Susceptibility to breast cancer is significantly increased in individuals with germ line mutations in RECQ1 (also known as RECQL or RECQL1), a gene encoding a DNA helicase essential for genome maintenance. We previously reported that RECQ1 expression predicts clinical outcomes for sporadic breast cancer patients stratified by estrogen receptor (ER) status.
KEYWORDS: DNA helicase, estrogen receptor, FOXA1, RECQ1, transcriptional regulation
ABSTRACT
Susceptibility to breast cancer is significantly increased in individuals with germ line mutations in RECQ1 (also known as RECQL or RECQL1), a gene encoding a DNA helicase essential for genome maintenance. We previously reported that RECQ1 expression predicts clinical outcomes for sporadic breast cancer patients stratified by estrogen receptor (ER) status. Here, we utilized an unbiased integrative genomics approach to delineate a cross talk between RECQ1 and ERα, a known master regulatory transcription factor in breast cancer. We found that expression of ESR1, the gene encoding ERα, is directly activated by RECQ1. More than 35% of RECQ1 binding sites were cobound by ERα genome-wide. Mechanistically, RECQ1 cooperates with FOXA1, the pioneer transcription factor for ERα, to enhance chromatin accessibility at the ESR1 regulatory regions in a helicase activity-dependent manner. In clinical ERα-positive breast cancers treated with endocrine therapy, high RECQ1 and high FOXA1 coexpressing tumors were associated with better survival. Collectively, these results identify RECQ1 as a novel cofactor for ERα and uncover a previously unknown mechanism by which RECQ1 regulates disease-driving gene expression in ER-positive breast cancer cells.
INTRODUCTION
Since the first demonstration that RECQ1 (also known as RECQL or RECQL1) is essential for chromosomal stability (1), emerging biochemical and cellular functions of RECQ1 have provided a strong rationale to investigate the roles of RECQ1 in cancer biology (2–5), ultimately leading to its discovery as a candidate breast cancer susceptibility gene (6–8). However, the underlying mechanisms of RECQ1 in breast cancer biology are not yet understood. Given the inarguable evidence on the diverse functions of RECQ1, unbiased genome-wide approaches could provide deeper mechanistic insights and uncover new molecular functions of RECQ1 in normal development and human disease.
RECQ1 is localized to chromosome 12p12 and encodes a 649-amino-acid protein RECQ1, a ubiquitous nuclear enzyme, and the most abundant homolog of the highly conserved RecQ helicase family (9–12). Biochemically, RECQ1 is a DNA-dependent ATPase, binds to single- and double-stranded DNA, unwinds DNA duplex in a 3′-to-5′ direction and promotes strand annealing (13, 14). Through these multiple catalytic activities, RECQ1 responds to oxidative DNA damage (15), restores productive replication following DNA damage (16–20), participates in DNA double-strand break repair (21), removes chemical alterations to DNA bases via base excision repair pathway (22), and maintains telomeres (23, 24).
DNA repair functions of RECQ1 are mediated through its interactions with critical protein partners, including PARP1, RPA, Top3α, MSH2/6, FEN1, and Ku70/80 (25). The essential role of RECQ1 in genome maintenance is underpinned by the fact that RECQ1 depletion in cells results in increased frequency of spontaneous sister chromatid exchanges, chromosomal instability, DNA damage accumulation, and increased sensitivity to cytotoxic chemotherapy (12, 26, 27). In addition to its DNA repair functions, and similar to its homologs in Neurospora (28) and rat (29), human RECQ1 also seems to have gene regulatory functions (30). The knockdown of RECQ1 in breast cancer cells has a significant effect on gene expression associated with tumorigenesis (31); however, the molecular mechanism of gene regulation by RECQ1 remains to be elucidated.
Both RECQ1 catalytic functions and expression levels are related to breast cancer. Whole-genome sequencing efforts revealed that rare, recurrent RECQ1 mutations in the catalytic domain increase the risk of breast cancer by 5-fold among unselected cases from Poland and by 16-fold among higher-risk cases in Quebec, Canada (7). The association of RECQ1 mutations with breast cancer was further confirmed in a Chinese population, suggesting that RECQ1 mutations are not limited to specific populations (8). Subsequently, we conducted an evaluation of RECQ1 mRNA and protein expression in the large METABRIC cohort of sporadic breast cancer patients (n = 1,977), providing the first clinical evidence that altered RECQ1 expression is significantly associated with patient survival (32). In this data set, high RECQ1 protein levels significantly (P = 0.021) correlate with better survival in estrogen receptor (ER)-positive tumors that received endocrine therapy, indicating a mechanistic link between RECQ1 and the estrogen response pathway (32).
ERα is a member of the nuclear receptor family of transcription factors and a master regulator of tumor biology in two-thirds of all human breast cancers (33). ERα expression is both necessary and sufficient to predict the responsiveness to anti-estrogen in a high proportion of breast tumors, and a low ERα expression level is generally associated with a poor prognosis (34). Understanding the mechanisms that regulate ERα in breast cancer is critical to understanding how ERα mediates gene transcription and what occurs during endocrine resistance (35–39).
Given the clinical significance of ERα and the significant correlation between RECQ1 expression levels in breast tumors with clinical outcome in ER-positive disease, we sought to determine whether RECQ1 regulates ERα signaling, contributing to breast cancer and response to therapy. Here, we describe a previously unrecognized regulation of ERα signaling by RECQ1 and uncover the mechanisms of RECQ1 target gene regulation in breast cancer cells.
RESULTS
RECQ1 depletion leads to downregulation of gene sets associated with estrogen response.
Our recently published results show that low RECQ1 expression in the METABRIC data set correlated with poor survival among ER-positive patients who received endocrine therapy (32). In our follow-up investigation, we also found that knockdown of RECQ1 in a breast cancer cell line results in reduced ERα protein levels (32). These initial observations prompted us to investigate a potential role of RECQ1 in ERα signaling. To begin this investigation, we used an unbiased approach by determining global changes in gene expression that occur upon RECQ1 depletion by performing RNA sequencing (RNA-seq) in biological triplicates following RECQ1 knockdown in MCF7 cells grown in regular complete medium. We also performed RNA-seq from MCF7 cells after transient knockdown of ESR1, the gene encoding ERα. This would allow us to compare the transcriptomes regulated by RECQ1 and ERα. As expected, we observed a strong decrease in RECQ1 and ESR1 mRNA levels upon knockdown of RECQ1 and ESR1, respectively (Fig. 1A). Interestingly, ESR1 mRNA levels were also decreased when RECQ1 was depleted, but RECQ1 mRNA levels did not change upon knockdown of ESR1 (Fig. 1A). We therefore examined the effect of small interfering RNA (siRNA)-mediated knockdown of RECQ1 on ERα expression in MCF7 and T47D breast cancer cell lines that are widely used models for ER-positive breast cancer. In both cell lines, knockdown of RECQ1 significantly reduced ESR1 mRNA and ERα protein levels (see Fig. S1A to D in the supplemental material). These experiments were performed using SMARTpool siRNAs against RECQ1. We next confirmed the role of RECQ1 as a positive regulator of ERα expression in MCF7 cells by using four individual siRNAs targeting RECQ1 (see Fig. S2A) and in MCF7 RECQ1-KO cells generated using CRISPR/Cas9 (see Fig. S2B and C). Because ERα mediates estrogen-stimulated cell proliferation (40), we next examined the effect of RECQ1 knockdown on cell proliferation in response to estrogen treatment. Unlike control cells where proliferation significantly increased upon estrogen treatment, RECQ1 knockdown cells did not show an increase in cell proliferation upon estrogen treatment in MCF7 and T47D cells (see Fig. S1E and F), indicating cross talk between RECQ1 and the estrogen response pathway.
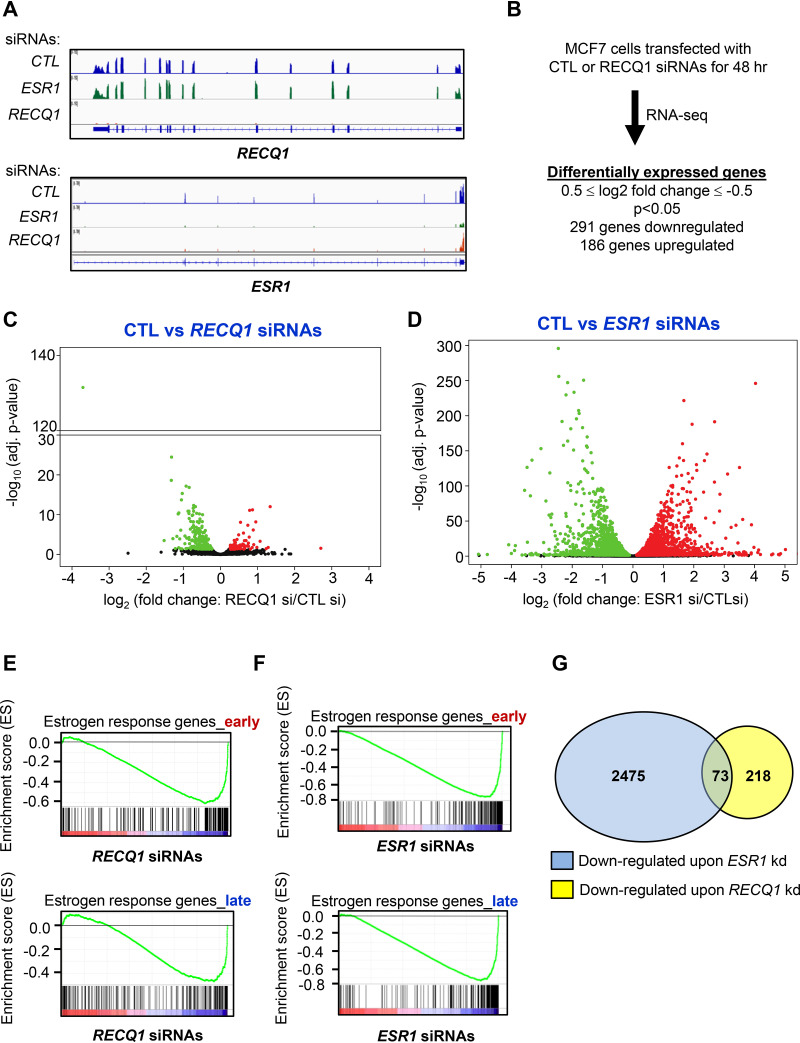
FIG 1.
Genome-wide RECQ1-regulated transcripts are significantly enriched for estrogen-responsive genes. (A and B) RNA-seq was performed from total RNA isolated from MCF7 cells transfected for 48 h with CTL, RECQ1, or ESR1 siRNAs. IGV snapshots for the RECQ1 and ESR1 loci (A) and the numbers of genes differentially expressed upon RECQ1 knockdown (B) are shown. (C and D) Volcano plots from the RNA-seq data showing significant differential gene expression upon knockdown of RECQ1 or ESR1 in MCF7 cells. Red and green dots correspond to genes up- or downregulated, respectively. (E and F) GSEA was performed using RNA-seq data for RECQ1 or ESR1 knockdown in MCF7 cells. The data show significant enrichment of estrogen response early and late genes in genes downregulated upon RECQ1 (E) or ESR1 (F) knockdown in MCF7 cells. (G) Venn diagram for the comparison of the number of genes downregulated upon knockdown of RECQ1 or ESR1 in the RNA-seq from MCF7 cells.
At the genome-wide level, 291 genes were significantly downregulated, and 119 genes were upregulated (0.5 < log2 fold change < –0.5; P < 0.05) upon knockdown of RECQ1 (Fig. 1B and C; see also Table S2 in the supplemental material). Using these statistical cutoffs, 1,355 genes were downregulated and 1,117 were upregulated upon ESR1 knockdown (Fig. 1D; see Table S3). Gene set enrichment analyses (GSEA) revealed the downregulation of many early and late estrogen response genes as most significant upon RECQ1 knockdown (Fig. 1E), and this pattern significantly overlapped with the expected enrichment of these pathways upon ESR1 knockdown (Fig. 1F). Given the observed similarity between the enrichment of gene sets that were downregulated following knockdown of RECQ1 or ESR1, we next looked at the intersection of genes whose expression was significantly decreased upon knockdown of RECQ1 or ESR1 in our RNA-seq analyses. We found a subset of genes that were commonly downregulated upon RECQ1 or ESR1 knockdown (Fig. 1G).
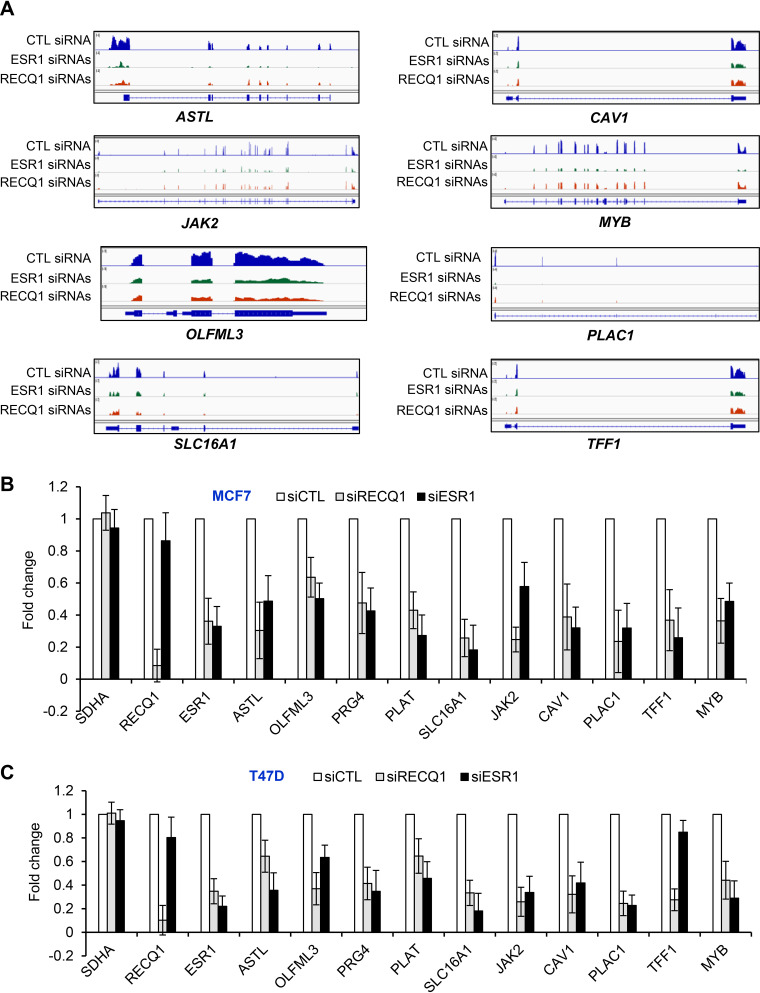
However, a majority of genes that were downregulated upon the ESR1 knockdown were not downregulated upon RECQ1 knockdown (Fig. 1G). This could be because RECQ1 may be regulating a subset of ERα targets and/or due to experimental variation in the extent of ESR1 downregulation upon RECQ1 knockdown. Importantly, as we had seen by reverse transcription-quantitative PCR (RT-qPCR) (see Fig. S1B and D), we observed a significant decrease in ESR1 mRNA levels (fold change = 0.75, P = 0.0028) in our RNA-seq analyses following RECQ1 knockdown (see Table S4). Of the 291 genes downregulated upon RECQ1 knockdown, 73 genes were also downregulated upon knockdown of ESR1 (Fig. 1G). Of these 73 genes, 26 were early and late estrogen response genes (see Table S5). As shown in the Integrative Genome Viewer (IGV; Broad Institute, http://software.broadinstitute.org/software/igv/) snapshot for 7 of these 26 genes (Fig. 2A), namely, ASTL, CAV1, JAK2, MYB, OLFLM3, PLAC1, SLC16A1, and TFF1, we observed a significant decrease in the expression of these genes upon knockdown of RECQ1 or ESR1. We validated the downregulation of select genes by RT-qPCR after knockdown of RECQ1 or ESR1 in both MCF7 and T47D cells, indicating that these genes are not cell-type-specific targets of RECQ1 and ERα (Fig. 2B and C). This result, together with the observed significant enrichment of estrogen response genes upon RECQ1 knockdown, indicates that RECQ1 has a direct or indirect role in regulating genes involved in estrogen signaling.
FIG 2.
A subset of ERα target genes is downregulated upon RECQ1 knockdown. (A) IGV snapshot for select genes downregulated in the RNA-seq data from MCF7 cells upon knockdown of RECQ1 or ESR1. (B and C) RT-qPCR was performed for ESR1, RECQ1, and a subset of ERα targets downregulated upon knockdown of RECQ1 or ESR1 in MCF7 (B) and T47D cells (C). The housekeeping gene SDHA was used as a negative control. Error bars represent standard deviations from three independent experiments.
RECQ1 ChIP-seq reveals significant genome-wide colocalization of RECQ1 and ERα.
To determine whether RECQ1 could play a direct role in the regulation of gene expression in ER-positive breast cancer cells, we performed chromatin immunoprecipitation sequencing (ChIP-seq) in duplicates for RECQ1 and for ERα in MCF7 cells grown in regular complete medium using a previously described method (41). Data from the replicates were pooled and peaks were called using model-based analysis for ChIP-seq (MACS). This resulted in the identification of thousands of RECQ1 binding events (see Table S6) distributed predominantly in intergenic regions and introns (Fig. 3A; see also Fig. S3A and Table S7). More than half (∼59%) of the RECQ1 peaks colocalized with transcription start sites (see Fig. S3B). We next validated our RECQ1 ChIP-seq data for 11 RECQ1-bound genes. We observed approximately 8- to 34-fold enrichment of RECQ1 at the target sites by ChIP-qPCR from MCF7 cells (Fig. 3B); knockdown of RECQ1 in MCF7 cells resulted in decreased expression of a majority of these genes (see Fig. S3C). Moreover, RECQ1 ChIP-seq peaks showed the most significant enrichment of estrogen response genes (Fig. 3C). Among the genes bound by RECQ1, a subset changed in expression upon RECQ1 knockdown (see Table S8).
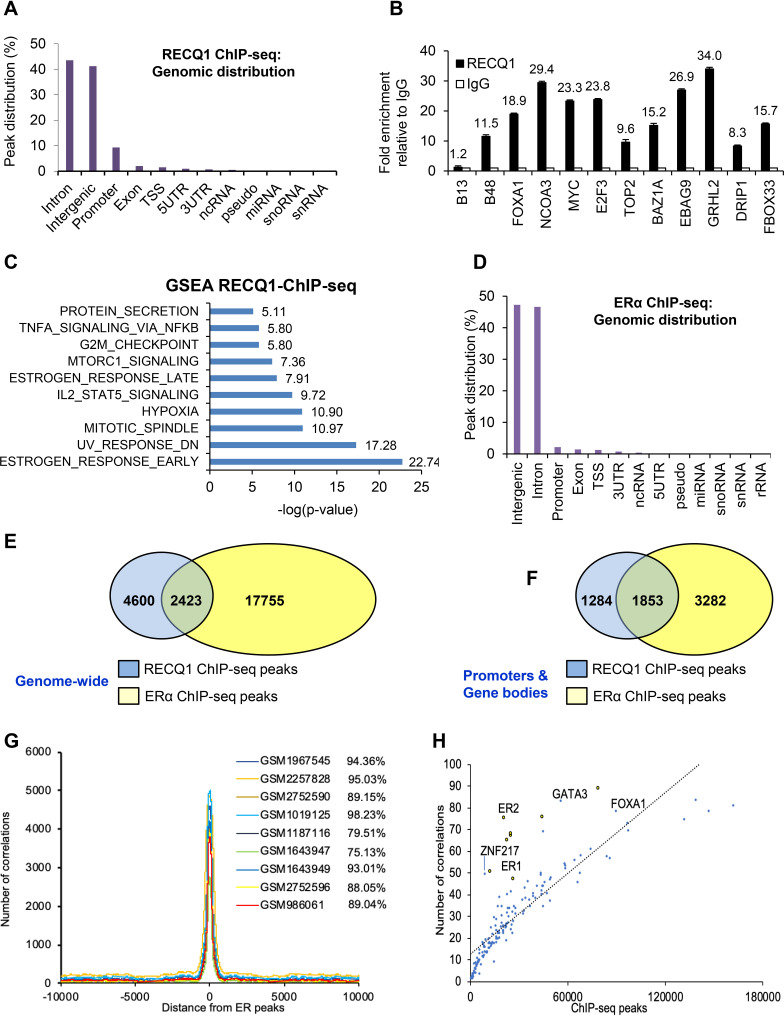
FIG 3.
Genome-wide identification of RECQ1 binding sites shows significant cooccupancy of RECQ1 and ERα-bound regions. (A) Genomic distribution of RECQ1 binding sites identified by ChIP-seq (replicate 2) in MCF7 cells is shown. The genomic distribution from both replicates is shown in Fig. S2 in the supplemental material. (B) Validation of a subset of RECQ1 targets in MCF7 cells using RECQ1 ChIP-qPCR. B48 primer targets lamin B2 origin of DNA replication and was used as a positive control since RECQ1 is known to bind to the lamin B2 origin in unperturbed cells (18). The B13 primers target a region 5 kb away from the origin and was used as a negative control. (C) The GSEA molecular signature database (https://www.gsea-msigdb.org/gsea/msigdb/annotate.jsp) was used for regions of the genome bound by RECQ1, as identified by ChIP-seq. The graph shows significant enrichment of estrogen response genes, together with other pathways. (D) Genomic distribution of ERα ChIP-seq (replicate 2). Data from both replicates are shown in Fig. S4 in the supplemental material. (E and F) A comparison of RECQ1 and ERα binding sites genome-wide (E) and in promoters and gene bodies (F) shows significant overlap. (G) Comparison of our ERα ChIP-seq data with nine previously published ERα ChIP-seq from MCF7 cells shows a strong overlap between our ERα ChIP-seq and previously published ERα ChIP-seq data. (H) Correlation between our ERα ChIP-seq (ER1 and ER2), RECQ1 ChIP-seq, and previously published ChIP-seq data for select transcription factors.
Given the central role of ERα as a transcription factor in ER-positive breast cancer and our data showing that RECQ1 enhances basal ERα expression and some ERα targets, we next performed ERα ChIP-seq in MCF7 cells. ChIP-seq for ERα revealed thousands of ChIP-seq peaks distributed predominantly in intergenic regions and introns (Fig. 3D; see also Fig. S4A and Table S9). The enrichment of the ERα binding events compared to the genome was strongest in the promoter regions (see Fig. S4B and Table S7). When we compared the genome-wide binding events of RECQ1 with that of ERα, we found that ∼35% (2,423 out of a total of 7,761 peaks) of RECQ1 peaks colocalize with ERα peaks (Fig. 3E); ∼12% of total ERα-bound sites overlapped with RECQ1 peaks (Fig. 3E). IGV snapshots for select genes that were bound by RECQ1 (Fig. 3B) are shown in Fig. S4C. Moreover, when we compared the RECQ1 peaks with ERα peaks in promoters and gene bodies, we found that ∼59% (1,853 out of a total of 3,137 peaks) of RECQ1 peaks colocalize with ERα peaks and ∼36% of ERα peaks colocalize with RECQ1 peaks (Fig. 3F; see also Table S10). Although our study is the first to report genome-wide RECQ1 binding sites via ChIP-seq, comparison of our ERα ChIP-seq data with nine previously published ERα ChIP-seq results from MCF7 cells that had ∼22,000 to 78,000 peaks showed approximately 75 to 95% colocalization (Fig. 3G). Moreover, comparing our RECQ1 ChIP-seq data with ChIP-seq data for a number of transcription factors, including our ERα ChIP-seq data, showed that RECQ1 genome-wide binding positively correlates with ERα, FOXA1, GATA3, TCF7L2, and ZNF217 (Fig. 3H).
To better understand the relationship between RECQ1 and its colocalized factors with associated chromatin features, we performed k-means clustering of ChIP-seq peaks into three clusters based on H3K4me1 (enhancer marker) and H3K4me3 (promoter marker) signals (Fig. 4A). Peaks in cluster 1 correspond to strong promoters with high H3K4me3 signal and low H3K4me1 signal, and peaks in cluster 2 correspond to strong enhancers with high H3K4me1 signal and low H3K4me3 signal, whereas peaks in cluster 3 correspond to weak and/or inactive enhancers and promoters. K-means clustering of the ChIP-seq data revealed that, in addition to active promoter regions, RECQ1 binding sites are associated with active enhancers similar to FOXA1 and GATA3 binding (ChIP-seq data from ENCODE) (Fig. 4A).
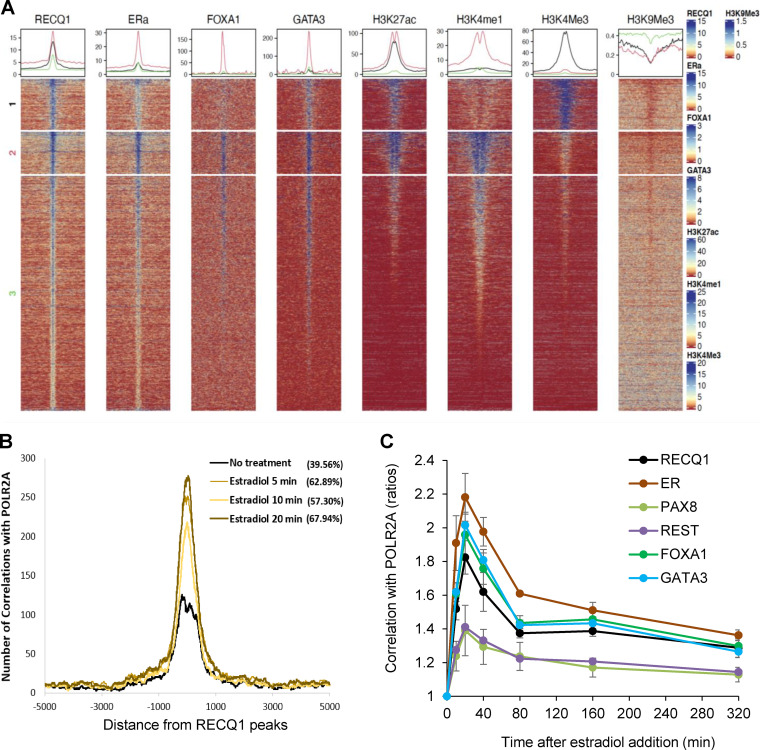
FIG 4.
Chromatin features of genome-wide RECQ1 binding. (A) Signal distribution of RECQ1, ERα, FOXA1, GATA3, H3K27ac, H3K4me1, H3K4me3, and H3K9me3 ChIP-seq data over RECQ1 peaks. The RECQ1 peaks were clustered into three groups based on H3K4me1 and H3K4me3 signal using K-means clustering. Peaks in cluster 1 correspond to strong promoters with a high H3K4me3 signal and a low H3K4me1 signal. Peaks in cluster 2 correspond to strong enhancers with a high H3K4me1 signal and a low H3K4me3 signal, with very strong RECQ1, ERα, FOXA1, and GATA3 occupancy. Cluster 3 corresponds to weak and/or inactive enhancers and promoters. (B) Colocalization of the RECQ1 binding sites (average of two replicates) with the RNA Pol II (POL2RA) binding sites. POL2RA binding sites were obtained over a time course of 0, 5, 10, and 20 min of estradiol treatment. The histogram x axis extends 5 kb upstream and 5 kb downstream from the center of the RECQ1 peaks. The extent of colocalization (percent) is measured as the fraction of RECQ1 peaks within the 5-kb window of the POLR2A peaks. (see Materials and Methods for details). (C) Graph depicting correlations between the binding sites of RECQ1, ERα, PAX8, REST, FOXA1, and GATA3 with the binding sites of POL2RA from cells collected over a time course of 0 to 320 min after estradiol treatment. Correlations were normalized using the time point t = 0. Correlations between peaks were calculated over 5-kb windows. (see Materials and Methods for details).
The colocalization of RECQ1 and ERα at the genome-wide level, combined with the significant enrichment of estrogen response pathway in the genes downregulated upon RECQ1 knockdown and RNA-seq overlap between the gene expression changes that occur upon knockdown of RECQ1 and ESR1, indicated a potential role of RECQ1 as a coregulator for ERα signaling. Therefore, we next compared RECQ1 ChIP-seq peaks with a previously published RNA Pol II (POLR2A) binding over a time course of estrogen treatment in MCF7 cells (42) (Fig. 4B and C). The correlation of RECQ1-peaks with estradiol-stimulated POLR2A occupancy onto chromatin is rapidly and significantly increased, as early as 5 min after treatment, and returns to an unstimulated level at 6 h after estradiol treatment indicating potentially dynamic recruitment of RECQ1 to the estrogen-responsive gene promoters (Fig. 4B). Similar analysis with the ERα ChIP-seq peaks revealed a stronger correlation with estrogen-stimulated POLR2A recruitment than RECQ1 ChIP-seq peaks; however, the trend was similar (Fig. 4C). The correlation of RECQ1 ChIP-seq with estrogen-stimulated POLR2A recruitment was comparable with the publicly available ChIP-seq data sets for FOXA1 and GATA3, the two proteins known to be critical in tethering ERα to the DNA (33) (Fig. 4C). In comparison to these well-established regulators of estrogen response, publicly available ChIP-seq data sets of REST and PAX8 showed significantly reduced correlation with POLR2A occupancy (Fig. 4C). Notably, estrogen treatment also upregulated RECQ1 mRNA levels (see Fig. S5 in the supplemental material), as reported previously (43). Taken together, these analyses suggest that the genome-wide binding of RECQ1 is correlated with estrogen-induced transcriptional dynamics in MCF7 cells and implicate RECQ1 in the estrogen response pathway.
RECQ1 associates with FOXA1 at the ESR1 locus to enhances ESR1 transcription.
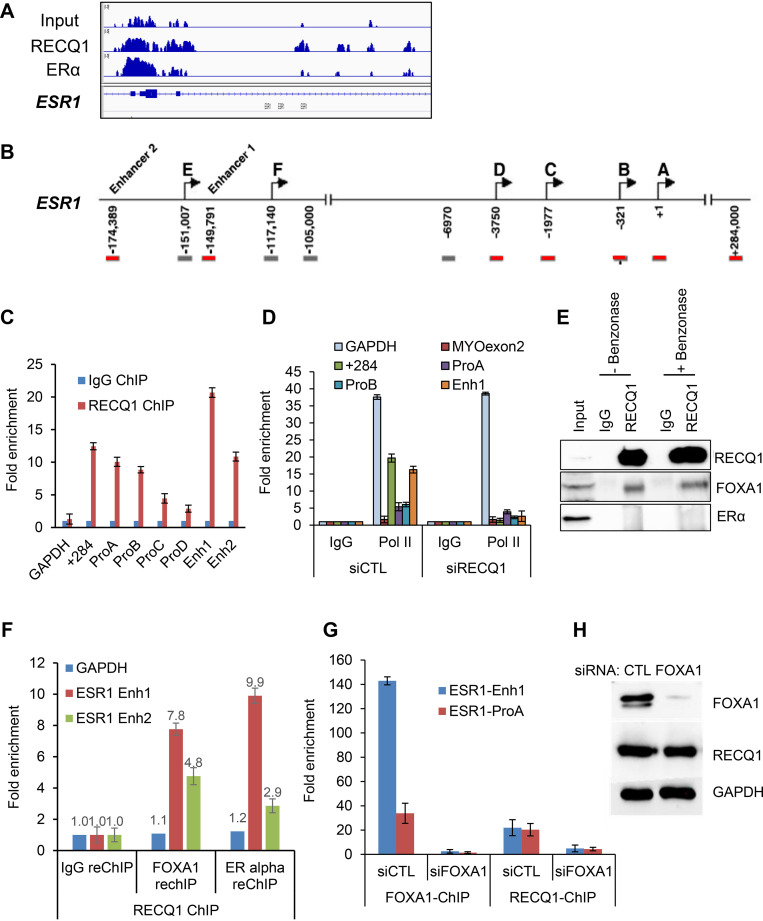
ERα is known to regulate its own expression by binding at the ESR1 gene locus (44). Therefore, in this study we decided to focus on a single RECQ1-bound gene locus and determining the molecular mechanism by which RECQ1 activates ESR1 transcription. RECQ1 ChIP-seq compared to input DNA (Fig. 5A) or compared to IgG control (see Table S6), and validation using ChIP-qPCR (Fig. 5B and C) demonstrated recruitment of RECQ1 at the ESR1 promoter and enhancer regions. RNA Pol II ChIP from control and RECQ1 knockdown MCF7 cells showed reduced occupancy of RNA Pol II at the ESR1 promoter and enhancer regions and exon 1 (+284) located near the transcription start site that has been previously shown to govern ERα expression in MCF7 cells (44, 45) (Fig. 5D). These results indicate that RECQ1 regulates ERα expression directly at the transcriptional level as reflected by the reduced RNA Pol II occupancy at regulatory regions of ESR1 upon RECQ1 knockdown. However, in coimmunoprecipitation (co-IP) experiments, we could not detect ERα in RECQ1 IPs (Fig. 5E).
FIG 5.
RECQ1 binding to the ESR1 locus is FOXA1-dependent. (A) An IGV snapshot of RECQ1 and ERα ChIP-seq peaks is shown for the ESR1 locus. (B) Location of primers used to amplify promoter and enhancer regions of ESR1 is shown. (C and D) ChIP-qPCR shows strong enrichment of RECQ1 at specific promoter and enhancer regions of ESR1. (C) MCF7 cells were transfected with control (siCTL) siRNA or RECQ1 siRNAs (siRECQ1) for 48 h, and ChIP-qPCR was performed using a control IgG antibody or RECQ1 antibody. (D) The data show that RECQ1 knockdown in MCF7 cells results in reduced RNA Pol II occupancy at specific promoter and enhancer regions. (E) RECQ1 associates with FOXA1 but not ERα protein, as assessed by immunoblotting after IP from MCF7 whole-cell extracts using a control IgG antibody or RECQ1 antibody. The RECQ-FOXA1 interaction is not DNA or RNA dependent because it is not sensitive to benzonase. (F) ChIP/re-ChIP assays show the colocalization of RECQ1, FOXA1, and ERα at ESR1 regulatory regions. ChIP/re-ChIP assays in MCF7 cells with RECQ1 as first ChIP, followed by a re-ChIP with either IgG or antibody against FOXA1 or ERα. Re-ChIP DNA was quantified by qPCR at ESR1 enhancer 1 and enhancer 2. (G) ChIP-qPCR was performed from MCF7 cells transfected with siCTL or FOXA1 siRNAs (siFOXA1) using a FOXA1 antibody or RECQ1 antibody. The data show that the binding of RECQ1 to a region in the ESR1 promoter and enhancer is abolished upon knockdown of FOXA1 with siRNAs in MCF7 cells. As expected, binding of FOXA1 to these regions was lost upon knockdown of FOXA1 in siRNAs in MCF7 cells. (H) Immunoblotting was performed from MCF7 whole-cell lysates prepared after transfection of MCF7 cells with control (CTL) or FOXA1 siRNAs for 48 h. Immunoblotting was used to confirm knockdown of FOXA1 in the ChIP-qPCR experiment in panel F; FOXA1 knockdown did not affect RECQ1 protein levels. GAPDH was used as a loading control.
Because FOXA1 is known to regulate ESR1 transcription (46, 47), we next sought to determine whether RECQ1 regulates ESR1 expression by associating with FOXA1. Indeed, in co-IP experiments, we pulled down FOXA1 in RECQ1 IPs indicating that RECQ1 associates with FOXA1 (Fig. 5E). The RECQ1-FOXA1 interaction was not disrupted in the presence of benzonase, an enzyme that degrades DNA and RNA, indicating that RECQ1 forms a protein-protein complex with FOXA1 (Fig. 5E). This observation is consistent with the reported association of FOXA1 with RECQ1 in chromatin and chromatin-free complexes (48). We next performed sequential ChIP to determine whether RECQ1, FOXA1, and ERα are bound at the same regulatory regions of ESR1. We found significant enrichment of ESR1 enhancer 1 and 2 in re-ChIP of RECQ1 ChIP material with FOXA1 or ERα antibodies (Fig. 5F), indicating that these proteins work together to regulate the ESR1 gene.
To further investigate the role of FOXA1 in binding of RECQ1 to the ESR1 enhancer regions, we performed ChIP-qPCR for RECQ1 after knockdown of FOXA1 in MCF7 cells with siRNAs. Although in cells transfected with CTL siRNA, RECQ1 was enriched ∼20-fold at the promoter and enhancer region of ESR1, knockdown of FOXA1 resulted in complete loss of binding of RECQ1 at these regions (Fig. 5G), suggesting that FOXA1 could recruit RECQ1 to these regions that are known to play important roles in ESR1 transcription. As expected, FOXA1 was more enriched at the ESR1 enhancer (∼140-fold) compared to the ESR1 promoter (∼30-fold), and this binding of FOXA1 to the ESR1 regulatory regions was dramatically reduced upon FOXA1 knockdown (Fig. 5G). Immunoblotting verified substantial decrease in FOXA1 expression in the FOXA1 siRNA-transfected cells (Fig. 5H); there was no change on RECQ1 mRNA or protein levels upon FOXA1 knockdown (Fig. 5H; see also Fig. S6), suggesting that the observed loss of RECQ1 binding at the ESR1 promoter and enhancer upon knockdown of FOXA1 was not due to decreased RECQ1 expression.
Helicase activity of RECQ1 is required for FOXA1-mediated ESR1 transcription.
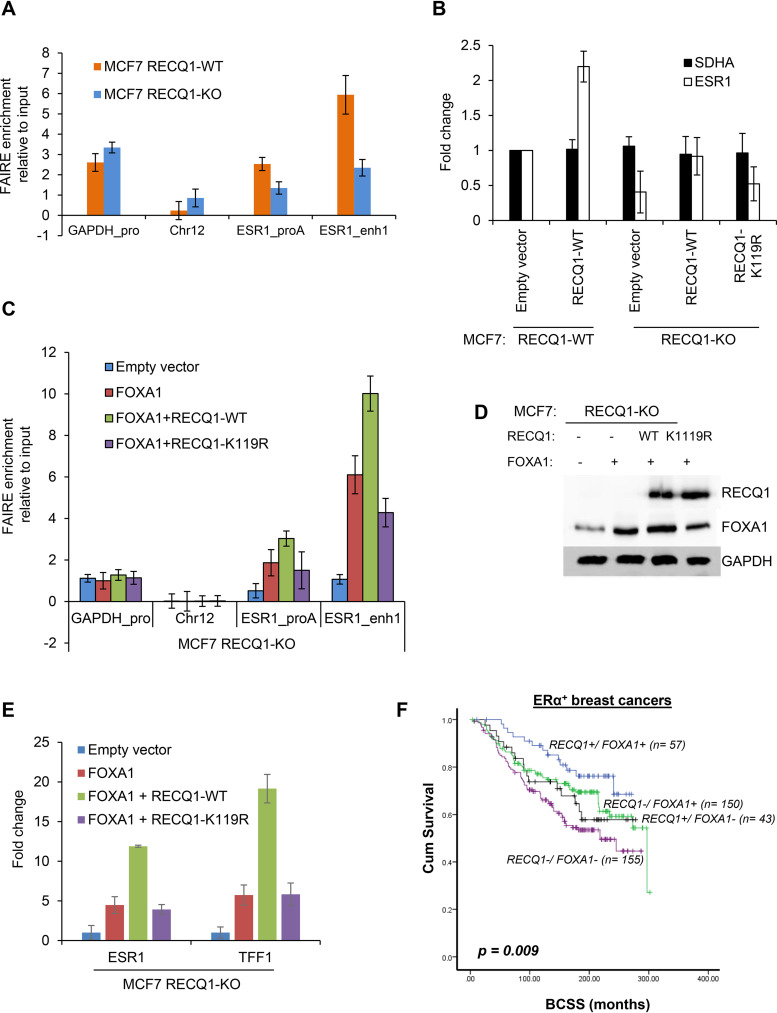
Given that FOXA1 is a known pioneer factor that can bind compact chromatin (39, 46, 49), we next assessed whether RECQ1 helicase activity might contribute to chromatin accessibility at ESR1 regulatory regions in MCF7 cells and hence facilitate ESR1 expression. Formaldehyde-assisted isolation of regulatory elements (FAIRE) is a method that assays for nucleosome-free regions of the genome with open chromatin, and higher values for enrichment correspond to more accessible DNA (50). We first performed FAIRE, followed by qPCR, for enrichment of ESR1 promoter and enhancer regions to determine the effect of loss of endogenous RECQ1 on chromatin accessibility at these specific ESR1 regulatory regions (Fig. 6A). Loss of endogenous RECQ1 induced a decrease in chromatin accessibility at ESR1 promoter and enhancer regions, as assessed by FAIRE-qPCR in RECQ1-WT and RECQ1-KO MCF7 cells (Fig. 6A). Consistent with the reduced chromatin accessibility at these ESR1 regulatory regions, RECQ1-KO cells transfected with an empty vector showed reduced ESR1 expression compared to RECQ1-WT cells transfected with an empty vector (Fig. 6B). Thus, the pattern of accessibility changes at the ESR1 regulatory regions in RECQ1-WT and RECQ1-KO is comparable to the ESR1 mRNA expression in these isogenic cells (Fig. 6A and B). Overexpression of wild-type RECQ1 in RECQ1-KO cells restored ESR1 expression to the RECQ1-WT level, whereas overexpression of a well-characterized helicase-dead RECQ1-K119R variant failed to restore ESR1 expression in RECQ1-KO cells (Fig. 6B).
FIG 6.
RECQ1 helicase cooperates with FOXA1 to regulate ESR1 expression. (A) FAIRE assays were performed using MCF7 RECQ1-WT or isogenic MCF7 RECQ1-KO cells, and the changes in chromatin accessibility at an ESR1 promoter (ESR1_proA) region and an ESR1 enhancer (ESR1_enh1) were assessed by FAIRE-qPCR. The promoter regions of GAPDH and a heterochromatin region in chromosome 12 were used as positive and negative controls, respectively, for open chromatin. (B) RT-qPCR assays for ESR1 and the housekeeping gene SDHA were performed with MCF7 RECQ1-WT transfected with empty vector or a vector that expresses WT RECQ1 (RECQ1-WT) for 48 h. In parallel, RT-qPCR assays for ESR1 and the housekeeping gene SDHA were performed with MCF7 RECQ1-KO cells transfected with empty vector or a vector that expresses WT RECQ1 (RECQ1-WT) or helicase-dead RECQ1 mutant (RECQ1-K119R) for 48 h. The fold change refers to the fold change in gene expression normalized to GAPDH. (C to E) MCF7 RECQ1-KO cells were transfected with empty vector, FOXA1-expressing vector (FOXA1), or FOXA1-expressing vector in combination with RECQ1-expressing vectors that were RECQ1 WT (RECQ1-WT) or helicase-dead RECQ1 mutant (RECQ1-K119R). (C) Changes in chromatin accessibility at ESR1 regulatory regions in these cells were determined by FAIRE-qPCR. An increase in FAIRE enrichments by FOXA1 was further enhanced by cotransfection with a helicase-active wild-type RECQ1. The relative enrichment of the FAIRE signal normalized to input chromatin is shown for the promoter of GAPDH (positive control), a heterochromatin region on chromosome 12 (negative control), and the ESR1 promoter A and enhancer 1. (D) The effect on FOXA1 and RECQ1 protein was determined by immunoblotting using GAPDH as a loading control. (E) The effect on ESR1 or TFF1 expression was determined by RT-qPCR at 48 h after transfection. Fold change refers to fold change in gene expression normalized to GAPDH. (F) Kaplan-Meier curves for RECQ1/FOXA1 coexpression and breast cancer-specific survival (BCSS) in ERα+ breast cancers.
We next sought to determine whether RECQ1 helicase may cooperate with FOXA1 in modulating DNA accessibility at ESR1 promoter and enhancer regions to restore ESR1 expression in RECQ1-KO cells. To test this, we performed FAIRE-qPCR assays in RECQ1-KO MCF7 cells overexpressing FOXA1 alone or in combination with wild-type RECQ1 or the helicase-dead RECQ1-K119R (Fig. 6C). Western blot analysis of transfected cells validated comparable expression of wild-type and K119R RECQ1 proteins in RECQ1-KO MCF7 cells (Fig. 6D). Compared to RECQ1-KO cells transfected with an empty vector, overexpression of FOXA1 in RECQ1-KO cells increased FAIRE enrichment at the ESR1 promoter (∼2-fold) and enhancer (∼6-fold) (Fig. 6C). Cotransfection of FOXA1- and wild-type RECQ1-expressing constructs resulted in further increase in FAIRE-enrichment at ESR1 promoter (∼3.5-fold) and enhancer (∼9-fold) compared to RECQ1-KO cells transfected with an empty vector (Fig. 6C). In contrast, cotransfection with RECQ1-K119R resulted in reduced FAIRE enrichment at the ESR1 promoter (∼1.5-fold) and enhancer (∼4-fold), indicating that the helicase activity of RECQ1 is essential to cooperate with FOXA1 and promote chromatin accessibility at ESR1 regulatory regions (Fig. 6C). Consistent with chromatin accessibility, as indicated by FAIRE enrichment at the ESR1 regulatory regions, cotransfection of FOXA1- and wild-type RECQ1-expressing constructs increased ESR1 and TFF1 expression in RECQ1-KO cells (Fig. 6E), whereas cotransfection with RECQ1-K119R had minimal effect.
These results indicate that RECQ1 acts as a partner for FOXA1 in its pioneer activity. MCF7 cells lacking RECQ1 have reduced accessibility at the ESR1 promoter, as well as at ESR1 enhancer, where FOXA1 has an impact on accessibility that is dependent on the helicase activity of RECQ1. This is consistent with the role of FOXA1 as a pioneer factor which are thought to act at the enhancers to initiate chromatin opening (51). Promoters for the most part maintain an open chromatin configuration (52). Therefore, our data suggest a more complex relationship of RECQ1 with regulatory chromatin. Collectively, these data provide evidence to support our hypothesis that RECQ1 helicase cooperates with FOXA1 to enhance chromatin accessibility and enhance ESR1 expression.
RECQ1-FOXA1 coexpression and survival outcomes in clinical ERα-positive breast cancers.
Preclinical evidence presented thus far suggests that RECQ1-FOXA1 interaction may influence ERα-positive breast cancer pathogenesis and impact survival outcomes in patients who received endocrine therapy. We have investigated the clinical significance of RECQ1 (32) or FOXA1 (53) in a large cohort of breast cancers. We observed nuclear staining only for RECQ1 and FOXA1. Here, we investigated the clinicopathological significance of RECQ1/FOXA1 protein coexpression in ERα-positive breast cancers (n = 1,406). Patient demographics are summarized in Tables S11 and S12 in the supplemental material. All patients received tamoxifen adjuvant endocrine therapy. As shown in Fig. 6D, breast cancer-specific survival (BCSS) was better in patients whose tumor had high RECQ1/high FOXA1 coexpression compared to those with low RECQ1/low FOXA1 coexpression (P = 0.009). Tumors with low RECQ1/low FOXA1 coexpression were associated with higher tumor stage, high tumor grade, dedifferentiation, pleomorphism, higher mitotic index, and high-risk Nottingham prognostic index (NPI; >3.4) compared to tumors with high RECQ1/high FOXA1 coexpression (P < 0.05; see Table S13). In ER– tumors (n = 244), RECQ1/FOXA1 coexpression did not influence survival outcomes (see Fig. S7 and Table S14). These data suggest that RECQ1/FOXA1 coexpression in ERα-positive breast cancers has clinicopathological and prognostic significance.
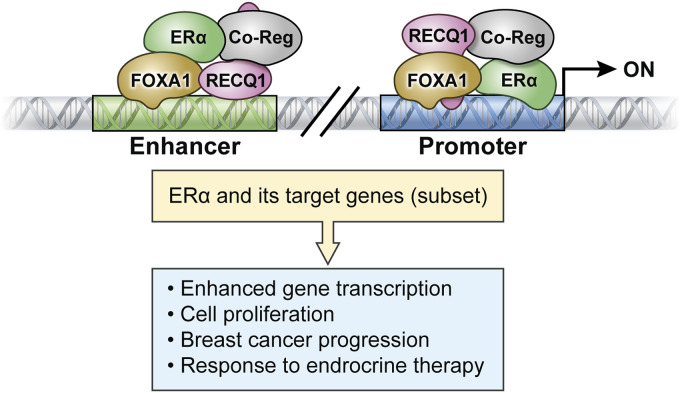
Based on our data, we propose a working model on the transcriptional circuitry linking RECQ1 to ERα and ultimately the biological response in ER-positive breast cancer cells (Fig. 7). According to this model, RECQ1 promotes transcription of ESR1, the gene encoding ERα. By regulating ERα levels and as a cofactor for ERα in cooperation with FOXA1 and other yet-to-be-identified factors, RECQ1 binding to promoter or enhancer regions regulates the expression of a subset of ERα target genes.
FIG 7.
Proposed model of the role of RECQ1 in regulating the expression of ERα and a subset of ERα target genes.
DISCUSSION
Previous work has described various roles of RECQ1 helicase in DNA repair and its requirement for genome maintenance. Here, we report an unexpected role of RECQ1 in the regulation of ERα signaling in breast cancer cells in the absence of estrogen. We demonstrate that RECQ1 directly regulates the expression of ESR1, a well-known therapeutic target that has been shown to determine clinical outcomes in breast cancer (33, 37). Using RNA-seq, we found that ESR1 and several of its downstream target genes are significantly downregulated in RECQ1 knockdown MCF7 cells. ChIP-seq analysis revealed a significant overlap of RECQ1 genomic binding sites with ERα, including binding to the ESR1. Mechanistically, RECQ1 is recruited to the regulatory regions of the ESR1 gene through its interaction with FOXA1 and cooperates with FOXA1 to facilitate chromatin accessibility and promote ESR1 expression. In addition, RECQ1 levels are significantly associated with clinical outcomes in ER-positive breast cancer patients, specifically in those receiving tamoxifen treatments. Taken together, our findings identify a novel mechanism of RECQ1 as an upstream regulator of ERα and strengthen the clinical relevance of the RECQ1 helicase in breast cancer.
Transcriptional regulation by RECQ1 through cooperating with other transcriptional regulators has not been studied before. Our findings begin to address this paucity using unbiased genome-wide approaches. The analysis of the transcriptome upon depletion of RECQ1 in ER-positive MCF7 cells allowed us to unveil the unanticipated function of RECQ1 as a regulator of ESR1 expression and a modulator of ERα-dependent gene expression. Indeed, we found that the siRECQ1 downregulated transcriptome is significantly enriched in early and late estrogen response genes. A comparison with siESR1 downregulated genes indicated a subset of genes commonly regulated by RECQ1 and ERα; however, a majority of differential gene expression in siRECQ1 cells are affected through an ERα-independent mechanism. ChIP-seq in MCF7 cells demonstrated significant enrichment of RECQ1 binding at a subset of estrogen-responsive genes. Comparative analysis of genome-wide binding of RECQ1 and ERα in MCF7 cells revealed that >35% of RECQ1-binding peaks overlap bona fide genomic ERα binding peaks independent of its ligand, estrogen. We note that a majority of ERα binding peaks were not enriched for RECQ1 binding, thus indicating that RECQ1 may not be a general regulator of ERα-driven transcription or that ERα-mediated gene expression change may not require a stable association of RECQ1. However, the fact that more than a third of the total RECQ1-binding peaks were cobound with ERα genome-wide and that ∼60% of the RECQ1-binding peaks were cobound by ERα at the promoters and gene bodies suggests a mechanistic cross talk between these two proteins for a wide range of genomic functions in breast cancer cells. The data presented here focused on a single RECQ1-bound gene locus and provide mechanistic insights into the role of RECQ1 in activating ESR1 transcription. Future studies utilizing a more global approach will establish the mechanisms by which RECQ1 regulates the transcription of specific genes important for genome stability and cancer.
ERα requires cofactors to assist with DNA binding and transcriptional regulation (54), and FOXA1 acts as a pioneer factor for ERα by modulating chromatin structure and promoter-enhancer interactions (47, 55). Our results suggest a novel role of RECQ1 in facilitating chromatin accessibility through a possible interaction with FOXA1 and in a helicase activity-dependent manner. Through its robust DNA-binding and unwinding activities, RECQ1 helicase cooperates with FOXA1 to promote ESR1 expression, as we have shown. RECQ1 in association with FOXA1 and other yet-to-be-identified cofactors may enhance the accessibility of transcriptional machinery and promote genomic binding of transcription factors such as ERα and its coregulators to control physiologically relevant gene expression programs (Fig. 7). Given that RECQ1 controls the transcription of ESR1, it may be difficult to separate out the changes in ERα-induced genes for which RECQ1 acts as a coactivator due to the fact that ERα levels decrease upon RECQ1 loss. Germ line mutations in RECQ1 that increase breast cancer risk inactivate its helicase activity (8) and thereby are also expected to impair its ability to regulate ERα expression and signaling in ER-positive breast cancer. Further studies using global approaches such as assay for transposase-accessible chromatin with sequencing (ATAC-seq) will determine how the displacement of nucleosomes by the pioneer factor FOXA1 is impacted by dysfunctional RECQ1 and affects ERα binding and activity genome-wide.
The implication of RECQ1 helicase activity in facilitating local chromatin states to modulate ERα occupancy at specific genomic loci is conceptually novel. However, transcriptional regulation by RECQ1 may also employ helicase-independent mechanisms through enhancer-binding and protein interactions. Whether RECQ1 acts by participation in a complex with ERα or by operating in a complex with its key coregulators such as FOXA1 (48) and other proteins remains to be elucidated in future studies. A critical interactor of RECQ1, PARP1, has been shown to interact with breast cancer related histone-modifying enzymes and regulate gene expression in breast cancer cells (56, 57). The results of genomic analyses conducted in the MCF7 human breast cancer cell line have localized RECQ1 at gene promoters as well as transcription start sites and intronic regions in the genome. Our integrative genomics approach provides important new, information about the chromatin properties (e.g., histone marks dictating chromatin state) of RECQ1 binding sites. Whether binding of RECQ1 to these sites modifies histones and thereby regulates chromatin openness is unknown. Additional factors and chromatin features that provide specificity to RECQ1 to be recruited at these subsets of genomic sites are unknown at this time.
What may be the role of RECQ1 in estrogen-dependent gene regulation in ERα-positive breast cancers? One key unifying feature of our RNA-Seq analysis upon RECQ1 knockdown and RECQ1 ChIP-seq analysis is the identification of estrogen response as the top enriched pathway. A functional role of RECQ1 in regulating a subset of estrogen response genes that underlie proliferation of breast cancer cells is evident by significantly compromised stimulation of proliferation of RECQ1-knockdown MCF7 and T47D cells upon estrogen treatment. Comparison of RECQ1 ChIP-seq peaks with RNA POLII occupancy after estrogen treatment shows a significant correlation with the dynamic transcriptional changes and suggests a role for RECQ1 as a mediator of early transcriptional response in estrogen signaling. This is consistent with our findings that RECQ1 cooperates with FOXA1 which is essential to reprogram the genome-wide occupancy of the ERα and mediate transcriptional response in estrogen signaling (58). However, we do not yet know whether RECQ1 is required for the implementation of an estrogen-regulated transcriptional program in an ERα-dependent manner.
A clear majority of the breast cancer patients express ERα in their tumors making the tumors amenable to endocrine therapies for ERα-positive breast cancer patients (33, 35). We have previously shown that RECQ1 deficiency was not only associated with aggressive breast cancer phenotypes but in ERα-positive tumors that received endocrine therapy, low RECQ1 expression was also linked with poor survival (32). High FOXA1 expression was previously shown to be associated with ERα-positive tumors, smaller tumor size, lower histological grade, and better survival (53). In another study of ERα-positive/HER-2 negative breast cancers high expression of FOXA1 was also linked with small tumor size and independently predicted long disease-free survival (59). This is further supported by a recent study suggesting that high FOXA1 may predict late recurrence in patients with ER-positive breast cancer (59). Although the mechanisms of endocrine resistance in breast cancer is complex (36), studies of response to selective estrogen receptor modulators (SERMs) like tamoxifen indicate that negative (corepressors) and positive (coactivators) coregulators of ERα may influence the balance of agonistic versus antagonistic activities of tamoxifen and determine endocrine sensitivity or resistance (36). Favorable survival outcomes in high RECQ1/high FOXA1 expressing ERα-positive tumors indicate that functional interaction between ERα, RECQ1, and FOXA1 may contribute to the anticancer activity of tamoxifen. It is well established that efficacy of ER blockade therapy is linked with wild-type ERα and downstream ER signaling pathway in tumors. On the other hand, the presence of ERα mutations or downstream overactivation such as those involving PIK3CA mutations are associated with endocrine resistance (60). In the present study, patients whose tumor had high RECQ1/FOXA1 coexpression had better breast cancer specific survival. The data would suggest that a functional ERα-FOXA1 interaction could predict response to endocrine therapy. However, further characterization will be required to determine whether ERα or FOXA1 mutational status would also influence endocrine therapy response in breast cancer. Therefore, the clinical data are largely hypothesis generating, and further prospective validation, including somatic ERα, RECQ1, and FOXA1 mutational analyses, will be required to confirm our initial observations.
In conclusion, we have demonstrated that RECQ1 can modulate a subset of ERα-driven gene expression by regulating the expression of ESR1 in a helicase activity-dependent and FOXA1-assisted manner. Cooperation of RECQ1 and FOXA1 is clinically relevant in response to endocrine therapy in ER-positive breast cancer. Together, this study provides the first mechanism by which RECQ1 could alter the progression and therapeutic response of ER-positive breast cancer. The notion that RECQ1 has a specific and direct role in gene expression control is novel and of potential significance to cancer biology. RECQ1 is a breast cancer susceptibility gene significantly correlated with clinical outcomes of sporadic breast cancer patients. Given that ERα is the major driving transcription factor in the mammary gland development, as well as breast cancer initiation and progression (61), evaluating the impact of breast cancer risk-associated RECQ1 variants on ER signaling may uncover the mechanisms by which RECQ1 helicase acts to suppress breast cancer development and progression. Elucidating the role of RECQ1 in coordinating genomic stability with transcriptomic networks could potentially predict cancer risk, achieve early diagnosis, track the prognosis of tumor fate, and ultimately provide valuable targets for novel therapeutic approaches.
MATERIALS AND METHODS
Cell culture, transfection, and treatment.
MCF7 and T47D human breast cancer cell lines were purchased from the American Type Culture Collection. All cells were grown as monolayers in Dulbecco modified Eagle medium (DMEM; Thermo Fisher) supplemented with 1% (vol/vol) sodium pyruvate, 10% (vol/vol) fetal bovine serum (FBS), and 10 IU/ml penicillin-streptomycin (Life Technologies) and maintained at 37°C and 5% CO2 in a humidified incubator. On-Target Plus SMARTpool siRNAs against RECQ1, ESR1, FOXA1, and control siRNAs were purchased from Dharmacon. All siRNA transfections were performed by reverse transfection at a final concentration of 20 nM the siRNA using Lipofectamine RNAiMAX (Invitrogen) as instructed by the manufacturer. Isogenic MCF7 RECQ1 knockout (RECQ1-KO) and its wild-type control (RECQ1-WT) cells were generated by using the CRISPR/Cas9 system (19).
Cell proliferation assays.
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies) was used to determine the effect of estrogen (17β-estradiol [E2]; Sigma) on RECQ1 knockdown MCF7 and T47D cell proliferation. At 48 h after siRNA transfection, MCF7 or T47D cells were plated in triplicates in a 96-well plate (8 × 103 cells/well), followed by incubation with phenol-red free DMEM containing 5% charcoal-stripped FBS (Gibco) for 24 h at 37°C in 5% CO2. The next day, the cells were treated with 10 nM E2 or 0.1% ethanol, followed by incubation for an additional 48 h at 37°C in 5% CO2. Cell proliferation was measured by adding 10 μl of CCK-8 reagent to each well containing 100 μl of growth medium. The plates were incubated at 37°C, and the absorbance was measured at 450 nm every hour for 4 h. The relative cell proliferation was calculated by normalizing the absorbance values to the untreated condition in each cell type.
RNA isolation and RT-qPCR.
Total RNA from MCF7 and T47D cells was extracted using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. A total of 0.5 μg of RNA was used for reverse transcription using an iScript reverse transcription supermix kit (Bio-Rad) according to the manufacturer’s instructions. The cDNA was subjected to real-time quantitative PCR using iTaq Universal SYBR green Supermix (Bio-Rad) in triplicate. Reactions were cycled at 95°C for 30 s, followed by 40 cycles of 94°C for 10 s and 60°C for 15 s with fluorescence data collection during the annealing/extension step on the CFX96 real-time PCR system (Bio-Rad). The relative transcript levels were normalized to the housekeeping gene GAPDH and differential expression measured using the 2–ΔΔCT method (62). The housekeeping gene SDHA served as a negative control in RT-qPCR experiments. The primer sequences are detailed in Table S1 in the supplemental material.
RNA-seq.
For RNA-seq, ∼10 million MCF7 cells transfected with CTL siRNA, ESR1 siRNAs (SmartPool) or RECQ1 siRNAs (SmartPool) for 48 h were harvested, and the total RNA was isolated by using an RNeasy Plus Micro kit (Qiagen). RNA integrity was checked with a Bioanalyzer (Agilent), and only samples with an RNA integrity number (RIN) of >9.5 were subsequently subjected to mRNA-seq. The mRNA-seq samples were pooled and sequenced on a HiSeq apparatus using an Illumina TruSeq mRNA Prep kit (RS-122-2101) and paired-end sequencing. The samples had ∼79 to 101 million pass filter reads with a base call quality of above ∼90% of bases with Q30 and above. Reads of the samples were trimmed for adapters and low-quality bases using Trimmomatic software before alignment with the reference genome (human, hg19) and the annotated transcripts using STAR. The average mapping rate of all samples was ∼95%. Unique alignment is above 89%. The mapping statistics are calculated using Picard software. The samples had ∼0.88% ribosomal reads. The percent coding base values were between 64 and 66%. The percent UTR base values were 29 to 31%, and mRNA base values were between 93 and 94% for all the samples. Library complexity was measured in terms of unique fragments in the mapped reads using Picard’s MarkDuplicate utility. The samples had 64 to 70% nonduplicate reads.
Read count per gene was calculated by HTSeq under the Gencode annotation and normalized by size factor implemented in the DESeq2 package. Regularized-logarithm transformation (rlog) values of gene expression were used to perform hierarchical clustering and principal-component analysis. To assess differential gene expression between different conditions (e.g., constructs versus mocks), we used a generalized linear model within DESeq2 that incorporates information from counts and uses negative binomial distribution with a fitted mean and a gene-specific dispersion parameter. DESeq2 used Wald statistics for significance testing and the Benjamini-Hochberg adjustment for multiple corrections.
Immunoblotting.
MCF7 and T47D cells were harvested after washing with phosphate-buffered saline (PBS), and whole-cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitor cocktail (Roche). The bicinchoninic acid (BCA) protein quantification kit (Thermo Scientific) was used to perform protein quantification. A total of 50 μg of the whole-cell lysate was mixed with Laemmli buffer (Bio-Rad), boiled at 95°C for 5 min, and subjected to SDS-PAGE, followed by transfer to a polyvinylidene difluoride membrane (Life Technologies). The membrane and primary antibodies—anti-RECQ1 (Bethyl lab), anti-ERα (Abcam), anti-FOXA1 (Abcam), and anti-glyceraldehyde dehydrogenase (anti-GAPDH; Cell Signaling)—were incubated on a rotating platform overnight at 4°C and followed by secondary antibodies conjugated to horseradish peroxidase (HRP; Jackson Immuno Research Laboratories). A chemiluminescent HRP substrate Immobilon Western kit (Invitrogen) was used to develop the immunoblots.
Coimmunoprecipitation.
Whole-cell lysate of MCF7 cells prepared in RIPA buffer (1 mg total protein) was incubated with Dynabeads protein A coupled with an antibody against human RECQ1 (Bethyl Lab) or normal rabbit IgG (Vector Labs) for overnight at 4°C in presence or absence of benzonase (Sigma, 50 U/ml). After four washes with 1× PBS, the immunocomplexes were eluted with 2× SDS sample buffer by boiling at 95°C for 5 min and resolved by 10% SDS-PAGE, followed by Western blotting detection with specific antibodies against RECQ1 (Bethyl Lab), FOXA1 (Abcam), and ERα (Santa Cruz Biotechnology).
Chromatin immunoprecipitation and re-ChIP.
A ChIP-IT High Sensitivity kit (Active Motif) was used to determine the associations of RECQ1, ERα, Pol II, and FOXA1 with the genomic regions on the promoter and enhancer of ERS1gene. MCF7 cells transfected with indicated siRNAs, as well as RECQ1-KO or RECQ1-WT cells, were grown at a density of 1 × 107 per 15-cm dish and subjected to fixation, chromatin sonication, and immunoprecipitation using 4 μg of anti-RECQ1 (Bethyl Lab), anti-ERα (Santa Cruz Biotechnology) anti-Pol II (Abcam), anti-FOXA1 (Abcam), or the same amount of rabbit IgG, followed by DNA purification according to manufacturer’s instructions. The immunoprecipitated fraction was analyzed by qPCR using iTaq Universal SYBR green Supermix (Bio-Rad) with technical triplicates, with an initial denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 30 s using the CFX96 real-time PCR system (Bio-Rad). Fold enrichments of the targeted genomic sequences were calculated over IgG as follows: fold enrichment = 2–(CtIP − CtIgG), where CtIP and CtIgG are the mean threshold cycles of PCR in triplicates on DNA samples immunoprecipitated with a specific antibody or IgG control, respectively. Melting-curve analyses and agarose gel electrophoresis were used to confirm the presence of a single specific product after the qPCR. The sequences of primers are listed in Table S1 in the supplemental material.
For ChIP and re-ChIP experiments, cell lysates were collected as described above, followed by incubation overnight with the first antibody, anti-RECQ1 (Bethyl Laboratories, Inc.) and protein G-Sepharose beads. Immunocomplexes were washed, followed by elution with 10 mM dithiothreitol at 37°C for 30 min. The eluted DNA was diluted 50-fold with lysis buffer and incubated with the second antibody, anti-FOXA1 or anti-ERα, for 3 h at 4°C. Protein G-Sepharose beads were then added, followed by incubation overnight with rotation at 4°C. The following day, the samples were processed according to the protocol of a ChIP-IT high-sensitivity kit (Active Motif), and subsequent qPCR analysis was carried out as described above.
ChIP-seq.
Chromatin from MCF7 cells was fragmented by sonication using a Bioruptor (Diagnode) at a 9 strength outcome (30 cycles, 30 s on plus 60 s off) to generate fragments <200 bp long and confirmed by agarose gel electrophoresis. Sheared DNA fragments were divided into two parts. One part was immunoprecipitated with specific RECQ1 and ERα antibodies as described above for ChIP-qPCR, and the second part was used as the corresponding input. After purification using the ChIP-IT high-sensitivity kit (Active Motif), immunoprecipitated DNA and input DNA were converted into sequencing libraries using the TruSeq ChIP sample preparation kit (Illumina), which were then sequenced in single-end 75-bp sequencing with the NextSeq 500 system (Illumina). The samples had ∼30 to 64 million pass filter reads with a base call quality of above ∼90% of bases with Q30 and above.
(i) Peak calling.
RECQ1 and ERα ChIP-seq peaks were called against IgG controls by using the MACS2 algorithm from the Genomatix genome analyzer (https://www.genomatix.de/solutions/genomatix-genome-analyzer.html), with the following parameters: broad region calling on, bandwidth = 300, q-value cutoff = 1.00e–02, and model fold = [5, 50].
(ii) Heatmap and colocalization analysis.
The normalized sequencing coverage for the RECQ1 and ERα ChIP-seq data was created using BAMscale (63). Colocalization of signal heatmap was created using the rtracklayer (v1.48.0) (64) and ComplexHeatmap (65) (v2.4.3) packages in R (v4.0.2, script available at https://github.com/ncbi/BAMscale) using the RECQ1 called peaks (n = 7,023) with the RECQ1 and ERα ChIP-seq data, along with FOXA1 (ENCFF255FPM), GATA3 (ENCFF477GZL), H3K27ac (ENCFF411FCW), H3K4me1 (ENCFF983TTS), H3K4me3 (ENCFF862CKA) and H3K9me3 (ENCFF688REP) data downloaded from ENCODE. Peaks were clustered using k-means clustering, setting the centers to 3, and using the H3K4me1 (enhancer mark) and H3K4me3 (promoter) histone marks for cluster identification. The RECQ1 peaks were extended by 5 kb upstream and downstream when creating the heatmaps.
Colocalization analyses between RECQ1 and other ChIP-seq bed files or transcription start regions were performed using the genome inspector program of the genomatix genome analyzer. Colocalization analysis were performed between the binding sites of POL2RA from cells collected over a time course of 0 to 320 min after estradiol treatment (42) and binding sites of RECQ1 (duplicates, this study), ERα (duplicates, this study), PAX8, REST, FOXA1, and GATA3. Publicly available ChIPseq bed files were obtained from Cistrome (http://cistrome.org/) (66) and ENCODE: POLR2A (GSM1091921 [control], GSM1091915 [10 min], GSM1091916 [20 min], GSM1091917 [40 min], GSM1091918 [80 min], GSM1091919 [160 min], and GSM1091920 [320 min]), PAX8 (GSM2828671 and GSM2828670), REST (GSM1010891), FOXA1 (ENCFF255FPM, GSM1534737, GSM3092505, GSM798437, and GSM798436), and GATA3 (ENCFF477GZL, GSM986068, GSM1241752, and GSM720423). The genome inspector program was used to quantify colocalization in 5-kb windows between anchor sets for RECQ1 (n = 2 data sets), ERα (n = 2), PAX8 (n = 2), REST (n = 1), FOXA1 (n = 5), and GATA3 (n = 4) and each of the partner sets POLR2A binding sites for 0, 10, 20, 40, 60, 160, and 320 min (n = 1 data set each). Averages of colocalization values for each anchor sets were normalized using the time point t = 0 for estradiol treatment. Colocalization analysis between the RECQ1 binding sites (7,023 peaks) and the transcriptional start regions (TSRs, 291,570 regions) was also performed. For the genomatix genome annotation and analysis project, TSRs are defined as regions of genomic sequence for which experimental evidence for transcription initiation is available. The extent of colocalization (%) was measured as the fraction of RECQ1 peaks within the 5-kb window of the TSRs.
(iii) Validation of ChIP-seq data.
Ten genes were selected for validation of ChIP-seq data by ChIP-qPCR. The selection of genes is based on three conditions: (i) the genomic sequence of the gene was commonly bound by RECQ1 and ERα, (ii) at least one binding site of RECQ1 or ERα was located in the promoter region of the gene, and (iii) the functional relationship of the gene with ESR1 had been previously reported.
FAIRE and FAIRE-qPCR.
Formaldehyde-assisted isolation of regulatory elements (FAIRE) and FAIRE-qPCR were carried out based on a protocol described by Rodríguez-Gil et al. (50). Briefly, 5 × 106 of MCF7 cells were cross-linked with 1% formaldehyde for 10 min at room temperature and then quenched with 125 mM glycine. Fixed cells were lysed in SDS lysis buffer and chromatin-bound DNA sheared by sonication to obtain fragments of approximately 200 to 300 bp in length, representing one to two nucleosomes. Each sample was divided into two aliquots. One aliquot was treated with proteinase K at 37°C for 4 h, followed by incubation at 65°C for 6 h to reverse the cross-link, and the other was left untreated. A 1% portion of the untreated aliquot was used as an input. All samples were then subjected to three consecutive phenol-chloroform extractions extracted by phenol-chloroform. The purified DNAs were subjected to qPCR analyses, and the primers used were the same as those used in ChIP qPCR, encompassing the promoter and enhancer of ESR1. The promoter of GAPDH, an actively transcribed housekeeping gene, served as a positive control, and a heterochromatin region on chromosome 12 was used as a negative control. The relative enrichment for each amplicon was calculated using the comparative CT method such that a ratio is calculated for the signal from the FAIRE sample relative to the signal from input control DNA (67).
Clinical study of RECQ1 and FOXA1 expression in ERα-positive human breast cancers.
(i) Patients. The study was performed in a consecutive series of patients with ERα+ primary invasive breast carcinomas who were diagnosed between 1986 and 1999 and entered into the Nottingham Tenovus Primary Breast Carcinoma series. Patient demographics are summarized in Table S11 in the supplemental material. This is a well-characterized series of patients with long-term follow-up that has been investigated in a wide range of biomarker studies (32). All patients were treated uniformly in a single institution with standard surgery (mastectomy or wide local excision), followed by radiotherapy. Prior to 1989, patients did not receive systemic adjuvant treatment (AT). After 1989, AT was scheduled based on prognostic and predictive factor status, including the Nottingham Prognostic Index (NPI), ERα status, and menopausal status. Patients with NPI scores of <3.4 (low risk) did not receive AT. For premenopausal patients with NPI scores of ≥3.4 (high risk), classical cyclophosphamide, methotrexate, and 5-fluorouracil (CMF) chemotherapy was administered; patients with ERα-positive tumors were also offered endocrine therapy. Postmenopausal patients with NPI scores of ≥3.4 and ERα positivity were offered endocrine therapy. Median follow-up was 111 months (range, 1 to 233 months). Survival data, including breast cancer-specific survival (BCSS), was maintained on a prospective basis. BCSS was defined as the number of months from diagnosis to the occurrence of breast cancer-related death. Survival was censored if the patient was still alive at the time of analysis, lost to follow-up, or died from other causes. Tumor Marker Prognostic Studies (REMARK) criteria, recommended by McShane et al. (68), were followed throughout this study. Ethical approval was obtained from the Nottingham Research Ethics Committee (C202313).
(ii) Tissue microarrays and immunohistochemistry. Tumors were arrayed in tissue microarrays (TMAs) constructed with 0.6-mm cores sampled from the periphery of the tumors. The TMAs were immunohistochemically profiled as described previously for RECQ1 (32), FOXA1 (53), and other biological antibodies (see Table S12 in the supplemental material). Immunohistochemical (IHC) staining was performed using the Thermo Scientific Shandon Sequenza chamber system (REF 72110017), in combination with the Novolink Max Polymer Detection System (RE7280-K: 1,250 tests) and the Leica Bond Primary Antibody Diluent (AR9352), each used according to the manufacturer’s instructions (Leica Microsystems). Leica Autostainer XL machine was used to dewax and rehydrate the slides. Pretreatment antigen retrieval was performed on the TMA sections using sodium citrate buffer (pH 6.0) and heated for 20 min at 95°C in a microwave (WhirlPool JT359 Jet Chef, 1,000 W). A set of slides were incubated for 60 min with the primary anti-RECQ1 antibody (Bethyl Laboratories, catalog no. A300-450A) at a dilution of 1:1,000. Negative and positive (by omission of the primary antibody and IgG-matched serum) controls were included in each run. The negative control ensured that all of the staining was produced from the specific interaction between antibody and antigen.
Mouse monoclonal antibody to FOXA1 (clone 2F83; ab40868; Abcam, Cambridge, UK) was optimized at a working dilution of 1:2,000 using full-face sections of mouse fetal lung tissue as a positive-control tissue. Immunohistochemical staining of FOXA1 was carried out using a Techmate500 Plus (Dako Cytomation, Cambridge, UK) automatic immune stainer with a linked streptavidin-biotin technique according to the manufacturer’s instructions after microwave antigen retrieval in citrate buffer (pH 6.0). Negative controls were performed by omitting the primary antibody. Sections were counterstained in hematoxylin and coverslipped using DPX mounting medium.
(iii) Evaluation of immune staining. Whole-field inspection of the core was scored, and the intensities of nuclear staining were grouped as follows: 0 = no staining, 1 = weak staining, 2 = moderate staining, and 3 = strong staining. The percentage of each category was estimated (0 to 100%). The H-score (range, 0 to 300) was calculated by multiplying the intensity of staining and percentage staining. RECQ1 expression was categorized based on the frequency histogram distributions. The tumor cores were evaluated by two scorers, and the concordance between the two scorers was excellent (k = 0.79). Xtile (v3.6.1) was used to identify a cutoff in protein expression values such that the resulting subgroups had significantly different survival courses. An H-score of ≥215 was taken as the cutoff for a high RECQ1 level (32). Not all cores within the TMA were suitable for IHC assessments since some cores were missing or containing inadequate invasive cancer (<15% tumor). The FOXA1 H-score cutoff point for determining positive and negative staining was chosen as the median H-score of the informative cases (H-score P10) (53). HER2 scoring was performed using the manufacturer recommendations (Ventana Medical Systems, Tucson, AZ). Breast carcinomas that were considered positive for HER2 protein overexpression met threshold criteria for the intensity and pattern of membrane staining (2+ or greater on a scale of 0 to 3+) and for the percentage of positive tumor cells (>10%).
(iv) Statistical analysis. Data analysis was performed using SPSS (v17; SPSS, Chicago, IL). Where appropriate, Pearson’s chi-square, Fisher exact, Student t, and one-way analysis of variance tests were used. Cumulative survival probabilities were estimated using the Kaplan-Meier method, and differences between survival rates were tested for significance using the log-rank test. Hazard ratios and 95% confidence intervals (95% CI) were estimated for each variable. All tests were two-sided with a 95% CI, and a P value of <0.05 was considered significant.
Data availability.
The RNA-seq data upon RECQ1 and ESR1 knockdown in MCF7 cells have been deposited in GEO under accession number GSE152323 (the reviewer token is qvepomgmvvobfel). The ChIP-seq data for RECQ1 and ERα have been deposited in GEO under accession number GSE153286.
Supplementary Material
ACKNOWLEDGMENTS
We thank the National Cancer Institute sequencing core facility for performing RNA-seq.
This study was supported by the National Institute of General Medical Sciences of the National Institutes of Health (under award SC1GM093999), the National Science Foundation (under award NSF1832163), the National Institute on Aging (under award 1R2 AG047843 [S.S.]), and the Intramural Research Program (A.L., S.A., J.K., and M.I.A.) of the National Cancer Institute, Center for Cancer Research, National Institutes of Health. B.B. was supported by a graduate student fellowship from Umm Al-Qura University, Saudi Arabia.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, Brosh RM, Jr, Blackshear PJ. 2007. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol Cell Biol 27:1784–1794. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma S. 2014. An appraisal of RECQ1 expression in cancer progression. Front Genet 5:426. doi: 10.3389/fgene.2014.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendoza-Maldonado R, Faoro V, Bajpai S, Berti M, Odreman F, Vindigni M, Ius T, Ghasemian A, Bonin S, Skrap M, Stanta G, Vindigni A. 2011. The human RECQ1 helicase is highly expressed in glioblastoma and plays an important role in tumor cell proliferation. Mol Cancer 10:83. doi: 10.1186/1476-4598-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li D, Liu H, Jiao L, Chang DZ, Beinart G, Wolff RA, Evans DB, Hassan MM, Abbruzzese JL. 2006. Significant effect of homologous recombination DNA repair gene polymorphisms on pancreatic cancer survival. Cancer Res 66:3323–3330. doi: 10.1158/0008-5472.CAN-05-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viziteu E, Klein B, Basbous J, Lin YL, Hirtz C, Gourzones C, Tiers L, Bruyer A, Vincent L, Grandmougin C, Seckinger A, Goldschmidt H, Constantinou A, Pasero P, Hose D, Moreaux J. 2017. RECQ1 helicase is involved in replication stress survival and drug resistance in multiple myeloma. Leukemia 31:2104–2113. doi: 10.1038/leu.2017.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowden AR, Tischkowitz M. 2019. Clinical implications of germline mutations in breast cancer genes: RECQL. Breast Cancer Res Treat 174:553–560. doi: 10.1007/s10549-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cybulski C, Carrot-Zhang J, Kluźniak W, Rivera B, Kashyap A, Wokołorczyk D, Giroux S, Nadaf J, Hamel N, Zhang S, Huzarski T, Gronwald J, Byrski T, Szwiec M, Jakubowska A, Rudnicka H, Lener M, Masojć B, Tonin PN, Rousseau F, Górski B, Dębniak T, Majewski J, Lubiński J, Foulkes WD, Narod SA, Akbari MR. 2015. Germline RECQL mutations are associated with breast cancer susceptibility. Nat Genet 47:643–646. doi: 10.1038/ng.3284. [DOI] [PubMed] [Google Scholar]
- 8.Sun J, Wang Y, Xia Y, Xu Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Lou H, Xie Y. 2015. Mutations in RECQL gene are associated with predisposition to breast cancer. PLoS Genet 11:e1005228. doi: 10.1371/journal.pgen.1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma S, Doherty KM, Brosh RM, Jr. 2006. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J 398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu WK, Hickson ID. 2009. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer 9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 11.Brosh RM, Jr. 2013. DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer 13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croteau DL, Popuri V, Opresko PL, Bohr VA. 2014. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem 83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM, Jr. 2005. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem 280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 14.Pike AC, Gomathinayagam S, Swuec P, Berti M, Zhang Y, Schnecke C, Marino F, von Delft F, Renault L, Costa A, Gileadi O, Vindigni A. 2015. Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: insights from DNA complex structures. Proc Natl Acad Sci U S A 112:4286–4291. doi: 10.1073/pnas.1417594112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma S, Phatak P, Stortchevoi A, Jasin M, Larocque JR. 2012. RECQ1 plays a distinct role in cellular response to oxidative DNA damage. DNA Repair (Amst) 11:537–549. doi: 10.1016/j.dnarep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popuri V, Croteau DL, Brosh RM, Jr, Bohr VA. 2012. RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures. Cell Cycle 11:4252–4265. doi: 10.4161/cc.22581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berti M, Chaudhuri AR, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, Marino F, Lucic B, Biasin V, Gstaiger M, Aebersold R, Sidorova JM, Monnat RJ, Jr, Lopes M, Vindigni A. 2013. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol 20:347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu X, Parvathaneni S, Hara T, Lal A, Sharma S. 2013. Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D. Mol Cancer 12:29. 29–4598-12–29. doi: 10.1186/1476-4598-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parvathaneni S, Sharma S. 2019. The DNA repair helicase RECQ1 has a checkpoint-dependent role in mediating DNA damage responses induced by gemcitabine. J Biol Chem 294:15330–15345. doi: 10.1074/jbc.RA119.008420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee T, Sommers JA, Huang J, Seidman MM, Brosh RM, Jr. 2015. Catalytic Strand Separation by RECQ1 is required for RPA-mediated response to replication stress. Curr Biol 25:2830–2838. doi: 10.1016/j.cub.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parvathaneni S, Stortchevoi A, Sommers JA, Brosh RM, Jr, Sharma S. 2013. Human RECQ1 interacts with Ku70/80 and modulates DNA end-joining of double-strand breaks. PLoS One 8:e62481. doi: 10.1371/journal.pone.0062481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodrick J, Gupta S, Camacho S, Parvathaneni S, Choudhury S, Cheema A, Bai Y, Khatkar P, Erkizan HV, Sami F, Su Y, Schärer OD, Sharma S, Roy R. 2017. A new sub-pathway of long-patch base excision repair involving 5′ gap formation. EMBO J 36:1605–1622. doi: 10.15252/embj.201694920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sami F, Lu X, Parvathaneni S, Roy R, Gary RK, Sharma S. 2015. RECQ1 interacts with FEN-1 and promotes binding of FEN-1 to telomeric chromatin. Biochem J 468:227–244. doi: 10.1042/BJ20141021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popuri V, Hsu J, Khadka P, Horvath K, Liu Y, Croteau DL, Bohr VA. 2014. Human RECQL1 participates in telomere maintenance. Nucleic Acids Res 42:5671–5688. doi: 10.1093/nar/gku200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debnath S, Sharma S. 2020. RECQ1 helicase in genomic stability and cancer. Genes (Basel) 11:622. doi: 10.3390/genes11060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S, Brosh RM, Jr. 2007. Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS One 2:e1297. doi: 10.1371/journal.pone.0001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sami F, Sharma S. 2013. Probing genome maintenance functions of human RECQ1. Comput Struct Biotechnol J 6:e201303014. doi: 10.5936/csbj.201303014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cogoni C, Macino G. 1999. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286:2342–2344. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- 29.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. 2006. Characterization of the piRNA complex from rat testes. Science 313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 30.Lu X, Parvathaneni S, Li XL, Lal A, Sharma S. 2016. Transcriptome guided identification of novel functions of RECQ1 helicase. Methods 108:111–117. doi: 10.1016/j.ymeth.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XL, Lu X, Parvathaneni S, Bilke S, Zhang H, Thangavel S, Vindigni A, Hara T, Zhu Y, Meltzer PS, Lal A, Sharma S. 2014. Identification of RECQ1-regulated transcriptome uncovers a role of RECQ1 in regulation of cancer cell migration and invasion. Cell Cycle 13:2431–2445. doi: 10.4161/cc.29419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arora A, Parvathaneni S, Aleskandarany MA, Agarwal D, Ali R, Abdel-Fatah T, Green AR, Ball GR, Rakha EA, Ellis IO, Sharma S, Madhusudan S. 2017. Clinicopathological and functional significance of RECQL1 helicase in sporadic breast cancers. Mol Cancer Ther 16:239–250. doi: 10.1158/1535-7163.MCT-16-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carroll JS. 2016. Mechanisms of oestrogen receptor (ER) gene regulation in breast cancer. Eur J Endocrinol 175:R41–R49. doi: 10.1530/EJE-16-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke R, Liu MC, Bouker KB, Gu Z, Lee RY, Zhu Y, Skaar TC, Gomez B, O’Brien K, Wang Y, Hilakivi-Clarke LA. 2003. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene 22:7316–7339. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- 35.Musgrove EA, Sutherland RL. 2009. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 36.Osborne CK, Schiff R. 2011. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med 62:233–247. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao L, Zhou S, Gustafsson JÅ. 2019. Nuclear receptors: recent drug discovery for cancer therapies. Endocr Rev 40:1207–1249. doi: 10.1210/er.2018-00222. [DOI] [PubMed] [Google Scholar]
- 38.Carroll JS, Brown M. 2006. Estrogen receptor target gene: an evolving concept. Mol Endocrinol 20:1707–1714. doi: 10.1210/me.2005-0334. [DOI] [PubMed] [Google Scholar]
- 39.Lupien M, Brown M. 2009. Cistromics of hormone-dependent cancer. Endocr Relat Cancer 16:381–389. doi: 10.1677/ERC-09-0038. [DOI] [PubMed] [Google Scholar]
- 40.Ciocca DR, Fanelli MA. 1997. Estrogen receptors and cell proliferation in breast cancer. Trends Endocrinol Metab 8:313–321. doi: 10.1016/S1043-2760(97)00122-7. [DOI] [PubMed] [Google Scholar]
- 41.Holmes KA, Brown GD, Carroll JS. 2016. Chromatin immunoprecipitation-sequencing (ChIP-seq) for mapping of estrogen receptor-chromatin interactions in breast cancer. Methods Mol Biol 1366:79–98. doi: 10.1007/978-1-4939-3127-9_8. [DOI] [PubMed] [Google Scholar]
- 42.Wa Maina C, Honkela A, Matarese F, Grote K, Stunnenberg HG, Reid G, Lawrence ND, Rattray M. 2014. Inference of RNA polymerase II transcription dynamics from chromatin immunoprecipitation time course data. PLoS Comput Biol 10:e1003598. doi: 10.1371/journal.pcbi.1003598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauss C, Kornowski M, Benvenisty A, Shahar A, Masury H, Ben-Porath I, Ravid T, Arbel-Eden A, Goldberg M. 2014. The DNA2 nuclease/helicase is an estrogen-dependent gene mutated in breast and ovarian cancers. Oncotarget 5:9396–9409. doi: 10.18632/oncotarget.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellison-Zelski SJ, Solodin NM, Alarid ET. 2009. Repression of ESR1 through actions of estrogen receptor alpha and Sin3A at the proximal promoter. Mol Cell Biol 29:4949–4958. doi: 10.1128/MCB.00383-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powers GL, Ellison-Zelski SJ, Casa AJ, Lee AV, Alarid ET. 2010. Proteasome inhibition represses ERα gene expression in ER+ cells: a new link between proteasome activity and estrogen signaling in breast cancer. Oncogene 29:1509–1518. doi: 10.1038/onc.2009.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 47.Jozwik KM, Carroll JS. 2012. Pioneer factors in hormone-dependent cancers. Nat Rev Cancer 12:381–385. doi: 10.1038/nrc3263. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Wang W, Wang J, Malovannaya A, Xi Y, Li W, Guerra R, Hawke DH, Qin J, Chen J. 2015. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol Syst Biol 11:775. doi: 10.15252/msb.20145504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glont SE, Chernukhin I, Carroll JS. 2019. Comprehensive genomic analysis reveals that the pioneering function of FOXA1 is independent of hormonal signaling. Cell Rep 26:2558–2565. doi: 10.1016/j.celrep.2019.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodríguez-Gil A, Riedlinger T, Ritter O, Saul VV, Schmitz ML. 2018. Formaldehyde-assisted isolation of regulatory elements to measure chromatin accessibility in mammalian cells. J Vis Exp 134:57272. doi: 10.3791/57272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. 2016. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol Cell 62:79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang SH, Kiefer CM, Yang TP. 2003. Role of the promoter in maintaining transcriptionally active chromatin structure and DNA methylation patterns in vivo. Mol Cell Biol 23:4150–4161. doi: 10.1128/mcb.23.12.4150-4161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Habashy HO, Powe DG, Rakha EA, Ball G, Paish C, Gee J, Nicholson RI, Ellis IO. 2008. Forkhead-box A1 (FOXA1) expression in breast cancer and its prognostic significance. Eur J Cancer 44:1541–1551. doi: 10.1016/j.ejca.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 54.Green KA, Carroll JS. 2007. Oestrogen-receptor-mediated transcription and the influence of cofactors and chromatin state. Nat Rev Cancer 7:713–722. doi: 10.1038/nrc2211. [DOI] [PubMed] [Google Scholar]
- 55.Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. 2011. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krishnakumar R, Kraus WL. 2010. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell 39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schiewer MJ, Knudsen KE. 2014. Transcriptional roles of PARP1 in cancer. Mol Cancer Res 12:1069–1080. doi: 10.1158/1541-7786.MCR-13-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. 2011. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horimoto Y, Sasahara N, Sasaki R, Hlaing MT, Sakaguchi A, Saeki H, Arakawa A, Himuro T, Saito M. 2020. High FOXA1 protein expression might predict late recurrence in patients with estrogen-positive and HER2-negative breast cancer. Breast Cancer Res Treat 183:41–48. doi: 10.1007/s10549-020-05751-x. [DOI] [PubMed] [Google Scholar]
- 60.Martínez-Sáez O, Chic N, Pascual T, Adamo B, Vidal M, González-Farré B, Sanfeliu E, Schettini F, Conte B, Brasó-Maristany F, Rodríguez A, Martínez D, Galván P, Rodríguez AB, Martinez A, Muñoz M, Prat A. 2020. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res 22:45. doi: 10.1186/s13058-020-01284-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chi D, Singhal H, Li L, Xiao T, Liu W, Pun M, Jeselsohn R, He H, Lim E, Vadhi R, Rao P, Long H, Garber J, Brown M. 2019. Estrogen receptor signaling is reprogrammed during breast tumorigenesis. Proc Natl Acad Sci U S A 116:11437–11443. doi: 10.1073/pnas.1819155116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 63.Pongor LS, Gross JM, Vera Alvarez R, Murai J, Jang SM, Zhang H, Redon C, Fu H, Huang SY, Thakur B, Baris A, Marino-Ramirez L, Landsman D, Aladjem MI, Pommier Y. 2020. BAMscale: quantification of next-generation sequencing peaks and generation of scaled coverage tracks. Epigenetics Chromatin 13:21. doi: 10.1186/s13072-020-00343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lawrence M, Gentleman R, Carey V. 2009. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 66.Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H, Chen CH, Brown M, Zhang X, Meyer CA, Liu XS. 2019. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res 47:D729–D735. doi: 10.1093/nar/gky1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simon JM, Giresi PG, Davis IJ, Lieb JD. 2012. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc 7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics . 2005. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data upon RECQ1 and ESR1 knockdown in MCF7 cells have been deposited in GEO under accession number GSE152323 (the reviewer token is qvepomgmvvobfel). The ChIP-seq data for RECQ1 and ERα have been deposited in GEO under accession number GSE153286.