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. ISME Journal, 8(3), 541–551. 10.1038/ismej.2013.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Sun, Y., Chen, K., Yu, N., Zhou, Z., Chen, L., Du, Z., & Li, E. (2014). Characterization of the intestinal microbiota in Pacific white shrimp, Litopenaeus vannamei, fed diets with different lipid sources. Aquaculture, 434, 449–455. 10.1016/j.aquaculture.2014.09.008 [DOI] [Google Scholar]
- Zhang, M., Sun, Y., Chen, L., Cai, C., Qiao, F., Du, Z., & Li, E. (2016). Symbiotic bacteria in gills and guts of Chinese mitten crab (Eriocheir sinensis) differ from the free‐living bacteria in water. PLoS One, 11(1), 10.1371/journal.pone.0148135 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence datasets generated during the current study are available at NCBI Sequence Read Archive (SRA) under BioProject PRJNA691201: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA691201. BioSample accession numbers are SAMN17283444 ‐ SAMN17283464.


