Under infection conditions, S. aureus needs to ensure survival when energy production via oxidative phosphorylation is not possible, e.g., either due to the lack of terminal electron acceptors or by the inactivation of components of the respiratory chain. Under these conditions, S. aureus can switch to mixed-acid fermentation to sustain ATP production by substrate-level phosphorylation.
KEYWORDS: Staphylococcus aureus, anaerobiosis, ccpA
ABSTRACT
To be a successful pathogen, Staphylococcus aureus has to adapt its metabolism to the typically oxygen- and glucose-limited environment of the host. Under fermenting conditions and in the presence of glucose, S. aureus uses glycolysis to generate ATP via substrate-level phosphorylation and mainly lactic acid fermentation to maintain the redox balance by reoxidation of NADH equivalents. However, it is less clear how S. aureus proceeds under anoxic conditions and glucose limitation, likely representing the bona fide situation in the host. Using a combination of proteomic, transcriptional, and metabolomic analyses, we show that in the absence of an abundant glycolysis substrate, the available carbon source pyruvate is converted to acetyl coenzyme A (AcCoA) in a pyruvate formate-lyase (PflB)-dependent reaction to produce ATP and acetate. This process critically depends on derepression of the catabolite control protein A (CcpA), leading to upregulation of pflB transcription. Under these conditions, ethanol production is repressed to prevent wasteful consumption of AcCoA. In addition, our global and quantitative characterization of the metabolic switch prioritizing acetate over lactate fermentation when glucose is absent illustrates examples of carbon source-dependent control of colonization and pathogenicity factors.
IMPORTANCE Under infection conditions, S. aureus needs to ensure survival when energy production via oxidative phosphorylation is not possible, e.g., either due to the lack of terminal electron acceptors or by the inactivation of components of the respiratory chain. Under these conditions, S. aureus can switch to mixed-acid fermentation to sustain ATP production by substrate level phosphorylation. The drop in the cellular NAD+/NADH ratio is sensed by the repressor Rex, resulting in derepression of fermentation genes. Here, we show that expression of fermentation pathways is further controlled by CcpA in response to the availability of glucose to ensure optimal resource utilization under growth-limiting conditions. We provide evidence for carbon source-dependent control of colonization and virulence factors. These findings add another level to the regulatory network controlling mixed-acid fermentation in S. aureus and provide additional evidence for the lifestyle-modulating effect of carbon sources available to S. aureus.
INTRODUCTION
The ability of Staphylococcus aureus to grow in the absence of oxygen is a prerequisite to infect and colonize its human host (1–3). When oxygen levels drop, S. aureus can fuel its metabolism with energy from anaerobic respiration, using nitrate or nitrite as terminal electron acceptors. Anaerobic respiration is controlled by the two-component systems NreBC and AirSR/YhcSR, which sense oxygen availability via reversible oxidation of an Fe-S cluster within their sensor kinases (4–7). Under these conditions, ATP is also generated by substrate-level phosphorylation and mixed-acid fermentation (8).
The ability of S. aureus to ferment is crucial to sustaining growth not only at low oxygen levels but also when its respiratory chain is nonfunctional, although oxygen might still be present. For instance, lactate dehydrogenase, the key enzyme in lactic acid fermentation, is essential for the metabolic adaptation of S. aureus toward inactivation of the respiratory chain by nitric oxide produced by cells of the innate immune response (9). Furthermore, S. aureus can acquire a slow-growing, difficult-to-eradicate, small-colony-variant (SCV) phenotype, which is associated with persistent infections. SCVs are often characterized by inhibition of respiration caused by inactivation of genes of the menadione or hemin biosynthesis pathways (10, 11). In these strains, energy production is maintained by fermentation, and mutations leading to an upregulation of fermentation pathways can result in fast-growing SCVs that are highly resistant to antimicrobial therapy (12).
Expression of mixed-acid fermentation pathways in S. aureus is regulated by the intracellular redox balance and thus linked to a functional respiratory chain rather than directly to oxygen availability as shown in Escherichia coli, where the oxygen level is sensed by the regulator FNR (fumarate and nitrate reductase regulation) (13). Fermentation gene expression in S. aureus is mainly controlled by the transcriptional repressor Rex, the mixed-acid fermentation master regulator, and, to a minor extent, by the pleiotropic two-component system SrrAB (13, 14). In vitro, the binding affinity of Rex to its operator sequence [TTGTGAA-W(4)-TTCACAA] is dependent on the intracellular NAD+/NADH ratio. In the presence of high NADH concentrations, NADH competes with NAD+ for Rex binding, lowering the affinity of Rex for its operator sites and consequently leading to derepression of pathways required for anaerobic NAD+ regeneration (13). The signal activating the SrrAB two-component system is most likely an impaired electron flow in the electron transport chain, sensed by a conserved cysteine in the SrrB PAS domain (14, 15).
Mixed-acid fermentation in S. aureus results in the secretion of 2,3-butanediol, ethanol, acetate, and lactate. Vitko and coworkers showed that S. aureus critically depends on a large number of glucose transporters to fuel glycolysis during invasive infection in hypoxic environments (16). During growth on glucose, lactate is the major fermentation product, and only small amounts of acetate are produced (8, 17–20). Acetate production from pyruvate is initiated by the pyruvate formate-lyase (Pfl) and subsequent oxidation of acetyl coenzyme A (AcCoA) to acetate by the concerted activities of phosphate acetyltransferase (Pta) and acetate kinase (AckA), yielding an additional ATP. Thus, if glycolysis remains active, a shift from lactate to acetate would result in a higher ATP gain. This situation is somewhat reminiscent of aerobic overflow metabolism, but the mechanisms balancing the carbon-to-ATP ratio under anaerobic conditions remain poorly understood (21).
Previous work has identified the Pfl-dependent conversion of pyruvate to AcCoA and formate as an important source of C1 building blocks under anaerobic conditions (22). Here, using a complementary global quantitative proteomic, transcriptional, and metabolomic approach, we show that in the absence of a glycolytic carbon source, the fermentation pathways of S. aureus are reorganized to maximize the activity of the Pfl branch in a catabolite control protein A (CcpA)-dependent manner, while reactions leading to wasteful consumption of AcCoA are repressed. We also provide additional evidence that carbon sources and oxygen availability impact the expression of colonization and virulence factors.
RESULTS
Availability of glycolytic versus nonglycolytic carbon sources influences the abundance and repertoire of fermentation and glycolytic enzymes.
To determine how S. aureus adapts to oxygen limitation in the absence of glucose, cells were cultivated in a chemically defined medium with pyruvate as the major carbon source. Samples were taken before (aerobic reference) and 3 h after a shift of the exponentially growing culture to hypoxic conditions (fermentation sample). For comparison, corresponding samples were harvested from a culture grown with glucose as the major carbon source. To ensure that cells are anaerobic at sampling points, we measured the concentration of dissolved oxygen in our experimental setup. We found that oxygen was depleted from the pyruvate and the glucose medium within 17.5 min and 22.5 min, respectively. This is in good agreement with previously published data showing oxygen depletion within 20 to 25 min under similar growth conditions (see Fig. S1 in the supplemental material) (8).
After the shift to hypoxic conditions, no increase in the optical density (OD) of the pyruvate culture could be determined, indicating that fermentative growth in the absence of glucose is not well supported. The OD of the glucose culture, however, continued to increase and approximately doubled during the experiment (Fig. S2).
After cell disruption, the soluble protein fraction was tryptic digested, and purified peptides were analyzed by liquid chromatography-ion mobility-mass spectrometry (LC-IMSE). This approach allowed generating quantitative data for 1,215 proteins across all conditions probed, corresponding to a coverage of 56% of the soluble S. aureus proteome (Table S1). A global overview summarizing carbon source- and oxygen-dependent changes is provided in Fig. 1. The results showed that about one-quarter of the proteome changed in a condition-specific manner. Three-quarters of the total protein amounts remained relatively constant over all tested conditions. The substantial changes in the proteome were also observed for the pyruvate culture, indicating that despite no obvious increase in biomass, the cells were still capable of allocating sufficient energy to restructure their proteome in response to fermentation conditions.
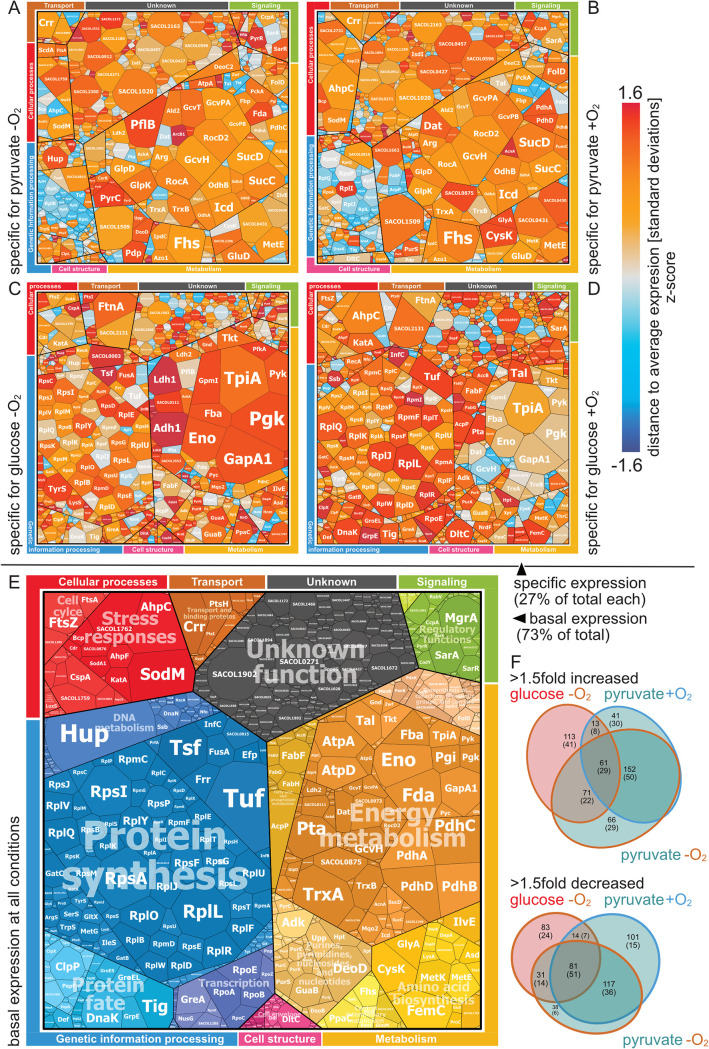
FIG 1.
Overview of global carbon source- and oxygen-dependent protein accumulation pattern. Voronoi treemaps showing the abundance of the dynamic proteome (27% of the total protein amount) during anaerobic growth with pyruvate (A) or glucose (C) and aerobic growth with pyruvate (B) or glucose (D). (E) Static or basal and unchanged fraction of the proteome (73%) at all tested growth conditions. The size of the tiles in the Voronoi treemaps is proportional to the relative protein abundance in the sample. Color code in panels A to D corresponds to Z-scores of protein abundance over all tested conditions to indicate condition specific up- and downregulation. (F) Venn diagrams summarize the numbers of regulated proteins showing an at least 1.5-fold up- or downregulation in abundance.
Following the shift to anaerobic conditions in the presence of glucose, a strong accumulation of proteins involved in mixed-acid fermentation and glycolysis was observed, which confirmed previously reported results (Fig. 2; Table S1) (8, 18). Of note, in particular, glycolytic enzymes catalyzing the committed steps (PfkA, PykA) increased significantly in abundance. In addition, Pgk was strongly induced, which, together with PykA, catalyzes the two ATP-generating reactions in glycolysis. Of the fermentation enzymes, l-lactate dehydrogenase 1 (Ldh1) and the potential alcohol dehydrogenases 1 (Adh1) were by far the proteins showing the strongest anaerobic induction. Together, both proteins accounted for almost 1% of the total accumulated soluble proteome. In particular, the massive accumulation of Adh1 was of interest, suggesting an important role in ethanol production, which, so far, lacks experimental confirmation. A comparison of Adh1 to alcohol dehydrogenases of known structures and substrate specificities, however, makes a function of Adh1 in ethanol production very likely (Fig. S3). Furthermore, l-lactate dehydrogenase 2 (Ldh2), acetate kinase (AckA), the B subunit of pyruvate formate-lyase (PflB), d-lactate dehydrogenase (LdhD), and the bifunctional acetaldehyde dehydrogenase/alcohol dehydrogenase (AdhE) accumulated significantly following the anaerobic shift (Fig. 2; Table S1).
FIG 2.
Road map of central carbon metabolism and mixed-acid fermentation. Metabolic pathways leading to the conversion of glucose to the various fermentation products, the citric acid cycle, and the pentose phosphate (PP) shunt are shown as road maps. Bar charts indicate absolute protein abundance. Values shown are mean with standard deviations calculated from three independent biological replicates, each measured as three technical replicates. The table on the right side of the road maps shows log2 values of fold changes in protein abundance, and a color code indicates up- (red) or downregulation (blue). Significant changes (significance level ≤ 0.05) are indicated by red circles for the different statistical test. Proteins completely absent from one of the compared samples are indicated as either “on” or “off” proteins.
Compared to glucose, a global reorganization of the protein repertoire was observed when cells were grown with pyruvate. This extensive reorganization of the proteome was already evident under aerobic conditions. The most prominent differences were a strong accumulation of proteins of the tricarboxylic acid cycle (CitC, OdhA, OdhB, SucC, SucD, FumC, SdhA, and SdhB), glycerol metabolism (GlpD and GlpK), and gluconeogenetic enzymes (Fbp, GapA2, and PckA) (Fig. 2; Table S1). In addition, enzymes involved in amino acid catabolism (RocA, RocD2, Ald2, and ArgF) were highly abundant during aerobic growth with pyruvate (Fig. 1; Table S1).
In response to oxygen depletion, pathways of mixed-acid fermentation were strongly induced in cells growing on pyruvate. However, compared to glucose-fed cells, a significant redistribution of the amounts of the different fermentation enzymes was observed (Fig. 2). For instance, in the presence of pyruvate, Ldh1 accumulated only 1.8-fold and thus 32 times lower than in cells grown on glucose. Furthermore, Adh1 was 18.3 times less abundant in cells growing anaerobically on pyruvate, while AdhE was not detectable. In contrast, abundance of PflB was 2.6-fold higher with pyruvate, and PflA, the PflB-activating enzyme, became only detectable under this condition (Fig. 2; Table S1). AckA, Pta, and Ldh2 protein levels, however, showed no significant carbon source-dependent differences in accumulation during anaerobiosis in the glucose versus pyruvate comparison (Fig. 2).
CcpA regulates pfl transcription during glucose limitation.
To further investigate the carbon source-dependent regulatory events controlling fermentation gene expression, we carried out transcriptional analysis of metabolic genes 30 min after shifting cells to hypoxic conditions (Fig. 3). As expected, based on our proteome data, we observed clear carbon source-dependent differences in the transcription of fermentation genes when oxygen was depleted from the culture medium. For instance, pflB transcription increased 3-fold in the medium with pyruvate compared to the medium with glucose, which correlates well with the proteome data (Fig. 2 and Fig. 3; Table S1). A slightly stronger anaerobic transcription with pyruvate was observed also for genes encoding l-lactate dehydrogenase 2 (Ldh2) and acetate kinase (AckA). However, induction at the transcriptional level was well below 2-fold and not reflected by major differences in the accumulation of both corresponding proteins (Fig. 2; Fig. 3).
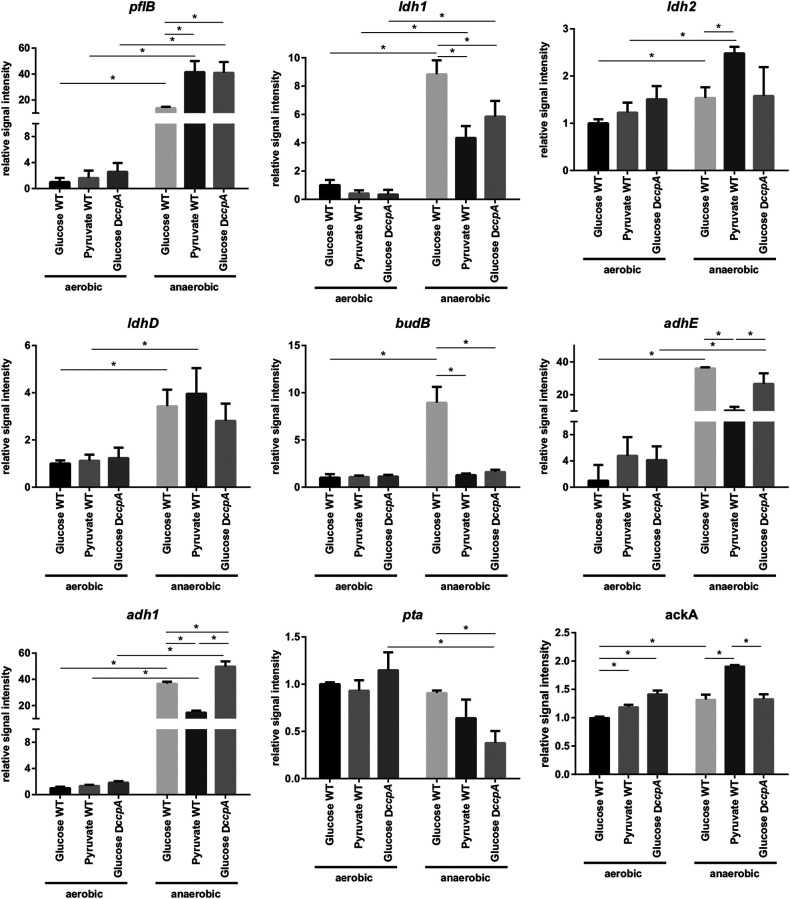
FIG 3.
Slot blot analysis of genes of mixed-acid fermentation. For RNA sampling, S. aureus COL WT or COL ccpA cells were grown in CDM supplemented with either glucose or pyruvate to an OD500 of 0.5 and shifted to hypoxic conditions. RNA was sampled directly before and 30 min after the shift to hypoxic conditions. Results shown are mean signal intensities and standard deviations from three independent biological replicates. For every gene signal intensity, the corresponding aerobic glucose sample was set to one. Significant differences (q ≤ 0.05, t test with Benjamini-Hochberg adjustment of P values) are indicated with a line and a star.
In contrast, anaerobic transcriptions of ldh1, the two alcohol dehydrogenases adhE and adh1, and budB, encoding catabolic acetolactate synthase, were reduced 2- to 4-fold in pyruvate medium compared to growth on glucose. Together, these genes encode proteins required for the regeneration of NAD+ during fermentation, resulting in the secretion of reduced fermentation products lactate (Ldh1), 2,3-butanediol (BudB), and ethanol (AdhE and possibly Adh1). Finally, we observed relatively similar transcript levels with both carbon sources for ldhD, contrasting the proteomic results showing a slightly but significantly increased accumulation (2.3-fold) of LdhD in glucose medium (Fig. 2; Fig. 3).
To elucidate the impact of carbon catabolite repression on the reorganization of fermentation pathways, transcription of different metabolic genes was also investigated in a ccpA mutant grown in glucose medium. Under anaerobic conditions, the most prominent effect in the ccpA mutant was a dramatically increased transcription of pflB, which reached the same transcription level as observed for cells growing on pyruvate. This observation indicates a strong negative effect of CcpA on pflB transcription in the presence of glucose (Fig. 3). CcpA dependency of pfl transcription is further supported by the presence of a potential catabolite-responsive element (cre) site (TATGAAAACGTTAACATA) identified 42 nucleotides (nt) upstream of the pflBA operon using Virtual Footprint 3.0 (23), the RSAT regulatory sequence analysis tools (24, 25), and the published cre consensus sequences (26).
A positive effect of CcpA under anaerobic conditions was observed for transcription of ldh1, budB, and the phosphate acetyltransferase gene, pta, but no well-defined cre sites were predicted for the corresponding upstream regions using our search criteria.
While transcription of pta was about 2-fold reduced in the ccpA mutant, anaerobic induction of budB transcription was completely abolished. Finally, anaerobic transcription of ldh2, ldhD, and ackA and of the gene encoding the alcohol dehydrogenase AdhE was independent from CcpA, while the small difference observed for adh1 transcription was deemed significant (Fig. 3).
Acetate is the major secretion product in pyruvate-fed cells under anaerobic conditions.
To analyze how carbon source-dependent changes in fermentation gene expression affect the exometabolome, we monitored the composition of the growth medium by proton nuclear magnetic resonance (1H NMR) (Fig. 4A).
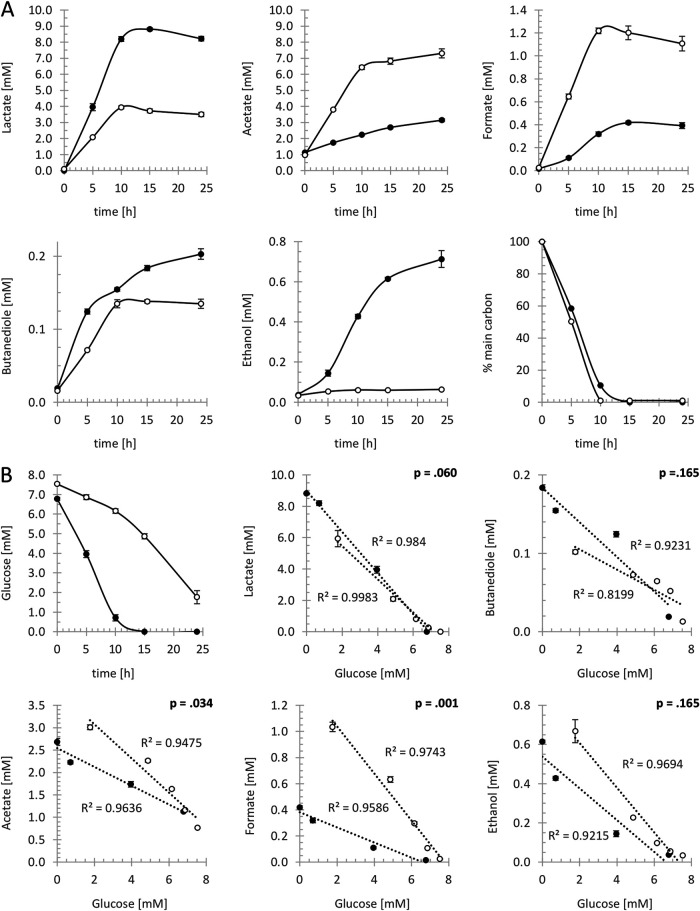
FIG 4.
Carbon source- and CcpA-dependent changes of mixed-acid fermentation metabolites. (A) Profiles of fermentation products secreted during growth with glucose (closed symbols) or pyruvate (open symbols). The bottom right graph shows time-dependent percentual consumption of the major carbon sources glucose (closed symbol) and pyruvate (open symbol). Values are means with standard deviations from four biological replicates. Statistics are summarized in Table S2 in the supplemental material. (B) The top left graph shows anaerobic glucose consumption in the S. aureus COL wild type (closed symbols) and the isogenic ccpA mutant (open symbols). The remaining graphs show changes in the concentration of key fermentation products plotted against glucose consumption to account for the reduced metabolic activity of the ccpA mutant compared to the wild type. Differences in the production of fermentation products were detected by comparing the slopes of linear regressions for the wild type (closed symbols) and ccpA mutant (open symbols) data. Corresponding P values are shown in the upper right corner of the graphs. Error bars indicate standard deviations from four biological replicates.
These analyses confirmed that during anaerobic growth with glucose, lactate is the major fermentation product (lactate-to-acetate ratio, 4:1). However, it was revealed that in the presence of pyruvate, S. aureus mainly secrets acetate (lactate-to-acetate ratio, 1:2), which indicates increased activity of the Pfl-Pta-AckA branch. In further agreement with increased Pfl activity is a 2.5-fold increased formate production in pyruvate medium. Finally, while the amount of secreted butanediol was carbon source independent, ethanol was only detected when cells were grown with glucose (Fig. 4A).
Of the 16 supplemented amino acids, threonine, serine, arginine, isoleucine, glutamate, and lysine were consumed in measurable amounts. Out of these, threonine, serine, arginine, lysine, and isoleucine were removed from the medium more quickly in the presence of glucose, while isoleucine and glutamate are consumed independently of the carbon source but at much lower quantities than threonine, serine, and arginine (Fig. S4).
To confirm a role of CcpA in the adjustment of fermentation pathways, the exometabolome of a ccpA mutant grown on glucose was investigated. Elevated acetate and formate production was observed in the ccpA mutant, aligning well with the derepressed transcription of pflB in this background. In the ccpA mutant, production of ethanol increased compared to the wild type, probably as a consequence of increased supply of AcCoA via Pfl. The rate of lactate production was not affected by a deletion of the ccpA gene. However, compared to wild-type cells, decreased secretion of butanediol was observed, aligning well with the strongly repressed transcription of budB in the ccpA mutant (Fig. 4B).
The importance of a precise carbon source-dependent adjustment of fermentation pathways is further supported by the significantly reduced growth of the ccpA mutant under anaerobic conditions compared to the wild type (Fig. S2).
Does Rex align transcription of fermentation pathways with carbon source availability?
The lower concentrations of reduced fermentation products secreted by S. aureus grown on pyruvate suggests a higher NAD+/NADH ratio with pyruvate than the more reduced glucose, as previously reported for E. coli (27, 28). Therefore, in addition to CcpA, a hierarchical derepression of fermentation pathways via the NAD+/NADH ratio-sensing regulator Rex could provide an additional mechanism to link expression of mixed-acid fermentation to the oxidation state of the fermented carbon source (13). In this context, it is interesting to note that two strong Rex-binding sites are present upstream of ldh1, and perfect, high-affinity Rex-binding sites are located in the adh1 and adhE promoter regions. Consequently, a very strong shift toward NADH might be required to achieve full derepression of ldh1, adh1, and adhE transcription. Contrary, the Rex-binding sites in front of the pfl and the lactate permease (lctP) genes show two mismatches relative to a perfect palindromic binding sequence, and in vitro analyses confirm a higher affinity of Rex for the ldh1, adhE, and adh1 upstream regions than the pfl and lctP promoter regions (13). Interestingly, the low-affinity binding site preceding lctP would fit well with the observation that cells growing on pyruvate still produced and need to secret relatively large amounts of lactate, even if Ldh1 expression is strongly reduced. Thus, the degree of Rex operator matching a perfect palindrome and the resulting affinities of Rex-binding sites align well with the different pathways used to oxidize NADH and therefore might have evolved to ensure that the efficiency of NADH oxidation matches carbon source-dependent NADH production (Fig. S5).
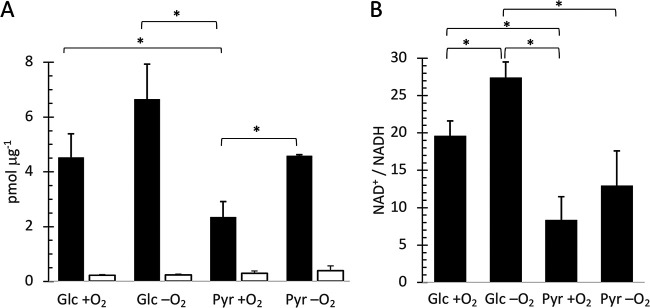
To test this hypothesis, we measured the NAD+/NADH ratio of cells 30 min after a shift to hypoxic growth conditions to correlate the NAD+/NADH ratio with the transcriptional data. We found that aerobic cells growing on pyruvate were generally characterized by lower NAD+ concentrations than the cells growing on glucose. A significant increase in total NAD+ was observed when pyruvate cells were shifted to hypoxic conditions, reaching levels in the range of the glucose culture (Fig. 5A). Furthermore, contrary to our hypothesis, the NAD+/NADH ratio was generally higher in the glucose culture than in the pyruvate culture. Finally, a significant shift in the NAD+/NADH ratio was observed only during cultivation with glucose, which, however, even increased upon anaerobiosis (Fig. 5B). Thus, the results do not provide support for the hypothesis that the oxidation state of the carbon source modulates the intracellular NAD+/NADH ratio in S. aureus and thereby triggers a hierarchical derepression of mixed-acid fermentation pathways.
FIG 5.
NAD+/NADH ratio during aerobic and fermentative growth with pyruvate or glucose. NAD+ (closed columns) and NADH (open columns) concentrations (A) and NAD+/NADH ratios (B) were measured in S. aureus cells grown aerobic (with O2) or after the shift to hypoxic conditions for 30 min (without O2) with glucose (Glc) or pyruvate (Pyr) as carbon source. Results shown are means from three biological replicates with standard deviations. Significant differences (P ≥ 0.05, two-sided t test) are indicated by a bracket and a star.
The carbon source modulates accumulation of extracellular and oxidative stress proteins.
It is conceivable that in addition to modulating the central carbon pathways, an oxygen-depleted and low-glucose environment might influence other aspects of the S. aureus physiology.
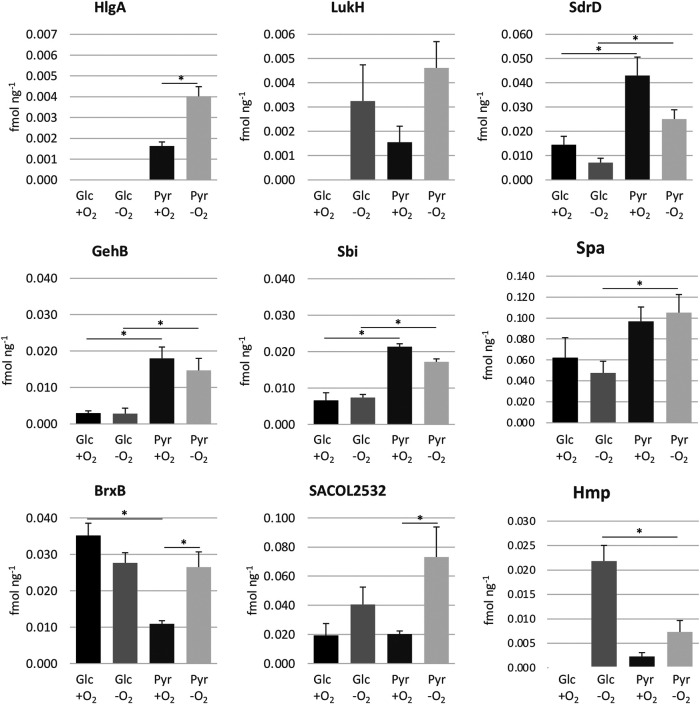
Although our proteomic approach did not specifically aim at the extracellular proteome, we detected clear medium-dependent differences in the expression of several exoproteins, which is in agreement with a recent study (29). For instance, gamma-hemolysin A (HlgA) was only detected in the pyruvate culture and showed a 2.4-fold stronger accumulation under anaerobic conditions (Fig. 6). Furthermore, the leucocidin LukH was detected only in the absence of oxygen or in the presence of pyruvate (30, 31). Other medium-regulated secreted virulence factors included the lipase GehB, two immunoglobulin-binding proteins (Spa and Sbi), and the Ser- and Asp-rich surface protein SdrD (Fig. 6).
FIG 6.
Pyruvate-regulated virulence and thiol stress response proteins. Protein abundance for selected virulence and stress protection factors showing pyruvate-dependent expression. Values shown are means and standard deviations from three biological replicates. Significant differences (q ≤ 0.05 [see Table S1 for details]) are indicated.
In addition to proteins related to host-pathogen interactions, we observed medium- and anaerobiosis-dependent expression of the bacilliredoxin BrxB (SACOL1558), which is central to the thiol stress response by reducing S-bacillithiolated proteins (32). We further observed upregulation of SACOL2532, a protein proposed to function as cysteine N-acetyltransferase in bile salt hydrolase (BSH)-dependent detoxification pathways of toxins (33, 34). Interestingly, Hmp, a conserved flavohemoprotein critical for the detoxification of NO˙ to nitrate (35) was significantly less expressed during anaerobiosis in pyruvate medium (Fig. 6).
DISCUSSION
In the absence of a glycolytic carbon source, CcpA regulation fosters the Pfl-Pta-AckA branch of mixed-acid fermentation.
Glucose availability may vary significantly with the specific host site. For instance, secretions produced in the nose, the major S. aureus host niche, contain different amino (10 to 200 μmol each) and organic acids, including pyruvate (70 μmol), but only small amounts of glucose (370 μmol) compared to levels in serum (6 mM) (36). Previous studies have shown that S. aureus can use different amino acids to grow and fuel its metabolism in aerobic environments but that a terminal electron acceptor is required to support anaerobic growth under these conditions (37). Here, we asked if and how fermentation pathways are organized in S. aureus to maintain redox balance in the absence of alternative electron acceptors and a glycolytic carbon source.
We used pyruvate as a model substrate to investigate the anaerobic S. aureus metabolism in the absence of glucose as an abundant glycolytic carbon source. During anaerobic growth on glucose, we measured a lactate-to-acetate ratio of 4:1, confirming that NAD+ is mainly regenerated by the reduction of pyruvate to lactate. When cells were grown on pyruvate, however, the lactate-to-acetate ratio reversed to 1:2, while the total amount of both secreted organic acids remained approximately the same (about 10 mM). This switch is in agreement with a study by Vitko and coworkers, showing that in the absence of glycolysis, ATP production in S. aureus is Pta and AckA dependent, resulting in an increased acetate secretion (38). We extend these observations by showing that differences in the expression of key fermentation enzymes (Ldh1, AdhE, Adh1, and PflAB) accompany the lactate-to-acetate switch, which is critically dependent on CcpA-dependent Pfl expression. This is reminiscent of the situation in Streptococcus pneumoniae where CcpA modulates Pfl expression in response to glucose and galactose availability. As a consequence, lactic acid fermentation is shifted to mixed-acid fermentation in S. pneumoniae, a step critical for full virulence of this bacterium (39). While Pfl abundance increased in pyruvate medium, enzymes that would drain large amounts of acetyl coenzyme A away from energy production, and thereby lead to wasteful synthesis of ethanol and lactate, are downregulated in S. aureus. An estimation of the produced ATP in the pyruvate-fed cells, based on the total amounts of secreted metabolites, suggests a yield of about 6.4 mM ATP (if only acetate is considered), which is about 38% below the theoretical yield in cells cultured in glucose medium (ca. 10.4 mM ATP). In addition, gluconeogenesis (which also requires carbon) provides another considerable energy burden for pyruvate-cultured cells. Approximately 6 mM high-energy phosphoryl groups, which are devoted to anabolism in the glucose culture, would be necessary to synthesize the 1 mM glucose from pyruvate. Consequently, although metabolic activity was still observed in cells cultured with pyruvate, growth was not well supported suggesting that pyruvate mainly serves as an energy source and only small amounts—if any—of pyruvate are diverted to anabolic processes.
In the ccpA mutant, where fermentation pathways can no longer be properly adjusted to the available glucose, we estimated an ATP yield of approximately 9 mM, which is close to the value estimated for the wild type. However, we observed a marked reduction in growth of the ccpA strain compared to the wild type. This illustrates the significance of a precise carbon source-dependent adjustment of anaerobic metabolism in S. aureus to avoid wasteful use of resources and to support growth.
Further mechanisms likely contribute to carbon source-dependent regulation of mixed-acid fermentation.
Although acetate was the major secretion product in pyruvate medium, lactate was still produced, suggesting that the cells still have to cope with excess NADH production under these conditions. Surprisingly, the about 3-fold decrease in total lactate secretion was accompanied by an over 30-fold decrease in the accumulation of the major lactate dehydrogenase, Ldh1, and only minor changes in the amounts of the much lower expressed Ldh2 and LdhD, suggesting that regulation of enzyme activity significantly contributes to the level of lactate production under these conditions. Of note, Garrard and Lascelles reported that in S. aureus, anaerobic lactate dehydrogenase activity was highest in the medium with pyruvate as carbon source, while addition of glucose to the growth medium resulted in a decrease of lactate dehydrogenase activity (40). The precise mechanism, however, of how the enzymatic activity of the lactate dehydrogenases is controlled in S. aureus remains to be elucidated. Furthermore, while the amounts of Ldh1 and Adh1 in cells grown on pyruvate decreased by 30- and 18-fold, respectively, compared to cultivation in glucose, only a 3-fold reduction in the mRNA levels was observed for both enzymes. Thus, posttranscriptional control by yet-to-be-identified factors likely regulates translation efficiencies of ldh1 and adh1 mRNAs or protein stability. Interestingly, we and others observed that ClpL, a poorly characterized HSP100 family AAA+ chaperone (41, 42), is strongly induced during anaerobiosis in glucose medium (8). At present, we cannot exclude that a CcpA-controlled mediator modulates translation of ldh1 or adh1, thereby linking expression of these enzymes to CcpA activity.
We were not able to provide support for a hierarchical derepression of mixed-acid fermentation in response to different intracellular NAD+/NADH ratios imposed by the redox state of the main catabolic carbon source. Although we cannot exclude limitations of the methodology used to determine intracellular NAD+ levels, it is interesting to note that transcription of fermentation genes can persist at high levels for at least 30 min after oxygen depletion (8). How this presumably NADH-dependent transcription is maintained in the presence of high levels of NAD+ remains unclear.
Potential significance during colonization and infection.
We show that under anaerobic conditions and in the absence of a glycolytic carbon source, mixed-acid fermentation pathways in S. aureus are reorganized in a CcpA-dependent manner to foster the Pfl-Pta-AckA pathway, which becomes an important source of ATP in addition to providing C1 building blocks for growth. The fact that S. aureus has evolved the regulatory capacity to control fermentation pathways in response to carbon source availability suggests that such scenarios are relevant for the in vivo situation as was shown for other Gram-positive cocci (43, 44). Although pyruvate, which we used to simulate a situation in which no ATP can be generated from glycolysis, did not support growth during anaerobiosis, it allowed for the maintenance of basic metabolic activities. Thus, an anaerobic metabolism geared toward the Pfl-Pta-AckA branch might not be sufficient for long-term survival if facing a highly active immune response encountered, e.g., during abscess formation, an environment characterized by a decrease in glucose and oxygen during progression of the abscess (45), but could become important during host colonization.
Interestingly, we and others (29) observed that two leucocidins were upregulated in a pyruvate- and anaerobiosis-dependent manner, which might represent a strategy to deal with the often hypoxic or microaerophilic environment associated with increased presence of leucocytes.
It is also interesting to note that the bacilliredoxin BrxB was upregulated during anaerobiosis in pyruvate medium, suggesting a possible oxidative shift of the bacillithiol redox potential and increased protein S-bacillithiolation as previously observed under oxidative stress (46).
Bacilliredoxins are critically involved in the response to hypochlorous acid, which is produced during the oxidative burst of activated neutrophils (47). Consistent with a reduced expression of many Rex-dependent proteins, Hmp, the major NO˙ detoxification enzyme, accumulated at significantly lower levels in pyruvate-grown cells. Thus, anaerobic accumulation of BrxB may represent a response to the lower Hmp levels under these conditions.
Finally, the pyruvate medium-dependent upregulation of SdrD, which specifically promotes adherence of bacteria to nasal epithelial cells (48, 49), suggests that this niche is characterized by low glucose. Indeed, a metabolomic analysis of nasal secretions indicates that amino acids and organic acids, including pyruvate, are prominent carbon sources in the nose (36). Additional surface proteins with a role in immune evasion (Sbi and Spa) were also upregulated in the absence of glucose, although not influenced by oxygen availability. Thus, the absence of high glucose initiates a specific host response in S. aureus, which is further modulated by oxygen depletion. To which extent this response might be directly modulated by a specific carbon source (e.g., pyruvate) or rather results from changes in metabolic flux or intracellular metabolite levels associated with glucose limitation and/or anaerobiosis remains to be determined. The latter possibility is supported by the observation that ArlRS, a central regulator of toxins, responds to changes in activity of the glycolytic pathway (50).
In summary, our data show that S. aureus specifically modulates the expression of the different routes of mixed-acid fermentation in response to available carbon sources to optimize ATP production. The data further highlight the intimate interaction between anaerobic metabolism and expression of host colonization and pathogenicity factors as well as the role of available carbon sources in this process.
MATERIALS AND METHODS
Growth conditions.
S. aureus COL wild-type (WT) and mutant strains were grown at 37°C under vigorous agitation in a chemically defined medium (CDM) according to Liebeke et al. (51) with 7.5 mM glucose or equimolar concentrations of pyruvate (15 mM) as a major carbon source. Overnight cultures in CDM were diluted to an optical density at 500 nm (OD500) of 0.075 in fresh, tempered CDM. To induce hypoxic conditions, at an OD500 of 0.5, an appropriate amount of cell culture was transferred to completely filled 50-ml Falcon tubes, leaving no residual air bubbles in the tubes. The filled Falcon tubes were subjected to static incubation at 37°C (8).
The Staphylococcus aureus COL markerless ΔccpA deletion mutant was constructed by allelic replacement with the help of the temperature‐sensitive shuttle vector pMAD as described previously (52, 53). Primer sequences for the amplification of the pMAD insert to disrupt the ccpA (SACOL_1786) gene were SACOL_1786_pMAD_F1, GCGAATTCGCCTTTAGCTGATTTATATAACGC; SACOL_1786_pMAD_R1, GAATGC CTAATTTTGTGAATAATTTCCTCCTTGTAAACG; SACOL_1786_pMAD_F2, CGTTTACAAGGAGGAAATTATT CACAAAATTAGGCATTC; and SACOL_1786_pMAD_R2, GCGAATTCCCTGATATTATTCTAAGCG.
RNA preparation.
Samples for total RNA isolation were taken at exponential phase (OD500, 0.5; aerobic reference) before and 30 min after the shift to hypoxic conditions (anaerobic sample). Twenty-five OD units of aerobic culture and approximately 30 OD units of anaerobic culture, respectively, were centrifuged for 3 min at 8,500 rpm at 4°C. The supernatant was completely removed, and the tubes containing the pellets were immediately flash frozen in liquid nitrogen and stored at −80°C until the next step. Total RNA was isolated using the acid phenol-chloroform method modified according to Fuchs et al. (8). The samples were extracted twice with acid phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) and twice with chloroform-isoamyl alcohol (24:1, vol/vol); the extraction step with diethyl ether was omitted. RNA was precipitated by adding 98% ethanol and 3 mol sodium acetate solution and following incubation at −20°C overnight. Precipitated RNA was centrifuged for 30 min at 4°C at 15,000 rpm, washed once with 70% ethanol, and finally resuspended in diethyl pyrocarbonate (DEPC) water. The quality of the RNA was ensured by gel electrophoresis.
Analysis of mRNA synthesis.
Digoxigenin (DIG)-labeled RNA probes were obtained by in vitro transcription with T7 RNA polymerase (Roche Diagnostics) and purified PCR fragments as the templates. PCR fragments were prepared using chromosomal DNA of S. aureus COL as a template, and appropriate primers are listed in Table S3 in the supplemental material.
Equal amounts of total RNA (10 μg each, diluted in 10× SSC [1× SSC is 0.15 mol NaCl, 0.015 mM sodium citrate]) were denatured at 65°C for 10 min and then transferred onto a positively charged nylon membrane (Roche Diagnostics), soaked first with aqua bidest and afterward with 20× SSC by slot blotting. After UV cross-linking (120 mJ/cm2, 5 min), the membranes were prehybridized for 1 h in hybridization buffer (5× SSC, 0.1% N-lauroylsarcosine, 7% sodium dodecyl sulfate [SDS], 1% blocking reagent [Roche Diagnostics], and 50% formamide) and hybridized overnight in hybridization buffer with DIG-labeled RNA probes. Membranes were washed twice at room temperature for 5 min with 2× SSC and 0.1% SDS and twice at 68°C for 15 min in 0.2× SSC and 0.1% SDS. After short incubation at room temperature in buffer I (0.1 mol maleine acid, 0.15 mol sodium chloride), the membranes were equilibrated for 30 min in buffer II (1% blocking reagent in buffer I). The following membranes were incubated for 30 min with anti-DIG antibodies coupled with alkaline phosphatase (Roche Diagnostics; diluted 1:10,000 in buffer II) and washed twice for 15 min with buffer I. Afterward, membranes were equilibrated in buffer III (0.1 mol diethanolamine) for 5 min and, finally, exposed for 5 min to CDP-Star (Roche Diagnostic; diluted 1:200 in buffer III). Chemiluminescence signals were detected using a ChemoCam imager (Intas Science Image Instruments GmbH). Experiments were carried out in biological triplicates. Statistical analysis was carried out using the two-sample t tests (two-sided, P = 0.05) implemented in Perseus (54). To account for multiple hypothesis testing, a Benjamini-Hochberg adjustment (false-discovery rate [FDR] q values set to 0.05) was applied. The following comparisons were tested: WT Glc with O2 versus WT Glc without O2, WT Pyr with O2 versus WT Pyr without O2, WT Glc without O2 versus ΔccpA Glc without O2, WT Glc without O2 versus WT Pyr without O2, and WT Pyr without O2 versus ΔccpA Glc without O2.
Sample preparation for global label-free quantification of protein levels.
Samples for analyses of cytoplasmic proteins were taken at the exponential phase (OD500, 0.5; aerobic reference) before and 3 h after the shift to hypoxic conditions (anaerobic sample). We centrifuged 50 ml bacterial cell culture (4°C, 8,500 rpm, 10 min), and cells were washed twice with 1 ml TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5). Cell pellets were resuspended in 1 ml TE buffer and transferred in a 2-ml screw cap microtube filled with 500-μl glass beads (0.10 to 0.11 mm diameter; Sartorius AG). Subsequently, cells were disrupted mechanically using a Precellys 24 homogenizator (PeqLab; 3× 30 s at 6,800 rpm). To separate cell debris and glass beads from the proteins, the suspension was centrifuged twice for 15 min at 4°C at 15,000 rpm. Protein concentration was determined by Roti-Nanoquant protein quantitation assay (Carl Roth, Germany). Afterward, 200 μg protein were digested with trypsin, desalted, and analyzed by mass spectrometry, as described earlier (18), for absolute quantification.
LC-IMSE was done with 3 technical and biological replicates for each time point. Statistical analysis of proteome data is based on calculated absolute protein amounts (femtomoles per picogram) summarized in Table S1. Statistical analysis was carried out in Perseus (54). Following sample normalization (median normalization) and data transformation (log10), two-way analysis of variance (ANOVA) was used to identify significant medium-, aeration-, or medium X aeration-dependent changes. A two-sample t test (two sided), corrected for multiple hypothesis testing (Benjamini-Hochberg), was used to identify conditions leading to significant changes in protein abundance. Nonadjusted (P values) and adjusted (q values) significance levels are reported. Changes in protein abundance of at least 1.5-fold associated with q values of ≤0.05 and were considered biologically relevant. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD017966.
Exometabolome analysis.
Exometabolome samples were taken immediately after inoculation (t0) and along the growth curve after 5, 10, 15, and 24 h of cultivation. At every sampling point, 2 ml cell suspension was filtered on ice using a 0.45-μm-pore-size filter (Sarstedt, Germany). Samples were stored at −20°C.
1H-NMR analysis and quantification of exometabolites were performed as previously described with further modifications (55). The Bruker Avance II 600 NMR spectrometer was operated by TopSpin 3.5 software (Bruker Biospin GmbH, Rheinstetten, Germany). Quantification was done by integration and comparison of designated peaks to the ERETIC signal, which was generated internally with the ERETIC quantification tool based on PULCON (56). The pyruvate concentration was determined using the pyruvate assay kit according to the manufacturer’s instructions (Abnova, Taiwan).
Statistical analysis of exometabolome data was carried out in Perseus using a two-sided two-sample t test corrected for multiple testing (Benjamini-Hochberg). P values for the t test and q value for the FDR correction were set to 0.05.
Measurement of NADH concentrations.
The NAD+ and NADH concentrations were calculated from measurements with a colorimetric assay (NAD+/NADH quantification kit; Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, cells were harvested using the same procedure as for the RNA samples. Lysates were prepared by resuspension of the cell pellet in 400 μl of NADH/NAD+ extraction buffer (provided in the kit). Resuspended cells were transferred to 2 ml cryotubes containing 0.5-ml glass beads (diameter, 0.1 mm; Roth). Cells were disrupted by three cycles in a ribolyser at 6,800 rpm for 30 s. To remove glass beads and cell debris, samples were centrifuged at 21,000 × g for 3 min at 4°C. The supernatant was transferred to new 1.5-ml tubes for a second centrifugation at 21,000 × g for 5 min at 4°C to remove residual aggregates. Finally, the supernatant was filtered through a 10-kDa cutoff filter, as suggested by the assay manufacturer, to remove enzymes which utilize NADH. Further processing of samples followed the protocol of the manufacturer. Samples were incubated for 4 h. Statistical analysis was performed with Microsoft Excel using a t test (two sided) and a significance level of 0.05.
Estimation of ATP yields.
To estimate the ATP yields generated by the cells during anaerobic cultivation, we considered the amounts of secreted fermentation products (acetate and lactate) produced within the first 10 h of anaerobic cultivation, during which the major carbon sources are consumed, as an approximation for the metabolic flow through the different mixed-acid fermentation routes. The lactate secreted in the pyruvate culture was considered solely as a sink for NADH, thus not contributing to ATP production. This assumption neglects the small amounts of ATP potentially produced from amino acid fermentation. Of note, the total amounts of consumed amino acids within 10 h of anaerobiosis were 1.0 mM and 1.6 mM in the pyruvate and glucose cultures, respectively, with serine and threonine consumption contributing to about 50% to these values. ATP requirements for gluconeogenesis were calculated as follows. Within 10 h of fermentation, the glucose culture consumes 6 mM glucose of which about 5 mM are converted to secreted fermentation products, leaving 1 mM for anabolism. The costs for synthesis of 1 mM glucose from pyruvate are 4 ATP, 2 GTP, and 2 NADH (57). Data for the extracellular metabolites are summarized in Table S2.
Identification of cre sites.
The bacterial regulon analyzer Virtual Footprint 3.0 (23) and the regulatory sequence analysis tool RSAT (“genome-scale dna-pattern”) were used to identify potential cre sites within the S. aureus COL genome using the published cre consensus sequence of Miwa and coworkers (WWTGNAARCGNWWWCAWW) with default settings, allowing no mismatches (26).
Supplementary Material
ACKNOWLEDGMENTS
This research received funding from the Deutsche Forschungsgemeinschaft (DFG) within the GRK 1870 (214214857) and the SFB/Transregio 34 (16524344). This research was further funded by the Mecklenburg-Pomerania Excellence Initiative (Germany) and European Social Fund (ESF) Grant KoInfekt (ESF_14-BM-A55-0005_16). Further support was provided by grants from the Deutsche Forschungsgemeinschaft (AN746/4-1 and AN746/4-2) within the DFG priority program SPP1710 to H.A.
We thank Silvia Dittmann and Christian Wolff for excellent technical assistance.
A.T. and J.P.-F. designed the experiments. A.T., V.V.L., K.M., D.Z., J.B., E.M.E., M.L., K.R., G.B., H.A., and J.P.-F. performed the experiments and analyzed the data. J.P.-F. conceived and supervised the study. All authors wrote, reviewed, and approved the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Coleman G, Garbutt IT, Demnitz U. 1983. Ability of a Staphylococcus aureus isolate from a chronic osteomyelitic lesion to survive in the absence of air. Eur J Clin Microbiol 2:595–597. 10.1007/BF02016574. [DOI] [PubMed] [Google Scholar]
- 2.Park MK, Myers RA, Marzella L. 1992. Oxygen tensions and infections: modulation of microbial growth, activity of antimicrobial agents, and immunologic responses. Clin Infect Dis 14:720–740. 10.1093/clinids/14.3.720. [DOI] [PubMed] [Google Scholar]
- 3.Chan PF, Foster SJ. 1998. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology (Reading, Engl) 144:2469–2479. 10.1099/00221287-144-9-2469. [DOI] [PubMed] [Google Scholar]
- 4.Kamps A, Achebach S, Fedtke I, Unden G, Götz F. 2004. Staphylococcal NreB: an O(2)-sensing histidine protein kinase with an O(2)-labile iron-sulphur cluster of the FNR type. Mol Microbiol 52:713–723. 10.1111/j.1365-2958.2004.04024.x. [DOI] [PubMed] [Google Scholar]
- 5.Fedtke I, Kamps A, Krismer B, Götz F. 2002. The nitrate reductase and nitrite reductase operons and the narT gene of Staphylococcus carnosus are positively controlled by the novel two-component system NreBC. J Bacteriol 184:6624–6634. 10.1128/jb.184.23.6624-6634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlag S, Fuchs S, Nerz C, Gaupp R, Engelmann S, Liebeke M, Lalk M, Hecker M, Götz F. 2008. Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J Bacteriol 190:7847–7858. 10.1128/JB.00905-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan M, Yu C, Yang J, Ji Y. 2011. The essential two-component system YhcSR is involved in regulation of the nitrate respiratory pathway of Staphylococcus aureus. J Bacteriol 193:1799–1805. 10.1128/JB.01511-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs S, Pané-Farré J, Kohler C, Hecker M, Engelmann S. 2007. Anaerobic gene expression in Staphylococcus aureus. J Bacteriol 189:4275–4289. 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson AR, Libby SJ, Fang FC. 2008. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319:1672–1676. 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- 10.Schaaff F, Bierbaum G, Baumert N, Bartmann P, Sahl H-G. 2003. Mutations are involved in emergence of aminoglycoside-induced small colony variants of Staphylococcus aureus. Int J Med Microbiol 293:427–435. 10.1078/1438-4221-00282. [DOI] [PubMed] [Google Scholar]
- 11.Dean MA, Olsen RJ, Long SW, Rosato AE, Musser JM. 2014. Identification of point mutations in clinical Staphylococcus aureus strains that produce small-colony variants auxotrophic for menadione. Infect Immun 82:1600–1605. 10.1128/IAI.01487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao S, Huseby DL, Brandis G, Hughes D. 2017. Alternative evolutionary pathways for drug-resistant small colony variant mutants in Staphylococcus aureus. mBio 8:e00358-17. 10.1128/mBio.00358-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pagels M, Fuchs S, Pané-Farré J, Kohler C, Menschner L, Hecker M, McNamarra PJ, Bauer MC, von Wachenfeldt C, Liebeke M, Lalk M, Sander G, von Eiff C, Proctor RA, Engelmann S. 2010. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol Microbiol 76:1142–1161. 10.1111/j.1365-2958.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinkel TL, Roux CM, Dunman PM, Fang FC. 2013. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio 4:e00696-13. 10.1128/mBio.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tiwari N, López-Redondo M, Miguel-Romero L, Kulhankova K, Cahill MP, Tran PM, Kinney KJ, Kilgore SH, Al-Tameemi H, Herfst CA, Tuffs SW, Kirby JR, Boyd JM, McCormick JK, Salgado-Pabón W, Marina A, Schlievert PM, Fuentes EJ. 2020. The SrrAB two-component system regulates Staphylococcus aureus pathogenicity through redox sensitive cysteines. Proc Natl Acad Sci U S A 117:10989–10999. 10.1073/pnas.1921307117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitko NP, Grosser MR, Khatri D, Lance TR, Richardson AR. 2016. Expanded glucose import capability affords Staphylococcus aureus optimized glycolytic flux during infection. mBio 7:e00296-16. 10.1128/mBio.00296-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J-L, Zhang S-K, Chen J-Y, Han B-Z. 2012. Metabolic profiling of Staphylococcus aureus cultivated under aerobic and anaerobic conditions with (1)H NMR-based nontargeted analysis. Can J Microbiol 58:709–718. 10.1139/w2012-046. [DOI] [PubMed] [Google Scholar]
- 18.Zühlke D, Dörries K, Bernhardt J, Maaß S, Muntel J, Liebscher V, Pané-Farré J, Riedel K, Lalk M, Völker U, Engelmann S, Becher D, Fuchs S, Hecker M. 2016. Costs of life - dynamics of the protein inventory of Staphylococcus aureus during anaerobiosis. Sci Rep 6:28172. 10.1038/srep28172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira MT, Manso AS, Gaspar P, Pinho MG, Neves AR. 2013. Effect of oxygen on glucose metabolism: utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLoS One 8:e58277. 10.1371/journal.pone.0058277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall DD, Sadykov MR, Thomas VC, Bayles KW, Powers R. 2016. Redox imbalance underlies the fitness defect associated with inactivation of the Pta-AckA pathway in Staphylococcus aureus. J Proteome Res 15:1205–1212. 10.1021/acs.jproteome.5b01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goel A, Eckhardt TH, Puri P, de Jong A, Branco Dos Santos F, Giera M, Fusetti F, de Vos WM, Kok J, Poolman B, Molenaar D, Kuipers OP, Teusink B. 2015. Protein costs do not explain evolution of metabolic strategies and regulation of ribosomal content: does protein investment explain an anaerobic bacterial Crabtree effect? Mol Microbiol 97:77–92. 10.1111/mmi.13012. [DOI] [PubMed] [Google Scholar]
- 22.Leibig M, Liebeke M, Mader D, Lalk M, Peschel A, Götz F. 2011. Pyruvate formate lyase acts as a formate supplier for metabolic processes during anaerobiosis in Staphylococcus aureus. J Bacteriol 193:952–962. 10.1128/JB.01161-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189. 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen NTT, Contreras-Moreira B, Castro-Mondragon JA, Santana-Garcia W, Ossio R, Robles-Espinoza CD, Bahin M, Collombet S, Vincens P, Thieffry D, van Helden J, Medina-Rivera A, Thomas-Chollier M. 2018. RSAT 2018: regulatory sequence analysis tools 20th anniversary. Nucleic Acids Res 46:W209–W214. 10.1093/nar/gky317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turatsinze J-V, Thomas-Chollier M, Defrance M, van Helden J. 2008. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc 3:1578–1588. 10.1038/nprot.2008.97. [DOI] [PubMed] [Google Scholar]
- 26.Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res 28:1206–1210. 10.1093/nar/28.5.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leonardo MR, Dailly Y, Clark DP. 1996. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol 178:6013–6018. 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alam KY, Clark DP. 1989. Anaerobic fermentation balance of Escherichia coli as observed by in vivo nuclear magnetic resonance spectroscopy. J Bacteriol 171:6213–6217. 10.1128/jb.171.11.6213-6217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harper L, Balasubramanian D, Ohneck EA, Sause WE, Chapman J, Mejia-Sosa B, Lhakhang T, Heguy A, Tsirigos A, Ueberheide B, Boyd JM, Lun DS, Torres VJ. 2018. Staphylococcus aureus responds to the central metabolite pyruvate to regulate virulence. mBio 9:e02272-17. 10.1128/mBio.02272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alonzo F, Torres VJ. 2014. The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol Mol Biol Rev 78:199–230. 10.1128/MMBR.00055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trstenjak N, Milić D, Graewert MA, Rouha h, Svergun D, Djinović-Carugo K, Nagy E, Badarau A. 2020. Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity. Proc Natl Acad Sci U S A 117:317–327. 10.1073/pnas.1913690116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaballa A, Chi BK, Roberts AA, Becher D, Hamilton CJ, Antelmann h, Helmann JD. 2014. Redox regulation in Bacillus subtilis: the bacilliredoxins BrxA(YphP) and BrxB(YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE. Antioxid Redox Signal 21:357–367. 10.1089/ars.2013.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newton GL, Fahey RC, Rawat M. 2012. Detoxification of toxins by bacillithiol in Staphylococcus aureus. Microbiology (Reading) 158:1117–1126. 10.1099/mic.0.055715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cort JR, Ramelot TA, Murray D, Acton TB, Ma L-C, Xiao R, Montelione GT, Kennedy MA. 2008. Structure of an acetyl-CoA binding protein from Staphylococcus aureus representing a novel subfamily of GCN5-related N-acetyltransferase-like proteins. J Struct Funct Genomics 9:7–20. 10.1007/s10969-008-9041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson AR, Somerville GA, Sonenshein AL. 2015. Regulating the intersection of metabolism and pathogenesis in Gram-positive bacteria. Microbiol Spectr 3:10. 10.1128/microbiolspec.MBP-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krismer B, Liebeke M, Janek D, Nega M, Rautenberg M, Hornig G, Unger C, Weidenmaier C, Lalk M, Peschel A. 2014. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog 10:e1003862. 10.1371/journal.ppat.1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halsey CR, Lei S, Wax JK, Lehman MK, Nuxoll AS, Steinke L, Sadykov M, Powers R, Fey PD. 2017. Amino acid catabolism in Staphylococcus aureus and the function of carbon catabolite repression. mBio 8:e01434-16. 10.1128/mBio.01434-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitko NP, Spahich NA, Richardson AR. 2015. Glycolytic dependency of high-level nitric oxide resistance and virulence in Staphylococcus aureus. mBio 6:e00045-15. 10.1128/mBio.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Bayati FAY, Kahya HFH, Damianou A, Shafeeq S, Kuipers OP, Andrew PW, Yesilkaya H. 2017. Pneumococcal galactose catabolism is controlled by multiple regulators acting on pyruvate formate lyase. Sci Rep 7:43587. 10.1038/srep43587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrard W, Lascelles J. 1968. Regulation of Staphylococcus aureus lactate dehydrogenase. J Bacteriol 95:152–156. 10.1128/JB.95.1.152-156.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jana B, Biswas I. 2020. Significance of individual domains of ClpL: a novel chaperone from Streptococcus mutans. Biochemistry 59:3368–3379. 10.1021/acs.biochem.0c00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim G, Lee S-G, Han S, Jung J, Jeong HS, Hyun J-K, Rhee D-K, Kim HM, Lee S. 2020. ClpL is a functionally active tetradecameric AAA+ chaperone, distinct from hexameric/dodecameric ones. FASEB J 34:14353–14370. 10.1096/fj.202000843R. [DOI] [PubMed] [Google Scholar]
- 43.Willenborg J, Goethe R. 2016. Metabolic traits of pathogenic streptococci. FEBS Lett 590:3905–3919. 10.1002/1873-3468.12317. [DOI] [PubMed] [Google Scholar]
- 44.Asanuma N, Yoshii T, Hino T. 2004. Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis. Appl Environ Microbiol 70:5244–5251. 10.1128/AEM.70.9.5244-5251.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thurlow LR, Joshi GS, Richardson AR. 2018. Peroxisome proliferator-activated receptor γ is essential for the resolution of Staphylococcus aureus skin infections. Cell Host Microbe 24:261–270.e4. 10.1016/j.chom.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Imber M, Huyen NTT, Pietrzyk-Brzezinska AJ, van Loi V, Hillion M, Bernhardt J, Thärichen L, Kolšek K, Saleh M, Hamilton CJ, Adrian L, Gräter F, Wahl MC, Antelmann H. 2018. Protein S-bacillithiolation functions in thiol protection and redox regulation of the glyceraldehyde-3-phosphate dehydrogenase gap in Staphylococcus aureus under hypochlorite stress. Antioxid Redox Signal 28:410–430. 10.1089/ars.2016.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beavers WN, Skaar EP. 2016. Neutrophil-generated oxidative stress and protein damage in Staphylococcus aureus. Pathog Dis 74:ftw060. 10.1093/femspd/ftw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corrigan RM, Miajlovic H, Foster TJ. 2009. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol 9:22. 10.1186/1471-2180-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Askarian F, Ajayi C, Hanssen A-M, van Sorge NM, Pettersen I, Diep DB, Sollid JUE, Johannessen M. 2016. The interaction between Staphylococcus aureus SdrD and desmoglein 1 is important for adhesion to host cells. Sci Rep 6:22134. 10.1038/srep22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Párraga Solórzano PK, Yao J, Rock CO, Kehl-Fie TE. 2019. Disruption of glycolysis by nutritional immunity activates a two-component system that coordinates a metabolic and antihost response by Staphylococcus aureus. mBio 10:e01321-19. 10.1128/mBio.01321-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liebeke M, Dörries K, Zühlke D, Bernhardt J, Fuchs S, Pané-Farré J, Engelmann S, Völker U, Bode R, Dandekar T, Lindequist U, Hecker M, Lalk M. 2011. A metabolomics and proteomics study of the adaptation of Staphylococcus aureus to glucose starvation. Mol Biosyst 7:1241–1253. 10.1039/c0mb00315h. [DOI] [PubMed] [Google Scholar]
- 52.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Müller M, Reiß S, Schlüter R, Mäder U, Beyer A, Reiß W, Marles-Wright J, Lewis RJ, Pförtner H, Völker U, Riedel K, Hecker M, Engelmann S, Pané-Farré J. 2014. Deletion of membrane-associated Asp23 leads to upregulation of cell wall stress genes in Staphylococcus aureus. Mol Microbiol 93:1259–1268. 10.1111/mmi.12733. [DOI] [PubMed] [Google Scholar]
- 54.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 55.Dörries K, Lalk M. 2013. Metabolic footprint analysis uncovers strain specific overflow metabolism and D-isoleucine production of Staphylococcus aureus COL and HG001. PLoS One 8:e81500. 10.1371/journal.pone.0081500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wider G, Dreier L. 2006. Measuring protein concentrations by NMR spectroscopy. J Am Chem Soc 128:2571–2576. 10.1021/ja055336t. [DOI] [PubMed] [Google Scholar]
- 57.Berg JM, Tymoczko JL, Stryer L. 2012. Stryer biochemie. Springer Spektrum, Berlin, Heidelberg, Germany. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.