PH domain leucine-rich repeat protein phosphatase 1 (PHLPP1) is a tumor suppressor that directly dephosphorylates a wide array of substrates, most notably the prosurvival kinase Akt. However, little is known about the molecular mechanisms governing PHLPP1 itself.
KEYWORDS: PHLPP, Cdk1, mitosis, phosphatase
ABSTRACT
PH domain leucine-rich repeat protein phosphatase 1 (PHLPP1) is a tumor suppressor that directly dephosphorylates a wide array of substrates, most notably the prosurvival kinase Akt. However, little is known about the molecular mechanisms governing PHLPP1 itself. Here, we report that PHLPP1 is dynamically regulated in a cell cycle-dependent manner and deletion of PHLPP1 results in mitotic delays and increased rates of chromosomal segregation errors. We show that PHLPP1 is hyperphosphorylated during mitosis by Cdk1 in a functionally uncharacterized region known as the PHLPP1 N-terminal extension (NTE). A proximity-dependent biotin identification (BioID) interaction screen revealed that during mitosis, PHLPP1 dissociates from plasma membrane scaffolds, such as Scribble, by a mechanism that depends on its NTE and gains proximity to kinetochore and mitotic spindle proteins such as KNL1 and TPX2. Our data are consistent with a model in which phosphorylation of PHLPP1 during mitosis regulates binding to its mitotic partners and allows accurate progression through mitosis. The finding that PHLPP1 binds mitotic proteins in a cell cycle- and phosphorylation-dependent manner may have relevance to its tumor-suppressive function.
INTRODUCTION
Dynamic changes in protein phosphorylation critically regulate accurate and timely progression through the cell cycle. Phosphorylation is particularly important during mitosis to ensure that the replicated genome is evenly divided between two daughter cells. Errors in this process can result in aneuploidy, a hallmark of cancer (1). Phospho-proteomic analyses have identified over 1,000 proteins whose phosphorylation increases during mitosis, with the majority of sites displaying S/T-P motifs that are recognized by cyclin-dependent kinases (Cdk) (2, 3). In addition to activation of kinases, most notably Cdk1 bound to cyclin B, inactivation of protein phosphatases such as protein phosphatase 2A (PP2A) drives the massive increase in protein phosphorylation. However, this cellular state of heightened protein phosphorylation is short-lived. Reactivation of protein phosphatases (4) and activation of the E3 ubiquitin ligase anaphase promoting complex (APC/C), which degrades key mitotic proteins such as cyclin B (5–7), occur during the final stages of mitosis. Phosphorylation of mitotic substrates by the Cdk1-cyclin B complex has diverse effects, including regulating the localization (8, 9), activity (10–12), stability (13), and interactome of substrate proteins (14). Given the massive phosphorylation changes that accompany mitosis (2, 3), it is increasingly apparent that many other mitotic kinases and phosphatases orchestrate mitotic responses, and many may do so downstream of Cdk1.
The PH domain leucine-rich repeat protein phosphatases 1 and 2 (PHLPP1 and PHLPP2) are emerging as key players opposing context-dependent signaling events (15). These two family members of the metal-dependent protein phosphatase (PPM) “shrub” of the phosphatome (16) were originally discovered for their role in dephosphorylating a key regulatory site on Akt, the hydrophobic motif (Ser473 on Akt1), to suppress the activity of this oncogenic kinase (17, 18). Supporting a tumor-suppressive role, the PHLPP1 gene locus is frequently deleted in cancer (19–22), and genetic deletion in a mouse model promotes tumor progression in both prostate (23) and colorectal cancer (24). While the importance of PHLPP1 signaling in the context of disease has mostly been attributed to its regulation of Akt and other AGC kinases (25–27), an increasing number of substrates involved in other biological pathways are being identified. Notably, PHLPP1 suppresses inflammatory signaling by dephosphorylating the transcription factor STAT1 (28), controls receptor tyrosine kinase transcription by suppressing histone phosphorylation (29), maintains regulatory T-cell development (30), and promotes bone morphogenesis (31–33). A role in mitosis was recently suggested in a study showing that PHLPP1 dephosphorylates and stabilizes the outer-kinetochore protein SGT1, resulting in proper kinetochore assembly (34).
PHLPP family members have low catalytic activity, and their scaffolding to protein substrates is essential for effective downstream signaling. This is achieved through specific regulatory modules that are part of the same polypeptide as the catalytic phosphatase domain, contrasting with most other Ser/Thr phosphatases, whose regulatory modules are distinct polypeptides. In addition to a catalytic protein phosphatase 2C (PP2C) phosphatase domain, both PHLPP1 and PHLPP2 have a pleckstrin homology (PH) domain, multiple leucine-rich repeats (LRRs), and an unstructured C-terminal extension (CTE) capped by a PDZ binding ligand (15). The main structural difference between the two family members is a unique, approximately 50-kDa N-terminal extension (NTE) on PHLPP1 which contains a bipartite arginine-rich nuclear localization signal (NLS) (28). This region has no known domain homology and is not required for targeting of shared PHLPP targets, such as Akt (17, 18), protein kinase C (PKC) (27), and ribosomal protein S6 kinase 1 (S6K1) (25). Each of these domains confers specificity required for substrate targeting. For example, Akt dephosphorylation in cells depends on an intact PDZ ligand (18), PKC dephosphorylation depends on the PH domain (27), and STAT1 binding and dephosphorylation require the NTE (28). Additionally, the binding of PHLPP1 to the plasma membrane scaffold Scribble (Scrib) depends on determinants in the CTE distinct from the PDZ ligand, and this interaction was shown to be necessary for the dephosphorylation of Akt Ser473 in epithelial cells (35). Identification of key binding partners to the NTE and CTE have opened up the possibility that these unstructured and understudied regions of the enzyme play critical roles in regulating PHLPP1 interactions and localization.
Here, we determined that the PHLPP1 NTE is a substrate of Cdk1 and that the NTE functions to switch the PHLPP1 protein interaction network during mitosis. Specifically, we report that endogenous PHLPP1 protein undergoes a distinct and reversible electrophoretic mobility shift in mitotic cells as a result of hyperphosphorylation on the NTE. Biochemical analysis and phospho-mass spectrometry uncovered 13 previously undescribed mitotic phospho-sites within the NTE, all exhibiting a minimal Cdk1 recognition motif, S/T-P. In vitro and cellular assays utilizing the Cdk1 inhibitor RO-3306 confirmed that the NTE is a Cdk1 substrate. A proximity-dependent biotin identification (BioID) screen revealed that the NTE regulates the interactome of PHLPP1 during mitosis, dampening PHLPP1 interactions with plasma membrane scaffolds such as Scrib and promoting interactions with the kinetochore and mitotic spindle assembly proteins. Importantly, mouse embryonic fibroblasts (MEFs) lacking PHLPP1 had increased errors in chromatin segregation and a mitotic delay phenotype, as assessed by fluorescence microscopy. Taken together, these results identify PHLPP1 as a Cdk1 substrate and a new player in the field of mitotic signaling.
RESULTS
PHLPP1 phosphorylation is dynamically regulated during the cell cycle.
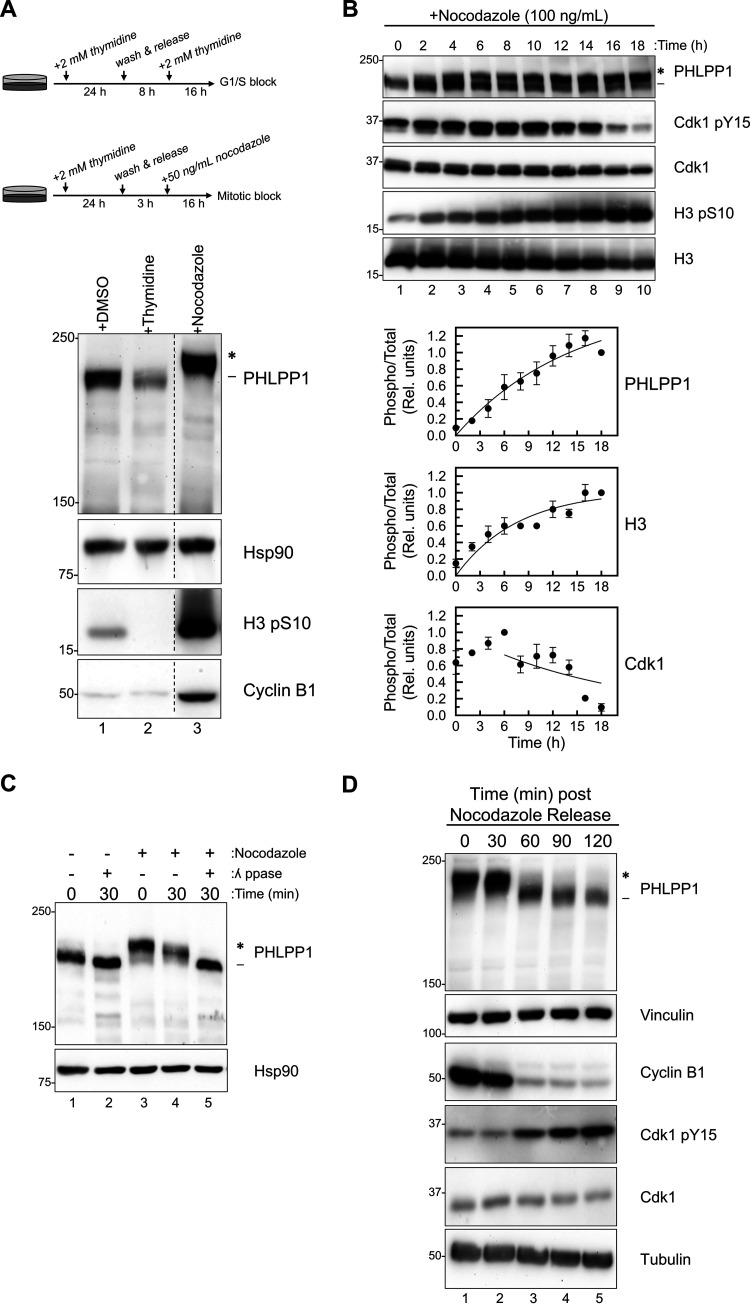
To determine how PHLPP1 is regulated during the cell cycle, we utilized two different cell cycle synchronization techniques. RPE1 cells were synchronized using either a double-thymidine block to enrich for G1/S cells or a thymidine/nocodazole block followed by a mitotic shake-off to isolate mitotic cells, and PHLPP1 levels were assessed (Fig. 1A). We observed a substantial electrophoretic mobility shift on PHLPP1 in nocodazole-treated cells. To determine whether this mobility shift tracked with cellular synchronization markers, HEK-293A cells were treated with nocodazole for various times and the PHLPP1 mobility shift, phosphorylation of Cdk1 on Tyr15 (which is dephosphorylated during mitotic exit), and phosphorylation of histone H3 on Ser10 were monitored (Fig. 1B). PHLPP1 transitioned from a faster-migrating (Fig. 1B, dash) to a slower-migrating (Fig. 1B, asterisk) species with a half-time of 10 ± 4 h. These kinetics were similar to those for Cdk1 dephosphorylation (which was half-maximally phosphorylated at 13 ± 2 h) and histone phosphorylation (half-time of 5 ± 2 h). These results indicate that PHLPP1 undergoes an electrophoretic mobility shift as a function of cell cycle synchronization.
FIG 1.
PHLPP1 phosphorylation is cell cycle dependent. (A) Western blot of RPE1 cells treated with either DMSO (asynchronous), double-thymidine block (G1/S), or thymidine/nocodazole block (M). Western blots are representative of four independent experiments. Lanes in the blot that were irrelevant to this experiment were removed (indicated by the dashed line). (B) Western blot of HEK-293A cells treated with nocodazole (100 ng/ml) for indicated amounts of time. Graphs show the quantification of three independent experiments for PHLPP1 and Cdk1 and two independent experiments for H3. Values are the ratio of the signal of phosphorylated protein to total protein (relative units) ± SEM. For PHLPP1, the ratio was calculated as the signal of the slower-mobility species (*) divided by the total PHLPP1 signal (* and -) normalized to the 18-h time point. For Cdk1, the ratio was calculated as the Cdk1 pY15 signal divided by the total Cdk1 signal normalized to the 6-h time point. For H3, the ratio was calculated as the signal of the H3 pS10 divided by total H3 signal normalized to the 18-h time point. Data analysis was performed using GraphPad Prism 8. (C) Lysates from asynchronous (lanes 1 and 2) or nocodazole-treated (lanes 3, 4, and 5) HEK-293A cells were incubated at 25°C with or without λ phosphatase (ppase) for indicated amounts of time. All blots are probed against endogenous proteins. Three independent experiments were performed. (D) Western blot of mitotic RPE1 cells that were collected, washed, and released from a thymidine/nocodazole block. Western blots are representative of three independent experiments. The asterisk shows the PHLPP1 electrophoretic mobility shift compared to faster-mobility species (dash).
To determine if the mitosis-dependent electrophoretic mobility shift of PHLPP1 resulted from phosphorylation, lysates from nocodazole-treated cells were incubated with either purified lambda phosphatase (Fig. 1C, lane 5) or buffer only (lane 4) for 30 min at 25°C. The shift observed in mitotic cells (Fig. 1C, lane 3) collapsed after 30 min of lambda phosphatase treatment (lane 5), indicating that the mobility shift resulted from phosphorylation. When asynchronous cells (Fig. 1C, lanes 1 and 2) were treated with lambda phosphatase (lane 2), a faster-mobility species was also observed, demonstrating that PHLPP1 is phosphorylated under basal conditions.
We next addressed whether the phosphorylation was reversed during mitotic exit. RPE1 cells were synchronized in mitosis using a thymidine/nocodazole block, followed by a mitotic shake-off (Fig. 1D). Mitotic cells were collected, washed, and released into nocodazole-free medium, allowing cells to resume and exit mitosis. Cyclin B1, which is degraded during mitotic exit, and Cdk1 Tyr15 phosphorylation, which occurs during mitotic exit, were analyzed to track mitotic progression. Exit from mitosis was accompanied by dephosphorylation of PHLPP1, assessed by the increase in electrophoretic mobility, with the appearance of the dephosphorylated species correlating with the increase in mitotic exit markers. Thus, PHLPP1 is reversibly phosphorylated during the cell cycle.
Western blotting revealed changes in endogenous PHLPP1 protein signal at different points in the cell cycle. Specifically, cells synchronized at mitosis had higher levels of PHLPP1 signal than asynchronous cells (Fig. 1A, lane 1) and thymidine-blocked cells (lane 2). Cells released from a nocodazole-induced mitotic block had decreasing levels of PHLPP1 protein signal (Fig. 1D, lanes 2 to 5). The changes in protein signal could indicate either that protein expression itself is dynamically regulated during the cell cycle or that the epitope for the PHLPP1 antibody may be better exposed when PHLPP1 is phosphorylated. Probing for overexpressed hemagglutinin (HA)-tagged PHLPP1 with an anti-HA antibody did not result in a large difference in signal between asynchronous and mitotic cells (Fig. 2B), which could reflect the latter possibility. Thus, our data suggest that PHLPP1 is dynamically regulated during the cell cycle at the posttranslational level.
FIG 2.
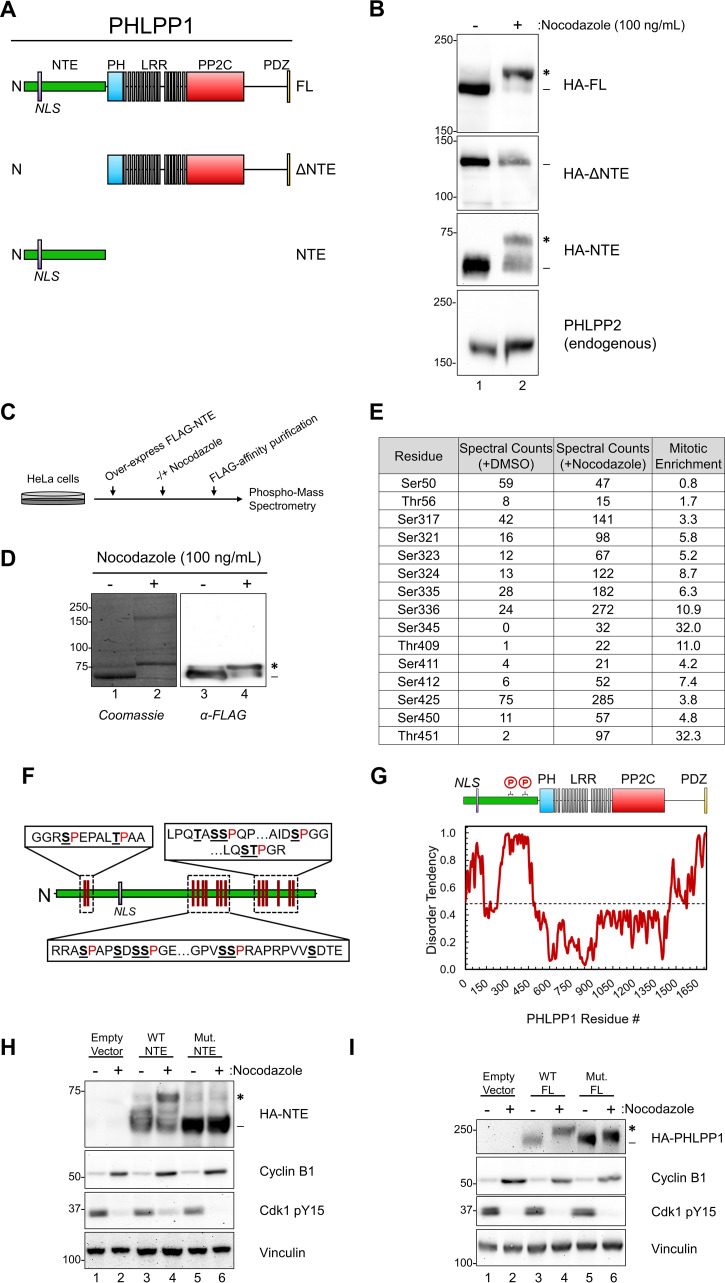
Mitotic PHLPP1 phosphorylation occurs within the unstructured N-terminal extension (NTE). (A) Schematic of HA-tagged PHLPP1 constructs used in this study: full-length PHLPP1 (amino acids 1 to 1717 [FL]), N-terminally truncated PHLPP1 (amino acids 513 to 1717 [ΔNTE]), and C-terminally truncated PHLPP1 (amino acids 1 to 512 [NTE]). (B) Western blot of DMSO- or nocodazole-treated HeLa cells overexpressing HA-tagged constructs from panel A using an anti-HA antibody. Samples were also analyzed for endogenous PHLPP2 using an anti-PHLPP2 antibody. (C) Schematic of experimental design for phospho-mass spectrometry. (D) FLAG-PHLPP1 NTE purified from DMSO or nocodazole-treated HeLa cells separated by SDS-PAGE (left, Coomassie blue staining; right, Western blot using FLAG antibody). (E) Table summarizing phospho-mass spectrometry results. The mitotic enrichment score was calculated as follows: (spectral countsnocodazole)/[1 + (spectral countsDMSO)]. (F) Schematic of mitotic phosphorylation sites (underlined) detected in the PHLPP1 NTE (proximal proline residues in red). (G) (Top) Diagram of PHLPP1 protein structure with mitotic phosphorylation clusters indicated. (Bottom) Predictions of intrinsic disorder tendency of PHLPP1 by IUPred2A long disorder (36). Scores above 0.5 (dotted line) indicate disordered regions. (H) Western blot of DMSO or nocodazole-treated HeLa cells overexpressing either an empty vector, HA-NTE wild type (WT NTE), or the HA-NTE containing Ser/Thr-to-Ala mutations at the mitotic phosphorylation sites identified in panel F (Mut. NTE) using an anti-HA antibody. Samples were also analyzed for endogenous cyclin B1 and Cdk1 pY15. These data are representative of three independent experiments. (I) Western blot of DMSO or nocodazole-treated HeLa cells overexpressing either an empty vector, full-length HA-PHLPP1 wild type (WT FL), or full-length HA-PHLPP1 containing Ser/Thr-to-Ala mutations at the mitotic phosphorylation sites identified in panel F (Mut. FL) using an anti-HA antibody. Samples were also analyzed for endogenous cyclin B1 and Cdk1 pY15. These data are representative of three independent experiments. Asterisks indicate PHLPP1 electrophoretic mobility shift compared to faster-mobility species (dash).
Mitotic PHLPP1 phosphorylation occurs at the PHLPP1 NTE.
To elucidate which region of PHLPP1 is phosphorylated during mitosis, truncated PHLPP1 mutants were expressed in asynchronous and mitotic HeLa cells (Fig. 2A). While full-length (FL) PHLPP1 exhibited a mobility shift following nocodazole treatment, a construct lacking the N-terminal extension (ΔNTE) did not (Fig. 2B). Additionally, the NTE alone had a mobility shift when expressed in nocodazole-treated cells. Endogenous PHLPP2 (the other PHLPP family member), which lacks the NTE that is present on PHLPP1, does not shift during mitosis (Fig. 2B), further suggesting that the NTE mediates the mitosis-induced electrophoretic mobility shift of PHLPP1.
We utilized phospho-mass spectrometry to determine which residues in the NTE are phosphorylated during mitosis. HeLa cells expressing FLAG-tagged NTE were treated with dimethyl sulfoxide (DMSO) or nocodazole, purified, and resolved on an SDS-PAGE gel (Fig. 2C). The faster-mobility species, corresponding to the unphosphorylated NTE, was excised from the DMSO-treated condition (Fig. 2D, lane 1 [dash]), and the slower-mobility species was excised from the nocodazole-treated condition (Fig. 2D, lane 2 [asterisk]). Gel slices were digested with trypsin and analyzed by ultra-high-pressure liquid chromatography (UPLC) coupled with tandem mass spectrometry (MS/MS) to identify phosphorylated peptides (see Data Set S1 in the supplemental material). A mitotic enrichment score was calculated by dividing the total spectral counts of the nocodazole-treated sample by that of the DMSO-treated samples for each phosphorylated residue. This analysis revealed that 13 residues had a ≥3-fold increase in spectral counts of phosphorylated peptides in the nocodazole-treated cells compared to the DMSO control, while two residues, Ser50 and Thr56, were phosphorylated to similar levels in both samples (Fig. 2E). Additionally, the residues modified were grouped in clusters rather than evenly spread out along the NTE (Fig. 2F). These clusters also appeared in regions of the NTE that are predicted to be intrinsically disordered (Fig. 2G). Intrinsic disorder prediction was completed by analysis of the PHLPP1 amino acid sequence using the IUPRED2 long disorder prediction tool using the default parameters (36).
To determine if the phosphorylated residues identified via mass spectrometry account for the PHLPP1 mobility shift observed during mitosis, mitotic phosphorylation site mutants were expressed in asynchronous and mitotic HeLa cells. Specifically, we mutated these Ser/Thr residues (Ser317, Ser321, Ser323, Ser324, Ser335, Ser336, Ser345, Thr409, Ser411, Ser412, Ser425, Ser450, and Thr451), as well as several nearby Ser residues that could function as compensatory phosphorylation sites (Ser313, Ser347, Ser349, Ser351, Ser353, and Ser355), to Ala both in the NTE alone (Fig. 2H) and in full-length PHLPP1 (Fig. 2I). Nocodazole treatment resulted in a mobility shift of wild-type NTE (Fig. 2H, lane 4, asterisk) but not of the mutated NTE construct (lane 6), indicating that phosphorylation of these residues mediates the mobility shift. Mutated full-length PHLPP1 (Fig. 2I, lane 6) also did not undergo the large mobility shift of wild-type PHLPP1 (lane 4); however, it did undergo an intermediate shift potentially resulting from compensatory phosphorylation sites that are present outside the NTE. These data reveal that PHLPP1 is hyperphosphorylated at the NTE following nocodazole treatment.
The PHLPP1 NTE is a substrate of Cdk1.
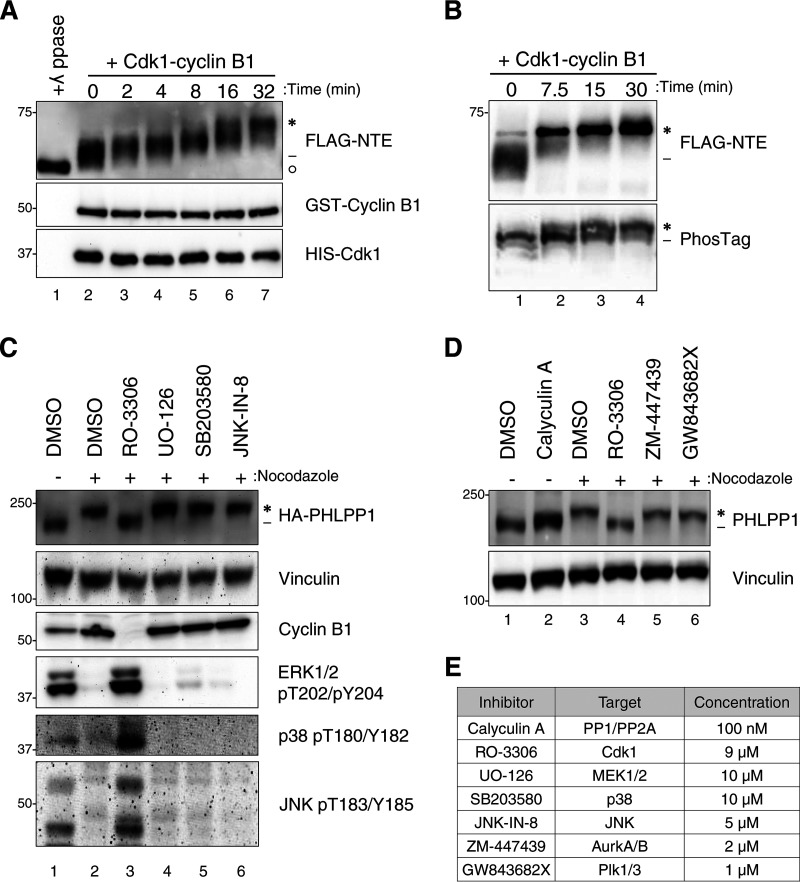
Almost all of the identified phosphorylation sites in the NTE occur on serine or threonine residues followed by a proline, the motif for a proline-directed kinase such as Cdk1, the master regulator of mitosis (37). Additionally, two of the residues, Ser450 and Thr451, are part of a canonical consensus motif for Cdk1, S/T-P-X-R/K, and previous studies have illustrated that Cdk1 can target a minimal sequence of S/T-P (38, 39). To determine if the PHLPP1 NTE is a Cdk1 substrate, we incubated FLAG-tagged PHLPP1 NTE, purified from asynchronous HEK-293T cells, with recombinant Cdk1-cyclin B1 and ATP. This resulted in an electrophoretic mobility shift of the NTE as assessed by Western blotting (Fig. 3A). Rather than accumulating all upper (asterisk) or all lower bands (dash), we observed a number of intermediate bands, supporting the multiple sites identified by mass spectrometry. Additionally, samples resolved on a PhosTag gel displayed several bands, further suggesting multiple phosphorylated residues (Fig. 3B). Treatment of the FLAG-NTE with lambda phosphatase resulted in a collapse of the slower-migrating species to a species migrating faster than untreated PHLPP1 (circle), likely reflecting basal phosphorylation on the Ser50 and Thr56 residues identified by mass spectrometry (Fig. 3A, lane 1).
FIG 3.
Cdk1 phosphorylates PHLPP1 NTE during mitosis. (A) In vitro phosphorylation of FLAG-tagged PHLPP1 NTE with recombinant Cdk1-cyclin B1 (0.01 μg/μl). Purified NTE was incubated with either λ phosphatase (lane 1) or recombinant Cdk1-cyclin B1 (lanes 2 to 7) at 37°C for the indicated times. PHLPP1 NTE mobility shift was evaluated by Western blotting using an anti-FLAG antibody. GST-cyclin B1 and HIS-Cdk1 were probed by antibodies against cyclin B1 and Cdk1, respectively. (B) Purified NTE treated with recombinant Cdk1-cyclin B1 (0.02 μg/μl) was probed by an anti-FLAG antibody on an SDS-PAGE (top) or PhosTag (bottom) gel. (C) HeLa cells expressing HA-tagged PHLPP1 (full length) were treated with nocodazole (100 ng/ml for 16 h), followed by the following kinase inhibitors for 30 min prior to lysis: RO-3306 (Cdk1 inhibitor; 9 μM), UO-126 (MEK1/MEK2 inhibitor; 10 μM), SB203580 (p38 inhibitor; 10 μM), or JNK-IN-8 (JNK inhibitor; 5 μM). Asynchronous cells were treated with DMSO for 30 min prior to lysis. Triton-soluble lysates were subjected to Western blot analysis and probed for HA-PHLPP1. (D) HeLa cells were treated with nocodazole (100 ng/ml for 16 h), followed by various mitotic kinase inhibitors for 30 min prior to lysis as indicated: RO-3306 (Cdk1 inhibitor; 9 μM), ZM-447439 (Aurora A/B inhibitor; 2 μM), and GW843682X (Polo-like kinase 1/3 inhibitor; 1 μM). Asynchronous cells were treated with the phosphatase inhibitor calyculin A (100 nM) for 30 min prior to lysis. Triton-soluble lysates were subjected to Western blotting and probed for endogenous PHLPP1. (E) List of inhibitors used in panels C and D. Asterisks indicate PHLPP1 electrophoretic mobility shift compared to faster-mobility species (dashes). The open circle indicates PHLPP1 species after phosphatase treatment.
We next employed a pharmacological approach to determine whether Cdk1 catalyzes the phosphorylation of PHLPP1 in cells. Treatment of mitotic HeLa cells with the Cdk1 active-site inhibitor RO-3306 for 30 min prior to lysis resulted in collapse of the endogenous PHLPP1 mobility shift (Fig. 3C, lane 3). Since mitogen-activated protein kinases (MAPK) also show a preference for substrates with the S/T-P motif, mitotic HeLa cells were incubated with inhibitors against the 3 subfamilies of MAPKs, specifically extracellular signal-regulated kinase (ERK), p38, and c-Jun N-terminal kinase (JNK) (Fig. 3C, lanes 4 to 6). None of these treatment conditions resulted in a collapse of the faster-mobility (phosphorylated) PHLPP1 species. Furthermore, a collapse was not observed when cells were treated with inhibitors against other mitotic kinases (Fig. 3D), specifically Aurora kinase A/B (Fig. 3D, lane 5) or Polo-like kinase 1/3 (lane 6). To determine whether this mobility shift could be induced in nonmitotic cells (when Cdk1 is not fully active), asynchronous cells were treated with the phosphatase inhibitor calyculin A (Fig. 3D, lane 2) for 30 min prior to lysis in order to preserve any transient phosphorylation events. Although phosphatase inhibition resulted in a small electrophoretic mobility shift, this species of PHLPP1 migrated below that of the hyperphosphorylated PHLPP1 observed in nocodazole-treated cells. These data reveal that Cdk1 activation is necessary for the hyperphosphorylation of PHLPP1 that results in a robust mobility shift.
Nocodazole treatment induces changes to the PHLPP1 interactome.
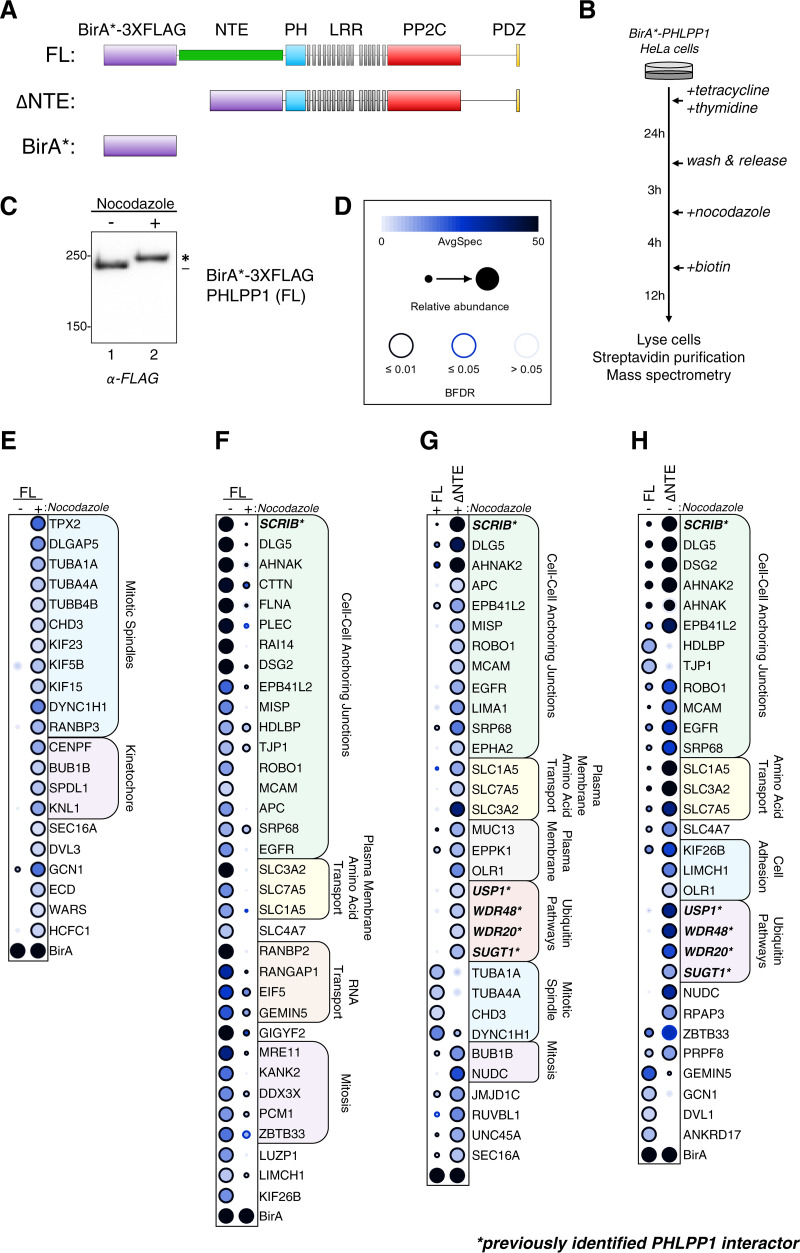
As mitotic phosphorylation events induce changes in protein-protein interactions (40), we hypothesized that the phosphorylation of the PHLPP1 NTE could alter the PHLPP1 interactome. To determine the differences in PHLPP1 protein-protein interactions between DMSO- and nocodazole-treated cells, we performed a proximity-dependent biotin identification (BioID) screen (41). A mutated form of the Escherichia coli biotin ligase BirA R118G (BirA*) (42) and a 3×FLAG tag were fused to the N terminus of full-length (FL) or truncated (ΔNTE) PHLPP1 and stably integrated into the tetracycline-inducible Flp-In T-REx HeLa cell system (Fig. 4A). As a negative control to determine background biotinylation, a cell line expressing just the BirA*-3×FLAG tag was generated. Cells were treated with tetracycline to induce expression of BirA*-3×FLAG fusion proteins and then either DMSO to obtain asynchronous cells or thymidine/nocodazole to block cells in mitosis (Fig. 4B). When lysates from cells expressing BirA*-3×FLAG-PHLPP1 (FL) were resolved on an SDS-PAGE gel and probed with an anti-FLAG antibody, a mobility shift of the construct was observed. Thus, fusion of BirA*-3×FLAG to PHLPP1 did not appear to prevent hyperphosphorylation of the NTE (Fig. 4C).
FIG 4.
BioID screen to determine changes in the mitotic PHLPP1 interactome. (A) Tetracycline-inducible Flp-IN T-REx HeLa cells expressing the following constructs were generated: full-length PHLPP1 (amino acids 1 to 1717 [FL]) and N-terminally truncated PHLPP1 (amino acids 513 to 1717 [ΔNTE]), fused with BirA* biotin ligase (R118G mutation) and a 3×FLAG tag. Additionally, cells expressing only the BirA*-3×FLAG tag were generated (BirA*). (B) BioID experimental workflow. (C) Western blot of BirA*-3×FLAG-PHLPP1 FL Flp-In T-REx HeLa cells treated or not treated with nocodazole. The blot was probed with an anti-FLAG antibody. (D) Legend for BioID dot plot analysis, where AvgSpec refers to averaged spectral counts across the biological duplicates (BFDR, false discovery rate). (E and F) Dot plots of prey that display either a ≥2-fold enrichment in nocodazole-treated (mitotic) versus DMSO-treated (asynchronous) cells expressing full-length (FL) PHLPP1 (E) or ≥2-fold enrichment in DMSO-treated (asynchronous) versus nocodazole-treated (mitotic) cells expressing FL PHLPP1 (F). (G) Dot plot of prey that display ≥2-fold enrichment or ≥2-fold depletion in nocodazole-treated ΔNTE cells compared to nocodazole-treated FL PHLPP1-expressing cells. (H) Dot plot of prey that display ≥2-fold enrichment or ≥2-fold depletion in DMSO-treated ΔNTE cells compared to FL cells. Boldface type and asterisks indicate previously identified PHLPP1-interacting proteins. The asterisk in panel C indicates PHLPP1 electrophoretic mobility shift compared to faster-mobility species (dash).
After synchronization, biotin was added to cells 12 h prior to harvest to allow for biotinylation of proteins proximal to the BirA*-3×FLAG-PHLPP1 fusion proteins. Following cell lysis, biotinylated proteins were purified using streptavidin-conjugated beads and analyzed by nano-high-pressure liquid chromatography (nano-HPLC) coupled to mass spectrometry. The data set was analyzed using the computational tool SAINTexpress (43) to define high-confidence proximity interactions and visualized by a dot plot (Fig. 4D to H; also, see Data Set S2). A number of previously identified PHLPP1-interacting proteins, such as Scrib (35, 44), USP1 (45–47), WDR48 (47–49), WDR20 (50), and SGT1 (gene name SUGT1) (34, 51), were identified in the BioID screen, indicative of the ability of this screen to identify known PHLPP1 interactors. To assess the PHLPP1 mitotic interactome, we determined which prey had a ≥2-fold increase in nocodazole-treated cells compared to DMSO-treated cells (Fig. 4E). Gene ontology (GO) analysis using g:Profiler (52) revealed an enrichment of proteins found at mitotic spindles, such as TPX2, tubulin (TUBA1A, TUBA4A, and TUBB4B), and kinesins (KIF23, KIF5B, and KIF15). Additionally, several kinetochore proteins, namely, Bub1B, Knl1, Spindly (SPDL1), and CENP-F, were identified, suggesting that PHLPP1 is in close proximity to a number of key mitotic signaling scaffolds. We also assessed which potential interactions were decreased ≥2-fold in nocodazole-treated compared to DMSO-treated cells (Fig. 4F). GO analysis identified an enrichment for prey involved in regulation of cell-cell anchoring junctions, such as Scrib, Dlg5, and cortactin (CTTN), and proteins involved in amino acid transport across the plasma membrane, such as 4F2hc (SL3A2), LAT1 (SLC7A5), and ASCT2 (SLC1A5). These data indicate that during interphase, PHLPP1 is in proximity to scaffolds at the plasma membrane, but mitosis disrupts these interactions, allowing PHLPP1 to switch binding partners and relocalize to key mitotic signaling complexes.
We next asked whether the NTE plays a role in this interactome change, considering that it is heavily modified during mitosis. To answer this question, we determined which prey displayed either a ≥2-fold enrichment or ≥2-fold depletion in nocodazole-treated cells expressing the ΔNTE construct compared to cells expressing full-length PHLPP1 (Fig. 4G). We observed that NTE deletion enhanced the interaction with proteins localized to the plasma membrane, such as Scrib, Dlg5, and 4F2hc (SLC3A2), and decreased the interaction with mitotic spindle proteins, such as tubulin (TUBA1A, TUBA4A, and TUBB4B) and dynein (DYNC1H1). These data suggest that the NTE is critical in releasing PHLPP1 from its plasma membrane-associated interactions. Furthermore, deletion of the NTE under basal conditions resulted in enhanced pulldown of plasma membrane-associated proteins (Fig. 4H), revealing that the NTE plays a general role in regulating PHLPP1 protein-protein interactions and localization outside mitosis.
Immortalized Phlpp1−/− mouse embryonic fibroblasts exhibit increased rates of mitotic defects compared to wild-type cells.
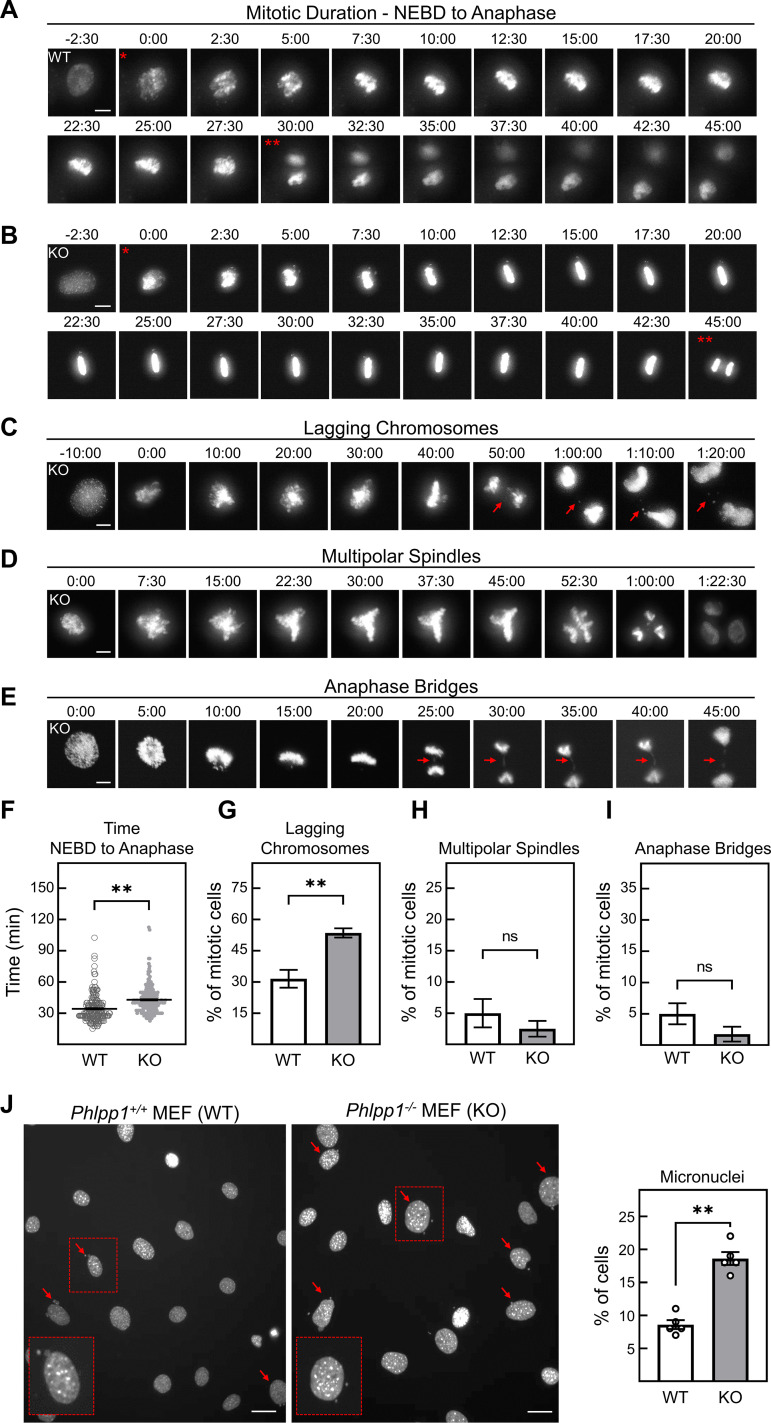
Since the BioID data suggested that PHLPP1 could be in proximity to mitotic signaling hubs, we hypothesized that PHLPP1 may function to regulate mitosis. To initially determine if PHLPP1 activity is required to maintain mitotic fidelity, we asked if the deletion of PHLPP1 resulted in any obvious mitotic defects. We performed live-cell imaging of Phlpp1−/− (knockout [KO]) and Phlpp1+/+ (wild-type [WT]) MEFs stably expressing a monomeric red fluorescent protein (mRFP)-tagged histone H2B to visualize chromatin dynamics during mitosis. Mitotic cells were scored on four individual parameters: (i) mitotic duration from nuclear envelope breakdown (NEBD) to anaphase (Fig. 5A and B), (ii) evidence of lagging chromosomes during anaphase (Fig. 5C), (iii) multipolar spindle formation (Fig. 5D), and (iv) anaphase bridge formation (Fig. 5E). While no significant difference was observed between WT and KO MEFs in the percentage of cells forming multipolar spindles (5% ± 2% versus 2% ± 1%, respectively [Fig. 5H]) or anaphase bridges (5% ± 2% versus 2% ± 1%, respectively [Fig. 5I]), lagging chromosomes were more prevalent in the KO MEFs than the WT MEFs (54% ± 2% versus 32% ± 4%, respectively [Fig. 5G]). Additionally, these lagging chromosomes can often form micronuclei, small extranuclear bodies containing chromatin that are a marker of genomic instability (53). DAPI (4′,6-diamidino-2-phenylindole) staining revealed that micronuclei were observed more frequently in KO than WT MEFs (19% ± 1% versus 8.6% ± 0.7%, respectively [Fig. 5J]). Live-cell imaging revealed that Phlpp1−/− MEFs spent an increased amount of time between NEBD and anaphase (43 min ± 1 min versus 34 min ± 1 min, respectively [Fig. 5A, B, and F]), suggesting that a mitotic delay occurs in these cells.
FIG 5.
Phlpp1−/− MEFs exhibit defects in mitosis. (A and B) Time-lapse fluorescence microscopy to determine mitotic duration in (A) Phlpp1+/+ (WT) and (B) Phlpp1−/− (KO) MEFs stably expressing mRFP-tagged H2B. The time when each frame was captured is noted in minutes and seconds, where 0:00 was set as the first frame indicating nuclear envelope breakdown (NEBD). The single asterisk indicates NEBD, and the double asterisks indicate the beginning of anaphase. (C to E) Representative images (from KO MEFs) of mitotic defects scored in WT and KO MEFs for lagging chromosomes (arrows) (C), multipolar spindle formation (D), and anaphase bridge formation (arrows) (E). Bars (A to E), 10 μm. (F to I) Quantification of parameters in panels A to E of WT (185 cells) and KO (211 cells) MEFs. Three clonal lines were imaged and analyzed separately; results were then averaged and used to calculate the SEM. Values are means ± SEM. **, P < 0.005; ns, not significant (Student's t test). (J) Representative image of DAPI-stained WT and KO MEFs. Arrows indicate cells containing micronuclei. Quantification of micronucleus-containing WT (n = 4,925) and KO (n = 5,302) cells from four independent experiments. Bars, 20 μm. Values are means ± SEM. **, P < 0.005 (Student's t test).
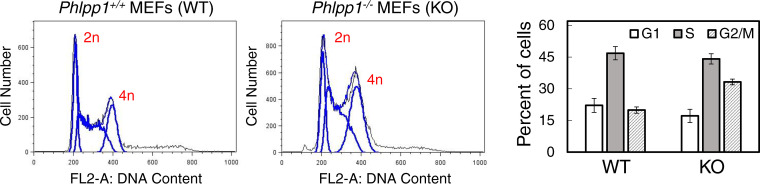
To further explore the effect of PHLPP1 loss on the cell cycle, we performed flow cytometry on propidium iodide-stained WT and Phlpp1−/− MEFs to determine DNA content. A higher percentage of cells were in G2/M in the Phlpp1−/− MEFs than in WT cells (33% ± 1% versus 20% ± 1%, respectively [Fig. 6]), complementing the results of the live-cell imaging experiments. Thus, cells lacking PHLPP1 exhibit subtle but measurable mitotic defects that could result in mitotic delays. Whether these defects result from the mitotic function of the PHLPP1 NTE remains to be elucidated.
FIG 6.
Cell cycle profile of WT and Phlpp1−/− MEFs. Ethanol fixed WT or Phlpp1−/− MEFs were treated with RNase, stained with propidium iodide, and analyzed by flow cytometry to determine their cell cycle profile. To determine the percentage of cells at various stages of the cell cycle (G1, S, or G2/M), DNA content (2n versus 4n) was assessed. The graph shows the percentage of cells in each phase of the cell cycle. Values are means ± SEM. Data are averages from four experiments performed independently.
DISCUSSION
Here, we identify the tumor suppressor phosphatase PHLPP1 as a component of the intricate machinery that maintains accuracy during the cell cycle, with its loss causing errors in cell division (Fig. 7). We show that PHLPP1 is a substrate for Cdk1, the master regulator of mitosis. Studies in vitro and in cells reveal that Cdk1 phosphorylates PHLPP1 at multiple S/T-P sites on the NTE as cells enter mitosis and that this region of the protein determines the cellular interactome of the phosphatase. The finding that PHLPP1 is involved in cell cycle regulation unveils a new player to oppose the plethora of phosphorylation events associated with cell division.
FIG 7.
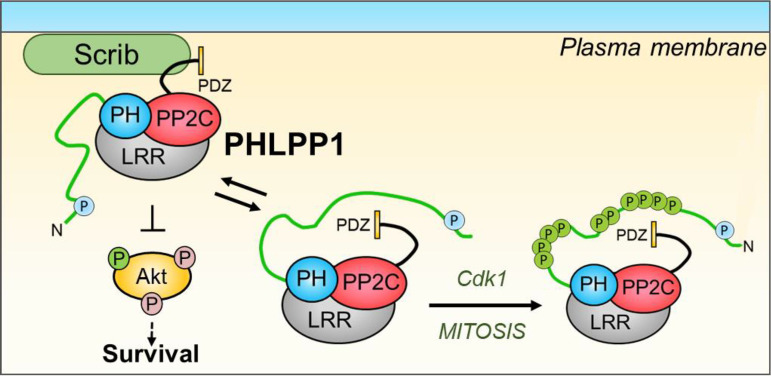
Proposed model. During interphase, cytoplasmic unphosphorylated PHLPP1 functions to regulate substrates such as Akt (left). PHLPP1 is in close proximity to membrane-associated scaffolds such as Scrib. Upon entry into mitosis, the PHLPP1 NTE is hyperphosphorylated by Cdk1 (right). Additionally, a diminished interaction with Scrib and an enhanced interaction with mitotic scaffolds are observed. Deletion of PHLPP1 results in mitotic defects, suggesting that this pathway is important for proper mitotic progression.
The phosphorylation of PHLPP1 by Cdk1 as cells enter mitosis suggests that the phosphatase may be one of the mitotic targets of Cdk1, whose activity is necessary for faithful cell division. Proper and timely activation of Cdk1 is essential for the accurate completion of cell division; deletion of the gene encoding Cdk1 results in early embryonic lethality in mice (54, 55), and expression of a Cdk1 lacking inhibitory phosphorylation sites (Thr14Ala/Tyr15Ala) in adult mice results in DNA damage and eventual death (55). Furthermore, increased Cdk1 expression and activity are associated with cancer progression and poor prognosis (56–60). Complete inhibition of Cdk1 by the active-site inhibitor RO-3306 arrests cells at the G2/M transition (61), while partial inhibition results in mitotic delays and errors in chromatin segregation, in turn resulting in formation of micronuclei (62, 63). The finding that cells lacking PHLPP1, which we establish as a Cdk1 substrate, display mitotic delays and increased chromatin segregation errors identifies the phosphatase as one of the downstream targets of Cdk1 whose phosphorylation ensures mitotic fidelity.
Cdk1 phosphorylates the relatively understudied PHLPP1 NTE. This region of the protein accounts for almost 20% of the entire polypeptide (residues 1 to 512 out of 1,717) and is predicted to be intrinsically disordered and unstructured, a desirable attribute for a potential kinase suitor. A recent study also identified a nuclear localization signal (NLS) within the PHLPP1 NTE (28). Interestingly, PHLPP2 lacks this NTE and does not undergo the dramatic electrophoretic mobility shift observed for PHLPP1. Both biochemical and mass spectrometry results indicate that the NTE is phosphorylated at multiple sites by Cdk1 during mitotic entry and that the phosphorylated species is removed during mitotic exit, either by dephosphorylation or degradation of the phospho-protein. Our data do not elucidate whether Cdk1 actively discriminates which sites it phosphorylates or the stoichiometry of the phosphorylation, but given the large electrophoretic mobility shift and detection of intermediate species, many sites are modified per molecule.
Of the 13 sites identified, only one (Thr451) adheres to the optimal Cdk1 consensus sequence of S/T-P-X-K/R (38, 39). Mutation of these 13 sites to alanine, a residue that cannot be phosphorylated, abolishes the electrophoretic mobility shift, validating the mass spectrometry results. Our data suggest that this phosphorylation event is specific to Cdk1, as inhibiting other mitotic kinases (Aurora A/B, Polo-like kinase 1/3) or other proline-directed kinases (ERK, p38, and JNK) did not result in a reduction of PHLPP1 phosphorylation. Interestingly, the phosphorylated residues are roughly clustered in two regions (residues 317 to 345 and 409 to 451), consistent with reports that many Cdk1 substrates are phosphorylated multiple times in clusters within regions predicted to be intrinsically disordered (64). Furthermore, these clusters are reported to occur in large numbers at rapidly evolving regions rather than on conserved residues (64), suggesting that the degree of phosphorylation of a substrate could be more significant than the specific sites phosphorylated. Whether these phosphorylation events on the NTE, which contains an NLS, regulate nuclear PHLPP1 is an intriguing possibility. Namely, does this phosphorylation event occur before the breakdown of the nuclear envelope, and if so, does it function to alter localization of nuclear PHLPP1? These data identify the unique NTE of PHLPP1 as a phosphorylation switch that coordinates with the cell cycle.
BioID analysis identified large changes in the interactome of PHLPP1 through its NTE and, through changes observed upon nocodazole treatment, the phosphorylation state of the NTE. For this analysis, we created stable cell lines encoding only one copy of BirA*-FLAG-tagged PHLPP1 (or a construct lacking the NTE). While this preliminary screen provides a wealth of potential targets to further explore, they await further validation using techniques such as coimmunoprecipitation or proximity ligation assays to better characterize these potential interactions. In the BioID screen, plasma membrane-associated functions, such as amino acid transport and regulation of cell-cell anchoring junctions, were the primary proteins identified in proximity to full-length PHLPP1. Several of the identified proteins are known Cdk1 substrates, including filamin A (FLNA) (65), plectin (PLEC) (66), and cortactin (CTTN) (67). It is of note that several known PHLPP1 binding partners, including Scrib, USP1, WDR48, WDR20, and SGT1 (gene name SUGT1), were identified as hits in this screen. Deletion of the NTE resulted in enrichment of these proteins in the screen, suggesting that the NTE functions to preclude interactions with other protein partners. This includes Scrib as well as WDR48 (48), another established target that interacts with the CTE of PHLPP1. These data suggest that the NTE functions to reversibly shield the CTE and other binding determinants from potential binding partners. It is not currently known if the other targets in this set of known PHLPP1 interactors, namely, USP1, WDR20, and SGT1, also interact with PHLPP1 through the CTE. If they do, it would strengthen the finding that the NTE functions to regulate protein interactions with the CTE. Conversely, deletion of the NTE abolished the interaction with another set of proteins, notably mitotic spindle proteins such as TUBA1A. Thus, the NTE controls the PHLPP1 interactome.
The Cdk1-mediated phosphorylation of PHLPP1 during mitosis may provide a mechanism to control the reversible shielding of binding determinants on PHLPP1. Consistent with this, BioID revealed that association with Scrib and WDR48 was reduced in cells expressing full-length BirA*-FLAG-PHLPP1 during mitosis (where a phosphorylated NTE is present) compared to nonmitotic cells (unphosphorylated NTE). Furthermore, pulldown of Scrib and WDR48 during mitosis was restored if the truncated form of PHLPP1 (ΔNTE) was expressed. Thus, the reversible regulation of binding partners mediated by phosphorylation of the NTE may be an important regulatory mechanism that defines PHLPP1 signaling.
Our finding that PHLPP1 KO MEFs exhibit mitotic delays and have increased mitotic errors resulting in formation of micronuclei compared to WT MEFs suggests that PHLPP1 plays a role in maintaining mitotic fidelity. Whether these defects are attributed to the inability for PHLPP1 to localize and function properly via the NTE is an open question. A recent study reported that knockdown of PHLPP1 in HeLa cells results in an increased rate of abnormal spindle formation and centrosomal defects (34), supporting the possibility that loss of PHLPP1 directly affects centrosomal maturation and spindle formation. Our BioID screen identified several targets that are in a complex with the mitotic kinase Aurora A, a known regulator of mitotic spindle maturation. These proteins included TPX2, Ch-TOG/XMAP215 (CKAP5), KIF11, and HURP (DLGAP5) (68). Importantly, increased spectral counts of these targets were observed in nocodazole-treated cells compared to untreated cells, suggesting a potential increase in colocalization of PHLPP1 with these targets. Whether PHLPP1 regulates Aurora A kinase activity to maintain mitotic fidelity is a question that has yet to be explored. PHLPP1 was also observed to be proximal to multiple outer kinetochore-associated proteins, such as Bub1B, Knl1, Spindly (SPDL1), and CENP-F. Since the kinetochore functions as the bridge between microtubule spindle fibers and chromosomes during mitosis (69), its proper formation and function are critical for accurate and timely chromatin segregation (70). Whether PHLPP1 directly interacts with, and regulates, these kinetochore proteins during mitosis awaits further experimentation. Thus, this BioID screen has opened up several potential avenues to better understand the role of PHLPP1 in mitosis and why its loss would result in an increase in mitotic errors.
Another mechanism that could explain the mitotic defect phenotype observed in the PHLPP1 KO MEFs is through PHLPP1-dependent regulation of Akt activity. Several studies have shown that Akt is active during mitosis and that activation of Akt can override the G2/M checkpoint, even in the presence of DNA damage (71–73). Furthermore, Akt inhibition can result in abnormal spindle formation (74). The release of PHLPP1 from the Scrib scaffold during mitosis could provide a potential mechanism for how Akt activity is increased during mitosis. Tethering of PHLPP1 to Scrib allows better access to Akt (35), so its release from the scaffold would be expected to enhance Akt activity during mitosis indirectly by removing PHLPP1 from its substrate. Whether the phosphorylations on the NTE additionally alter the catalytic phosphatase activity of PHLPP1 by allosteric mechanisms has yet to be determined. Taken together, our data are consistent with a model in which Cdk1 phosphorylates the NTE of PHLPP1, inducing a conformational change that releases PHLPP1 from plasma membrane-bound substrates and unmasks determinants that promote binding to centrosomal proteins.
The identification of PHLPP1 as a novel mitotic phosphatase unveils novel insight into how PHLPP1 functions as a tumor suppressor, as errors in mitosis can result in aneuploidy, a hallmark of cancer. Thus, in addition to its previously characterized functions of suppressing signaling pathways, it plays a role in the fundamental process of cell division. Furthermore, the identification of the unstructured NTE as a key regulator of the PHLPP1 interactome suggests that allosteric modulators have the potential to control deregulated PHLPP1 signaling in disease. Our findings provide a new lens through which to view PHLPP1 function during cancer.
MATERIALS AND METHODS
Materials and antibodies.
The pharmacological reagents used in this study include biotin (BB0078; BioBasic), calyculin A (9902; Cell Signaling), GW843682X (sc-203202; Santa Cruz Biotechnology), JNK-IN-8 (HY-13319; MedChemExpress), RO-3306 (270-463; Enzo Life Sciences), SB203580 (559389; Calbiochem), tetracycline (TB0504; BioBasic), UO-126 (662005; Calbiochem), and ZM-447439 (sc-200696; Santa Cruz Biotechnology). The pCDNA3-HA-tagged PHLPP1 construct was described previously (75). The pCDNA3-HA-tagged PHLPP1 N-terminal extension (NTE) construct was described previously (28). The pCDNA3-HA-tagged PHLPP1 ΔNTE construct was described previously (17). The PHLPP1 NTE was subcloned into the pCMV-3XFLAG vector (E4401; Sigma-Aldrich). The pCDNA3-HA-tagged PHLPP1 mitotic phosphorylation site mutant was generated by synthesizing a transgene of the NTE (amino acid residues 1 to 512) containing Ser/Thr-to-Ala mutations at mitotic phosphorylation sites (Ser50, Ser317, Ser321, Ser323, Ser324, Ser335, Ser336, Ser345, Thr409, Ser411, Ser412, Ser425, Ser450, and Thr451), as well as Ser-to-Ala mutations of nearby Ser residues that could function as compensating phosphorylation sites (Ser313, Ser347, Ser349, Ser351, Ser353, and Ser355). This transgene was then subcloned into either the pCDNA3-HA-PHLPP1 (full-length) vector or the pCDNA3-HA-NTE vector, and sequences were verified. Full-length PHLPP1 and the PHLPP1 NTE were subcloned via Gateway cloning (Thermo Fisher Scientific) into pDEST 5′ BirA*-FLAG pcDNA5 FRT TO (76). All DNA plasmid transfections were done using Effectene (301425; Qiagen), following the manufacturer’s instructions. Antibodies used in this study include Cdk1 (cdc2) total (77055; Cell Signaling), Cdk1 (cdc2) pY15 (9111; Cell Signaling), cyclin B1 (4138; Cell Signaling), ERK1/2 pT202/pY204 (9101; Cell Signaling), FLAG M2 (F3165; Sigma-Aldrich), H3 total (39163; Active Motif), H3 pS10 (39253; Active Motif), HA clone 3F10 (11867423001; Roche), Hsp90 (610418; BD Transductions), JNK pT183/Y185 (9251; Cell Signaling), p38 pT180/Y182 (9216; Cell Signaling), PHLPP1 (22789-1-AP; Proteintech), PHLPP2 (A300-661A; Bethyl Laboratories), tubulin (T6074; Sigma-Aldrich), and vinculin (13901; Cell Signaling).
Cell culture.
MEFs from WT or Phlpp1−/− mice stably expressing shp53 and green fluorescent protein (GFP) were a kind gift from the group of Lloyd Trotman (Cold Spring Harbor Laboratory [CSHL]) and were described previously (23). MEFs, HeLa cells (ATCC), HEK-293T cells, and HEK-293A cells (a kind gift from the lab of Kun-Liang Guan, University of California, San Diego [UCSD]) were grown in Dulbecco’s modified Eagle medium (DMEM; 10-013-CV; Corning), and RPE1 cells (a kind gift from the lab of Karen Oegema, UCSD) were grown in DMEM–Ham’s F-12 50/50 mix (10-092-CV; Corning). All cell lines were supplemented with 10% fetal bovine serum (S11150; Atlanta Biologicals) and 1% penicillin-streptomycin (15140-122; Gibco) and cultured at 37°C in 5% (vol/vol) CO2. Cells were periodically tested for Mycoplasma contamination using a PCR-based protocol (77) and showed no evidence of contamination.
Cell cycle synchronization.
To block cells in G1/S, a double-thymidine block was used. Thymidine (T9250; Sigma-Aldrich) was added to cells to a final concentration of 2 mM for 24 h. Thymidine-containing medium was then removed, and cells were washed four times with PBS and incubated with fresh medium for 8 h, after which thymidine was added to cells to a final concentration of 2 mM for 16 h, after which cells were lysed. To block cells in mitosis, a thymidine/nocodazole block was used. Following a single thymidine block, cells were washed and incubated in fresh medium for 3 h, followed by the addition of nocodazole (2190; Cell Signaling) to a final concentration of 50 ng/ml (RPE1 cells) or 100 ng/ml (HEK-293A and HeLa cells). After 16 h, mitotic cells were harvested by a mitotic shake-off. For mitotic release experiments, mitotic cells were pooled by mitotic shake-off, centrifuged at 150 × g for 3 min, and then resuspended in 10 ml phosphate-buffered saline (PBS). This step was repeated three times to ensure removal of nocodazole-containing medium. Cells were then resuspended in fresh growth medium, replated, and lysed at various time points.
Cell lysis and Western blotting.
Cells were lysed in buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μM microcystin, 20 μM benzamidine, and 40 μg/ml leupeptin and sonicated briefly. Following a 10-min incubation at 4°C, lysates were centrifuged, and soluble fractions were separated on SDS-PAGE gels or PhosTag gels containing 100 μM PhosTag reagent (a kind gift from the lab of Dario Alessi, University of Dundee) and 100 μM MnCl2, followed by transfer to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Membranes were blocked with 5% milk for 1 h at room temperature and analyzed by immunoblotting with specific antibodies. Detection and quantification of Western blots was conducted by chemiluminescence on a FluorChem Q imaging system (ProteinSimple).
Lambda phosphatase assay.
Cells were lysed in buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1% Triton X-100, 1 mM DTT, 1 mM PMSF, 20 μM benzamidine, and 40 μg/ml leupeptin. Following a 10-min incubation at 4°C, lysates were centrifuged, and soluble fractions were incubated with 1× NEBuffer for protein metallophosphatases (PMP), 1 mM MnCl2, and either 1 μl (400 U) lambda phosphatase (P0753; NEB) or 1 μl NEBuffer for PMP. Samples were incubated at room temperature for 30 min, after which reactions were terminated by the addition of protein sample buffer containing 250 mM Tris-HCl, 8% (wt/vol) SDS, 40% (vol/vol) glycerol, 80 μg/ml bromophenol blue, and 2.86 M β-mercaptoethanol.
FLAG-PHLPP1 NTE purification for phospho-mass spectrometry.
pCMV-3XFLAG-PHLPP1-NTE was transfected into HeLa cells using Effectene. Medium was replaced 24 h later with fresh medium containing either 100 ng/ml nocodazole or DMSO for 16 h, after which cells were lysed in a buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4, 1 mM DTT, 1 mM PMSF, 1 mM microcystin, 20 mM benzamidine, and 40 mg/ml leupeptin. The detergent-solubilized cell lysates were incubated with anti-FLAG M2 affinity gel (30 ml per plate; A2220; Sigma-Aldrich) for 1 h at 4°C, washed four times in lysis buffer, and then eluted using protein sample buffer. Eluted samples were analyzed by Western blotting to determine the presence of the FLAG epitope. The remainder of the samples were separated on an 8% SDS-PAGE gel. Following staining with Coomassie blue, relevant bands were excised from the gel and processed further for phospho-mass spectrometry.
Phospho-mass spectrometry of PHLPP1 NTE.
For in-gel digests, colloidal Coomassie gel slices were washed three times with 100 mM ammonium bicarbonate in 5 to 15% acetonitrile (ACN) for 15 min. The gel pieces were dried in a SpeedVac and subsequently reduced by mixing with 10 mM DTT in 100 mM ammonium bicarbonate at 56°C for 30 min. Gel pieces were dehydrated by incubation with 55 mM iodoacetamide in 100 mM ammonium bicarbonate, followed by increasing concentrations of acetonitrile–100 mM ammonium bicarbonate (50%, 75%, and 95%). For digestion, samples were incubated with trypsin (0.01 μg/μl) in 100 mM ammonium bicarbonate at 37°C overnight. Peptides were extracted twice by addition of 50 μl 0.2% formic acid and 65% ACN and vortexed at room temperature for 30 min. The supernatant was dried in a SpeedVac, after which a total of 50 μl 50% ACN–0.2% formic acid was added to the sample, followed by a 30-min vortexing at room temperature.
Trypsin-digested peptides were subsequently analyzed by ultra-high-pressure liquid chromatography (UPLC) coupled with tandem mass spectroscopy (LC-MS/MS) using nanospray ionization. The nanospray ionization experiments were performed using a TripleTOF 5600 hybrid mass spectrometer (AB SCIEX) interfaced with nanoscale reverse-phase UPLC (nanoACQUITY system; Waters Corporation) using a 20- to 75-μm ID glass capillary )packed with 2.5-μm C18 (130-Å) CSH beads (186007038; Waters Corporation) Peptides were eluted from the C18 column into the mass spectrometer using a linear gradient (5% to 80%) of ACN at a flow rate of 250 μl/min for 1 h. The buffers used to create the ACN gradient were buffer A (98% H2O, 2% ACN, 0.1% formic acid, and 0.005% trifluoroacetic acid [TFA]) and buffer B (100% ACN, 0.1% formic acid, and 0.005% TFA). MS/MS data were acquired in a data-dependent manner in which the MS1 data were acquired for 250 ms at m/z of 400 to 1,250 Da and the MS/MS data were acquired at m/z of 50 to 2,000 Da. The independent data acquisition (IDA) parameters were as follows; MS1-TOF (time of flight) acquisition time of 250 ms, followed by 50 MS2 events of 48 ms acquisition time for each event. The threshold to trigger the MS2 event was set to 150 counts when the ion had the charge state +2, +3, and +4. The ion exclusion time was set to 4 s.
The collected data were analyzed using Protein Pilot 5.0 (AB SCIEX) for peptide identifications. MS/MS spectra were searched against only human PHLPP1 (UniProtKB accession no. O60346 [PHLPP1_HUMAN]). Database parameters were set to allow 1 missed cleavage site per peptide with a mass tolerance of ±10 ppm, a maximum charge of 4+, and a minimum allowed peptide length of 6 amino acids. The following variable modifications were selected for: acetylation, amidation, carbamidomethylation, carboxylation, deamidation, dehydration, demethylation, formylation, formylation, oxidation, phosphorylation, and sodium adducts. Search parameters were set based on all possible posttranslationally modified peptides generated for this specific protein. To determine mitotic enrichment, the number of peptides identified that include the phosphorylated residue were counted (total spectral counts). The following formula was used to calculate the mitotic enrichment: (spectral countsnocodazole)/[1 + (spectral countsDMSO)]. Any phosphorylated residue with an enrichment score of ≥3 was designated a mitotic phosphorylation. Data analysis and scoring parameters can be found in Data Set S1 in the supplemental material.
Intrinsic disorder prediction.
To predict regions of intrinsic disorder in the PHLPP1 amino acid sequence, IUPred2A prediction software was used (https://iupred2a.elte.hu/) (36). The full-length PHLPP1 amino acid sequence was queried using the IUPred2 long disorder setting.
In vitro Cdk1 kinase assay.
pCMV-3XFLAG-PHLPP1-NTE was transfected into HEK-293T cells using Effectene. Cells were collected 72 h posttransfection and lysed in a buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4, 1 mM DTT, 1 mM PMSF, 1 mM microcystin, 20 mM benzamidine, and 40 mg/ml leupeptin. The detergent-solubilized cell lysates were incubated with anti-FLAG M2 affinity gel (30 ml per plate; A2220; Sigma-Aldrich) for 1 h at 4°C and washed four times in lysis buffer. FLAG-tagged NTE was eluted from the beads for 1 h at 4°C in a buffer containing 0.5 mg/ml 3×FLAG peptide (F4799; Sigma), 50 mM HEPES (pH 7.4), 100 mM NaCl, 1 mM DTT, 0.01% Igepal, and 10% glycerol. Samples were centrifuged, and the eluate was transferred to a new tube. Protein concentration and purity were measured by Bradford assay (5000006; Bio-Rad) and SDS-PAGE. Each reaction mixture contained 2 μg/ml purified FLAG-NTE, 0.01 μg/μl recombinant human Cdk1-cyclin B1 (14-450; EMD Millipore), and 1 mM ATP in a buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, and 0.01% Brij 35 (NEBuffer for protein kinases; B6022; NEB). Samples were incubated at 30°C for various times, and reactions were terminated by the addition of protein sample buffer, followed by Western blot analysis.
BioID. (i) Cell line generation and cell cycle synchronization.
BirA*-FLAG constructs were generated via Gateway cloning (Thermo Fisher Scientific) into pDEST 5′ BirA*-FLAG pcDNA5 FRT TO and used to generate stable cell lines in HeLa Flp-In T-REx cell pools, as described in reference 76. To obtain a mitotic population, cells were plated on a 150-mm plate and treated with thymidine (2 mM final concentration) and tetracycline (1 μg/ml final concentration) to induce expression of BirA*-tagged proteins. After 24 h, cells were washed twice with PBS and incubated in cell culture medium containing tetracycline (1 μg/ml final concentration). After 3 h, nocodazole (100 ng/μl final concentration) was added to the cells. After 4 h, biotin was added to the cells at a final concentration of 50 μM. Cells were harvested 12 h later. For asynchronous cells, the same treatment protocol was used, but the addition of thymidine and nocodazole was omitted.
(ii) Streptavidin affinity purification and data acquisition.
Cell pellets were resuspended in a 1:4 ratio of pellet weight to radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 1 mM EGTA, 4.5 mM MgCl2 and 0.4% SDS) supplemented with Benzonase (250 U/ml; E1014; Sigma-Aldrich) and 1× protease inhibitor cocktail (P8340; Sigma-Aldrich). Cells were lysed by one freeze-thaw cycle and then gently agitated on a nutating mixer at 4°C for 30 min, followed by centrifugation at 16,000 × g for 20 min at 4°C. The supernatant was transferred to 1.5-ml microcentrifuge tubes, and 25 μl of 60% streptavidin-Sepharose bead slurry (17-5113-01; GE Healthcare) was added. Following overnight incubation at 4°C with rocking, beads were washed with 2% SDS buffer (2% SDS, 50 mM Tris [pH 7.5]), twice with RIPA buffer, and three times with 50 mM ammonium bicarbonate, pH 8.0 (ABC). After removal of the last wash solution, beads were resuspended in 50 mM ABC (100 μl) with 1 μg of trypsin (T6567; Sigma-Aldrich) and rocked at 37°C for 4 h. Then, an additional 1 μg of trypsin (in 2 μl of 50 mM ABC) was added to each sample, and samples were incubated at 37°C with rocking overnight. Beads were centrifuged at 500 × g for 1 min, and the supernatant (pooled peptides) was transferred to a new tube. Beads were rinsed with MS-grade H2O (100 μl), and the rinse solution was combined with the pooled peptide sample. Formic acid (10%) was added to the supernatant to a final concentration of 2%. The pooled supernatant was centrifuged at 16,000 × g for 5 min to pellet remaining beads. A 230-μl portion of the pooled supernatant was transferred to a new 1.5-ml microcentrifuge tube, and samples were dried using a vacuum concentrator. Tryptic peptides were resuspended in 10 μl of 5% formic acid, and 2.5 μl was used for each analysis.
Peptides were analyzed by nano high-pressure liquid chromatography (nano-HPLC) coupled to MS. Nanospray emitters were generated from fused silica capillary tubing (100-μm internal diameter, 365-μm outer diameter) using a laser puller (Sutter Instrument Co. model P-2000; heat = 280, filament = 0, velocity = 18, delay = 2,000). Nanospray emitters were packed with C18 reverse-phase material (Reprosil-Pur 120 C18-AQ; 3 μm) in methanol using a pressure injection cell. A 5-μl portion of sample (2.5 μl of each sample with 3.5 μl of 5% formic acid) was directly loaded at 800 nl/min for 20 min onto a 100-μm by 15-cm nanospray emitter. Peptides were eluted from the column with an acetonitrile gradient generated by an Eksigent ekspert nanoLC 425 system and analyzed on a TripleTOF6600 instrument (AB SCIEX, Concord, Ontario, Canada). The gradient was delivered at 400 nl/min from 2% acetonitrile with 0.1% formic acid to 35% acetonitrile with 0.1% formic acid using a linear gradient of 90 min. This was followed by a 15-min wash with 80% acetonitrile with 0.1% formic acid and equilibration for another 15 min to 2% acetonitrile with 0.1% formic acid, resulting in a total of 120 min for the data-dependent acquisition (DDA) protocol. The first MS1 scan had an accumulation time of 250 ms within a mass range of 400 to 1,800 Da. This was followed by 10 MS/MS scans of the top 10 peptides identified in the first DDA scan, with an accumulation time of 100 ms for each MS/MS scan. Each candidate ion was required to have a charge state from 2 to 5 and a minimum threshold of 300 cps, isolated using a window of 50 mDa. Previously analyzed candidate ions were dynamically excluded for 7 s.
(iii) MS data analysis.
Mass spectrometry data generated were stored, searched, and analyzed using ProHits laboratory information management system (LIMS) platform (78). Within ProHits, WIFF files were converted to an MGF format using the WIFF2MGF converter and to a mzML format using ProteoWizard (V3.0.10702) and the AB SCIEX MS data converter (V1.3 beta). The DDA data were then searched using Mascot (V2.3.02) (79) and Comet (V2016.01 rev.2) (80). The spectra were searched with the human and adenovirus sequences in the RefSeq database (version 57, 30 January 2013) acquired from NCBI, supplemented with “common contaminants” from the Max Planck Institute (http://maxquant.org) and the Global Proteome Machine (GPM; ftp://ftp.thegpm.org/fasta/cRAP/crap.fasta), forward and reverse sequences (labeled gi|9999 or DECOY), sequence tags (BirA, glutathione S-transferase 26 [GST26], mCherry, and GFP), and streptavidin, for a total of 72,481 entries. Database parameters were set to search for tryptic cleavages, allowing up to 2 missed cleavage sites per peptide with a mass tolerance of 35 ppm for precursors with charges of 2+ to 4+ and a tolerance of 0.15 amu for fragment ions. Variable modifications were selected for deamidated asparagine and glutamine and for oxidized methionine. Results from each search engine were analyzed through TPP (the Trans-Proteomic Pipeline, v.4.7, POLAR VORTEX rev 1) via the iProphet pipeline (81). Data analysis can be found in Data Set S2.
(iv) SAINT analysis.
SAINTexpress (version 3.6.1) was used as a statistical tool to calculate the probability of potential protein-protein interactions from background contaminants using default parameters (43). SAINT analysis was performed using two biological replicates per bait for DDA. Four negative-control experiments were used for the SAINT analysis: 2 biological replicates each of empty-3×FLAG and empty-BirA*FLAG stable cell lines, treated with DMSO. Controls were not compressed, and two unique peptide ions and a minimum iProphet probability of 0.99 were required for protein identification. SAINT probabilities were calculated independently for each sample, averaged (AvgP) across biological replicates, and reported as the final SAINT score. Only SAINT scores with a false discovery rate (FDR) of ≤1% were considered high-confidence protein interactions. Data were visualized using ProHitz-viz (82). Data analysis can be found in Data Set S2.
Time-lapse imaging of H2B-mRFP-expressing MEFs.
WT or Phlpp1−/− KO MEFs stably expressing H2B-mRFP were generated using lentivirus. Briefly, HEK-293T cells were transfected with H2B-mRFP cloned into the pCDH-EF1 vector (a kind gift from the lab of Karen Oegema, UCSD), as described in reference 83, and the packaging plasmids psPAX2 (12260; Addgene) and pMD2.G (12259; Addgene), using Fugene 6 (E2691; Promega). Forty-eight hours after transfection, virus-containing culture was added to WT and KO MEFs along with 8 μg/ml Polybrene. Fluorescence-activated cell sorting (FACS) of RFP-expressing cells was performed to isolate single clones to generate clonal lines. One day prior to imaging, cells were plated in 8-well μ-slides (80826; Ibidi). Time-lapse imaging was performed on a deconvolution microscope (DeltaVision Elite; Applied Precision) equipped with a charge-coupled device camera (pco.edge 5.5 sCMOS; PCO) and a 60× 1.42 numerical aperture (NA) PlanApo N objective (Olympus). z-stacks (5 by 5 μm) without binning were acquired at 2 min 30 s intervals over a 14-h period using an environmental control chamber set to 37°C in 5% (vol/vol) CO2. For mRFP-tagged H2B, 5% illumination intensity and 50 ms exposure were used. Images were analyzed using ImageJ (NIH) and scored on four separate parameters: (i) time (in minutes) from nuclear envelope breakdown (NEBD) to anaphase, (ii) percentage of mitotic cells exhibiting chromatin segregation errors, (iii) percentage of mitotic cells exhibiting multipolar spindle formation, and (iv) percentage of mitotic cells that formed an anaphase bridge. For both the WT and KO MEFs, three clonal lines were imaged and analyzed separately, and the results from each analysis were combined to calculate an average and standard error of the mean (SEM). Statistical significance was determined using Student's t test.
Micronucleus quantification.
WT and Phlpp1−/− KO MEFs were plated on glass coverslips in a 6-well plate. Medium was removed 24 h after plating, and cells were washed with PBS, stained, and fixed with DAPI (10236276001; Roche) diluted in 100% methanol (1-μg/ml final concentration) for 15 min at 37°C. Cover slips with methanol-fixed cells were washed with PBS, and images were acquired on a Zeiss Axiovert 200M microscope (Carl Zeiss Microimaging Inc.) using an iXon Ultra 888 electron-multiplying charge-coupled device (EMCCD) camera (Andor) controlled by MetaFluor software (Molecular Devices). Imaging analysis was performed on ImageJ software (NIH). For each slide, multiple fields of view were quantified. The number of cells with micronuclei were divided by the total number of cells counted, generating the percentage of cells with micronuclei. Data over three separate experiments were averaged, and SEM were calculated. Statistical significance was determined using Student's t test.
Cell cycle analysis by flow cytometry.
WT and Phlpp1−/− KO MEFs were cultured at low confluence in 10-cm2 plates (100,000 cells/plate). As a separate control, a pool of WT and Phlpp1−/− KO MEFs were serum starved for 48 h to arrest cells in G1. The following day, cells were trypsinized and collected by centrifugation (200 × g, 8 min, 4°C). Cells were then washed with PBS and centrifuged (200 × g, 8 min, 4°C), followed by fixation with 70% (vol/vol) cold ethanol with gentle vortexing to ensure proper fixation of the cells and to minimize aggregation. Following 48 h of incubation in 70% ethanol at 4°C, fixed cells were centrifuged (200 × g, 8 min, 4°C), washed with PBS, and treated with RNase (100 μg/ml) for 30 min at 37°C. Cells were washed with PBS to remove RNase, centrifuged, resuspended in PBS containing propidium iodide (50 μg/ml; P3566; Thermo Scientific), and incubated for 1 h at room temperature in the dark prior to running samples on a FACSCalibur (BD) 4-color, dual-laser benchtop flow cytometer. Data were analyzed with FlowJo software (BD) using the Watson pragmatic approach (84).
Data availability.
The BioID data have been deposited as a complete submission in the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) and assigned accession number MSV000085628. The ProteomeXchange accession number is PXD020003 (http://www.proteomexchange.org).
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Newton and Dixon laboratories for helpful discussion. We kindly thank Hannah Tovell for assistance with revisions and Sourav Banerjee (UCSD) for performing FACS for live-cell imaging experiments. We thank Lloyd Trotman’s lab (CSHL) for the WT and Phlpp1−/− MEFs, Kun Liang Guan’s lab (UCSD) for the HEK-293A cells, and Karen Oegema’s lab (UCSD) for the RPE1 cells and the pCDH-EF1 vector. We thank Dario Alessi’s lab (University of Dundee) for the PhosTag reagent, Robert Tukey’s lab (UCSD) for the SB203580 inhibitor, and Michael Karin’s lab (UCSD) for the JNK-IN-8 inhibitor and the pJNK antibody. We thank Dennis Young (Moore’s Cancer Center, UCSD) for help with analysis of flow cytometry data.
This work was supported by NIH R35 GM122523 (A.C.N.), NIH GM067946 (A.C.N.), NIH R01 GM074215 (A.D.) and the Canadian Institutes of Health Research (CIHR FDN 144301 to A.-C.G.). BioID experiments were performed at the Network Biology Collaborative Centre at the Lunenfeld-Tanenbaum Research Institute, a facility supported by Canada Foundation for Innovation funding, by the Ontarian Government and by Genome Canada and Ontario Genomics (OGI-139). Flow cytometry was performed at the Flow Cytometry Shared Resource Service at the Moore’s Cancer Center (UCSD). A.T.K. was supported in part by the University of California, San Diego Graduate Training Grant in Cellular and Molecular Pharmacology through National Institutes of Health Institutional Training Grant T32 GM007752 from the NIGMS. A.-C.G. is the Canada Research Chair (Tier 1) in Functional Proteomics.
Author contributions are as follows: A.T.K., C.W., G.L., and C.C.K. performed the experiments. C.W. designed and performed the BioID screen under the supervision of A.-C.G. C.C.K. performed the phospho-mass spectrometry. G.L. performed experiments whose results are shown in Fig. 2H and I, Fig. 3C, and Fig. 6. A.T.K. performed the remaining experiments and generated the figures. P.L.-G. and A.D. provided assistance and equipment for time-lapse fluorescence microscopy. A.T.K. and A.C.N. wrote the manuscript, and all authors edited the manuscript. A.T.K. and A.C.N. conceived the project.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Naylor RM, van Deursen JM. 2016. Aneuploidy in cancer and aging. Annu Rev Genet 50:45–66. doi: 10.1146/annurev-genet-120215-035303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. 2011. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal 4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. 2008. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A 105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wurzenberger C, Gerlich DW. 2011. Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol 12:469–482. doi: 10.1038/nrm3149. [DOI] [PubMed] [Google Scholar]
- 5.King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. 1995. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 6.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. 1995. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell 6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irniger S, Piatti S, Michaelis C, Nasmyth K. 1995. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 8.Dou Z, von Schubert C, Körner R, Santamaria A, Elowe S, Nigg EA. 2011. Quantitative mass spectrometry analysis reveals similar substrate consensus motif for human Mps1 kinase and Plk1. PLoS One 6:e18793. doi: 10.1371/journal.pone.0018793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayward D, Alfonso-Pérez T, Cundell MJ, Hopkins M, Holder J, Bancroft J, Hutter LH, Novak B, Barr FA, Gruneberg U. 2019. CDK1-CCNB1 creates a spindle checkpoint-permissive state by enabling MPS1 kinetochore localization. J Cell Biol 218:1182–1199. doi: 10.1083/jcb.201808014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gharbi-Ayachi A, Labbé J-C, Burgess A, Vigneron S, Strub J-M, Brioudes E, Van-Dorsselaer A, Castro A, Lorca T. 2010. The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science 330:1673–1677. doi: 10.1126/science.1197048. [DOI] [PubMed] [Google Scholar]
- 11.Blake-Hodek KA, Williams BC, Zhao Y, Castilho PV, Chen W, Mao Y, Yamamoto TM, Goldberg ML. 2012. Determinants for activation of the atypical AGC kinase Greatwall during M phase entry. Mol Cell Biol 32:1337–1353. doi: 10.1128/MCB.06525-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vigneron S, Gharbi-Ayachi A, Raymond A-A, Burgess A, Labbé J-C, Labesse G, Monsarrat B, Lorca T, Castro A. 2011. Characterization of the mechanisms controlling Greatwall activity. Mol Cell Biol 31:2262–2275. doi: 10.1128/MCB.00753-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mailand N, Podtelejnikov AV, Groth A, Mann M, Bartek J, Lukas J. 2002. Regulation of G(2)/M events by Cdc25A through phosphorylation-dependent modulation of its stability. EMBO J 21:5911–5920. doi: 10.1093/emboj/cdf567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z, Jiang Q, Clarke PR, Zhang C. 2013. Phosphorylation of Crm1 by CDK1–cyclin-B promotes Ran-dependent mitotic spindle assembly. J Cell Sci 126:3417–3428. doi: 10.1242/jcs.126854. [DOI] [PubMed] [Google Scholar]
- 15.Grzechnik AT, Newton AC. 2016. PHLPPing through history: a decade in the life of PHLPP phosphatases. Biochem Soc Trans 44:1675–1682. doi: 10.1042/BST20160170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen MJ, Dixon JE, Manning G. 2017. Genomics and evolution of protein phosphatases. Sci Signal 10:eaag1796. doi: 10.1126/scisignal.aag1796. [DOI] [PubMed] [Google Scholar]
- 17.Gao T, Furnari F, Newton AC. 2005. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell 18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Brognard J, Sierecki E, Gao T, Newton AC. 2007. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell 25:917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Goel A, Arnold CN, Niedzwiecki D, Chang DK, Ricciardiello L, Carethers JM, Dowell JM, Wasserman L, Compton C, Mayer RJ, Bertagnolli MM, Boland CR. 2003. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res 63:1608–1614. [PubMed] [Google Scholar]
- 20.Johnson-Pais TL, Nellissery MJ, Ammerman DG, Pathmanathan D, Bhatia P, Buller CL, Leach RJ, Hansen MF. 2003. Determination of a minimal region of loss of heterozygosity on chromosome 18q21.33 in osteosarcoma. Int J Cancer 105:285–288. doi: 10.1002/ijc.11070. [DOI] [PubMed] [Google Scholar]
- 21.Rakha EA, Green AR, Powe DG, Roylance R, Ellis IO. 2006. Chromosome 16 tumor-suppressor genes in breast cancer. Genes Chromosomes Cancer 45:527–535. doi: 10.1002/gcc.20318. [DOI] [PubMed] [Google Scholar]
- 22.Tørring N, Borre M, Sørensen KD, Andersen CL, Wiuf C, Ørntoft TF. 2007. Genome-wide analysis of allelic imbalance in prostate cancer using the Affymetrix 50K SNP mapping array. Br J Cancer 96:499–506. doi: 10.1038/sj.bjc.6603476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, O'Neill A, Castillo-Martin M, Nowak DG, Naguib A, Grace DM, Murn J, Navin N, Atwal GS, Sander C, Gerald WL, Cordon-Cardo C, Newton AC, Carver BS, Trotman LC. 2011. Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell 20:173–186. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Stevens PD, Liu J, Yang H, Wang W, Wang C, Zeng Z, Schmidt MD, Yang M, Lee EY, Gao T. 2014. PHLPP is a negative regulator of RAF1, which reduces colorectal cancer cell motility and prevents tumor progression in mice. Gastroenterology 146:1301–1312.E10. doi: 10.1053/j.gastro.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Stevens PD, Li X, Schmidt MD, Gao T. 2011. PHLPP-mediated dephosphorylation of S6K1 inhibits protein translation and cell growth. Mol Cell Biol 31:4917–4927. doi: 10.1128/MCB.05799-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baffi TR, Van AN, Zhao W, Mills GB, Newton AC. 2019. Protein kinase C quality control by phosphatase PHLPP1 unveils loss-of-function mechanism in cancer. Mol Cell 74:378–392.E5. doi: 10.1016/j.molcel.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao T, Brognard J, Newton AC. 2008. The phosphatase PHLPP controls the cellular levels of protein kinase C. J Biol Chem 283:6300–6311. doi: 10.1074/jbc.M707319200. [DOI] [PubMed] [Google Scholar]
- 28.Cohen Katsenelson K, Stender JD, Kawashima AT, Lordén G, Uchiyama S, Nizet V, Glass CK, Newton AC. 2019. PHLPP1 counter-regulates STAT1-mediated inflammatory signaling. Elife 8:e48609. doi: 10.7554/eLife.48609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyes G, Niederst M, Cohen-Katsenelson K, Stender JD, Kunkel MT, Chen M, Brognard J, Sierecki E, Gao T, Nowak DG, Trotman LC, Glass CK, Newton AC. 2014. Pleckstrin homology domain leucine-rich repeat protein phosphatases set the amplitude of receptor tyrosine kinase output. Proc Natl Acad Sci U S A 111:E3957–E3965. doi: 10.1073/pnas.1404221111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patterson SJ, Han JM, Garcia R, Assi K, Gao T, O'Neill A, Newton AC, Levings MK. 2011. Cutting edge: PHLPP regulates the development, function, and molecular signaling pathways of regulatory T cells. J Immunol 186:5533–5537. doi: 10.4049/jimmunol.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang SM, Feigenson M, Begun DL, Shull LC, Culley KL, Otero M, Goldring MB, Ta LE, Kakar S, Bradley EW, Westendorf JJ. 2018. Phlpp inhibitors block pain and cartilage degradation associated with osteoarthritis. J Orthop Res 36:1487–1497. doi: 10.1002/jor.23781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradley EW, Carpio LR, Westendorf JJ. 2013. Histone deacetylase 3 suppression increases PH domain and leucine-rich repeat phosphatase (Phlpp)1 expression in chondrocytes to suppress Akt signaling and matrix secretion. J Biol Chem 288:9572–9582. doi: 10.1074/jbc.M112.423723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradley EW, Carpio LR, Newton AC, Westendorf JJ. 2015. Deletion of the PH-domain and leucine-rich repeat protein phosphatase 1 (Phlpp1) increases fibroblast growth factor (Fgf) 18 expression and promotes chondrocyte proliferation. J Biol Chem 290:16272–16280. doi: 10.1074/jbc.M114.612937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gangula NR, Maddika S. 2017. Interplay between the phosphatase PHLPP1 and E3 ligase RNF41 stimulates proper kinetochore assembly via the outer-kinetochore protein SGT1. J Biol Chem 292:13947–13958. doi: 10.1074/jbc.M117.782896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Yang H, Liu J, Schmidt MD, Gao T. 2011. Scribble-mediated membrane targeting of PHLPP1 is required for its negative regulation of Akt. EMBO Rep 12:818–824. doi: 10.1038/embor.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mészáros B, Erdos G, Dosztányi Z. 2018. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res 46:W329–W337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malumbres M. 2014. Cyclin-dependent kinases. Genome Biol 15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. 1994. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol 4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 39.Mok J, Kim PM, Lam HYK, Piccirillo S, Zhou X, Jeschke GR, Sheridan DL, Parker SA, Desai V, Jwa M, Cameroni E, Niu H, Good M, Remenyi A, Ma J-LN, Sheu Y-J, Sassi HE, Sopko R, Chan CSM, De Virgilio C, Hollingsworth NM, Lim WA, Stern DF, Stillman B, Andrews BJ, Gerstein MB, Snyder M, Turk BE. 2010. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal 3:ra12. doi: 10.1126/scisignal.2000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, Barnes G. 2003. Kinetochore protein interactions and their regulation by the aurora kinase Ipl1p. Mol Biol Cell 14:3342–3355. doi: 10.1091/mbc.e02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roux KJ, Kim DI, Burke B, May DG. 2018. BioID: a screen for protein-protein interactions. Curr Protoc Protein Sci 91:19.23.1–19.23.15. doi: 10.1002/cpps.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi-Rhee E, Schulman H, Cronan JE. 2004. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci 13:3043–3050. doi: 10.1110/ps.04911804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras AC, Choi H. 2014. SAINTexpress: improvements and additional features in significance analysis of INTeractome software. J Proteomics 100:37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan S, Meyer AS, Weiler SME, Rupp C, Tóth M, Sticht C, Singer S, Thomann S, Roessler S, Schorpp-Kistner M, Schmitt J, Gretz N, Angel P, Tschaharganeh DF, Marquardt J, Schirmacher P, Pinna F, Breuhahn K. 2018. Cytoplasmic localization of the cell polarity factor scribble supports liver tumor formation and tumor cell invasiveness. Hepatology 67:1842–1856. doi: 10.1002/hep.29669. [DOI] [PubMed] [Google Scholar]
- 45.Zhiqiang Z, Qinghui Y, Yongqiang Z, Jian Z, Xin Z, Haiying M, Yuepeng G. 2012. USP1 regulates AKT phosphorylation by modulating the stability of PHLPP1 in lung cancer cells. J Cancer Res Clin Oncol 138:1231–1238. doi: 10.1007/s00432-012-1193-3. [DOI] [PubMed] [Google Scholar]
- 46.Goldbraikh D, Neufeld D, Eid-Mutlak Y, Lasry I, Gilda JE, Parnis A, Cohen S. 2020. USP1 deubiquitinates Akt to inhibit PI3K-Akt-FoxO signaling in muscle during prolonged starvation. EMBO Rep 21:e48791. doi: 10.15252/embr.201948791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, Lu X, Akhter S, Georgescu MM, Legerski RJ. 2016. FANCI is a negative regulator of Akt activation. Cell Cycle 15:1134–1143. doi: 10.1080/15384101.2016.1158375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gangula NR, Maddika S. 2013. WD repeat protein WDR48 in complex with deubiquitinase USP12 suppresses Akt-dependent cell survival signaling by stabilizing PH domain leucine-rich repeat protein phosphatase 1 (PHLPP1). J Biol Chem 288:34545–34554. doi: 10.1074/jbc.M113.503383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gagnon D, Lehoux M, Archambault J. 2015. Artificial recruitment of UAF1-USP complexes by a PHLPP1-E1 chimeric helicase enhances human papillomavirus DNA replication. J Virol 89:6227–6239. doi: 10.1128/JVI.00560-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McClurg UL, Summerscales EE, Harle VJ, Gaughan L, Robson CN. 2014. Deubiquitinating enzyme Usp12 regulates the interaction between the androgen receptor and the Akt pathway. Oncotarget 5:7081–7092. doi: 10.18632/oncotarget.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao G, Kun T, Sheng Y, Qian M, Kong F, Liu X, Yu Z, Zhang H, Zhang Q, Gu J, Zhang X. 2013. SGT1 regulates Akt signaling by promoting beta-TrCP-dependent PHLPP1 degradation in gastric cancer cells. Mol Biol Rep 40:2947–2953. doi: 10.1007/s11033-012-2363-8. [DOI] [PubMed] [Google Scholar]
- 52.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. 2007. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res 35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalsbeek D, Golsteyn RM. 2017. G2/M-phase checkpoint adaptation and micronuclei formation as mechanisms that contribute to genomic instability in human cells. Int J Mol Sci 18:2344. doi: 10.3390/ijms18112344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M, Barbacid M. 2007. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 55.Szmyd R, Niska-Blakie J, Diril MK, Renck Nunes P, Tzelepis K, Lacroix A, van Hul N, Deng L-W, Matos J, Dreesen O, Bisteau X, Kaldis P. 2019. Premature activation of Cdk1 leads to mitotic events in S phase and embryonic lethality. Oncogene 38:998–1018. doi: 10.1038/s41388-018-0464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piao J, Zhu L, Sun J, Li N, Dong B, Yang Y, Chen L. 2019. High expression of CDK1 and BUB1 predicts poor prognosis of pancreatic ductal adenocarcinoma. Gene 701:15–22. doi: 10.1016/j.gene.2019.02.081. [DOI] [PubMed] [Google Scholar]
- 57.Ravindran Menon D, Luo Y, Arcaroli JJ, Liu S, KrishnanKutty LN, Osborne DG, Li Y, Samson JM, Bagby S, Tan A-C, Robinson WA, Messersmith WA, Fujita M. 2018. CDK1 interacts with Sox2 and promotes tumor initiation in human melanoma. Cancer Res 78:6561–6574. doi: 10.1158/0008-5472.CAN-18-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasukawa M, Ando Y, Yamashita T, Matsuda Y, Shoji S, Morioka MS, Kawaji H, Shiozawa K, Machitani M, Abe T, Yamada S, Kaneko MK, Kato Y, Furuta Y, Kondo T, Shirouzu M, Hayashizaki Y, Kaneko S, Masutomi K. 2020. CDK1 dependent phosphorylation of hTERT contributes to cancer progression. Nat Commun 11:1557. doi: 10.1038/s41467-020-15289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heo J, Noh B-J, Lee S, Lee H-Y, Kim Y, Lim J, Ju H, Yu HY, Ryu C-M, Lee PCW, Jeong H, Oh Y, Kim K, Kim S-Y, Son J, Hong B, Kim JS, Cho YM, Shin D-M. 2020. Phosphorylation of TFCP2L1 by CDK1 is required for stem cell pluripotency and bladder carcinogenesis. EMBO Mol Med 12:e10880. doi: 10.15252/emmm.201910880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong S, Huang F, Zhang H, Chen Q. 2019. Overexpression of BUB1B, CCNA2, CDC20, and CDK1 in tumor tissues predicts poor survival in pancreatic ductal adenocarcinoma. Bioscience Rep 39:BSR20182306. doi: 10.1042/BSR20182306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. 2006. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A 103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voets E, Marsman J, Demmers J, Beijersbergen R, Wolthuis R. 2015. The lethal response to Cdk1 inhibition depends on sister chromatid alignment errors generated by KIF4 and isoform 1 of PRC1. Sci Rep 5:14798. doi: 10.1038/srep14798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCloy RA, Rogers S, Caldon CE, Lorca T, Castro A, Burgess A. 2014. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holt LJ, Tuch BB, Villén J, Johnson AD, Gygi SP, Morgan DO. 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cukier IH, Li Y, Lee JM. 2007. Cyclin B1/Cdk1 binds and phosphorylates Filamin A and regulates its ability to cross-link actin. FEBS Lett 581:1661–1672. doi: 10.1016/j.febslet.2007.03.041. [DOI] [PubMed] [Google Scholar]
- 66.Foisner R, Malecz N, Dressel N, Stadler C, Wiche G. 1996. M-phase-specific phosphorylation and structural rearrangement of the cytoplasmic cross-linking protein plectin involve p34cdc2 kinase. Mol Biol Cell 7:273–288. doi: 10.1091/mbc.7.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blethrow JD, Glavy JS, Morgan DO, Shokat KM. 2008. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci U S A 105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koffa MD, Casanova CM, Santarella R, Köcher T, Wilm M, Mattaj IW. 2006. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol 16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 69.Musacchio A, Desai A. 2017. A molecular view of kinetochore assembly and function. Biology 6:5. doi: 10.3390/biology6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levine MS, Holland AJ. 2018. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev 32:620–638. doi: 10.1101/gad.314351.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kandel ES, Skeen J, Majewski N, Di Cristofano A, Pandolfi PP, Feliciano CS, Gartel A, Hay N. 2002. Activation of Akt/protein kinase B overcomes a G(2)/m cell cycle checkpoint induced by DNA damage. Mol Cell Biol 22:7831–7841. doi: 10.1128/mcb.22.22.7831-7841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shtivelman E, Sussman J, Stokoe D. 2002. A role for PI 3-kinase and PKB activity in the G2/M phase of the cell cycle. Curr Biol 12:919–924. doi: 10.1016/s0960-9822(02)00843-6. [DOI] [PubMed] [Google Scholar]
- 73.Dangi S, Cha H, Shapiro P. 2003. Requirement for phosphatidylinositol-3 kinase activity during progression through S-phase and entry into mitosis. Cell Signal 15:667–675. doi: 10.1016/s0898-6568(03)00002-0. [DOI] [PubMed] [Google Scholar]
- 74.Liu X, Shi Y, Woods KW, Hessler P, Kroeger P, Wilsbacher J, Wang J, Wang JY, Li C, Li Q, Rosenberg SH, Giranda VL, Luo Y. 2008. Akt inhibitor a-443654 interferes with mitotic progression by regulating aurora a kinase expression. Neoplasia 10:828–837. doi: 10.1593/neo.08408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Warfel NA, Niederst M, Stevens MW, Brennan PM, Frame MC, Newton AC. 2011. Mislocalization of the E3 ligase, β-transducin repeat-containing protein 1 (β-TrCP1), in glioblastoma uncouples negative feedback between the pleckstrin homology domain leucine-rich repeat protein phosphatase 1 (PHLPP1) and Akt. J Biol Chem 286:19777–19788. doi: 10.1074/jbc.M111.237081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kean MJ, Couzens AL, Gingras A-C. 2012. Mass spectrometry approaches to study mammalian kinase and phosphatase associated proteins. Methods 57:400–408. doi: 10.1016/j.ymeth.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 77.Uphoff CC, Drexler HG. 2004. Detecting Mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol Med 88:319–326. doi: 10.1385/1-59259-406-9:319. [DOI] [PubMed] [Google Scholar]
- 78.Liu G, Knight JD, Zhang JP, Tsou CC, Wang J, Lambert JP, Larsen B, Tyers M, Raught B, Bandeira N, Nesvizhskii AI, Choi H, Gingras AC. 2016. Data independent acquisition analysis in ProHits 4.0. J Proteomics 149:64–68. doi: 10.1016/j.jprot.2016.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. doi:. [DOI] [PubMed] [Google Scholar]
- 80.Eng JK, Jahan TA, Hoopmann MR. 2013. Comet: an open-source MS/MS sequence database search tool. Proteomics 13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 81.Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. 2011. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics 10:M111.007690. doi: 10.1074/mcp.M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Knight JDR, Choi H, Gupta GD, Pelletier L, Raught B, Nesvizhskii AI, Gingras AC. 2017. ProHits-viz: a suite of web tools for visualizing interaction proteomics data. Nat Methods 14:645–646. doi: 10.1038/nmeth.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meitinger F, Anzola JV, Kaulich M, Richardson A, Stender JD, Benner C, Glass CK, Dowdy SF, Desai A, Shiau AK, Oegema K. 2016. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J Cell Biol 214:155–166. doi: 10.1083/jcb.201604081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Watson JV, Chambers SH, Smith PJ. 1987. A pragmatic approach to the analysis of DNA histograms with a definable G1 peak. Cytometry 8:1–8. doi: 10.1002/cyto.990080101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The BioID data have been deposited as a complete submission in the MassIVE repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) and assigned accession number MSV000085628. The ProteomeXchange accession number is PXD020003 (http://www.proteomexchange.org).