MicroRNAs (miRNAs) are small regulatory RNAs of relatively long half-life in non-proliferative human cells. However, in cancer cells the half-lives of miRNAs are comparatively short.
KEYWORDS: miRNA export, GW182, translation repression by miRNA, miRNA turnover, P-bodies, cell senescence, target-mediated miRNA regulation, extracellular vesicles, HuR, P-body, miRNA
ABSTRACT
MicroRNAs (miRNAs) are small regulatory RNAs of relatively long half-life in non-proliferative human cells. However, in cancer cells the half-lives of miRNAs are comparatively short. To understand the mechanism of rapid miRNA turnover in cancer cells, we explored the effect of target mRNAs on the abundance of the miRNAs that repress them. We have noted an accelerated extracellular vesicle (EV)-mediated export of miRNAs in presence of their target mRNAs in mammalian cells, and this target-driven miRNA-export process is retarded by Ago2-interacting protein GW182B. The GW182 group of proteins are localized to GW182 bodies or RNA processing bodies in mammalian cells, and GW182B-dependent retardation of miRNA export depends on GW body integrity and is independent of the HuR protein-mediated auxiliary pathway of miRNA export. Our data thus support the existence of a HuR-independent pathway of miRNA export in human cells that can be targeted in MDA-MB-231 cancer cells, to increase the level of cellular let-7a, a known negative regulator of cancer growth.
INTRODUCTION
MicroRNAs (miRNAs), noncoding RNAs of 22 nucleotides (nt) in length, form a complex with Argonaute group of proteins and regulate the majority of genes by imperfect base pairing to the 3′ untranslated region (UTR) of target messages (1). Most of the human genes are regulated by one or more miRNAs, and this regulation is contextual. Impairment of miRNA function has been documented in human diseases, and abnormal expression of miRNAs has been associated with several diseases, including different types of cancers (2–4). Microribonucleoproteins (miRNPs) repress target genes by inhibiting protein translation and affecting target mRNA stability (5, 6). Expressions and activities of miRNAs have been noted to be sensitive to cellular context (7, 8). But despite the fact that miRNAs are important regulators of gene expression in eukaryotes, factors controlling miRNA stability and turnover in mammalian cells are only partly explored.
The mechanism to regulate miRNA level and activity, for the most part, remains limited to discoveries related to specific miRNAs (9). Recent reports suggest that the RNAs with miRNA binding sites not only influence their activity by altering stability of the corresponding miRNAs in animal cells but also affect their de novo biogenesis (10). The claim of miRNA stabilization due to its base pairing with target mRNA in Caenorhabditis elegans (11) is opposite to what has been reported for mammalian cells, in which cellular and viral target messages can downregulate corresponding miRNAs (12–14). Target RNA-directed microRNA degradation (TDMD) is another pathway described in recent times in mammalian neuronal cells where 3′-end tailing of miRNA by the addition of A/U nontemplated nucleotides, trimming, or shortening from the 3′ end, and highly specific microRNA loss happens in the presence of target mRNAs (15). It has also been reported that extracellular miRNA export could be a key mechanism of miRNA lowering in mammalian cells (16–18).
In animal cells, multivesicular bodies (MVBs) play an important role in restricting miRNP recycling. Inhibition of MVB formation decreases miRISC activity, whereas blockage of MVB maturation into lysosomes leads to increase in miRNP activity (19, 20). MVBs can also fuse with cell membranes to form extracellular vesicles (EVs) or exosomes in animal cells (21). Exosomes, small (30 to 100 nm) EVs secreted by the animal cells to the extracellular space, are considered means of intercellular communication of cargos like proteins and RNAs (22). As miRNAs are shown to be present in EVs, they serve as intercellular carriers of miRNAs (23).
Extracellular export of excess miRNAs is another way of regulating miRNA levels in human cells. Several posttranscriptional modifications of miRNAs are also noted to be linked with their EV-mediated export. Several RNA binding proteins have been identified as miRNA exporters, as they promote export of miRNAs via EVs (18, 24). Human ELAV protein HuR was identified earlier as a derepressor of miRNA activity that inhibits the action of miRNA on cis-bound target mRNAs primarily by uncoupling the miRNPs from the target messages and mobilizing them from P-bodies (PBs) in amino acid-starved human hepatoma cells (25). HuR also acts as an miRNA sponge, and by its reversible binding with miRNAs in stressed cells, it can accelerate miRNA loss by promoting its extracellular export (18).
Phase separation of proteins and other components contributes to regulation of biochemical pathways in mammalian cells, and miRNA regulation has also been found to be compartmentalized in mammalian cells (26). The membranes of endoplasmic reticulum (ER) and P-bodies are the compartments where the repression and storage of miRNA targets happen, respectively (25, 26). The process of miRNA or target RNA compartmentalization could therefore be the primary mechanism of reversible translation repression and miRNA turnover regulation in mammalian cells.
In mammalian cells, repressed messages can be stored in P-bodies, as the phase-separated compartments. P-bodies have increased abundances of miRNA and mRNA-catabolizing enzymes there. However, mRNAs can also be stored in P-bodies in a reversible manner (25). The GW182 family of proteins, also known as the TNRC6 family, has three members in human cells that interact with Ago proteins (27). GW182 proteins are components of mammalian P-bodies and are the effectors of miRNP-mediated gene repression. They can also act as stabilizers of miRNPs (28). GW182 proteins, by recruiting PABP protein, prevent the initiation step of protein translation from the target mRNAs (29).
This work shows how phase separation of Ago2, by interacting with the GW182 group of proteins, ensures retardation of miRNA export in mammalian cells. The process depends on the integrity of the separated phase and it opposes the miRNA export that happens otherwise, specifically for miRNAs having their target mRNAs in abundance. We observed that the levels of GW182 do not have an effect on HuR-driven export of miRNA, and thus, it acts as the main pathway for miRNA export in human cancer cells other than the known HuR-mediated auxiliary miRNA export path (18).
RESULTS
Lowering of miRNAs in human cells in the presence of their substrate mRNAs.
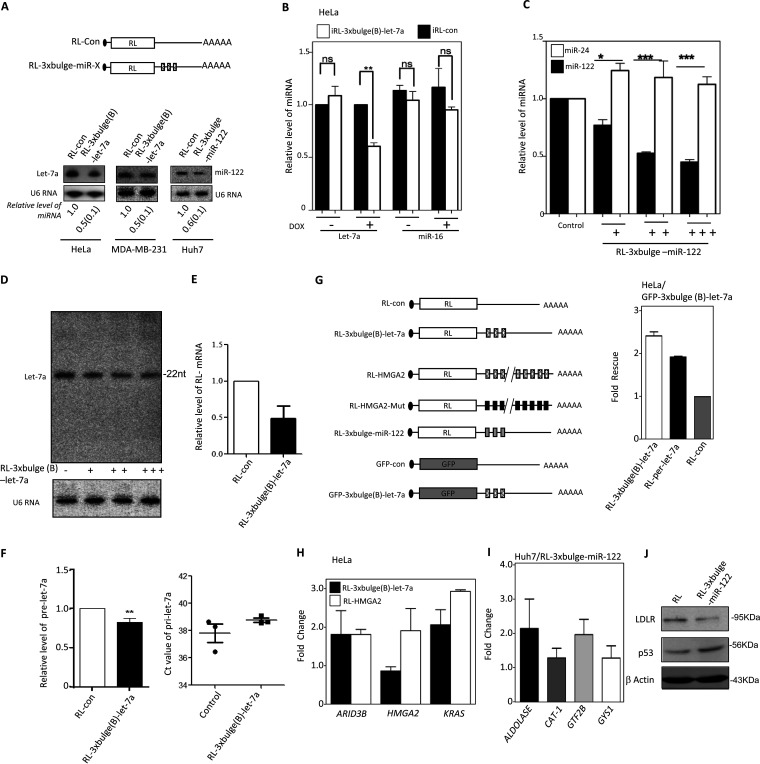
miRNAs, in complex with Ago proteins, find their targets and repress the target mRNAs with complementary sequences to induce translation suppression and degradation. Can the translation-repressive activity of a miRNA and miRNP make them different from the resting miRNA/miRNP pool that is yet to go for a repression act? To find out how the target repression by miRNA is linked to miRNA turnover, we expressed reporter mRNAs with three imperfect miRNA binding sites for let-7a in HeLa or MDA-MB-231 cells, in which let-7a miRNA is also expressed. The effects of expression of reporter mRNAs on existing let-7a pools of the respective cells were compared. Quantitative estimation of the remaining let-7a after 48 h of reporter mRNA expression was performed. A decrease in existing let-7a level was noted in the presence of the target. A similar decrease of miR-122 in hepatic Huh7 cells was also documented in the presence of miR-122 target reporter mRNA expressed there (Fig. 1A). A similar observation was also documented for HeLa cells when an inducible Renilla luciferase (RL) reporter with three let-7a binding sites was expressed. The reporters were expressed in an inducible manner, and after 24 h of induction with doxycycline, cellular levels of miRNAs were quantified. A similar observation was made with an miR-122 reporter in Huh7 cells expressing miR-122. In both cases, the levels of the nontarget miRNAs miR-16 and miR-24 were not changed with let-7a and miR-122 reporter mRNA, respectively (Fig. 1B and C). The reduction of the mature miRNA content was also dependent on the amount of target mRNA. With increasing amounts of target reporter mRNA for miR-122, we could see specific reduction of the cognate miRNA-122 in Huh7 cells, while nonspecific miRNA miR-24 did not change (Fig. 1C).
FIG 1.
Engagement of miRNA in target RNA repression affects miRNA level in human cells. (A) Downregulation in cellular miRNA levels in the presence of their substrate mRNAs. Shown is a scheme of reporters used for expression of a target message in excess. Representative blots show let-7a or miR-122 and U6 snRNA levels (endogenous control) in human cell lines expressing RL reporters. Quantifications of miRNA levels from multiple experiments are shown (means ± SEM; n = 3 to 5). (B) Relative levels of let-7a and miR-16 in HeLa cells expressing a control or let-7a reporter from the tetracycline-inducible constructs. Real-time quantifications of endogenous miRNA were done after 24 h of induction and were normalized against U6 snRNA. The value obtained with RL-con was set as the unit (means ± SEM; n = 3). (C) Level of miR-122 and miR-24 in Huh7 cells transfected with increasing concentrations (100 ng to 1,000 ng for 106 cells) of in vitro-transcribed RL reporter mRNA with three miR-122 binding sites. Levels of miRNAs in RL-con (1000 ng) (means ± SEM; n = 3) are used as units. (D) No effect of target mRNA on let-7a miRNA length. Effect of transfection of HeLa cells with increasing concentrations of target mRNA expression plasmid on let-7a miRNA levels evident by Northern blotting done with 32P-labeled anti-let-7a oligonucleotide. No change in miRNA length was observed. The Northern blot data of U6 snRNA were used as loading control. (E and F) Level of reporter mRNAs in HeLa cells expressing control or let-7a target containing mRNAs (E). In same cells, relative levels of pre-let-7a (left) and pri-let-7a (right) were also measured. All estimation was done using qRT-PCR. RL-con-expressing cells were used as a control (means ± SEM; n = 3) (F). (G) Scheme of target mRNA expression constructs used for reversal of miRNA action of endogenous targets (left) and derepression of let-7a Renilla reporters in HeLa cells expressing GFP let-7a reporter (right). Normalized expression levels of different RL reporters were measured in cells coexpressing GFP mRNAs with or without three let-7a target sites. Fold rescue represents the fold increase in activity of each RL reporter estimated by RL protein expressed in GFP-3×bulge-let-7a expressing cells than GFP-con. (H and I) Expression of target mRNA leads to upregulation of endogenous miRNA targets. Fold increases of endogenous target mRNAs in HeLa (H) and Huh7 (I) cells expressing let-7 a (RL-3×bulgeB-let7a or RL-HMGA2) and miR-122 (RL-3×bulge-miR-122) reporter were estimated by real-time quantification against their expression levels in control reporter-transfected cells. For RL-HMGA2, the control was RL-HMGA2-Mut reporter with mutated let-7a binding sites, while RL-con served as the control for the rest. (J) Reduction in miRNA level by its substrate leads to derepression of endogenous targets at the protein level. Expression of a reciprocally regulated indirect miR-122 target, LDL receptor, and the direct target p53 in Huh7 cells expressing reporters with or without miR-122 sites is shown. β-Actin served as a loading control. ns, nonsignificant. *, P < 0.05; **, P < 0.01; ***, P < 0.0001. P values were determined by paired t test.
It has been reported earlier that posttranscriptional modification of miRNA, associated with change of length of miRNAs, may cause the accelerated export of the modified miRNA in mammalian cells (30). To evaluate this possibility, we checked the length of the miRNA by Northern blotting of let-7a in the presence of increasing concentrations of its substrate. Interestingly, we did not observe any change in its length (Fig. 1D).
The levels of primary miRNA (pri-miRNA) do not change in the presence of target mRNA. However, there is a slight decrease in pre-miRNA levels that may account for an accelerated biogenesis of the miRNA in the presence of its target mRNAs, which corroborates our previous findings (10, 26). In this context, the target mRNA levels also drop compared to that of control mRNA without the let-7a sites (Fig. 1E and F).
As expected, substrate-dependent lowering of a miRNA resulted in higher expression of other endogenous or reporter mRNA targets of the cognate let-7a or miR-122 in respective cases, confirming lowering of effective miRNP pools in the presence of the substrate in HeLa or Huh7 cells. Using a green fluorescent protein (GFP)–let-7a reporter RNA, we found that luciferase activity of RL–let-7a reporters was rescued (Fig. 1G). Using two other let-7a reporters, one having 3 imperfect let-7a sites and another containing the 3′ UTR of HMGA2 genes (endogenous let-7a substrate), the expression levels of the endogenous mRNA target in HeLa cells were rescued (Fig. 1H). RL-3×bulge-miR-122 reporter mRNA also rescued the expression of the endogenous miR-122 target mRNA (Fig. 1I) and was associated with changes in protein levels of miR-122-controlled LDLR and p53 genes, which are known to be regulated positively and negatively by miR-122, respectively (Fig. 1J).
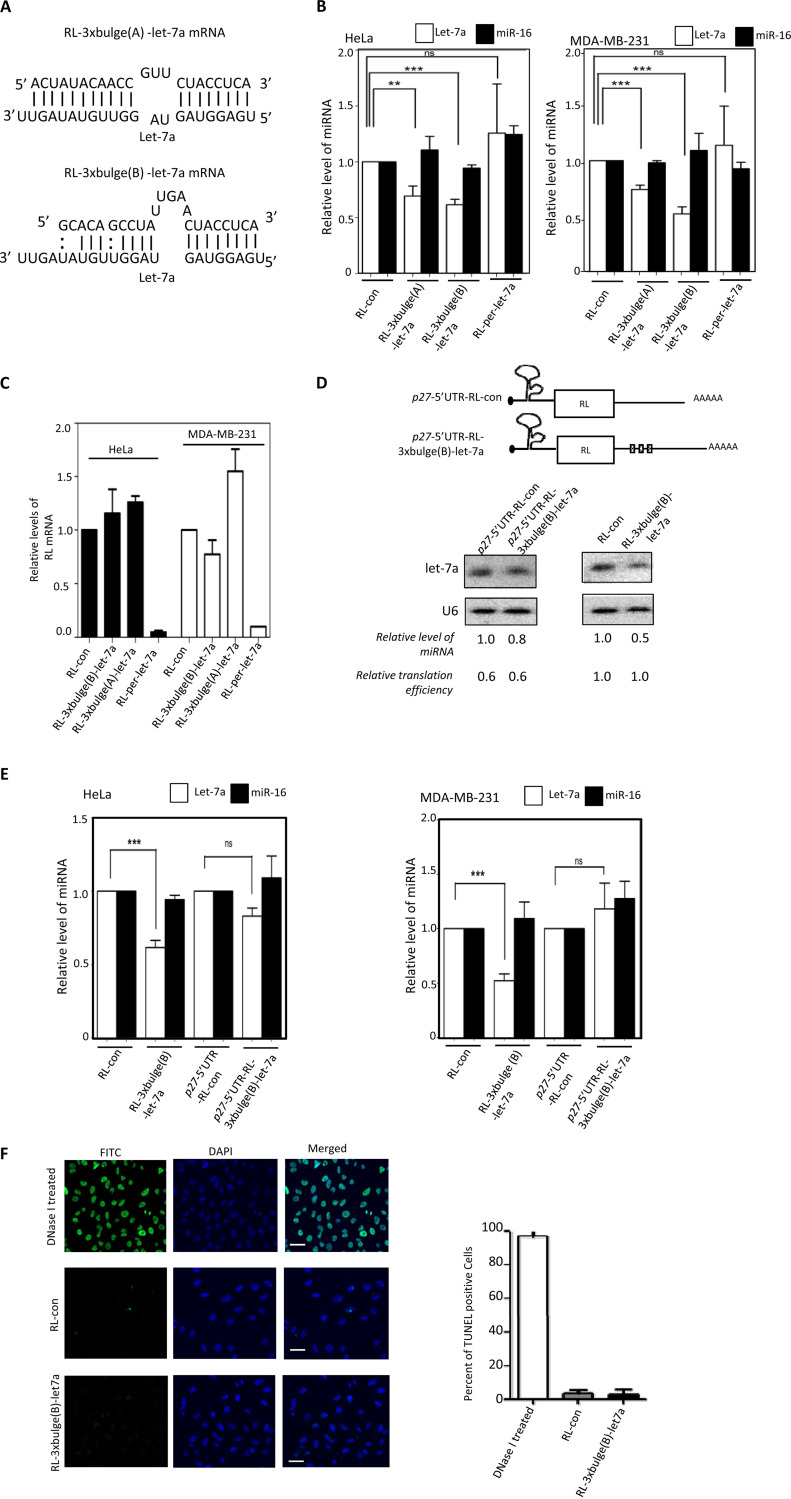
The miRNAs binding to the target sites could be strong or weak depending on the complementarities of target mRNAs to the respective miRNAs. We have used three let-7a reporters; one has one perfect let-7a site (RL-per), and the other two have either A- or B-type bulges and have different complementarities to let-7a in a nonseed region. The RL-per reporter should undergo a RISC-mediated cleavage upon expression in cells expressing let-7a. The A- or B-type bulges containing reporters are known to get repressed in HeLa cells but are also known to undergo different levels of degradation of the target mRNA (31). We found the target RNA-dependent miRNA decrease to be specific and dependent on target mRNA availability and on the nature of the target sites (Fig. 2A to C). The target mRNA with perfect miRNA sites undergoes RNA interference (RNAi)-like degradation, and availability of target mRNA becomes less. This target mRNA thus failed to induce lowering of miRNA content, although the respective miRNPs were engaged in RISC cleavage (Fig. 2A to C). This suggests that target-dependent miRNA lowering very much depends on active translation of the target messages. We found in subsequent experiments that mRNA with a translation-inhibitory secondary structure at its 5′ UTR (32) caused a reduced downregulation of corresponding miRNAs compared to the same target site containing reporter mRNA without the translation-inhibitory sequence in its 5′ UTR. This has been tested in two human cell lines (Fig. 2D and E) in which the let-7a reporter mRNA with the p27 5′ UTR failed to show downregulation of let-7a. This reporter also showed reduced translatability compared to those of respective mRNAs without the p27 5′ UTR. The observed effect of target mRNA on miRNA loss was not due to an increased apoptosis of the transfected cells, as transfection of plasmids expressing the control or reporter mRNA did not induce apoptosis in cells as evident the result obtained from a TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assay (Fig. 2F).
FIG 2.
Target site complementarities and target mRNA translatability affect target-driven miRNA lowering. (A) Target site complementarities for two let-7a reporters used to downregulate cellular let-7a level. Sequences and complementarities of two reporter (A and B types) mRNAs with let-7a are shown. (B) Level of let-7a and miR-16 in HeLa or MDA-MB-231 cells expressing control or reporter mRNAs with let-7a binding sites. RL-per-let-7a has one perfect let-7a site. miRNA levels normalized against U6 snRNA and values obtained with RL-con reporter were set as 1 (means ± SEM; n = 3). (C) Relative RL reporter mRNA levels in HeLa and MDA-MB-231 cells were estimated by real-time quantification, and values were normalized against GAPDH mRNA level. Values for RL-con were set as the unit. All experiments were performed in triplicates, and values are means ± SEM. (D and E) Target mRNA containing a translation-inhibitory element from the 5′ UTR of p27 mRNA reduces substrate dependent lowering of let-7a miRNA. Schematic representation of RL reporter construct containing the p27 5′ UTR element (D, top). The level of let-7a was measured in HeLa cells expressing either p27-RL-control or p27-RL-3×bulge(B)-let-7a mRNA (D, bottom). Luciferase assays for reporter gene and real-time quantification of reporter mRNA levels were performed to calculate the relative translation efficiencies, defined by amount of protein per unit of mRNA present in reporter mRNA-expressing cells (D, bottom left). Similar experiments were also done with RL-3×bulge(B)-let-7a and RL-con reporters. let-7a levels in RL-con reporter-transfected cells were considered 1 (D, bottom right). Northern blot data in panel D were substantiated with more quantitative data measured by real-time-based quantification of let-7a in HeLa and MDA-MB-231 cells and shown in panel E (means ± SEM; n = 3). (F) Effect of let-7a target RNA expression on cell apoptosis. TUNEL-positive cells (green) in the fluorescein isothiocyanate (FITC) channel were visualized and counted in nontransfected control and RL-con- or RL-3×bulge(B)-let-7a-expressing plasmid-transfected HeLa cells. DNase I-treated cells were used as the positive control. Estimation was done on the basis of data obtained from multiple measurements (means ± SEM; n = 10; right). Scale bars, 20 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.0001. P values were determined by paired t test.
Enhanced extracellular export of miRNA accounts for the lowering of miRNA content in human cancer cells.
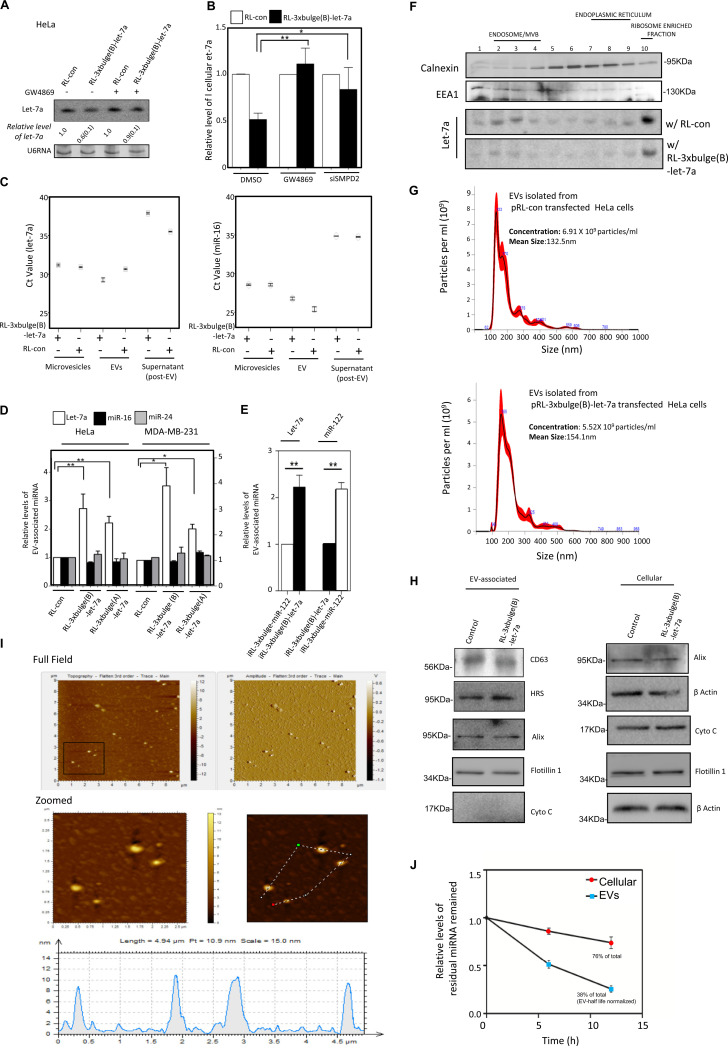
We have explored the effect of GW4869, a neutral sphingomyelinase inhibitor that is known for its inhibitory effect on EV-mediated miRNA export (33, 34). GW4869 blocks EV-mediated export of miRNA from mammalian cells by preventing biogenesis of EVs. The effect of GW4869 on target-driven miRNA export was evident, as the lowering of cellular miRNA in the presence of its target mRNAs was retarded when GW4869 was applied (Fig. 3A). SMPD2 is the target of GW4869. Confirming the role of EV biogenesis in target-dependent miRNA lowering, a similar effect was observed on cellular miRNA content when small interfering RNA (siRNA)-mediated knockdown of the protein SMPD2 was targeted (Fig. 3B).
FIG 3.
Substrate-driven extracellular export regulates cellular miRNA level in mammalian cells. (A) Effect of GW4869 treatment on target-mediated downregulation of let-7a in HeLa cells. U6 RNA serves as a loading control. Quantification was done from three different sets of Northern blot data. SD is shown in parentheses. (B) Real-time estimation of let-7a levels in MDA-MB-231 cells upon depletion of SMPD2 by RNAi or treatment of control siRNA-transfected cells with either dimethyl sulfoxide (DMSO) or GW4869 (means ± SEM; n = 4). (C) Relative levels of let-7a and miR-16 (used as control) present in different extracellular components like microvesicles, exosomes, or EVs and also in EV-free culture supernatant. For the same amount of mRNAs, CT values were plotted. (D) Real-time estimations of miRNA levels in EVs isolated from culture supernatants of cells expressing reporter mRNAs with or without let-7a miRNA binding sites (means ± SEM; n = 3). Two different reporters with differences in let-7a complementarities (see Fig. 2A) were used. (E) RL control and reporter mRNAs were induced in HeLa cells expressing miR-122, and cellular levels of let-7a or miR-122 were measured in EVs in each case after 24 h of induction (means ± SEM; n = 3). (F) Effect of target RNA on subcellular localization of miRNA in HeLa cells. HeLa cells expressing RL-con or RL-3×bulge(B)-let-7a were harvested, and isotonic lysates prepared from those cells were analyzed on a 3 to 30% OptiPrep density gradient. The individual fractions were analyzed for the presence of let-7a miRNA by Northern blotting. The presence of calnexin and EEA1 was used as a marker of endoplasmic reticulum and early endosome, respectively. (G) Nanoparticle tracking analysis of EVs obtained after isolation using a sucrose cushion. Average size and number obtained from cells expressing RL control and reporter mRNAs were compared. (H) Level of different marker proteins known to be present and absent in EVs isolated from HeLa cells expressing RL control and reporter mRNAs. Both cellular and EV contents were analyzed by Western blotting. (I) Amplitude and tomographic pictures of the EVs obtained from let-7a reporter RL-3×bulge(B)-let-7a-expressing cells by atomic force microscopy (top). The zoomed part was tracked (middle) for the particles for their size estimation (bottom). (J) Stability of miR-122 measured in EVs and at the cellular level. HeLa cells which do not express miR-122 were transfected with miRNA-122 mimic, and EVs were isolated 24 h posttransfection. Isolated EVs were incubated in a cell-free medium for another 24 h at 37°C. At regular intervals, intact miRNA content was measured. Similarly, cellular content of miR-122 was also measured. Values were compared against the amount of miRNA present at the beginning and plotted as percent residual miRNA remaining. The value of residual miRNA in the EV fraction was normalized against the amount of residual EV that had remained intact at 37°C after the incubation (36). *, P < 0.05; **, P < 0.01; ***, P < 0.0001. P values were determined by paired t test.
These data suggest the involvement of extracellular export in regulation of miRNA content by their target mRNAs. Comparing RNA from the EV, microvesicles, and EV-free supernatant of cell culture, the increase of let-7a level in the presence of substrate RNA was found only in the EV-associated RNA pool (Fig. 3C). Control miR-16 did not show similar changes in its EV content in the presence of let-7a reporter mRNA.
To further strengthen the concept that the reduction of miRNA in the presence of its substrate occurs due to a selective export of those miRNAs in human cells, we performed experiments with different target mRNAs. We noted that the extracellular content of miRNA let-7a was increased specifically both in HeLa and MDA-MB-231 cells, in the presence of abundant amounts of translatable target mRNAs. The nonspecific miRNAs miR-16 and miR-24 did not show any increase in the presence of let-7a target mRNAs in HeLa or MDA-MB-231 cells. The amounts of miRNAs exported from HeLa and MDA-MB-231 cells were also dependent on the availability of the target messages (Fig. 3D). In this context, using ectopic expression of a hepatic miRNA in nonhepatic cells, we documented a substrate-specific export of miR-122 from HeLa cells when its target mRNA was expressed (Fig. 3E). It has also been reported earlier that miRNA-repressed mRNAs relocalize from the endoplasmic reticulum (ER)-attached polysomal compartment to the endosomal compartment for their uncoupling from miRNPs and degradation (26). Therefore, the miRNPs present in the endosomal fraction can be considered the target-uncoupled miRNA pool that has completed one round of target repression. In the presence of target RNA, we documented a loss of miRNA association with polysome-enriched fractions which were resolved on a 3 to 30% OptiPrep density gradient (Fig. 3F) (26, 35). This supports our hypothesis that target RNA favors the miRNA export process by mobilizing miRNAs from ribosomal fractions, where they are engaged in repression (17, 26). Thus, we conclude that miRNAs that have completed one round of target RNA repression are possibly preferentially exported out from mammalian cells.
The EVs with the exported miRNAs were characterized to determine the effect of target RNA on EV number and size. We performed nanoparticle tracking analysis (NTA) and documented no major alteration in EV number and size (Fig. 3G). We also did not find any significant change in protein contents of EVs when several marker proteins were analyzed in EVs and cellular fractions prepared from HeLa cells expressing control and let-7a reporter mRNAs. The absence of cytochrome c also ruled out the presence of apoptotic bodies in EVs used in this analysis (Fig. 3H). Atomic force microscopic analysis of the isolated EVs also showed the integrity and shape of the particle carrying the exported let-7a from RL-3×bulge(B)-let-7a reporter mRNA-expressing cells (Fig. 3I).
We measured the stability of miRNA in cellular and EV-associated pools. We transfected HeLa cells with an exogenous mature and preformed double-stranded miR-122 mimic, and compared residual levels of miR-122 present in cellular and EV-associated fractions. As HeLa cells do not express miR-122, it allowed us to measure the turnover of miR-122 in HeLa cells without applying a transcriptional inhibitor. EVs are noted to be stable at 37°C, and considering that there is no significant turnover of EVs at 37°C (36), the reduction of miR-122 there represents the rate of degradation of miR-122 in EVs. A higher degradation rate of miR-122 in EVs than for the cellular miR-122 was noted (Fig. 3J). These data suggest effectiveness of EV-mediated miRNA export as a key mechanism of miRNA turnover in human cells. As the exported miRNAs are less stable in EVs, they might be degraded within EVs or in the extracellular matrix upon lysis of the EVs by matrix proteases; EV-mediated miRNA export thus is an effective way of reducing cellular miRNA content under specific conditions.
Substrate-mediated export of miRNA is ESCRT independent.
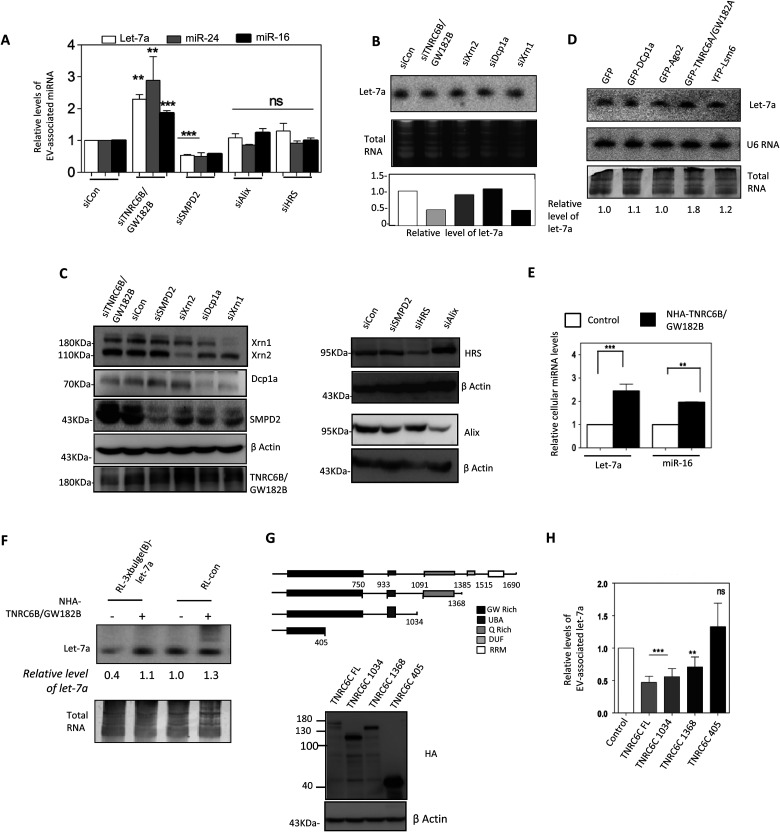
MVBs play an important role in restricting miRNP recycling and activity (19, 20). These vesicles can fuse with cell membranes to form EVs in animal cells (37). Maturation of early endosomes to MVBs is dependent on ESCRT, a multiprotein complex associated with MVBs. Depletion of ALIX or HRS proteins, the ESCRT components, had no considerable effect on EV-associated miRNA levels. siRNA-mediated knockdown of SMPD2 increased cellular miRNA levels considerably in MDA-MB-231 cells. RNAi for ESCRT components and SMPD2 revealed that the substrate-dependent EV-mediated export of miRNA is ESCRT independent but requires ceramide biosynthesis (Fig. 4A and B).
FIG 4.
TNRC6/GW182 proteins restrict EV-mediated export of miRNA. (A) EV-mediated export of miRNA is ESCRT independent but requires TNRC6B/GW182B. Shown is real-time estimation of miRNA content in EVs from MDA-MB-231 cells depleted of ESCRT components (ALIX and HRS), SMPD2, or TNRC6B/GW182B by RNAi (means ± SEM; n = 3). (B) HeLa cells were transfected with siRNAs against different P-body components, and levels of let-7a were determined by Northern blotting. Relative levels of let-7a in cells transfected with siRNAs were measured by real-time quantification and are shown below each lane. siRNA against RL luciferase was used as a control. An ethidium bromide (EtBr)-stained RNA gel used for blotting served as an RNA loading control. (C) Western blots for different P-body components were prepared with extracts isolated from respective siRNA-treated cells to confirm effective knockdown. Levels of SMPD2 and ESCRT components ALIX and HRS in cells depleted of these factors by RNAi were detected. β-Actin was used as a loading control. (D) Expression of P-body components alters let-7a level in HeLa cells. GFP fusion constructs of different P-body components were transfected individually, and levels of let-7a in cells expressing these constructs were measured by Northern blot analysis. Total RNA was used as a control. Relative levels of let-7a present under each condition are shown below the lanes. (E) Effect of NHA-TNRC6B/GW182B expression on cellular levels of two miRNAs, miR-16 and let-7a, in HeLa cells. (F) TNRC6B/GW182B restricts target-dependent lowering of miRNA in MDA-MB-231 cells. Levels of cellular let-7a in the presence and absence of target reporter mRNA in cells also expressing NHA-TNRC6B/GW182B are shown. Total RNA was used as a loading control, and relative values of let-7a present are shown below each lane. (G and H) Ago2-interacting motif of TNRC6C/GW182C is required for restricting miRNA export in human cells. (G) Schematic diagram and Western blot analysis to check the expression of the full-length and truncated versions of NHA-TNRC6C/GW182C in HeLa cells. (H) Effect of expression of TNRC6C/GW182C and its truncated mutants on EV-associated let-7a level in MDA-MB-231 cells (means ± SEM; n = 4). *, P < 0.05; **, P < 0.01; ***, P < 0.0001. P values were determined by paired t test.
GW182 proteins prevent target-mediated lowering of miRNA.
Ago-interacting TNRC6/GW182 proteins are the effectors of miRNP-mediated gene repression that can accelerate the decay of Ago-bound mRNAs (27, 38–40) and may act as stabilizers of miRNPs in animal cells (28). RNAi against TNRC6B/GW182B enhanced EV-mediated export of miRNA (Fig. 4A to C). Moreover, ectopic expression of TNRC6B or GW182B restricted the extracellular export of miRNA, thereby increasing the cellular miRNA level (Fig. 4D and E). A hemagglutinin (HA)-tagged version of GW182B or TNRC6B having a box B RNA element-interacting N protein in its N terminus was used for GW182 expression. Target-dependent miRNA lowering also was restricted in cells expressing TNRC6B/GW182B (Fig. 4F). Expression of TNRC6C/GW182C (another member of the TNRC6 protein family) also decreased the EV-associated miRNA level (Fig. 4G and H). The C-terminal motif of TNRC6C, necessary for miRNA-mediated gene repression, was dispensable for its inhibitory role on EV-mediated export, but the middle Ago2-interacting motif (41) was found to be important (Fig. 4G and H). Depletion or exogenous expression of the majority of other GFP-tagged P-body components did not have any prominent effect on cellular miRNA levels in HeLa cells except GFP-GW182A (Fig. 4B to D).
GW182 proteins prevent EV-mediated export of miRNAs by retaining the miRNPs within the P-body.
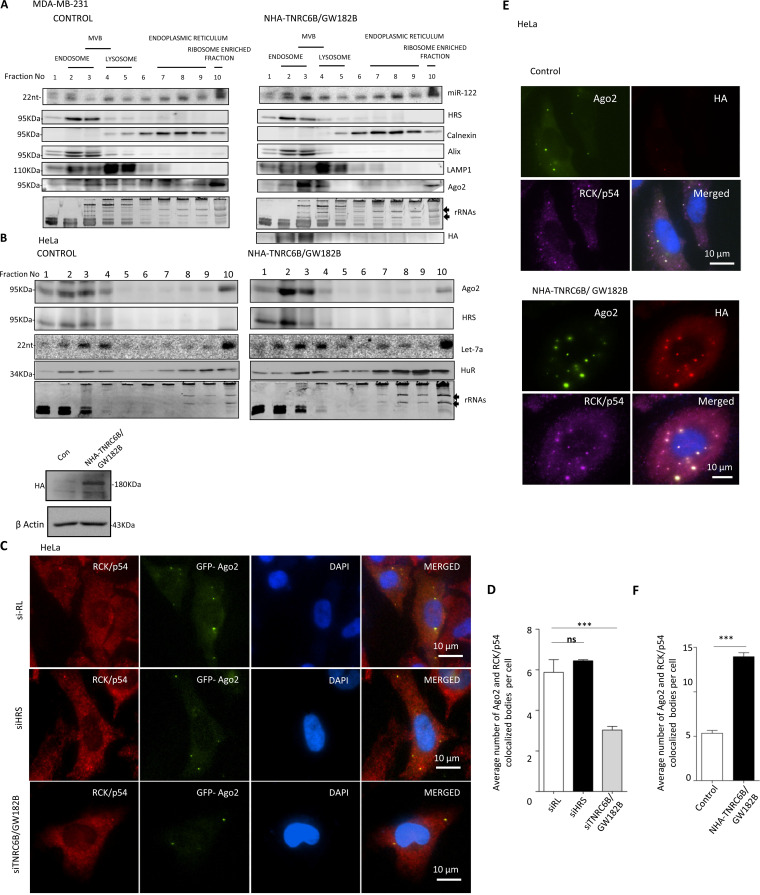
To understand the mechanism by which the TNRC6/GW182 proteins restrict EV-mediated export of miRNAs, we separated the subcellular organelles on a 3 to 30% OptiPrep density gradient to identify the fractions with which miRNPs remained associated in cells expressing NHA-TNRC6B/GW182B. In animal cells, miRNAs primarily remain associated with MVBs (19, 20) and the ER (42). With NHA-GW182B (with the 22-amino-acid RNA-binding domain of the lambda bacteriophage antiterminator protein N in the N terminusl) expression we documented increased association of Ago2 and both exogenously expressed (miR-122) and endogenous (let-7a) miRNAs with endosome/MVB-enriched fractions in MDA-MB-231 and HeLa cells, respectively (Fig. 5A and B).
FIG 5.
GW182 proteins promote P-body targeting of miRNPs to restrict the miRNA export process. (A and B) Distribution of Ago2 and miRNA in MDA-MB-231 and HeLa cells expressing TNRC6B/GW182B. Increased miRNP is associated with endosome fraction positive for HRS marker protein in NHA-TNRC6B/GW182B-expressing MDA-MB-231 (A) and HeLa (B) cells. A 3 to 30% OptiPrep gradient analysis and distribution of Ago2 in cells expressing NHA-TNRC6B/GW182B are shown. Expression and distribution of NHA-TNRC6B/GW182B were determined by Western blotting using anti-HA antibody in panel A. Levels of let-7a or exogenously expressed miR-122 were determined by Northern blot analysis. Calnexin served as an ER marker. Alix was also an endosomal marker, while Lamp1 was used as a lysosomal marker. Distribution of stress response protein HuR was also determined in HeLa cells. (C) Inhibition of TNRC6B/GW182B by RNAi reduces Ago2-positive bodies in HeLa cells. Cells were transfected with GFP-Ago2 and siRNAs against TNRC6B/GW182B or HRS. GFP-Ago2 bodies were visualized in cells where they colocalized with RCK/p54. siRL was used as a control (means ± SEM; n = 20). (D) Quantification of Ago2 bodies showing colocalization with P-body marker RCK/p54 under different conditions (means ± SEM; n = 10 cells; P = 0.0005). (E) Larger numbers of P-bodies in NHA-TNRC6B/GW182B-expressing cells. HeLa cells were cotransfected with GFP-Ago2-encoding plasmid along with either the control or the NHA-TNRC6B/GW182B-encoding plasmids. GFP-positive Ago2 bodies were visualized and their colocalization with P-body marker RCK/p54 was documented. Increased size of Ago2 bodies showing colocalization with TNRC6B/GW182B was visualized in cells expressing NHA-TNRC6B/GW182B. (F) Relative quantification of colocalization of Ago2 and RCK/p54 from the experiments described for panel E was done and plotted (means ± SEM; n = 23 cells; P < 0.0001).
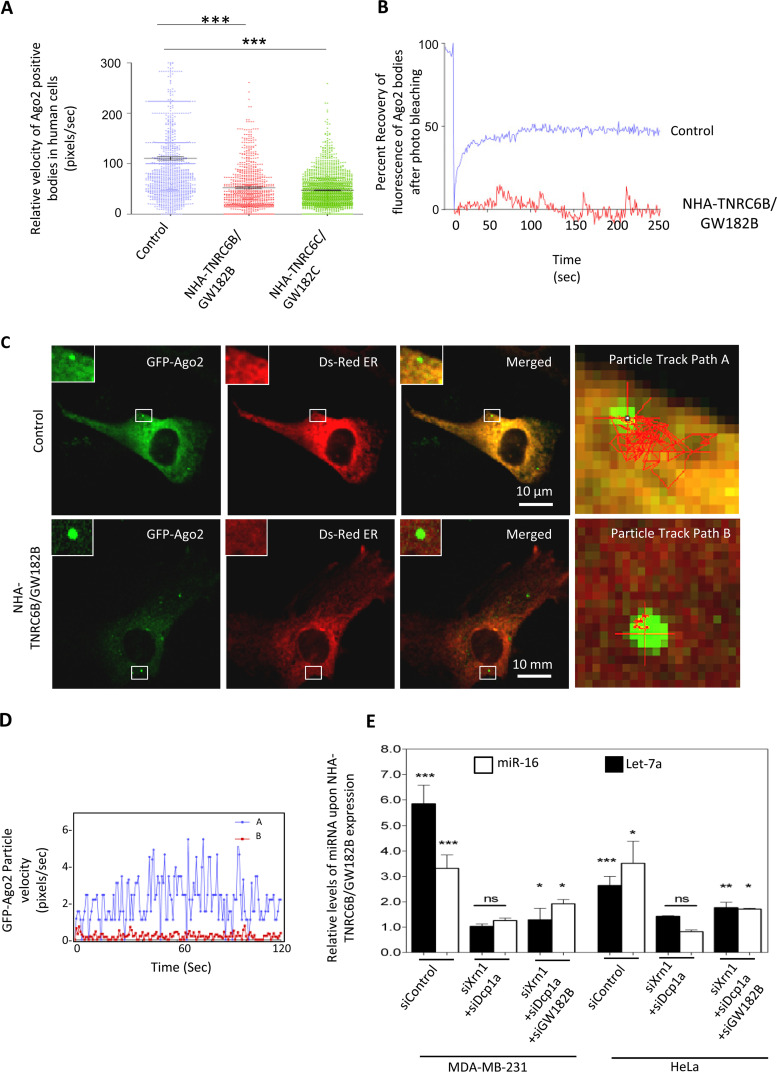
Relative subcellular distribution of miRNPs may get altered with changes in cell physiology and on availability of excess substrate RNA. Depletion of TNRC6B/GW182B by siRNA reduced P-body localization of Ago2 (Fig. 5C and D). With NHA-TNRC6B/GW182B expression, we found an increased association of miRNA let-7a with the MVB-associated pool (Fig. 5B). TNRC6B/GW182B expression increased the size of Ago2-positive P-bodies that showed a retarded rate of exchange of Ago2 molecules localized in bodies with the cytosolic pool (Fig. 5E and F). Therefore, P-body compartmentalization of miRNPs in human cells, regulated by TNRC6/GW182 proteins, may control extracellular export and hence stability of miRNAs in mammalian cells. Additionally, in NHA-TNRC6B/GW182B-expressing human cells, Ago2 bodies were found to be less dynamic, with reduced mobility (Fig. 6A). A fluorescence recovery after photobleaching (FRAP) experiment also suggested less mobility of P-body-localized Ago2 in the presence of NHA-TNRC6B/GW182B expression (Fig. 6B). Individual particle tracking analysis also revealed lateral and vertical movement restriction of the Ago2-positive bodies in three dimensions (3D) in the presence of NHA-TNRC6B/GW182B (Fig. 6C and D). Do the Ago2 movement and miRNA export restriction facilitated by NHA-TNRC6B/GW182B depend upon P-body integrity? The dependence of miRNA stabilization by GW182 was found to be governed by the integrity of PBs. Therefore, depletion of PBs by siRNAs against specific components like Xrn1 and Dcp1a resulted in reduced stabilization of miRNA in HeLa and MDA-MB-231 cells expressing NHA-TNRC6B/GW182B (Fig. 6E).
FIG 6.
NHA-TNRC6B/GW182B controls miRNA export by restricting miRNP mobility in a P-body-dependent manner. (A) From live-cell time-lapse imaging, the instantaneous velocity of multiple GFP-Ago2 particles was measured and plotted for both control and NHA-TNRC6B/GW182B plasmid-transfected cells (means ± SEM; n = 600; P < 0.0001). (B) Impaired recovery of GFP fluorescence of GFP-Ago2 in bodies after photobleaching in cells expressing NHA-TNRC6B/GW182B with respect to control. (C and D) Lower mobility of Ago2 bodies. Localization of GFP-Ago2 bodies was followed in cells expressing NHA-GW182B. Shown is change in particle velocity with time for two GFP-Ago2-positive bodies in control (path A [C]) and NHA-TNRC6B/GW182B-expressing (path B [C]) HeLa cells (D). (E) Effect of depletion of PBs on miRNA levels in cells expressing NHA-TNRC6B/GW182B. The protein was expressed in HeLa cells pretargeted with siRNAs against different P-body components. The cellular miRNA levels in individual cases were measured against control cells transfected with control siRNA (siRL) (means ± SEM; n = 4). For statistical analysis, the values with pCIneo vector plasmid-expressing cells were taken as a unit. *, P < 0.05; **, P < 0.01; ***, P < 0.0001. P values were determined by paired t test.
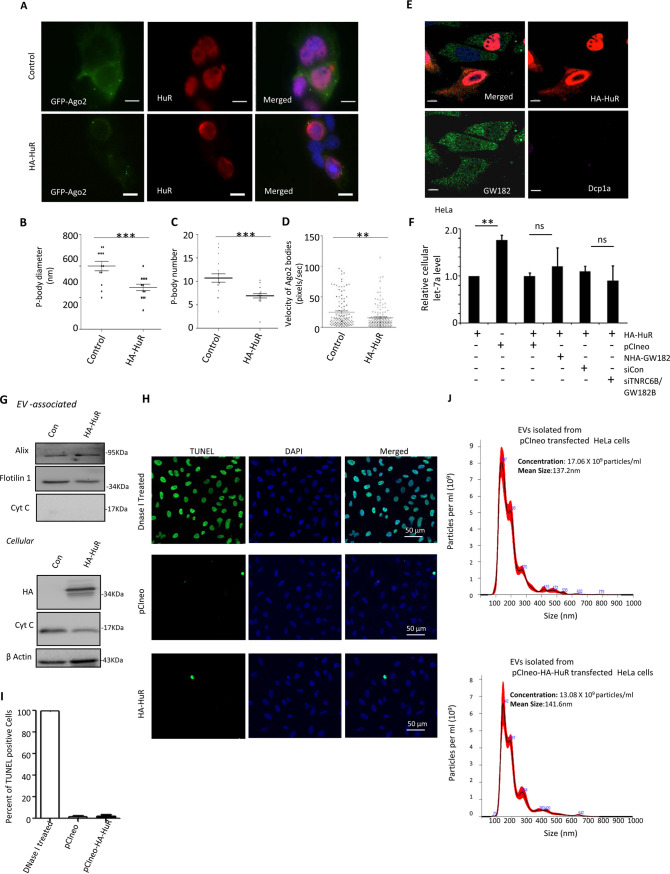
In this context, HuR is known to relocalize miRNA-repressed messages from PBs and ensure their translation in stressed human cells without an effect on PBs (25). HuR is also known to facilitate the export of miRNAs under stress conditions or when expressed ectopically in human cells by reversibly binding the miRNA in hepatic cells; ubiquitination of HuR is the key step happening on endosomes that allows its miRNA unbinding and loading of miRNA to MVBs for EV-mediated export (18). Now the question here was could GW182 proteins counteract the miRNA export mediated by HuR? GW182 was found to facilitate the relocalization of miRNPs to PBs, and these phase-separated miRNPs are unavailable for export. In this process of target RNA-driven export, the interaction of GW182 with Ago2 occurs primarily on the endosomal fraction (26). Therefore, an event most likely happens upon relocalization of the repressed mRNAs along with miRNPs to the endosome. HuR, in contrast, works upon ER-attached polysomes, first to disengage the miRNPs from the targets and then to uncouple miRNAs from Ago2 (18). Therefore, these two processes are likely to be independent of each other. We have observed a limited effect of HuR overexpression on P-body size and number (Fig. 7A to C) and has little effect on Ago2 mobility in mammalian cells (Fig. 7D). However, expression of HA-HuR did not alter GW182 or Dcp1a localization in HeLa cells (Fig. 7E). The cellular distribution pattern of HuR also was not altered in the presence of NHA-TNRC6B/GW182B expression in HeLa cells (Fig. 5B). HuR overexpression accelerated miRNA export, and siRNA-mediated GW182 depletion or overexpression of NHA-TNRC6B/GW182B did not have any significant effect on the cellular miRNA content already lowered in HA-HuR-expressing cells. These results confirmed the uncoupling of GW182-mediated miRNA stabilization from HuR-mediated export of miRNA. Therefore, in mammalian cells, HuR-mediated miRNA export is a predetermined step that cannot be reversed by Ago2-interacting GW182 proteins that work downstream of miRNP coupling of target message and happens in a spatiotemporally distinct manner compared to HuR-driven miRNA export, which is a target RNA-independent event (Fig. 7F). The effect of HuR expression on levels of EV-associated proteins suggests no major alteration in EV content, which was further substantiated by NTA data (Fig. 7G and J). HuR expression was also not associated with apoptosis induction, as no change in TUNEL-positive cell numbers in HA-HuR-expressing cells compared to control vector-transfected cells was noted (Fig. 7H and I). The lack of detection of cytochrome c in the EVs and no change in cellular cytochrome c level with HA-HuR expression also signify unaltered apoptosis status of cells expressing HA-HuR (Fig. 7G).
FIG 7.
HuR-mediated miRNA export remains unperturbed by cellular GW182 levels. (A) Effect of HA-HuR on Ago2-positive bodies in Huh7 cells. P-body localization of GFP-Ago2 in HA-HuR-expressing cells is shown. Cells were transfected with GFP-Ago2-encoding plasmid along with either the HA-HuR expression plasmid or control vector, and GFP-positive bodies were visualized. Anti-HuR antibody was used to detect HuR, and DAPI was used to stain nucleus. Scale bars, 10 μm. (B to D) Scatterplots depicting HA-HuR-mediated changes in diameter (means ± SEM; n = 18; P = 0.0007) (B), number (means ± SEM; n = 14; P = 0.0004) (C), and mobility (D) of GFP-Ago2-positive bodies in Huh7 cells (means ± SEM; n = 108; P = 0.0016). (E) Effect of HA-HuR expression (red) on Dcp1a (purple) and GW182 (green) in HeLa cells. HA signal was detected with indirect immunofluorescence using anti-HA antibody. Antibodies against GW182 and Dcp1a were used to detect endogenous proteins. Scale bars, 10 μm. (F) Effect of GW182 depletion or NHA-TNRC6B/GW182B expression on cellular miRNA export by HuR in HeLa cells. Levels of cellular miRNA in pCIneo- or HA-HuR-expressing HeLa cells were compared. In the context of HA-HuR expression, siGW182B treatment or NHA-GW182B expression seems to have no effect on cellular miRNA levels compared to control cells treated with siCon or expressing an equivalent amount of pCIneo control plasmid (means ± SEM; n = 4). (G) Effect of ectopic expression of HA-HuR on EV and cellular contents of marker proteins. Western blotting for different marker proteins and cytochrome c was performed for both EV and cell extract to check their levels in control and HA-HuR-expressing cells. (H and I) HA-HuR expression does not induce apoptosis in HeLa cells. Numbers of apoptotic cells were estimated in a TUNEL assay done for pCIneo and HA-HuR expression plasmid-transfected cells. Apoptotic cells positive for FITC signals (panel H) were estimated from different fields and quantified (I). DNase I-treated cells were used as a positive control (means ± SEM; n = 10). (J) Effect of HA-HuR expression on EV number and size. Nanoparticle tracking analysis was performed with EVs isolated by ultracentrifugation from HeLa cells expressing HA-HuR or control plasmid-transfected cells. The EV numbers and diameters were calculated in each case. *, P < 0.05; **, P < 0.01; ***, P < 0.0001. P values were determined by paired t test.
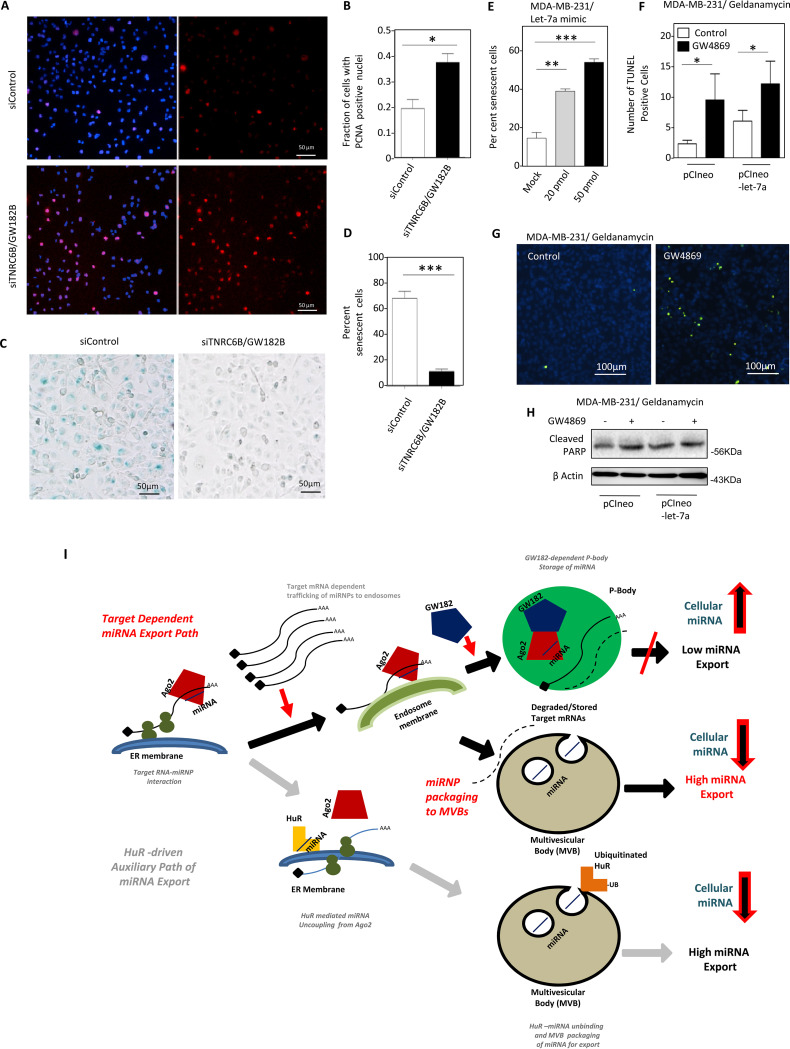
It has been shown before that EV-mediated export of miRNA-let-7a is impaired under HuR-compromised condition or in growth-retarded human cells. This increase of let-7a induces cell senescence in MDA-MB-231 human breast cancer cells (18). Is GW182B required for inducing senescence in MDA-MB-231 cells by changing the cellular content of let-7a? As expected, we have noted enhanced cell proliferation and decreased senescence in MDA-MB-231 cells depleted for GW182B. This suggests that GW182 protein has a role to play in cell growth regulation (Fig. 8A to D). To confirm let-7a as the key molecule behind the increased senescence in MDA-MB-231 cells through an increase in let-7a level, we tried to express let-7a in MDA-MB-231 cells by transfecting cells with increasing concentrations of let-7a mimic. Increased levels of let-7a have a positive effect on increase of senescence in MDA-MB-231 cells (Fig. 8E). Blockage of EV-mediated export of let-7a also altered the efficacy of drugs to induce apoptosis in treated cells. Cotreatment of MDA-MB-231 cells with GW4869 and geldanamycin, an inhibitor of HSP90, increased TUNEL-positive cells (Fig. 8F and G). This cotreatment also increased cleaved PARP levels, which indicates increased apoptosis in MDA-MB-231 cells blocked for EV export (Fig. 8H).
FIG 8.
Decreased senescence and increased proliferation of MDA-MB-231 cells in the absence of GW182B. (A to D) Increased numbers of proliferating cells with PCNA-positive nuclei in siTNRC6B/GW182B-treated cells were visualized (A) and plotted (means ± SEM; n = 2 independent experiments; P = 0.0212) (B). Reduced senescence in cells treated with siTNRC6B was detected (C) and quantified (means ± SEM; n = 2 independent experiments; P = 0.007) (D). (E) Increased let-7a causes elevated senescence in MDA-MB-231 cells. Percent senescence in total population of cells transfected with different concentrations of let-7a mimic (20 or 50 pmol/ml) was plotted (means ± SEM, n = 10 independent observations; P < 0.0001). Mock-transfected cells served as controls. (F and G) Increased sensitivity of GW4869-treated MDA-MB-231 cells to apoptosis induction by geldanamycin. Numbers of TUNEL-positive cells transfected with pCIneo (means ± SEM; n = 5 independent observations; P = 0.0313) or pCIneo-let-7a (expressing pre-let-7a) (means ± SEM; n = 5 independent observations; P = 0.0408) were estimated after treatment with geldanamycin in the presence or absence of GW4869 (F). Representative pictures of pCIneo-transfected and geldanamycin-treated cells with or without GW4869 cotreatment are shown in panel G. (H) Cotreatment with geldanamycin and GW4869 increases PARP cleavage in MDA-MB-231 cells transfected with pCIneo or pCIneo-let-7a plasmids. (I) Two counteractive pathways, controlled by GW182 and HuR, buffer cellular miRNA levels by negative and positive regulation of cellular miRNA export process in mammalian cells. In this model, the engagement of miRNA in translational repression of target messages accelerates miRNA movement to endosomes before its export. The process is regulated negatively by GW182, which by interacting with Ago2 miRNPs retains them with P-bodies to prevent their target-driven export. However, the HuR-mediated miRNA export is classified by binding of the target mRNAs with HuR and subsequent reversible miRNA binding by the protein HuR to augment their export that operates to help the cells to adjust to the stress (18). *, P < 0.05; **, P < 0.01; ***, P < 0.0001. P values were determined by paired t test.
DISCUSSION
miRNAs are otherwise considered stable molecules with low turnover rates in mammalian cells. Our findings provide evidence that miRNAs engaged in translational repression could be selected for export in a target RNA-dependent manner in human cells. In this study, we focused on how the target-driven export of miRNA controls gene expression in mammalian cells. Our data also provide an important molecular link between engagement of miRNA in translation repression of its targets and its EV-mediated export that can explain why export and hence turnover of miRNAs are selective in animal cells. In this work, we have been able to document EV-mediated miRNA export as a mechanism used by human cells to regulate cellular miRNA content and have delineated the molecular mechanism underlying this phenomenon (Fig. 8I).
In the presence of their substrates, miRNAs are more susceptible to extracellular export and their levels are reduced in human cells. Previous observation in C. elegans suggests that let-7a miRNA stability is dependent on its binding to the substrate (43). This rationale may be valid for certain exceptional cases, but it is more common that target mRNAs and miRNAs are usually present in animal cells either in a mutually exclusive manner or with their expressions inversely correlated (44). Furthermore, miRNAs have been reported to be “sponged” with mRNAs having multiple miRNA sites and are used to titrate out endogenous miRNAs to rescue their target genes, accompanied by reduction of respective miRNAs (45). Competing endogenous RNAs (ceRNAs) act as natural miRNA sponges and can attenuate the function of specific miRNAs by binding in a competitive manner to rescue the expression of the endogenous target genes in animals and plants (46, 47). It is also interesting to test whether ceRNAs can accelerate the export of miRNAs in animal cells. Besides noncoding RNAs, developmental stages of lymphopoiesis have also been reported to modulate miRNA abundance brought about by changes in mRNA profiles during ontogeny (14). Modification of miRNA with additional bases at the 3′ end is also known to play important role to modulate miRNA stability and activity in animal cells (48, 49). Whether such a modification in a substrate-dependent manner can modulate miRNA export will be a subject of interest for future exploration.
Target RNA is also known to upregulate the biogenesis of cognate miRNAs; that apparent contradiction is important for maintaining miRNA homeostasis (10). The target RNA-driven biogenesis and export of miRNA may happen in a mutually coordinated manner to ensure an effective way of balancing a “new” pool of miRNA available for the next round or in-line mode of miRNA repression where target RNA not only drives the biogenesis of a miRNA in its presence but also ensures a rapid turnover of the miRNA pool. The exact reason and consequence of this de novo biogenesis of miRNA are not clear.
In this context, the data published by Squadrito et al. (50) suggest an apparent opposite effect of targets on the miRNAs having the binding sites. They have argued in favor of retention of miRNAs in PBs in the presence of the target mRNAs and thus exclusion of respective miRNAs from the EV- associated miRNA pool. This could be a cell-type-specific event, and considering an abundant amount of GW182 present in the macrophage system, miRNP storage in PBs could happen in a target-dependent manner, but this should not hold true for the proliferative cancer cells where rapid turnover of transcripts is also ensured (Fig. 8I).
Alternatively to extracellular degradation of EV-associated miRNAs, miRNA-loaded EVs are used for intercellular communications where the retaken EV contents are reused in recipient cells (16, 21). The EV release of miRNAs may ensure the homogeneity of miRNA expression and activities among neighboring cells. In tissues, the spiked expression of target mRNAs in individual cells in a population may be tamed by the extra miRNAs received via EVs until the balanced expressions of target genes and miRNAs are achieved. This possibly ensures robustness of the biological processes in tissue, as discussed by Ebert and Sharp elsewhere (51).
We have identified TNRC6/GW182 and HuR as inverse regulators of the EV-mediated miRNA export process, and it would be interesting to identify additional factors that may act as regulators of miRNA export. miRNA homeostasis is controlled by several factors that contribute to buffering of the cellular miRNA level and stability in animal cells. From this study, it also seems that P-bodies can effectively act as a reservoir for miRNAs that otherwise get exported or recycled via MVBs/EVs (Fig. 8I). An increase of let-7a in breast cancer cells impaired EV-mediated export, increased senescence, and sensitized cells to an anticancer drug. Therefore, the manipulation of miRNA turnover machineries may lead to a better management of antiproliferative drugs in cancer treatment.
MATERIALS AND METHODS
Expression plasmids, cell culture, and transfection.
Plasmids bearing humanized Renilla luciferase (RL) coding sequences (RL-con), three miR-122 binding sites downstream of the RL coding region (RL-3×bulge-miR-122), three let-7a binding sites (RL-3×bulge-let-7a), and firefly luciferase (FL) under the control of a simian virus 40 (SV40) promoter (pGL3FF) were kind gifts from Witold Filipowicz. Plasmid expressing GFP-3×bulge-let-7a was made by replacing the RL fragment with GFP in pRL-3×bulge-let-7a. Downstream of the pRL-con plasmid the 3′ UTR of the HMGA2 gene with intact let-7a binding sites was cloned (RL-HMGA2 3′UTR), and for the mutated one, let-7 a binding sites were mutated (RL-HMGA2 3′UTR mut). Both the constructs were a kind gift from Anindya Dutta. FLAG- and hemagglutinin (HA)-tagged human Ago2 expression plasmid (FH-Ago2) was a kind gift from Tom Tuschl. DsRed2-expressing and ER target sequence-bearing plasmid pDsRed2-ER was purchased from Clontech. Different versions of NHA-tagged TNRC6/GW182 isoforms and their mutants for expression in mammalian cells, i.e., NHA-TNRC6B, NHA-TNRC6C, and truncated forms, were kind gifts from Witold Filipowicz. A 472-bp product of the full-length (cyclin-dependent kinase inhibitor 1B) p27 5′ UTR (NM_004064.3) fragment was cloned upstream of RL-con and RL- 3×bulgeB-let-7a plasmids to make p27-5′UTR-RL-con and p27-5′UTR-RL-3×bulge-let-7a, respectively. Plasmids bearing human Ago2 cloned in frame with GFP in peGFP-C2 (Clontech) (GFP-Ago2) were kind gifts from Witold Filipowicz. Plasmid bearing an N-terminally truncated version of TNRC6A isoform in frame with GFP in a mammalian expression vector (GFP-GW182) was a kind gift from M. J. Fritzler. The miR-122 expression plasmid, pmiR-122, was described previously (16). Full-length and truncated HuR without hinge region (HNS) with HA coding sequence has been cloned into pCIneo vector (HA-HuR) was also a kind gift from Witold Filipowicz.
SMARTpool ON-TARGETplus human HGS (HRS) siRNA (siHRS; L-016835-00-0005), human SMPD2 (neutral sphingomyelinase 2) siRNA (siSMPD2; L-006677-01-0005), human ALIX (PDCD6IP) siRNA (siAlix; L-004233-00-0005), human TNRC6B siRNA (siTNRC6B; L-024575-00-0005), human XRN2 siRNA (siXRN2; L-017622-01-0005), and human XRN1 siRNA (siXRN1; L-013754-01-0005) were purchased from Dharmacon. For silencing of RL expression, siRNA against RL was synthesized by (siRL-sense strand, GCGAGAUCCCUCUCGUUAATT, and siRL-antisense strand, UUAACGAGAGGGAUCUCGCGG) and purchased from Eurogentech. si All Star Negative (siCon) was purchased from Qiagen.
Luciferase assay and Northern and Western blotting.
RL and FL activities were measured using a dual-luciferase assay kit (Promega) following the supplier’s protocol on a Victor X3 plate reader with injectors (Perkin Elmer). FL-normalized RL expression levels for reporter and control were used to calculate fold repression as the ratio of control to reporter normalized RL values.
Northern blotting of total cellular RNA (5 to 15 μg) was performed as described earlier (23, 28). For miRNA detection, 32P-labeled 22-nt antisense DNA or locked nucleic acid (LNA) modified probes specific for respective miRNAs or siRL or U6 snRNA were used. Phosphorimaging of the blots was performed in a Cyclone Plus storage phosphor system (Perkin Elmer), and Optiquant software (Perkin Elmer) was used for quantification.
Western analyses of different miRNP components were performed as described previously (20). After running the respective samples on SDS-PAGE, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane overnight. After blocking of the membrane with 3% bovine serum albumin (BSA) in Tris-buffered saline with Tween 20 (TBST), probing was done with the following sets of primary antibodies at 4°C for 16 h: rabbit anti-p53 (Biovision), 1:1,000; rabbit anti-LDL receptor (Cayman Chemical), 1:1,000; rabbit anti-PCNA (Bethyl Laboratories), 1:1000, rabbit anti-Hsp90 (Cell Signaling), 1:1,000; horseradish peroxidase (HRP)-conjugated β-actin (Sigma), 1:10,000; rat anti-HA (Roche), 1:1,000; mouse anti-GFP (Roche), 1:1,000; mouse anti-Ago2 (Novus Biologicals), 1:500; rabbit anti-Xrn1 (Bethyl Laboratories), 1:10,000; rabbit anti-RCK/p54 (Bethyl Laboratories), 1:10,000; mouse anti-Dcp1a (Novus Biologicals), 1:1,000; rabbit anti-HRS (Bethyl Laboratories), 1:1,000; mouse anti-β-tubulin (Sigma), 1:1,000; rabbit anticalnexin (Cell Signaling), 1:1,000; mouse anti-ALIX (Santa Cruz), 1:500; rabbit anti-GW182 (Bethyl Laboratories), 1:1,000; rabbit anti-Xrn2 (Bethyl Laboratories), 1:10,000; mouse anti-SMPD (Abnova), 1:2,000; rabbit anti-flotilin 1 (Cell Signaling), 1:1,000; rabbit cytochrome c (Cell Signaling), 1:1,000; and mouse anti-HuR (Santa Cruz), 1:1,000. Imaging of all Western blots was performed using an UVP Bio Imager 600 system equipped with VisionWorks Life Science software (UVP) V6.80.
Preparation of Microvesicles, EVs, and treatments.
Extracellular vesicles (EVs) were isolated based on the published protocols (18, 52). Briefly, for isolation of EVs, cells were grown in medium prepared with EV-free fetal bovine serum (FBS) and the culture supernatants were clarified for cellular debris and other contaminants by centrifugation at 400 × g for 5 min, 2,000 × g for 10 min, and 10,000 × g for 30 min and filtration through a 0.22-μm filter. The 10,000 × g pellet fraction was treated as microvesicles, and RNA was isolated from this fraction by Trizol. Extracellular vesicles designated as EVs were separated by centrifugation at 100,000 × g for 75 min and were resuspended in 200 μl of 1× passive lysis buffer (Promega) for isolation of EV-associated proteins and RNA. All operations were done at 4oC. Characterization of isolated EVs and its purity check were done by procedures adopted elsewhere (18).
NTA and atomic force microscopy.
For characterization by atomic force microscopy, EVs isolated on a 30% sucrose cushion were resuspended in phosphate-buffered saline (PBS) after being washed by ultracentrifugation. EV suspension was placed onto a mica sheet and dried for 15 min before being washed with autoclaved Milli-Q water and dried again. AAC mode atomic force microscopy was performed using a Pico Plus 5500 ILM atomic force microscope (Agilent Technologies, USA) with a piezo scanner (18). Microfabricated silicon cantilevers from Nano Sensors, USA, were used. Images were processed by flatten using Pico view1.1 version software (Agilent Technologies). For nanoparticle tracking analysis (NTA), EVs resuspended in 1 ml of PBS were diluted 5-fold before injection into the sample chamber of a nanoparticle tracker (Nanosight NS300) and analyzed as recommended (18).
Fractionation and separation of endosomes and ER on OptiPrep gradients.
OptiPrep (Sigma-Aldrich, USA) was used to prepare a 3 to 30% continuous gradient in a buffer containing 78 mM KCl, 4 mM MgCl2, 8.4 mM CaCl2, 10 mM EGTA, and 50 mM HEPES-NaOH (pH 7.0) for separation of subcellular organelles as described previously, with minor modifications described below (17). Cells were trypsinized, washed, and homogenized with a Dounce homogenizer in a buffer containing 0.25 M sucrose, 78 mM KCl, 4 mM MgCl2, 8.4 mM CaCl2, 10 mM EGTA, and 50 mM HEPES-NaOH (pH 7.0) supplemented with 100 μg/ml of cycloheximide, 5 mM vanadyl ribonucleoside complex (VRC; Sigma-Aldrich), 0.5 mM dithiothreitol (DTT), and 1× protease inhibitor. The lysate was clarified by centrifugation at 1,000 × g for 5 min and layered on top of the prepared gradient. The tubes were centrifuged for separation of gradient using established protocols, and 10 fractions were collected.
Immunofluorescence.
For immunofluorescence, cells were transfected with 250 ng of GFP-Ago2 alone or cotransfected with 2 μg of either NHA-TNRC6B/GW182B or HA-HuR expression plasmid per well of a six-well plate. pCIneo plasmid was used as a control. The cells were split after 24 h of transfection and subjected to specific experimental conditions. siRNAs were transfected at 100 nM concentration. For immunofluorescence analysis, cells were fixed with 4% paraformaldehyde for 30 min, permeabilized, and blocked with PBS containing 1% BSA, 0.1% Triton X-100, and 10% goat serum (GIBCO) for 30 min. Secondary anti-rabbit or anti-mouse antibodies labeled with Alexa Fluor 488 dye (green), Alexa Fluor 594 dye (red), or Alexa Fluor 647 dye (far red) fluorochrome (Molecular Probes) were used at a 1:500 dilution. Cells were observed either under a Plan Apo VC 60×/1.40 oil or Plan Fluor 10×/0.30 objective on an inverted Eclipse Ti Nikon microscope equipped with a Nikon Qi1MC or QImaging-Rolera EMC2 camera for image capture, or the images were captured using a Zeiss LSM800 confocal microscope.
For live-cell analysis, 250 ng of GFP-Ago2-encoding plasmid used for transfection of a six-well plate was further transfected with NHA-GW182 or HA HuR after overnight durations. pCIneo was used as the control plasmid. The cells were split at 24 h posttransfection for analysis. For FRAP experiments, cells were photobleached, and recovery was monitored for a total duration of less than 4 min. Glass bottom petri dishes precoated with gelatin were used for cell growth for live-cell imaging. Cells were observed with a 60×/numerical aperture 1.42 Plan Apo N objective. Images were captured with an IXON3 electron microscopy charge-coupled-device (EMCCD) camera in an ANDOR spinning-disc confocal imaging system on an Olympus IX81 inverted microscope.
Analyses of mRNA and miRNA by qRT-PCR.
Real-time analysis by two-step reverse transcription-PCR (RT-PCR) was performed for quantification of miRNA and mRNA levels on a 7500 real-time PCR system (Applied Biosystems) or Bio-Rad CFX96 real-time system using an Applied Biosystems TaqMan chemistry-based miRNA assay system. mRNA real-time quantification was generally performed in a two-step format using a Eurogentec reverse transcriptase core kit and MESA GREEN quantitative PCR (qPCR) Master Mix Plus for SYBR Assay with Low Rox kit from Eurogentec following the supplier’s protocols. The comparative cycle threshold (CT) method, which typically included normalization by the 18S rRNA or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) level for each sample, was used for relative quantification.miRNA assay by real-time PCR was performed with 25 ng of cellular RNA and 200 ng of EV-derived RNA unless specified otherwise, using specific primers for human let-7a (assay 000377), human miR-122 (assay 000445), or human miR-16 (assay 000391). U6 snRNA (assay 001973) was used as an endogenous control. One-third of the reverse transcription mix was subjected to PCR amplification with TaqMan Universal PCR Master Mix No AmpErase (Applied Biosystems) and the respective TaqMan reagents for target miRNA. Samples were analyzed in triplicates from a minimum of two biological replicates. The miRNA levels were defined from the cycle threshold values for representation of EV-associated miRNA or immunoprecipitated miRNA levels. The comparative CT method, which typically included normalization by the U6 snRNA or a nonrelevant miRNA for each sample, was used for all other instances. For real-time quantification and gene amplification studies, the following primers were used: ARID3B (forward, 5′-GTCTGGCTGTGCCCGTGACC-3′; reverse, 5′-TGGGCAAACAGCACACCTGC-3′), HMGA2 (forward, 5′-AGCAGCAGCAAGAACCAACCG-3′; reverse, 5′-TCGAACGTTGGCGCCCCCTA-3′), KRAS (forward, 5′-GCAAAGACAAGACAGAGAGTGGAGG-3′; reverse, 5′-AACTGCATGCACCAAAAACCCCA-3′), ALDOLASE (forward, 5′-TGGACCTAGCTTGGCGCGGA-3′; reverse, 5′-CCTGGGCCAGCAGGCAGTTC-3′), GYS1 (forward, 5′-GGTGGCTAACAAGGTGGGTGGC-3′; reverse, 5′-CGATCAGCCAGCGCCCGAAA-3′), CAT1 (forward, 5′-GCCGCCGGCTTGGATTCTGA-3′; reverse, 5′-CCCCGAGGGCCACCAGATCA-3′), GTF2B (forward, 5′-TGCGCGTCTCTTCGCCACAT-3′; reverse, 5′-AGCTCCTGTGCCCTTGCCAAT-3′), GAPDH (forward, 5′-CAGGGGGGAGCCAAAAGGG-3′; reverse, 5′-CTTGGCCAGGGGTGCTAAGC-3′), and 18S rRNA (forward, 5′-TGACTCTAGATAACCTCGGG-3′; reverse, 5′-GACTCATTCCAATTACAGGG-3′).
Cell senescence and TUNEL assay.
Cells were assayed for senescence using a senescence cell histochemical staining kit from Sigma after fixation of the cells in a buffer containing 20% formaldehyde, 2% glutaraldehyde, 70.4 mM Na2HPO4, 14.7 mM KH2PO4, 1.37 M NaCl, and 26.8 mM KCl as a stock 10% solution for 7 min. TUNEL assays were performed with a DeadEnd fluorometric TUNEL system kit (Promega) as per the manufacturer’s protocol. Cells were mounted on a cover slide with Vectashield 4′,6-diamidino-2-phenylindole (DAPI) for observation with a 10× Plan Fluor 10×/0.30 objective on a Nikon Eclipse Ti microscope and imaged with a Nikon Qi1MC camera.
Postimaging and other analyses.
All Western blot and Northern blot images were processed with Adobe Photoshop CS4 for all linear adjustments and cropping. All images captured on a Nikon Eclipse Ti microscope or ANDOR spinning-disc microscope were analyzed and processed with Nikon NIS Element AR 3.1 software, including P-body tracking and velocity calculations. Image cropping was done using Adobe Photoshop CS4. Statistical analysis of data was done by performing paired, nonparametric, two-tailed t tests with a confidence interval of 95%, and four significant digits were considered in all cases.
ACKNOWLEDGMENTS
We thank W. Filipowicz, G. Meister, R. Pillai, and E. Bertrand for their generous help with reagents and plasmid constructs. We also thank B. Barman, who helped us with plasmid construction. We thank T. Muruganandan for atomic force microscopy analysis.
This work was primarily funded by the Swarnajayanti Fellowship (DST/SJF/LSA-03/2014-15) and CEFIPRA (6003-2) grant to S.N.B. S.N.B. also acknowledge a High Risk High Reward grant (HRR/2016/000093) from the Department of Science and Technology, Government of India. All the authors except S.N.B. and K.M. were supported by a fellowship from CSIR.
The work was conceived and analyzed by S.N.B., and he was helped by other authors in formulating, analyzing, and critical reading of the text and figures. S.G., K.M., Y.C., S.C. and B.G. did all the experiments and data analysis. K.M. and S.G. also contributed to conceptualization of the idea.
We declare no conflict of interest related to this article.
REFERENCES
- 1.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iorio MV, Croce CM. 2012. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4:143–159. 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendell JT, Olson EN. 2012. MicroRNAs in stress signaling and human disease. Cell 148:1172–1187. 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834–838. 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 5.Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114. 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 6.Huntzinger E, Izaurralde E. 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12:99–110. 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 7.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. 2005. Stem cell division is regulated by the microRNA pathway. Nature 435:974–978. 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 8.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. 2006. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A 103:2746–2751. 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, Oertner TG, Schratt G, Bibel M, Roska B, Filipowicz W. 2010. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141:618–631. 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 10.Bose M, Bhattacharyya SN. 2016. Target-dependent biogenesis of cognate microRNAs in human cells. Nat Commun 7:12200. 10.1038/ncomms12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee S, Grosshans H. 2009. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461:546–549. 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 12.Baccarini A, Chauhan H, Gardner TJ, Jayaprakash AD, Sachidanandam R, Brown BD. 2011. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr Biol 21:369–376. 10.1016/j.cub.2011.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cazalla D, Yario T, Steitz JA, Steitz J. 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328:1563–1566. 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, Hu-Li J, Lu K, Dubois W, Kitamura Y, Charles N, Sun HW, Muljo S, Schwartzberg PL, Paul WE, O’Shea J, Rajewsky K, Casellas R. 2010. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32:828–839. 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Mata M, Gaidatzis D, Vitanescu M, Stadler MB, Wentzel C, Scheiffele P, Filipowicz W, Großhans H. 2015. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep 16:500–511. 10.15252/embr.201540078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu S, Bhattacharyya SN. 2014. Insulin-like growth factor-1 prevents miR-122 production in neighbouring cells to curtail its intercellular transfer to ensure proliferation of human hepatoma cells. Nucleic Acids Res 42:7170–7185. 10.1093/nar/gku346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh S, Bose M, Ray A, Bhattacharyya SN. 2015. Polysome arrest restricts miRNA turnover by preventing exosomal export of miRNA in growth-retarded mammalian cells. Mol Biol Cell 26:1072–1083. 10.1091/mbc.E14-11-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee K, Ghoshal B, Ghosh S, Chakrabarty Y, Shwetha S, Das S, Bhattacharyya SN. 2016. Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep 17:1184–1203. 10.15252/embr.201541930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. 2009. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11:1143–1149. 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 20.Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim do H, Cho IS, Nakahara K, Preall JB, Bellare P, Sontheimer EJ, Carthew RW. 2009. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol 11:1150–1156. 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller S, Sanderson MP, Stoeck A, Altevogt P. 2006. Exosomes: from biogenesis and secretion to biological function. Immunol Lett 107:102–108. 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Simons M, Raposo G. 2009. Exosomes—vesicular carriers for intercellular communication. Curr Opin Cell Biol 21:575–581. 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 24.Temoche-Diaz MM, Shurtleff MJ, Nottingham RM, Yao J, Fadadu RP, Lambowitz AM, Schekman R. 2019. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife 8:e47544. 10.7554/eLife.47544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125:1111–1124. 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 26.Bose M, Barman B, Goswami A, Bhattacharyya SN. 2017. Spatiotemporal uncoupling of microRNA-mediated translational repression and target RNA degradation controls MicroRNP recycling in mammalian cells. Mol Cell Biol 37:e00464-16. 10.1128/MCB.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. 2011. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol 18:1218–1226. 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao B, La LB, Chen YC, Chang LJ, Chan EK. 2012. Defining a new role of GW182 in maintaining miRNA stability. EMBO Rep 13:1102–1108. 10.1038/embor.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, Chen CY, Shyu AB, Yates JR, III, Hannon GJ, Filipowicz W, Duchaine TF, Sonenberg N. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35:868–880. 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX, Wurdinger T, Meijer GA, Pegtel DM. 2014. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep 8:1649–1658. 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 31.Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. 2006. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res 34:4801–4815. 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kullmann M, Gopfert U, Siewe B, Hengst L. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5'UTR. Genes Dev 16:3087–3099. 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. 2010. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 285:17442–17452. 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319:1244–1247. 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 35.Bose M, Chatterjee S, Chakrabarty Y, Barman B, Bhattacharyya SN. 2020. Retrograde trafficking of Argonaute 2 acts as a rate-limiting step for de novo miRNP formation on endoplasmic reticulum-attached polysomes in mammalian cells. Life Sci Alliance 3:e201800161. 10.26508/lsa.201800161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng Y, Zeng Q, Han Q, Xia W. 2019. Effect of pH, temperature and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell 10:295–299. 10.1007/s13238-018-0529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bobrie A, Colombo M, Raposo G, Thery C. 2011. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12:1659–1668. 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 38.Braun JE, Huntzinger E, Fauser M, Izaurralde E. 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44:120–133. 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. 2009. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA 15:1067–1077. 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eulalio A, Tritschler F, Izaurralde E. 2009. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA 15:1433–1442. 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. 2009. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA 15:781–793. 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cikaluk DE, Tahbaz N, Hendricks LC, DiMattia GE, Hansen D, Pilgrim D, Hobman TC. 1999. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol Biol Cell 10:3357–3372. 10.1091/mbc.10.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatterjee S, Fasler M, Bussing I, Grosshans H. 2011. Target-mediated protection of endogenous microRNAs in C. elegans. Dev Cell 20:388–396. 10.1016/j.devcel.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. 2005. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123:1133–1146. 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 45.Ebert MS, Neilson JR, Sharp PA. 2007. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4:721–726. 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147:358–369. 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358. 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kai ZS, Pasquinelli AE. 2010. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol 17:5–10. 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. 2010. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328:1534–1539. 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, Ibberson M, De Palma M. 2014. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep 8:1432–1446. 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 51.Ebert MS, Sharp PA. 2012. Roles for microRNAs in conferring robustness to biological processes. Cell 149:515–524. 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thery C, Amigorena S, Raposo G, Clayton A. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]