Besides the ubiquitin-proteasome system, autophagy is a major degradation pathway within cells. It delivers invading pathogens, damaged organelles, aggregated proteins, and other macromolecules from the cytosol to the lysosome for bulk degradation. This so-called canonical autophagy activity contributes to the maintenance of organelle, protein, and metabolite homeostasis as well as innate immunity. Over the past years, numerous studies rapidly deepened our knowledge on the autophagy machinery and its regulation, driven by the fact that impairment of autophagy is associated with several human pathologies, including cancer, immune diseases, and neurodegenerative disorders.
KEYWORDS: canonical autophagy, noncanonical autophagy, LC3-associated endocytosis, LC3-associated phagocytosis, microglia, neurodegeneration
ABSTRACT
Besides the ubiquitin-proteasome system, autophagy is a major degradation pathway within cells. It delivers invading pathogens, damaged organelles, aggregated proteins, and other macromolecules from the cytosol to the lysosome for bulk degradation. This so-called canonical autophagy activity contributes to the maintenance of organelle, protein, and metabolite homeostasis as well as innate immunity. Over the past years, numerous studies rapidly deepened our knowledge on the autophagy machinery and its regulation, driven by the fact that impairment of autophagy is associated with several human pathologies, including cancer, immune diseases, and neurodegenerative disorders. Unexpectedly, components of the autophagic machinery were also found to participate in various processes that do not involve lysosomal delivery of cytosolic constituents. These functions are defined as noncanonical autophagy. Regarding neurodegenerative diseases, most research was performed in neurons, while for a long time, microglia received considerably less attention. Concomitant with the notion that microglia greatly contribute to brain health, the understanding of the role of autophagy in microglia expanded. To facilitate an overview of the current knowledge, here we present the fundamentals as well as the recent advances of canonical and noncanonical autophagy functions in microglia.
AUTOPHAGY PROCESSES
Maintaining cellular homeostasis involves controlled protein, organelle, and metabolite degradation systems, which are conserved among all eukaryotic cells. Autophagy, Greek for “self-eating,” is an intracellular recycling process by which cytoplasmic material is targeted for lysosomal degradation (1). Depending on the type of cargo delivery to the lysosome, three different autophagy processes can be classified. (i) Microautophagy describes the internalization of smaller cytosolic portions by inward-budding vesicles from the lysosomal membrane (2), whereas in (ii) chaperone-mediated autophagy, cytosolic proteins are specifically recognized by chaperones and taken up by the lysosome in a transporter-dependent manner (2). Finally, (iii) macroautophagy (here abbreviated autophagy) is characterized by the engulfment of cytosolic constituents by double-membrane structures, termed autophagosomes, which fuse with lysosomes (Fig. 1). Regardless of the delivery route, autophagic cargo is eventually digested by lysosomal enzymes, and the degradation products are released to the cytosol as new building blocks (3).
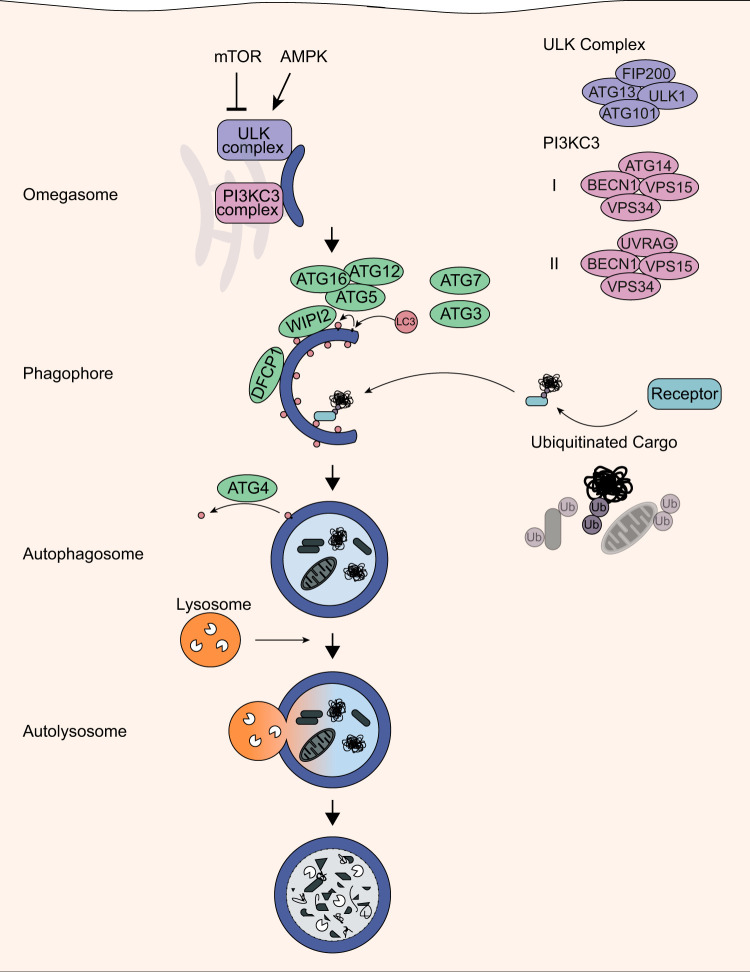
FIG 1.
Autophagy pathway. The energy sensors mTORC1 and AMPK control autophagy activation via the ULK1 complex. Following activation, the ULK1 and PI3KC3 complex regulate the formation of the omegasome, and cargo is then recruited by autophagic receptors to the phagophore. Finally, the mature autophagosome fuses with lysosomes to autolysosomes, and the cargo is degraded.
During autophagy, autophagy-related (ATG) proteins regulate the formation of the autophagosome in a hierarchical manner through different phosphorylating and ubiquitin-like conjugating events (Fig. 1). Initiation of autophagy requires assembly of the unc-51-like kinase 1 (ULK1) complex, including its subunits ULK1, ATG13, ATG101, and RB1-inducible coiled-coil protein 1 (RB1CC1), with dephosphorylation of ATG13 being a main triggering factor (4). Following activation, ULK1 phosphorylates and recruits the transmembrane protein ATG9 and class III phosphatidylinositol 3-kinase (PI3KC3) complex I, which in turn promote autophagosome biogenesis (5–7). Mammalian PI3KC3 complex I consists of a core composed of hVps34, hVps15, beclin-1 (BECN1), and ATG14 as well as associated factors such as activating molecule in beclin-1-regulated autophagy (AMBRA1) and nuclear receptor-binding factor 2 (NRBF2) (8, 9). PI3KC3 produces phosphatidylinositol 3-phosphate (PI3P), which then induces the formation of the omegasome, a membrane platform tightly associated with the endoplasmic reticulum (ER) that gives rise to a preautophagosomal structure termed phagophore or isolation membrane (10, 11). Enrichment of PI3P leads to the recruitment of WD repeat domain phosphoinositide-interacting protein 2 (WIPI2) and FYVE domain-containing protein 1 (DFCP1) to the omegasome (11, 12). Subsequently, WIPI2 orchestrates the conjugation of ubiquitin-like human ATG8 (hATG8) proteins to phosphatidylethanolamine (PE), which anchors them to the nascent phagophore (12). The family of hATG8 proteins comprises seven members which are categorized in two protein subfamilies: (i) the microtubule-associated proteins 1A/1B light chain 3 (LC3A, LC3B, LC3B2, and LC3C) and (ii) the γ-aminobutyric acid receptor-associated proteins (GABARAP, GABARAPL1, and GABARAPL2) (13). This conjugation is exerted by a ubiquitin-like machinery, where ATG7 is equivalent to an E1 activating enzyme, ATG3 to an E2 conjugating enzyme, and the ATG12-ATG5-ATG16 complex to an E3 scaffolding ligase (14). Upon lipidation to the concave phagophore membrane, hATG8 proteins are retained in the autophagosome and degraded along with its engulfed cargo, whereas hATG8-PE conjugates on the outer membrane of autophagosomes are eventually removed by the family of ATG4 hydrolases (15). The dynamics of LC3 and GABARAP conjugation play a central role in autophagosome formation by controlling the tethering and hemifusion of membranes (16, 17). In addition, LC3 was found to participate in transporting autophagosomes to lysosomes by forming a complex with Rab7 and its effector protein FYCO1, which facilitates microtubule transport of vesicles (18).
Importantly, the hierarchical orchestration of ATG proteins is not absolutely mandatory for autophagic degradation of cytosolic content. BECN1-independent autophagy, for example, can be induced, among other conditions, by resveratrol and was shown to be insensitive to the knockdown of PI3KC3 complex components such as BECN1 or hVps34 (19). In the following years, other independent forms were discovered, including the bypassing of ATG3, ATG5, ATG7, or ULK1 and ULK2 (20–22).
Most cell types exert basal levels of autophagy but can upregulate the pathway in response to a variety of stimuli, including nutrient deprivation. Thereby, autophagy provides the cell with metabolic building blocks for the synthesis of proteins, carbohydrates, and lipids. Key energy sensors are the mammalian target of rapamycin complex I (mTORC1) and AMP-activated protein kinase (AMPK), which induce bulk autophagic degradation of cytosolic constituents according to the nutrient status of the cell (23). As a negative autophagy regulator, mTORC1 couples sensing of amino acids, growth factors, and metabolic stress to the phosphorylation-mediated repression of the ULK1 complex at high-nutrient stages (24, 25). Decline of intracellular ATP levels triggers the activation of AMPK, which in turn positively controls ULK1 complex activation by direct phosphorylation of ULK1 and suppression of mTOR activity (26). The urge of the cell to overcome starvation and to promote cell survival requires autophagic activity to be elevated above basal levels and represents a rather nonselective pathway.
In contrast, selective autophagic degradation pathways exert cellular quality and quantity control through a number of soluble and membrane-bound receptors which mediate autophagic engulfment of specific substrates, including protein aggregates, damaged organelles, or pathogens (27–29). The ability of these autophagy receptors to recognize and tether their targets to the forming autophagosome is mediated by distinct functional domains. Here, polyubiquitination of cargo is a major signal for selective autophagy, allowing the binding of cargo by receptors via their ubiquitin-binding domain (UBD) (30, 31). It remains unclear how different polyubiquitin chains confer specificity for different autophagy receptors. Several studies demonstrated lysine-63 ubiquitination to be predominantly associated with autophagic turnover. However, in autophagy-deficient mice, several different types of polyubiquitin chains accumulate, suggesting that substrate oligomerization rather than ubiquitin topology is the driving signal for receptor selectivity (32–34).
Apart from target recognition, autophagy receptors directly interact with members of the hATG8 family, causing the autophagosome to zip around specific cargo (35). This interaction is mediated by the so-called LC3 interaction region (LIR), a characteristic linear motif among autophagy receptors and other hATG8-interacting proteins (36). Several autophagy receptors have been identified to carry both the UBD and LIR, including sequestosome 1 (SQSTM1; also known as p62), next to BRCA1 gene 1 (NBR1), optineurin (OPTN), calcium-binding and coiled-coil domain-containing protein 2 (CALCOCO2; alias, NDP52), Tax1-binding protein 1 (TAX1BP1), and Toll-interacting protein (TOLLIP) (37). Genetic mutations within some of these autophagy receptor domains are linked to neurodegenerative diseases, thus assigning selective autophagy as a contributor to preserving neuronal homeostasis (38).
Throughout the brain and spinal cord, tissue maintenance and removal of cellular debris is accomplished by parenchymal macrophages, the microglia (39). Given the vital role of microglia in the central nervous system (CNS), this article discusses the latest findings regarding microglial autophagy in context of the inflammatory response and different degradation pathways. Besides the degradation of cytosolic components by canonical autophagy, recently identified noncanonical pathways participate in the clearance of extracellular entities. Here, we focus on the molecular aspects of both autophagy pathway variants in microglia.
MICROGLIA
Microglia are the major resident immune cells of the central nervous system (CNS) that constitute up to 15% of all CNS cells. They arise early in development and originate from a pool of primitive macrophages derived from the embryonic yolk sac. During development as well as during adulthood, they contribute in multiple ways to overall brain function (40). Microglia are capable of orchestrating tissue homeostasis by releasing inflammatory and neurotrophic factors as well as by phagocytosis (41, 42). Phagocytosis is the ingestion and digestion of extracellular particles, for example, bacteria or dead cells. In an intact brain, microglial processes undergo rapid extensions and retractions, thereby scanning their environment for tissue abnormalities (43). They can sense disturbances due to a variety of neurotransmitter and immune receptors as well as ion channels (44–50). As a response, microglia can undergo multiple morphological and functional changes depending on microenvironmental factors and migrate to the site of injury. These transformations include the retraction of processes accompanied by a more amoeboid appearance as well as changes in signaling transduction, receptor expression, and phagocytic capacity. When microglia are faced with potentially harmful entities, they can recognize these through Toll-like receptors (TLRs), triggering receptor expressed on myeloid cells 2 (TREM2) or other receptors such as the mannose receptor (51–53). The various activated receptors trigger different signaling pathways, which initiate reorganization of the actin cytoskeleton and phagosome formation (54, 55). First, the target is engulfed by the plasma membrane, leading to interiorization, also called ingestion (56). Thereby, a membrane vesicle containing the particle, termed phagosome, buds inwards and fuses with lysosomes (Fig. 2A). This phagocytic mechanism is needed for the clearance of all kinds of harmful targets. Furthermore, microglial activation can result in the release of a broad range of pro- and anti-inflammatory factors such as cytokines or chemokines (57, 58). Interestingly, accumulating evidence indicates that autophagy plays a crucial role in the immune functions of microglia.
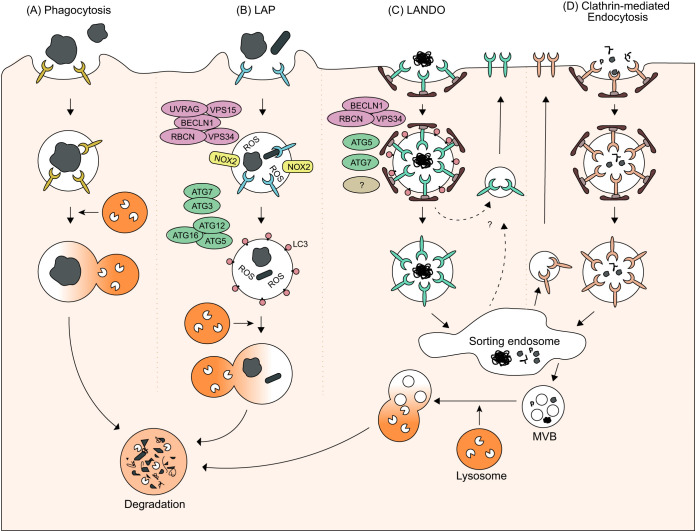
FIG 2.
Overview of phagocytosis, LC3-associated phagocytosis (LAP), LC3-associated endocytosis (LANDO), and clathrin-mediated endocytosis. (A) During phagocytosis, large extracellular particles are recognized by specific receptors, engulfed by the plasma membrane, and finally internalized by phagosomes. (B) When LC3 is recruited via the PI3KC3 II complex and other autophagy proteins to the phagosome before its fusion with the lysosome, the pathway is referred to as LAP. (C) When conjugation of LC3 by autophagy proteins to Rab5- and clathrin-positive endosomes is necessary for receptor recycling, the pathway is called LANDO. (D) Clathrin-mediated endocytosis describes the uptake of smaller extracellular cargo into clathrin-coated vesicles. After uncoating, the nascent vesicle is further transported within the cell, for example, to the sorting endosome.
CANONICAL AUTOPHAGY IN MICROGLIA
Canonical autophagy pathways follow a stepwise variable assembly of the autophagic machinery. While in most cells, there is no strict dependency on distinct components, autophagy induction often includes the association of the ULK1 kinase complex, the recruitment of the PI3KC3 complex, ATG9, PI3P effector proteins, and the conjugation of hATG8s to PE on the expanding phagophore. In this way, constituents from the cytosol are engulfed and delivered to lysosomes. Whether these autophagy components are compulsory for microglial autophagy is unknown. Within the CNS, autophagic activity has been mainly examined in neurons, leaving unanswered questions regarding microglia as the key players in neuronal immune responses in the brain (59).
Microglial autophagy and intracellular aggregates.
Accumulation of protein aggregates is a pathological hallmark of neurodegenerative diseases (60). Intracellular aggregates are predominantly found in neurons, whereas they are remarkably less abundant in microglia. Interestingly, recent studies showed that extracellular aggregates, which reach the cytosol of microglia, can be cleared by autophagy.
Synucleinphagy, the degradation of neuron-released α-synuclein by selective autophagy in microglia, was discovered by Choi and colleagues studying in vivo and in vitro models (61). α-Synuclein is thought to function in vesicular trafficking and is predominantly known due to its pathological contribution to Parkinson’s disease (PD), where it aggregates in neuronal inclusions called Lewy bodies (62, 63). Following overexpression of human α-synuclein in transgenic mice, microglia are activated and engulf extracellular α-synuclein deposits (61). Binding of α-synuclein to TLR4 on the microglial cell surface is accompanied by a significant increase of p62, whereas levels of other autophagy receptors remain unchanged (61). This process is mediated by nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), which regulates the transcription of p62 (61). Upregulated p62 is thought to associate with internalized α-synuclein to promote its sequestration in autophagosomes (61). This selective binding is assumed to be mediated by p62’s recognition of ubiquitinated α-synuclein (64). The mechanism of how α-synuclein is taken up by microglia and released into the cytosol remains unclear. Different from other TLR4 signaling, binding of α-synuclein does not stimulate TLR4 endocytosis, thus excluding phagocytosis or receptor-mediated endocytosis as an internalization mechanism (61, 65). Alternatively, α-synuclein might permeate the microglial cell membrane in a lipid raft-dependent manner (66). When exposed to neuronal SH-SY5Y cells, biochemically generated α-synuclein fibrils were shown to escape intracellular vesicles by rupturing their membrane after endocytosis (67). Consistent with this study, exogenous fibrillary α-synuclein is able to trigger lysosomal rupture following internalization by microglia-derived BV2 cells and primary microglia (68). In response to this damage, TANK-binding kinase 1 (TBK1), OPTN, and LC3 are recruited to ubiquitin earmarked rupture sites and drive the removal of irreparable lysosomes by autophagy (68).
Moreover, Cho and colleagues described a function of autophagy in the clearance of extracellular β-amyloid (Aβ) fibrils using primary microglia and BV2 cells (69). Here, autophagy initiation is thought to be dependent on AMPK and serine/threonine-protein kinase 11 (STK11) signaling (69, 70). OPTN and LC3B were found to coimmunoprecipitate with Aβ, and deletion of OPTN resulted in higher intracellular Aβ concentrations (69). Accordingly, OPTN was suggested to be an autophagy receptor that recruits the autophagy machinery to cytosolic Aβ via binding to LC3B (69). How Aβ aggregates are selectively targeted by OPTN is not yet understood. However, OPTN was also found to associate with protein aggregates in a ubiquitin-independent manner and to mediate their autophagic clearance (71). While the uptake of extracellular Aβ can be accomplished by receptor-mediated phagocytosis (72, 73), Aβ might leak through the phagosomal membrane, causing its release into the cytosol and thereby allowing the binding of OPTN for autophagic degradation (69).
Understanding the molecular dependencies between internalization of extracellular Aβ and microglial autophagy requires further investigations. For instance, TREM2 is capable of sensing Aβ through lipoproteins and mediates the phagocytic uptake of Aβ in microglia (74). Deletion of TREM2, in turn, has a severe impact on mTOR signaling and causes increased activation of autophagy (75).
Microglial autophagy and pathogen defense.
Autophagy is able to modulate the immune response, as it functions as an intracellular defense mechanism by capturing cytosol-invading pathogens and delivering them for lysosomal degradation (76), a selective autophagy pathway termed xenophagy. In macrophages, recognition of pathogen-associated molecular patterns and damage-associated molecular patterns stimulates phagocytosis and subsequent elimination of pathogens by autophagy (77). Comparable to selective degradation of other cytosolic cargo, xenophagy involves the core autophagic machinery, autophagosome formation, and ubiquitination as an “eat-me” signal. Several autophagy receptors (p62, NDP52, and OPTN) can target ubiquitinated pathogens or damaged pathogen-containing phagosomes to the autophagosome via binding to LC3 (29, 78–80). In addition, NDP52 interacts with galectin-8, a cytosolic lectin and danger receptor that recognizes cytosol-accessible β-galactosides on damaged phagosomes (81, 82). This specific interaction represents an alternative way to selectively target invading bacteria for autophagic degradation.
As host cell invasion serves for microbe replication, several bacterial pathogens possess strategies to escape xenophagy. For instance, after infection of epithelial MDCK cells with the Gram-negative bacterium Shigella flexneri, binding of the Shigella virulence factor VirG to ATG5 triggers autophagy induction (83). By secreting the bacterial effector IscB, Shigella flexneri is capable of concealing the presence of its virulence factor VirG (83). IscB binds VirG in a competitive manner, thereby ultimately preventing the autophagic capture of Shigella flexneri (83). Disrupting lysosomal degradation by inhibiting components of the xenophagy machinery is another strategy of pathogens to ensure bacterial multiplication and survival. During invasion of HEK 293T cells and primary macrophages, Legionella pneumophila utilizes the bacterial effector RavZ, a cysteine protease, to hydrolyze the linkage of LC3 to PE (84). This irreversible cleavage results in a conjugation-deficient LC3 protein and inhibits autophagy in the host cell (84, 85). Furthermore, the Salmonella enterica serovar Typhimurium virulence factors SseF and SseG inhibit autophagy initiation by interfering with Rab1A signaling (86). Rab1A belongs to the family of small GTPases and is involved in translocation of the ULK1 kinase complex to the phagophore (87). In murine macrophage-like cells infected with SseF- or SseG-deficient Salmonella variants, bacterial replication was impaired, whereas downregulation of Rab1A restored this effect (86).
To what extent microglia deploy autophagy as a pathogen defense mechanism and in what way bacterial effectors impact microglial autophagy remain largely elusive. Given that several bacterial pathogens are able to cross the blood-brain barrier and infect cells in the CNS, microglial xenophagy could serve as an important neuroprotective mechanism.
Microglial autophagy and proinflammatory response.
Microglia-dependent secretion of cytokines is a key event in regulating the stimulation of proinflammatory responses after brain injury. Evidence shows that autophagy proteins modulate microglial inflammation by exhibiting either inductive or suppressive effects (69, 88, 89). In general, proinflammatory signaling is executed by the activation of the inflammasome, a cytoplasmic tripartite protein complex consisting of a sensory component for recognizing ligands, an adaptor component for the binding of caspases, and caspases for proteolytic processing of cytokines (90).
Belonging to the inflammasome sensors, NACHT, LRR, and PVD domain-containing protein 3 (NLRP3) can be activated in the presence of Aβ followed by oligomerization of the adaptor component apoptosis-associated speck-like protein containing a CARD (ASC) and the intrinsic cleavage of caspase-1 (CASP1) (91–93). These molecular events, in turn, promote the secretion of the cytokine interleukin-1β (IL-β) and the proinflammatory response to Aβ plaques (Fig. 3) (91). Studies on the Aβ-induced activation of the NLRP3 inflammasome indicate that dysfunctional autophagy enhances the inflammasomal activation state (69). For example, primary microglia treated with Aβ showed intensified cleavage of CASP1 and increased release of IL-1β when the autophagy proteins LC3B and ATG7 were depleted (69). A constitutive and abnormally high inflammatory response can also lead to neuronal toxicity and cell damage. In this context, elevated autophagic activity points toward a reduction of Aβ-induced inflammation, thereby possibly promoting cell survival (69, 94). Which autophagy proteins directly participate in the regulation of microglial inflammation is poorly understood. Houtman and colleagues described a novel function of microglial autophagy in regulating the NLRP3 inflammasome via BECN1 (88). In comparison to microglia from wild-type mice, the activation of BECN1-deficient microglia resulted in larger amounts of IL-1β secretion and higher NLRP3 protein levels (88). Furthermore, NLRP3 colocalizes with LC3 and NDP52 at autophagosomes (88). It is hypothesized that NDP52 recruits NLRP3 for autophagosomal turnover, since downregulation of NDP52 leads to elevated levels of IL-1β (Fig. 3) (88). In addition, Shi and colleagues showed p62-dependent colocalization of ubiquitinated inflammasomes with autophagosomes in macrophages (95). Hence, microglia presumably modulate the activity of inflammasomes via autophagosomal degradation of NLRP3. Saitoh and colleagues demonstrated ATG16L1-dependent regulation of endotoxin-induced inflammation in macrophages (89). ATG16L1 is essential for the formation and phagophore localization of the ATG12-ATG5-ATG16L1 E3 ligase scaffold and, thus, for the lipidation of LC3 to the autophagosomal membrane (96). Upon endotoxic activation, ATG16L1-depleted macrophages elicit higher expression of IL-1β than wild-type macrophages (89). In what way this observation translates to microglial function needs to be further elucidated. However, clearly, these findings suggest that autophagy has an important role in balancing microglial proinflammatory responses.
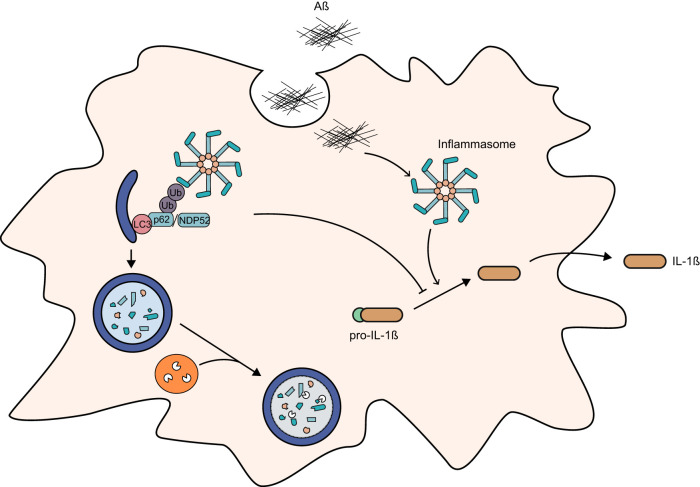
FIG 3.
Selective autophagy as a possible regulator of inflammasome activity in microglia. Presence of Aβ causes activation of proinflammatory response and release of the cytokine IL-1β. Autophagy receptors p62 and NDP52 might recognize and target ubiquitinated inflammasomes for lysosomal degradation, thereby controlling Aβ-induced inflammation and survival of the cell.
Given the morphological changes accompanied by frequent encounters with invading pathogens and toxic aggregates, activation of microglia is in urgent need of high energy levels in order to maintain microglial function. Therefore, autophagy not only is critical for the regulation of proinflammatory responses and elimination of extracellular aggregates but also needs to be considered a catabolic process for permanently supplying microglia with essential nutrients and controlling intracellular protein and organelle quality.
NONCANONICAL AUTOPHAGY IN MICROGLIA
In noncanonical autophagy processes, components of the autophagy machinery are deployed to fulfill functions which do not involve lysosomal delivery of cytosolic entities. Initial indicators for these pathways in microglia were provided by Lucin and colleagues (97), who were able to show that BECN1 is required for phagocytic uptake and subsequent degradation of Aβ. In ex vivo studies, deletion of BECN1 in microglia-derived BV2 cells exposed to APP transgenic mouse brain slices resulted in insufficient phagocytosis of Aβ (97). More importantly, isolated microglia from postmortem human Alzheimer’s disease (AD) brains exhibit reduced levels of BECN1, suggesting a link between autophagic regulation and phagocytic uptake of Aβ deposits (97). Recently, two pathways with key roles in myeloid cells deepened the understanding of noncanonical autophagy: LC3-associated phagocytosis (LAP) and LC3-associated endocytosis (LANDO) (98, 99). During LAP and LANDO autophagy, machinery components are used to conjugate LC3 to the membranes of phagosomes and endosomes, respectively. Due to their important function in myeloid cells, these two pathways will be discussed below in more detail.
LC3-associated phagocytosis.
The LAP pathway makes use of components of the canonical autophagy machinery to conjugate LC3 to the phagosome (Fig. 2B). Several membrane receptors can initiate this process, which is mainly studied in macrophages. Among others, these receptors include TLRs (1/2, 2/6, and 4) that are able to detect pathogen-associated patterns, immunoglobulin receptors that recognize antigens and pathogens opsonized with IgG, and T-cell immunoglobulin mucin receptor 4 that binds to phosphatidylserines on the surfaces of dead cells (100–103). Binding to the receptor mediates the phagocytosis of the extracellular target. Upon complete engulfment of the cargo and the sealing of the phagosome, LAP effectors are recruited. However, it remains unclear how exactly this recruitment is triggered. The most upstream autophagy complex present in LAP is the PI3KC3 complex II, which shares the core subunits hVps34 and hVps15 with PI3KC3 complex I, but instead of ATG14, it contains UV radiation resistance-associated gene protein (UVRAG) and Rubicon (RUBCN) (8, 9). While RUBCN is a known negative regulator of PI3KC3 during canonical autophagy (104), it is also compulsory for LAP (99). During LAP, RUBCN is, on one side, essential for PI3KC3-mediated PI3P generation and, on the other side, it recruits and stabilizes the NADPH oxidase 2 (NOX2) complex at the phagosome (99, 105). NOX2 activity itself as well as NOX-induced reactive oxygen species (ROS) production are crucial for the following LC3 lipidation. However, the underlying mechanism remains unknown (99, 101, 106). PI3P generation and NOX2 association to the phagosome result in conjugation of LC3 to PE on the phagosomal membrane. Notably, lipidation of LC3 and GABARAP proteins during LAP differs in two important mechanistic aspects from its use in canonical autophagy. First, WIPI2 is not necessary but probably substituted by another, yet-undiscovered effector protein (107). Second, the WD40 domain of ATG16L1 is indispensable (107, 108). One other main difference between canonical autophagy and LAP is the timing and the location of LC3 lipidation. In canonical autophagy, LC3 lipidation occurs during autophagosome formation on the inner and outer autophagosomal membranes (109). In contrast, LC3 conjugation during LAP occurs after formation of the single-membrane phagosome, leading to LC3 lipidation on the outer leaflet only. The resulting structure is called a LAPosome (103). Consequently, the function of conjugated LC3 does not entirely overlap between canonical autophagy and LAP. However, a potential common role of LC3-PE in both pathways is to facilitate the fusion of the respective transient organelle with lysosomes, leading to the degradation of the engulfed material (99, 110, 111).
Due to the targeting of several pathogens, LAP is critical for fighting bacterial as well as fungal infection (106, 112–114). Martinez et al. highlighted an additional importance of LAP by showing ineffective efferocytosis, the phagocytosis of dead cells, accompanied by an increase in proinflammatory cytokines in LAP-deficient mice (115). Furthermore, LAP plays also a role in major histocompatibility complex (MHC) class II-restricted antigen presentation. MHC class II proteins present antigens from extracellular proteins to CD4+ T cells and are found on a number of professional antigen-presenting cells, including macrophages and microglia. Antigenic peptides derived from partially degraded exogenous particles taken up by phagocytosis or endocytosis are loaded onto MHC class II molecules in late endosomes. Intriguingly, it was shown that recruitment of LC3 to the phagosome facilitates MHC class II presentation of fungal antigens in a murine macrophage cell line (116). This process is conserved in human macrophages, in which compromised fungal antigen presentation was reported when LAP was inhibited (117). Mechanistically, the coupling of LC3 to phagosomes is thought to result in prolonged antigen presentation by MHC class II molecules in human macrophages.
So far, LAP has mainly been studied in macrophages. Although microglia are distinct from other macrophages, they share many features such as the ability to phagocytose and to release cytokines. Therefore, it was generally assumed that LAP occurs in a similar or even the same manner in microglia. This was recently confirmed by two independent studies, which observed a role of LAP in the uptake of zymosan in microglia (98, 118). However, the physiological targets of microglial LAP still remain unknown.
LC3-associated endocytosis.
Remarkably, in microglia, another uptake pathway was recently demonstrated to employ autophagy proteins and therefore, in analogy to LAP, termed LANDO (98). Endocytosis is a ubiquitous cellular process that is defined by the active uptake of extracellular materials as well as components of the plasma membrane into the cytoplasm. It is important for many physiological processes, including nutrient uptake and cell signaling. Generally, endocytic pathways are differentiated as clathrin mediated and independent, with clathrin-independent endocytosis processes classified as macropinocytosis and phagocytosis. In comparison to phagocytosis, clathrin-mediated endocytosis (CME) is common in all eukaryotic cells and describes the uptake of smaller cargo into clathrin-coated vesicles (Fig. 2D) (119, 120). The early endosome matures to a late multivesicular endosome which then finally fuses with lysosomes, resulting in the degradation of the cargo (121). This process is driven by constant fusion and fission with other vesicles. Importantly, a considerable subset of the cargo escapes lysosomal degradation. For example, a large fraction of membrane components, including receptors, is often recycled back to the plasma membrane, while other cargo is targeted to the trans-Golgi network (122–124).
LANDO is characterized by LC3 conjugation to clathrin- and Rab5-positive endosomes (Fig. 2C). As in LAP, the conjugation process is dependent on BECN1, Vps34, ATG5, ATG7, and RUBCN. In contrast to LAP, the absence of LC3 conjugation does not result in defects in cargo degradation but in reduced receptor recycling. Therefore, secondary uptake of the cargo is diminished. So far, roles for LANDO have been observed for the recycling of the putative Aβ receptors TREM2, CD36, and TLR4 in microglia (98). It is conceivable that this noncanonical function of autophagic proteins plays a role not only for other receptors but also in other cell types. Notably, previous studies reporting a link between autophagy, retromer trafficking, and receptor recycling are in line with the proposed role of LANDO. The retromer complex sorts endosomal cargo back to the plasma membrane, the trans-Golgi network, or other compartments. Popovic et al. reported that induction of autophagy results in dissociation of the retromer-associated Rab GTPase-activating protein TBC1D5 from the retromer complex (125). This partition was observed to be due to an activation of Rab7a and resulted in enhanced retromer receptor recycling (126, 127). Furthermore, Lucin et al. described a dependency of TREM2 as well as CD36 receptor recycling on BECN1 and Vps34 in microglia (97). Taken together, LANDO is a newly discovered noncanonical pathway in microglia involved in receptor recycling. Although, so far, evidence is limited to one report, several previous studies are in accordance with such a pathway. Clearly, there is a need for further investigation to dissect the mechanisms of LANDO, in microglia and other cell types, in detail.
RELEVANCE IN DISEASE
The human brain comprises only 2% of total body weight but consumes 20% of total body oxygen (128). This highlights its relevance for the human body. Among others, it senses and coordinates information and controls body movements and personality as well as thoughts. Accordingly, disruption of brain functions has severe consequences such as impaired mental abilities, memory damage, and loss of muscle coordination and strength or vision and language impairment. Therefore, CNS repair and protection strategies are indispensable. The brain is protected not only by the skull and the blood-brain barrier but also by a multitude of molecular mechanisms. Among others, these include enzymatic antioxidant systems, the ubiquitin-proteasome pathway, neuroinflammation, and autophagy. Due to the role of autophagy in organelle and protein quality control, its impairment results in accumulation of aggregated proteins and damaged organelles, common hallmarks of neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS), Alzheimer’s disease (AD), and PD. The essentiality of autophagic processes for the CNS is substantiated by the multiplicity of autophagic genes which are known to be linked to neurological diseases, ranging from those encoding autophagic receptors, regulators, and adaptors (including OPTN, SQSTM1, TBK1, C9orf72, UBQLN2, HTT, and PICALM), which are risk genes for neurodegenerative diseases, to that encoding AMBRA1, which has been linked to autism and schizophrenia (129–139). Furthermore, mutations in the hATG8-binding protein tectonin β-propeller containing protein 2 (TECPR2) are known to result in hereditary spastic paraplegias, while absence of the autophagic protein WD repeat domain phosphoinositide-interacting protein 4 (WIPI4) causes beta-propeller protein-associated neurodegeneration (140, 141). Although the value of autophagic processes for brain health is widely appreciated, they were mainly studied in neurons. However, the contribution of canonical as well as noncanonical autophagy functions in microglia increasingly receives more attention in several CNS diseases (Fig. 4).
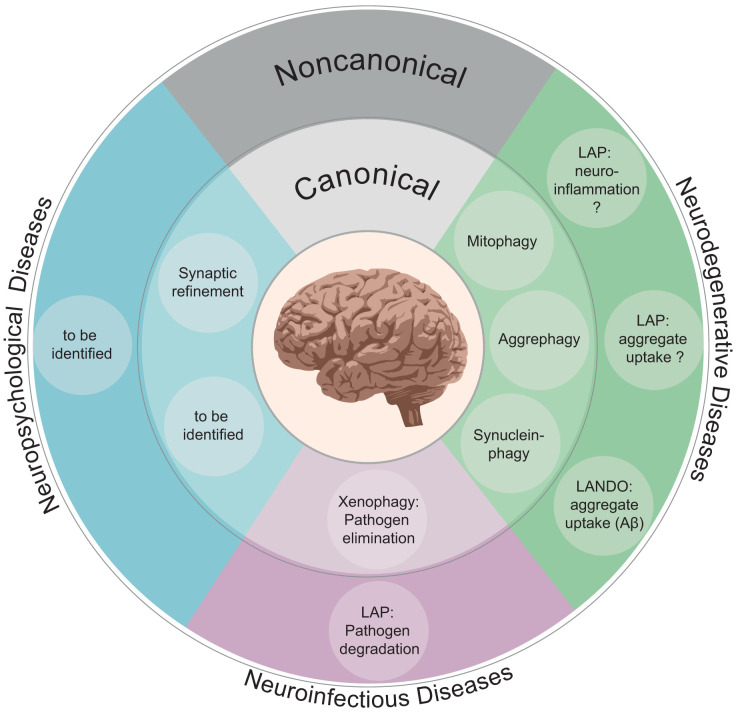
FIG 4.
Canonical and noncanonical autophagy functions associated with neurological diseases. The inner and outer circles present canonical and noncanonical autophagy processes, respectively. So far, these pathways were most extensively studied in neurodegenerative diseases. However, only little is known about their functions in neuropsychological diseases. Further aspects remain to be identified. Concerning neuroinfectious diseases, either canonical xenophagy or LAP can eliminate the invading particle, depending on its pathogenic characteristics.
Relevance of microglial autophagy in diseases.
As aging progresses, loss of protein homeostasis is a generic consequence (142). The tight regulation of inflammatory responses and degradation of extracellular aggregates by microglial autophagy supports a link between disease development and impaired autophagic processes at different stages. During AD progression, alterations in AMPK signaling are a central issue (143, 144). As a homeostasis sensor, AMPK activates microglial autophagy in the presence of Aβ plaques, consequently leading to their lysosomal degradation (69). Inactivation of AMPK with compound C decreases Aβ clearance, thereby making AMPK a potential target for AD treatment. In PD, neuron-released α-synuclein and its accumulation in Lewy bodies result in degeneration of the dopaminergic system (145). Intriguingly, impaired microglial autophagy provokes a decrease of dopaminergic neurons when α-synuclein is expressed in mice (61). Hence, removal of extracellular α-synuclein by microglial autophagy is suggested to have a vital role in maintaining neuronal protection and function.
Modulation of proinflammatory responses by autophagy was found to be crucial in Crohn’s disease, where excessive production of cytokines results in chronic inflammation, compromising the entire gastrointestinal tract (146, 147). Among patients suffering from Crohn’s disease, a genetic missense variant of ATG16L1 was identified (148). This disease-related polymorphism shows an elevated sensitivity to cleavage by caspase-3, which eventually leads to the degradation of ATG16L1 (146, 149). Following ATG16L1 loss of function in macrophages, autophagic activity is reduced and, as a consequence, levels of cytokines increase (146). Therefore, microglial autophagy could own a similar role in controlling proinflammatory responses within the CNS.
Despite neurodegeneration and inflammatory diseases, microglia were shown to have a critical impact on neuropsychological behaviors (150). In autism spectrum disorders, impaired microglial autophagy pathways studied in mice led to defective synaptic pruning, which becomes visible by an abnormal high dendritic spine density (151). Synapsis formation and elimination is fundamental, as they ensure proper brain development (152). Here, autophagy could have a modulatory role in neuronal connectivity by clearing excessive and dysfunctional synapses (151). Uncovering the molecular basis of autophagy pathways in microglia is an essential objective in order to provide a wide-ranging platform for neurotherapeutic approaches.
Relevance of microglial noncanonical autophagy in diseases.
LANDO contributes to the clearance of Aβ plaques. Furthermore, an upregulation of proinflammatory cytokines was detected in LANDO-deficient cells, which resulted in microglial hyperactivation in mice harboring several mutations associated with human familial AD. These mice also presented accelerated tau phosphorylation. In this model, the deficiency of microglial LANDO caused neuronal cell death accompanied by behavioral and memory impairments, emphasizing the role of microglia as well as LANDO in AD (98). Although the relevance of microglial LANDO has so far only been reported for AD, it is likely that this process targets other molecules or particles besides Aβ, especially since LANDO supports the recycling of the receptors CD36, TLR4, and TREM2, which can bind a broad range of ligands besides Aβ. Although LANDO is suggested to be a major player in Aβ clearance, it is not known to what extent this process is physiologically relevant. Still, it is tempting to speculate that stimulation of LANDO could be a promising target for therapeutic interventions in neurodegenerative diseases, especially in AD. However, it needs to be clarified to what extend increased LANDO is beneficial for microglia and at what point negative effects arise. In contrast to LANDO, to date, LAP was not investigated in detail in microglia. Interestingly, LAP is firmly entangled with inflammation. Mice, which lack LAP-obligatory proteins, release more proinflammatory cytokines and are defective in the clearance of apoptotic cells (115). Since microglia are responsible for the phagocytosis of apoptotic cells in the CNS and neuroinflammation is a common feature of neurodegenerative disorders, it is plausible that defective LAP could play an important role in the onset or progression of neurodegenerative diseases (153, 154). Another known function of LAP in macrophages is the elimination of pathogens. Several of these LAP-targeted pathogens, including Streptococcus pneumoniae and Listeria monocytogenes, can penetrate the blood brain barrier to enter the CNS and are common causes for bacterial meningitis (155, 156). While it remains to be firmly established that microglial LAP is part of the defense against these pathogens in the CNS, increasing LAP activity in microglia could represent a potential target for antibacterial therapeutic approaches. However, due to the two-sided role of neuroinflammation in neurodegenerative diseases, beneficial as well as detrimental impacts could result from an overactivation of LAP.
CONCLUSION
As resident immune cells of the brain and spinal cord, microglia accomplish vital functions, including synaptic pruning, neurogenesis, and immune responses. Growing evidence indicates that microglia utilize autophagy to meet CNS homeostasis, thereby strictly controlling proinflammatory responses and removal of protein aggregates as well as damaged organelles from the cytosol. Compared to canonical autophagy, noncanonical pathways allow microglia to target also noncytosolic entities for lysosomal degradation. LAP and LANDO are two noncanonical pathways which were shown to be present in microglia. While it was observed that microglial LANDO is crucial for the uptake of Aβ, the exact role of LAP in microglia remains unknown. Regarding the decline of protein function during aging and various neuronal diseases, these noncanonical features contribute to the maintenance of CNS homeostasis, and their defect can have detrimental consequences. With regard to the energetic aspect, microglia may omit and exploit specific signaling pathways to activate autophagy, thereby recycling nutrients and sustaining energy. Taken together, canonical as well as noncanonical autophagy pathways play key parts in microglia functioning, most likely in complementary ways. Yet, several questions as to the role of LAP in microglia and the detailed interplay between canonical and noncanonical autophagy remain unanswered and should be elucidated by further research.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) within the frameworks of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy identifier [ID] 390857198) and of the Collaborative Research Center 1177 (ID 259130777).
J.J., L.S., and C.B. contributed to the drafting of the article. J.J. and L.S. wrote the manuscript and designed the figures. L.S. realized digital illustrations. All authors reviewed the final manuscript. C.B. acquired funding.
REFERENCES
- 1.Ohsumi Y. 1999. Molecular mechanism of autophagy in yeast, Saccharomyces cerevisiae. Philos Trans R Soc Lond B Biol Sci 354:1577–1580. doi: 10.1098/rstb.1999.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. 2008. Autophagy fights disease through cellular self-digestion. Nature 451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky DJ, Ohsumi Y. 1999. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol 15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Shang L, Wang X. 2011. AMPK and mTOR coordinate the regulation of Ulk1 and mammalian autophagy initiation. Autophagy 7:924–926. doi: 10.4161/auto.7.8.15860. [DOI] [PubMed] [Google Scholar]
- 5.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. 2013. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou C, Ma K, Gao R, Mu C, Chen L, Liu Q, Luo Q, Feng D, Zhu Y, Chen Q. 2017. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res 27:184–201. doi: 10.1038/cr.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Long YC, Shen HM. 2015. Differential regulatory functions of three classes of phosphatidylinositol and phosphoinositide 3-kinases in autophagy. Autophagy 11:1711–1728. doi: 10.1080/15548627.2015.1043076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young LN, Goerdeler F, Hurley JH. 2019. Structural pathway for allosteric activation of the autophagic PI 3-kinase complex I. Proc Natl Acad Sci U S A 116:21508–21513. doi: 10.1073/pnas.1911612116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 11.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. 2014. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell 55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slobodkin MR, Elazar Z. 2013. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem 55:51–64. doi: 10.1042/bse0550051. [DOI] [PubMed] [Google Scholar]
- 14.Sakoh-Nakatogawa M, Matoba K, Asai E, Kirisako H, Ishii J, Noda NN, Inagaki F, Nakatogawa H, Ohsumi Y. 2013. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol 20:433–439. doi: 10.1038/nsmb.2527. [DOI] [PubMed] [Google Scholar]
- 15.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. 2000. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakatogawa H, Ichimura Y, Ohsumi Y. 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 17.Yu ZQ, Ni T, Hong B, Wang HY, Jiang FJ, Zou S, Chen Y, Zheng XL, Klionsky DJ, Liang Y, Xie Z. 2012. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 8:883–892. doi: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. 2008. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ 15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 20.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. 2009. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 21.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. 2011. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci U S A 108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang TK, Shravage BV, Hayes SD, Powers CM, Simin RT, Wade Harper J, Baehrecke EH. 2013. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol 15:1067–1078. doi: 10.1038/ncb2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Kundu M, Viollet B, Guan KL. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganley IG, Lam Du H, Wang J, Ding X, Chen S, Jiang X. 2009. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 26.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanki T, Klionsky DJ. 2008. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem 283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. 2009. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol 183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 30.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. 2011. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell 44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 32.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. 2004. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol 24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP, Ho MW, Troncoso J, Gygi SP, Lee MK, Dawson VL, Dawson TM, Lim KL. 2008. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet 17:431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- 34.Riley BE, Kaiser SE, Shaler TA, Ng AC, Hara T, Hipp MS, Lage K, Xavier RJ, Ryu KY, Taguchi K, Yamamoto M, Tanaka K, Mizushima N, Komatsu M, Kopito RR. 2010. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J Cell Biol 191:537–552. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 36.Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F. 2008. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 37.Wild P, McEwan DG, Dikic I. 2014. The LC3 interactome at a glance. J Cell Sci 127:3–9. doi: 10.1242/jcs.140426. [DOI] [PubMed] [Google Scholar]
- 38.Deng Z, Purtell K, Lachance V, Wold MS, Chen S, Yue Z. 2017. Autophagy receptors and neurodegenerative diseases. Trends Cell Biol 27:491–504. doi: 10.1016/j.tcb.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Li Q, Barres BA. 2018. Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol 18:225–242. doi: 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- 40.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyamoto A, Wake H, Ishikawa AW, Eto K, Shibata K, Murakoshi H, Koizumi S, Moorhouse AJ, Yoshimura Y, Nabekura J. 2016. Microglia contact induces synapse formation in developing somatosensory cortex. Nat Commun 7:12540. doi: 10.1038/ncomms12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hagemeyer N, Hanft KM, Akriditou MA, Unger N, Park ES, Stanley ER, Staszewski O, Dimou L, Prinz M. 2017. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol 134:441–458. doi: 10.1007/s00401-017-1747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB. 2005. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci 8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 44.Noda M, Nakanishi H, Nabekura J, Akaike N. 2000. AMPA-kainate subtypes of glutamate receptor in rat cerebral microglia. J Neurosci 20:251–258. doi: 10.1523/JNEUROSCI.20-01-00251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biber K, Laurie DJ, Berthele A, Sommer B, Tolle TR, Gebicke-Harter PJ, van Calker D, Boddeke HW. 1999. Expression and signaling of group I metabotropic glutamate receptors in astrocytes and microglia. J Neurochem 72:1671–1680. doi: 10.1046/j.1471-4159.1999.721671.x. [DOI] [PubMed] [Google Scholar]
- 46.Kuhn SA, van Landeghem FK, Zacharias R, Farber K, Rappert A, Pavlovic S, Hoffmann A, Nolte C, Kettenmann H. 2004. Microglia express GABA(B) receptors to modulate interleukin release. Mol Cell Neurosci 25:312–322. doi: 10.1016/j.mcn.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 47.Town T, Jeng D, Alexopoulou L, Tan J, Flavell RA. 2006. Microglia recognize double-stranded RNA via TLR3. J Immunol 176:3804–3812. doi: 10.4049/jimmunol.176.6.3804. [DOI] [PubMed] [Google Scholar]
- 48.Bianco F, Fumagalli M, Pravettoni E, D'Ambrosi N, Volonte C, Matteoli M, Abbracchio MP, Verderio C. 2005. Pathophysiological roles of extracellular nucleotides in glial cells: differential expression of purinergic receptors in resting and activated microglia. Brain Res Brain Res Rev 48:144–156. doi: 10.1016/j.brainresrev.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 49.Lv M, Liu Y, Zhang J, Sun L, Liu Z, Zhang S, Wang B, Su D, Su Z. 2011. Roles of inflammation response in microglia cell through Toll-like receptors 2/interleukin-23/interleukin-17 pathway in cerebral ischemia/reperfusion injury. Neuroscience 176:162–172. doi: 10.1016/j.neuroscience.2010.11.066. [DOI] [PubMed] [Google Scholar]
- 50.Kettenmann H, Hoppe D, Gottmann K, Banati R, Kreutzberg G. 1990. Cultured microglial cells have a distinct pattern of membrane channels different from peritoneal macrophages. J Neurosci Res 26:278–287. doi: 10.1002/jnr.490260303. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi K, Rochford CD, Neumann H. 2005. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med 201:647–657. doi: 10.1084/jem.20041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blander JM, Medzhitov R. 2004. Regulation of phagosome maturation by signals from Toll-like receptors. Science 304:1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 53.Sung SS, Nelson RS, Silverstein SC. 1983. Yeast mannans inhibit binding and phagocytosis of zymosan by mouse peritoneal macrophages. J Cell Biol 96:160–166. doi: 10.1083/jcb.96.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao H, Coppola K, Schweig JE, Crawford F, Mullan M, Paris D. 2019. Distinct signaling pathways regulate TREM2 phagocytic and NFkappaB antagonistic activities. Front Cell Neurosci 13:457. doi: 10.3389/fncel.2019.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koizumi S, Shigemoto-Mogami Y, Nasu-Tada K, Shinozaki Y, Ohsawa K, Tsuda M, Joshi BV, Jacobson KA, Kohsaka S, Inoue K. 2007. UDP acting at P2Y6 receptors is a mediator of microglial phagocytosis. Nature 446:1091–1095. doi: 10.1038/nature05704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rabinovitch M. 1967. The dissociation of the attachment and ingestion phases of phagocytosis by macrophages. Exp Cell Res 46:19–28. doi: 10.1016/0014-4827(67)90405-3. [DOI] [PubMed] [Google Scholar]
- 57.Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, Gulati G, Bennett ML, Sun LO, Clarke LE, Marschallinger J, Yu G, Quake SR, Wyss-Coray T, Barres BA. 2019. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101:207.e10–223.e10. doi: 10.1016/j.neuron.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, Marsh SE, Saunders A, Macosko E, Ginhoux F, Chen J, Franklin RJM, Piao X, McCarroll SA, Stevens B. 2019. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50:253.e6–271.e6. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hickey WF, Kimura H. 1988. Perivascular microglial cells of the CNS are bone marrow-derived and present antigen in vivo. Science 239:290–292. doi: 10.1126/science.3276004. [DOI] [PubMed] [Google Scholar]
- 60.Ross CA, Poirier MA. 2004. Protein aggregation and neurodegenerative disease. Nat Med 10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 61.Choi I, Zhang Y, Seegobin SP, Pruvost M, Wang Q, Purtell K, Zhang B, Yue Z. 2020. Microglia clear neuron-released alpha-synuclein via selective autophagy and prevent neurodegeneration. Nat Commun 11:1386. doi: 10.1038/s41467-020-15119-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, Su LJ, Caldwell KA, Caldwell GA, Rochet JC, McCaffery JM, Barlowe C, Lindquist S. 2008. The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci U S A 105:145–150. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. 1997. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science 276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 64.Kuusisto E, Salminen A, Alafuzoff I. 2001. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport 12:2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- 65.Liang Y, Chu H, Jiang Y, Yuan L. 2016. Morphine enhances IL-1beta release through Toll-like receptor 4-mediated endocytic pathway in microglia. Purinergic Signal 12:637–645. doi: 10.1007/s11302-016-9525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park JY, Kim KS, Lee SB, Ryu JS, Chung KC, Choo YK, Jou I, Kim J, Park SM. 2009. On the mechanism of internalization of alpha-synuclein into microglia: roles of ganglioside GM1 and lipid raft. J Neurochem 110:400–411. doi: 10.1111/j.1471-4159.2009.06150.x. [DOI] [PubMed] [Google Scholar]
- 67.Flavin WP, Bousset L, Green ZC, Chu Y, Skarpathiotis S, Chaney MJ, Kordower JH, Melki R, Campbell EM. 2017. Endocytic vesicle rupture is a conserved mechanism of cellular invasion by amyloid proteins. Acta Neuropathol 134:629–653. doi: 10.1007/s00401-017-1722-x. [DOI] [PubMed] [Google Scholar]
- 68.Bussi C, Peralta Ramos JM, Arroyo DS, Gallea JI, Ronchi P, Kolovou A, Wang JM, Florey O, Celej MS, Schwab Y, Ktistakis NT, Iribarren P. 2018. Alpha-synuclein fibrils recruit TBK1 and OPTN to lysosomal damage sites and induce autophagy in microglial cells. J Cell Sci 131:jcs226241. doi: 10.1242/jcs.226241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cho MH, Cho K, Kang HJ, Jeon EY, Kim HS, Kwon HJ, Kim HM, Kim DH, Yoon SY. 2014. Autophagy in microglia degrades extracellular beta-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy 10:1761–1775. doi: 10.4161/auto.29647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, Slingerland JM, Mills GB. 2007. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol 9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 71.Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, Terzic J, Dikic I. 2013. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci 126:580–592. doi: 10.1242/jcs.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bamberger ME, Harris ME, McDonald DR, Husemann J, Landreth GE. 2003. A cell surface receptor complex for fibrillar beta-amyloid mediates microglial activation. J Neurosci 23:2665–2674. doi: 10.1523/JNEUROSCI.23-07-02665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koenigsknecht J, Landreth G. 2004. Microglial phagocytosis of fibrillar beta-amyloid through a beta1 integrin-dependent mechanism. J Neurosci 24:9838–9846. doi: 10.1523/JNEUROSCI.2557-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yeh FL, Wang Y, Tom I, Gonzalez LC, Sheng M. 2016. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron 91:328–340. doi: 10.1016/j.neuron.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 75.Ulland TK, Song WM, Huang SC, Ulrich JD, Sergushichev A, Beatty WL, Loboda AA, Zhou Y, Cairns NJ, Kambal A, Loginicheva E, Gilfillan S, Cella M, Virgin HW, Unanue ER, Wang Y, Artyomov MN, Holtzman DM, Colonna M. 2017. TREM2 maintains microglial metabolic fitness in Alzheimer's disease. Cell 170:649.e13–663.e13. doi: 10.1016/j.cell.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 77.Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT. 2012. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol Rev 249:158–175. doi: 10.1111/j.1600-065X.2012.01146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. 2009. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 79.Ravenhill BJ, Boyle KB, von Muhlinen N, Ellison CJ, Masson GR, Otten EG, Foeglein A, Williams R, Randow F. 2019. The cargo receptor NDP52 initiates selective autophagy by recruiting the ULK complex to cytosol-invading bacteria. Mol Cell 74:320.e6–329.e6. doi: 10.1016/j.molcel.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. 2011. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. 2012. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim BW, Hong SB, Kim JH, Kwon DH, Song HK. 2013. Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8. Nat Commun 4:1613. doi: 10.1038/ncomms2606. [DOI] [PubMed] [Google Scholar]
- 83.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. 2005. Escape of intracellular Shigella from autophagy. Science 307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 84.Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, Roy CR. 2012. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park SW, Jeon P, Jun YW, Park JH, Lee SH, Lee S, Lee JA, Jang DJ. 2019. Monitoring LC3- or GABARAP-positive autophagic membranes using modified RavZ-based probes. Sci Rep 9:16593. doi: 10.1038/s41598-019-53372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feng ZZ, Jiang AJ, Mao AW, Feng Y, Wang W, Li J, Zhang X, Xing K, Peng X. 2018. The Salmonella effectors SseF and SseG inhibit Rab1A-mediated autophagy to facilitate intracellular bacterial survival and replication. J Biol Chem 293:9662–9673. doi: 10.1074/jbc.M117.811737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, Myszczynska MA, Higginbottom A, Walsh MJ, Whitworth AJ, Kaspar BK, Meyer K, Shaw PJ, Grierson AJ, De Vos KJ. 2016. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J 35:1656–1676. doi: 10.15252/embj.201694401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Houtman J, Freitag K, Gimber N, Schmoranzer J, Heppner FL, Jendrach M. 2019. Beclin1-driven autophagy modulates the inflammatory response of microglia via NLRP3. EMBO J 38:e99430. doi: 10.15252/embj.201899430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 90.Sharma D, Kanneganti TD. 2016. The cell biology of inflammasomes: mechanisms of inflammasome activation and regulation. J Cell Biol 213:617–629. doi: 10.1083/jcb.201602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ, Golenbock DT. 2008. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol 9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vajjhala PR, Mirams RE, Hill JM. 2012. Multiple binding sites on the pyrin domain of ASC protein allow self-association and interaction with NLRP3 protein. J Biol Chem 287:41732–41743. doi: 10.1074/jbc.M112.381228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martinon F, Burns K, Tschopp J. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 94.Block ML, Zecca L, Hong JS. 2007. Microglia-mediated neurotoxicity: uncovering the molecular mechanisms. Nat Rev Neurosci 8:57–69. doi: 10.1038/nrn2038. [DOI] [PubMed] [Google Scholar]
- 95.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. 2012. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. 2003. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci 116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 97.Lucin KM, O'Brien CE, Bieri G, Czirr E, Mosher KI, Abbey RJ, Mastroeni DF, Rogers J, Spencer B, Masliah E, Wyss-Coray T. 2013. Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer's disease. Neuron 79:873–886. doi: 10.1016/j.neuron.2013.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Heckmann BL, Teubner BJW, Tummers B, Boada-Romero E, Harris L, Yang M, Guy CS, Zakharenko SS, Green DR. 2019. LC3-associated endocytosis facilitates beta-amyloid clearance and mitigates neurodegeneration in murine Alzheimer's disease. Cell 178:536.e14–551.e14. doi: 10.1016/j.cell.2019.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, Orchard R, Guan JL, Tan H, Peng J, Kanneganti TD, Virgin HW, Green DR. 2015. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol 17:893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100.Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, Coyle AJ, Kolbeck R, Green DR, Sanjuan MA. 2012. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity 37:986–997. doi: 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. 2009. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A 106:6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. 2011. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A 108:17396–17401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 104.Kim YM, Jung CH, Seo M, Kim EK, Park JM, Bae SS, Kim DH. 2015. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell 57:207–218. doi: 10.1016/j.molcel.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang CS, Lee JS, Rodgers M, Min CK, Lee JY, Kim HJ, Lee KH, Kim CJ, Oh B, Zandi E, Yue Z, Kramnik I, Liang C, Jung JU. 2012. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe 11:264–276. doi: 10.1016/j.chom.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lam GY, Cemma M, Muise AM, Higgins DE, Brumell JH. 2013. Host and bacterial factors that regulate LC3 recruitment to Listeria monocytogenes during the early stages of macrophage infection. Autophagy 9:985–995. doi: 10.4161/auto.24406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fletcher K, Ulferts R, Jacquin E, Veith T, Gammoh N, Arasteh JM, Mayer U, Carding SR, Wileman T, Beale R, Florey O. 2018. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J 37:e97841. doi: 10.15252/embj.201797840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lystad AH, Carlsson SR, de la Ballina LR, Kauffman KJ, Nag S, Yoshimori T, Melia TJ, Simonsen A. 2019. Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat Cell Biol 21:372–383. doi: 10.1038/s41556-019-0274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Manil-Segalen M, Lefebvre C, Jenzer C, Trichet M, Boulogne C, Satiat-Jeunemaitre B, Legouis R. 2014. The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev Cell 28:43–55. doi: 10.1016/j.devcel.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 111.Wang H, Sun HQ, Zhu X, Zhang L, Albanesi J, Levine B, Yin H. 2015. GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proc Natl Acad Sci U S A 112:7015–7020. doi: 10.1073/pnas.1507263112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Masud S, Prajsnar TK, Torraca V, Lamers GEM, Benning M, Van Der Vaart M, Meijer AH. 2019. Macrophages target Salmonella by Lc3-associated phagocytosis in a systemic infection model. Autophagy 15:796–812. doi: 10.1080/15548627.2019.1569297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oikonomou V, Moretti S, Renga G, Galosi C, Borghi M, Pariano M, Puccetti M, Palmerini CA, Amico L, Carotti A, Prezioso L, Spolzino A, Finocchi A, Rossi P, Velardi A, Aversa F, Napolioni V, Romani L. 2016. Noncanonical fungal autophagy inhibits inflammation in response to IFN-gamma via DAPK1. Cell Host Microbe 20:744–757. doi: 10.1016/j.chom.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Herb M, Gluschko A, Schramm M. 2018. LC3-associated phagocytosis initiated by integrin ITGAM-ITGB2/Mac-1 enhances immunity to Listeria monocytogenes. Autophagy 14:1462–1464. doi: 10.1080/15548627.2018.1475816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martinez J, Cunha LD, Park S, Yang M, Lu Q, Orchard R, Li QZ, Yan M, Janke L, Guy C, Linkermann A, Virgin HW, Green DR. 2016. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 533:115–119. doi: 10.1038/nature17950. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 116.Ma J, Becker C, Lowell CA, Underhill DM. 2012. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem 287:34149–34156. doi: 10.1074/jbc.M112.382812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Romao S, Gasser N, Becker AC, Guhl B, Bajagic M, Vanoaica D, Ziegler U, Roesler J, Dengjel J, Reichenbach J, Munz C. 2013. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J Cell Biol 203:757–766. doi: 10.1083/jcb.201308173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee JW, Nam H, Kim LE, Jeon Y, Min H, Ha S, Lee Y, Kim SY, Lee SJ, Kim EK, Yu SW. 2019. TLR4 (Toll-like receptor 4) activation suppresses autophagy through inhibition of FOXO3 and impairs phagocytic capacity of microglia. Autophagy 15:753–770. doi: 10.1080/15548627.2018.1556946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brodsky FM. 1988. Living with clathrin: its role in intracellular membrane traffic. Science 242:1396–1402. doi: 10.1126/science.2904698. [DOI] [PubMed] [Google Scholar]
- 120.Pearse BM. 1976. Clathrin: a unique protein associated with intracellular transfer of membrane by coated vesicles. Proc Natl Acad Sci U S A 73:1255–1259. doi: 10.1073/pnas.73.4.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mullock BM, Perez JH, Kuwana T, Gray SR, Luzio JP. 1994. Lysosomes can fuse with a late endosomal compartment in a cell-free system from rat liver. J Cell Biol 126:1173–1182. doi: 10.1083/jcb.126.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Goldstein JL, Brown MS, Anderson RG, Russell DW, Schneider WJ. 1985. Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annu Rev Cell Biol 1:1–39. doi: 10.1146/annurev.cb.01.110185.000245. [DOI] [PubMed] [Google Scholar]
- 123.Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR. 1993. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J 12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mayor S, Presley JF, Maxfield FR. 1993. Sorting of membrane components from endosomes and subsequent recycling to the cell surface occurs by a bulk flow process. J Cell Biol 121:1257–1269. doi: 10.1083/jcb.121.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Popovic D, Akutsu M, Novak I, Harper JW, Behrends C, Dikic I. 2012. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol 32:1733–1744. doi: 10.1128/MCB.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roy S, Leidal AM, Ye J, Ronen SM, Debnath J. 2017. Autophagy-dependent shuttling of TBC1D5 controls plasma membrane translocation of GLUT1 and glucose uptake. Mol Cell 67:84.e5–95.e5. doi: 10.1016/j.molcel.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Seaman MNJ, Mukadam AS, Breusegem SY. 2018. Inhibition of TBC1D5 activates Rab7a and can enhance the function of the retromer cargo-selective complex. J Cell Sci 131:jcs217398. doi: 10.1242/jcs.217398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mink JW, Blumenschine RJ, Adams DB. 1981. Ratio of central nervous system to body metabolism in vertebrates: its constancy and functional basis. Am J Physiol 241:R203–R212. doi: 10.1152/ajpregu.1981.241.3.R203. [DOI] [PubMed] [Google Scholar]
- 129.Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E. 2001. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem 276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 130.Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, Eng C. 2005. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet 42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T. 2011. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. 2007. Ambra1 regulates autophagy and development of the nervous system. Nature 447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 133.Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Muller K, Marroquin N, Nordin F, Hubers A, Weydt P, Pinto S, Press R, Millecamps S, Molko N, Bernard E, Desnuelle C, Soriani MH, Dorst J, Graf E, Nordstrom U, Feiler MS, Putz S, Boeckers TM, Meyer T, Winkler AS, Winkelman J, de Carvalho M, Thal DR, Otto M, Brannstrom T, Volk AE, Kursula P, Danzer KM, Lichtner P, Dikic I, Meitinger T, Ludolph AC, Strom TM, Andersen PM, Weishaupt JH. 2015. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci 18:631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 134.Kwok CT, Morris A, de Belleroche JS. 2014. Sequestosome-1 (SQSTM1) sequence variants in ALS cases in the UK: prevalence and coexistence of SQSTM1 mutations in ALS kindred with PDB. Eur J Hum Genet 22:492–496. doi: 10.1038/ejhg.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. 2010. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci 13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mitjans M, Begemann M, Ju A, Dere E, Wustefeld L, Hofer S, Hassouna I, Balkenhol J, Oliveira B, van der Auwera S, Tammer R, Hammerschmidt K, Volzke H, Homuth G, Cecconi F, Chowdhury K, Grabe H, Frahm J, Boretius S, Dandekar T, Ehrenreich H. 2017. Sexual dimorphism of AMBRA1-related autistic features in human and mouse. Transl Psychiatry 7:e1247. doi: 10.1038/tp.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rietschel M, Mattheisen M, Degenhardt F, Mühleisen TW, Kirsch P, Esslinger C, Herms S, Demontis D, Steffens M, Strohmaier J, Haenisch B, Breuer R, Czerski PM, Giegling I, Strengman E, Schmael C, Mors O, Mortensen PB, Hougaard DM, Ørntoft T, Kapelski P, Priebe L, Basmanav FF, Forstner AJ, Hoffman P, Meier S, Nikitopoulos J, Moebus S, Alexander M, Mössner R, Wichmann H-E, Schreiber S, Rivandeneira F, Hofman A, Uitterlinden AG, Wienker TF, Schumacher J, Hauser J, Maier W, Cantor RM, Erk S, Schulze TG, Craddock N, Owen MJ, O'Donovan MC, Børglum AD, Rujescu D, Walter H, Meyer-Lindenberg A, Nöthen NM, SGENE-plus Consortium, et al. 2012. Association between genetic variation in a region on chromosome 11 and schizophrenia in large samples from Europe. Mol Psychiatry 17:906–917. doi: 10.1038/mp.2011.80. [DOI] [PubMed] [Google Scholar]
- 138.Tian Y, Chang JC, Fan EY, Flajolet M, Greengard P. 2013. Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer's APP-CTF for terminal degradation via autophagy. Proc Natl Acad Sci U S A 110:17071–17076. doi: 10.1073/pnas.1315110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wong YC, Holzbaur EL. 2014. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A 111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stadel D, Millarte V, Tillmann KD, Huber J, Tamin-Yecheskel BC, Akutsu M, Demishtein A, Ben-Zeev B, Anikster Y, Perez F, Dotsch V, Elazar Z, Rogov V, Farhan H, Behrends C. 2015. TECPR2 cooperates with LC3C to regulate COPII-dependent ER export. Mol Cell 60:89–104. doi: 10.1016/j.molcel.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 141.Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K, Kasai-Yoshida E, Sawaura N, Nishida H, Hoshino A, Ryujin F, Yoshioka S, Nishiyama K, Kondo Y, Tsurusaki Y, Nakashima M, Miyake N, Arakawa H, Kato M, Mizushima N, Matsumoto N. 2013. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet 45:445–449. doi: 10.1038/ng.2562. [DOI] [PubMed] [Google Scholar]
- 142.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 143.Vingtdeux V, Giliberto L, Zhao H, Chandakkar P, Wu Q, Simon JE, Janle EM, Lobo J, Ferruzzi MG, Davies P, Marambaud P. 2010. AMP-activated protein kinase signaling activation by resveratrol modulates amyloid-beta peptide metabolism. J Biol Chem 285:9100–9113. doi: 10.1074/jbc.M109.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zimmermann HR, Yang W, Kasica NP, Zhou X, Wang X, Beckelman BC, Lee J, Furdui CM, Keene CD, Ma T. 2020. Brain-specific repression of AMPKalpha1 alleviates pathophysiology in Alzheimer's model mice. J Clin Invest 130:3511–3527. doi: 10.1172/JCI133982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xu J, Kao SY, Lee FJ, Song W, Jin LW, Yankner BA. 2002. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat Med 8:600–606. doi: 10.1038/nm0602-600. [DOI] [PubMed] [Google Scholar]
- 146.Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR, van Lookeren Campagne M. 2014. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506:456–462. doi: 10.1038/nature13044. [DOI] [PubMed] [Google Scholar]
- 147.Xavier RJ, Podolsky DK. 2007. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 148.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, Gunther S, Prescott NJ, Onnie CM, Hasler R, Sipos B, Folsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M, Schreiber S. 2007. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 149.Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, Villablanca EJ, Norman JM, Liu TC, Heath RJ, Becker ML, Fagbami L, Horn H, Mercer J, Yilmaz OH, Jaffe JD, Shamji AF, Bhan AK, Carr SA, Daly MJ, Virgin HW, Schreiber SL, Stappenbeck TS, Xavier RJ. 2014. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A 111:7741–7746. doi: 10.1073/pnas.1407001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Koyama R, Ikegaya Y. 2015. Microglia in the pathogenesis of autism spectrum disorders. Neurosci Res 100:1–5. doi: 10.1016/j.neures.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 151.Kim HJ, Cho MH, Shim WH, Kim JK, Jeon EY, Kim DH, Yoon SY. 2017. Deficient autophagy in microglia impairs synaptic pruning and causes social behavioral defects. Mol Psychiatry 22:1576–1584. doi: 10.1038/mp.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, Gross CT. 2011. Synaptic pruning by microglia is necessary for normal brain development. Science 333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 153.Guzman-Martinez L, Maccioni RB, Andrade V, Navarrete LP, Pastor MG, Ramos-Escobar N. 2019. Neuroinflammation as a common feature of neurodegenerative disorders. Front Pharmacol 10:1008. doi: 10.3389/fphar.2019.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Witting A, Muller P, Herrmann A, Kettenmann H, Nolte C. 2000. Phagocytic clearance of apoptotic neurons by microglia/brain macrophages in vitro: involvement of lectin-, integrin-, and phosphatidylserine-mediated recognition. J Neurochem 75:1060–1070. doi: 10.1046/j.1471-4159.2000.0751060.x. [DOI] [PubMed] [Google Scholar]
- 155.Gluschko A, Herb M, Wiegmann K, Krut O, Neiss WF, Utermohlen O, Kronke M, Schramm M. 2018. The β2 integrin Mac-1 induces protective LC3-associated phagocytosis of Listeria monocytogenes. Cell Host Microbe 23:324.e5–337.e5. doi: 10.1016/j.chom.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 156.Ogawa M, Takada N, Shizukuishi S, Tomokiyo M, Chang B, Yoshida M, Kakuta S, Tanida I, Ryo A, Guan JL, Takeyama H, Ohnishi M. 2020. Streptococcus pneumoniae triggers hierarchical autophagy through reprogramming of LAPosome-like vesicles via NDP52-delocalization. Commun Biol 3:25. doi: 10.1038/s42003-020-0753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]