Abstract
Carotenoid cleavage dioxygenases (CCDs) are a group of enzymes that catalyze the selective oxidative cleavage steps from carotenoids to apocarotenoids, which are essential for the synthesis of biologically important molecules such as retinoids, and the phytohormones abscisic acid (ABA) and strigolactones. In addition, CCDs play important roles in plant biotic and abiotic stress responses. Till now, a comprehensive characterization of the CCD gene family in the economically important crop cotton (Gossypium spp.) is still missing. Here, we performed a genome-wide analysis and identified 33, 31, 16 and 15 CCD genes from two allotetraploid Gossypium species, G. hirsutum and G. barbadense, and two diploid Gossypium species, G. arboreum and G. raimondii, respectively. According to the phylogenetic tree analysis, cotton CCDs are classified as six subgroups including CCD1, CCD4, CCD7, CCD8, nine-cis-epoxycarotenoid dioxygenase (NCED) and zaxinone synthase (ZAS) sub-families. Evolutionary analysis shows that purifying selection dominated the evolution of these genes in G. hirsutum and G. barbadense. Predicted cis-acting elements in 2 kb promoters of CCDs in G. hirsutum are mainly involved in light, stress and hormone responses. The transcriptomic analysis of GhCCDs showed that different GhCCDs displayed diverse expression patterns and were ubiquitously expressed in most tissues; moreover, GhCCDs displayed specific inductions by different abiotic stresses. Quantitative reverse-transcriptional PCR (qRT-PCR) confirmed the induction of GhCCDs by heat stress, salinity, polyethylene glycol (PEG) and ABA application. In summary, the bioinformatics and expression analysis of CCD gene family provide evidence for the involvement in regulating abiotic stresses and useful information for in-depth studies of their biological functions in G. hirsutum.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02805-9.
Keywords: CCD genes, Cotton, Genome-wide analysis, Phylogenetic tree, Abiotic stresses, Expression analysis
Introduction
Cotton (Gossypium spp.) is the most important crop for natural textile fiber and one of the main sources for editable oil in the world (Chen et al. 2007). The genus Gossypium consists of about 45 diploid (2n = 2× = 26) and five allotetraploid (2n = 4× = 52) species. Among them, two allotetraploid cotton, G. hirsutum (Upland cotton) and G. barbadense (Sea Island cotton) are two main domesticated cultivars that are used for fiber production. G. hirsutum dominates more than 90% of worldwide cotton production due to its high yield, and G. barbadense, with superior fiber quality, accounts for less than 10%. It is assumed that allotetraploid Gossypium arises 1–1.5 million years ago (MYA) through one hybridization event between one extant progenitor of G. herbaceum (Marion-Poll) or G. arboreum (A2) and another progenitor, G. raimondii Ulbrich (D5) (Endrizzi et al. 1985; Wendel 1989; Zhang et al. 2015). Cotton is a moderately salt-tolerant crop; however, it is usually planted on saline-alkali or dry land due to insufficient farm land. Abiotic stresses, such as high salt, drought, heat, seriously affect the productivity and quality of cotton fiber (Sexton and Gerard 1982). Thus, developing stress-resistant cotton varieties is important to improve cotton fiber quality and yield performance.
Carotenoids are a class of C40 isoprenoid pigments that play as accessory photosynthetic pigments and constituents of photosynthetic apparatus, and play essential roles in photosynthesis and photoprotection (Hashimot 2016). Apart from that, carotenoids act as precursors of a series of biologically important regulatory molecules in plants, such as β-cyclocitral (Ramel et al. 2012), the phytohormones abscisic acid (ABA) and strigolactone (Schwartz et al. 1997; Alder et al. 2012), and the recently identified plant regulatory metabolites, anchorene, iso-anchorene and zaxinone (Jia et al. 2019b, 2021; Wang et al. 2019). These derivatives are produced from the selective oxidative cleavage of carotenoids, mainly catalyzed by carotenoid cleavage dioxygenases (CCDs). CCDs are a family of non-heme FeII-dependent enzymes that cleave the conjugated C–C double bonds in carotenoids to produce different apocarotenoids. CCDs differ in their substrate specificity that covers the type of carotenoid, e.g., cyclic or acyclic, its stereo configuration (cis/trans) and the cleavage site, i.e., regio-specificity (Giuliano et al. 2003; Walter and Strack2011; Nisar et al. 2015; Jia et al. 2017). Moreover, several CCDs have been shown to cleave apocarotenoids rather than carotenoids (Alder et al. 2012; Nisar et al. 2015).
In Arabidopsis (Arabidopsis thaliana), there are five sub-families of CCDs, which are designated as CCD1, CCD4, CCD7, CCD8 and nine-cis-epoxycarotenoid dioxygenase (NCED) (Ohmiya 2009). Among them, CCD1 enzymes showed a wide substrate and relaxed double-bond specificity by cleaving various cyclic and acyclic all-trans-carotenoids as well as apocarotenoids in vitro, suggesting CCD1 enzymes are likely involved in scavenging unnecessary apocarotenoid metabolites (Ilg et al. 2009, 2014). In addition, CCD1s have been shown to be involved in the formation of flavor volatiles in different species (Sun et al. 2008; Ilg et al. 2009). CCD4 enzymes mainly cleave bicyclic carotenoids either at the C7′–C8′ to produce a C30 apocarotenal and C10 volatile, or at C9′–C10′ position to produce a C27 apocarotenal and C13 volatile, respectively (Ma et al. 2013; Rodrigo et al. 2013; Bruno et al. 2015; Zheng et al. 2019). The carotenoid cleavage activity of CCD4 determines carotenoid profiles in non-photosynthetic tissues of different plant species (Ohmiya et al. 2006; Gonzalez-Jorge et al. 2013; Zheng et al. 2019). CCD7 and CCD8 sub-families are mainly involved in the biosynthesis of strigolactones, a class of phytohormones that regulate plant architecture, as well as rhizospheric interactions with symbiotic arbuscular fungi and root parasitic plants (Alder et al. 2012; Al-Babili and Bouwmeester 2015; Jia et al. 2019a). The Arabidopsis NCED sub-family consists of five genes, including NCED2, NCED3, NCED5, NCED6, and NCED9 (Tan et al. 2003). NCEDs mainly catalyze the stereospecific cleavage of 9-cis-epoxycarotenoids into apo-12′-epoxycarotenal (C25) and the ABA precursor xanthoxin (C15), which is a key rate-limiting step in ABA biosynthesis (Schwartz et al. 1997; Giuliano et al. 2003; Felemban et al. 2019). ABA plays a key role in plant response to various abiotic stresses, such as cold, heat, and drought (Nambara et al. 2010; Peleg and Blumwald 2011). In addition to the above-mentioned groups common present in Arabidopsis, rice (Oryza sativa) and other species contain a further CCD sub-family that has been recently identified and called zaxinone synthase (ZAS) (Wang et al. 2019). A rice ZAS was reported to catalyze the cleavage of 3-OH-β-apo-10′-carotenal into zaxinone, a novel carotenoid-derived plant growth regulator required for normal rice growth and a determinant of strigolactone content in both rice and Arabidopsis (Wang et al. 2019; Ablazov et al. 2020).
Albeit their importance for plant growth, development and response to environmental abiotic stresses, a comprehensive study on the CCD gene family in Gossypium spp. is still missing. Such a study has become possible through the recent release of high quality of third generation genome sequencing of diploid and allotetraploid Gossypium species (Wang et al. 2012; Li et al. 2014; Fang et al. 2017; Hu et al. 2019). Here, we performed genome-wide analysis to identify CCD family genes in two diploid and two allotetraploid Gossypium species, and comprehensively analyzed phylogeny, gene structure, gene duplications, cis-acting elements, molecular evolution and gene expression for the cotton CCD gene family. Our study demonstrates the role of cotton CCDs in regulating abiotic stress response and provides important insights for investigating their biological functions in the future.
Methods
Identification of the CCD gene family members in Gossypium spp.
Two approaches were adopted to identify cotton CCD genes in different Gossypium genomes. First, the whole genome translation protein sequences of G. arboretum (A2, CRIv1.0), G. raimondii (D5, JGI_v2.0), G. hirsutum (AD1, ZJUv2.1), and G. barbadense (AD2, H7124_ZJUv1.1) were downloaded from the CottonGen database (https://www.cottongen.org/home). Nine Arabidopsis and 13 rice CCD proteins downloaded from (https://phytozome.jgi.doe.gov/pz/portal.html) were used as a query to search for CCD proteins in four Gossypium species protein databases by local blast tool with default setting(e-value < 1e−10). Then, the hidden Markov model (HMM) profiles with RPE65 (PF03055) domains (http://Pfam.sanger.ac.uk/), and the HMMER 3.0 software (http://hmmer.org/) (Potter et al. 2018) were used to perform local HMM searches in the four protein databases. All candidate sequences were submitted to domain analysis using InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan5/) and SMART (http://smart.embl-heidelberg.de/) tools with default parameters to substantiate the existence of the two conserved domains and determine the exact location of them. Protein sequences without any one domain were rejected.
Comparative phylogenetic analysis of the cotton CCD proteins
CCD protein sequences from G. arboretum, G. raimondii, G. hirsutum, G. barbadense, Arabidopsis and rice were performed multiple alignments in MUSCLE program of Molecular Evolutionary Genetics Analysis (MEGA 7) software (Kumar et al. 2016). An unrooted phylogenetic tree was constructed based on the NJ method, with a 1000 bootstrap replicates with a Jones–Taylor–Thornton (JTT) model. The phylogenetic tree was further visualized and annotated using Evolview (https://www.evolgenius.info) (Subramanian et al. 2019).
Gene exon/intron architecture and promoter cis-acting element analysis of the cotton CCD genes
The gene exon/intron architecture of CCD genes from the four Gossypium species was determined using the Gene Structure Display Server (GSDS: http://gsds.cbi.pku.edu.cn/) based on the coding and corresponding genomic sequences (Hu et al. 2015). For determining cis-acting elements of GhCCDs promoters, 2 kb upstream sequences were analyzed in PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/search_CARE.html) (Lescot et al. 2002 ) and the cis-acting elements were categorized based on their predicted functions.
Chromosome location and synteny analysis
Chromosomal positions of the CCD genes were obtained from the annotation information of cotton genome database (https://www.cottongen.org/home). The chromosomal distributions of genes were drafted on the cotton chromosomes according to the gene positions by Mapchart software (http://www.plantbreeding.wur.nl/UK/software_mapinspect.html) (Voorrips 2002). Meanwhile, the orthologous/paralogous gene pairs of the CCD gene in A, D genomes, At- and Dt-subgenomes were searched by InParanoid software (http://inparanoid.sbc.su.se/cgi-bin/index.cgi) (Sonnhammer and Ostlund 2015). Additionally, the evolutionary rates Ka, Ks, and Ka/Ks ratio were estimated by KaKs_Calculator package (https://kakscalculator.herokuapp.com/) (Wang et al. 2010). On the basis of the synonymous substitutions per year (λ) of 2.6 × 10–9 for cotton, the divergent time of CCD orthologous gene pairs were estimated by T = Ks/2 λ × 10–6 MYA (Zhang et al. 2015).
Transcriptome analysis and quantitative reverse-transcriptional PCR (qRT-PCR) confirmation of GhCCD gene expression
All raw RNA-seq data were downloaded from the NCBI Sequence Read Archive (SRA: PRJNA490626). TopHat2 (Kim et al. 2013), and cufflinks (Trapnell et al. 2010) were used to analyze transcriptome data. Gene expression was measured in fragments per kilobase million (FPKM). The TBtools program (Chen et al. 2018) was used to display the heat map of gene expressions.
Four-week-old G. hirsutum L. acc. Texas Marker-1 (TM-1) seedlings grown in a climate-controlled chamber (light/dark cycle: 16 h at 28 °C/8 h at 22 °C) were used for ABA and different abiotic stresses treatments. 100 μM ABA, 42 °C, 200 mM NaCl and 5% PEG6000 were used as ABA, heat, salt and PEG treatment conditions, and normal-grown TM-1 seedlings were used as a control group. Leaves collected at 3 h or 6 h after treatments onset were immediately frozen in liquid nitrogen and stored at – 80 °C. Total RNA was extracted using an RNAprep Pure Plant Kit (Tiangen, Beijing, China), according to the manufacturer’s instructions. The first-strand cDNA was synthesized using a PrimeScript RT reagent kit (Takara, Dalian, China). Oligo 7.0 software was used to design the gene-specific primers for qRT-PCR (Table S5). Cotton Histone 3 (GenBank accession no. AF094716) was used as an internal control. The qRT-PCR (Promega, Madison, WI, USA) on an ABI 7500 real-time PCR system (Applied Biosystems, USA) with three replicates. The 2−ΔΔCT method was used to calculate the relative expression levels of GhCCDs, and t-tests were used for statistical analysis.
Results
Identification and characterization of CCD genes in Gossypium spp.
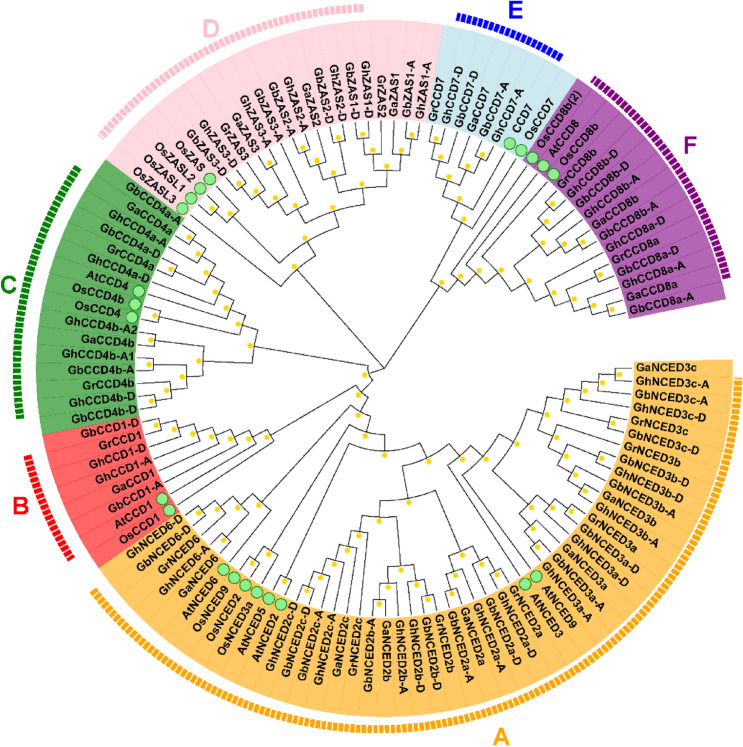
CCDs were characterized as a RPE65 (retinal pigment epithelial membrane protein) domain which is key for their enzymatic oxidative carotenoid cleavage activity (Ohmiya 2009). In the present study, we employed two approaches to identify CCD family genes in two allotetraploid Gossypium species, G. hirsutum and G. barbadense, and two diploid Gossypium species, G. arboreum and G. raimondii, respectively. First, we used the CCD protein sequences from the dicot model plant Arabidopsis and the monocot model plant rice as query to perform a protein blast analysis in the four corresponding Gossypium genome databases, respectively. Then, we did another Pfam search and SMART programs analysis using the RPE65 domain in the same corresponding cotton genome database. By combination of the results from the two approaches and removal of the reduplicating proteins, we identified a total of 16, 15, 33 and 31 putative CCD members in G. arboreum, G. raimondii, G. hirsutum and G. barbadense, respectively (Table S1). The cotton CCD genes were then named according to their chromosomal location and homologs in Arabidopsis and rice (Fig. 1 and Table S1).
Fig. 1.
Phylogenetic tree of CCD proteins from Arabidopsis, rice, G. hirsutum, G. barbadense, G. arboreum and G. raimondii. Full-length CCD proteins were aligned using MUSCLE program in MEGA 7.0. The phylogenetic tree was generated by the Neighbor-Joining (NJ) method with 1,000 bootstrap replicates. CCD proteins were divided into A-F sub-families distinguished by different colors. CCD proteins of Arabidopsis and rice were marked by green dots
We next investigated the basic features of all CCD genes and their corresponding encoding polypeptides from the four Gossypium species, e.g., the gene ID, chromosomal location, gene length, open reading frames (ORF) and deduced protein length, predicted protein molecular weights (MW) and isoelectric points (pI), predicted chloroplast transit peptide (cTP) presence and length, and predicted subcellular localization. As shown in Table S2, the predicted protein lengths of 90.5% of cotton CCDs (86 out of 95) range from 508 to 667 amino acids (aa), with molecular weight (MW) from 57.553 to 74.461 Da. There are four cotton CCDs (GhCCD4b-A1, GhCCD4b-A2, GhNCED2c-D, GhZAS2-A) with a predicted encoded protein length of less than 400 aa, suggesting the occurrence of deletions or truncations. The gene length of all CCDs ranges from 351 to 9070 bp, indicating a large diversity in the gene structure. CCD proteins usually, with the exception of CCD1s, contain an N-terminal cTP domain which targets them to the plastid, where carotenoids are synthesized and metabolized (Al-Babili and Bouwmeester 2015). We used the online cTP analysis software “ChloroP 1.1 Server” (http://www.cbs.dtu.dk/services/ChloroP/) to predict the presence of cTP in the identified CCDs. As shown in Table S2, most CCDs contain a CTP domain with the exception of CCD1s and CCD8as from all the four Gossypium species which were predicted without a cTP domain presence.
Classification and phylogenetic analysis of the cotton CCD genes
To investigate the evolutionary relationships of the 117 CCD genes from 6 different plant species including A. thaliana, O. sativa, G. arboreum, G. raimondii, G. hirsutum and G. barbadense, we generated a phylogenetic tree using the Neighbor Joint (NJ) method based on multiple sequences alignment with 1000 bootstrap replicates. As shown in Fig. 1, all CCDs from the four Gossypium species were clearly divided into 6 sub-families, CCD1, CCD4, CCD7, CCD8, ZAS and NCED, which is similar to the case in rice. CCD4 sub-family is divided into two sub-branches, CCD4a and CCD4b (Fig. 1). CCD4a sub-branch shows high similarity with OsCCD4s and AtCCD4; while, CCD4b branch is divergent from OsCCD4s and AtCCD4 in the phylogenetic tree. A multi-member NCED sub-family (NCED2a, NCED2b, NCED2c, NCED3a, NCED3b, NCED3c and NCED6) was identified in all the four Gossypium species (Fig. 1). In addition, Gossypium species contain a multi-member ZAS sub-family which showed high similarity with rice (Fig. 1). The phylogenetic tree indicated that G. hirsutum experienced significant gene family expansion, because it has more than double the number of CCD genes compared to A. thaliana and O. sativa. The numbers of CCD sub-family members in the two allotetraploid Gossypium species are almost double of those in the two diploid species, except CCD4b in G. hirsutum and NCED6 in G. barbadense. In G. hirsutum, we noticed that GhCCD4b-A was broken into two genes, GhCCD4b-A1 and GhCCD4b-A2, likely due to gene truncation. In G. barbadense, GbNCED6 is only present in the D-subgenome but not in the A-subgenome. These results suggest that most cotton CCD family genes likely retain conservative biological functions during evolution. In addition, the clusters provide evidence that G. hirsutum and G. barbadense are the result of hybridization of two diploid cotton species.
Gene structure and promoter analysis of the cotton CCD family genes
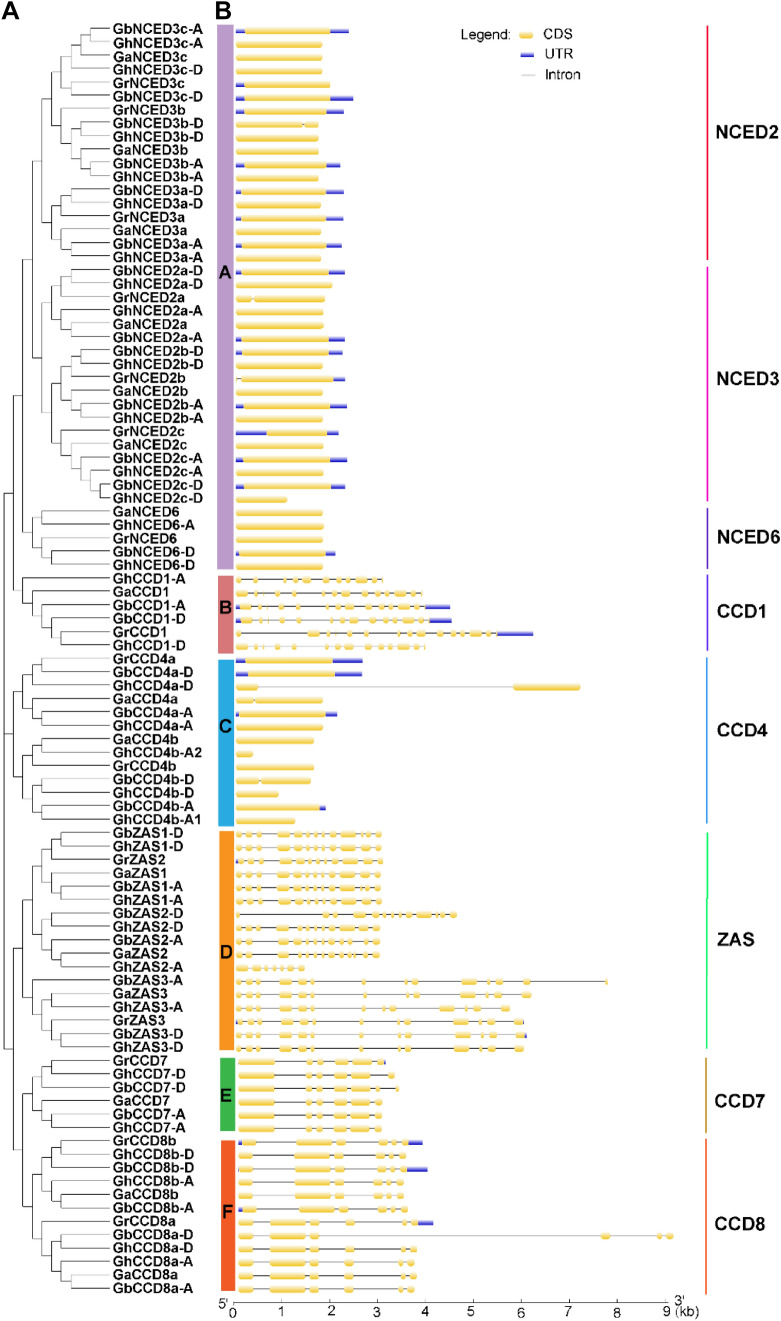
The gene structure, e.g., number of introns, plays an important role in the evolution of gene function (Roy and Gilbert 2006). Therefore, we compared the exon/intron number of different CCDs from the four Gossypium species using GSDS online tools (http://gsds.cbi.pku.edu.cn/). As shown in Fig. 2, the structure of the exon/intron displayed a similar pattern among the same CCD sub-family; while, different CCD sub-families displayed quite divergent exon/intron structures. Specifically, no more than one intron is present in all members of both NCED and CCD4 sub-families; while all members of the CCD1 and the ZAS sub-families contain no less than 10 introns except GhZAS2-A (Fig. 2). All CCD7 and CCD8 genes from the four Gossypium species contain five introns except GhCCD7-D (Fig. 2). These results suggest the common origin of the gene members from same sub-family and are in line with the functional diversity from different CCD sub-families.
Fig. 2.
Gene structures of CCD genes from different Gossypium spp.. a The phylogenetic tree was constructed using the Neighbor-Joining (NJ) method, with 1000 bootstrap replicates. Sub-families are distinguished by different colors. b Exon/intron architectures of CCD genes from different Gossypium spp.. Yellow and blue boxes indicate exons and UTRs, respectively. Black lines represent introns. The size of exons and introns can be calculated according to the scale at the bottom
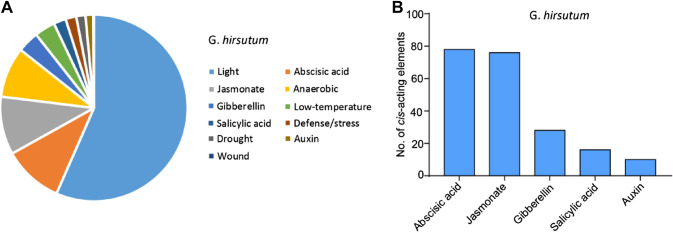
The cis-acting elements located in gene promoters are binding sites of various transcription factors which are key to regulate gene expression (Bilas et al. 2016). To predict the gene regulation behavior of different CCDs in G. hirsutum, we analyzed the cis-acting elements in the 2 kb promoter region of GhCCDs and categorized them based on their functional relevance. As shown in Fig. 3a, the identified cis-acting elements were mainly classified into light, hormones, and stress response-related elements, suggesting complex regulation of GhCCDs by various environmental cues and hormonal signals. We further investigated the cis-acting elements that are related to hormones, which showed that most of them are targeted by ABA- and jasmonate acid-responsive genes (Fig. 3b). The other hormone related cis-acting elements were predicted to be targeted by salicylic acid-, gibberellins- and auxin-responsive genes (Fig. 3b). ABA, jasmonate acid and salicylic acid are important regulators of biotic and abiotic stress responses (Ku et al. 2018), suggesting the expression of GhCCDs are involved in these stress response processes.
Fig. 3.
Classification of cis-acting elements in GhCCD gene promoters. a The percentage of different cis-acting elements in the GhCCD promoters. b Numbers of cis-acting elements related to hormone response. The 2 kb promoter region of the GhCCD genes was used for cis-acting elements analysis
Gene duplication and syntenic analysis of the CCD family genes in Gossypium spp.
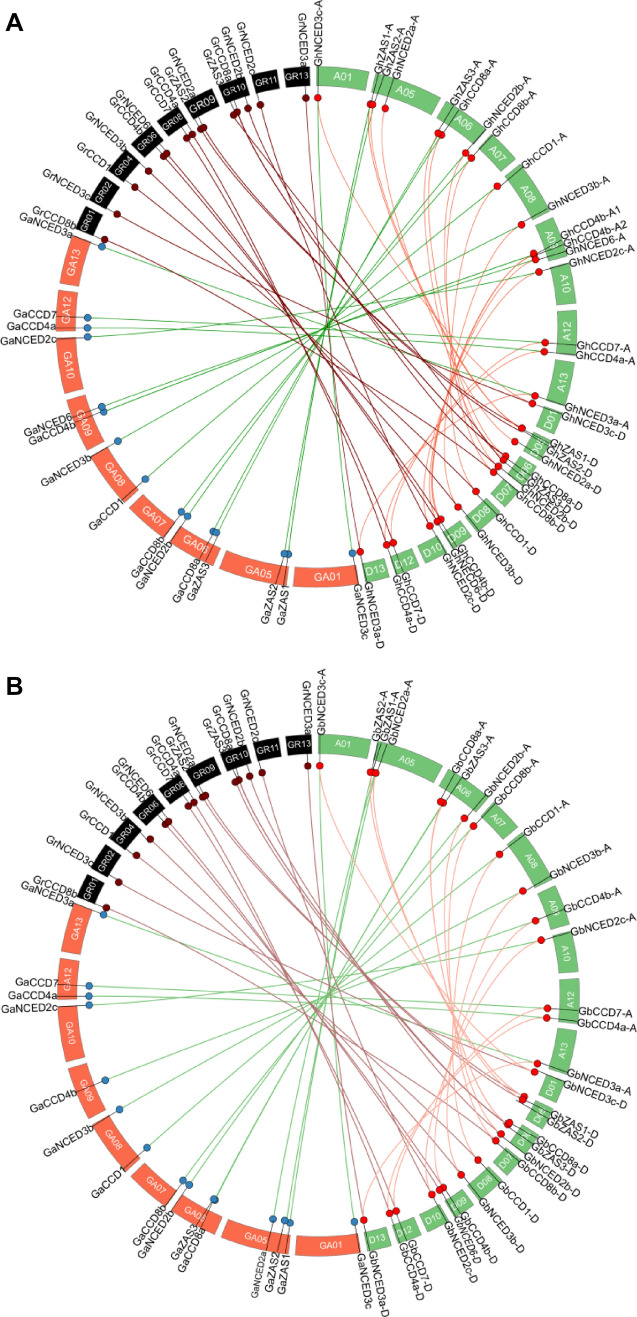
Gene duplication is a common event during evolution, which is important to generate functional divergence. To study the locus relationships among the CCD genes in Gossypium spp., we identified the orthologous/paralogous gene pairs in the subgenomes of the two allotetraploid cottons, G. hirsutum and G. barbadense. 47 and 44 orthologous/paralogous gene pairs were identified for G. hirsutum and G. barbadense, respectively (Fig. 4, Table S3 and S4). Most CCDs in the A and D subgenomes of G. hirsutum and G. barbadense have corresponding orthologous/paralogous in the A (G. arboreum) or D (G. raimondii) genomes. As shown in Fig. 4, the gene syntenic analysis indicates that the gene duplication in the two allotetraploid Gossypium species is mainly due to hybridization. This provides evidence that cotton CCD genes likely have not experienced large-scale genomic arrangements during polyploidization.
Fig. 4.
Genome-wide synteny analysis for CCD genes among allopolyploid and diploid Gossypium species. a Synteny analysis between G. hirsutum and two diploid species, G. arboreum and G. raimondii. b Synteny analysis between G. barbadense and two diploid species, G. arboreumand G. raimondii. Green lines link gene pairs between G. arboreum and G. hirsutum or G. barbadense; dark red lines connect gene pairs between G. raimondii and G. hirsutum or G. barbadense; light red lines bridge gene pairs between At- and Dt-subgenome in allopolyploid cotton. Red boxes indicate chromosomes of G. arboreum, dark boxes indicate chromosomes of G. raimondii, and green boxes indicate chromosomes of G. hirsutum or G. barbadense
To investigate whether the duplicated orthologous/paralogous gene pairs of G. hirsutum and G. barbadense gained functional divergence during evolution, the Non-synonymous (Ka) and synonymous (Ks) divergence values were calculated. Usually, Ka/Ks ratios less than 1 indicate that the genes have undergone purifying selection, while ratios above 1 point to positive selection. For G. hirsutum, out of the Ka/Ks ratios of the 49 duplicated gene pairs, 36 were < 0.5, nine were between 0.5 and 1.0, and five gene pairs were > 1.0 (Table S3). For G. barbadense, out of the Ka/Ks ratios of the 44 duplicated gene pairs, 30 were < 0.5, 11 were between 0.5 and 1.0, and three gene pairs were > 1.0 (Table S4). These results suggest that most CCDs of both G. hirsutum and G. barbadense have been subjected to a strong purifying selection, with exception of five CCDs (GhCCD4b, GhNCED2b-A, GhNCED3c-A, GhNCED6-A and GhZAS1-A) in G. hirsutum and three CCDs (GbNCED2b-A, GbZAS1-A, GbCCD4b-D) in G. barbadense that likely experienced positive selection (Table S3 and S4).
Expression patterns of the CCD genes in G. hirsutum
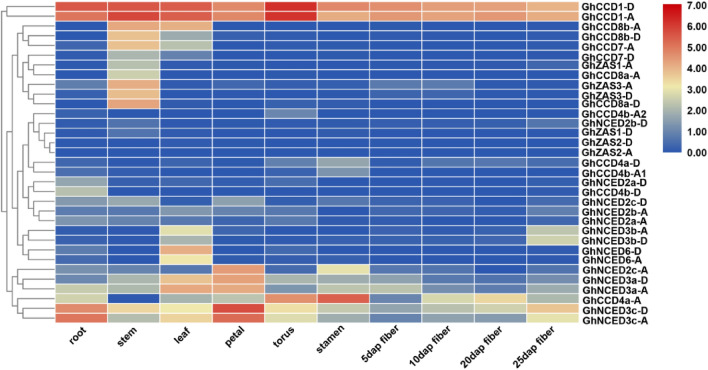
Spatial expression pattern of genes is important for the study of their biological functions. Therefore, we analyzed the tissue expression pattern of different CCDs in G. hirsutum, using the published transcriptome data downloaded from NCBI (SRA: PRJNA490626) for 10 different tissues mirroring key developmental stages in cotton growth and development. These tissues include roots, stems, leaves, petals, torus, stamens, and fibers sampled at 5, 10, 20 and 25 days post anthesis (DPA). As shown in the Heat Map in Fig. 5, CCD1 sub-family showed ubiquitous high expression in all selected tissues. Both CCD7 and CCD8 sub-families exhibited high expression in stems and leaves (Fig. 5). All members of the ZAS sub-family were expressed at low levels in all tissues, with exception of GhZAS1-A, GhZAS3-A and GhZAS3-D, which showed a stem-specific expression pattern (Fig. 5). For CCD4 sub-family, GhCCD4a-A exhibited a ubiquitous expression with high expression rates in torus and stamens, while all the other members showed a general low expression in all tissues (Fig. 5). Members of the NCED sub-family were characterized by different expression patterns, with four and eight members showing ubiquitous and tissue specific expression pattern, respectively (Fig. 5).
Fig. 5.
Tissue expression pattern of GhCCD genes. Heat map of the tissue expression pattern of 33 GhCCD genes in 10 different tissues, including roots, stems, leaves, petals, torus, stamens, and fibers sampled at 5, 10, 20 and 25 DPA. The expression data were analyzed using the published transcriptome data downloaded from NCBI (SRA: PRJNA490626)
CCD genes in G. hirsutum are induced by different abiotic stressors
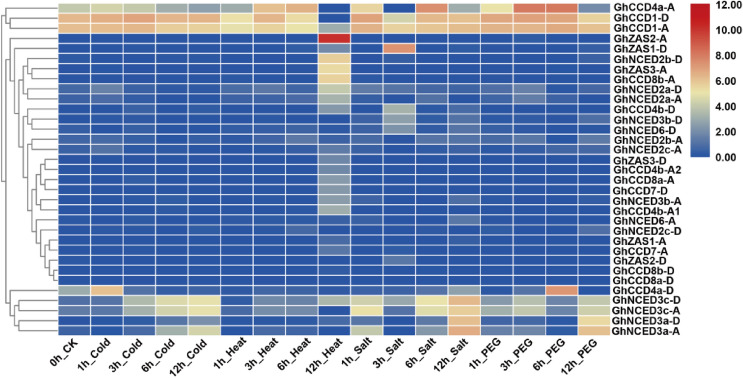
To get insight into the responses of Gossypium CCDs to abiotic stressors, we analyzed the related transcriptome data from NCBI (SRA: PRJNA490626). The analyzed data include four different abiotic stress factors, i.e., cold, heat, salt and PEG treatment at time points 1, 3, 6, and 12 h. As shown in Fig. 6, GhCCD4a-A/D and GhNCED3s were induced under most abiotic stress conditions. Most members of the CCD7, CCD8, GhNCED2s and several ZAS sub-family members showed specific induction by heat stress (Fig. 6). These results suggest that GhCCDs likely contribute to abiotic stress responses.
Fig. 6.
Heat map of the expression of GhCCDs under different abiotic stressors. The abiotic stressors include cold, heat, salt and PEG treatment for 1, 3, 6, 12 h. The expression data were analyzed using the published transcriptome data downloaded from NCBI (SRA: PRJNA490626)
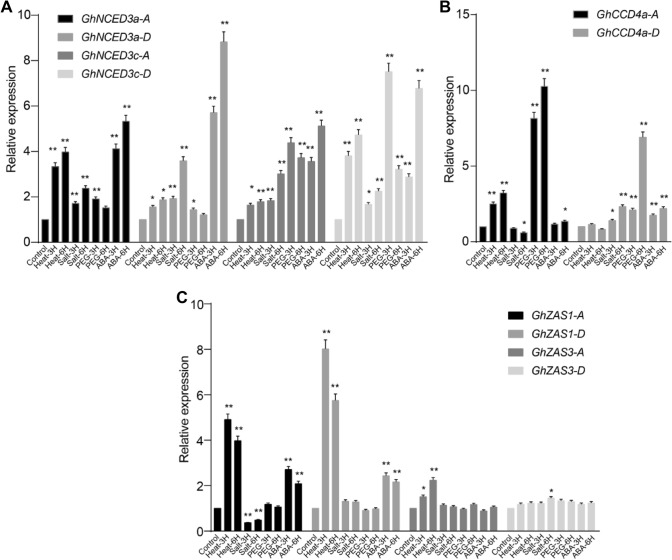
To confirm the published transcriptome data and to investigate the effect of ABA application on the expression of CCD genes in G. hirsutum, we performed a qRT-PCR analysis from seedling exposed to heat-, salt-, PEG- and ABA-treatment. Four NCEDs (GhNCED3a-A/D, GhNCED3c-A/D), two CCD4a (GhCCD4a-A/D) genes and four ZAS genes (GhZAS1-A/D, GhZAS3-A/D), which were obviously induced by different abiotic stress factors in the transcriptome data, were selected for the qRT-PCR investigation. As shown in Fig. 7, the four NCEDs, GhCCD4a-D and GhZAS1-A were clearly induced by ABA application. The transcript levels of GhNCED3a-A, GhNCED3c-D, GhCCD4a-A, and GhZAS1-A/D increased significantly upon heat treatment, while salt treatment for 6 h enhanced the expression of GhNCED3a-D, GhNCED3c-A, and GhCCD4a-D. In addition, NCEDs and CCD4as, especially GhNCED3c-D and GhCCD4a-A, were strongly induced by PEG treatment. Most of these results are consistent with the transcriptomic data analysis (Fig. 6) and support the importance of GhCCDs in the reaction to abiotic stresses.
Fig. 7.
qRT-PCR expression analysis of selected GhCCD genes under different abiotic stresses treatments or upon exposure to exogenous ABA for 3 h and 6 h. Relative expression level of selected GhNCEDs (a), GhCCD4s (b) and GhZASs (c) under different conditions. Relative gene expression levels were normalized to the reference gene (GhHistone 3) and were re-adjusted to the expression levels in the control, which were set as 1. Average fold change ± SE (n = 3). Two-tailed Student’s t-test, *P < 0.05, **P < 0.01
Discussion
CCDs are key enzymes involved in carotenoid metabolism and the production of phytohormones, plant bioactive molecules, as well as plant pigmentations (Ohmiya 2009; Felemban et al. 2019; Wang et al. 2021 ). In this study, we comprehensively characterized the CCD gene family in four Gossypium species, including two allotetraploid cotton, G. hirsutum and G. barbadense, and two diploid cotton, G. arboreum and G. raimondii. CCDs from all four Gossypium species are classified into six sub-families corresponding to the CCD1, CCD4, CCD7, CCD8, ZAS and NCED clades, similar to rice and other mycorrhizal plants; in contrast, Arabidopsis lacks ZAS sub-family (Fig. 1) (Wang et al. 2019). In addition, the gene structural analysis showed that the pattern of exons/introns distribution was highly conservative among the same sub-family and quite diverse in different sub-families (Fig. 2). Introns number is an important factor for the evolution of gene function and regulation, and exon/intron structure differences usually result from insertion/deletion events in genes and are useful for investigating gene evolution (Lecharny et al. 2003). Furthermore, introns are considered to be under weak selection pressure, and gene families with fewer or no introns are usually thought to be evolutionarily advanced (Roy and Gilbert 2005; Roy and Penny 2007). Thus, it seems that the CCD4 and NCED sub-family likely evolved faster than the other CCD sub-families (Fig. 2). This is consistent with reported biological functions of CCD4s and NCEDs, which are involved in secondary metabolism and abiotic stresses, respectively, and are highly adapted the environmental cues (Ohmiya 2009). Moreover, it is supposed that the intron-free structure of NCED genes is related with the rapid induction of ABA production and ABA-mediated stress response in plants (Fig. 2) (Wang et al. 2017).
It is assumed that allotetraploid cotton arose due to one hybridization event between A-genome of G. arboreum (A2) and D-genome of G. raimondii Ulbrich (D5) about 1–1.5 million years ago (MYA) (Endrizzi et al. 1985; Wendel 1989). The genome-wide syntenic analysis for CCD genes among allopolyploid and diploid Gossypium species well supported the above assumption (Fig. 4). Gene duplication generates functional divergence, which is essential for environmental adaptability and speciation (Conant and Wolfe 2008). The similar distribution pattern of CCDs on the chromosomes of A- and D-subgenome of G. hirsutum and G. barbadense indicates that CCDs likely have not experienced large-scale genomic arrangements during polyploidization (Fig. 4). However, the unequal CCD gene number between G. hirsutum and G. barbadense suggests that gene loss or addition through segmental events may have occurred. Nevertheless, an incomplete genome assembly or inaccurate gene annotation could also be a reason for this observation. We speculate that the CCD gene family underwent a quite conservative evolution after polyploidy. Consistently, evolutionary analysis showed that purifying selection dominated the evolution of these genes (Table S3 and Table S4). However, four CCDs (GhCCD4b, GhNCED2b-A, GhNCED3c-A, GhNCED6-A and GhZAS1-A) in G. hirsutum and three CCDs (GbNCED2b-A, GbZAS1-A, GbCCD4b-D) in G. barbadense, which likely experienced positive selection during evolution, might acquire new biological functions (Table S3 and Table S4).
Cotton CCDs are classified into 6 sub-families according to the phylogenetic analysis. CCD1 sub-family is highly conservative among different species. CCD1 enzymes are characterized by a wide substrate and low double-bond specificity by cleaving various cyclic and acyclic all-trans-carotenoids as well as apocarotenoids (Ilg et al. 2009, 2014). Cotton CCD1s are predicted to be localized in cytoplasm and are ubiquitously expressed in all tissues (Figs. 1 and 5), suggesting Gossypium CCD1s likely have conservative biological functions, e.g., to scavenger unnecessary carotenoids and apocarotenoids similar as AtCCD1 (Ilg et al. 2014). CCD4 sub-family members were showed to affect carotenoid profile in a tissue-dependent manner in different species (Gonzalez-Jorge et al. 2013; Ohmiya et al. 2006; Zheng et al. 2019). Cotton CCD4 sub-family seems have two sub-branches, CCD4a and CCD4b based on the phylogenetic analysis (Fig. 1). The CCD4a sub-branch is highly similar to OsCCD4s rice and AtCCD4, but CCD4b seems represent a different sub-branch (Fig. 1). In G. hirsutum, the GhCCD4a-A is ubiquitously expressed with high levels in stamen, and GhCCD4a-D showed a specific expression in torus and stamen, suggesting GhCCD4a might have important roles in stamen (Fig. 5). GhCCD4b-A1 and GhCCD4b-A2 supposedly originate from the truncation of GhCCD4b-A, which was likely accompanied by the loss of function of both genes, as supported by the fact that both genes are almost not expressed in all tissues (Fig. 5). In addition, GhCCD4b-D is predicted to be cytoplasm localized different with the plastid localization of GhCCD4as, suggesting this gene might evolve new biological functions (Fig. 5).
CCD7 and CCD8 are mainly involved in the biosynthesis of strigolactones (Alder et al.2012). Strigolactones are quite conservative plant hormones which are synthesized even in moss (Decker et al. 2017). Members of cotton CCD8 and CCD7 sub-families showed high similarity to CCD8 and CCD7 from both rice and Arabidopsis based on the phylogenetic analysis, indicating conservative biological functions of them (Fig. 1). As we know, CCD7 and CCD8 catalyzed strigolactone biosynthesis steps that occur in plastids; however, CCD8as from all the four Gossypium species were predicted as cytoplasm localized, suggesting CCD8as might acquire unknown biological functions rather than involvement in strigolactone biosynthesis. In the other side, CCD7s and CCD8bs from all the four Gossypium species were predicted as plastid localized and have similar tissue expression patterns, suggesting they are likely involved in strigolactone biosynthesis (Figs. 1 and 5).
NCEDs catalyze the stereospecific cleavage of 9-cis-epoxycarotenoids into the ABA precursor xanthoxin (C15) (Schwartz et al. 1997; Giuliano et al. 2003). The gene numbers of NCED sub-family dominated the CCD gene family In Arabidopsis and the four Gossypium species, suggesting the gene expansion and important roles of this sub-family (Fig. 1). The transcriptome analysis of NCEDs in G. hirsutum shows that GhNCED3c-A/D and GhNCED3a-A/D were generally high expressed in most tissues (Fig. 6). Furthermore, GhNCED3c-A/D and GhNCED3a-A/D were all strongly induced by ABA application (Fig. 7a), suggesting their involvement in ABA biosynthesis and abiotic stresses (Wan and Li 2006).
ZASs belong to a CCD sub-family which is not present in Brassicacea species. In rice, OsZAS catalyzes the cleavage of apo-10′-zeaxanthinal to produce zaxinone (Wang et al. 2019). Although Arabidopsis doesn't have ZAS homologs, zaxinone was detected in this specie and exogenously applied zaxinone was shown to promote strigolactone and ABA biosynthesis (Ablazov et al. 2020). Cotton ZASs showed high similarity to OsZAS based on phylogenetic analysis, suggesting a similar enzymatic activity (Fig. 1). However, Gossypium spp. are dicotyledonous plants, different from O. sativa. Thus, the biological functions of ZASs in Gossypium species are still open and interesting questions.
Abiotic stress is becoming a major constraint for cotton fiber yield, due to insufficient farm land and climate change. CCD genes have been reported to play key roles in regulating biotic and abiotic stress responses in different species, (Wang et al. 2013; Zhou et al. 2019). The cis-acting elements analysis of the promoter regions in GhCCDs showed that most elements are related to light, hormones, and response to different abiotic stresses, such as drought, wound, and low temperature (Fig. 3). Most identified hormone-regulated cis-acting elements in the promoters of GhCCDs are related to ABA and jasmonate acid, the major two hormones regulating stress responses, suggesting GhCCDs are likely involved in various biotic and abiotic stresses (Fig. 3). This assumption was substantiated by transcriptome data of GhCCDs under various abiotic stress conditions, including cold, heat, salt and PEG treatments (Fig. 6) and by qRT-PCR analysis performed in this study (Fig. 7). Consistently, it was previously reported that the expression of NCEDs in Gossypium is induced by drought stress and H2O2 application (Kong et al. 2016). CCD4 genes were also shown to be enhanced by various abiotic stress factors in different species (Wang et al. 2013; Rubio-Moraga et al. 2014). In the present study, we show that members of GhNCED, GhZAS, GhCCD7 and GhCCD8 sub-families are specifically promoted by heat stress (Figs. 6 and 7). GhNCEDs and GhCCD4as were strongly induced by PEG treatment, suggesting possible roles of these genes in drought stress response (Fig. 7). Taken together, our study identifies the CCD gene family in four Gossypium species and suggests key roles of GhCCDs in regulating abiotic stress response, which paves the way for investigating their biological functions in the future.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgement
We thank Dr. Salim Al-Babili and Dr. Xiongjie Zheng for valuable discussions and critical reading for the manuscript. We thank Dr. Dingding Zhu for technical support.
Author contributions
KJ, YM and SZ designed the research. KJ and YG did the qRT-PCR experiments. KJ, YM, SZ, YG, YZ, JG, KL, WF, ZJ and WL analyzed the data with the input of L-SPT. KJ and SZ wrote the manuscript. YM, L-SPT and WL revised the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Grant No. 31900245 and 31770300), the key research and promotion projects (202102110002), Open Fund of State Key Laboratory of Cotton Biology (CB2019A19) and 111 project of China.
Availability of data and materials
Nine Arabidopsis and 13 rice CCD proteins downloaded from (https://phytozome.jgi.doe.gov/pz/portal.html). All raw RNA-seq data were downloaded from the NCBI Sequence Read Archive (SRA: PRJNA490626).
Declarations
Conflicts of interest
The authors declare that they have no conflict of interest in the publication.
Footnotes
Shulin Zhang and Yutao Guo authors contribute equally to this work.
Contributor Information
Kun-Peng Jia, Email: Kunpengjia_0515@126.com.
Yuchen Miao, Email: miaoych@henu.edu.cn.
References
- Ablazov A, Mi J, Jamil M, Jia K-P, Wang JY, Feng Q, et al. The apocarotenoid zaxinone is a positive regulator of strigolactone and abscisic acid biosynthesis in arabidopsis roots. Front Plant Sci. 2020 doi: 10.3389/fpls.2020.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Babili S, Bouwmeester HJ. Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol. 2015;66:161–186. doi: 10.1146/annurev-arplant-043014-114759. [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, et al. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335(6074):1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- Bilas R, Szafran K, Hnatuszkokonka K, Kononowicz AK. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult. 2016;127(2):269–287. doi: 10.1007/s11240-016-1057-7. [DOI] [Google Scholar]
- Bruno M, Beyer P, Al-Babili S. The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of β-ionone ring-containing carotenes and non-epoxidated xanthophylls. Arch Biochem Biophys. 2015;572:126–133. doi: 10.1016/j.abb.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Scheffler BE, Dennis ES, Triplett BA, Zhang T, Guo W, et al. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2007;145(4):1303–1310. doi: 10.1104/pp.107.107672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, He Y, Xia R. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv. 2018 doi: 10.1101/289660. [DOI] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: How duplicated genes find new functions. Nat Rev Genet. 2008;9(12):938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Decker EL, Alder A, Hunn S, Ferguson J, Lehtonen MT, Scheler B, et al. Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss. Physcomitrella patens New Phytol. 2017;216(2):455–468. doi: 10.1111/nph.14506. [DOI] [PubMed] [Google Scholar]
- Endrizzi JE, Turcotte EL, Kohel RJ (1985) "Genetics, cytology, and evolution of gossypium," in Advances in Genetics, (eds) Caspari EW, Scandalios JG (Academic Press), 271–375. 10.1016/S0065-2660(08)60515-5
- Fang L, Gong H, Hu Y, Liu C, Zhou B, Huang T, et al. Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Bio. 2017;18(1):33. doi: 10.1186/s13059-017-1167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felemban A, Braguy J, Zurbriggen MD, Al-Babili S. Apocarotenoids involved in plant development and stress response. Front Plant Sci. 2019;10:1168. doi: 10.3389/fpls.2019.01168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Al-Babili S, Von Lintig J. Carotenoid oxygenases: cleave it or leave it. Trends Plant Sci. 2003;8(4):145–149. doi: 10.1016/S1360-1385(03)00053-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Jorge S, Ha S-H, Magallanes-Lundback M, Gilliland LU, Zhou A, Lipka AE, et al. Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell. 2013;25(12):4812–4826. doi: 10.1105/tpc.113.119677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimot H, Cogdell RJ. Carotenoids and photosynthesis. Subcell Biochem. 2016;79:111–139. doi: 10.1007/978-3-319-39126-7_4. [DOI] [PubMed] [Google Scholar]
- Hu B, Jin J, Guo A, Zhang H, Luo J, Gao G. GSDS 2.0 an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet. 2019;51(4):739–748. doi: 10.1038/s41588-019-0371-5. [DOI] [PubMed] [Google Scholar]
- Ilg A, Beyer P, Al-Babili S. Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J. 2009;276(3):736–747. doi: 10.1111/j.1742-4658.2008.06820.x. [DOI] [PubMed] [Google Scholar]
- Ilg A, Bruno M, Beyer P, Al-Babili S. Tomato carotenoid cleavage dioxygenases 1A and 1B: Relaxed double bond specificity leads to a plenitude of dialdehydes, mono-apocarotenoids and isoprenoid volatiles. FEBS Open Biol. 2014;4:584–593. doi: 10.1016/j.fob.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia KP, Baz L, Al-Babili S. From Carotenoids to Strigolactones. J Exp Bot. 2017;69(9):2189–2204. doi: 10.1093/jxb/erx476. [DOI] [PubMed] [Google Scholar]
- Jia KP, Li C, Bouwmeester HJ, Al-Babili S (2019a) Strigolactone biosynthesis and signal transduction. (eds) Strigolactones - Biology and Applications. 1–45. Springer, Cham. 10.1007/978-3-030-12153-2_1
- Jia KP, Dickinson AJ, Mi J, Cui G, Xiao TT, Kharbatia NM, et al. Anchorene is a carotenoid-derived regulatory metabolite required for anchor root formation in Arabidopsis. Sci Adv. 2019 doi: 10.1126/sciadv.aaw6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia KP, Mi J, Ablazov A, Ali S, Yang Y, Balakrishna A, Berqdar L, Feng Q, Blilou I, Al-Babili S. Iso-anchorene is an endogenous metabolite that inhibits primary root growth in Arabidopsis. Plant J. 2021;10(10):15271. doi: 10.1111/tpj.15271. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Luo Z, Dong H, Eneji AE, Li W. H2O2 and ABA signaling are responsible for the increased Na+ efflux and water uptake in Gossypium hirsutum L. roots in the non-saline side under non-uniform root zone salinity. J Exp Bot. 2016;67(8):2247–2261. doi: 10.1093/jxb/erw026. [DOI] [PubMed] [Google Scholar]
- Ku YS, Sintaha M, Cheung MY, Lam HM. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int J Mol Sci. 2018;19(10):3206. doi: 10.3390/ijms19103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecharny A, Boudet N, Gy I, Aubourg S, Kreis M. Introns in, introns out in plant gene families: a genomic approach of the dynamics of gene structure. J Struct Funct Genomics. 2003;3:111–116. doi: 10.1023/A:1022614001371. [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46(6):567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- Ma G, Zhang L, Matsuta A, Matsutani K, Yamawaki K, Yahata M, et al. Enzymatic formation of β-citraurin from β-cryptoxanthin and zeaxanthin by carotenoid cleavage dioxygenase 4 in the flavedo of citrus fruit. Plant Physiol. 2013;163(2):682–695. doi: 10.1104/pp.113.223297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y. Abscisic acid and the control of seed dormancy and germination. Seed Sci Res. 2010;20(2):55–67. doi: 10.1017/S0960258510000012. [DOI] [Google Scholar]
- Nisar N, Li L, Lu S, Khin NC, Pogson BJ. Carotenoid metabolism in plants. Mol Plant. 2015;8(1):68–82. doi: 10.1016/j.molp.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Ohmiya A. Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnol. 2009;26(4):351–358. doi: 10.5511/plantbiotechnology.26.351. [DOI] [Google Scholar]
- Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 2006;142(3):1193–1201. doi: 10.1104/pp.106.087130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol. 2011;14(3):290–295. doi: 10.1016/j.pbi.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylides C, Havaux M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA. 2012;109(14):5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo MJ, Alquézar B, Alós E, Medina V, Carmona L, Bruno M, et al. A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. J Exp Bot. 2013;64(14):4461–4478. doi: 10.1093/jxb/ert260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. Complex early genes. Proc Natl Acad Sci USA. 2005;102(6):1986–1991. doi: 10.1073/pnas.0408355101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet. 2006;7(3):211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- Roy SW, Penny D. A very high fraction of unique intron positions in the intron-rich diatom thalassiosira pseudonana indicates widespread intron gain. Mol Biol Evol. 2007;24(7):1447–1457. doi: 10.1093/molbev/msm048. [DOI] [PubMed] [Google Scholar]
- Rubio-Moraga Á, Rambla J, Fernández-de-Carmen A, Trapero A, Ahrazem O, Orzáez D, et al. New target carotenoids for CCD4 enzymes are revealed with the characterization of a novel stress-induced carotenoid cleavage dioxygenase gene from Crocus sativus. Plant Mol Biol. 2014 doi: 10.1007/s11103-014-0250-5. [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276(5320):1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- Sexton PD, Gerard CJ. Emergence force of cotton seedlings as influenced by salinity. Agron J. 1982;74(4):699–702. doi: 10.2134/agronj1982.00021962007400040025x. [DOI] [Google Scholar]
- Sonnhammer ELL, Ostlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43:234–239. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian B, Gao S, Lercher MJ, Hu S, Chen W. Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019;47:270–275. doi: 10.1093/nar/gkz357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Hans J, Walter MH, Matusova R, Beekwilder J, Verstappen FWA, et al. Cloning and characterisation of a maize carotenoid cleavage dioxygenase (ZmCCD1) and its involvement in the biosynthesis of apocarotenoids with various roles in mutualistic and parasitic interactions. Planta. 2008;228(5):789. doi: 10.1007/s00425-008-0781-6. [DOI] [PubMed] [Google Scholar]
- Tan B, Joseph LM, Deng W, Liu L, Li Q, Cline K, et al. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35(1):44–56. doi: 10.1046/j.1365-313x.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Walter MH, Strack D. Carotenoids and their cleavage products: biosynthesis and functions. Nat Prod Rep. 2011;28(4):663–692. doi: 10.1039/c0np00036a. [DOI] [PubMed] [Google Scholar]
- Wan XR, Li L. Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem Biophys Res Commun. 2006;347(4):1030–1038. doi: 10.1016/j.bbrc.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteom Bioinform. 2010;8(1):77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang Z, Li F, Ye W, Wang J, Song G, et al. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 2012;44(10):1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- Wang R, Wang C, Fei Y, Gai J, Zhao T. Genome-wide identification and transcription analysis of soybean carotenoid oxygenase genes during abiotic stress treatments. Mol Biol Rep. 2013;40(8):4737–4745. doi: 10.1007/s11033-013-2570-y. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ding G, Gu T, Ding J, Li Y. Bioinformatic and expression analyses on carotenoid dioxygenase genes in fruit development and abiotic stress responses in Fragaria vesca. Mol Genet Genomics. 2017;292(4):895–907. doi: 10.1007/s00438-017-1321-5. [DOI] [PubMed] [Google Scholar]
- Wang JY, Haider I, Jamil M, Fiorilli V, Saito Y, Mi J, et al. The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat Commun. 2019;10(1):810. doi: 10.1038/s41467-019-08461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Lin PY, Al-Babili S. On the biosynthesis and evolution of apocarotenoid plant growth regulators. Semin Cell Dev Biol. 2021;109:3–11. doi: 10.1016/j.semcdb.2020.07.007. [DOI] [PubMed] [Google Scholar]
- Wendel JF. New World tetraploid cottons contain Old World cytoplasm. Proc Natl Acad Sci USA. 1989;86(11):4132–4136. doi: 10.1073/pnas.86.11.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- Zheng X, Zhu K, Sun Q, Zhang W, Wang X, Cao H, et al. Natural variation in CCD4 promoter underpins species-specific evolution of red coloration in citrus peel. Mol Plant. 2019;12(9):1294–1307. doi: 10.1016/j.molp.2019.04.014. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li Q, Li P, Zhang S, Liu C, Jin J, et al. Carotenoid cleavage dioxygenases: identification, expression, and evolutionary analysis of this gene family in tobacco. Int J Mol Sci. 2019;20(22):5796. doi: 10.3390/ijms20225796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nine Arabidopsis and 13 rice CCD proteins downloaded from (https://phytozome.jgi.doe.gov/pz/portal.html). All raw RNA-seq data were downloaded from the NCBI Sequence Read Archive (SRA: PRJNA490626).