ABSTRACT
Transcription factor (TF)-based biosensors have proven useful for increasing biomanufacturing yields, large-scale functional screening, and in environmental monitoring. Most yeast TF-based biosensors are built from natural promoters, resulting in large DNA parts retaining considerable homology to the host genome, which can complicate biological engineering efforts. There is a need to explore smaller, synthetic biosensors to expand the options for regulating gene expression in yeast. Here, we present a systematic approach to improving the design of an existing oxidative stress sensing biosensor in Saccharomyces cerevisiae based on the Yap1 transcription factor. Starting from a synthetic core promoter, we optimized the activity of a Yap1-dependent promoter through rational modification of a minimalist Yap1 upstream activating sequence. Our novel promoter achieves dynamic ranges of activation surpassing those of the previously engineered Yap1-dependent promoter, while reducing it to only 171 base pairs. We demonstrate that coupling the promoter to a positive-feedback-regulated TF further improves the biosensor by increasing its dynamic range of activation and reducing its limit of detection. We have illustrated the robustness and transferability of the biosensor by reproducing its activity in an unconventional probiotic yeast strain, Saccharomyces boulardii. Our findings can provide guidance in the general process of TF-based biosensor design.
Keywords: synthetic biology, cell-based biosensor, promoter engineering, oxidative stress, reactive oxygen species, Saccharomyces boulardii
The paper presents the design and optimization of a novel yeast biosensor using an engineered promoter to sense oxidative stress, and by extension, the presence of reactive oxygen species.
INTRODUCTION
Biosensors are a broad class of detection devices in which a biological molecule or system is employed to detect a molecule or chemical state of interest. In recent years, there has been increasing interest in cell-based or ‘living’ biosensors, in which the detection is accomplished by a living cell (often a microbe). Such biosensors offer key advantages: (i) since the detection mechanism is encoded in a replicating cell, creating additional copies of the sensor is easily accomplished; (ii) cells are robust to environmental perturbations and can repair themselves when damaged; (iii) some types of cell can be easily introduced into environments that are otherwise inconvenient to sample (the human intestine, for example) and (iv) cells can be engineered to use an internal biosensor's output to drive a metabolic (or other) response, allowing cells to act as individual or collective feedback systems, sensing a state and generating a response without external intervention. Cell-based biosensors have been successfully used to dynamically regulate metabolic pathways in microorganisms with the aim of optimizing chemical production yields (Farmer and Liao 2000; Zhang, Carothers and Keasling 2012; Dahl et al. 2013; Xu et al. 2014; Venayak et al. 2015), for large-scale screening of mutant cell strains for functionality (Zhang, Jensen and Keasling 2015; Dietrich et al. 2013; Umeyama, Okada and Ito 2013; Siedler et al. 2014; Lee and Oh 2015), for environmental monitoring of pollutants (Wang, Barahona and Buck 2013; Cai et al. 2018; Wan et al. 2019), and for diagnostic and therapeutic applications (Archer, Robinson and Süel 2012; Daeffler et al. 2017; Landry and Tabor 2017; Riglar et al. 2017; Riglar and Silver 2018; Woo et al. 2020).
One common method for creating cellular biosensors is to couple a transcription factor (TF) with a target promoter. The TF is selected for its ability to change its activity level or function upon exposure to the chemical signal of interest; the modified TF then activates or represses the promoter's transcriptional activity, modulating the production of a protein that serves as the biosensor's output. In both yeast and bacterial cells, TF-based biosensors have been successfully designed to detect a variety of molecules and environmental states, including, but not limited to, arsenic and mercury (Wan et al. 2019), fatty acids (Zhang, Carothers and Keasling 2012), malonyl-CoA (Xu et al. 2014), xylose (Teo and Chang 2015), naringenin (Wang et al. 2019), S-adenosylmethionine (SAM) (Umeyama, Okada and Ito 2013), tetrathionate and thiosulfate (Daeffler et al. 2017; Riglar et al. 2017), nitrate (Daeffler et al. 2017), and oxidative stress (Zhang et al. 2016). Equipping cells with the capacity to detect the oxidative stress induced by increased intracellular reactive oxygen species (ROS) concentrations could be important in a number of contexts, including in microbial bioproduction (where responding to changes in the redox state can help optimize lipid biosynthesis (Qiao et al. 2017; Xu, Qiao and Stephanopoulos 2017) or the yields of secreted proteins (Raimondi et al. 2008)) and disease diagnosis or surveillance (where the presence of ROSs is strongly correlated with intestinal inflammation (Grisham 1994; Tian, Wang and Zhang 2017)). Here, we will describe how we have improved the design of a biosensor that uses the oxidative stress response pathways in yeast to respond to ROSs.
Beyond specificity for the target molecule, there are other parameters to be tuned and optimized in a cell-based biosensor. The ability to control the biosensor's response curve is key to the process of matching a biosensor to the requirements of a particular application; this includes the magnitude of the response (or dynamic range of activation), the slope (or sensitivity) of the response, and the linear range of detection (or operational range). One attractive candidate for facilitating such tuning is the output promoter itself. Well-established molecular cloning techniques make it straightforward and inexpensive to synthesize, assemble, and modify the DNA sequence of the promoter. Previously-designed promoters in yeast have often been engineered by adding TF-binding sites inside various regions of truncated natural yeast promoters (Bovee et al. 2007; Khalil et al. 2012; McIsaac et al. 2014; Ottoz, Rudolf and Stelling 2014; Mukherjee, Bhattacharyya and Peralta-Yahya 2015; Teo and Chang 2015; David, Nielsen and Siewers 2016; Skjoedt et al. 2016; Ikushima and Boeke 2017; Rantasalo et al. 2018). However, this can result in relatively large promoters that retain substantial regions homologous to the natural promoter sequences, negatively affecting chromosomal integration efficiency. Furthermore, if assembling a multiple-gene heterolgous pathway, the accumulated size of each promoters necessarily increases the amount of additional DNA required to synthetize and introduce within the cells. The desire for reducing regulatory DNA size was one of the principal motivation in the development of synthetic minimal core promoter sequences, a number of which have recently been designed in yeast (Redden and Alper 2015; Portela et al. 2017). These promoters contain only the necessary DNA sequences required to recruit the eukaryotic RNA polymerase II and initiate transcription, while also offering the potential to add TF-binding sites to regulate the expression of a reporter gene; their small size and minimalist construction made them an ideal starting point for the design of our synthetic oxidative stress responsive biosensor in yeast.
In this work, we have optimized the activity of a TF-based yeast biosensor constructed from a small synthetic core yeast promoter. Our starting point was a previously designed oxidative stress sensor activated by the natural yeast TF Yap1 (Zhang et al. 2016). Yap1 is an evolutionarily conserved TF in eukaryotes responsible for the intracellular response to oxidative stress caused by a redox imbalance (Morano, Grant and Moye-Rowley 2012). Starting from a minimal core promoter, we have created a Yap1-dependent oxidative stress biosensor and confirmed that it has a considerable dynamic range of activation in response to oxidative stress induced by several distinct ROSs. During the design and optimization process, we found that the spacing between binding sites, number of Yap1 binding sites, and especially the sequence of the binding site itself can affect the dynamic range of activation of the biosensor. We have been able to achieve biosensor response levels to oxidative stress that, importantly, are far greater than previous oxidative stress sensing biosensors, while also reducing the size of the promoter to only 171 base pairs (including the 5'UTR). We have demonstrated that the addition of a positive feedback loop can significantly improve the dynamic range of activation of the biosensor. Finally, we have established initial evidence for the ability of the biosensor to operate in multiple biological contexts, by demonstrating that it functions efficiently in the non-conventional probiotic yeast strain Saccharomyces boulardii. Beyond the utility of the biosensor itself, our results offer useful guidance in the design of other TF-based biosensors in yeast.
MATERIALS AND METHODS
Media, reagents and chemicals
For yeast cultures, we used standard YPD broth (1% yeast extract, 2% peptone, 2% dextrose). Plasmids encoding the oxidative stress sensors were maintained in S. cerevisiae with yeast synthetic drop-out (SD-URA) media (0.174% yeast nitrogen base, 0.5% ammonium sulfate, 0.2% amino acid mix minus uracil, 2% dextrose). Plasmids for S. boulardii were maintained in synthetic complete (SC) media with 200 µg/mL G-418 (Wisent). For CRISPR-Cas9 mediated integration, YED media (1% yeast extract, 2% dextrose) with 200 µg/mL G-418 was used to reduce the amount of small background colonies after transformation. To grow E. coli strains with cloned plasmids, we used standard LB Miller broth (1% yeast extract, 0.5% Tryptone, 1% NaCl) with 100 µg/mL of ampicillin. Agar plates were made by adding 2% agar to the media before autoclaving. Hydrogen peroxide (30% w/v) was diluted in water and added to yeast cultures at desired concentrations. Other oxidative stress inducing chemicals include diamide (TCI Chemicals) and tert-butyl hydroperoxide (Thermo Fisher Scientific), which were diluted in water prior to use, as well as diethyl maleate (Thermo Fisher Scientific), which was diluted in dimethyl sulfoxide prior to use.
Yeast strains, oligos and plasmids
Yeast strains used for this study are listed in Table S1 (Supporting Information). The yeast strain Saccharomyces cerevisiae (mating type A, S288C background; ATCC:4040002) was used for all initial sensor construction, characterization, and optimization steps. The probiotic strain Saccharomyces boulardii CNCM I-745 was purchased and isolated from the commercially-available Fluorastor® product. Oligonucleotides (for genomic integrations), plasmids, and gene/DNA sequences used for this study are listed in Tables S2, S3 and S4 (Supporting Information), respectively. All primers were ordered unmodified and salt-free from Eurofins Genomics. Plasmids for testing the oxidative stress sensor in S. cerevisiae were cloned into the pRS416 backbone or pRS415 backbone (Sikorski and Hieter 1989). Original CRISPR-Cas9 plasmid and gRNA expressing plasmids were purchased from Addgene, deposited by the Borodina lab ( Addgene:83946, 83947). The previously published oxidative stress sensing biosensor (5XUAS-pTRX2) was purchased from Addgene, deposited by the Jensen lab (Addgene: 124708). The synthetic minimal promoter sequence from the previously characterized core promoter, including the neutral A/T region and the core promoter sequence (Redden and Alper 2015) number 1, was ordered as a gene fragment from Eurofins Genomics. Yap1 binding sites, spacer sequences, and 5’UTR sequences were added or modified by standard PCR cloning protocols and/or NEB HIFI assembly protocols (New England Biolabs Canada). For standard PCR cloning, primers which included insertion or deletion were used to amplify the plasmid following the NEB Q5 protocol. The resulting PCR product was then digested with DpnI at 37°C for 15 min at and gel extracted using the NEB gel extraction kit. Eluted PCR product was then co-treated with T4 polynucleotide kinase and T4 ligase in 1X ligase reaction buffer for 2 hours at room temperature. The resulting reaction mix was subsequently transformed into the DH5 alpha E. coli strain and incubated at 37°C for 16 hours. Three to ten colonies were then picked for plasmid extraction and sequencing. For NEB HIFI assembly, primers were designed to include 20- to 25-nucleotides overhangs homologous to the desired insertion sites. NEB Q5 protocols were used to amplify all backbones and inserts. Amplified backbones were then treated with DpnI at 37°C for 15 min then gel extracted using the NEB gel extraction kit. Amplified inserts were purified using the NEB PCR clean-up kit. Resulting fragments and/or single-stranded oligos were then assembled following the NEB HIFI master mix assembly protocol. Resulting reaction mix was subsequently transformed in DH5 alpha E. coli strain and incubated at 37°C for 16 h. Three to ten colonies were then picked for plasmid extraction and sequencing. Isolates were confirmed by sequencing using primers to span all recombination regions.
Yeast transformation
Plasmid transformation into both S. cerevisiae and S. boulardii followed the standard LiAc/PEG protocol. Briefly, cells were grown to logarithmic growth phase, harvested, washed once with 100 mM LiAc, then resuspended in transformation mix (240 µL 50% w/v P.E.G. (M.M. 3350), 36 µL 1M LiAc, 50 μL 2 mg/mL salmon sperm DNA). Roughly 100 ng of plasmid were added to each reaction mix and dH2O was used to complete volume to 360 µL. The reaction mixes were vortexed for 30 sec, incubated at 30°C for 20 min, then heat-shocked at 42°C for 20 min. Afterwards, cells were harvested and resuspended in YPD. If plasmid selection used auxotrophic markers, transformed cells were immediately plated unto selective media (synthetic drop-out agar media minus the selective amino acid). If plasmids were selected for G-418 resistance, transformed cells were left to recover in 1–2 mL of YPD at 30°C for 4–24 h (longer recovery time (24 h) was required for S. boulardii) with shaking (200-250 rpm) before plating on selective media (YED media plus G-418 (200 µg/mL).
CRISPR-Cas9 mediated integration into yeast chromosome
Cas9 and gRNA expression cassettes were initially on two separate plasmids derived from a previous publication (Stovicek, Borodina and Forster 2015). To simplify the transformation process, the gRNA cassette, including the structural gRNA gene expressed from the SNR52 promoter, was cloned into the Cas9 plasmid at the BsiWI cut site. Expressing gRNA from this plasmid (low-copy CEN plasmid) rather than its original plasmid (high-copy 2 micron plasmid) considerably reduced integration efficiency due to increased background colony formation. However, it was discovered that by screening three or four different gRNA sequences in the region of the desired integration site, we could find at least one gRNA that caused very efficient cutting, resulting in low background colony formation and over 80% integration efficiency of a 1400 bp GFP cassette. For integrating Yap1 cassettes into both S. cerevisiae and S. boulardii, gRNA was designed for efficient cutting and integration in the region adjacent to the previously characterized chromosomal integration site 20, (YPRCΔ15, Chromosome XVI), site 21 (YPRCτ3, chromosome XVI), and site 19 (YORWΔ22, chromosome XV) (Table S4) (Flagfeldt et al. 2009). The Yap1 cassette was amplified by PCR using primers with 50 bp overhangs homologous to the sequence immediately surrounding the gRNA target region. For transformation, we added between 4 µg of Cas9-gRNA plasmid with 3 to 6 µg of purified PCR product into the reaction mix and proceeded with the same protocol as described in the previous section. After recovery in YPD media, cells were plated on YED + G-418 agar plates (using YED media instead of YPD media reduced the amount of small background colonies). Genomic DNA was extracted from resulting colonies and verified for integration by PCR. Resulting PCR product was also verified by sequencing to confirm there weren't any mutations. After PCR confirmation, positive colonies were cured of their plasmid by successive passaging in YDP media for 1–2 days and selection for loss of G-418 resistance. For multiplex genome editing in S. boulardii, GFP reporters (pTDH3_GFP_tADH1) were successively integrated at sites 20, 21, and 19 in the genome making strains with either 1, 2, or 3 GFP reporters. After which, a gRNA was designed and tested for efficient cutting within the GFP gene. To integrate Yap1 cassettes in these strains, healing fragments were created by PCR as described above with 50 bp overhangs homologous to the integration sites. For transformation, the cells were prepared as above before adding 4 µg of the CRISPR-Cas9 plasmid with the GFP-specific gRNA and roughly 3 µg of purified PCR products of the healing fragments and proceeding with the transformation protocol. Colonies were screened for loss of fluorescence using a blue-light illuminator (Safe Imager, Invitrogen) as well as confirmed by PCR and sequencing.
Testing the oxidative stress responsive sensor
Three separate colonies containing the oxidative stress responsive reporter were inoculated in a 96 deep well plate (VWR Canada) in 1 mL of SD-URA (S. cerevisiae) or SC + 100 µg/mL of G-418 (S. boulardii) and grown at 30°C overnight on a plate shaker (600-700 rpm) (VWR Canada). In the morning, cultures were diluted 1/60 in 1 mL of fresh media and grown for 4–5 hours under the same conditions. After which, the oxidative stress inducing chemicals were prepared at 100X concentration in its appropriate vehicle and 10 µL were added to the cultures. Cultures were incubated for another 3 hours under the same growth conditions. At the end of the induction, 200 µL aliquots of each samples were transferred into a 96 black well plate (Thermo) and measured on a Tecan M1000 Pro plate reader: optical density (600 nm), mCherry fluorescence (Exc. 587 ±5 nm/Em. 610 ±5 nm). For the data analysis, the average OD600 and mCherry fluorescence of the media without cells were subtracted from the values of the samples. Then, fluorescence of each sample was normalized to its corresponding OD600 and averaged between its three biological replicates. Fold activation was calculated based on the ratio of the OD-normalized fluorescence between the treated samples and the untreated samples: Fold activation = ((mCherrytreatment—mCherrymedia)/(OD600treatment—OD600media))/((mCherrycontrol—mCherrymedia)/(OD600control—OD600media)). Dose response curves were fitted by nonlinear regression (curve fit) analysis (log[agonist] vs. response- variable slope (four parameters), least squares fit) using GraphPad Prism (version 9.0.0 for Mac OS X, GraphPad Software, San Diego, California USA, www.graphpad.com) with the ‘Bottom’ value fixed to 1.0. Curves were fitted up to an ROS concentration beyond which the response levels of one of the biosensor variants dropped significantly from peak activation (In the case of hydrogen peroxide, for example, the observed decrease in fluorescence became apparent at 600 µM for pSynOs_4(alt1)+, and thus defined as the cut-off concentration for the curve fitting for all curves; see Fig. S1A, Supporting Information) The dynamic range of activation for each curve was calculated from the relative fold activation of the fitted curve at the highest ROS concentration used in the curve fitting. Limit of detection was arbitrarily defined as the lowest ROS concentration needed to induce 2-fold relative expression above the baseline (zero concentration of ROS). Sensitivity was defined as the estimated Hill coefficient of the fitted curves.
RESULTS AND DISCUSSION
Design and characterization of the oxidative stress responsive TF-based biosensor
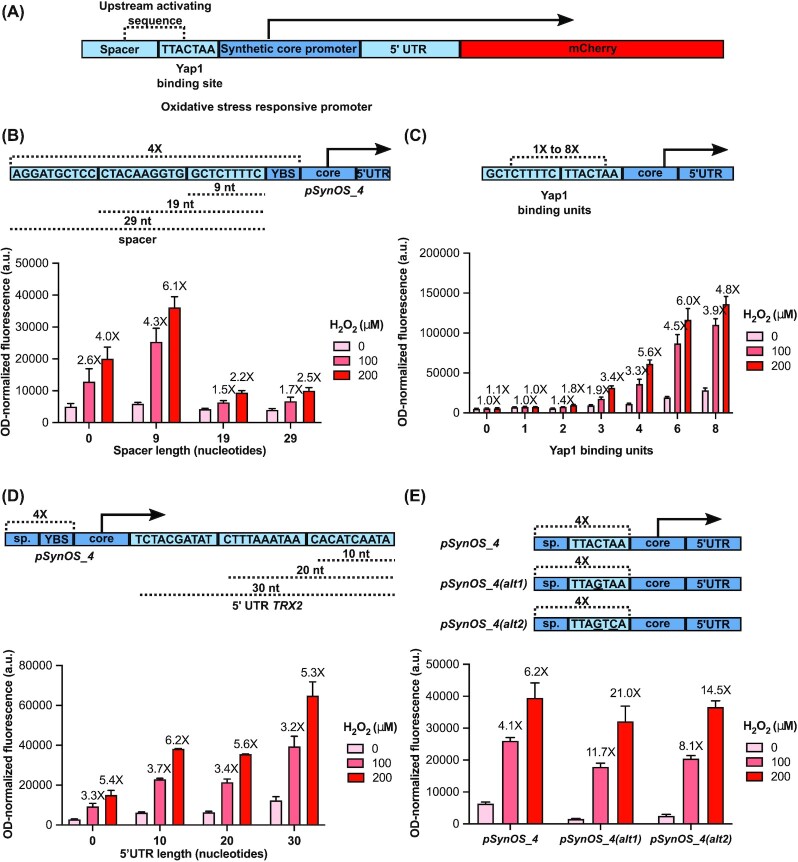
The yeast TRX2 promoter is a natural Yap1-dependent oxidative stress sensing promoter of approximately 275 base pairs (bp) long containing two Yap1 binding sites within its upstream activating sequence (UAS) (Kuge and Jones 1994). A previously published oxidative stress TF-based biosensor was designed by adding multiple tandem repeats of a Yap1 responsive element upstream to the existing TRX2 promoter (as well as other natural core promoters), creating hybrid promoters with sizes reaching over 700 bp (Zhang et al. 2016). To improve on this design, we assembled a promoter using minimal elements, which consisted of: (i) a minimal upstream activating sequence to recruit Yap1; (ii) a synthetic core promoter containing elements required for the recruitment of the RNA polymerase and (iii) an untranslated sequence of nucleotides directly upstream of the start codon of the reporter gene (5’UTR) (Fig. 1A). For the UAS, we added multiple copies of the preferred Yap1 binding site of Yap1 (TTACTAA) (Fernandes, Rodrigues-Pousada and Struhl 1997) while also including a spacer element between each Yap1 binding site, taken from the spacer element between the two Yap1 binding sites in the native TRX2 promoter (nucleotides -182 to -210 of pTRX2; see Table S4, Supporting Information). For the core promoter, we chose a previously characterized synthetic minimal yeast promoter containing a consensus TATA box and transcription start site separated by optimal spacing and sequences (Redden and Alper 2015). Finally, to complete our promoter, we added a 5’UTR whose sequence was taken from the 5’UTR of the TRX2 promoter directly upstream of the start codon (see Table S4, Supporting Information). These promoters were cloned in front of a mCherry reporter on a low-copy vector and tested in S. cerevisiae for activation upon addition of hydrogen peroxide, a reactive oxygen species commonly used to induce oxidative stress in yeast. Initially, we characterized the activity of an earlier version of the synthetic promoter (pSynOS_4) to determine optimal growth and fluorescence measurement conditions (Fig. S1A, Supporting Information), while also confirming it was completely dependent on Yap1 (Fig. S1B, Supporting Information) and that it could be activated by other oxidative stress inducing chemicals (diethyl maleate, tert-butyl hydroperoxide and diamide (Kuge and Jones 1994)) (Fig. S1C, Supporting Information). Altogether, this confirms that the base, initial design of the oxidative stress sensing synthetic promoter can achieve similar function to the previously designed hybrid promoter (Zhang et al. 2016), but it is much smaller in size (171 bp) and modular, which simplified the next steps in the process of optimization. Two major factors were studied to improve biosensor activity and ultimately surpass the function of the previously published biosensor: (i) elements within the promoter sequence itself and (ii) positive-feedback-regulated Yap1 expression.
Figure 1.
Optimizing the synthetic oxidative stress responsive promoter through promoter modifications. A) Schematic of the synthetic oxidative stress biosensor. Various copies of the Yap1 binding site were added in the UAS region of a synthetic core promoter separated by a spacer element. A 30 bp 5’UTR was added after the core promoter.Upon increasing levels of reactive oxygen species, Yap1 will be recruited to the promoter and increase expression of the fluorescent reporter (mCherry). B) The effect of different spacer region length on promoter activity. Synthetic promoters were designed with either 0, 9, 19, or 29 nucleotides spacer length between 4 Yap1 binding sites. Spacer sequences were derived from the TRX2 promoter in yeast directly upstream of the first Yap1 binding site. C) The effect of increasing Yap1 binding units on promoter activity. 0 to 8 copies of a minimal Yap1 binding element were added upstream of the synthetic core promoter. D) The effect of different 5’UTR length on promoter activity. Synthetic promoters were created with 5’UTRs of either 0, 10, 20, or 30 nucleotides in length. Sequences of the 5’UTR were derived from the TRX2 promoter in yeast directly upstream of the start codon. E) The effect of alternative Yap1 binding sites on promoter activity. Synthetic promoters were created using two alternative Yap1 binding sites (pSynOS_4(alt1): TTAGTAA and pSynOS_4(alt2): TTAGTCA). Bar graph illustrates the mean OD-normalized mCherry fluorescence plus standard deviation of three biological replicates under indicated concentration of the hydrogen peroxide. Fold fluorescent change of biosensor compared to basal, non-stressed conditions (0 µM) is reported over each bar.
Modifying the promoter sequence to improve the dynamic range of the biosensor
The minimal design of our biosensor facilitated the process of modifying the promoter to improve its dynamic range of activation. Increasing the dynamic range (that is, the ratio between the maximal response and the no-ROS response) serves to reduce the detection limit of the biosensor, allowing lower ROS concentrations to be reliably distinguished from the basal zero-concentration response. There were four parameters that were varied in the promoter design: (i) the length of the spacer region between the Yap1 binding sites; (ii) the number of Yap1 binding sites; (iii) the length of the 5’UTR and (iv) the sequence of the Yap1 binding site. To optimize the oxidative stress biosensor, we chose to investigate the effect for each of these parameters on the sensor activity under oxidative stress. We first examined the effect of the spacer element in between Yap1 sites on promoter activity. In the TRX2 promoter of S. cerevisiae, the two Yap1 binding sites within its UAS are separated by a 29 nucleotide spacer region (Kuge and Jones 1994). To determine the optimal spacer length for our sensor, we designed promoters with 0, 9, 19 and 29 nucleotides between 4 Yap1 binding sites. The 29 nucleotide spacer is drawn directly from the spacing region between the two Yap1 binding sites of the TRX2 promoter (nucleotides -182 to -210 of pTRX2), and the 9 and 19 nucleotide spacers are truncated versions of this spacer element starting from the -182 position (see Table S4, Supporting Information) (Fig. 1B). Surprisingly, we found that a 9 nucleotide spacer, and not the original 29 nucleotide spacer, was not only functional but in fact necessary for promoter activation, since increasing the spacing region beyond 9 nucleotides prevented significant activation of our reporter upon addition of hydrogen peroxide. On the other hand, adding four consecutive Yap1 binding sites without spacing was still functional but did not respond as well as the promoter with 9 nucleotide spacing. This suggests that there is a previously unknown cooperation between closely separated consecutive Yap1 binding sites that helps to activate transcription upon oxidative stress. The 9 nucleotide spacer sequence was selected for subsequent promoter design.
The second modification we employed was to vary the number of copies of Yap1 binding sites in the upstream activating sequence. We made variant promoters that included up to eight copies of the Yap1 binding site with the 9 nucleotide spacer. Through this approach, we found that we could achieve an oxidative stress sensitive promoter with the addition of at least three Yap1 responsive elements, but the promoter with four Yap1 sites (pSynOS_4) was more responsive to hydrogen peroxide (Fig. 1C). Using a two-way ANOVA test with multiple comparisons (on all pairs of numbers of binding sites), we confirmed that the differences at 100 μM were statistically significant between four and six Yap1 binding sites, while the differences at 200 μM were not. Adding eight Yap1 binding sites decreased the fold activation of the promoter due to higher basal level of expression. A positive correlation between the number of Yap1 TF-binding sites and biosensor activity had been previously reported (Zhang et al. 2016); however, the trade-off we encountered between the number of TF binding sites and the basal level of activation of our biosensor underscores that in TF-based biosensor designs, more TF binding sites is not always better. Not only does it increase the size of the promoter, but it might also lead to an undesirable decrease in fold activation. To achieve the best available results while minimizing the size of the system, the promoter with four Yap1 binding sites was selected for subsequent optimization.
The third modification we employed was to vary the length of the 5’UTR. We originally added a 30 nucleotide 5’UTR upstream of the mCherry reporter with the assumption that it would help with translation of the protein due to the inclusion of potentially necessary elements for efficient ribosomal translation. To determine the effect of the 5’UTR on the sensor activity, we designed promoters with 0, 10, 20 and 30 nucleotide 5’UTRs drawn from the sequence directly preceding the start codon of TRX2 (Fig. 1D). In general, addition of a 5’UTR improved protein expression levels but did not sustantially affect the fold activation. Compared to the 10 and 20 nucleotide 5’UTR, the longer 30 nucleotide 5’UTR achieved higher absolute protein levels without compromising the fold activation of the sensor. Therefore, the 30 nucleotide 5’UTR was kept in the promoter.
Lastly, we examined the effect of modifying the Yap1 binding sequences in our promoter. The original Yap1 binding site (TTACTAA) was used because it was previously determined to be the preferred Yap1 binding site in the native TRX2 promoter (Fernandes, Rodrigues-Pousada and Struhl 1997). To determine how the Yap1 binding sequence affected fold activation of the sensor, we designed promoters with two alternative Yap1 binding sequences: TTAGTAA (pSynOS_4(alt1)) and TTAGTCA (pSynOS_4(alt2)). The first alternative binding site is the less efficient Yap1 binding site within the TRX2 promoter and is simply the inverse (reverse complement) sequence of the preferred binding site, effectively reversing the orientation of the binding site. The second alternative binding site is the inverse sequence of the AP-1 protein (a mammalian analog of Yap1) recognition motif. The original assumption was that these alternative binding sites would reduce basal level of expression of the sensor by changing the optimal orientation and binding affinity of the sites. Surprisingly, although using alternative Yap1 binding sites did reduce basal level of expression under control conditions, it also maintained relatively high levels of expression upon addition of hydrogen peroxide comparable to the preferred Yap1 binding site, thus leading to a substantially improved fold activation (Fig. 1E). Specifically, the promoter with the alternative site TTAGTAA (pSynOS_4(alt1)) improved the fold activation of the sensor from 4.1X to 11.7X and 6.2X to 21.0X at 100 μM and 200 μM of hydrogen peroxide, respectively. This effect of reducing basal level of expression while also retaining much of its activity under stressed conditions is likely the result of the improved spacing and cooperativity between adjacent Yap1 binding sites. The alternative Yap1 binding site TTAGTAA was therefore included in the final design of the oxidative stress sensor.
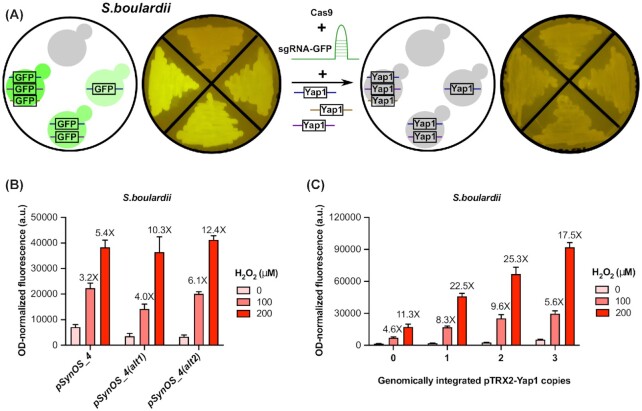
Controlling Yap1 expression with a positive feedback mechanism improves biosensor activation
Another method we used to improve the dynamic range of our biosensor was through the transcriptional regulation the TF Yap1. Positive feedback loops have been used in the past to control the expression of a metabolite-responsive TF to build an organic acid biosensor in yeast (Williams et al. 2017). In that work, an organic acid responsive promoter, pPDR12, was used to control the expression of War1, a transcriptional regulator responsive to organic acids, as well as control the expression of the output signal of the biosensor. This positive feedback loop design improved the dynamic range of their output signal, and we applied this method to improve our oxidative stress biosensor (Fig. 2A). In this design, an additional copy of Yap1 is placed under the control of an oxidative stress responsive promoter (either pTRX2 or pSynOS_4). This way, Yap1 activation under oxidative stress would lead to increased Yap1 expression, leading in turn to increased binding and activation of the downstream oxidative stress sensor. In an initial experiment, we compared the activity of pSynOS_4 expressed in S. cerevisiae with a second plasmid expressing Yap1 from one of the oxidative stress responsive promoters on a low-copy plasmid (Fig. 2B). However, this design led to a substantial increase in basal level of expression of our reporter, while not increasing the high-end expression enough to result in an improvement in the fold activation of the sensor. Additionally, growth was severally impaired for cells containing two plasmid expression vectors. To improve this design, we instead created strains of S. cerevisiae containing one genomically-integrated copy of Yap1 under an oxidative stress responsive promoter (Fig. 2C). Using this design, expressing Yap1 from the native TRX2 promoter did in fact improve the activation of the biosensor from 3.3X to 5.9X and from 5.8X to 9.4X at 100 μM and 200 μM of hydrogen peroxide, respectively. However, expressing Yap1 from the synthetic promoter (pSynOS_4) proved to be ineffective. This seems to indicate that Yap1 expression levels need to surpass a certain threshold before having an effect on the biosensor. This also indicates that there is a trade-off between the number of Yap1 copies we can use in our positive feedback loop and the basal level of expression of our sensor's reporter. To try to maximize the fold induction capability of our positive feedback design, we created strains with up to three integrated copies of Yap1 under the control of the pTRX2 promoter (Fig. 2D). Although increasing the copy number of the pTRX2-Yap1 did lead to a slight increase in reporter expression, the basal level of expression under non-stressed conditions also increased, yielding a decreased fold activation. In summary, controlling Yap1 expression using a positive feedback loop improved the fold activation of the oxidative stress biosensor. However, its effectiveness depends on the oxidative stress responsive promoter regulating Yap1 expression, and on copy numbers.
Figure 2.
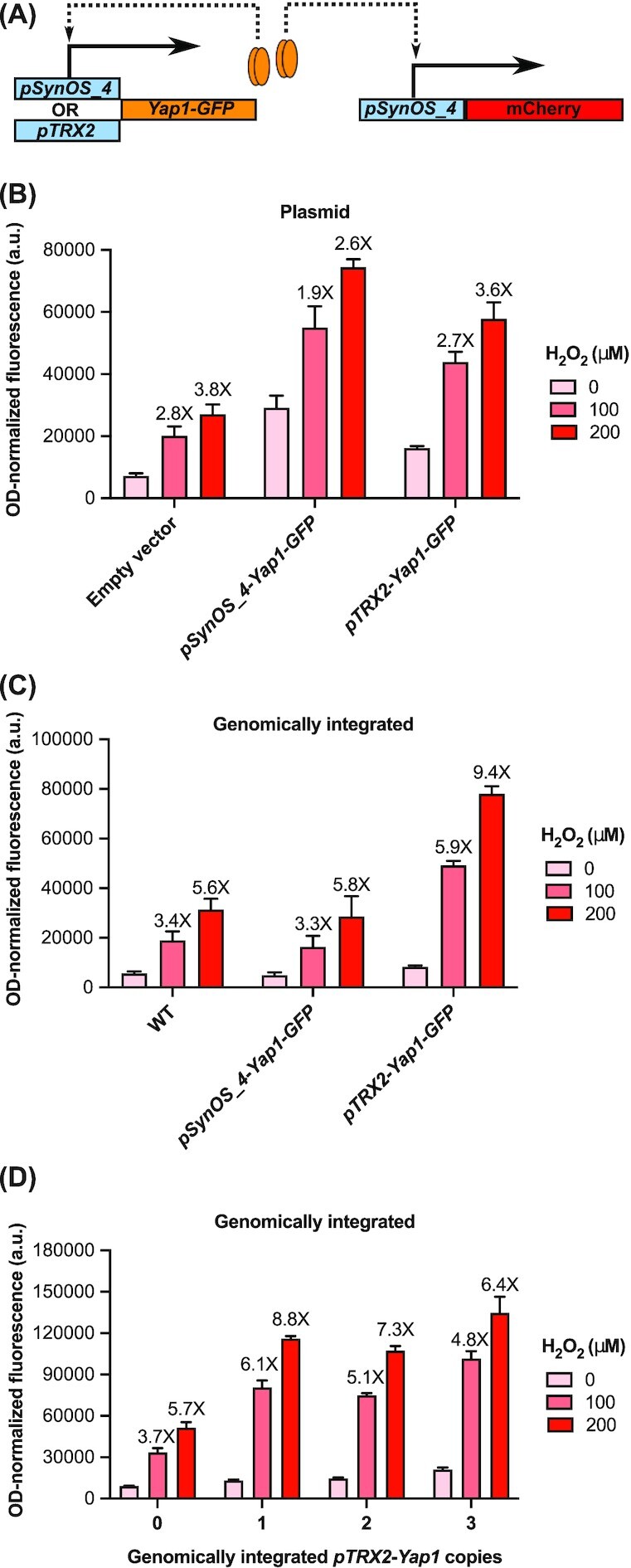
Implementing a positive feedback loop to improve the fold activation of the oxidative stress biosensor. A) Schematic of the positive feedback loop for controlling the expression of Yap1 with two different oxidative stress responsive promoters: pTRX2 and pSynOS_4. We tested the activity of the oxidative stress biosensor pSynOS_4-mCherry with the positive feedback loop design using either B) plasmids or C) genomically integrated Yap1 reporters under two different oxidative stress responsive promoters. D) Testing the effect of integrated copy numbers of the positive feedback loop on the activity of the oxidative stress biosensor. Bar graph illustrates the mean OD-normalized mCherry fluorescence, with error bars indicating one standard deviation over three biological replicates. Fold fluorescent change of biosensor output compared to basal, non-stressed conditions (0 µM) is reported over each bar.
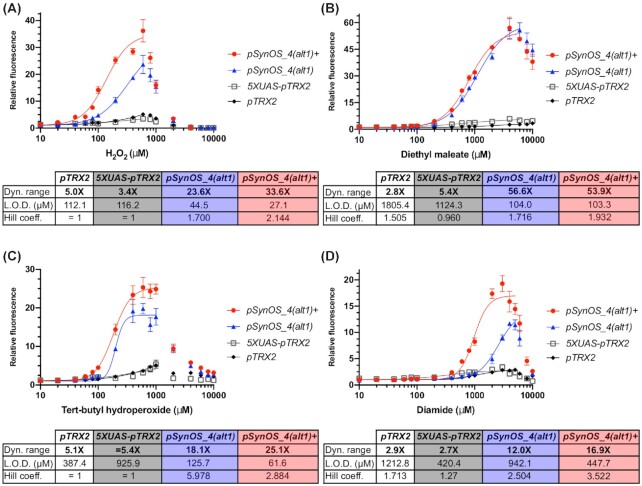
Comparing the activity of the fully optimized synthetic biosensor against a previously published oxidative stress sensing biosensor
To summarize the results from the optimization steps, maximal fold activation of the oxidative stress biosensor is achieved by adding four tandem repeats of a Yap1 binding element in front of the synthetic core promoter which includes a 30 nucleotide 5’UTR. The Yap1 binding element consists of the sub-optimal Yap1 binding sequence (TTAGTAA) separated by a 9 nucleotide spacer element. Finally, the sensor is completed by integrating a Yap1 reporter under the TRX2 promoter into the genome. Within the context of the S. cerevisiae strain, a single copy of the positive-feedback regulated Yap1 reporter is sufficient to achieve the best fold activation of our biosensor.
The fully optimized oxidative stress sensor was compared to the activity of the natural yeast promoter (pTRX2) and to the previously published oxidative stress sensing promoter (5XUAS-pTRX2) (Zhang et al. 2016). Both promoters were first cloned within the same expression vector as the one used with our synthetic promoter. The relative expression of the fluorescent reporter under 5XUAS-pTRX2 in the new expression vector was comparable to that of the original expression vector despite using different fluorescent reporters and different normalization procedures (Fig. S2, Supporting Information). We were therefore able to directly compare activity of these two promoters against the synthetic promoter alone (pSynOS_4(alt1)) and the synthetic promoter with the positive feedback regulated Yap1 (pSynOS_4(alt1)+). The absolute protein expression levels of pSynOS_4(alt1) and pSynOS_4(alt1)+ were lower compared to 5XUAS-pTRX2, which was expected given our use of a synthetic core promoter (Fig. S3, Supporting Information). But, at its peak, pSynOS_4(alt1) and pSynOS_4(alt1)+ reaches protein levels 24% and 58% of that of 5XUAS-pTRX2, respectively. Additionally, protein levels achieved by pSynOS_4(alt1)+ exceed those of the native pTRX2 promoter. This demonstrates that the newly developed biosensors can still achieve relatively high expression levels. The greatest improvement, however, was in terms of the fold activation in response to ROSs. To demonstrate this, we generated dose response curves against all four ROSs to compare the relative responses of each biosensor (Fig. 3A-D). Hill functions could be properly fitted on the dose responses for both pSynOS_4(alt1) and pSynOS_4(alt1)+, which were used to estimate the dynamic range of activation, the limit of detection, and the sensitivity of the biosensors. Hill functions could not be fitted to all of the pTRX2 or the 5XUAS-pTRX2 datasets, but approximate values could be estimated by fixing the Hill coefficient at n = 1, yielding a non-sigmoidal function to be used to compare with the other curves. Even without the positive feedback loop, the synthetic promoter (pSynOS_4(alt1)) outperformed the natural yeast promoter (pTRX2) and the previous published promoter (5XUAS-pTRX2) by a wide margin, with dynamic ranges of activation reaching 21.9X (vs. 5.0X and 3.4X), 56.6X (vs. 2.8X and 5.4X), 18.1X (vs. 5.1X and 5.4X), and 12.0X (vs. 2.9X and 2.7X) with hydrogen peroxide, diethyl maleate, tert-butyl hydroperoxide, and diamide, respectively. Moreover, these improvements also generally led to decreased limits of detection (the lowest concentration of ROS needed to induce a 2X induction of the output) and increased sensitivity (related to the calculated Hill coefficient of the fitted curves) to the ROSs. When we also include the positive feedback loop, the responses to hydrogen peroxide, tert-butyl hydroperoxide and diamide are further improved (with dynamic ranges of activation reaching 33.6X, 25.1X and 18.0X, respectively), and the limit of detection is lowered. Interestingly, the positive feedback loop did not affect the dose response curves to diethyl maleate: the dynamic range of activation of the completed biosensor and that of the synthetic promoter without positive feedback are comparable. With or without the positive feedback, our biosensor does significantly exceed the response of the previous oxidative stress sensing promoter towards diethyl maleate.
Figure 3.
Comparing the dose response of the fully optimized oxidative stress sensing biosensor with previous biosensors in Saccharomyces cerevisiae. Comparison of the dose response curves of the fully optimized biosensor (pSynOS_4(alt1)+) with the synthetic promoter (pSynOS_4(alt1)), the natural yeast promoter (pTRX2), and the previously published Yap1-dependent promoter (5XUAS-pTRX2) against the ROSs A) hydrogen peroxide, B) diethyl maleate, C) tert-butyl hydroperoxide, and D) diamide. 16 to 18 different concentrations of each ROS were used ranging from 10^1 to 10^4 µM. Mean fluorescence levels of the output (mCherry) from three biological replicates is normalized to cell concentration (OD-normalized) and relative to basal levels of expression at 0 µM (±S.D.). Nonlinear regression models (lines) were fitted to the datasets up to an appropriately chosen concentration. If nonlinear regression could not be appropriately fitted, certain variables (Hill coefficient, top value) were fixed ( = ) to allow an approximation of the other values. Table beneath the graph summarizes the calculated maximal dynamic range of activation, limit of detection (L.O.D.), and Hill coefficient from the fitted curves.
To show how versatile our promoter can be, we fused the smaller optimized Yap1-dependent upstream activating sequence to non-synthetic core promoters (pCYC-core, pTRX2-core, pTDH3-core,). And, when also coupled with the positive feedback regulated Yap1 (pCYC-core+, pTRX2-core+, pTDH3-core+), we were able to create different versions of the sensors with variable expression levels (Fig. S4, Supporting Information). Each version of the sensor were activated by hydrogen peroxide although not to the same extent as with the synthetic core promoter. This was not all too surprising since all previous optimization steps were completed using the synthetic core promoter; however, each sensor can still achieve dynamic ranges of activation over ten-fold, well above what could be previously acheived. Expression levels of this small library of biosensors ranges from as low as 3% of basal expression and up to 72% of peak expression relative to what was achieved with 5XUAS-pTRX2 allowing for more precise protein expression control. This demonstrates that, much like the previously designed promoter, the smaller synthetic UAS is not necessarily context-dependent and can be fused to any core promoter to tune for an appropriate expression level.
Transferring the oxidative stress responsive biosensor into Saccharomyces boulardii
To demonstrate the transferability of our biosensor, we next tried translating the different components into the probiotic yeast strain S. boulardii. Implementing the biosensor in the probiotic strain posed certain challenges. First, unlike S. cerevisiae, S. boulardii is strictly diploid as it does not have the ability to sporulate (McCullough et al. 1998; Edwards-Ingram et al. 2007). Furthermore, because it is a wild, non-laboratory yeast strain, S. boulardii does not have specific auxotrophic mutations for use as selective markers in plasmid transformation. Some modifications to the sensor were therefore required to simplify its implementation in the probiotic strain. To facilitate genomic integration of the positive feedback loop into S. boulardii, we modified a CRISPR-Cas9 method for multiplex genome editing using a single gRNA (Fig. 4A). We first generated strains with one to three integrated GFP reporters at three previously established genomic sites suitable for heterologous gene expression (Flagfeldt et al. 2009). Under a blue light illuminator, one can visually distinguish the GFP gradient between the different strains (Fig. 4A). Next, a gRNA was designed and tested for efficient cutting within the GFP gene. This gRNA was cloned within a Cas9 expression vector and co-transformed with healing fragments containing 50 bp overhangs homologous to the different integration sites. This technique allowed for the simultaneous integration of one to three Yap1 copies using a single gRNA, with screening easily accomplished by visual inspection for loss of fluorescence (Fig. 4A). The use of a single gRNA saves the trouble of having to co-express multiple gRNAs at the same time and the pre-integrated GFP reporter facilitates the confirmation of genomic integration at the different sites.
Figure 4.
Translating the oxidative stress biosensor into Saccharomyces boulardii. A) Pictures of plated S. boulardii strains to illustrate how we created S. boulardii strains containing one to three Yap1 cassettes at different integration sites. Three separate GFP reporters were integrated at three different sites (see Methods) then used as visual markers for successful integration of the Yap1 cassettes using a CRISPR-Cas9 system with a single gRNA designed to efficiently and specifically cut within the GFP gene. B) Testing oxidative stress responsive promoters in S. boulardii. The three different oxidative stress sensing promoters with variant Yap1 binding sites (pSynOS_4,pSynOS_4(alt1), pSynOS_4(alt2)) were transformed in S. boulardii and tested for their response to two concentrations of hydrogen peroxide, as described in the Methods section. Bar graph illustrates the mean OD-normalized mCherry fluorescence plus standard deviation of three biological replicates. Fold fluorescent change of biosensor compared to basal, non-stressed conditions (0 μM) is reported over each bar. C) Testing the effect of the positive feedback loop to control Yap1 expression on the activity of the oxidative stress biosensor in S. boulardii. Three strains were generated from S. boulardii with one to three Yap1 copies under the control of the pTRX2 promoter. Oxidative stress biosensor pSynOS_4(alt1) was transformed in each strains and tested for its response to two concentrations of hydrogen peroxide, as described above.
Initially, we cloned the biosensors with different Yap1 binding sites (pSynOS_4, pSynOS_4(alt1), pSynOS_4(alt2)) into a high-copy plasmid conferring G-418 resistance and tested its activity in wildtype S. boulardii (Fig. 4B). As expected, the biosensors were activated upon induction with hydrogen peroxide, although their fold activation was lower than when it was tested in the S. cerevisiae background. Despite this, we were able to demonstrate that the oxidative stress sensor using the alternative Yap1 binding sites, pSynOS_4(alt1) and pSynOS_4(alt2), achieved a better fold activation than the original sensor, pSynOS_4, lending support to the idea that the optimization steps completed in S. cerevisiae could be at least partially transferred to S. boulardii. We then generated three S. boulardii strains that included multiple copies of Yap1 under the TRX2 promoter and compared the activity of pSynOS_4(alt1) within these strains (Fig. 4C). The fold activation of our biosensor improved significantly up to the addition of two Yap1 copies but decreased in the strain with three Yap1 copies. With two copies of Yap1, the sensor's fold activation was improved from 4.6X to 9.6X and from 11.3X to 25.3X at 100 μM and 200 μM of hydrogen peroxide, respectively, which greatly exceeded the fold activity of just the synthetic promoter.
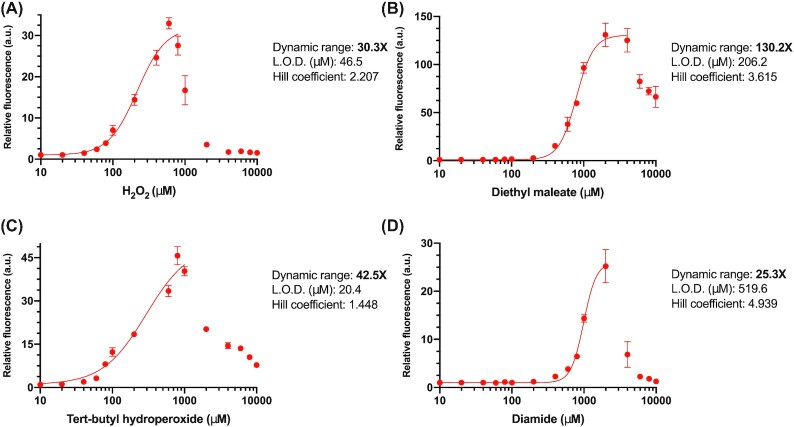
To finalize our biosensor, the oxidative stress sensing reporter (pSynOS_4(alt1)) was genomically integrated into the S. boulardii strain with two Yap1 cassettes. We tested the integrated biosensor against all four ROSs spanning a range of concentrations from 10 µM to 10 mM (Fig. 5). The dose response curves have features comparable to the optimized sensor in S. cerevisiae, achieving high dynamic ranges of activation (30.3X, 130.2X, 42.5X, and 25.3X) in response to hydrogen peroxide, diethyl maleate, tert-butyl hydroperoxide, and diamide, respectively. In conclusion, our results clearly demonstrate a highly responsive and robust oxidative stress biosensor built from a minimal synthetic core promoter that functions within different yeast strains.
Figure 5.
Evaluating the dose response of the fully optimized oxidative stress sensing biosensor in Saccharomyces boulardii. Dose response curves of the fully optimized biosensor (pSynOS_4(alt1)+) integrated into S. boulardii against the ROSs A) hydrogen peroxide, B) diethyl maleate, C) tert-butyl hydroperoxide, and D) diamide. 16 different concentrations of each ROSs were used ranging from 10^1 to 10^4 µM. Mean fluorescence levels of the output (mCherry) from three biological replicates is normalized to cell concentration (OD-normalized) and relative to basal levels of expression at 0 µM (±S.D.). Nonlinear regression models (lines) were fitted to each dataset up to an appropriately chosen concentration. Calculated dynamic range of activation, limit of detection (L.O.D.), and Hill coefficient from the fitted curves are presented next to each graph.
DISCUSSION
We set out to design a new synthetic oxidative stress sensitive promoter not only to reduce its size, but also to find the elements most responsible for the promoter's activity which we could then modify to optimize its performance. The natural Yap1-dependent oxidative stress sensing promoter, pTRX2, is 275 bp long and shows only modest activity towards various ROSs (Fig. 3, pTRX2). This promoter was previously redesigned to improve its function, but the optimization steps led to the creation of a relatively long promoter of 700 bp plus the size of the core promoter (Fig. 3, 5XUAS-pTRX2) (Zhang et al. 2016). The synthetic promoter we have designed was reduced in size to 171 bp while also creating a highly responsive oxidative stress promoter outperforming the activity of both the natural yeast promoter and the previously optimized promoter by a large margin (Fig. 3, pSynOS_4(alt1)). Predictably, increasing the dynamic range of activation of our promoter concurrently widened the effective dosage range allowing us to distinguish changes in activity for lower concentrations of ROSs. The only drawback with the new synthetic promoter appears to have been the reduction of absolute protein levels to a quarter of what could be achieved with the previous promoter (Fig. S3, Supporting Information). However, with the inclusion of the positive feedback loop, we could raise protein expression levels to over half of what the previous promoter could achieve while maintaining a considerably higher dynamic range of activation (Fig. 3 and Fig. S3, Supporting Information; pSynOS_4(alt1)+).
Three factors were found to have a significant effect on promoter activation, and we believe these might be useful when designing other TF-based biosensors. First, optimizing the size of the spacer element between the TF binding sites proved to be important for promoter activation. Using the natural 29 nucleotide spacer element between the two Yap1 binding sites of the TRX2 promoter completely inactivated promoter activation with hydrogen peroxide (Fig. 1B). The Yap1 binding sites only effectively recruited the transcription once they were closer together demonstrating a previously undiscovered cooperativity between adjacent Yap1 binding sites. The fact that the natural spacer element could not effectively recruit Yap1 suggests that the TRX2 promoter has other elements in its upstream activating sequence that either favor the recruitment of the TF or enhance the process of transcriptional activation. For example, TRX2 expression upon oxidative stress is dependent not only on Yap1 but also another stress responsive TF called Skn7 (Morgan 1997). The Skn7 binding site lies within the sites -164 and -142 of the TRX2 promoter and was not included in our synthetic promoter. The lack of the Skn7 binding site and potentially other elements within the UAS of TRX2 might be the reason our promoter needs over three Yap1 binding sites to become sensitive to oxidative stress rather than only two. Conversely, removing the spacer element between the Yap1 binding sites also substantially reduced the activity of our promoter (Fig. 1B). We assume this is because of steric hindrance between the TF that would prevent cooperative binding. The original 9 bp spacer element used turned out to be the ideal spacing required, even though its sequence was arbitrarily chosen. The promoter could be even further optimized by changing the size of the spacer element between 0 bp and 19 bp as well as changing the sequence of the spacer itself to improve cooperativity between the TF binding sites.
The second factor we discovered to be crucial for promoter activity was the TF binding sequence. The original sequence used (TTACTAA) was previously described as the most effective for recruiting the yeast TF. Other binding sequences have also been shown to recruit Yap1, albeit with different affinities, like the mammalian AP-1 recognition motif (TGACTAA) (Harshman, Moye-Rowley and Parker 1988) and the Gcn4 recognition motif (TGACTCA) (Fernandes, Rodrigues-Pousada and Struhl 1997). The binding sites all have in common a central C·G pair and two half-sites (TTA-AAT for the preferred binding site). Alternative binding sequences were used on the basis that they would lower the activation of the promoter under non-stressed conditions by altering the orientation of the binding sites and lowering the binding affinity. The first alternative binding site (TTAGTAA) is the second Yap1 binding site within the TRX2 promoter and has an inverted central C-G pair compared to the preferred Yap1 binding site; this is also simply the reverse complement of the preferred Yap1 binding sequence. Using this binding site likely changes the preferred orientation of the TF without necessarily affecting its binding affinity. The second alternative binding site (TTAGTCA) also changes one of the half-sites to the sequence used by the mammalian AP-1 recognition motif; this is also simply the reverse complement of the same motif. Changing one of the half site sequences would likely reduce its binding affinity towards Yap1. As seen in Fig. 1C, both these modifications caused a substantial reduction in basal level of activation compared to the preferred Yap1 binding sites. What was unexpected was that the promoter with alternative binding sites also retained a very similar level of activation as the promoter with preferred Yap1 binding sequence. We believe this might be the result of the newly found cooperation between closely separated Yap1 binding sites. The Yap1 binding sequence itself turned out to be the most important factor for achieving higher activity. It is possible that introducing small, rational modifications to an existing TF binding sequence could improve the activity of other biosensors.
Thirdly, we found that using a positive feedback loop to control the expression of an additional copy of the TF improved the dynamic range of activation of the biosensor, at least for some of the ROSs (Fig. 2). However, this does come at the cost of using larger DNA components, which we recognize works against our original proposal of reducing the total amount of regulatory DNA in the system. If improvements to the dynamic range or the operational range of detection are required for an application, then the positive feedback loop could serve as a useful tool for tuning the parameters of the dose response curves beyond what is possible from promoter optimization alone. It should also be noted that the positive feedback loop not only improves the fold activation of the biosensor, but also increases the total output levels relative to the synthetic promoter (Fig. 2C and D, Fig. S4, Supporting Information). Inclusion of the positive feedback loop could therefore also serve as a tool to increase expression levels of the reporter without negatively affecting the biosensor's fold activation.
Importantly, we showed that all these optimizations could translate into a different yeast strain, the probiotic Saccharomyces boulardii. Both the alternative Yap1 binding sites and the positive feedback loop improved the activity of the oxidative stress sensing biosensor within S. boulardii which, when combined, achieved dose response profiles comparable to those observed in S. cerevisiae (Fig. 5). Transferring the biosensor into S. boulardii also led to the development of genomic editing tools that could be used to engineer the probiotic for therapeutic purposes. One future application of this biosensor would be to detect ROSs in the intestine, where such species serve as a recurrent marker for colitis (Keshavarzian et al. 1992; Simmonds et al. 1992; Lih-Brody et al. 1996; Bronsart et al. 2016); development in this direction would likely require the biosensor to generate an output that would be conveniently detected in stool samples. Overall, we believe that the optimization steps detailed here could be applied towards improving other TF-based biosensors and their applications.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kristin Baetz for providing yeast strain (BY4741) and plasmids (pRS415, pRS416).
Contributor Information
Louis C Dacquay, Dept of Cell and Systems Biology, University of Toronto, 25 Harbord St, Toronto, ON M5S 3G5, Canada; Department of Chemical and Physical Sciences, University of Toronto Mississauga, 3359 Mississauga Rd, Mississauga ON L5L 1C6, Canada.
David R McMillen, Dept of Cell and Systems Biology, University of Toronto, 25 Harbord St, Toronto, ON M5S 3G5, Canada; Department of Chemical and Physical Sciences, University of Toronto Mississauga, 3359 Mississauga Rd, Mississauga ON L5L 1C6, Canada; Departments of Chemistry and Physics, University of Toronto, 80 St. George St., Toronto ON M5S 3H6, Canada.
FUNDING
This work was supported by the Canada First Research Excellence Fund (Medicine by Design program) and the Natural Sciences and Engineering Research Council (NSERC) Discovery grant program. Stipend support for Louis Dacquay has been provided by the Government of Ontario (Ontario Graduate Scholarship) and the Government of Canada (NSERC PhD Graduate Scholarship).
Conflict of interest
None declared.
REFRENCES
- Archer EJ, Robinson AB, Süel GM. Engineered E. Coli That Detect and Respond to Gut Inflammation through Nitric Oxide Sensing. ACS Synthetic Biology. 2012;1:451–7. [DOI] [PubMed] [Google Scholar]
- Bovee TFH, Helsdingen RJR, Hamers ARMet al. A New Highly Specific and Robust Yeast Androgen Bioassay for the Detection of Agonists and Antagonists. Anal Bioanal Chem. 2007;389:1549–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronsart L, Nguyen L, Habtezion Aet al. Reactive Oxygen Species Imaging in a Mouse Model of Inflammatory Bowel Disease. Mol Imaging Biol. 2016;18:473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Shen Y, Zou Yet al. Engineering Highly Sensitive Whole-Cell Mercury Biosensors Based on Positive Feedback Loops from Quorum-Sensing Systems. Analyst. 2018;143:630–4. [DOI] [PubMed] [Google Scholar]
- Daeffler KN, Galley JD, Sheth RUet al. Engineering Bacterial Thiosulfate and Tetrathionate Sensors for Detecting Gut Inflammation. Mol Syst Biol. 2017;13:923. 10.15252/msb.20167416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl RH, Zhang F, Alonso-Gutierrez Jet al. Engineering Dynamic Pathway Regulation Using Stress-Response Promoters. Nat Biotechnol. 2013;31:1039–46. [DOI] [PubMed] [Google Scholar]
- David F, Nielsen J, Siewers V. Flux Control at the Malonyl-CoA Node through Hierarchical Dynamic Pathway Regulation in Saccharomyces Cerevisiae. ACS Synthetic Biology. 2016;5:224–33. [DOI] [PubMed] [Google Scholar]
- Dietrich JA, Shis DL, Alikhani Aet al. Transcription Factor-Based Screens and Synthetic Selections for Microbial Small-Molecule Biosynthesis. ACS Synthetic Biology. 2013;2:47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards-Ingram L, Gitsham P, Burton Net al. Genotypic and Physiological Characterization of Saccharomyces Boulardii, the Probiotic Strain of Saccharomyces Cerevisiae. Appl Environ Microbiol. 2007;73:2458–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer WR, Liao JC. Improving Lycopene Production in Escherichia Coli by Engineering Metabolic Control. Nat Biotechnol. 2000;18:533–7. [DOI] [PubMed] [Google Scholar]
- Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a Novel Family of Eight BZIP Proteins in Saccharomyces Cerevisiae with Distinct Biological Functions. Mol Cell Biol. 1997;17:6982–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagfeldt DB, Siewers V, Huang Let al. Characterization of Chromosomal Integration Sites for Heterologous Gene Expression in Saccharomyces Cerevisiae. Yeast. 2009;26:545–51. [DOI] [PubMed] [Google Scholar]
- Grisham MB. Oxidants and Free Radicals in Inflammatory Bowel Disease. Lancet North Am Ed. 1994;344:859–61. [DOI] [PubMed] [Google Scholar]
- Harshman KD, Moye-Rowley WS, Parker CS. Transcriptional Activation by the SV40 AP-1 Recognition Element in Yeast Is Mediated by a Factor Similar to AP-1 That Is Distinct from GCN4. Cell. 1988;53:321–30. [DOI] [PubMed] [Google Scholar]
- Ikushima S, Boeke JD. New Orthogonal Transcriptional Switches Derived from Tet Repressor Homologues for Saccharomyces Cerevisiae Regulated by 2,4-Diacetylphloroglucinol and Other Ligands. ACS Synthetic Biology. 2017;6:497–506. [DOI] [PubMed] [Google Scholar]
- Keshavarzian A, Sedghi S, Kanofsky Jet al. Excessive Production of Reactive Oxygen Metabolites by Inflamed Colon: analysis by Chemiluminescence Probe. Gastroenterology. 1992;103:177–85. [DOI] [PubMed] [Google Scholar]
- Khalil AS, Lu TK, Bashor CJet al. A Synthetic Biology Framework for Programming Eukaryotic Transcription Functions. Cell. 2012;150:647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S, Jones N. YAP1 Dependent Activation of TRX2 Is Essential for the Response of Saccharomyces Cerevisiae to Oxidative Stress by Hydroperoxides. EMBO J. 1994;13:655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry BP, Tabor JJ. Engineering Diagnostic and Therapeutic Gut Bacteria. Microbiol Spectr. 2017;5:333–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-W, Oh M-K. A Synthetic Suicide Riboswitch for the High-Throughput Screening of Metabolite Production in Saccharomyces Cerevisiae. Metab Eng. 2015;28:143–50. [DOI] [PubMed] [Google Scholar]
- Lih-Brody L, Powell SR, Collier KPet al. Increased Oxidative Stress and Decreased Antioxidant Defenses in Mucosa of Inflammatory Bowel Disease. Dig Dis Sci. 1996;41:2078–86. [DOI] [PubMed] [Google Scholar]
- McCullough MJ, Clemons KV, McCusker JHet al. Species Identification and Virulence Attributes Of Saccharomyces Boulardii (Nom. Inval.). J Clin Microbiol. 1998;36:2613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac RS, Gibney PA, Chandran SSet al. Synthetic Biology Tools for Programming Gene Expression without Nutritional Perturbations in Saccharomyces Cerevisiae. Nucleic Acids Res. 2014;42:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano KA, Grant CM, Moye-Rowley WS. The Response to Heat Shock and Oxidative Stress in Saccharomyces Cerevisiae. Genetics. 2012;190:1157–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA. The Skn7 Response Regulator Controls Gene Expression in the Oxidative Stress Response of the Budding Yeast Saccharomyces Cerevisiae. EMBO J. 1997;16:1035–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Bhattacharyya S, Peralta-Yahya P. GPCR-Based Chemical Biosensors for Medium-Chain Fatty Acids. ACS Synthetic Biology. 2015;4:1261–9. [DOI] [PubMed] [Google Scholar]
- Ottoz DSM, Rudolf F, Stelling J. Inducible, Tightly Regulated and Growth Condition-Independent Transcription Factor in Saccharomyces Cerevisiae. Nucleic Acids Res. 2014;42:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela RMC, Vogl T, Kniely Cet al. Synthetic Core Promoters as Universal Parts for Fine-Tuning Expression in Different Yeast Species. ACS Synthetic Biology. 2017;6:471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao K, Wasylenko TM, Zhou Ket al. Lipid Production in Yarrowia Lipolytica Is Maximized by Engineering Cytosolic Redox Metabolism. Nat Biotechnol. 2017;35:173–7. [DOI] [PubMed] [Google Scholar]
- Raimondi S, Zanni E, Talora Cet al. SOD1, a New Kluyveromyces Lactis Helper Gene for Heterologous Protein Secretion. Appl Environ Microbiol. 2008;74:7130 LP–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantasalo A, Kuivanen J, Penttilä Met al. Synthetic Toolkit for Complex Genetic Circuit Engineering in Saccharomyces Cerevisiae. ACS Synthetic Biology. 2018;7:1573–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redden H, Alper HS. The Development and Characterization of Synthetic Minimal Yeast Promoters. Nat Commun. 2015;6:7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riglar DT, Giessen TW, Baym Met al. Engineered Bacteria Can Function in the Mammalian Gut Long-Term as Live Diagnostics of Inflammation. Nat Biotechnol. 2017;35:653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riglar DT, Silver PA. Engineering Bacteria for Diagnostic and Therapeutic Applications. Nat Rev Microbiol. 2018;16:214–25. [DOI] [PubMed] [Google Scholar]
- Siedler S, Schendzielorz G, Binder Set al. SoxR as a Single-Cell Biosensor for NADPH-Consuming Enzymes in Escherichia Coli. ACS Synthetic Biology. 2014;3:41–7. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A System of Shuttle Vectors and Yeast Host Strains Designed for Efficient Manipulation of DNA in Saccharomyces Cerevisiae. Genetics. 1989;122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds NJ, Allen RE, Stevens TRJet al. Chemiluminescence Assay of Mucosal Reactive Oxygen Metabolites in Inflammatory Bowel Disease. Gastroenterology. 1992;103:186–96. [DOI] [PubMed] [Google Scholar]
- Skjoedt ML, Snoek T, Kildegaard KRet al. Engineering Prokaryotic Transcriptional Activators as Metabolite Biosensors in Yeast. Nat Chem Biol. 2016;12:951–8. [DOI] [PubMed] [Google Scholar]
- Stovicek V, Borodina I, Forster J. CRISPR–Cas System Enables Fast and Simple Genome Editing of Industrial Saccharomyces Cerevisiae Strains. Metabolic Engineering Communications. 2015;2:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo WS, Chang MW. Bacterial XylRs and Synthetic Promoters Function as Genetically Encoded Xylose Biosensors in Saccharomyces Cerevisiae. Biotechnol J. 2015;10:315–22. [DOI] [PubMed] [Google Scholar]
- Tian T, Wang Z, Zhang J. Pathomechanisms of Oxidative Stress in Inflammatory Bowel Disease and Potential Antioxidant Therapies. Oxid Med Cell Longev. 2017;2017:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeyama T, Okada S, Ito T. Synthetic Gene Circuit-Mediated Monitoring of Endogenous Metabolites: identification of GAL11 as a Novel Multicopy Enhancer of S-Adenosylmethionine Level in Yeast. ACS Synthetic Biology. 2013;2:425–30. [DOI] [PubMed] [Google Scholar]
- Venayak N, Anesiadis N, Cluett WRet al. Engineering Metabolism through Dynamic Control. Curr Opin Biotechnol. 2015;34:142–52. [DOI] [PubMed] [Google Scholar]
- Wang B, Barahona M, Buck M. A Modular Cell-Based Biosensor Using Engineered Genetic Logic Circuits to Detect and Integrate Multiple Environmental Signals. Biosens Bioelectron. 2013;40:368–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Cress BF, Yang Zet al. Design and Characterization of Biosensors for the Screening of Modular Assembled Naringenin Biosynthetic Library in Saccharomyces Cerevisiae. ACS Synthetic Biology. 2019;8:2121–30.. 10.1021/acssynbio.9b00212. [DOI] [PubMed] [Google Scholar]
- Wan X, Volpetti F, Petrova Eet al. Cascaded Amplifying Circuits Enable Ultrasensitive Cellular Sensors for Toxic Metals. Nat Chem Biol. 2019;15:540–8.. 10.1038/s41589-019-0244-3. [DOI] [PubMed] [Google Scholar]
- Williams TC, Xu X, Ostrowski Met al. Positive-Feedback, Ratiometric Biosensor Expression Improves High-Throughput Metabolite-Producer Screening Efficiency in Yeast. Synth Biol. 2017;2. 10.1093/synbio/ysw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S-G, Moon S-J, Kim SKet al. A Designed Whole-Cell Biosensor for Live Diagnosis of Gut Inflammation through Nitrate Sensing. Biosens Bioelectron. 2020;168:112523. 10.1016/j.bios.2020.112523. [DOI] [PubMed] [Google Scholar]
- Xu P, Li L, Zhang Fet al. Improving Fatty Acids Production by Engineering Dynamic Pathway Regulation and Metabolic Control. Proc Natl Acad Sci. 2014;111:11299 LP–11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Qiao K, Stephanopoulos G. Engineering Oxidative Stress Defense Pathways to Build a Robust Lipid Production Platform in Yarrowia Lipolytica. Biotechnol Bioeng. 2017;114:1521–30. [DOI] [PubMed] [Google Scholar]
- Xu P, Wang W, Li Let al. Design and Kinetic Analysis of a Hybrid Promoter–Regulator System for Malonyl-CoA Sensing in Escherichia Coli. ACS Chem Biol. 2014;9:451–8. [DOI] [PubMed] [Google Scholar]
- Zhang F, Carothers JM, Keasling JD. Design of a Dynamic Sensor-Regulator System for Production of Chemicals and Fuels Derived from Fatty Acids. Nat Biotechnol. 2012;30:354–9. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jensen MK, Keasling JD. Development of Biosensors and Their Application in Metabolic Engineering. Curr Opin Chem Biol. 2015;28:1–8. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sonnenschein N, Pihl TPBet al. Engineering an NADPH/NADP + Redox Biosensor in Yeast. ACS Synthetic Biology. 2016;5:1546–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.