Bacteria commonly live in dense polymicrobial communities and compete for scarce resources. Consequently, they employ a diverse array of mechanisms to harm, inhibit, and kill their competitors. The cell wall is essential for bacterial survival by providing mechanical strength to resist osmotic stress. Because peptidoglycan is the major component of the cell wall and its synthesis is a complex multistep pathway that requires the coordinate action of several enzymes, it provides a target for rival bacteria, which have developed a large arsenal of antibacterial molecules to attack the peptidoglycan of competitors.
KEYWORDS: peptidoglycan, antibiotic, antimicrobial peptide, bacteriocin, effector, interbacterial competition, bacterial warfare, microbial ecology
ABSTRACT
Bacteria commonly live in dense polymicrobial communities and compete for scarce resources. Consequently, they employ a diverse array of mechanisms to harm, inhibit, and kill their competitors. The cell wall is essential for bacterial survival by providing mechanical strength to resist osmotic stress. Because peptidoglycan is the major component of the cell wall and its synthesis is a complex multistep pathway that requires the coordinate action of several enzymes, it provides a target for rival bacteria, which have developed a large arsenal of antibacterial molecules to attack the peptidoglycan of competitors. These molecules include antibiotics, bacteriocins, and contact-dependent effectors that are either secreted into the medium or directly translocated into a target cell. In this minireview, we summarize the diversity of these molecules and highlight distinct mechanisms to disrupt the peptidoglycan, giving special attention to molecules that are known or have the potential to be used during interbacterial competitions.
BACTERIAL WARFARE
Because bacteria live in densely populated polymicrobial communities and compete over limited resources, they deploy a broad arsenal of antibacterial weapons, including both contact-independent and contact-dependent mechanisms (1). Diffusible toxins such as small molecule antibiotics and proteinaceous bacteriocins, ranging in size from peptides to proteins, are secreted into the medium and can target cells at a distance (2–4). Contact-dependent antagonism is mediated by specialized protein secretion systems, including the type I, IV, V, and VI pathways in Gram-negative organisms and the type VII secretion system of Gram-positive bacteria (5–9). Secreted/translocated antibacterial toxins attack components in target cell’s periplasm or cytoplasm, acting as lipases, pore-forming proteins, peptidoglycan hydrolases, nucleases, protein-modifying enzymes, and protein synthesis inhibitors. One could arguably say that attacking the peptidoglycan is one of the most effective ways to render rival cells vulnerable. The location of the peptidoglycan layer makes it more accessible to antagonistic attacks, which do not have to cross the cytoplasmic membrane to exert toxicity. In the following sections, we discuss the vast diversity of antimicrobials that target this conserved structure.
PEPTIDOGLYCAN STRUCTURE, SYNTHESIS, AND DIVERSITY
Peptidoglycan is composed of glycan chains formed by alternating β-(1→4)-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc), which are later cross-linked by short peptide stems (10–12). In Gram-negative bacteria, the peptide stems are usually made of l-alanine (l-Ala1), followed by d-isoglutamic acid (d-iGlu2), meso-diaminopimelic acid (mDAP3), d-alanine (d-Ala4), and d-Ala5. Conversely, in Gram-positive bacteria, the peptide stems often contain d-iso-glutamine (d-iGln) at position 2 and a diamino acid residue at position 3 (13). These peptide stems are cross-linked in two manners: (i) 4→3 cross-link, made between d-Ala4 of a donor pentapeptide and mDAP3 of an acceptor tetrapeptide, and (ii) at a lower frequency, 3→3 cross-link, produced between two mDAP3 residues (10). The cross-link can be either direct (Gram-negative bacteria) or indirect (Gram-positive bacteria), mediated by a short interpeptide bridge (13).
Peptidoglycan synthesis begins in the cytoplasm with the precursors UDP-N-acetylglucosamine (UDP-GlcNAc) and UDP-N-acetylmuramic acid (UDP-MurNAc) (Fig. 1). The enzymes MurA to MurF, alanine racemase (Alr), and d-Ala–d-Ala ligase (DdlA) contribute to the synthesis of UDP-MurNAc-pentapeptide. The enzyme UDP-MurNAc-pentapeptide phosphotransferase (MraY) links UDP-MurNAc-pentapeptide to the lipid transporter undecaprenyl phosphate, forming the intermediate lipid I (undecaprenyl pyrophosphate-MurNAc-pentapeptide) (14). Next, UDP-GlcNAc is coupled by the enzyme MurG to the muramyl moiety of lipid I to form lipid II, which is flipped across the cytoplasmic membrane to the periplasm by the flippase MurJ (15). The precursor lipid II is incorporated into a nascent glycan chain by glycosyltransferases (GTases). The undecaprenyl pyrophosphate is converted by the enzyme undecaprenyl pyrophosphate phosphatase (UppP) to undecaprenyl phosphate, which is recycled to the cytoplasm for the next cycle (16). Transpeptidases (TPases) are responsible for cross-linking the newly polymerized glycan chain via their peptide stems to previously synthesized chains. Transpeptidation can be performed either by d,d-TPases or l,d-TPases, which form 4→3 and 3→3 cross-links, respectively (13).
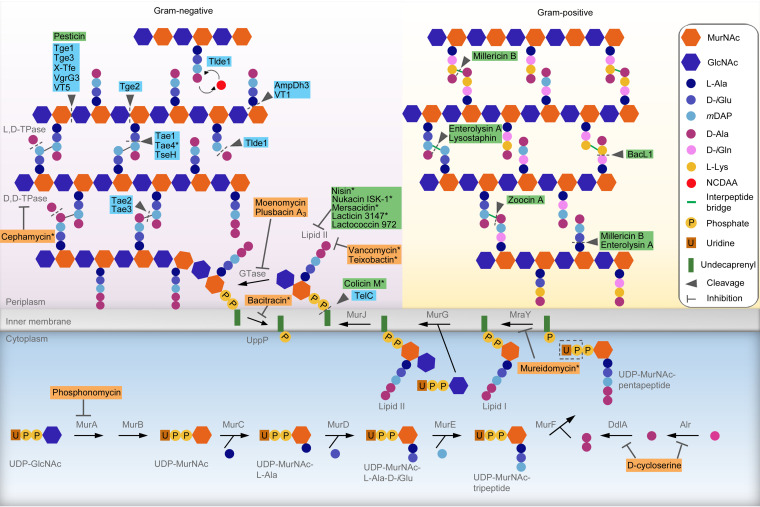
FIG 1.
Antibiotics, bacteriocins, and effectors targeting peptidoglycan synthesis and structure. Peptidoglycan precursors UDP-N-acetylglucosamine (UDP-GlcNAc) and UDP-N-acetylmuramic acid (UDP-MurNAc) are synthesized in the cytoplasm. The enzymes MurA to MurF, Alr, and DdlA are responsible for the synthesis of UDP-MurNAc-pentapeptide, which are linked to the lipid transporter undecaprenyl phosphate, forming the intermediate lipid I. Next, UDP-GlcNAc is coupled by the enzyme MurG to form lipid II, which is flipped across the cytoplasmic membrane by the flippase MurJ. The precursor lipid II is incorporated into a glycan chain by glycosyltransferases (GTases), and the undecaprenyl pyrophosphate is recycled by the enzyme UppP. Transpeptidases (TPases) are responsible for cross-linking peptide stems of the newly polymerized glycan chain to previously synthesized chains. Antibiotics (orange boxes), bacteriocins (green boxes), and contact-dependent effectors (blue boxes) targeting the peptidoglycan either by binding and inhibition or by enzymatic cleavage are indicated. Representative molecules with similar activities are indicated by asterisks, and the complete list is described in Table 1. GlcNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid; MurA, UDP-GlcNAc enolpyruvyl transferase; MurB, UDP-MurNAc dehydrogenase; MurC, UDP-MurNAc-l-Ala ligase; MurD, UDP-MurNAc-l-Ala-d-Glu ligase; MurE, UDP-MurNAc-l-Ala-d-Glu-mesoDAP ligase; MurF, UDP-MurNAc-tripeptide-d-alanyl-d-Ala ligase; Alr, alanine racemase; DdlA, d-Ala–d-Ala ligase A; MraY, UDP-MurNAc-pentapeptide-phosphotransferase; MurG, UDP-GlcNAc-undecaprenyl-pyrophosphoryl-MurNAc-pentapeptide transferase.
The overall peptidoglycan structure is conserved among different species, but there is variation in the glycan chains and peptide stems. In the Gram-positive bacterium Staphylococcus aureus, the glycan chains terminate with a MurNAc or GlcNAc reducing end (17), while in Gram-negative bacteria, the glycan chains terminate with a 1,6-anydroMurNAc (13). In addition, there is variability in the length of the chains, with most Gram-negative bacteria (e.g., Escherichia coli) displaying a mean length of 25 to 35 disaccharide units and Gram-positive bacteria (e.g., Bacillus subtilis) showing an average of 500 disaccharide units. However, some Gram-positive bacteria such as S. aureus have short glycan chains with 3 to 10 disaccharide units (13). Furthermore, modifications of the sugar residues such as N-glycosylation, O-acetylation, and N-deacetylation are present in many Gram-positive bacteria, and O-acetylation in Gram-negative bacteria (13, 18). Regarding the peptide stems, there is variation in the amino acids at positions 2 and 3. In Gram-negative bacteria, d-iGlu at position 2 remains unmodified, but in most Gram-positive bacteria, it can be amidated to d-iGln (19). The most common amino acid at position 3 is mDAP in Gram-negative bacteria and l-Lys in Gram-positive bacteria; however, other amino acids such as l-ornithine, d-Lys, meso-lanthionine, l-homoserine, l-Ala, and l-Glu have been found at this position (10). Such variability in peptidoglycan composition, which changes according to the species and growth conditions, may explain the vast array of molecules developed by bacteria to antagonize different competitors in distinct environments.
ANTIBIOTICS
In 1928, Alexander Fleming accidentally discovered penicillin as he noticed that a fungus, Penicillium notatum, contaminated a plate containing Staphylococcus and created bacterium-free zones. Penicillin is an antibiotic that contains a β-lactam ring in its chemical structure and targets d,d-TPases of the peptidoglycan (20). In polymicrobial communities, fungi, viruses, bacteria, and other unicellular eukaryotes produce molecules that disrupt the peptidoglycan; however, in this section, we will only discuss antibiotics produced by bacteria.
Bacteria produce several classes of antibiotics that target the peptidoglycan: β-lactams, glycopeptides, cyclic peptides and depsipeptides, phosphoglycolipids, peptidyl nucleosides, and phosphonic antibiotics (Fig. 2; Table 1). These antibiotics are mostly non-ribosomally synthesized peptides, which often contain unusual amino acids and are conjugated to carbohydrate and lipid moieties. The structural diversity of these peptides provides distinct mechanisms for inhibition of peptidoglycan synthesis.
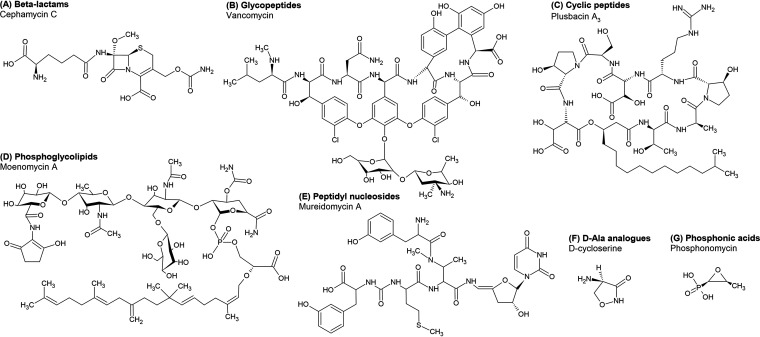
FIG 2.
Chemical structures of representative classes of antibiotics that affect the peptidoglycan. (A) β-Lactams are represented by cephamycin C. (B) Glycopeptides are represented by vancomycin. (C) Cyclic peptides are depicted by plusbacin A3. (D) Phosphoglycolipids are represented by moenomycin A. (E) Peptidyl nucleosides are represented by mureidomycin A. (F) d-Cycloserine is an analogue of d-Ala. (G) Phosphonic acids are represented by phosphonomycin. Chemical structures were drawn using ChemSketch software (Advanced Chemistry Development, Inc.).
TABLE 1.
Antibiotics, bacteriocins, and effectors that target peptidoglycan synthesis and structurea
| Antibiotic(s), bacteriocin(s), or effector(s) (reference[s]) | Class or secretion system | Activity or target |
|---|---|---|
| Antibiotics | ||
| Cephamycin A to C (22); cephabacins (23); nocardicin A (29, 30); monobactams (31–33); thienamycin (24); epithienamycin (25); 1-carbapen-2-em-3-carboxylic acid (26, 27) | β-Lactam | d,d-Transpeptidases |
| Mannopeptimycins α to ε (55) | Glycopeptide | Lipid II |
| Vancomycin (49); balhimycin (50); chloroeremomycin (51); actinoidin A (48); ristocetin A (48); teicoplanin (52); A40926 (53); A47934 (54); complestatin (48) | Glycopeptide | d-Ala–d-Ala of lipid II |
| Amphomycin (70); friulimicin (71) | Cyclic peptide | Undecaprenyl phosphate |
| Bacitracin (74, 75) | Cyclic peptide | Undecaprenyl pyrophosphate |
| Plusbacin A3 (61) | Cyclic peptide | Glycosyltransferases |
| Teixobactin (62); katanosin B (59, 60); enduracidin A and B (63); empedopeptin (66); lysocin E (68); ramoplanins (69) | Cyclic peptide | Lipid II |
| Moenomycins (77) | Phosphoglycolipid | Glycosyltransferases |
| Mureidomycins (79); pacidamycins (80); tunicamycins (81); capuramycins (82); napsamycins (83); liposidomycins (84); muraymycins (85) | Peptidyl nucleoside | MraY |
| d-Cycloserine (89) | Analogue of d-Ala | Alr and DdlA |
| Phosphonomycin (90) | Phosphonic acid | MurA |
| Bacteriocins from Gram-positive bacteria | ||
| Nisin (103); gallidermin (104); epidermin (105); subtilin (106); mutacin B-Ny266 (107); mutacin III/1140 (108, 109); mutacin I (110); streptin (111); ericin A (112); ericin S (112); bovicin HC5 (113); microbisporicin (114); clausin (115); Bsa (116) | Type AI lantibiotic | Lipid II |
| Nukacin ISK-1 (117); lacticin 481 (118); mutacin II (119); variacin (120); salivaricin A2 (121); salivaricin B (121); bovicin HJ50 (122); nukacin IVK45 (123) | Type AII lantibiotic | Lipid II |
| Mersacidin (132); plantaricin C (133); actagardine (134); Ala(0)-actagardine (135) | Type B lantibiotic | Lipid II |
| Lacticin 3147 (137); lichenicidin VK21 (138); Sh-lantibiotic-α/β (139); staphylococcin C55 (140); plantaricin W (141); haloduracin (142) | Type C lantibiotic | Lipid II |
| Lactococcin 972 (143) | Class II | Lipid II |
| Enterolysin A (151) | Class III | Amidase (l-Ala¹ and d-iGlu²;| l-Lys3 and d-Asp interpeptide bridge) |
| BacL1 (153) | Class III | Amidase (d-iGln² and l-Lys3) |
| Lysostaphin (145) | Class III | Amidase (Gly3 and Gly4 interpeptide bridge) |
| Millericin B (150) | Class III | Amidase (l-Ala1 and d-iGlu2; l-Thr and l-Ala interpeptide bridge) |
| Zoocin A (148) | Class III | Amidase (d-Ala4 and l-Ala1 interpeptide bridge) |
| Bacteriocins from Gram-negative bacteria | ||
| Colicin M (156); PaeM (42); PsyM (42); PflM (42) | Lipid II | |
| Pesticin (160) | Muramidase | |
| Contact-dependent effectors | ||
| Tae1 (165, 170); Tae4 (170); TseH (171); Ssp1 (173); Ssp2 (173) | T6SS | Amidase (d-iGlu2 and mDAP3) |
| Tae2 (170); Tae3 (170) | T6SS | Amidase (mDAP3 and d-Ala4| within cross-link) |
| VT1 (168); AmpDh3 (175) | T6SS | Amidase (MurNAc and l-Ala¹) |
| Tge1 (165); Tge3 (167); VT5 (168); VgrG3 (169) | T6SS | Muramidase |
| Tge2 (167) | T6SS | Glucosaminidase |
| Tlde1 (176) | T6SS | l,d-Carboxypeptidase and l,d-transpeptidase |
| X-TfeXAC2609 (5) | T4SS | Muramidase |
| TelC (164) | T7SS | Lipid II |
MurNAc, N-acetylmuramic acid; d-iGlu, d-isoglutamic acid; mDAP, meso-diaminopimelic acid; d-Ala, d-alanine; l-Ala, l-alanine; d-Asp, d-aspartic acid; l-Thr, l-threonine.
β-Lactams are structural analogues of the terminal d-Ala4–d-Ala5 moiety of the pentapeptide stems and act as suicide substrates for d,d-TPases, preventing the formation of 4→3 cross-links (Fig. 1; Table 1) (21). Bacteria synthesize a variety of β-lactams, including cephems (cephamycins and cephabacins), carbapenems, and monocyclic β-lactams, all of them containing the characteristic β-lactam ring. Cephamycins have a methoxyl group in the d-α-aminoadipic acid of the cephem nucleus, which is a β-lactam ring fused to a six-member sulfur-containing dihydrothiazine ring (Fig. 2A). A variety of species of Streptomyces produce cephamycin A and B, and Streptomyces clavuligerus, Streptomyces cattleya, and Nocardia lactamdurans produce cephamycin C (22). Cephabacins display a 7-formylamino group or 7-hydrogen and an oligopeptide as a side chain of the cephem nucleus (23) and are produced by Lysobacter lactamgenus, Xanthomonas lactamgena, and Flavobacterium sp. Carbapenems are characterized by an unsaturated five-membered carbon ring fused to the β-lactam ring. Examples of this group comprise thienamycin (24), epithienamycin (25), and 1-carbapen-2-em-3-carboxylic acid (Car) (26, 27). Car is produced by the phytopathogen Pectobacterium carotovorum under the control of quorum-sensing mechanisms and is associated with growth inhibition of competing species in planta (28). Monocyclic β-lactams, such as nocardicines and monobactams, display only a single β-lactam ring. Nocardicin A has a p-hydroxy-l-phenylglycine unit and is produced by Nocardia uniformis (29) and Actinosynnema mirum (30). Monobactams have a 2-oxoazetine-1-sulfonic acid moiety and are produced by species of Pseudomonas, Gluconobacter, Flexibacter, and Acetobacter and by Chromobacterium violaceum and Agrobacterium radiobacter (31–33).
Both Gram-positive and Gram-negative bacteria rely on an important and ancient defense mechanism against β-lactam antibiotics, the production of β-lactamases (34, 35). Additional resistance mechanisms to β-lactams comprise changes in the active site of d,d-TPases (36, 37), the presence of efflux pumps, changes in membrane permeability to reduce antibiotic uptake (38), and changes in the type of peptidoglycan cross-link from 4→3 to 3→3, which is made by l,d-TPases (39, 40).
Lipid II is a common cell wall component targeted by many classes of antibiotics, as well as bacteriocins and contact-dependent effectors (discussed below) (Fig. 3). Some characteristics of this intermediate may explain why it is prone to attack. (i) The amount of lipid II that can be synthetized is limited due to the small amount of undecaprenyl phosphate present in the cell (∼2 × 105 molecules per cell) (41, 42), which is considered to be the bottleneck for cell wall synthesis (43). (ii) Molecules that attack lipid II do not have to cross the target cell cytoplasmic membrane (44). (iii) The development of resistance to inhibitors that target complex nonprotein intermediates such as lipid II requires alteration of several enzymes in the pathway of peptidoglycan synthesis (45, 46).
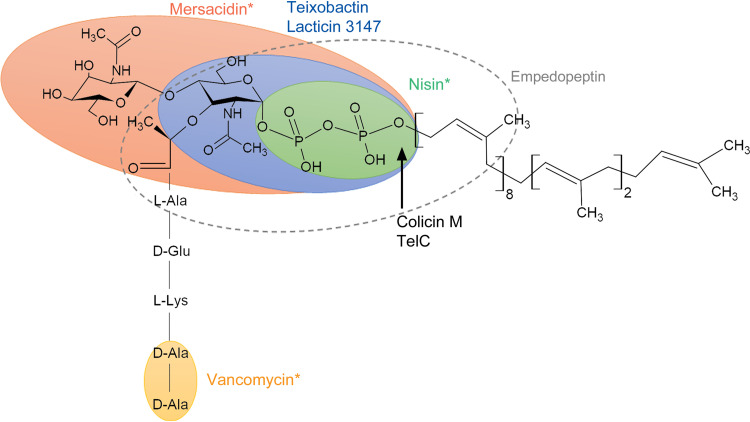
FIG 3.
The peptidoglycan intermediate lipid II is the most common target of antimicrobials. The chemical structure of lipid II was drawn using ChemSketch software (Advanced Chemistry Development, Inc.) and is represented by GlcNAc-MurNAc-pentapeptide linked to undecaprenyl pyrophosphate. The bacteriocin mersacidin, which is representing type B lantibiotics shown in Table 1, binds to GlcNAc-MurNAc and the pyrophosphate (orange ellipse). The cyclic peptide antibiotic teixobactin and the bacteriocin lacticin 3147 (type C lantibiotic) bind to MurNAc and pyrophosphate (blue ellipse). The bacteriocin nisin, which is representing type AI lantibiotics shown in Table 1, binds to the pyrophosphate moiety of lipid II (green ellipse). The cyclic lipodepsipeptide antibiotic empedopeptin binds to the pyrophosphate moiety, MurNAc, and a portion of the peptide stem (dashed ellipse). The binding site of glycopeptide antibiotics, which are represented by vancomycin, is shown by the yellow ellipse. The arrow indicates the cleavage site of the bacteriocin colicin M and the T7SS effector TelC.
Glycopeptides bind to the terminal d-Ala4–d-Ala5 of lipid II (Fig. 3), blocking transglycosylation and transpeptidation by preventing its incorporation into the glycan chain (Fig. 1) (47). Glycopeptides are divided into five structural subtypes (I to V), and representatives of each subtype are vancomycin (I), actinoidin A (II), ristocetin A (III), teicoplanin (IV), and complestatin (V) (48). There are numerous examples of glycopeptides, but their mechanism was proposed to be the same (48). Vancomycin from Amycolatopsis orientalis was the first glycopeptide to be discovered (Fig. 2B) (49). Vancomycin-type glycopeptides, such as balhimycin produced by Amycolatopsis balhimycina (50) and chloroeremomycin by A. orientalis (51), have the same heptapeptide backbone but differ in the glycosylation pattern. Teicoplanin produced by Actinoplanes teichomyceticus (52) is distinguished from vancomycin by the presence of a fatty acid moiety attached to one of the sugars, and it is often called lipoglycopeptide (53). Teicoplanin-type glycopeptides are A40926 from Nonomuraea sp. (53) and A47934 from Streptomyces toyocaensis (54). A novel class of glycopeptides, named mannopeptimycins, produced by Streptomyces hygroscopicus is composed of a cyclic hexapeptide glycosylated with mannose residues (55). Mannopeptimycins α to ε were proposed to interact with lipid II differently from vancomycin, mannopeptimycins bind to the disaccharide unit MurNAc-GlcNAc or the pyrophosphate moiety, while vancomycin binds to the terminal d-Ala4–d-Ala5 of the pentapeptide (55).
Resistance to glycopeptides is attributed to modification of the pentapeptide sequence. In enterococci, there are six types of vancomycin resistance named VanA to VanG (56). The VanA type depends on a dehydrogenase (VanH), which reduces pyruvate to d-lactate, and the VanA ligase, which catalyzes the formation of an ester bond between d-Ala and d-Lac. The resulting d-Ala–d-Lac depsipeptide replaces the d-Ala–d-Ala dipeptide in peptidoglycan synthesis, a substitution that decreases the affinity for glycopeptides. The VanC type of resistance is similar to VanA, but it replaces d-Ala5 for d-serine (d-Ser) (56, 57).
Cyclic peptides and depsipeptides (the last one containing an ester/depside bond as part of their backbone) may contain a fatty acid (cyclic lipodepsipeptides) and a carbohydrate moiety (cyclic glycolipodepsipeptides) (58). Katanosin B (also known as lysobactin) from Cytophaga sp. and Lysobacter sp. is a cyclic depsipeptide of 11 amino acids composed of a d-leucine–d-leucine (d-Leu–d-Leu) dipeptide and nine amino acids forming a macrocycle (59, 60). Katanosin B binds to lipid II, blocking transglycosylation and the following steps of peptidoglycan synthesis (61). Teixobactin is a 11-amino-acid cyclic depsipeptide, produced by the previously uncultured Eleftheria terrae, containing a methylphenylalanine, four d-amino acids, and the unusual amino acid l-allo-enduracididine (62). Teixobactin binds to the pyrophosphate and MurNAc moieties of lipid II (Fig. 1 and 3) (62). Enduracidin A and B are cyclic lipodepsipeptides composed of 17 amino acids linked to a fatty acid and are produced by Streptomyces fungicidicus (63), which were proposed to bind to lipid II and block transglycosylation (Fig. 1) (64). Plusbacin A3 is a cyclic lipodepsipeptide from Pseudomonas sp. composed of a cyclic peptide head of eight amino acids and a lipophilic side chain tail (Fig. 2C) (61); its cyclic peptide head was proposed to insert near the interpeptide cross-link bridge of Gram-positive bacteria (S. aureus), while the tail would displace the glycan chain to impair transglycosylation (Fig. 1) (65). Empedopeptin is a cyclic lipodepsipeptide from Empedobacter haloabium (66) that was shown to bind to lipid II in a Ca2+-dependent manner, the lipid II-interacting region comprises the pyrophosphate moiety, MurNAc, and a portion of the peptide stem (Fig. 3) (67). Another cyclic lipodepsipeptide is lysocin E from Lysobacter sp., which disrupts the bacterial membrane and interacts with lipid II (68). Ramoplanins are a family of cyclic glycolipodepsipeptides, produced by Actinoplanes sp., structurally and functionally related to enduracidins that block transglycosylation upon binding to MurNAc and the pyrophosphate moieties of lipid II (Fig. 3) (69). Amphomycin from Streptomyces canus (70) and friulimicin from Actinoplanes friuliensis (71) are cyclic lipopeptides sharing a peptide core with 10 amino acids but differing in their exocyclic amino acids (asparagine for friulimicin and aspartic acid for amphomycin) and hydrophobic tail; both antibiotics form complexes with undecaprenyl phosphate and prevent its recycling (72, 73). Bacitracin is a cyclic peptide of 12 amino acids produced by B. subtilis and Bacillus licheniformis (74, 75) that binds and sequesters the undecaprenyl pyrophosphate, impairing its conversion to the monophosphate form by UppP and its recycling (Fig. 1) (76).
Moenomycins comprise a family of phosphoglycolipid antibiotics produced by Streptomyces sp. in which a pentasaccharide is linked to a short polycaprenol chain via a phosphoglycerate linkage (Fig. 2D) (77). Moenomycins are analogues of the disaccharide pyrophosphate moiety of lipid II and bind to the active site of glycosyltransferases, blocking transglycosylation (Fig. 1) (78).
In addition to periplasmic targets, there are a few examples of antibiotics that affect precursors of peptidoglycan in the cytoplasm or at the inner face of the cytoplasmic membrane (44), these molecules usually require an specific transport system to reach the cytoplasm. An example are antibiotics known as peptidyl nucleosides, which share a structure containing a 3′-deoxyuridine nucleoside attached to N-methyl-2,3-diaminobutyric acid (Fig. 2E). Mureidomycins (79), pacidamycins (80), tunicamycins (81), capuramycins (82), napsamycins (83), liposidomycins (84), and muraymycins from Streptomyces sp. (85) affect the enzyme MraY by competitive inhibition (Fig. 1) (86, 87). Another example is a cyclic small molecule analogue of d-Ala, named d-cycloserine (Fig. 2F), which acts as competitive inhibitor of the enzymes Alr and DdlA (Fig. 1) (88, 89). The CycA (d-serine/d-alanine/glycine transporter) system is responsible for the d-cycloserine uptake (44). Phosphonomycin (fosfomycin) from Streptomyces sp. belongs to the class of phosphonic acid antibiotics and is a phosphoenolpyruvate analogue (Fig. 2G) that acts on peptidoglycan by inactivating MurA (Fig. 1) (90). Phosphonomycin is actively transported to the cytoplasm by the GlpT (glycerol-3-phosphate) or UhpT (hexose-6-phosphate) transport systems (90). Bacteria that produce phosphonomycin, such as Streptomyces wedmorensis and Streptomyces fradiae, harbor an immunity mechanism involving kinases that inactivate the antibiotic by phosphorylation (91).
Despite the knowledge about the diversity of antibiotics, their mechanism of action, and applications, there is a lack of information about their role in natural settings (92, 93). According to their therapeutic activity, antibiotics have been inferred to act as growth inhibitors of competitors in natural habitats. However, some studies proposed that antibiotics could have a role in communication rather than antibiosis (94, 95). One of the arguments against the antagonistic role of antibiotics is the low concentration of antibiotics found in nature compared to the concentration in therapeutic applications (92, 94). Studies revealed that some antibiotics at a nonlethal concentration may increase the expression of bacterial virulence determinants (94, 96). Further work will be necessary to understand the ecological roles of naturally occurring peptidoglycan-targeting antibiotics.
BACTERIOCINS
Bacteriocin is a broad term used to describe natural peptides and proteins synthesized by bacterial ribosomes that display antimicrobial activity and provide a competitive advantage against closely related bacteria (3). Bacteriocins are made up of molecules of various sizes, structures, and mechanisms and are produced by Gram-negative and Gram-positive bacteria (4).
Bacteriocins from Gram-positive bacteria.
These bacteriocins have self-regulated synthesis. Some have evolved a bacteriocin-specific transport system, whereas others employ the Sec-dependent export pathway (3). Bacteriocins produced by Gram-positive bacteria are either peptides or proteins and are divided into three classes. Class I comprises polycyclic posttranslationally modified peptides called lantibiotics or lanthipeptides. Class II includes small heat-stable minimally modified peptides (<10 kDa), and class III contains larger and heat-labile proteins (>25 kDa) (97).
Lactic acid bacteria produce lantibiotics that contain posttranslational modifications such as dehydration of serine to 2,3-dehydroalanine and dehydration of threonine to 2,3-dehydrobutyrine, which are covalently bound to the sulfur of neighboring cysteines forming lanthionines and/or 3-methyllanthionine rings (98). Lantibiotics have an N-terminal signal peptide for secretion via ATP-binding cassette (ABC) transporters or via Sec-dependent pathway (99). Immunity to lantibiotics is provided by two mechanisms. (i) A specific immunity protein binds to the antimicrobial peptide. (ii) A specialized ABC transporter pumps the peptide out of the cytoplasmic membrane (100, 101). Lantibiotics are divided into three major groups: type A lantibiotics are elongated, flexible, and positively charged; type B lantibiotics are globular peptides; and type C lantibiotics comprise two peptides that act synergistically (102). Type A lantibiotics are further subdivided into type AI, which is modified by the sequential action of two enzymes (LanB and LanC), and type AII, which is modified by a bifunctional enzyme (LanM) (102). Examples of type AI lantibiotics that target the peptidoglycan include nisin (103), gallidermin (104), epidermin (105), subtilin (106), mutacin B-Ny266 (107), mutacin III/1140 (108, 109), mutacin I (110), streptin (111), ericin A and S (112), bovicin HC5 (113), microbisporicin (114), clausin (115), and Bsa (116) (Table 1). Type AII lantibiotics comprise nukacin ISK-1 (117), lacticin 481 (118), mutacin II (119), variacin (120), salivaricin A2 and B (121), bovicin HJ50 (122), and nukacin IVK45 (123) (Table 1).
The food preservative nisin, from Lactococcus lactis, is the most well studied lantibiotic and has a dual mode of action: (i) binding to the pyrophosphate moiety of lipid II and inhibition of peptidoglycan synthesis (Fig. 1 and 3); (ii) binding to lipid II to induce membrane pore formation (124, 125). The conserved N-terminal A/B ring of nisin is the lipid II-binding motif (126), and most type AI lantibiotics that have the A/B ring were proposed to display a similar mechanism (127)—an exception is clausin that might bind to the first amino acids of the pentapeptide stem rather than the pyrophosphate moiety (115). It has been suggested that the binding of nisin to lipid II could explain the low resistance levels detected for nisin compared to antibiotics such as vancomycin. It is unlikely that bacteria would change the highly conserved pyrophosphate configuration or reduce the cellular amount of lipid II (41). Nevertheless, resistance to nisin has been reported and is associated with changes in the cell wall such as incorporation of positive charges, which restrict the access of the positively charged nisin molecule (128).
Experimental evidence pointing to a role of type A lantibiotics in interbacterial antagonism has been reported. A nukacin-related lantibiotic produced by Staphylococcus epidermidis strain IVK45, named nukacin IVK45, was purified from the culture supernatant and coincubated with commensal bacterial species from the nasal microbiota, leading to a decrease in target cell viability (123). Furthermore, the contribution of mutacin I was studied during interbacterial competition between Streptococcus mutans and Streptococcus sanguinis in dental biofilm (129). Mutacin I is similar to epidermin and binds to lipid II to inhibit transglycosylation (127). S. mutans defective in mutacin I production can no longer inhibit the growth of S. sanguinis (129). Bovicin HC5, from Streptococcus bovis, binds to lipid II like nisin, and bovicin producers outcompete sensitive strains (130). S. aureus produces an epidermin-like lantibiotic named Bsa (116). Isolates of methicillin-resistant S. aureus (MRSA) subjected to a biofilm formation assay diversify spontaneously into two distinct sequentially arising strains. The first strain acquires mutations in regulatory genes and hyperactivate a quorum-sensing system, which upregulates the production of surfactants, the bacteriocin Bsa, and its resistance machinery, providing a competitive advantage over the parental strain. After a while, a second strain emerges from the parental strain containing resistance to Bsa after acquiring point mutations in regulatory genes that lead to the thickening of the cell wall, thus reducing access to lipid II (131). Interestingly, this second strain resistant to Bsa also has intermediate resistance to the glycopeptide antibiotic vancomycin (131).
Examples of type B lantibiotics are mersacidin (132), plantaricin C (133), actagardine (or gardimycin) (134), and Ala(0)-actagardine (135) (Table 1). These lantibiotics bind to a different region of lipid II that requires the GlcNAc residue (Fig. 3) (132). Unlike type A lantibiotics, mersacidin acts only by inhibiting peptidoglycan synthesis and does not recruit lipid II to form membrane pores (132, 136). Type C lantibiotics comprise two peptides that act synergistically. The two-component lantibiotic lacticin 3147 from L. lactis is composed of the A1 peptide that binds to lipid II (Fig. 1 and 3) and the A2 peptide that forms membrane pores (137). Similarly, lichenicidin VK21 from B. licheniformis is composed of the peptide Lchα that binds to lipid II and Lchβ that forms membrane pores leading to target cell death (138). The commensal skin bacteria Staphylococcus hominis produces Sh-lantibiotic-α and Sh-lantibiotic-β, which were shown to inhibit S. aureus growth and participate in host defense (139). Staphylococcin C55 (140), plantaricin W (141), and haloduracin (142) are also included in the two-component type C lantibiotic group (Table 1).
Most class II bacteriocins cause membrane pore formation, the only example targeting the peptidoglycan is lactococcin 972, which is produced by L. lactis and binds to lipid II, inhibiting septum formation and cell division (Fig. 1 and Table 1) (143, 144).
Class III bacteriocins are enzymes that degrade the peptidoglycan by cleaving at different sites (Table 1). Lysostaphin from Staphylococcus simulans is a zinc-containing metallopeptidase that cleaves between the third and fourth glycine residues within the cross-link interpeptide bridge of target cells (Fig. 1) (145, 146). Lysostaphin producers have a modified peptidoglycan in which the interpeptide bridge is made of serine instead of glycine to avoid cleavage (147). Zoocin A from Streptococcus zooepidemicus cleaves the peptide stems between d-Ala4 and the first l-Ala of the interpeptide bridge (Fig. 1) (148, 149). Streptococcus milleri produces millericin B that cleaves the peptidoglycan at two sites: (i) between l-Ala1 and d-iGlu2 in the same peptide stem and (ii) between the l-threonine (l-Thr) and l-Ala within the interpeptide bridge (Fig. 1) (150). Enterolysin A from Enterococcus faecalis cleaves between l-Ala1 and d-iGlu2 within the same peptide stem and between l-Lys3 and d-aspartic acid (d-Asp) of the interpeptide bridge of Lactobacillaceae (Fig. 1) (151, 152). Bac41 from E. faecalis is composed of two subunits: BacL1 that cleaves between d-iGln2 and l-Lys3 (Fig. 1) and an accessory factor BacA (153).
Bacteriocins from Gram-negative bacteria.
Colicins from E. coli were the first bacteriocins from Gram-negative bacteria to be discovered. Colicin operons encode the toxic protein followed by an immunity protein, which confers resistance by binding and inactivating the colicin. In some operons, there is also a lysis gene responsible for the release of colicins after lysis of the producer cell under stress conditions (154). Colicins are usually composed by an N-terminal translocation domain, a central receptor-binding domain, and a C-terminal toxic domain (155). Specific outer membrane receptors are required for target cell recognition by colicins, which explains their narrow killing spectrum (155). Translocation to the periplasm or cytoplasm of target cells occurs in a Tol- or TonB-dependent manner (155). Colicin M binds to the ferrichrome-iron receptor FhuA (ferric hydroxamate uptake) for translocation (156, 157) and affects peptidoglycan synthesis by targeting lipid II and cleaving between the lipid moiety and the pyrophosphoryl group (Fig. 1 and 3) (158). Given the low cellular pool of undecaprenyl phosphate, cleavage of lipid II impairs recycling of undecaprenyl phosphate and peptidoglycan synthesis (158). Genes encoding proteins with similarity to the C-terminal domain of colicin M were identified in the genomes of Pseudomonas aeruginosa (PaeM), Pseudomonas syringae (PsyM), Pseudomonas fluorescens (PflM), Burkholderia spp., and P. carotovorum species (159). The three colicin M homologs from Pseudomonas species were expressed as recombinant proteins, and in vitro assays revealed that they cleave lipid II as colicin M (159). Pesticin from Yersinia pestis binds to the ferric yersinia bactin receptor FyuA and is translocated in a TonB-dependent manner (160). Pesticin displays muramidase activity, cleaving between MurNAc and GlcNAc (Fig. 1) (160). The expression of both FhuA and FyuA receptors in E. coli and Y. pestis, respectively, is repressed by the Fur regulator (161, 162), and it was suggested that an increase in sensitivity to bacteriocin-mediated killing could occur in iron-deprived conditions where the expression of these receptors is induced (163).
CONTACT-DEPENDENT ANTIBACTERIAL EFFECTORS
Besides releasing peptides and proteins into the extracellular medium, bacteria also translocate effector proteins directly into target bacterial cells via contact-dependent protein secretion systems. Type I, IV, V, and VI secretion systems (T1SS, T4SS, T5SS, and T6SS) from Gram-negative bacteria and type VII (T7SS) from Gram-positive bacteria are involved in this process (5–9), but effectors targeting the peptidoglycan were described so far for only T4SS, T6SS, and T7SS (Table 1) (5, 164, 165).
The T6SS is a contractile nanomachine evolutionarily related to bacteriophage tails. Effectors are translocated fused to structural proteins such as Hcp (hemolysin coregulated protein), VgrG (valine-glycine repeat protein G), and PAAR (proline-alanine-alanine-arginine) as C-terminal extension domains (specialized effectors) or associated via noncovalent interaction with these proteins (cargo effectors) (166). T6SS effectors targeting the peptidoglycan can act on the glycan backbone (glycoside hydrolases) or within peptide stems and cross-links (amidases). Effectors with glycosidase activity were divided into three families and named Tge1 to Tge3 (type VI secretion glycoside hydrolase effectors) (167). P. aeruginosa carries a gene that encodes the muramidase Tge1 (Tse3) that contains a 70-kDa soluble lytic transglycosylase motif (Slt70) and cleaves between MurNAc and GlcNAc (Fig. 1) (165). Pseudomonas protegens secretes Tge2, a predicted glucosaminidase that cleaves between GlcNAc and MurNAc and confers a competitive advantage against Pseudomonas putida (167). Tge3 was suggested to act as muramidase (Fig. 1) (167). Enterotoxigenic E. coli carries a gene that encodes a predicted muramidase named VT5 (Fig. 1) (168). The specialized effector VgrG-3 from Vibrio cholerae contains a C-terminal domain with muramidase activity (Fig. 1) (169).
T6SS amidase effectors were divided into four families named Tae1 to Tae4 (type VI amidase effectors) (170). Tae1 and Tae4 are gamma-glutamyl-d,l-endopeptidases that cleave between d-iGlu2 and mDAP3 within the same peptide stem, while Tae2 and Tae3 are d,d-endopeptidases that cleave the cross-link bridge between mDAP3 and d-Ala4 (Fig. 1) (170). P. aeruginosa secretes Tae1 (Tse1) and induces death of competitor species (7). TseH from V. cholerae has structural similarity with Tae1 (171, 172). Serratia marcescens carries genes that encode two Tae4 homologs, named Ssp1 and Ssp2 (173). Salmonella enterica serotype Typhimurium carries a gene that encodes Tae4 (170), which was proposed to participate during competition with the gut microbiota (174). In addition, enterotoxigenic E. coli encodes VT1 (TaeX) that cleaves the bond between MurNAc and l-Ala1 (Fig. 1) (168). The zinc protease AmpDh3 from P. aeruginosa is secreted in a T6SS-dependent manner and cleaves between MurNAc and l-Ala1 (Fig. 1), increasing the fitness during competition with E. coli and Yersinia pseudotuberculosis (175). A recently characterized effector, Tlde1 (type VI l,d-transpeptidase effector 1) from S. Typhimurium, is evolutionarily related to l,d-transpeptidases (176). Tlde1 exhibits both l,d-carboxypeptidase activity, cleaving between mDAP3 and d-Ala4 of the acceptor tetrapeptide stem, and l,d-transpeptidase d-amino acid exchange activity, replacing the d-Ala4 by a noncanonical d-amino acid (NCDAA) (Fig. 1) (176).
Immunity to T6SS-dependent killing is conferred by expression of specific immunity proteins that usually bind and inactivate the cognate effector (7, 170). However, immunity gene-independent protection was also shown to provide immunity to T6SS (172). Production of exopolysaccharides during biofilm formation provides defense against T6SS attacks by acting as a physical barrier (177). In addition, modifications in the peptidoglycan confer resistance to T6SS effectors. Acinetobacter baumannii incorporates the NCDAA d-Lys into its peptidoglycan during stationary phase. This activity confers immunity against the amidase Tae1 and increases the survival of A. baumannii during competition with P. aeruginosa, S. marcescens, and Acinetobacter nosocomialis (178). Amidation of mDAP in the peptidoglycan of Gluconobacter frateurii reduces the cleavage efficacy of T6SS effectors that target the d-Ala–mDAP cross-link in vitro, and such modification was proposed to work as a protective mechanism against competitor bacteria (179). In addition, O-acetylation of MurNAc may protect bacteria against glycoside hydrolases (180, 181).
T4SSs are involved in bacterial conjugation and secretion of effector proteins into both eukaryotic and prokaryotic cells (182–184). Antibacterial T4SSs were described for the plant pathogen Xanthomonas citri (5) and the opportunistic bacterium Stenotrophomonas maltophilia (185). Among the T4SS effectors of X. citri, X-TfeXAC2609 (Xanthomonadaceae-T4SS effector) has a lysozyme-like activity and degrades peptidoglycan, conferring a competitive advantage to the attacker (Fig. 1) (5).
The T7SS/Esx system is present in both pathogenic and nonpathogenic Gram-positive bacteria (e.g., Actinobacteria and Firmicutes) (186, 187). This system was first associated with the export of the protein ESAT-6 (early secreted antigen target 6; also called EsxA) (188). The antibacterial function of T7SS was demonstrated in an environmental strain of S. aureus, which kills competing bacteria using a nuclease effector (8). Proteins with an N-terminal LXG domain contain a conserved [LF]XG sequence motif (189) and are often associated with the T7SS/Esx system. TelC (toxin exported by Esx with LXG domain C) is a LXG-containing protein secreted by Streptococcus intermedius, which cleaves lipid II between the lipid moiety and pyrophosphoryl group (Fig. 1 and 3) (164).
DISTRIBUTION AND EVOLUTION OF PEPTIDOGLYCAN-TARGETING MOLECULES
Given the importance of the cell wall for bacterial survival, it is not surprising that peptidoglycan-targeting molecules are widespread across nature. Lysozymes (muramidases) are a good example as they are encoded by phages, bacteria, fungi, plants, and animals (190, 191). Bacteriophages use lysozymes in order to infect host cells and to release the progeny (192, 193). Phagocytes of the immune system such as dendritic cells, neutrophils, and macrophages produce lysozymes that work as a defense mechanism against pathogenic bacteria (190). Environmental amoebae, such as Dictyostelium discoideum, also encode lysozymes that play a role in digestion of phagocytosed bacteria for nutrition (194).
The presence of some lysozyme families (e.g., glycosyl hydrolase 25 [GH25]) in all domains of life indicates the occurrence of horizontal gene transfer events from bacteria to archaea and to eukaryotes (195). A typical example of horizontal gene transfer are β-lactam genes, which are shared between Actinobacteria and the fungi Acremonium chrysogenum (cephalosporin) and Penicillium sp. (penicillin) (196, 197). Production of β-lactams allows fungi to antagonize a myriad of bacteria in the soil. Interestingly, a gene for the β-lactam synthetic pathway was also found in the soil arthropod Folsomia candida (198), for which the most likely donor is a soil-living bacteria or a fungus associated with its diet. The production of β-lactams by this arthropod might be important to control its microbiota (198).
T6SS effectors Tae have also been horizontally transferred to eukaryotes at least six times during evolution resulting in what was called domesticated amidase effectors (Dae) (199). Dae can be found in unicellular protozoans (Naegleria gruberi, Oxytricha trifallax, and Monosiga brevicollis) and in multicellular metazoans (Daphnia sp., Capitella teleta, mollusks, mites, and ticks). It was suggested that Dae2 from the tick Ixodes scapularis acts as an immune factor that can kill S. epidermidis, which is a skin commensal of mammalian hosts, thus protecting the tick from opportunistic infections during the long feeding period (200).
CONCLUDING REMARKS
Bacteria have evolved different warfare mechanisms over millions of years. These strategies range from contact-dependent weapons to diffusible molecules that target cells at a distance (1, 201). Given the great abundance of molecules targeting the peptidoglycan found in nature, it seems evident that carrying a set of these weapons is fundamental for bacterial warfare (Fig. 1 and Table 1). Likewise, the diversity of molecules targeting the peptidoglycan also represents the variety of competitors encountered. As the peptidoglycan structure and composition changes according to the species and environmental conditions, a diverse array of poisonous molecules increases the probability of an effective attack.
The large number of toxic molecules targeting the peptidoglycan, and the number of resistance mechanisms that promote immunity to these molecules, call our attention to the fact that antibiotic resistance is an ancient and naturally occurring phenomenon widespread in the environment. Bacteria encoding β-lactamases and proteins conferring vancomycin resistance precede the modern use of clinical antibiotics (35). In addition, experimental data confirmed that antibiotic resistance can arise solely by competitive interactions between bacteria without previous antibiotic exposure (131). Increased understanding of bacterial immunity mechanisms against natural antimicrobials might help us anticipate the emergence of new resistance mechanisms in clinical settings. Peptidoglycan continues to be the Achilles’ heel of bacteria, and the more we know about molecules targeting this structure and its intrinsic resistance mechanisms, the better equipped we will be to design new antimicrobial strategies and fight infections.
ACKNOWLEDGMENTS
This work was supported by the Sao Paulo Research Foundation (FAPESP) to E.B.-S. (2017/02178-2). FAPESP fellowships were awarded to S.S.-S. (2019/27644-1), J.T.H. (2018/25316-4), and E.B.-S. (2018/04553-8).
REFERENCES
- 1.Peterson SB, Bertolli SK, Mougous JD. 2020. The central role of interbacterial antagonism in bacterial life. Curr Biol 30:R1203–R1214. doi: 10.1016/j.cub.2020.06.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh C. 2003. Antibiotics: actions, origins, resistance. American Society for Microbiology, Washington, DC. [Google Scholar]
- 3.Riley MA, Wertz JE. 2002. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 4.Simons A, Alhanout K, Duval RE. 2020. Bacteriocins, antimicrobial peptides from bacterial origin: overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 8:639. doi: 10.3390/microorganisms8050639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, Hobeika L, Cavalcante NS, Alegria MC, Barbosa LR, Salinas RK, Guzzo CR, Farah CS. 2015. Bacterial killing via a type IV secretion system. Nat Commun 6:6453. doi: 10.1038/ncomms7453. [DOI] [PubMed] [Google Scholar]
- 6.Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 7.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T. 2016. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol 2:16183. doi: 10.1038/nmicrobiol.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Bayona L, Guo MS, Laub MT. 2017. Contact-dependent killing by Caulobacter crescentus via cell surface-associated, glycine zipper proteins. Elife 6:e24869. doi: 10.7554/eLife.24869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schleifer KH, Kandler O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477. doi: 10.1128/BR.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vollmer W, Joris B, Charlier P, Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 12.Vollmer W, Blanot D, De Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 13.Vollmer W, Born P. 2009. Bacterial cell envelope peptidoglycan, p 15–28. In Moran A, Brennan P, Holst O, von Itszstein M (ed), Microbial glycobiology: structures, relevance and applications. Elsevier, London, United Kingdom. [Google Scholar]
- 14.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sham L-T, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N. 2014. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Ghachi M, Bouhss A, Blanot D, Mengin-Lecreulx D. 2004. The bacA gene of Escherichia coli encodes an undecaprenyl pyrophosphate phosphatase activity. J Biol Chem 279:30106–30113. doi: 10.1074/jbc.M401701200. [DOI] [PubMed] [Google Scholar]
- 17.Boneca IG, Huang Z-H, Gage DA, Tomasz A. 2000. Characterization of Staphylococcus aureus cell wall glycan strands, evidence for a new β-N-acetylglucosaminidase activity. J Biol Chem 275:9910–9918. doi: 10.1074/jbc.275.14.9910. [DOI] [PubMed] [Google Scholar]
- 18.Bernard E, Rolain T, Courtin P, Guillot A, Langella P, Hols P, Chapot-Chartier MP. 2011. Characterization of O-acetylation of N-acetylglucosamine: a novel structural variation of bacterial peptidoglycan. J Biol Chem 286:23950–23958. doi: 10.1074/jbc.M111.241414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siewert G, Strominger JL. 1968. Biosynthesis of the peptidoglycan of bacterial cell walls. XI. Formation of the isoglutamine amide group in the cell walls of Staphylococcus aureus. J Biol Chem 243:783–790. doi: 10.1016/S0021-9258(19)81734-9. [DOI] [PubMed] [Google Scholar]
- 20.Kelly JA, Moews PC, Knox JR, Frère JM, Ghuysen JM. 1982. Penicillin target enzyme and the antibiotic binding site. Science 218:479–481. doi: 10.1126/science.7123246. [DOI] [PubMed] [Google Scholar]
- 21.Tipper DJ, Strominger JL. 1965. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci U S A 54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stapley E, Jackson M, Hernandez S, Zimmerman S, Currie S, Mochales S, Mata J, Woodruff H, Hendlin D. 1972. Cephamycins, a new family of β-lactam antibiotics I. Production by Actinomycetes, including Streptomyces lactamdurans sp. n. Antimicrob Agents Chemother 2:122–131. doi: 10.1128/aac.2.3.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada S, Tsubotani S, Ono H, Okazaki H. 1984. Cephabacins, new cephem antibiotics of bacterial origin. II. Isolation and structural elucidation. J Antibiot 37:1536–1545. doi: 10.7164/antibiotics.37.1536. [DOI] [PubMed] [Google Scholar]
- 24.Williamson JM, Inamine E, Wilson KE, Douglas AW, Liesch JM, Albers-Schönberg G. 1985. Biosynthesis of the beta-lactam antibiotic, thienamycin, by Streptomyces cattleya. J Biol Chem 260:4637–4647. doi: 10.1016/S0021-9258(18)89118-9. [DOI] [PubMed] [Google Scholar]
- 25.Stapley EO, Cassidy PJ, Tunac J, Monaghan RL, Jackson M, Hernandez S, Zimmerman SB, Mata JM, Currie SA, Daoust D, Hendlin D. 1981. Epithienamycins–novel β-lactams related to thienamycin. J Antibiot (Tokyo) 34:628–636. doi: 10.7164/antibiotics.34.628. [DOI] [PubMed] [Google Scholar]
- 26.Derzelle S, Duchaud E, Kunst F, Danchin A, Bertin P. 2002. Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Appl Environ Microbiol 68:3780–3789. doi: 10.1128/aem.68.8.3780-3789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker WL, Rathnum ML, Wells JS, Trejo WH, Principe PA, Sykes RB. 1982. SQ 27, 860, a simple carbapenem produced by species of Serratia and Erwinia. J Antibiot (Tokyo) 35:653–660. doi: 10.7164/antibiotics.35.653. [DOI] [PubMed] [Google Scholar]
- 28.Shyntum DY, Nkomo NP, Shingange NL, Gricia AR, Bellieny-Rabelo D, Moleleki LN. 2019. The impact of type VI secretion system, bacteriocins and antibiotics on bacterial competition of Pectobacterium carotovorum subsp. brasiliense and the regulation of carbapenem biosynthesis by iron and the ferric-uptake regulator. Front Microbiol 10:2379. doi: 10.3389/fmicb.2019.02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aoki H, Sakai H-I, Kohsaka M, Konomi T, Hosoda J, Kubochi Y, Iguchi E, Imanaka H. 1976. Nocardicin A, a new monocyclic β-lactam antibiotic. I. Discovery, isolation and characterization. J Antibiot (Tokyo) 29:492–500. doi: 10.7164/antibiotics.29.492. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe K, Okuda T, Yokose K, Furumai T, Maruyama HB. 1983. Actinosynnema mirum, a new producer of nocardicin antibiotics. J Antibiot (Tokyo) 36:321–324. doi: 10.7164/antibiotics.36.321. [DOI] [PubMed] [Google Scholar]
- 31.Imada A, Kitano K, Kintaka K, Muroi M, Asai M. 1981. Sulfazecin and isosulfazecin, novel β-lactam antibiotics of bacterial origin. Nature 289:590–591. doi: 10.1038/289590a0. [DOI] [PubMed] [Google Scholar]
- 32.Singh PD, Johnson JH, Ward PC, Wells JS, Trejo WH, Sykes RB. 1983. SQ 28, 332, a new monobactam produced by a Flexibacter sp. J Antibiot (Tokyo) 36:1245–1251. doi: 10.7164/antibiotics.36.1245. [DOI] [PubMed] [Google Scholar]
- 33.Sykes R, Bonner D, Bush K, Georgopapadakou N, Wells J. 1981. Monobactams—monocyclic β-lactam antibiotics produced by bacteria. J Antimicrob Chemother 8(Suppl E):1–16. doi: 10.1093/jac/8.suppl_E.1. [DOI] [PubMed] [Google Scholar]
- 34.Ogawara H. 1975. Production and property of beta-lactamases in Streptomyces. Antimicrob Agents Chemother 8:402–408. doi: 10.1128/aac.8.4.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. 2011. Antibiotic resistance is ancient. Nature 477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 36.Rybkine T, Mainardi JL, Sougakoff W, Collatz E, Gutmann L. 1998. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of β-lactam resistance. J Infect Dis 178:159–163. doi: 10.1086/515605. [DOI] [PubMed] [Google Scholar]
- 37.Contreras-Martel C, Dahout-Gonzalez C, Martins ADS, Kotnik M, Dessen A. 2009. PBP active site flexibility as the key mechanism for beta-lactam resistance in pneumococci. J Mol Biol 387:899–909. doi: 10.1016/j.jmb.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 38.Rumbo C, Gato E, Lopez M, Ruiz de Alegria C, Fernandez-Cuenca F, Martinez-Martinez L, Vila J, Pachon J, Cisneros JM, Rodriguez-Bano J, Pascual A, Bou G, Tomas M, Spanish Group of Nosocomial Infections and Mechanisms of Action and Resistance to Antimicrobials (GEIH-GEMARA), Spanish Society of Clinical Microbiology and Infectious Diseases (SEIMC), Spanish Network for Research in Infectious Diseases (REIPI). 2013. Contribution of efflux pumps, porins, and beta-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 57:5247–5257. doi: 10.1128/AAC.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mainardi JL, Fourgeaud M, Hugonnet JE, Dubost L, Brouard JP, Ouazzani J, Rice LB, Gutmann L, Arthur M. 2005. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J Biol Chem 280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- 40.Hugonnet JE, Mengin-Lecreulx D, Monton A, den Blaauwen T, Carbonnelle E, Veckerle C, Brun YV, van Nieuwenhze M, Bouchier C, Tu K, Rice LB, Arthur M. 2016. Factors essential for l,d-transpeptidase-mediated peptidoglycan cross-linking and beta-lactam resistance in Escherichia coli. Elife 5:e19469. doi: 10.7554/eLife.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kramer NE, Smid EJ, Kok J, de Kruijff B, Kuipers OP, Breukink E. 2004. Resistance of Gram-positive bacteria to nisin is not determined by lipid II levels. FEMS Microbiol Lett 239:157–161. doi: 10.1016/j.femsle.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 42.Barreteau H, Magnet S, El Ghachi M, Touze T, Arthur M, Mengin-Lecreulx D, Blanot D. 2009. Quantitative high-performance liquid chromatography analysis of the pool levels of undecaprenyl phosphate and its derivatives in bacterial membranes. J Chromatogr B Analyt Technol Biomed Life Sci 877:213–220. doi: 10.1016/j.jchromb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Breukink E, de Kruijff B. 2006. Lipid II as a target for antibiotics. Nat Rev Drug Discov 5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 44.Silver LL. 2013. Viable screening targets related to the bacterial cell wall. Ann N Y Acad Sci 1277:29–53. doi: 10.1111/nyas.12006. [DOI] [PubMed] [Google Scholar]
- 45.Silver LL. 2007. Multi-targeting by monotherapeutic antibacterials. Nat Rev Drug Discov 6:41–55. doi: 10.1038/nrd2202. [DOI] [PubMed] [Google Scholar]
- 46.Silver LL. 2011. Challenges of antibacterial discovery. Clin Microbiol Rev 24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarkar P, Yarlagadda V, Ghosh C, Haldar J. 2017. A review on cell wall synthesis inhibitors with an emphasis on glycopeptide antibiotics. Medchemcomm 8:516–533. doi: 10.1039/c6md00585c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicolaou K, Boddy CN, Bräse S, Winssinger N. 1999. Chemistry, biology, and medicine of the glycopeptide antibiotics. Angew Chem Int Ed 38:2096–2152. doi:. [DOI] [PubMed] [Google Scholar]
- 49.McCormick MH. 1956. Vancomycin, a new antibiotic. I. Chemical and biologic properties. Antibiot Annu 3:606–611. [PubMed] [Google Scholar]
- 50.Nadkarni SR, Patel MV, Chatterjee S, Vijayakumar EK, Desikan KR, Blumbach J, Ganguli BN, Limbert M. 1994. Balhimycin, a new glycopeptide antibiotic produced by Amycolatopsis sp. Y-86, 21022. J Antibiot (Tokyo) 47:334–341. doi: 10.7164/antibiotics.47.334. [DOI] [PubMed] [Google Scholar]
- 51.van Wageningen AM, Kirkpatrick PN, Williams DH, Harris BR, Kershaw JK, Lennard NJ, Jones M, Jones SJ, Solenberg PJ. 1998. Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem Biol 5:155–162. doi: 10.1016/s1074-5521(98)90060-6. [DOI] [PubMed] [Google Scholar]
- 52.Parenti F, Beretta G, Berti M, Arioli V. 1978. Teichomycins, new antibiotics from Actinoplanes teichomyceticus nov. sp. I. Description of the producer strain, fermentation studies and biological properties. J Antibiot 31:276–283. doi: 10.7164/antibiotics.31.276. [DOI] [PubMed] [Google Scholar]
- 53.Goldstein BP, Selva E, Gastaldo L, Berti M, Pallanza R, Ripamonti F, Ferrari P, Denaro M, Arioli V, Cassani G. 1987. A40926, a new glycopeptide antibiotic with anti-Neisseria activity. Antimicrob Agents Chemother 31:1961–1966. doi: 10.1128/aac.31.12.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boeck LD, Mertz FP. 1986. A47934, a novel glycopeptide-aglycone antibiotic produced by a strain of Streptomyces toyocaensis taxonomy and fermentation studies. J Antibiot (Tokyo) 39:1533–1540. doi: 10.7164/antibiotics.39.1533. [DOI] [PubMed] [Google Scholar]
- 55.Ruzin A, Singh G, Severin A, Yang Y, Dushin RG, Sutherland AG, Minnick A, Greenstein M, May MK, Shlaes DM, Bradford PA. 2004. Mechanism of action of the mannopeptimycins, a novel class of glycopeptide antibiotics active against vancomycin-resistant Gram-positive bacteria. Antimicrob Agents Chemother 48:728–738. doi: 10.1128/aac.48.3.728-738.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Courvalin P. 2006. Vancomycin resistance in gram-positive cocci. Clin Infect Dis 42:S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 57.Marshall C, Lessard I, Park I-S, Wright G. 1998. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob Agents Chemother 42:2215–2220. doi: 10.1128/AAC.42.9.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davies JS. 2003. The cyclization of peptides and depsipeptides. J Pept Sci 9:471–501. doi: 10.1002/psc.491. [DOI] [PubMed] [Google Scholar]
- 59.O’Sullivan J, McCullough JE, Tymiak AA, Kirsch DR, Trejo WH, Principe PA. 1988. Lysobactin, a novel antibacterial agent produced by Lysobacter sp. I. Taxonomy, isolation and partial characterization. J Antibiot (Tokyo) 41:1740–1744. doi: 10.7164/antibiotics.41.1740. [DOI] [PubMed] [Google Scholar]
- 60.Shoji J, Hinoo H, Matsumoto K, Hattori T, Yoshida T, Matsuura S, Kondo E. 1988. Isolation and characterization of katanosins A and B. J Antibiot (Tokyo) 41:713–718. doi: 10.7164/antibiotics.41.713. [DOI] [PubMed] [Google Scholar]
- 61.Maki H, Miura K, Yamano Y. 2001. Katanosin B and plusbacin A(3), inhibitors of peptidoglycan synthesis in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 45:1823–1827. doi: 10.1128/AAC.45.6.1823-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higashide E, Hatano K, Shibata M, Nakazawa K. 1968. Enduracidin, a new antibiotic. I Streptomyces fungicidicus NO B.5477, an enduracidin producing organism. J Antibiot (Tokyo) 21:126–137. doi: 10.7164/antibiotics.21.126. [DOI] [PubMed] [Google Scholar]
- 64.Fang X, Tiyanont K, Zhang Y, Wanner J, Boger D, Walker S. 2006. The mechanism of action of ramoplanin and enduracidin. Mol Biosyst 2:69–76. doi: 10.1039/b515328j. [DOI] [PubMed] [Google Scholar]
- 65.O’Connor RD, Singh M, Chang J, Kim SJ, VanNieuwenhze M, Schaefer J. 2017. Dual mode of action for Plusbacin A3 in Staphylococcus aureus. J Phys Chem B 121:1499–1505. doi: 10.1021/acs.jpcb.6b11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Konishi M, Sugawara K, Hanada M, Tomita K, Tomatsu K, Miyaki T, Kawaguchi H, Buck RE, More C, Rossomano VZ. 1984. Empedopeptin (Bmy-28117), a new depsipeptide antibiotic. J Antibiot (Tokyo) 37:949–957. doi: 10.7164/antibiotics.37.949. [DOI] [PubMed] [Google Scholar]
- 67.Muller A, Munch D, Schmidt Y, Reder-Christ K, Schiffer G, Bendas G, Gross H, Sahl HG, Schneider T, Brotz-Oesterhelt H. 2012. Lipodepsipeptide empedopeptin inhibits cell wall biosynthesis through Ca2+-dependent complex formation with peptidoglycan precursors. J Biol Chem 287:20270–20280. doi: 10.1074/jbc.M112.369561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santiago M, Lee W, Fayad AA, Coe KA, Rajagopal M, Do T, Hennessen F, Srisuknimit V, Muller R, Meredith TC, Walker S. 2018. Genome-wide mutant profiling predicts the mechanism of a lipid II binding antibiotic. Nat Chem Biol 14:601–608. doi: 10.1038/s41589-018-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamburger JB, Hoertz AJ, Lee A, Senturia RJ, McCafferty DG, Loll PJ. 2009. A crystal structure of a dimer of the antibiotic ramoplanin illustrates membrane positioning and a potential lipid II docking interface. Proc Natl Acad Sci U S A 106:13759–13764. doi: 10.1073/pnas.0904686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heinemann B, Kaplan M, Muir R, Hooper I. 1953. Amphomycin, a new antibiotic. Antibiot Chemother (Northfield) 3:1239–1242. [PubMed] [Google Scholar]
- 71.Aretz W, Meiwes J, Seibert G, Vobis G, Wink J. 2000. Friulimicins: novel lipopeptide antibiotics with peptidoglycan synthesis inhibiting activity from Actinoplanes friuliensis sp. nov. J Antibiot (Tokyo) 53:807–815. doi: 10.7164/antibiotics.53.807. [DOI] [PubMed] [Google Scholar]
- 72.Rubinchik E, Schneider T, Elliott M, Scott WR, Pan J, Anklin C, Yang H, Dugourd D, Muller A, Gries K, Straus SK, Sahl HG, Hancock RE. 2011. Mechanism of action and limited cross-resistance of new lipopeptide MX-2401. Antimicrob Agents Chemother 55:2743–2754. doi: 10.1128/AAC.00170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schneider T, Gries K, Josten M, Wiedemann I, Pelzer S, Labischinski H, Sahl HG. 2009. The lipopeptide antibiotic friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob Agents Chemother 53:1610–1618. doi: 10.1128/AAC.01040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnson BA, Anker H, Meleney FL. 1945. Bacitracin: a new antibiotic produced by a member of the B. subtilis group. Science 102:376–377. doi: 10.1126/science.102.2650.376. [DOI] [PubMed] [Google Scholar]
- 75.Bernlohr RW, Novelli G. 1960. Some characteristics of bacitracin production by Bacillus licheniformis. Arch Biochem Biophys 87:232–238. doi: 10.1016/0003-9861(60)90166-1. [DOI] [Google Scholar]
- 76.Stone KJ, Strominger JL. 1971. Mechanism of action of bacitracin: complexation with metal ion and C55-isoprenyl pyrophosphate. Proc Natl Acad Sci U S A 68:3223–3227. doi: 10.1073/pnas.68.12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huber G, Nesemann G. 1968. Moenomycin, an inhibitor of cell wall synthesis. Biochem Biophys Res Commun 30:7–13. doi: 10.1016/0006-291x(68)90704-3. [DOI] [PubMed] [Google Scholar]
- 78.Lovering AL, de Castro LH, Lim D, Strynadka NCJ. 2007. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science 315:1402–1405. doi: 10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]
- 79.Isono F, Inukai M. 1991. Mureidomycin A, a new inhibitor of bacterial peptidoglycan synthesis. Antimicrob Agents Chemother 35:234–236. doi: 10.1128/aac.35.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen RH, Buko AM, Whittern DN, McAlpine JB. 1989. Pacidamycins, a novel series of antibiotics with anti-Pseudomonas aeruginosa activity II. Isolation and structural elucidation. J Antibiot (Tokyo) 42:512–520. doi: 10.7164/antibiotics.42.512. [DOI] [PubMed] [Google Scholar]
- 81.Tamura G, Sasaki T, Matsuhashi M, Takatsuki A, Yamasaki M. 1976. Tunicamycin inhibits the formation of lipid intermediate in cell-free peptidoglycan synthesis of bacteria. Agric Biol Chem 40:447–449. doi: 10.1080/00021369.1976.10862071. [DOI] [Google Scholar]
- 82.Hotoda H, Furukawa M, Daigo M, Murayama K, Kaneko M, Muramatsu Y, Ishii MM, Miyakoshi S-I, Takatsu T, Inukai M, Kakuta M, Abe T, Harasaki T, Fukuoka T, Utsui Y, Ohya S. 2003. Synthesis and antimycobacterial activity of capuramycin analogues. Part 1: substitution of the azepan-2-one moiety of capuramycin. Bioorg Med Chem Lett 13:2829–2832. doi: 10.1016/S0960-894X(03)00596-1. [DOI] [PubMed] [Google Scholar]
- 83.Chatterjee S, Nadkarni S, Vijayakumar E, Patel M, Gangul B, Fehlhaber H-W, Vértesy L. 1994. Napsamycins, new Pseudomonas active antibiotics of the mureidomycin family from Streptomyces sp. HIL Y-82, 11372. J Antibiot (Tokyo) 47:595–598. doi: 10.7164/antibiotics.47.595. [DOI] [PubMed] [Google Scholar]
- 84.Muroi M, Kimura K-I, Osada H, Inukai M, Takatsuki A. 1997. Liposidomycin B inhibits in vitro formation of polyprenyl (pyro) phosphate N-acetylglucosamine, an intermediate in glycoconjugate biosynthesis. J Antibiot (Tokyo) 50:103–104. doi: 10.7164/antibiotics.50.103. [DOI] [PubMed] [Google Scholar]
- 85.McDonald LA, Barbieri LR, Carter GT, Lenoy E, Lotvin J, Petersen PJ, Siegel MM, Singh G, Williamson RT. 2002. Structures of the muraymycins, novel peptidoglycan biosynthesis inhibitors. J Am Chem Soc 124:10260–10261. doi: 10.1021/ja017748h. [DOI] [PubMed] [Google Scholar]
- 86.Brandish PE, Kimura K-I, Inukai M, Southgate R, Lonsdale JT, Bugg T. 1996. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob Agents Chemother 40:1640–1644. doi: 10.1128/AAC.40.7.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winn M, Goss RJ, Kimura K, Bugg TD. 2010. Antimicrobial nucleoside antibiotics targeting cell wall assembly: recent advances in structure-function studies and nucleoside biosynthesis. Nat Prod Rep 27:279–304. doi: 10.1039/b816215h. [DOI] [PubMed] [Google Scholar]
- 88.Lambert MP, Neuhaus FC. 1972. Mechanism of D-cycloserine action: alanine racemase from Escherichia coli W. J Bacteriol 110:978–987. doi: 10.1128/JB.110.3.978-987.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Neuhaus FC, Lynch JL. 1964. The enzymatic synthesis of D-alanyl-D-alanine. III. On the inhibition of D-alanyl-D-alanine synthetase by the antibiotic D-cycloserine. Biochemistry 3:471–480. doi: 10.1021/bi00892a001. [DOI] [PubMed] [Google Scholar]
- 90.Kahan FM, Kahan JS, Cassidy PJ, Kropp H. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann N Y Acad Sci 235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 91.Castañeda-García A, Blázquez J, Rodríguez-Rojas A. 2013. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics (Basel) 2:217–236. doi: 10.3390/antibiotics2020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davies J. 2006. Are antibiotics naturally antibiotics? J Ind Microbiol Biotechnol 33:496–499. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 93.Aminov RI. 2009. The role of antibiotics and antibiotic resistance in nature. Environ Microbiol 11:2970–2988. doi: 10.1111/j.1462-2920.2009.01972.x. [DOI] [PubMed] [Google Scholar]
- 94.Linares JF, Gustafsson I, Baquero F, Martinez J. 2006. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci U S A 103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fajardo A, Martinez JL. 2008. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol 11:161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 96.Herold S, Siebert J, Huber A, Schmidt H. 2005. Global expression of prophage genes in Escherichia coli O157:H7 strain EDL933 in response to norfloxacin. Antimicrob Agents Chemother 49:931–944. doi: 10.1128/AAC.49.3.931-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alvarez-Sieiro P, Montalban-Lopez M, Mu D, Kuipers OP. 2016. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100:2939–2951. doi: 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nes IF, Diep DB, Holo H. 2007. Bacteriocin diversity in Streptococcus and Enterococcus. J Bacteriol 189:1189–1198. doi: 10.1128/JB.01254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zheng S, Sonomoto K. 2018. Diversified transporters and pathways for bacteriocin secretion in gram-positive bacteria. Appl Microbiol Biotechnol 102:4243–4253. doi: 10.1007/s00253-018-8917-5. [DOI] [PubMed] [Google Scholar]
- 100.Draper LA, Ross RP, Hill C, Cotter PD. 2008. Lantibiotic immunity. Curr Protein Pept Sci 9:39–49. doi: 10.2174/138920308783565750. [DOI] [PubMed] [Google Scholar]
- 101.Geiger C, Korn SM, Häsler M, Peetz O, Martin J, Kötter P, Morgner N, Entian K-D. 2019. LanI-mediated lantibiotic immunity in Bacillus subtilis: functional analysis. Appl Environ Microbiol 85:e00534-19. doi: 10.1128/AEM.00534-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barbour A, Wescombe P, Smith L. 2020. Evolution of lantibiotic salivaricins: new weapons to fight infectious diseases. Trends Microbiol 28:578–593. doi: 10.1016/j.tim.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 103.Linnett PE, Strominger JL. 1973. Additional antibiotic inhibitors of peptidoglycan synthesis. Antimicrob Agents Chemother 4:231–236. doi: 10.1128/aac.4.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kellner R, Jung G, Horner T, Zahner H, Schnell N, Entian KD, Gotz F. 1988. Gallidermin: a new lanthionine-containing polypeptide antibiotic. Eur J Biochem 177:53–59. doi: 10.1111/j.1432-1033.1988.tb14344.x. [DOI] [PubMed] [Google Scholar]
- 105.Schnell N, Engelke G, Augustin J, Rosenstein R, Ungermann V, Gotz F, Entian KD. 1992. Analysis of genes involved in the biosynthesis of lantibiotic epidermin. Eur J Biochem 204:57–68. doi: 10.1111/j.1432-1033.1992.tb16605.x. [DOI] [PubMed] [Google Scholar]
- 106.Klein C, Kaletta C, Schnell N, Entian KD. 1992. Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol 58:132–142. doi: 10.1128/AEM.58.1.132-142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mota-Meira M, Lacroix C, LaPointe G, Lavoie MC. 1997. Purification and structure of mutacin B-Ny266: a new lantibiotic produced by Streptococcus mutans. FEBS Lett 410:275–279. doi: 10.1016/s0014-5793(97)00425-0. [DOI] [PubMed] [Google Scholar]
- 108.Hillman JD, Novak J, Sagura E, Gutierrez JA, Brooks TA, Crowley PJ, Hess M, Azizi A, Leung K, Cvitkovitch D, Bleiweis AS. 1998. Genetic and biochemical analysis of mutacin 1140, a lantibiotic from Streptococcus mutans. Infect Immun 66:2743–2749. doi: 10.1128/IAI.66.6.2743-2749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qi F, Chen P, Caufield PW. 1999. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl Environ Microbiol 65:3880–3887. doi: 10.1128/AEM.65.9.3880-3887.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qi F, Chen P, Caufield PW. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl Environ Microbiol 67:15–21. doi: 10.1128/AEM.67.1.15-21.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Karaya K, Shimizu T, Taketo A. 2001. New gene cluster for lantibiotic streptin possibly involved in streptolysin S formation. J Biochem 129:769–775. doi: 10.1093/oxfordjournals.jbchem.a002918. [DOI] [PubMed] [Google Scholar]
- 112.Stein T, Borchert S, Conrad B, Feesche J, Hofemeister B, Hofemeister J, Entian KD. 2002. Two different lantibiotic-like peptides originate from the ericin gene cluster of Bacillus subtilis A1/3. J Bacteriol 184:1703–1711. doi: 10.1128/jb.184.6.1703-1711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mantovani HC, Russell JB. 2003. Inhibition of Listeria monocytogenes by bovicin HC5, a bacteriocin produced by Streptococcus bovis HC5. Int J Food Microbiol 89:77–83. doi: 10.1016/s0168-1605(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 114.Castiglione F, Lazzarini A, Carrano L, Corti E, Ciciliato I, Gastaldo L, Candiani P, Losi D, Marinelli F, Selva E, Parenti F. 2008. Determining the structure and mode of action of microbisporicin, a potent lantibiotic active against multiresistant pathogens. Chem Biol 15:22–31. doi: 10.1016/j.chembiol.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 115.Bouhss A, Al-Dabbagh B, Vincent M, Odaert B, Aumont-Nicaise M, Bressolier P, Desmadril M, Mengin-Lecreulx D, Urdaci MC, Gallay J. 2009. Specific interactions of clausin, a new lantibiotic, with lipid precursors of the bacterial cell wall. Biophys J 97:1390–1397. doi: 10.1016/j.bpj.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Daly KM, Upton M, Sandiford SK, Draper LA, Wescombe PA, Jack RW, O’Connor PM, Rossney A, Gotz F, Hill C, Cotter PD, Ross RP, Tagg JR. 2010. Production of the Bsa lantibiotic by community-acquired Staphylococcus aureus strains. J Bacteriol 192:1131–1142. doi: 10.1128/JB.01375-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sashihara T, Kimura H, Higuchi T, Adachi A, Matsusaki H, Sonomoto K, Ishizaki A. 2000. A novel lantibiotic, nukacin ISK-1, of Staphylococcus warneri ISK-1: cloning of the structural gene and identification of the structure. Biosci Biotechnol Biochem 64:2420–2428. doi: 10.1271/bbb.64.2420. [DOI] [PubMed] [Google Scholar]
- 118.Piard JC, Muriana PM, Desmazeaud MJ, Klaenhammer TR. 1992. Purification and partial characterization of lacticin 481, a lanthionine-containing bacteriocin produced by Lactococcus lactis subsp. lactis CNRZ 481. Appl Environ Microbiol 58:279–284. doi: 10.1128/AEM.58.1.279-284.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chikindas ML, Novak J, Driessen AJ, Konings WN, Schilling KM, Caufield PW. 1995. Mutacin II, a bactericidal antibiotic from Streptococcus mutans. Antimicrob Agents Chemother 39:2656–2660. doi: 10.1128/aac.39.12.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pridmore D, Rekhif N, Pittet AC, Suri B, Mollet B. 1996. Variacin, a new lanthionine-containing bacteriocin produced by Micrococcus varians: comparison to lacticin 481 of Lactococcus lactis. Appl Environ Microbiol 62:1799–1802. doi: 10.1128/AEM.62.5.1799-1802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hyink O, Wescombe PA, Upton M, Ragland N, Burton JP, Tagg JR. 2007. Salivaricin A2 and the novel lantibiotic salivaricin B are encoded at adjacent loci on a 190-kilobase transmissible megaplasmid in the oral probiotic strain Streptococcus salivarius K12. Appl Environ Microbiol 73:1107–1113. doi: 10.1128/AEM.02265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang J, Feng Y, Teng K, Lin Y, Gao Y, Wang J, Zhong J. 2014. Type AII lantibiotic bovicin HJ50 with a rare disulfide bond: structure, structure-activity relationships and mode of action. Biochem J 461:497–508. doi: 10.1042/BJ20131524. [DOI] [PubMed] [Google Scholar]
- 123.Janek D, Zipperer A, Kulik A, Krismer B, Peschel A. 2016. High frequency and diversity of antimicrobial activities produced by nasal Staphylococcus strains against bacterial competitors. PLoS Pathog 12:e1005812. doi: 10.1371/journal.ppat.1005812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hasper HE, Kramer NE, Smith JL, Hillman JD, Zachariah C, Kuipers OP, de Kruijff B, Breukink E. 2006. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313:1636–1637. doi: 10.1126/science.1129818. [DOI] [PubMed] [Google Scholar]
- 125.Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 126.Hsu ST, Breukink E, Tischenko E, Lutters MA, de Kruijff B, Kaptein R, Bonvin AM, van Nuland NA. 2004. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol 11:963–967. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 127.Bonelli RR, Schneider T, Sahl HG, Wiedemann I. 2006. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies. Antimicrob Agents Chemother 50:1449–1457. doi: 10.1128/AAC.50.4.1449-1457.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Crandall AD, Montville TJ. 1998. Nisin resistance in Listeria monocytogenes ATCC 700302 is a complex phenotype. Appl Environ Microbiol 64:231–237. doi: 10.1128/AEM.64.1.231-237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187:7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xavier BM, Russell JB. 2006. Bacterial competition between a bacteriocin-producing and a bacteriocin-negative strain of Streptococcus bovis in batch and continuous culture. FEMS Microbiol Ecol 58:317–322. doi: 10.1111/j.1574-6941.2006.00160.x. [DOI] [PubMed] [Google Scholar]
- 131.Koch G, Yepes A, Forstner KU, Wermser C, Stengel ST, Modamio J, Ohlsen K, Foster KR, Lopez D. 2014. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell 158:1060–1071. doi: 10.1016/j.cell.2014.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brotz H, Bierbaum G, Reynolds PE, Sahl HG. 1997. The lantibiotic mersacidin inhibits peptidoglycan biosynthesis at the level of transglycosylation. Eur J Biochem 246:193–199. doi: 10.1111/j.1432-1033.1997.t01-1-00193.x. [DOI] [PubMed] [Google Scholar]
- 133.Gonzalez B, Glaasker E, Kunji E, Driessen A, Suarez JE, Konings WN. 1996. Bactericidal mode of action of plantaricin C. Appl Environ Microbiol 62:2701–2709. doi: 10.1128/AEM.62.8.2701-2709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zimmermann N, Metzger JW, Jung G. 1995. The tetracyclic lantibiotic actagardine. 1H-NMR and 13C-NMR assignments and revised primary structure. Eur J Biochem 228:786–797. doi: 10.1111/j.1432-1033.1995.tb20324.x. [DOI] [PubMed] [Google Scholar]
- 135.Vertesy L, Aretz W, Bonnefoy A, Ehlers E, Kurz M, Markus A, Schiell M, Vogel M, Wink J, Kogler H. 1999. Ala(0)-actagardine, a new lantibiotic from cultures of Actinoplanes liguriae ATCC 31048. J Antibiot (Tokyo) 52:730–741. doi: 10.7164/antibiotics.52.730. [DOI] [PubMed] [Google Scholar]
- 136.Brotz H, Josten M, Wiedemann I, Schneider U, Gotz F, Bierbaum G, Sahl HG. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol 30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 137.Wiedemann I, Bottiger T, Bonelli RR, Wiese A, Hagge SO, Gutsmann T, Seydel U, Deegan L, Hill C, Ross P, Sahl HG. 2006. The mode of action of the lantibiotic lacticin 3147–a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol Microbiol 61:285–296. doi: 10.1111/j.1365-2958.2006.05223.x. [DOI] [PubMed] [Google Scholar]
- 138.Shenkarev ZO, Finkina EI, Nurmukhamedova EK, Balandin SV, Mineev KS, Nadezhdin KD, Yakimenko ZA, Tagaev AA, Temirov YV, Arseniev AS, Ovchinnikova TV. 2010. Isolation, structure elucidation, and synergistic antibacterial activity of a novel two-component lantibiotic lichenicidin from Bacillus licheniformis VK21. Biochemistry 49:6462–6472. doi: 10.1021/bi100871b. [DOI] [PubMed] [Google Scholar]
- 139.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, Latif H, Kim JN, Lockhart A, Artis K, David G, Taylor P, Streib J, Dorrestein PC, Grier A, Gill SR, Zengler K, Hata TR, Leung DY, Gallo RL. 2017. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med 9:eaah4680. doi: 10.1126/scitranslmed.aah4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Navaratna MA, Sahl HG, Tagg JR. 1998. Two-component anti-Staphylococcus aureus lantibiotic activity produced by Staphylococcus aureus C55. Appl Environ Microbiol 64:4803–4808. doi: 10.1128/AEM.64.12.4803-4808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Holo H, Jeknic Z, Daeschel M, Stevanovic S, Nes IF. 2001. Plantaricin W from Lactobacillus plantarum belongs to a new family of two-peptide lantibiotics. Microbiology (Reading) 147:643–651. doi: 10.1099/00221287-147-3-643. [DOI] [PubMed] [Google Scholar]
- 142.McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. 2006. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc Natl Acad Sci U S A 103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Martinez B, Rodriguez A, Suarez JE. 2000. Lactococcin 972, a bacteriocin that inhibits septum formation in lactococci. Microbiology 146:949–955. doi: 10.1099/00221287-146-4-949. [DOI] [PubMed] [Google Scholar]
- 144.Martinez B, Bottiger T, Schneider T, Rodriguez A, Sahl HG, Wiedemann I. 2008. Specific interaction of the unmodified bacteriocin lactococcin 972 with the cell wall precursor lipid II. Appl Environ Microbiol 74:4666–4670. doi: 10.1128/AEM.00092-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schindler CA, Schuhardt VT. 1964. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc Natl Acad Sci U S A 51:414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Grundling A, Schneewind O. 2006. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J Bacteriol 188:2463–2472. doi: 10.1128/JB.188.7.2463-2472.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bastos MD, Coutinho BG, Coelho ML. 2010. Lysostaphin: a staphylococcal bacteriolysin with potential clinical applications. Pharmaceuticals (Basel) 3:1139–1161. doi: 10.3390/ph3041139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Simmonds RS, Pearson L, Kennedy RC, Tagg JR. 1996. Mode of action of a lysostaphin-like bacteriolytic agent produced by Streptococcus zooepidemicus 4881. Appl Environ Microbiol 62:4536–4541. doi: 10.1128/AEM.62.12.4536-4541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gargis SR, Heath HE, Heath LS, Leblanc PA, Simmonds RS, Abbott BD, Timkovich R, Sloan GL. 2009. Use of 4-sulfophenyl isothiocyanate labeling and mass spectrometry to determine the site of action of the streptococcolytic peptidoglycan hydrolase zoocin A. Appl Environ Microbiol 75:72–77. doi: 10.1128/AEM.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Beukes M, Bierbaum G, Sahl HG, Hastings JW. 2000. Purification and partial characterization of a murein hydrolase, millericin B, produced by Streptococcus milleri NMSCC 061. Appl Environ Microbiol 66:23–28. doi: 10.1128/aem.66.1.23-28.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nilsen T, Nes IF, Holo H. 2003. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl Environ Microbiol 69:2975–2984. doi: 10.1128/aem.69.5.2975-2984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Khan H, Flint SH, Yu PL. 2013. Determination of the mode of action of enterolysin A, produced by Enterococcus faecalis B9510. J Appl Microbiol 115:484–494. doi: 10.1111/jam.12240. [DOI] [PubMed] [Google Scholar]
- 153.Kurushima J, Hayashi I, Sugai M, Tomita H. 2013. Bacteriocin protein BacL1 of Enterococcus faecalis is a peptidoglycan D-isoglutamyl-L-lysine endopeptidase. J Biol Chem 288:36915–36925. doi: 10.1074/jbc.M113.506618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Riley M. 2009. Bacteriocins, biology, ecology and evolution, p 32–44. In Schaechler M (ed), Encyclopedia of microbiology, 3rd ed. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 155.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schaller K, Holtje JV, Braun V. 1982. Colicin M is an inhibitor of murein biosynthesis. J Bacteriol 152:994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Harkness RE, Braun V. 1989. Colicin M inhibits peptidoglycan biosynthesis by interfering with lipid carrier recycling. J Biol Chem 264:6177–6182. doi: 10.1016/S0021-9258(18)83329-4. [DOI] [PubMed] [Google Scholar]
- 158.El Ghachi M, Bouhss A, Barreteau H, Touze T, Auger G, Blanot D, Mengin-Lecreulx D. 2006. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J Biol Chem 281:22761–22772. doi: 10.1074/jbc.M602834200. [DOI] [PubMed] [Google Scholar]
- 159.Barreteau H, Bouhss A, Fourgeaud M, Mainardi JL, Touze T, Gerard F, Blanot D, Arthur M, Mengin-Lecreulx D. 2009. Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors. J Bacteriol 191:3657–3664. doi: 10.1128/JB.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Patzer SI, Albrecht R, Braun V, Zeth K. 2012. Structural and mechanistic studies of pesticin, a bacterial homolog of phage lysozymes. J Biol Chem 287:23381–23396. doi: 10.1074/jbc.M112.362913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hantke K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet 182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 162.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. 1993. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron‐repressible outer membrane polypeptide of 65 000 Da and pesticin sensitivity. Mol Microbiol 8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 163.Nedialkova LP, Denzler R, Koeppel MB, Diehl M, Ring D, Wille T, Gerlach RG, Stecher B. 2014. Inflammation fuels colicin Ib-dependent competition of Salmonella serovar Typhimurium and E. coli in enterobacterial blooms. PLoS Pathog 10:e1003844. doi: 10.1371/journal.ppat.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Whitney JC, Peterson SB, Kim J, Pazos M, Verster AJ, Radey MC, Kulasekara HD, Ching MQ, Bullen NP, Bryant D, Goo YA, Surette MG, Borenstein E, Vollmer W, Mougous JD. 2017. A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife 6:e26938. doi: 10.7554/eLife.26938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jana B, Salomon D. 2019. Type VI secretion system: a modular toolkit for bacterial dominance. Future Microbiol 14:1451–1463. doi: 10.2217/fmb-2019-0194. [DOI] [PubMed] [Google Scholar]
- 167.Whitney JC, Chou S, Russell AB, Biboy J, Gardiner TE, Ferrin MA, Brittnacher M, Vollmer W, Mougous JD. 2013. Identification, structure, and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J Biol Chem 288:26616–26624. doi: 10.1074/jbc.M113.488320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ma J, Sun M, Pan Z, Lu C, Yao H. 2018. Diverse toxic effectors are harbored by vgrG islands for interbacterial antagonism in type VI secretion system. Biochim Biophys Acta Gen Subj 1862:1635–1643. doi: 10.1016/j.bbagen.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 169.Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. 2013. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem 288:7618–7625. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, Mougous JD. 2012. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11:538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Altindis E, Dong T, Catalano C, Mekalanos J. 2015. Secretome analysis of Vibrio cholerae type VI secretion system reveals a new effector-immunity pair. mBio 6:e00075. doi: 10.1128/mBio.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hersch SJ, Watanabe N, Stietz MS, Manera K, Kamal F, Burkinshaw B, Lam L, Pun A, Li M, Savchenko A, Dong TG. 2020. Envelope stress responses defend against type six secretion system attacks independently of immunity proteins. Nat Microbiol 5:706–714. doi: 10.1038/s41564-020-0672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.English G, Trunk K, Rao VA, Srikannathasan V, Hunter WN, Coulthurst SJ. 2012. New secreted toxins and immunity proteins encoded within the type VI secretion system gene cluster of Serratia marcescens. Mol Microbiol 86:921–936. doi: 10.1111/mmi.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, Monack DM. 2016. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A 113:E5044–E5051. doi: 10.1073/pnas.1608858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang T, Hu Z, Du X, Shi Y, Dang J, Lee M, Hesek D, Mobashery S, Wu M, Liang H. 2020. A type VI secretion system delivers a cell wall amidase to target bacterial competitors. Mol Microbiol 114:308–321. doi: 10.1111/mmi.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sibinelli-Sousa S, Hespanhol JT, Nicastro GG, Matsuyama BY, Mesnage S, Patel A, de Souza RF, Guzzo CR, Bayer-Santos E. 2020. A family of T6SS antibacterial effectors related to l,d-transpeptidases targets the peptidoglycan. Cell Rep 31:107813. doi: 10.1016/j.celrep.2020.107813. [DOI] [PubMed] [Google Scholar]
- 177.Toska J, Ho BT, Mekalanos JJ. 2018. Exopolysaccharide protects Vibrio cholerae from exogenous attacks by the type 6 secretion system. Proc Natl Acad Sci U S A 115:7997–8002. doi: 10.1073/pnas.1808469115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Le NH, Peters K, Espaillat A, Sheldon JR, Gray J, Di Venanzio G, Lopez J, Djahanschiri B, Mueller EA, Hennon SW, Levin PA, Ebersberger I, Skaar EP, Cava F, Vollmer W, Feldman MF. 2020. Peptidoglycan editing provides immunity to Acinetobacter baumannii during bacterial warfare. Sci Adv 6:eabb5614. doi: 10.1126/sciadv.abb5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Espaillat A, Forsmo O, El Biari K, Bjork R, Lemaitre B, Trygg J, Canada FJ, de Pedro MA, Cava F. 2016. Chemometric analysis of bacterial peptidoglycan reveals atypical modifications that empower the cell wall against predatory enzymes and fly innate immunity. J Am Chem Soc 138:9193–9204. doi: 10.1021/jacs.6b04430. [DOI] [PubMed] [Google Scholar]
- 180.Yadav AK, Espaillat A, Cava F. 2018. Bacterial strategies to preserve cell wall integrity against environmental threats. Front Microbiol 9:2064. doi: 10.3389/fmicb.2018.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bera A, Herbert S, Jakob A, Vollmer W, Gotz F. 2005. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol 55:778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 182.Christie PJ, Whitaker N, Gonzalez-Rivera C. 2014. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta 1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Grohmann E, Christie PJ, Waksman G, Backert S. 2018. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol 107:455–471. doi: 10.1111/mmi.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sgro GG, Oka GU, Souza DP, Cenens W, Bayer-Santos E, Matsuyama BY, Bueno NF, Dos Santos TR, Alvarez-Martinez CE, Salinas RK, Farah CS. 2019. Bacteria-killing type IV secretion systems. Front Microbiol 10:1078. doi: 10.3389/fmicb.2019.01078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bayer-Santos E, Cenens W, Matsuyama BY, Oka GU, Di Sessa G, Mininel IDV, Alves TL, Farah CS. 2019. The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing. PLoS Pathog 15:e1007651. doi: 10.1371/journal.ppat.1007651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. 2007. Type VII secretion–mycobacteria show the way. Nat Rev Microbiol 5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 187.Unnikrishnan M, Constantinidou C, Palmer T, Pallen MJ. 2017. The enigmatic Esx proteins: looking beyond mycobacteria. Trends Microbiol 25:192–204. doi: 10.1016/j.tim.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 188.Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, Eisenberg D, Russell RG, Derrick SC, Collins FM, Morris SL, King CH, Jacobs WR. 2003. The primary mechanism of attenuation of bacillus Calmette–Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A 100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang D, Iyer LM, Aravind L. 2011. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res 39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Callewaert L, Michiels CW. 2010. Lysozymes in the animal kingdom. J Biosci 35:127–160. doi: 10.1007/s12038-010-0015-5. [DOI] [PubMed] [Google Scholar]
- 191.Jollès P, Jollès J. 1984. What's new in lysozyme research? Always a model system, today as yesterday. Mol Cell Biochem 63:165–189. [DOI] [PubMed] [Google Scholar]
- 192.Kanamaru S, Ishiwata Y, Suzuki T, Rossmann MG, Arisaka F. 2005. Control of bacteriophage T4 tail lysozyme activity during the infection process. J Mol Biol 346:1013–1020. doi: 10.1016/j.jmb.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 193.Pritchard DG, Dong S, Baker JR, Engler JA. 2004. The bifunctional peptidoglycan lysin of Streptococcus agalactiae bacteriophage B30. Microbiology (Reading) 150:2079–2087. doi: 10.1099/mic.0.27063-0. [DOI] [PubMed] [Google Scholar]
- 194.Lamrabet O, Jauslin T, Lima WC, Leippe M, Cosson P. 2020. The multifarious lysozyme arsenal of Dictyostelium discoideum. Dev Comp Immunol 107:103645. doi: 10.1016/j.dci.2020.103645. [DOI] [PubMed] [Google Scholar]
- 195.Metcalf JA, Funkhouser-Jones LJ, Brileya K, Reysenbach AL, Bordenstein SR. 2014. Antibacterial gene transfer across the tree of life. Elife 3:e04266. doi: 10.7554/eLife.04266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Brakhage AA, Thon M, Sprote P, Scharf DH, Al-Abdallah Q, Wolke SM, Hortschansky P. 2009. Aspects on evolution of fungal beta-lactam biosynthesis gene clusters and recruitment of trans-acting factors. Phytochemistry 70:1801–1811. doi: 10.1016/j.phytochem.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 197.Buades C, Moya A. 1996. Phylogenetic analysis of the isopenicillin-N-synthetase horizontal gene transfer. J Mol Evol 42:537–542. doi: 10.1007/BF02352283. [DOI] [PubMed] [Google Scholar]
- 198.Roelofs D, Timmermans MJ, Hensbergen P, van Leeuwen H, Koopman J, Faddeeva A, Suring W, de Boer TE, Marien J, Boer R, Bovenberg R, van Straalen NM. 2013. A functional isopenicillin N synthase in an animal genome. Mol Biol Evol 30:541–548. doi: 10.1093/molbev/mss269. [DOI] [PubMed] [Google Scholar]
- 199.Chou S, Daugherty MD, Peterson SB, Biboy J, Yang Y, Jutras BL, Fritz-Laylin LK, Ferrin MA, Harding BN, Jacobs-Wagner C, Yang XF, Vollmer W, Malik HS, Mougous JD. 2015. Transferred interbacterial antagonism genes augment eukaryotic innate immune function. Nature 518:98–101. doi: 10.1038/nature13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hayes BM, Radkov AD, Yarza F, Flores S, Kim JR, Zhao Z, Lexa KW, Marnin L, Biboy J, Bowcutt V, Vollmer W, Pedra JHF, Chou S. 2020. Immune factor of bacterial origin protects ticks against host microbial commensals. bioRxiv 10.1101/2020.04.10.036376. [DOI] [PMC free article] [PubMed]
- 201.Garcia-Bayona L, Comstock LE. 2018. Bacterial antagonism in host-associated microbial communities. Science 361:eaat2456. doi: 10.1126/science.aat2456. [DOI] [PubMed] [Google Scholar]