A resident of the human oral biofilm, Streptococcus mutans is one of the major bacterial pathogens associated with dental caries. This report highlights a spontaneously occurring mutation within the laboratory strain S. mutans UA159 found in the coding region of perR, a gene encoding a transcriptional repressor associated with peroxide tolerance.
KEYWORDS: S. mutans, PerR, oxidative stress, mutation
ABSTRACT
The ability of bacteria, such as the dental pathogen Streptococcus mutans, to coordinate a response against damage-inducing oxidants is a critical aspect of their pathogenicity. The oxidative stress regulator SpxA1 has been demonstrated to be a major player in the ability of S. mutans to withstand both disulfide and peroxide stresses. While studying spontaneously occurring variants of an S. mutans ΔspxA1 strain, we serendipitously discovered that our S. mutans UA159 host strain bore a single-nucleotide deletion within the coding region of perR, resulting in a premature truncation of the encoded protein. PerR is a metal-dependent transcriptional repressor that senses and responds to peroxide stress such that loss of PerR activity results in activation of oxidative stress responses. To determine the impact of loss of PerR regulation, we obtained a UA159 isolate bearing an intact perR copy and created a clean perR deletion mutant. Our findings indicate that loss of PerR activity results in a strain that is primed to tolerate oxidative stresses in the laboratory setting. Interestingly, RNA deep sequencing (RNA-Seq) and targeted transcriptional expression analyses reveal that PerR offers a minor contribution to the ability of S. mutans to orchestrate a transcriptional response to peroxide stress. Furthermore, we detected loss-of-function perR mutations in two other commonly used laboratory strains of S. mutans, suggesting that this may be not be an uncommon occurrence. This report serves as a cautionary tale regarding the so-called domestication of laboratory strains and advocates for the implementation of more stringent strain authentication practices.
IMPORTANCE A resident of the human oral biofilm, Streptococcus mutans is one of the major bacterial pathogens associated with dental caries. This report highlights a spontaneously occurring mutation within the laboratory strain S. mutans UA159 found in the coding region of perR, a gene encoding a transcriptional repressor associated with peroxide tolerance. Though perR mutant strains of S. mutans showed a distinct growth advantage and enhanced tolerance toward H2O2, a ΔperR deletion strain showed a small number of differentially expressed genes compared to the parent strain, suggesting few direct regulatory targets. In addition to characterizing the role of PerR in S. mutans, our findings serve as a warning to laboratory researchers regarding bacterial adaptation to in vitro growth conditions.
INTRODUCTION
Transition metals are essential micronutrients for cell life. However, when found in excess, these same metals can be extremely harmful in part because of the deleterious effects of Fenton chemistry, by which intermingling of H2O2 and soluble ferrous iron (Fe2+) or copper (Cu2+) results in formation of the highly reactive hydroxyl radical (HO·) (1). To maintain metal homeostasis and avoid reactive oxygen species (ROS) intoxication, bacteria employ a number of metal-sensing regulators (metalloregulators) that are used to maintain metal homeostasis and coordinate oxidative stress responses. Among the most extensively studied metal-sensing regulators are the ferric uptake regulator (Fur) protein family (2). Members of this family function as homodimers and include sensors for iron (Fur), zinc (Zur), manganese (Mur), nickel (Nur), and peroxide (PerR). PerR is an oxidative stress regulator that senses peroxide stress when bound to a metal cofactor that, depending on the bacterial species, can be iron, manganese, or both (2, 3). PerR has been shown to function as a transcriptional repressor by binding directly to operator sites within the promoter region of a gene target (4–6). Among Gram-positive bacteria, PerR is associated with repression of oxidative stress-detoxification genes, such as dpr (encoding an iron-binding peroxide resistance protein), sodA (superoxide dismutase), and ahpCF (alkyl hydroperoxidase), though the composition of regulons governed by PerR varies among bacterial species and even between strains within a species (7–11). In general, the genes regulated by PerR can be divided into two major groups as follows: genes directly involved in ROS detoxification and genes involved in metal homeostasis (12). Of note, PerR homologs are widespread in Firmicutes but are also occasionally found in Gram-negative bacteria (13–15).
Among Firmicutes, Streptococcus mutans is a resident of the dental biofilm shown to be a major contributor to the development of dental caries and, occasionally, associated with systemic infections, such as infective endocarditis (16). The association of S. mutans with dental caries is strongly linked with its ability to tolerate environmental stresses encountered in its environmental niche, namely, acidic and oxidative stresses (17). Multiple sources contribute to the oxidative stresses found in dental plaque, including the capacity of the oral microbial community as a whole to reduce oxygen into ROS and the direct production of H2O2 by peroxigenic bacteria. The transcriptional regulators SpxA1 and SpxA2 have been demonstrated to function as central regulators of the oxidative stress response of S. mutans, with SpxA1 serving as a direct activator of the genes classically associated with the oxidative stress response, whereas SpxA2, whose major role has recently been linked to cell envelope homeostasis, serves as a backup system (18, 19). As part of our ongoing characterization of S. mutans spx mutant strains, we found that the S. mutans ΔspxA1 strain was hypersensitive to arginine, a phenotype that was linked to alkaline stress tolerance (P. Zuber, unpublished data). As we observed the emergence of stable ΔspxA1 phenotypic revertants on plates containing arginine, we subjected those revertants to whole-genome sequencing (WGS) for the purpose of identifying the locus or loci linked to arginine tolerance. In addition to linking loss-of-function mutations in an iron transporter to the rescue of arginine tolerance in the ΔspxA1 strain, we unexpectedly discovered that all isolates subjected to WGS, including the original arginine-sensitive ΔspxA1 isolate, bore a mutation in the perR (NCBI annotation SMU_RS02830, formerly SMU_593) gene that likely renders PerR inactive. In light of this unexpected finding, we sought to characterize the impact of PerR on S. mutans physiology. Despite displaying a rather modest role in the transcriptional regulation of oxidative stress genes, we showed that loss of PerR regulation primed S. mutans for growth in the presence of oxidative stresses under in vitro conditions. In silico and PCR sequencing analyses of UA159 laboratory stocks as well as other widely used S. mutans strains revealed that the occurrence of spontaneous perR mutations may not be uncommon in laboratory strains.
RESULTS AND DISCUSSION
Loss-of-function mutations of an iron transporter in the ΔspxA1 strain lead to serendipitous identification of a spontaneous perR mutation that increases oxidative stress tolerance.
The work described here began as an exploration of the initial observation that our ΔspxA1 strain was hypersensitive to arginine stress. Though the ΔspxA1 strain showed enhanced sensitivity to growth in the presence of arginine compared to the parent strain, a number of revertant colonies were identified that regained the ability to grow in the presence of arginine (see Fig. S1A in the supplemental material). Four stable revertant isolates and the original ΔspxA1 strain were subjected to WGS analysis to identify the locus in which the mutation(s) that restored arginine tolerance to the ΔspxA1 strain resided. When compared to the annotated S. mutans UA159 genome (GenBank accession number NC_004350.2), all four colony revertants (numbered R1, R4, R6, and R7) harbored single nucleotide polymorphisms (SNPs) in SMU_RS04590 (previously annotated as SMU_997) or SMU_RS04595 (SMU_998) genes, which encode the ATP-binding and substrate-binding proteins of an iron transport system (Fig. S1B). Coincidently, we have previously identified the SMU_RS04580 to SMU_RS04595 (SMU_995 to SMU_998) operon as the site of suppressor mutations that reverted the hypersensitivity of a Δdpr strain to oxidative stresses (20). Notably, Dpr is an iron-sequestering intracellular protein that plays a major role in bacterial oxidative stress tolerance (21). While a mechanistic explanation for the association of the SMU_995 to SMU_998 operon with restoration of arginine tolerance in the ΔspxA1 strain is not the focus of this report, we expect that reductions in intracellular iron pools in the suppressor mutants diminish Fenton reactivity at elevated pH, which compensates for the loss of SpxA1-dependent antioxidant activity.
In addition to SNPs within the smu995-998 operon, all sequenced isolates, including the original arginine-sensitive ΔspxA1 isolate, bore a single base deletion at position 393 within the coding region of the SMU_RS02830 (previously SMU_593) gene (Fig. 1A), which is predicted to code for the peroxide regulator PerR. This single base deletion introduced a frameshift mutation that resulted in a premature truncation of the PerR protein (Fig. 1B). In light of this unexpected finding, we used PCR sequencing to determine if this perR mutation was present in our freezer stocks of strain UA159 and the ΔspxA1 strain. To our surprise, the frameshift mutation discovered in the ΔspxA1 strain was found in our original UA159 freezer stocks. We contacted six other labs that routinely work with S. mutans UA159 and found that UA159 stocks from some of these labs had the same single base deletion at position 393 while one other lab had a PerR C106F codon substitution mutation. We suspect that the perR mutation at position 393 occurred early on and was passed among different labs over the years. Nonetheless, the perR gene sequence was intact in stocks from two labs as well as in a lyophilized stock that was purchased from ATCC.
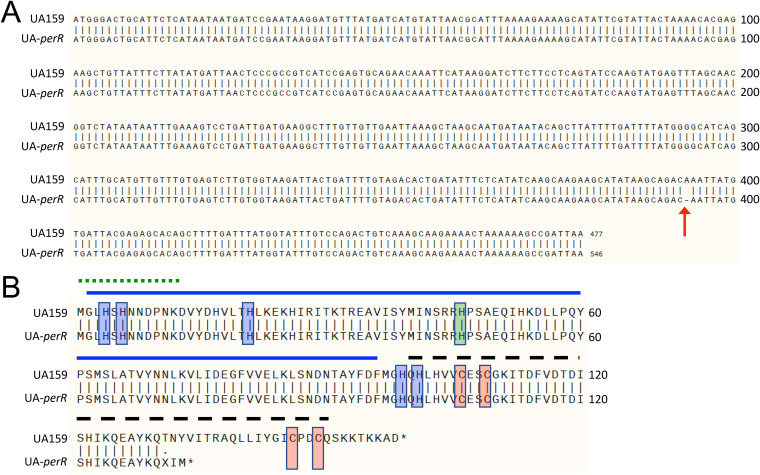
FIG 1.
Whole genomic sequencing reveals a perR SNP that results in a prematurely truncated protein in S. mutans UA159. (A) Alignment of nucleotide sequences of perR from UA159 from the NCBI (perR+) and UA-perR (perR SNP strain). The arrow at base 393 indicates a 1-bp deletion in the sequence of UA-perR. (B) Alignment of amino acid sequences of PerR from UA159 and UA-perR, indicating a premature termination of the latter. Red shading indicates zinc-coordinating cysteine residues critical for PerR function. Blue shading indicates histidine residues involved in binding of a regulatory Fe molecule. Green shading indicates a histidine residue believed to be important in DNA binding. The solid blue line indicates the N-terminal DNA-binding domain. The dashed black line indicates the C-terminal dimerization domain. The dotted green line indicates an N-terminal extension unique to PerR members of the Fur family and conserved among streptococci. Structural domains were based on homologies to those described in PerR of S. pyogenes, whose crystal structure has been solved (22).
Similar to most members of the Fur family, the PerR proteins of Bacillus subtilis (PerRBS) and Streptococcus pyogenes (PerRSP) require a zinc molecule for structural stability (8, 22). This zinc is tightly coordinated by cysteines that compose two highly conserved CXXC redox switch domains found in each member of the Fur family. The zinc molecule that is bound cooperatively by these 4 cysteine residues contributes to the structural stability and to the capacity for dimerization of PerR (23). The premature perR stop codon identified in some of the UA159 stocks indicates that the second CXXC motif is missing in the truncated PerR protein while the C106F codon substitution in the UA159 stock from one other lab directly disrupts the first CXXC motif. Thus, both the frameshift mutation at codon 393 and C106F amino acid substitution are predicted to affect the structure of the zinc-binding domain that constitutes part of the dimer interface of PerR. For the remaining parts of this manuscript, we renamed our UA159 isolate containing the truncated PerR as UA-perR, maintaining the UA159 designation for the strain harboring the intact perR gene.
Next, we used the intact UA159 PerR amino acid sequence to BLAST against sequenced S. mutans genomes. Out of the dozens of S. mutans PerR sequences currently available at the NCBI, the overwhelming majority were found to have no important changes at the amino acid level compared to the original UA159 sequence. We also amplified and sequenced the S. mutans perR open reading frame from several fresh clinical isolates available in the lab and found that the perR gene was intact in all of these strains (data not shown). Interestingly, we found that two other “laboratory strains,” NG8 and GS-5, also displayed perR SNPs. The GS-5 strain displayed an A36D amino acid substitution while the NG8 strain carried a 7-bp deletion that resulted in a truncation of the PerR protein even more premature than that of our UA159 stock (see Fig. S2 in the supplemental material). Based on this analysis, we conclude that the PerR protein sequence is fairly conserved in S. mutans, though possibly a genetic hot spot of spontaneous mutations in laboratory strains.
Disruption of PerR enhances the tolerance of S. mutans to peroxide stress.
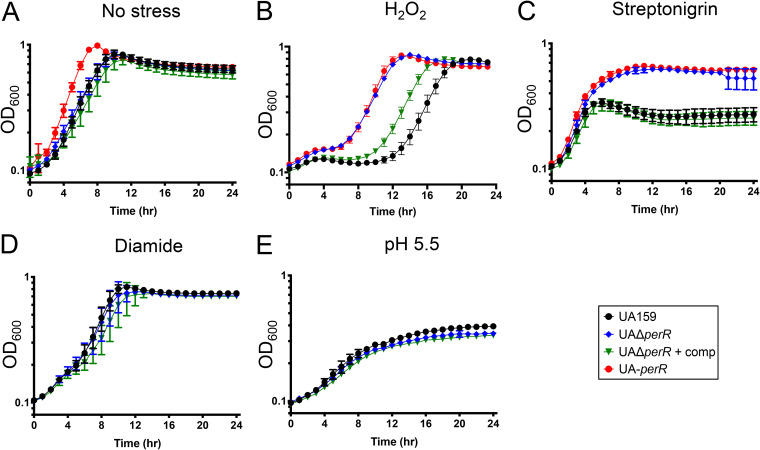
Next, we sought to determine the impact of the PerR truncation in our UA159 stock (UA-perR) and more broadly the importance of PerR to S. mutans physiology by replacing the entire perR gene in UA159 with an erythromycin resistance cassette to generate the UAΔperR strain. When grown in a 5% CO2 atmosphere using brain heart infusion (BHI), the UAΔperR and UA-perR strains displayed a slight growth advantage compared to that of UA159 of the complemented UAΔperR (see Fig. S3 in the supplemental material). Next, we grew the strains in BHI at 37°C in an automated growth reader containing an oil overlay on top of each well to create a semianaerobic environment. When grown in BHI alone in this environment, UA-perR displayed a slight growth advantage compared to that of UA159 and UAΔperR (Fig. 2A) similar to the differences observed in the 5% CO2 atmosphere (Fig. S3). When grown in medium containing 0.4 mM H2O2, the UA159 strain displayed an extended lag (∼10 h) while UA-perR and UAΔperR showed a much shorter adaptation phase (<4 h) (Fig. 2B). Of note, the extended lag phase of S. mutans when grown in subinhibitory concentrations of H2O2 likely occurs due to the combination of a cell-mediated adaptive process and the unstable nature of H2O2. To ensure that growth after an extended lag phase was not caused by emergence of spontaneous peroxide-resistant mutants, we reinoculated peroxide-grown cultures in H2O2 and observed the same extended lag phase that was observed when cells were grown in H2O2 for the first time (data not shown). Because peroxide stress and iron homeostasis are intertwined due to their role in hydroxyl radical generation via Fenton chemistry, we also tested the sensitivity of the different strains to streptonigrin, an iron-dependent antibiotic (24) used as a proxy to determine intracellular iron availability. Here, both UA-perR and UAΔperR grew significantly better than UA159, reaching a significantly higher final growth yield and indicating that PerR also controls intracellular iron homeostasis (Fig. 2C). Importantly, the phenotypes of the ΔperR strain were fully (Fig. 2A and C) or partially (Fig. 2B) restored in the genetically complemented UAΔperRComp strain, which was engineered by integrating the perR gene under the control of its native promoter elsewhere in the chromosome. Of interest, overexpression of PerR through a constitutive strong promoter in either parent or ΔperR strains severely inhibited their growth, indicating that PerR levels must be tightly controlled (data not shown).
FIG 2.
Disruption of perR enhances growth of S. mutans in the presence of stresses. Growth of UA159 (perR+), UA-perR (perR SNP strain), the perR deletion mutant strain (UAΔperR), and complemented strain (UAΔperRComp). Strains were grown in BHI (A), BHI containing 0.4 mM H2O2 (B), BHI containing 0.2 μg ml−1 streptonigrin (C), BHI containing 0.8 mM diamide (D), or BHI adjusted to pH 5.5 (E). Data represent averages and standard deviations of results from four independent experiments.
The enhanced streptonigrin tolerance of the perR mutants can be further supported by evidence that PerR controls expression of iron and manganese transporters of other streptococcal species (7, 25, 26). To further investigate this connection, we compared intracellular iron and manganese content of BHI-grown UA159, UAΔperR, and UA-perR strains. However, total intracellular metal quantifications revealed similar profiles for intracellular iron and manganese among the different strains (see Fig. S4A in the supplemental material), suggesting that the streptonigrin sensitivity of PerR-negative strains might be due to differences in freely available iron. To investigate this possibility, we took advantage of the availability of an anti-Dpr antibody (20) to determine whether the Dpr protein was more abundant in strains lacking a functional PerR. Indeed, we observed greater quantities of Dpr protein in UA-perR and in UAΔperR than in the UA159 and UAΔperRComp strains (Fig. S4B). In streptococci, Dpr is the primary iron-binding protein within the cell (21), and its ability to sequester iron molecules minimizes the generation of ROS through Fenton chemistry. Thus, the increased abundance of Dpr in perR-defective strains may help explain, at least in part, why the UA-perR and UAΔperR strains were more tolerant to streptonigrin despite showing similar intracellular iron concentrations when compared to those of PerR+ strains.
Given the phenotypic similarities of UA-perR and UAΔperR, we opted to continue our investigations only with UAΔperR and its genetic complemented version. Different than H2O2 or streptonigrin, a growth advantage for UAΔperR was not observed in the presence of the thiol-stressor diamide (Fig. 2D) or at low pH conditions (Fig. 2E). Thus, the relationship of PerR regulation, oxidative stress, and metal homeostasis appears to be rather complex, with our results reinforcing that PerR has a primary role in (if not restricted to) the peroxide stress response.
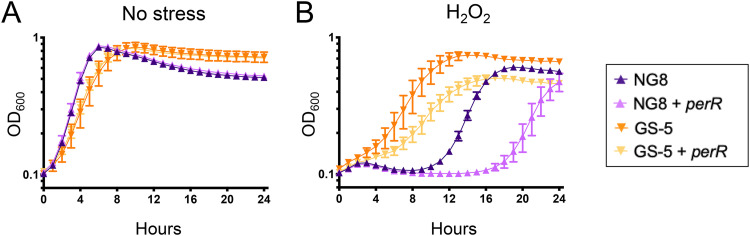
While the premature protein truncation in S. mutans NG8 almost certainly rendered PerR inactive, the consequence of the PerR A36D amino acid substitution in the GS-5 strain is unclear (Fig. S2). To begin to understand the consequence of these mutations to each respective strain, we transformed NG8 and GS-5 with the pMC340B-perR integration plasmid that was used to complement UAΔperR. Genetic complementation of perR in NG8 and GS-5 did not affect growth kinetics in BHI medium; however, both complemented strains became more sensitive to peroxide stress, showing an extended lag phase when grown in media containing 0.3 mM H2O2 (Fig. 3). Thus, it appears that the emergence of perR loss-of-function mutations may not be an uncommon phenomenon in laboratory strains of S. mutans.
FIG 3.
Chromosomal integration of an intact copy of the perR gene diminishes H2O2 tolerance in S. mutans NG8 or GS-5. Growth curves in BHI (A) or BHI containing 0.3 mM H2O2 (B) are shown. Data represent averages and standard deviations of results from four independent experiments.
Exploration of the regulatory reach of PerR.
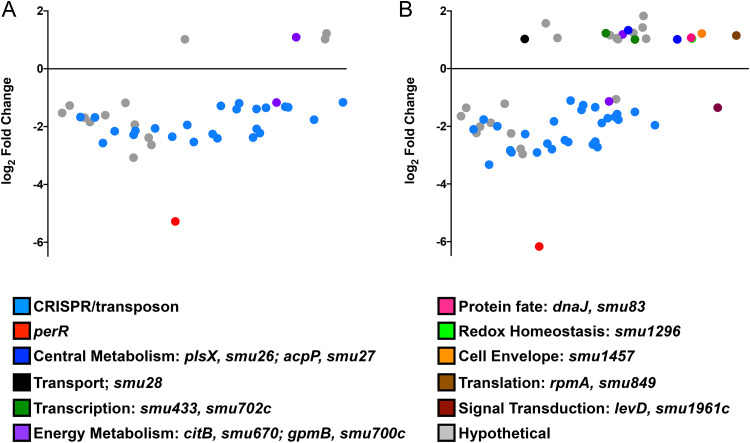
Based on extensive evidence that PerR functions as a peroxide stress regulator in Gram-positive bacteria (4, 27–29), including S. mutans (30), we next used RNA-Seq to determine the scope of the PerR regulon. As UA-perR and UA159ΔperR showed similar phenotypes, we chose to compare the transcriptome of UAΔperR to that of UA159. Briefly, UA159 and UAΔperR were grown to mid-exponential phase and exposed to 0.5 mM H2O2 for 5 min, a condition that we had previously demonstrated as optimal for activation of the peroxide stress response (19). As we have shown before (19), peroxide stress resulted in a strong and rapid induction of SpxA1-regulated genes, including archetypal oxidative stress genes, such as ahpC-ahpF, dpr, tpx, and sodA (see Table S1 in the supplemental material). Surprisingly, the peroxide stress transcriptome of UAΔperR (data not shown) was nearly identical to the transcriptional profile of UA159 subjected to H2O2 stress. Yet, when directly compared to UA159, 40 genes (4 upregulated and 36 downregulated) in the unstressed condition and 60 genes (19 upregulated and 41 downregulated) following exposure to H2O2 were differentially expressed in the UAΔperR strain (false discovery rate, 0.05; 2-fold cutoff) (see Table S2 in the supplemental material). Notably, all genes differentially expressed by UAΔperR in the unstressed condition showed the same expression trends following H2O2 exposure. Unexpectedly, none of the typical PerR-regulated genes (e.g., dpr, tpx, and sodA) were differentially expressed in the UAΔperR strain when compared to that in UA159 under any given growth condition. A number of the differentially expressed genes belong to two large polycistronic transcriptional units (SMU_RS04595-SMU_RS01120, formerly SMU_193c-SMU_217; and SMU_RS09955-SMU_RS08050, formerly SMU_1750c-SMU_1764c) coding, respectively, for hypothetical proteins of unknown function and the CRISPR2-cas system, a defense mechanism believed to provide immunity against phage invasion (31). In S. mutans UA159, the two CRISPR systems are also linked to stress responses, though the reason for this association remains to be determined (32). For visualization purposes, a dot plot graph showing all genes differently expressed in UAΔperR is provided (Fig. 4). It is worth noting that the changes in gene expression in most cases were rather modest as can be visualized by the large number of genes clustered closely below or above the x axis. While we expected that at least a subset of oxidative stress genes would be differently expressed in the ΔperR strain, a previous transcriptome analysis of an S. pyogenes ΔperR strain revealed that only 6 genes were strongly regulated by PerR, with only the iron efflux pmtA gene later confirmed to be under direct PerR control (7). Also in S. pyogenes, PerR was found to bind specifically to the ahpC promoter region, but transcriptional analysis failed to confirm a role for PerR in ahpC transcriptional expression (33). Finally, PerR was also shown to exert fairly modest transcriptional control of a small number of genes in Enterococcus faecalis (29). Collectively, these observations indicate that PerR does not exert major transcriptional control over the oxidative stress genes of these bacteria, supporting previous evidence that activation of the peroxide stress response of S. mutans is primarily controlled by SpxA1 (19).
FIG 4.
Dot plot of genes differentially expressed in UAΔperR compared to those in UA159 as determined by Degust (http://degust.erc.monash.edu/) under control (A) or H2O2 stress (B) conditions. The y axis indicates the log2 fold change in expression, while the x axis indicates the average expression level of each gene compared to that of all other genes. The details of the expression trends are shown in Table S2 in the supplemental material.
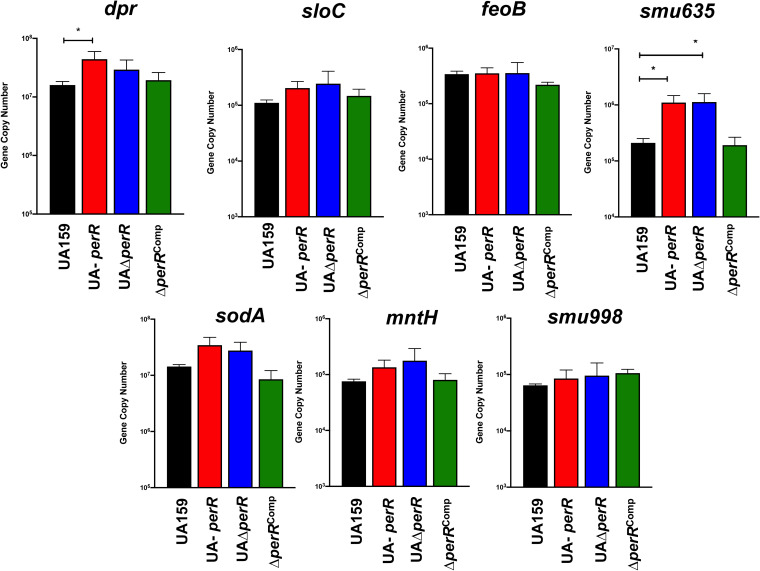
Because global transcriptional approaches are not as sensitive as targeted approaches and may overlook small, yet meaningful, changes in transcription, we also used real-time quantitative PCR (RT-qPCR) to compare the expression of a subset of oxidative stress genes, with or without putative PerR-binding DNA motifs, in the UA159, UA-perR, UAΔperR, and UAΔperRComp strains. Starting with the premise that SpxA1-mediated activation during peroxide stress overrules PerR-mediated effects on genes controlled by both regulators, this analysis was focused on cells grown in the absence of stress. PerR is reported to bind to a 15-bp palindrome motif (TTANAATNATTNTAA) within the promoter region of its targeted genes (33). This motif is similar to that recognized by Fur, maintaining the 7-1-7 arrangement, though it has been noted that a number of PerR-regulated genes do not have perfect repeats (6). Despite the similarity of PerR and Fur operators, there is little overlap of these regulators and their targets (12). For example, Fur binding sites can be distinguished from PerR boxes by guanine and cytosine residues that are recognized by a conserved arginine residue present in the Fur protein but not in PerR (34). To identify putative PerR-regulated genes in the S. mutans genome, we accessed the RegPrecise database (35) and manually scanned the promoter regions of selected oxidative stress and metal transport genes. RegPrecise listed five putative PerR-regulated genes (feoA, dpr, sloA, SMU_635, and perR itself) that contained one or two mismatches compared to the consensus sequence (Table 1). Our manual searches identified four additional genes (mntH, spxA1, sodA, and SMU_995) with putative PerR-binding boxes, albeit these additional motifs contained four mismatches from the consensus with the exception of sodA, which contained only one mismatch (Table 1). Of note, we recently showed that spxA1 is corepressed by PerR and SloR in a redundant manner, as only the simultaneous inactivation of perR and sloR increased spxA1 transcription (36). Overall, a trend was observed showing that expression levels of most of the genes tested were elevated in UA-perR and UAΔperR compared to those in UA159; the iron transport systems feoAB and smu998 were the notable exceptions (Fig. 5). Interestingly, SMU_RS03000 (formerly SMU_635) coding for a hypothetical protein of unknown function, seemingly unique to the streptococci, was the most upregulated gene in the absence of PerR. Work is under way to determine the function of Smu635. We also used, for control purposes, genes that are not (spxA2, [36]) or are not expected (pbp2A) to be regulated by PerR. In both cases, expression of these genes was not affected by the loss of perR (see Fig. S5 in the supplemental material).
TABLE 1.
Putative PerR-binding motifs in the S. mutans UA159 genome
| Gene, name | Distance from ATG | Sequencea |
|---|---|---|
| smu182, sloA | 26 | TTATATTACTTATAA |
| smu540, dpr | 57 | TTAGAATCGTTCTAA |
| smu569, feoA | 118 | TTATAATGTTTCTCA |
| smu593, perR | 157 | TTTGAATGATTTTTA |
| smu635 | 37 | TTGTAATCATTCTAA |
| smu629, sodA | 121 | TTAGAATTATTTTAC |
| smu770c, mntH | 79 | TTTTAACGATACTTA |
| smu995 | 63 | ATAAAATGGTTTTGT |
| smu1142, spxA1 | 52 | TTTAAAATCTTGTAT |
| Consensus | TTANAATNATTNTAA |
Nucleotides that vary from the consensus sequence are identified by bold type.
FIG 5.
RT-qPCR analysis of genes with putative PerR-binding motifs reveals a trend of PerR transcriptional repression. Bars represent the gene copy number and represent averages and standard deviations (SDs) from three replicate samples. ANOVA was performed to verify significance of the data; expression levels of each gene were compared to those of UA159. *, P ≤ 0.03.
Concluding remarks.
The accidental discovery of spontaneously occurring perR gene mutations in laboratory stocks of UA159, the first fully sequenced and (by far) the most thoroughly characterized lab strain of S. mutans, supports anecdotal observations of bacterial adaptation to in vitro growth conditions. As shown in this report, loss of PerR regulation conferred a growth advantage to S. mutans under conditions (i.e., in the presence of air) that are routinely encountered in the laboratory setting. Interestingly, loss-of-function perR mutations were also identified in strains GS-5 and NG8, two of the most commonly used laboratory strains after the UA159 strain, suggesting that routine conditions used to cultivate S. mutans in the laboratory may facilitate the emergence of perR-inactive strains. However, how the loss of PerR regulation increased peroxide tolerance without making a significant impact in oxidative stress gene expression remains to be understood.
It has been proposed that tight regulation of stress genes is important because expression of these genes in the absence of stress may decrease overall fitness (37, 38). In both S. pyogenes and E. faecalis, the ability of perR mutant strains to tolerate peroxide stress in vitro did not correlate with the ability of those strains to cause disease (10, 29, 33), indicating that PerR is required to fine-tune gene expression during infection. Thus, the growth benefit acquired by S. mutans perR mutants under in vitro conditions is unlikely to arise during host colonization, providing a possible explanation as to why S. mutans has retained a functional perR. Studies to determine the significance of S. mutans perR during host colonization and disease progression are still warranted.
This report must also serve as a cautionary tale for conclusions broadly based on a single bacterial strain and the importance of keeping culture passages to a bare minimum and of routine (re)assessment of strains’ genetic and phenotypic traits. While these precautions may not completely eliminate the emergence of spontaneous mutants, especially those that confer a growth advantage, the accessibility and cost reductions of WGS technologies have made identification of these occurrences a manageable undertaking.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 2. Strains of S. mutans were routinely grown in brain heart infusion (BHI) at 37°C under anaerobic conditions. For physiologic analysis, overnight cultures were subcultured 1:20 in BHI medium in a 5% CO2 atmosphere, and growth was monitored over time. To evaluate the ability of the different strains to grow in the presence of stress conditions, cultures were grown to an optical density at 600 nm (OD600) of 0.25 and then diluted 1:50 into the appropriate medium. Growth was then monitored in a microtiter plate with an overlay of sterile mineral oil to minimize oxygen exposure using an automated growth reader (Bioscreen C) at 37°C. For growth in the presence of arginine, overnight cultures were serially diluted and 10 μl spotted onto tryptone-yeast extract-glucose (TYG) agar with or without arginine supplementation. For gene or protein expression analysis, overnight cultures were subcultured 1:20 in BHI as described above and grown to an optical density at 600 nm of 0.4, at which point control samples were harvested by centrifugation. For RNA isolation, harvested pellets were resuspended in 1 ml RNAprotect bacteria reagent (Qiagen), incubated for 5 min at ambient temperature, harvested by centrifugation, and the pellets stored at −80°C until use.
TABLE 2.
Bacterial strains used in this study
| Strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| S. mutans UA159 | Wild type | ATCC |
| S. mutans UA-perR | UA159 variant with perR mutation | Spontaneously occurring/laboratory stock |
| S. mutans ΔspxA1 | spxA1::Spec | 42 |
| S. mutans UAΔperR | perR::Erm | This study |
| S. mutans UAΔperR + perR | perR::Erm, mtl insertion, perR+ | This study |
| S. mutans NG8 | Wild type, truncated PerR | Laboratory stock |
| S. mutans NG8 + perR | mtl insertion, perR+ | This study |
| S. mutans GS-5 | Wild type, PerRA36D | Laboratory stock |
| S. mutans GS-5+ perR | mtl insertion, perR+ | This study |
DNA purification for whole-genomic sequencing.
For DNA isolation, cells from 25-ml stationary-phase cultures of S. mutans were harvested by centrifugation (3,000 × g, 15 min); suspended in a mixture of 50 mM NaCl, 25 mM EDTA, and lysozyme (1 mg ml−1); and incubated for 3 h at 37°C. Then, 0.1-mm glass beads and 1% sodium lauryl sulfate (SDS) were added to the suspension, which was subjected to high-speed vortexing for 5 min, followed by an additional 30-min incubation at 37°C. Nucleic acid was extracted with 1 volume of buffer-saturated phenol. The aqueous phase was collected and extracted with phenol-chloroform-isoamyl alcohol followed by another centrifugation step. The aqueous phase was precipitated with 0.3 M sodium acetate, pH 5.5, and an equal volume of cold absolute ethanol. The precipitated DNA was washed with 70% ethanol and then resuspended in sterile deionized H2O containing RNase. After 30 min incubation at room temperature, the DNA solution was again extracted with phenol-chloroform-isoamyl alcohol and ethanol precipitated as described above. The DNA was examined by agarose gel electrophoresis to ensure that the preparation contained high-molecular-weight DNA free of RNA. The DNA was used for WGS with SNPs and indels detected using CLC Genomics Workbench at Tufts University Core Facility (Boston, MA) using S. mutans UA159 (GenBank accession number NC_004350) as the reference sequence.
Construction of mutant and complemented strains.
A UA159 (perR+) strain lacking the perR gene was constructed using a PCR ligation mutagenesis approach (39). Briefly, PCR fragments flanking the perR gene were ligated to an erythromycin resistance cassette after digestion with the same restriction enzymes. This ligation mixture was used to transform S. mutans UA159; the recombinant strain was isolated on BHI agar supplemented with 10 μg ml−1 erythromycin. The gene deletion was confirmed by sequencing the amplicon containing the antibiotic cassette insertion site and flanking region. The mutant strain was complemented by cloning the full-length perR gene (and native promoter) into the S. mutans integration vector pMC340B (40) to yield plasmid pMC340B-perR. The plasmid was used to transform the S. mutans ΔperR strain for integration at the mtl locus. This same strategy was also used to transform S. mutans GS-5 and NG8 strains. All primers used in this study are listed in Table 3.
TABLE 3.
Primers used in this study
| Primer | Sequence (5′–3′)a | Application |
|---|---|---|
| perRdeletion5’Arm1 | CGACGAGGACCAACATTTTTACC | perR deletion |
| perRdeletion5’ERI | CCGGAATTCAGAATGCAGTCCCATTTTTGAC | perR deletion |
| perRdeletion3’HiIII | CCCAAGCTTAAGCCGATTAATTTGCTTATTG | perR deletion |
| perRdeletion3’Arm2 | AGAGCACGGATAGAGTTACCATGTG | perR deletion |
| perRcomp5’BamHI | CGCGGATCCCGATGAATACGAGCAGGTTCC | perR complement |
| perRcomp3’XhoI | CCGCTCGAGGATAATCAATAAGCAAATTAATCGGC | perR complement |
| sodART Fwd | GATGCTGAAACGATGACCCTTC | qPCR, sodA |
| sodART Rev | CACATCAGCCAAAAGCACTTCC | qPCR, sodA |
| dprRT Fwd | GAAGAAACAGTTGGCACATGGG | qPCR, dpr |
| dprRT Rev | TTCCGTTTGAGCTGCTGTAAAG | qPCR, dpr |
| feoBRT Fwd | GGCTTATCAGATGGGAGTGC | qPCR, feoB |
| feoBRT Rev | GCAATCGCTGCTTCAAATTTGG | qPCR, feoB |
| smu998RT Fwd | GAGCAACAATTGGTCAGGATGC | qPCR, smu.998 |
| smu998RT Rev | GCTTTGGTTGTTCTAATACCACCTG | qPCR, smu.998 |
| smu635RT Fwd | GCAGGTGTCTTAGGAGCTAATGATGG | qPCR, smu.635 |
| smu635RT Rev | GGCCTCTTCAGTATCCTTTTGCG | qPCR, smu.635 |
| mntHRT Fwd | AATGCCCAGTTTACCAGCCA | qPCR, mntH |
| mntHRT Rev | TCAGCGAGGTCAATCAGAGC | qPCR, mntH |
| sloCRT Fwd | CAGTGAGCGATAGTGTTAAGAC | qPCR, sloC |
| sloCRT Rev | CTCTTGTTTTTAGGATCTTTTTCAGATAG | qPCR, sloC |
| spxA2RT Fwd | CCGCTATGCTAAGGCTCTCCA | qPCR, spxA2 |
| spxA2RT Rev | TCAACATTACGAATAGAACGCGGTA | qPCR, spxA2 |
| pbp2ART Fwd | AGCCAAGCCTCTGACAACC | qPCR, pbp2A |
| pbp2ART Rev | CCGCAATCAGCAAAATCTTAC C | qPCR, pbp2A |
Restriction sites used to facilitate cloning are indicated in bold.
ICP-MS analysis.
The total metal content within bacterial cells was determined using inductively coupled plasma mass spectrometry (ICP-MS) performed at the University of Florida Analytical Toxicology Core Laboratory. Briefly, cultures were grown to mid-exponential phase (OD600 = 0.4) in BHI, harvested by centrifugation at 4°C for 15 min at 4,000 rpm, and washed first in phosphate-buffered saline (PBS) supplemented with 0.1 mM EDTA to chelate extracellular divalent cations followed by a wash in PBS alone. Cell pellets were resuspended in HNO3, and metal composition was quantified using a 7900 ICP mass spectrometer (Agilent). Metal concentrations were then normalized to total protein content determined by the bicinchoninic acid (BCA) assay (Pierce).
Western blot analysis.
Whole-cell protein lysates of mid-log-grown cultures of S. mutans UA159, UA-perR, UAΔperR, and UAΔperRComp strains were obtained by homogenization in the presence of 0.1-mm glass beads using a bead beater. The protein concentration was determined using the BCA assay. Equal amounts of the protein lysates were separated by 13.5% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Prometheus). Dpr detection was performed using rabbit anti-rDpr polyclonal antibody (20) diluted 1:1,000 in phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBS-T) followed by anti-rabbit horseradish peroxidase (HRP)-coupled antibody (Sigma-Aldrich) diluted 1:1,000 in PBS-T. Immune reactivity was visualized by incubation of the blot with 3,3-diaminobenzidine (Sigma-Aldrich) substrate solution for horseradish peroxidase conjugates. As a loading control, duplicate gels were stained with SYPRO Ruby protein gel stain (Thermo Fisher Scientific) and visualized with UV light.
RNA analysis.
Total RNA was isolated by the acid-phenol chloroform extraction method from homogenized cell lysates that was modified from a previously described protocol (41). Briefly, harvested cells were resuspended in Tris-EDTA (50 mM:10 mM) in the presence of acid phenol-chloroform (5:1) and SDS detergent, mixed with 0.1-mm glass beads and subjected to homogenization in a BeadBeater (BioSpec Products) for three cycles of 30 s, with 3 min on ice between cycles. The cell lysates were subjected to centrifugation (14,000 × g, 10 min), and the upper aqueous phase containing nucleic acid transferred to a new tube. An equal volume of acid phenol-chloroform was vigorously mixed with the aqueous phase by vortexing followed by incubation for 5 min at ambient temperature, and the upper aqueous phase was collected after another centrifugation cycle. This nucleic acid-containing aqueous phase was digested with Ambion DNase I (Thermo Fisher) according to the manufacturer’s instructions. Then, RNA was purified using an RNeasy kit (Qiagen) including an on-column DNase digestion according to the manufacturer’s instructions. The final RNA concentration was determined by measuring on a NanoDrop One spectrophotometer (Thermo Fisher). Quantifications of mRNA were obtained by reverse transcriptase quantitative real-time PCR (RT-qPCR) according to protocols described elsewhere (41) using gene-specific primers listed in Table 3. Analysis of variance (ANOVA) was performed to verify significance of the RT-qPCR results using Dunnett’s test to compare expression to that of S. mutans UA159.
For RNA-Seq analysis, samples were prepared as previously described (32). Sample quality and quantity were assessed on an Agilent 2100 Bioanalyzer at the University of Florida Interdisciplinary Center for Biotechnology Research (UF-ICBR). RNA (5 μg per sample) was subjected to two rounds of mRNA enrichment using a MICROBExpress bacterial mRNA purification kit (Thermo Fisher). cDNA libraries with unique barcodes were generated from 100 ng enriched mRNA using NEBNext Ultra II directional RNA library prep kit for Illumina (New England Biolabs) and then assessed for quality and quantity by Qubit. Pooled cDNA libraries were subjected to RNA deep sequencing (RNA-Seq) using the Illumina NextSeq 500 platform. Read mapping was performed on a Galaxy server hosted by the UF research computer using Map with Bowtie for Illumina and the S. mutans UA159 genome (GenBank accession number NC_004350.2) as a reference. The reads per open reading frame were tabulated with htseq-count. Final comparisons were performed with Degust (http://degust.erc.monash.edu/), with a false-discovery rate (FDR) of 0.05 and a 1.5-fold change cutoff.
Data availability.
Gene expression data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) under GEO Series accession number GSE158080.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH-NIDCR award DE019783.
We thank our colleagues from the “S. mutans community” for sharing strains and helpful discussions.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499. 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 3.Lee JW, Helmann JD. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363–367. 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 4.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol 29:189–198. 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 5.Fuangthong M, Herbig AF, Bsat N, Helmann JD. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J Bacteriol 184:3276–3286. 10.1128/jb.184.12.3276-3286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herbig AF, Helmann JD. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol Microbiol 41:849–859. 10.1046/j.1365-2958.2001.02543.x. [DOI] [PubMed] [Google Scholar]
- 7.Brenot A, Weston BF, Caparon MG. 2007. A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol Microbiol 63:1185–1196. 10.1111/j.1365-2958.2006.05577.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Keramati L, Helmann JD. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci U S A 92:8190–8194. 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grifantini R, Toukoki C, Colaprico A, Gryllos I. 2011. Peroxide stimulon and role of PerR in group A Streptococcus. J Bacteriol 193:6539–6551. 10.1128/JB.05924-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gryllos I, Grifantini R, Colaprico A, Cary ME, Hakansson A, Carey DW, Suarez-Chavez M, Kalish LA, Mitchell PD, White GL, Wessels MR. 2008. PerR confers phagocytic killing resistance and allows pharyngeal colonization by group A Streptococcus. PLoS Pathog 4:e1000145. 10.1371/journal.ppat.1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun 69:3744–3754. 10.1128/IAI.69.6.3744-3754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinochet-Barros A, Helmann JD. 2018. Redox sensing by Fe2+ in bacterial Fur family metalloregulators. Antioxid Redox Signal 29:1858–1871. 10.1089/ars.2017.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kebouchi M, Saul F, Taher R, Landier A, Beaudeau B, Dubrac S, Weber P, Haouz A, Picardeau M, Benaroudj N. 2018. Structure and function of the Leptospira interrogans peroxide stress regulator (PerR), an atypical PerR devoid of a structural metal-binding site. J Biol Chem 293:497–509. 10.1074/jbc.M117.804443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JC, Oh E, Hwang S, Ryu S, Jeon B. 2015. Non-selective regulation of peroxide and superoxide resistance genes by PerR in Campylobacter jejuni. Front Microbiol 6:126. 10.3389/fmicb.2015.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Vliet AH, Baillon ML, Penn CW, Ketley JM. 1999. Campylobacter jejuni contains two Fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J Bacteriol 181:6371–6376. 10.1128/JB.181.20.6371-6376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pant S, Patel NJ, Deshmukh A, Golwala H, Patel N, Badheka A, Hirsch GA, Mehta JL. 2015. Trends in infective endocarditis incidence, microbiology, and valve replacement in the United States from 2000 to 2011. J Am Coll Cardiol 65:2070–2076. 10.1016/j.jacc.2015.03.518. [DOI] [PubMed] [Google Scholar]
- 17.Lemos JA, Burne RA. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology (Reading) 154:3247–3255. 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kajfasz JK, Rivera-Ramos I, Abranches J, Martinez AR, Rosalen PL, Derr AM, Quivey RG, Lemos JA. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol 192:2546–2556. 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajfasz JK, Rivera-Ramos I, Scott-Anne K, Gregoire S, Abranches J, Lemos JA. 2015. Transcription of oxidative stress genes is directly activated by SpxA1 and, to a lesser extent, by SpxA2 in Streptococcus mutans. J Bacteriol 197:2160–2170. 10.1128/JB.00118-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganguly T, Kajfasz JK, Miller JH, Rabinowitz E, Galvao LCC, Rosalen PL, Abranches J, Lemos JA. 2018. Disruption of a novel iron transport system reverses oxidative stress phenotypes of a dpr mutant strain of Streptococcus mutans. J Bacteriol 200:e00062-18. 10.1128/JB.00062-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto Y, Poole LB, Hantgan RR, Kamio Y. 2002. An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J Bacteriol 184:2931–2939. 10.1128/jb.184.11.2931-2939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makthal N, Rastegari S, Sanson M, Ma Z, Olsen RJ, Helmann JD, Musser JM, Kumaraswami M. 2013. Crystal structure of peroxide stress regulator from Streptococcus pyogenes provides functional insights into the mechanism of oxidative stress sensing. J Biol Chem 288:18311–18324. 10.1074/jbc.M113.456590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JW, Helmann JD. 2006. Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J Biol Chem 281:23567–23578. 10.1074/jbc.M603968200. [DOI] [PubMed] [Google Scholar]
- 24.Yeowell HN, White JR. 1982. Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob Agents Chemother 22:961–968. 10.1128/aac.22.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner AG, Ong CY, Djoko KY, West NP, Davies MR, McEwan AG, Walker MJ. 2017. The PerR-regulated P1B-4-type ATPase (PmtA) acts as a ferrous iron efflux pump in Streptococcus pyogenes. Infect Immun 85:e00140-17. 10.1128/IAI.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Tong H, Dong X. 2014. PerR-regulated manganese ion uptake contributes to oxidative stress defense in an oral streptococcus. Appl Environ Microbiol 80:2351–2359. 10.1128/AEM.00064-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King KY, Horenstein JA, Caparon MG. 2000. Aerotolerance and peroxide resistance in peroxidase and PerR mutants of Streptococcus pyogenes. J Bacteriol 182:5290–5299. 10.1128/jb.182.19.5290-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricci S, Janulczyk R, BjöRck L. 2002. The regulator PerR is involved in oxidative stress response and iron homeostasis and is necessary for full virulence of Streptococcus pyogenes. Infect Immun 70:4968–4976. 10.1128/IAI.70.9.4968-4976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verneuil N, Rince A, Sanguinetti M, Posteraro B, Fadda G, Auffray Y, Hartke A, Giard JC. 2005. Contribution of a PerR-like regulator to the oxidative-stress response and virulence of Enterococcus faecalis. Microbiology 151:3997–4004. 10.1099/mic.0.28325-0. [DOI] [PubMed] [Google Scholar]
- 30.Fujishima K, Kawada-Matsuo M, Oogai Y, Tokuda M, Torii M, Komatsuzawa H. 2013. dpr and sod in Streptococcus mutans are involved in coexistence with S. sanguinis, and PerR is associated with resistance to H2O2. Appl Environ Microbiol 79:1436–1443. 10.1128/AEM.03306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 32.Kajfasz JK, Katrak C, Ganguly T, Vargas J, Wright L, Peters ZT, Spatafora GA, Abranches J, Lemos JA. 2020. Manganese uptake, mediated by SloABC and MntH, is essential for the fitness of Streptococcus mutans. mSphere 5:e00764-19. 10.1128/mSphere.00764-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brenot A, King KY, Caparon MG. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol Microbiol 55:221–234. 10.1111/j.1365-2958.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- 34.Fuangthong M, Helmann JD. 2003. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J Bacteriol 185:6348–6357. 10.1128/jb.185.21.6348-6357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, Kazanov MD, Riehl W, Arkin AP, Dubchak I, Rodionov DA. 2013. RegPrecise 3.0–a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14:745. 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ganguly T, Kajfasz JK, Abranches J, Lemos JA. 2020. Regulatory circuits controlling Spx levels in Streptococcus mutans. Mol Microbiol 114:109–126. 10.1111/mmi.14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cotter PA, Miller JF. 1998. In vivo and ex vivo regulation of bacterial virulence gene expression. Curr Opin Microbiol 1:17–26. 10.1016/S1369-5274(98)80138-0. [DOI] [PubMed] [Google Scholar]
- 38.Mahan MJ, Heithoff DM, Sinsheimer RL, Low DA. 2000. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu Rev Genet 34:139–164. 10.1146/annurev.genet.34.1.139. [DOI] [PubMed] [Google Scholar]
- 39.Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49:193–205. 10.1016/s0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- 40.Chen PM, Chen YY, Yu SL, Sher S, Lai CH, Chia JS. 2010. Role of GlnR in acid-mediated repression of genes encoding proteins involved in glutamine and glutamate metabolism in Streptococcus mutans. Appl Environ Microbiol 76:2478–2486. 10.1128/AEM.02622-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J Bacteriol 188:3748–3756. 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kajfasz JK, Martinez AR, Rivera-Ramos I, Abranches J, Koo H, Quivey RG, Jr, Lemos JA. 2009. Role of Clp proteins in expression of virulence properties of Streptococcus mutans. J Bacteriol 191:2060–2068. 10.1128/JB.01609-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo) under GEO Series accession number GSE158080.