Abstract
This work introduces the Fourier-Bessel series expansion-based decomposition (FBSED) method, which is an implementation of the wavelet packet decomposition approach in the Fourier-Bessel series expansion domain. The proposed method has been used for the diagnosis of pneumonia caused by the 2019 novel coronavirus disease (COVID-19) using chest X-ray image (CXI) and chest computer tomography image (CCTI). The FBSED method is used to decompose CXI and CCTI into sub-band images (SBIs). The SBIs are then used to train various pre-trained convolutional neural network (CNN) models separately using a transfer learning approach. The combination of SBI and CNN is termed as one channel. Deep features from each channel are fused to get a feature vector. Different classifiers are used to classify pneumonia caused by COVID-19 from other viral and bacterial pneumonia and healthy subjects with the extracted feature vector. The different combinations of channels have also been analyzed to make the process computationally efficient. For CXI and CCTI databases, the best performance has been obtained with only one and four channels, respectively. The proposed model was evaluated using 5-fold and 10-fold cross-validation processes. The average accuracy for the CXI database was 100% for both 5-fold and 10-fold cross-validation processes, and for the CCTI database, it is 97.6% for the 5-fold cross-validation process. Therefore, the proposed method may be used by radiologists to rapidly diagnose patients with COVID-19.
Keywords: COVID-19, CT images, FBSED method, Image decomposition, X-ray image
Graphical abstract
1. Introduction
The first noted case of the disease caused by the 2019 novel coronavirus (COVID-19) was detected in Wuhan (Hubei province of China) and was described as a case of pneumonia [1]. Subsequently, the virus was named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the disease it causes is called COVID-19. The World Health Organization (WHO) declared COVID-19 as a pandemic on 11 March 2020 [2]. Patients with COVID-19 often present with dry cough, sore throat, fever, dyspnea, and ageusia, which may rapidly deteriorate to pneumonia and organ failure [1,3]. The SARS-CoV-2 can be transmitted from human-to-human, thus the main challenge in controlling its transmission is to test every person rapidly, accurately, and without any contact.
Currently, the most common way of testing for COVID-19 is the real-time reverse transcription-polymerase chain reaction (RT-PCR) test. The RT-PCR test takes approximately 10–15 h to declare the result which makes the testing process very slow. In addition, countries with large population face the problem of insufficient test kits due to their global shortage. Rapid diagnostic test (RDT) is another way of testing for COVID-19. Although it is faster (as it takes approximately 30 min) than the RT-PCR, it is less reliable [1]. The reported sensitivity (SEN) and specificity (SPE) of the RT-PCR test are 100% and 67%, respectively. Contrarily, the RDT has moderate SEN of around 50% and high SPE. In practice, that means if you test 100 COVID-19 patients, it will provide positive results for only 50 of them [4]. Although the RT-PCR test provides good performance, it requires a high level of skill to handle samples collected from the patients. It was noted in March 2020 that US Centers for Disease Control and Prevention withdrew testing kits due to contamination of samples [5]. Hence, there is a pressing need for fast, accurate, and contamination-free diagnostic techniques to test for COVID-19.
SARS-CoV-2 commonly affects the lungs of the patient and often presents as pneumonia. So chest X-ray image (CXI) and chest computer tomography image (CCTI) can be directly used for diagnosis of COVID-19 and can also be used as assistive tools along with RT-PCR test and RDT [6,7]. Even at the beginning of the pandemic, the Chinese Clinical Center and Turkey used CCTI results to diagnose COVID-19 due to an acute shortage of test kits [8,9].
Signal/image processing techniques with machine learning algorithm can be used for the automated diagnosis of COVID-19 from CXI and CCTI. Researchers from all around the world have proposed many models for the diagnosis of COVID-19 using CXI and CCTI. Hemdan et al. [10] diagnosed COVID-19 from CXI using deep learning (DL) model COVIDX-Net. It comprises seven convolutional neural network (CNN) models. Sethy and Behera [11] used CXI to train several CNN models along with a support vector machine (SVM) classifier to detect COVID-19. Their study showed that the ResNet-50 model with SVM provides the best result. Similar work was done by Nayak et al. [12] in which they compared the performance of eight pre-trained CNN models for the diagnosis of COVID-19. The best performance is obtained by the ResNet-34 model with an accuracy (ACC) of 98.33%. Ozturk et al. [3] proposed the DL model DarkCovidNet which is based on the DarkNet CNN model. The CXI with DarkCovidNet was used for the diagnosis of COVID-19. The backbone of the model is DarkNet-19 and the ACC obtained by the model was 98.08%. Wang and Wong [13] proposed a model COVID-Net based on a residual deep architecture that uses CXI for COVID-19 detection. The COVID-Net achieved an ACC of 83.5%. Similarly, the CCTIs were also studied by several authors for the diagnosis of COVID-19. It has been observed that CCTI is preferable to CXI for the diagnosis of COVID-19 [14]. Ni et al. [15] proposed NiNet for the diagnosis of COVID-19 using CCTI. The NiNet is a CNN model utilizing both 3D U-Net and multi-viewpoint regression network (MVP-Net). The proposed model offers a SEN of 100% and an F1-score of 0.97 in detecting lesions from CTIs. Li et al. [16] proposed COVNet which uses ResNet-50 as the backbone network. The CCTI with COVNet is used for COVID-19 diagnosis. The SEN and SPE obtained from COVNet model were 90% and 96%, respectively.
In our previous work [17], the Fourier-Bessel dyadic decomposition (FBD) method-based ensemble ResNet-50 model was used for the diagnosis of COVID-19 from CXI. Motivated by the results and advantages of using FBD, we extended our previous work in this paper. The main contributions of this work are as follows:
-
•
Fourier-Bessel series expansion-based decomposition (FBSED) has been introduced for image decomposition. The best level of decomposition is selected based on classification performance.
-
•
Five different pre-trained CNN (which recently won ImageNet challenge [18]) are used for feature extraction. The best CNN model is also examined for different optimizers.
-
•
The performance of five different classifiers is compared for the features, obtained from CNN.
-
•
Sub-band image (SBI) channel based analysis is proposed to select the combination of channels to get better performance.
-
•
A study of the proposed method was carried out on both CCTI and CXI.
The paper is organized as follows: Database and FBSED method are described in Section 2. The proposed methodology for the diagnosis of COVID-19 is explained in Section 3. The experiment results are illustrated in Section 4, the discussion of the obtained results is presented in Section 5, and Section 6 concludes the study.
2. Database and proposed FBSED method
2.1. Database
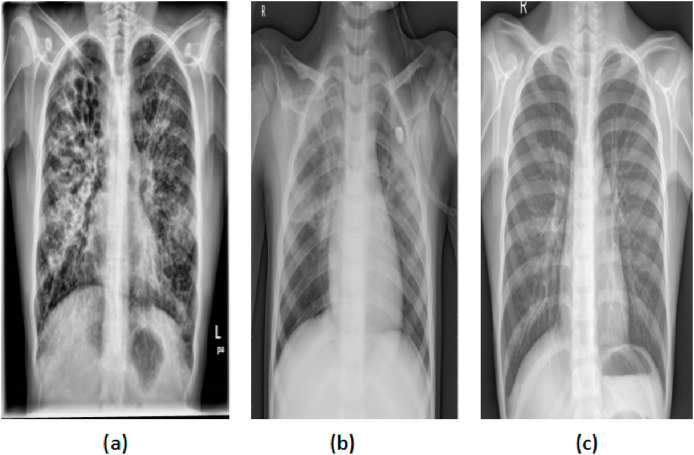
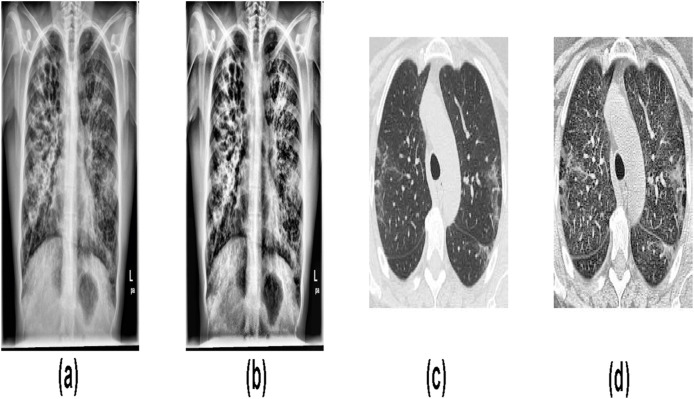
In this work, both CXI and CCTI were used. For CXI, a total of 1,446 images were collected from two databases. The 785 CXIs were downloaded from https://github.com/ieee8023/covid-chestxray-dataset. Out of 785 CXIs, 482 images are of pneumonia caused by COVID-19, 285 images were of other pneumonia subjects, and the remaining 18 images were of normal subjects [19]. These images were collected from various open sources. Most of the studies on COVID-19 were carried out on the same source, and in order to balance the database, the remaining images were collected from other sources [3,[10], [11], [12], [13]]. For balancing the CXI database, 661 CXIs were downloaded from the Kaggle repository database called “Chest X-Ray Images” [20]. From the Chest X-ray Image database, 197 images of pneumonia and 464 images of normal subjects were downloaded. So the CXI database had a total of 1,446 images, and each of the three classes having 482 images. Fig. 1 shows the CXI of COVID-19, pneumonia, and normal subjects.
Fig. 1.
CXI of (a) COVID-19, (b) Pneumonia, and (c) Normal subjects.
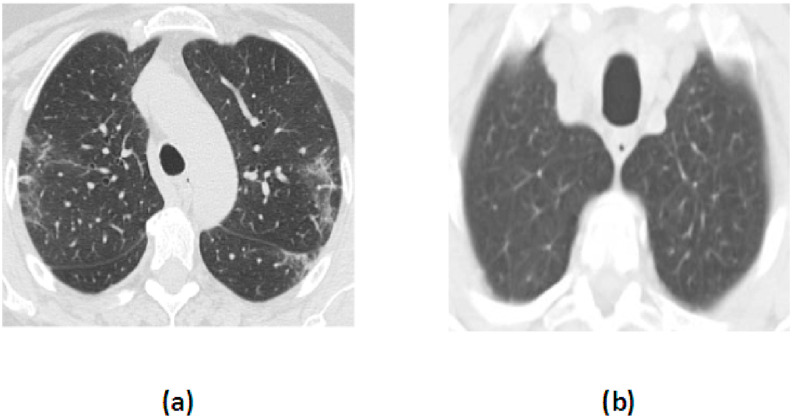
For the CCTI database, a total of 2,481 images have been downloaded from website https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset. The 1,229 CCTIs are of non-COVID-19 subjects and 1,252 CCTIs are of COVID-19 subjects [21]. Fig. 2 shows the CCTI of COVID-19 and non-COVID-19 subjects. So, the CXI database has three classes: COVID-19, pneumonia, and normal. The CCTI database has two classes: COVID-19 and non-COVID-19.
Fig. 2.
CCTI of (a) COVID-19 and (b) Non-COVID-19 subjects.
2.2. FBSED method
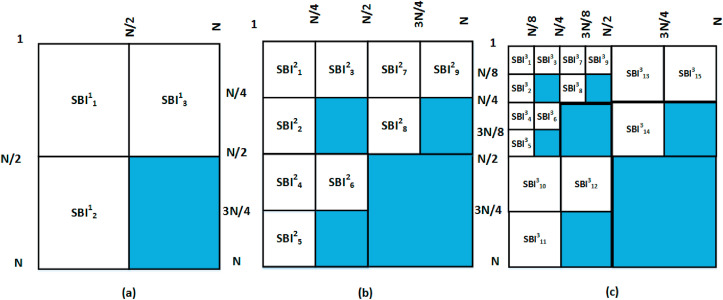
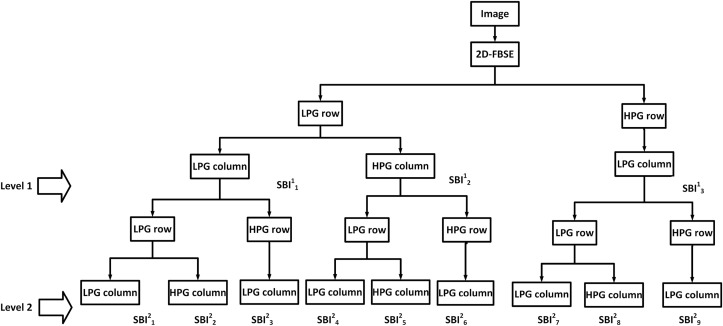
The FBSED is an extension of the FBD method [17]. The FBD is a dyadic decomposition method motivated by the two-dimensional discrete wavelet transform (2D-DWT) [22,23] and multi-frequency scale two-dimensional Fourier-Bessel series expansion-based empirical wavelet transform (2D-FBSE-EWT) method [24]. The 2D-DWT and FBD provide four SBIs for level-1 decomposition. These four SBIs include one approximation component and three detailed components (vertical, horizontal, and diagonal). For a higher level of decomposition, iterative filtering (or grouping) is performed only on the approximation component. Hence, these decomposed components often fail to accurately capture high-frequency information present in the image. On the other hand, the wavelet packet decomposition (WPD) can provide better high-frequency information present in the image [25]. It has been observed that the diagonal detail component of each level of decomposition degrades the classification performance, as these components contain the majority of noise or irrelevant information in the image [26,27]. In Ref. [26], the authors have proposed three ways of SBI grouping for 2D-DWT: 10-channel, 7-channel, and 4-channel for level-3 decomposition. The best performance was obtained by a 7-channel SBI grouping operation where the diagonal component was removed in each level of decomposition. The authors have extended the 7-channel SBI grouping operation for WPD in Ref. [27]. Being motivated from this concept, 7-channel SBI grouping has been used on FBSED SBIs. The SBI grouping used in our work for level-1, level-2, and level-3 decompositions for an image of size N N are shown in Fig. 3 (a), (b), and (c), respectively. Superscript in SBI represents the decomposition level and subscript represents SBI. The shaded portion shown in Fig. 3 is the diagonal detail component which is ignored in the grouping process.
Fig. 3.
FBSED at (a) Level-1, (b) Level-2, and (c) Level-3.
In FBSED, the WPD concept is implemented in the Fourier-Bessel series expansion (FBSE) domain. The mathematical expression of order-zero 1D-FBSE [28,29] of signal of length N is shown in Eq. (1).
| (1) |
where denotes order-zero 1D-FBSE coefficients which can be expressed as follows [28,30]:
| (2) |
In the above-mentioned expressions, and are order-zero and order-one Bessel functions, respectively. The variable denotes the positive root of equation [28]. By using FBSE, the signal is transformed from the l (spatial or time) domain to the x (order) domain.
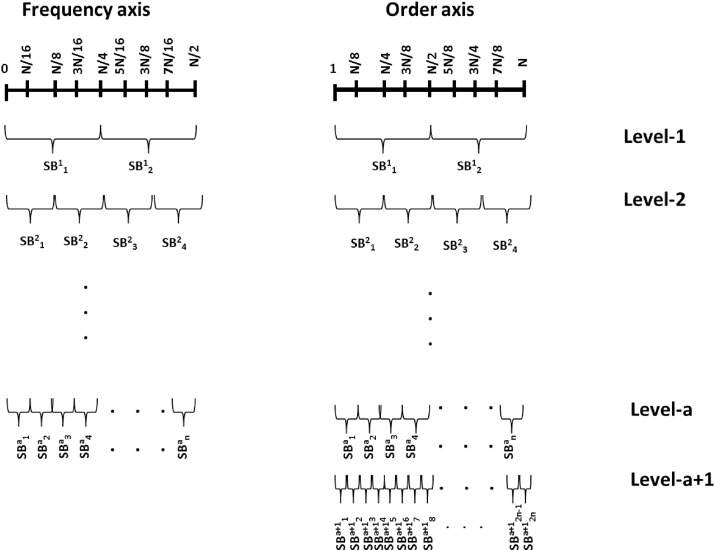
While implementing FBD using FBSE, one extra level of decomposition can be obtained as compared to DWT. Because for a signal with a length of N samples, the FBSE provides N unique FBSE coefficients, whereas discrete Fourier transform (DFT) provides number of coefficients. The same is true for FBSED, i.e., implementation of WPD using FBSE can provide a better multiresolution analysis (see Fig. 4 ). In Fig. 4, ‘a’ represents the level of decomposition, SB represents the sub-band signal, and ‘n’ is the number of SB signals at each level of decomposition. For 1D-WPD, n is equal to .
Fig. 4.
WPD in frequency axis and order axis.
Block diagram for implementation of FBSED for level-1 and level-2 decomposition (analysis part) for proposed decomposition scheme in Fig. 3 has been shown in Fig. 5 . In Fig. 5, the LPG row and HPG row represent low pass grouping and high pass grouping row-wise. Similarly, the LPG column and HPG column represent low pass grouping and high pass grouping column-wise.
Fig. 5.
Block diagram of the analysis part of FBSED.
Authors have shown the effect of pass band filtering on multi-component, amplitude modulated and frequency modulated (AM-FM) signals [31]. The changes were noted in the amplitude and frequency functions of the filtered signal. To address this concern, the authors used the FBSE spectrum and grouping of the coefficients to separate components. As the FBSE coefficients are real, each component can be reconstructed directly from the coefficients without affecting the amplitude and frequency functions of the filtered signal. Consequently, the grouping of FBSE coefficients is used for the implementation of the FBSED method. Using the concept of grouping, any level of decomposition can be obtained in a single step and it makes the implementation of FBSED easier. For level-1 FBSED (shown in Fig. 3, Fig. 5), the 2D-FBSE method is applied to the image. In order to have an approximation component (), the 2D-FBSE coefficients are grouped from x = 1 to row-wise (LPG row) and from to column-wise (LPG column) and 2D-inverse FBSE (2D-IFBSE) is applied to the grouped coefficients. The 2D-FBSE coefficients are grouped from to row-wise (LPG row) and from to N column-wise (HPG column) and 2D-IFBSE is applied to the grouped coefficients in order to obtain vertical detail component (). The horizontal detail component () requires grouping of 2D-FBSE coefficients from to N row-wise (HPG row) and from to column-wise (LPG column), and 2D-IFBSE is carried out on these grouped coefficients. For diagonal detail component (shaded block in Fig. 3), 2D-FBSE coefficients grouping is performed from to N row-wise (HPG row) and from to N column-wise (HPG column) and 2D-IFBSE has been performed to the grouped coefficients. The diagonal detail component is not used for classification. The 2D-FBSE can be obtained by applying 1D-FBSE using Eq. (2) row-wise followed by 1D-FBSE column-wise. On the other hand, the 2D-IFBSE can be obtained by applying 1D-IFBSE using Eq. (1) column-wise followed by 1D-IFBSE row-wise. Similarly, a higher level of decomposition can be obtained, as shown in Fig. 3, Fig. 5 (b), and (c).
3. Proposed methodology for COVID-19 diagnosis
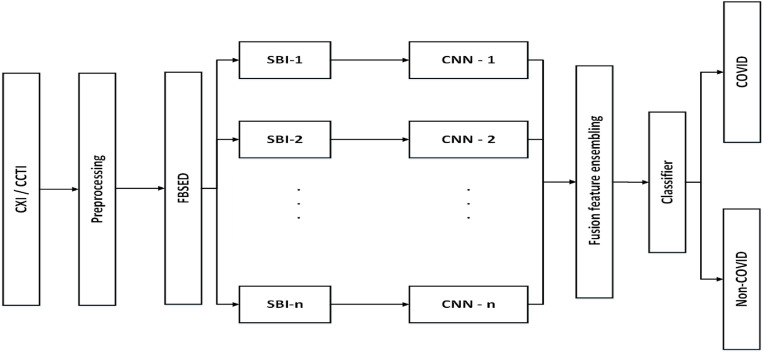
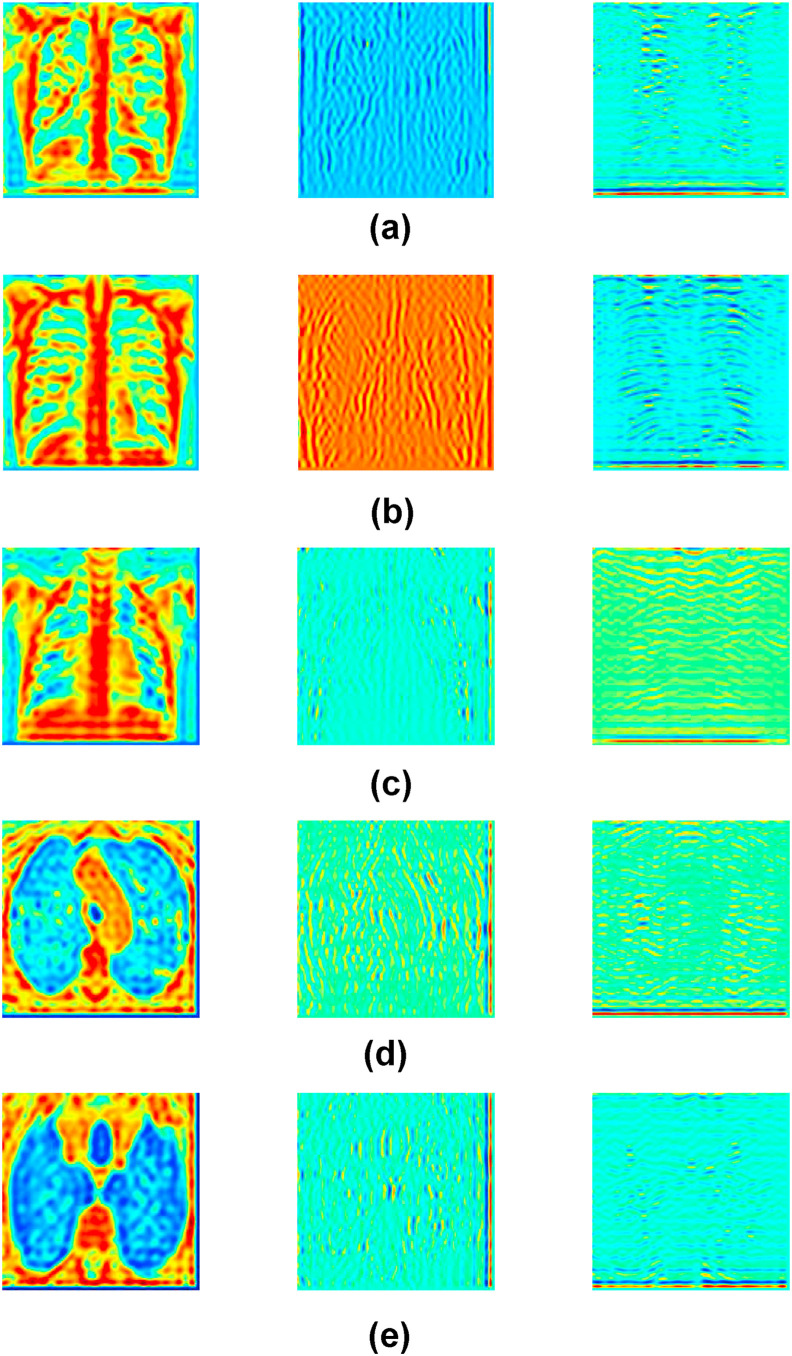
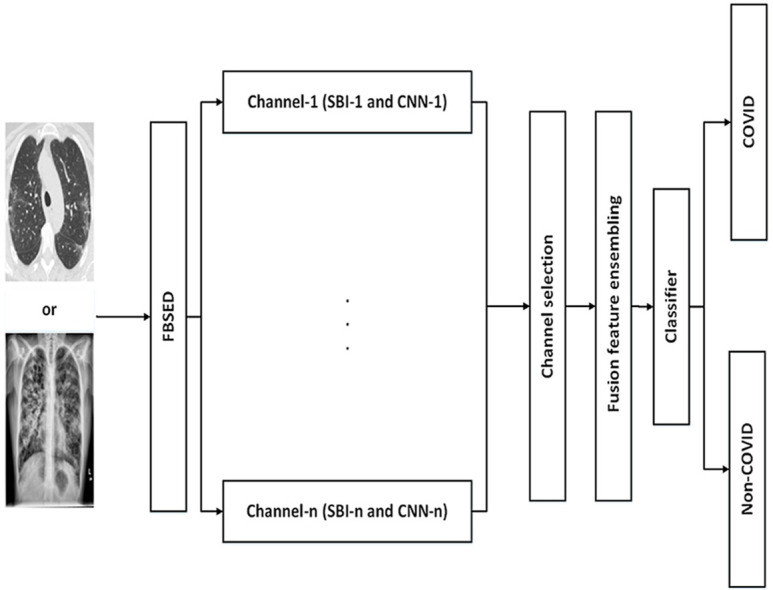
The framework of the proposed methodology for the diagnosis of COVID-19 from CXI and CCTI is shown in Fig. 6 . In the preprocessing step, the input images undergo normalization followed by contrast enhancement. In the normalization step, the pixel values of input images have been normalized in the range 0–1. Normalization of input image brings numerical stability in the CNN model [12]. For contrast enhancement, the contrast limited adaptive histogram equalization (CLAHE) method [32] is applied to the normalized image. The CLAHE performs histogram equalization on each tile of an input image. The small regions in the image are termed as tiles. After equalization, the CLAHE combines neighboring tiles using bilinear interpolation to eliminate artificially induced boundaries. Fig. 7 (a) and (b) show the raw and preprocessed CXI, respectively. Similarly, Fig. 7 (c) and 7 (d) illustrate raw and preprocessed CCTI, respectively. Contrast enhanced image is then resized according to the input layer size of the pre-train model that is used in this work. Then, the FBSED is used to decompose preprocessed images into SBIs. The number of SBIs depends on the level of decomposition. There are 3, 9, and 15 SBIs for level-1, -2, and -3 decompositions, respectively. Fig. 8 shows the first three components (pseudo color images) of level-3 decomposed FBSED. Fig. 8 (a), (b), and (c) show SBIs of COVID-19, pneumonia, and normal CXI. Fig. 8 (d) and (e) show SBIs of CCTI of COVID-19 and non-COVID-19. First row shows the approximation SBI () at level-3 decomposition and other two rows show vertical and horizontal detail SBIs ( and ) at level-3 decomposition. From each SBI, deep features are extracted from the last fully connected (FC) layer of the pre-trained CNN. Extracted features from each channel (SBI-n with CNN-n) are then ensembled using fusion operation. Fusion of features is achieved by concatenating features of each channels in order to get one feature vector.
Fig. 6.
Proposed framework for automated diagnosis of COVID-19 using CXI and CCTI.
Fig. 7.
Preprocessing of CXI and CCTI, where (a) Raw CXI, (b) CLAHE CXI, (c) Raw CCTI, and (d) CLAHE CCTI.
Fig. 8.
First three components of level-3 FBSED of (a) COVID-19 CXI, (b) Pneumonia CXI, (c) Normal CXI, (d) COVID-19 CCTI, and (e) Non-COVID-19 CCTI.
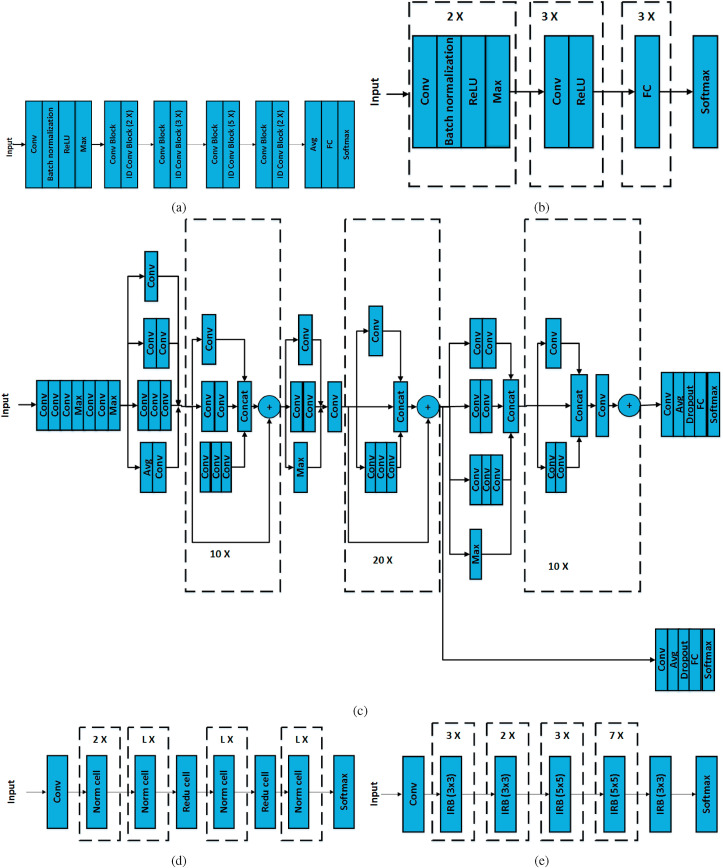
For classification, we have used different pre-trained networks, namely ResNet-50 [33], AlexNet [34], Inception-ResNet-v2 [35], NASNet [36], and EfficientNet [37]. ResNet-50 consists of five stages. The first stage consists of the convolution (Conv) layer, batch normalization layer, rectified linear unit (ReLU) activation layer, and maximum pooling (Max) layer. The second to fifth stage consists of a set convolution block (Conv Block) and identity convolution block (ID Conv Block). Both blocks have three Conv layers in each. Each block uses skip connections i.e., it adds output of earlier layer to output of present layer. ID Conv Block is used when there is no change in dimension of input and output. Conv Block uses Conv layer in skip connection to match the dimension of input and output. The output layer consists of the average pooling (Avg) layer, FC layer, and Softmax layer. Fig. 9 (a) shows the block diagram of ResNet-50 architecture. The AlexNet consists of five Conv and three FC layers. Fig. 9 (b) depicts the block diagram of AlexNet architecture. Inception-ResNet-v2 is a combination of InceptionNet and the residual connection. InceptionNet uses multiple Conv layers at the same level. In Inception-ResNet such InceptionNet networks are used along with the residual network. The architecture of Inception-ResNet-v2 is shown in Fig. 9 (c). In Fig. 9 (c), Max, Concat, and Dropout represent the maximum pooling, concatenation, and dropout layer, respectively. Fig. 9 (d) shows the basic architecture of NASNet. The number of repetitions ‘L’ and number of Conv layers are searched by the reinforcement learning search method. The NASNet model consists of two main cells: normal cell (Norm cell) and reduction cell (Redu cell). The Norm cell consists of Conv layer and returns the feature map of the same size as the input. The Redu cell is used when the feature map size is less than the input size. This concept is the same as of Conv Block of ResNet-50. The EfficientNet is a CNN that uses a scaling method, which uniformly scales all dimensions like depth, width, and resolution. The EfficientNet model extracts feature using multiple Conv layers and inverse residual block (IRB). The IRB connects narrow layers, while wider layers are in between skip connections. Fig. 9 (e) shows the architecture of EfficientNet. In Fig. 9 and 20 X, 10 X, 7 X, 3 X, 2 X, and L X mean repetition of the same block 20, 10, 7, 3, 2, and L times, respectively.
Fig. 9.
Architecture of (a) ResNet-50, (b) AlexNet, (c) Inception-ResNet-v2, (d) NASNet, and (e) EfficientNet CNN.
All the pre-trained CNNs mentioned above were trained using ImageNet database [18] which consists of more than a million images belonging to 1,000 classes. Transfer learning was applied to transfer the trained feature by freezing the weights of the previously trained layers and replacing the last trainable layer (usually the last FC layer) with the new trainable layer [38]. The number of outputs from the new trainable layer was set according to the number of classes in the new database. The obtained network was then trained with the new database. Table 1 shows the size of the input layer, number of layers, total parameters, trainable parameter (two-class and three-class) of CNNs used in the study, and size of feature vector from single SBI or channel (i.e., size of the deep feature of last FC layer). Size of a deep feature vector depends on the level of decomposition (number of SBIs) and the type of pre-trained network. For example, if level of decomposition is 2 (9 SBIs) and pre-trained network is ResNet-50 (2,048 features from single SBI or channel), then number of deep features will be equal to 18,432 (i.e., 2,0489).
Table 1.
Description of architecture used in the study.
| Model | Input layer size | Number of layers | Parameter (in millions) | Trainable parameter |
Feature vector size from single SBI | |
|---|---|---|---|---|---|---|
| two-class | three-class | |||||
| ResNet-50 | (224, 224) | 50 | 25.6 | 4,098 | 6,147 | 2,048 |
| AlexNet | (227, 227) | 8 | 61 | 8,192 | 12,291 | 4,096 |
| Inception-ResNet-v2 | (299, 299) | 169 | 55.9 | 3,074 | 4,611 | 1,536 |
| NASNet | (331, 331) | * | 88.9 | 8,066 | 12,099 | 4,032 |
| EfficientNet | (224, 224) | 82 | 5.3 | 2,562 | 3,843 | 1,280 |
Note: * Number of layers of NASNet networks is not known because it is not a linear sequence model.
Deep features are then fed to the classifier for three and two classes classification. In this work, random forest (RF) [39], J48 [40], Naive Bayes [41], AdaBoost [42], and Softmax [43] classifiers were used. Measuring parameters for evaluation models are, namely precision (PRE), SEN, SPE, F1-score, ACC, and area under the receiver operating characteristic curve (AUC) [[44], [45], [46]]. PRE indicates how accurately the model classifies positive class out of all positive cases. SEN indicates how correctly the model is able to classify positive class. SPE indicates how correctly the model is able to classify the negative class. ACC indicates how correctly the model can classify positive class image to positive class and negative class image to negative class. The F1-score is the harmonic mean of PRE and Recall. AUC indicates the extent to which the model can capable of distinguish between classes. The mathematical expression of these measures are represented by Eqs. (3)–(7).
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
where,
| (8) |
The notations NTP, NTN, NFP, and NFN represent the number of true positives, number of true negatives, number of false positives, and number of false negatives, respectively. The CXI database has three different classes. Overall performance measure of a model can be computed by taking weighted average of performance measures of each class. In this case, the weighted average is equivalent to the normal average as the number of images in each class is same. For example: SEN for CXI model can be calculated by taking the average of SEN of individual class [47].
Experiments were conducted on a Dell OptiPlex 790 computer equipped with an Intel Core i7 processor running at 3.60 GHz, RAM of 32 GB, and Windows 10 operating system. The entire algorithm was implemented on MATLAB 2020b platform.
4. Results
Earlier studies [17,24] showed, that fusion ensemble based ResNet-50 CNN model gives a better performance out of maxima, minima, average, and fusion ensemble operations. In this work, the deep features obtained from SBIs are ensembled using fusion operations. Both CXI and CCTI databases were used to evaluate the proposed framework. The CXI database had three classes: COVID-19, pneumonia, and normal. The CCTI database had two classes: COVID-19 and non-COVID-19. The database was randomly divided into three parts: 85% for training purposes, 5% for validation purposes, and the remaining 10% for testing purposes. Total 1,446 CXIs (482 for each class) and 248 CCTIs (125 images for COVID-19 and 123 images for non-COVID-19) were employed for testing the model. Three major experiments were carried out in the current study. The first experiment was performed to find the best level of decomposition of FBSED method for diagnosis of COVID-19. Second experiment was performed to find the best CNN model, optimizer, and classifier. The best combination of channels was also analyzed in the second experiment. In the third experiment, the best proposed model was studied using 5-fold and 10-fold cross-validation processes. The proposed model was also used for classification of two-class CXI database, namely pneumonia class and pneumonia due to COVID-19 class.
4.1. Level of decomposition selection
In this experiment, the best level of decomposition was selected out of level-1, -2, and -3 decompositions based on hit-and-trial basis. Computational time increases with an increase in level of decomposition. If decomposition level-i provides better performance than decomposition level-, stopping further decomposition at level-i reduces the computational complexity. The classification results of the model implemented using level-i FBSED SBIs were compared with the results shown by the proposed model based on level-1 FBD SBIs (which was found best in Ref. [17]) and the model based on full images (images without decomposition). In this experiment, we used ResNet-50 along with Softmax classifier. During the training of the CNN model, number of epochs, initial learning rate, and optimizer are set to 20, 0.0003, and stochastic gradient descent with momentum (SGDM) [48], respectively. Table 2 shows the performance of level-1, -2, and -3 FBSEDs, level-1 FBD, and full image based COVID-19 diagnosis model for both CXI and CCTI databases.
Table 2.
Performance comparison of level-1, -2, and -3 FBSEDs.
| Channels |
PRE (%) |
SEN (%) |
SPE (%) |
F1-score |
ACC (%) |
AUC |
PRE (%) |
SEN (%) |
SPE (%) |
F1-score |
ACC (%) |
AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CXI database | CCTI database | |||||||||||
| Level-1 | ||||||||||||
| Channel-1 | 72.92 | 92.11 | 87 | 0.81 | 88 | 0.86 | 88 | 93.22 | 88.46 | 0.9 | 90.73 | 0.90 |
| Channel-2 | 79.17 | 90.48 | 90.20 | 0.84 | 90.28 | 0.9 | 80 | 92.59 | 82.14 | 0.85 | 86.69 | 0.87 |
| Channel-3 | 77.08 | 84.09 | 89 | 0.80 | 87.50 | 0.86 | 76.8 | 90.57 | 79.58 | 0.83 | 84.27 | 0.86 |
| Fusion | 89.58 | 87.76 | 94.74 | 0.88 | 92 | 0.98 | 91.20 | 94.21 | 91.34 | 0.92 | 92.74 | 0.98 |
| FBD | 79.17 | 82.61 | 89.80 | 0.80 | 87.50 | 0.95 | 86.40 | 85.71 | 86.67 | 0.86 | 85.89 | 0.92 |
| Level-2 | ||||||||||||
| Channel-1 | 87.50 | 91.30 | 93.88 | 0.89 | 93.06 | 0.96 | 80 | 95.24 | 82.52 | 0.86 | 87.96 | 0.88 |
| Channel-2 | 100 | 100 | 100 | 1 | 100 | 1 | 77.6 | 74.62 | 76.27 | 0.76 | 75.4 | 0.86 |
| Channel-3 | 100 | 100 | 100 | 1 | 100 | 1 | 70.40 | 73.33 | 71.09 | 0.71 | 72.18 | 0.81 |
| Channel-4 | 100 | 100 | 100 | 1 | 100 | 1 | 68.80 | 74.78 | 70.68 | 0.71 | 72.58 | 0.80 |
| Channel-5 | 100 | 100 | 100 | 1 | 100 | 1 | 68 | 69.1 | 68 | 0.68 | 68.55 | 0.80 |
| Channel-6 | 100 | 100 | 100 | 1 | 100 | 1 | 62.90 | 82.11 | 69.74 | 71.23 | 74.49 | 0.78 |
| Channel-7 | 100 | 100 | 100 | 1 | 100 | 1 | 72 | 70.31 | 70.83 | 0.71 | 70.56 | 0.80 |
| Channel-8 | 100 | 100 | 100 | 1 | 100 | 1 | 77.60 | 74.62 | 76.27 | 0.76 | 75.40 | 0.85 |
| Channel-9 | 100 | 100 | 100 | 1 | 100 | 1 | 88 | 95.65 | 88.72 | 0.91 | 91.94 | 0.94 |
| Fusion | 100 | 100 | 100 | 1 | 100 | 1 | 98.40 | 97.60 | 98.36 | 0.98 | 97.6 | 0.98 |
| Level-3 | ||||||||||||
| Channel-1 | 97.92 | 92.16 | 98.92 | 0.94 | 96.53 | 0.98 | 76.80 | 90.57 | 79.58 | 0.83 | 84.27 | 0.92 |
| Channel-2 | 89.58 | 97.73 | 95 | 0.93 | 95.83 | 0.97 | 76 | 63.76 | 69.7 | 0.69 | 66.13 | 0.78 |
| Channel-3 | 66.67 | 96.67 | 85.56 | 0.79 | 88.19 | 0.97 | 75.2 | 59.5 | 65.56 | 0.66 | 61.69 | 0.76 |
| Channel-4 | 83.33 | 80 | 91.49 | 0.81 | 87.50 | 0.97 | 82.40 | 57.2 | 67.65 | 0.67 | 60.08 | 0.78 |
| Channel-5 | 83.33 | 88.89 | 91.92 | 0.86 | 90.97 | 0.97 | 88.88 | 58.42 | 75.86 | 0.70 | 62.50 | 0.78 |
| Channel-6 | 75 | 85.71 | 88.24 | 0.8 | 87.50 | 0.95 | 27.2 | 80.95 | 55.83 | 40.72 | 60.08 | 0.66 |
| Channel-7 | 93.75 | 88.24 | 96.77 | 0.90 | 93.75 | 0.97 | 77.06 | 72.93 | 75.65 | 0.75 | 74.19 | 0.81 |
| Channel-8 | 90 | 91.84 | 94.85 | 0.90 | 93.84 | 0.97 | 80.80 | 74.26 | 78.57 | 0.77 | 76.21 | 0.83 |
| Channel-9 | 81.25 | 88.64 | 91 | 0.84 | 90.28 | 0.97 | 89.6 | 64 | 82.19 | 74.67 | 69.35 | 0.80 |
| Channel-10 | 87.5 | 89.36 | 93.81 | 0.88 | 92.36 | 0.97 | 72.8 | 80.53 | 74.81 | 0.76 | 77.42 | 0.80 |
| Channel-11 | 100 | 100 | 100 | 1 | 100 | 1 | 68 | 69.1 | 68 | 0.68 | 68.55 | 0.80 |
| Channel-12 | 100 | 100 | 100 | 1 | 100 | 1 | 62.90 | 82.11 | 69.74 | 71.23 | 74.49 | 0.78 |
| Channel-13 | 100 | 100 | 100 | 1 | 100 | 1 | 72 | 70.31 | 70.83 | 0.71 | 70.56 | 0.80 |
| Channel-14 | 100 | 100 | 100 | 1 | 100 | 1 | 77.60 | 74.62 | 76.27 | 0.76 | 75.40 | 0.81 |
| Channel-15 | 100 | 100 | 100 | 1 | 100 | 1 | 88 | 95.65 | 88.72 | 0.91 | 91.94 | 0.94 |
| Fusion | 100 | 100 | 100 | 1 | 100 | 0.99 | 92.80 | 89.92 | 92.44 | 0.91 | 91.13 | 0.97 |
| Full | 68.75 | 78.57 | 85.29 | 0.73 | 83.3 | 0.94 | 88 | 82.71 | 86 | 0.85 | 84.68 | 0.91 |
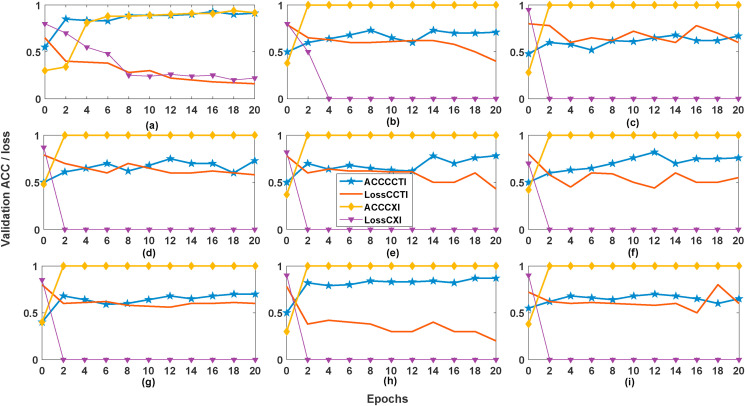
Table 2 shows that level-2 FBSED SBI model demonstrates better performance as compared to level-1 and -3 FBSED models. Fig. 10 shows the plot of validation ACC and validation loss versus epochs of level-2 FBSED SBI model for both CXI and CCTI databases. In Fig. 10, the abbreviations ACCCCTI and lossCCTI represent validation ACC and validation loss for the CCTI database. Similarly, the abbreviations ACCCXI and lossCXI represent validation ACC and validation loss for the CXI database. Table 2, also illustrates that level-1 FBSED SBI model provides better performance than level-1 FBD SBI model (or including diagonal component), and any of level-1, -2, and -3 FBSED based fusion ensemble models have provided better performance than the model based on full images.
Fig. 10.
Validation ACC and validation loss of CXI and CCTI database of different channels:(a) Channel-1, (b) Channel-2, (c) Channel-3, (d) Channel-4, (e) Channel-5, (f) Channel-6, (g) Channel-7, (h) Channel-8, (i) Channel-9.
4.2. Best CNN model, optimizer, classifier, and channel selection
In the experiment, level-2 FBSED SBIs were used to train different pre-trained CNN (preferred model which recently won the ImageNet Large Scale Visual Recognition Challenge), namely AlexNet, Inception-ResNet-v2, NASNet, and EfficientNet along with ResNet-50. Table 3 tabulates the performance of different pre-trained CNN models for level-2 FBSED. ResNet-50 and AlexNet perform better for the CXI database and ResNet-50 performs better for the CCTI database.
Table 3.
Performance comparison with different pre-trained CNN models.
| Model |
PRE (%) |
SEN (%) |
SPE (%) |
F1-score |
ACC (%) |
AUC |
PRE (%) |
SEN (%) |
SPE (%) |
F1-score |
ACC (%) |
AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CXI databse | CCTI database | |||||||||||
| ResNet-50 | 100 | 100 | 100 | 1 | 100 | 1 | 98.40 | 97.60 | 98.36 | 0.98 | 97.6 | 0.98 |
| AlexNet | 100 | 100 | 100 | 1 | 100 | 1 | 92.8 | 97.48 | 93.02 | 0.95 | 95.2 | 0.97 |
| Inception-ResNet-v2 | 89.58 | 89.58 | 94.79 | 0.89 | 93.06 | 0.97 | 84.17 | 85.59 | 84.17 | 0.84 | 84.87 | 0.88 |
| NASNet | 93.62 | 81.48 | 96.63 | 0.87 | 90.91 | 0.98 | 84 | 85.37 | 84 | 0.84 | 84.68 | 0.87 |
| EfficientNet | 79.17 | 82.61 | 89.80 | 0.80 | 87.50 | 0.94 | 81.60 | 83.61 | 82.17 | 0.82 | 82.87 | 0.87 |
For both the databases, the ResNet-50 offered the best results. Hence, the ResNet-50 model was further studied with different optimizers: root mean square propagation (RMSprop) [49], adaptive moment estimation (ADAM) [50], and SGDM. Table 4 shows performance of ResNet-50 CNN model using RMSprop, ADAM, and SGDM optimizers for both databases. The SGDM optimizer was found to be the best optimizer for ResNet-50.
Table 4.
Performance comparison with different optimizers.
| Optimizer |
PRE (%) |
SEN (%) |
SPE (%) |
F1-score |
ACC (%) |
AUC |
PRE (%) |
SEN (%) |
SPE (%) |
F1-score |
ACC (%) |
AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CXI database and ResNet-50 model | CCTI database and ResNet-50 model | |||||||||||
| SGDM | 100 | 100 | 100 | 1 | 100 | 1 | 98.40 | 97.60 | 98.36 | 0.98 | 97.6 | 0.98 |
| ADAM | 100 | 100 | 100 | 1 | 100 | 1 | 90.98 | 90.98 | 91.06 | 0.90 | 91.02 | 0.94 |
| RMSprop | 100 | 100 | 100 | 1 | 100 | 1 | 88 | 88 | 87.80 | 0.88 | 87.96 | 0.95 |
Feature extracted from level-2 FBSED SBIs with ResNet-50 (SGDM optimizer) was examined with different classifiers (RF, J48, Naive Bayes, AdaBoost, and Softmax). Table 5 shows the performance of different classifiers and highlights that the best classification performance was provided by Softmax classifier.
Table 5.
Performance comparison with different classifiers.
| Classifier |
PRE (%) |
SEN (%) |
SPE (%) |
F1-score |
ACC (%) |
AUC |
PRE (%) |
SEN (%) |
SPE (%) |
F1-score |
ACC (%) |
AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CXI database | CCTI database | |||||||||||
| RF | 100 | 100 | 100 | 1 | 100 | 1 | 90.83 | 90.83 | 92.13 | 0.9 | 91.53 | 0.96 |
| J48 | 100 | 100 | 100 | 1 | 100 | 1 | 87.04 | 86.24 | 88.98 | 0.86 | 87.71 | 0.88 |
| Naive Bayes | 100 | 100 | 100 | 1 | 100 | 1 | 86.92 | 85.32 | 88.98 | 0.86 | 87.29 | 0.95 |
| AdaBoost | 100 | 100 | 100 | 1 | 100 | 1 | 90.82 | 81.65 | 92.91 | 0.85 | 87.71 | 0.94 |
| Softmax | 100 | 100 | 100 | 1 | 100 | 1 | 98.40 | 97.60 | 98.36 | 0.98 | 97.6 | 0.98 |
Though the ensembling of features from channels shows a good performance, it is computationally expensive. In order to reduce the computational complexity, fewer number of channels were selected which also give better performance. Table 2 showed that for the CXI database, any single channel out of channel-2 and channel-9 could produce the same performance.
For CCTI database, on the basis of performance parameters like ACC and SEN, channels were arranged in decreasing order (Channel-9, -1, -2, -8, -3, -7, -5, and -6). Performance comparison for the combination of the different channels is shown in Table 6 . Combination of channel-9, -1, -2, and -8 outperformed the performance obtained by fusion of all channels. Since FBSED offers advantage of obtaining any level of SBIs independently, all such SBIs can be computed in a single step.
Table 6.
Performance comparison of CCTI database with different channels.
| Channels | PRE (%) | SEN (%) | SPE (%) | F1-score | ACC (%) | AUC |
|---|---|---|---|---|---|---|
| 9 | 88 | 95.65 | 88.72 | 0.91 | 91.94 | 0.94 |
| 9 and 1 | 94 | 95 | 94 | 0.95 | 95.16 | 0.96 |
| 9, 1, and 2 | 93.61 | 95.12 | 93.60 | 0.94 | 94.35 | 0.96 |
| 9, 1, 2, and 8 | 99.20 | 97.64 | 99.17 | 0.98 | 98.39 | 0.98 |
| 9, 1, 2, 8, and 3 | 96 | 93.02 | 95.8 | 0.94 | 94.35 | 0.96 |
| 9, 1, 2, 8, 3, and 7 | 95.02 | 93.02 | 95.8 | 0.94 | 94.35 | 0.96 |
| 9, 1, 2, 8, 3, 7, and 4 | 96 | 96 | 95.93 | 0.96 | 95.97 | 0.96 |
| 9, 1, 2, 8, 3, 7, 4, and 5 | 96.80 | 97.58 | 96.77 | 0.97 | 97.18 | 0.97 |
| 9, 1, 2, 8, 3, 7, 4, 5, and 6 | 98.40 | 97.60 | 98.36 | 0.98 | 97.6 | 0.98 |
The time taken for extracting a single channel from CXI and four channels from CCTI using FBSED was 0.06 and 0.15 s, respectively. Time taken to extract features from four SBIs of single CCTI from ResNet-50, AlexNet, Inception-ResNet-v2, NASNet, and EfficientNet model were 0.79, 0.86, 0.84, 0.88, and 0.62 s, respectively. Furthermore, time taken to extract features from one SBI of CXI database from ResNet-50, AlexNet, Inception-ResNet-v2, NASNet, and EfficientNet were 0.60, 0.66, 0.63, 0.67, and 0.56 s, respectively. Total time taken to test single image from CCTI database using proposed method was 0.943 s, which can be expressed as sum of image decomposition time (0.150 s), feature extraction time (0.790 s), and Softmax classifier time (0.003 s). Similarly, for CXI database total time required for testing a single image proposed method was 0.663 s. The time taken for testing one CCTI and CXI using full image model was 0.700 and 0.603 s, respectively. Time taken by full image model for classification of a single CXI and CCTI was slightly less than that of the proposed model, but the proposed model showed more ACC than full image model. The enhanced ACC provides sufficient justification to prefer our proposed method for the diagnosis of COVID-19 despite of the increased computational time.
4.3. Performance of proposed model at different K-fold cross-validation
The best combination of channels with ResNet-50 (which was trained using SGDM optimizer) and Softmax classifier were evaluated using 5-fold and 10-fold cross-validation processes. K-fold cross-validations were used in order to obtain reliable and unbiased classification performance of the model. In K-fold cross-validation, whole database was randomly divided into K equal sets. (K-1) sets were used to train CNN and remaining 1 set was used to validate the trained model. Same process was performed K number of times and then average of performance measure for all K experiments give final performance measure. Table 7 shows performance measures of proposed model for 5-fold and 10-fold cross-validation processes. Table 7 shows the average value and standard deviation of performance parameters. Performance measure of proposed method for classification of pneumonia due to COVID-19 from pneumonia is shown in Table 8 . Table 9 shows the performance comparison of the proposed method with the methods available in the literature for diagnosis of COVID-19 from CXI and CCTI databases.
Table 7.
Performance of proposed model at different K-fold cross-validation.
| K- Fold |
PRE (%) |
SEN (%) |
SPE (%) |
F1-score |
ACC (%) |
AUC |
PRE (%) |
SEN (%) |
SPE (%) |
F1-score |
ACC (%) |
AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CXI databse | CCTI database | |||||||||||
| 5-fold | 100 0 | 100 0 | 100 0 | 1 0 | 100 0 | 1 0 | 97.40 1.2 | 97 0.5 | 96.5 2 | 0.97 0.01 | 97.6 0.4 | 0.98 0 |
| 10-fold | 100 0 | 100 0 | 100 0 | 1 0 | 100 0 | 1 0 | 93.05 0.5 | 98.25 0.8 | 92.02 1 | 0.95 0 | 95 0.7 | 0.97 0 |
Table 8.
Performance of proposed model for CXI database for pneumonia cases and pneumonia due to COVID-19 classification.
| K- Fold | PRE (%) | SEN (%) | SPE (%) | F1-score | ACC (%) | AUC |
|---|---|---|---|---|---|---|
| 5-fold | 100 0 | 100 0 | 100 0 | 1 0 | 100 0 | 1 0 |
| 10-fold | 100 0 | 100 0 | 100 0 | 1 0 | 100 0 | 1 0 |
Table 9.
Performance comparison of proposed method with different works used for identification of COVID-19 using CXI and CCTI databases.
| Reference | Number of classes | Number of images | Train: Test: Valid | K-fold | PRE (%) | SEN (%) | SPE (%) | F1-score | ACC (%) | AUC |
|---|---|---|---|---|---|---|---|---|---|---|
| CCTI database | ||||||||||
| [21] | 2 | 2482 | 80:0:20 | – | 99.16 | – | – | 0.97 | 97.38 | 0.97 |
| [51] | 2 | 2945 | – | 5-fold | 99.20 | 98.80 | – | – | 98.99 | – |
| [52] | 2 | 2945 | – | 4-fold | 95.75 | – | – | 0.90 | 90.83 | 0.96 |
| [46] | 2 | 2482 | 60:0:40 | – | 98.74 | – | – | 98.14 | 0.98 | 0.98 |
| [53] | 2 | 2482 | 64:20:16 | – | 95 | – | – | 0.95 | 95 | – |
| Proposed method | 2 | 2482 | 85:10:5 | – | 98.40 | 97.60 | 98.36 | 0.98 | 97.6 | 0.98 |
| Proposed method | 2 | 2482 | – | 5-fold | 97.40 | 97 | 96.5 | 0.97 | 97.6 | 0.98 |
| CXI database | ||||||||||
| [3] | 3 | 1127 | – | 5-fold | 89.96 | 85.35 | – | 87.37 | 87.02 | – |
| [47] | 3 | 1251 | – | 4-fold | 90 | 96.4 | – | 0.87 | 89.06 | – |
| [12] | 2 | 406 | 70:0:30 | – | 96.77 | 100 | – | 0.98 | 98.33 | 0.98 |
| Proposed method | 3 | 1446 | 85:10:5 | – | 100 | 100 | 100 | 1 | 100 | 1 |
| Proposed method | 3 | 1446 | – | 5-fold and 10-fold | 100 | 100 | 100 | 1 | 100 | 1 |
5. Discussions
A total three experiments were performed in this study. In the first experiment, the best level of decomposition was selected. Performance of proposed method was compared with FBD based method and full image at each level. The results of the first experiment show that level-2 decomposition provides the best performance. In second experiment, level-2 SBIs obtained from FBSED were used to identify the best CNN model out of ResNet-50, AlexNet, Inception-ResNet-v2, NASNet, and EfficientNet, the best optimizer out of SGDM, ADAM, and RMSprop, the best classifiers out of RF, J48, Naive Bayes, AdaBoost, and Softmax classifier. The comparison revealed that ResNet-50 trained with SGDM optimizer and Softmax classifier gives the best results for both CXI and CCTI databases. For the proposed method, the channels were arranged to get better results with fewer number of SBIs which also reduces computation time. For the CXI database, any single channel between channel-2 to channel-9 is enough to get 100% classification performance. For the CCTI database, combination of channel-9, -1, -2, and -8 yields the best results with PRE, SEN, SPE, F1-score, ACC, and AUC of 98.40%, 97.60%, 98.36%, 0.98, 97.6%, and 0.98, respectively. The time required to classify one image from CXI and CCTI databases was 0.663 and 0.943 s, respectively. Finally in the third experiment, the model obtained from the previous experiments was evaluated using both 5-fold and 10-fold cross-validations. For CXI database, both 5-fold and 10-fold cross-validation processes provide the same results with 100% classification performance. For CCTI database 5-fold cross-validation provides the best results (average standard deviation) of PRE, SEN, SPE, F1-score, ACC, and AUC are 97.40 1.2%, 97 0.5%, 96.5 2%, 0.97 0.01%, 97.6 0.4%, and 0.98 0%, respectively. In this experiment, the classification performance of the proposed model for classification of pneumonia caused by COVID-19 from the broader pneumonia class of CXI database was studied. The study demonstrated that the classification performance of 5-fold and 10-fold cross-validation processes is 100%.
Generally CCTIs are found to be more suitable than CXIs for COVID-19 because a computer tomography (CT) scan offers significant level of detail by creating a 360 ∘ view of the body. This makes a CT scan preferable for emergency situations and for diagnostic purposes. In the proposed work, the results obtained from CXI database were better than the results obtained from the CCTI database. This may be due to the fact that the proposed framework was studied on several publicly available databases of two different imaging models (X-ray and CT), made up of different number of classes (two for CCTI and three for CXI), and of different patients. The deviation of our results from literature may be attributed to fact that X-ray scans are only able to detect COVID-19 in later stages of the disease in comparison with CT-scans because the imaging field of the lung is limited by the ribcage. Hence, the X-ray images classified as having COVID-19 will have more pronounced features than CT scans, thereby increasing the perceived effectiveness of X-rays.
6. Conclusion
FBSED is proposed in this paper for image analysis. FBSED uses grouping of FBSE coefficients for implementation of WPD where diagonal detail components are neglected. As the FBSE provides higher spectral resolution, one extra level of decomposition could be obtained using FBSED as compared to WPD. The FBSED uses grouping operation so any level of decomposition (or SBI) can be obtained in a single step. The ongoing COVID-19 pandemic has devastated the entire world and caused immense loss of life. Countries with more population are suffering due to the shortage of test kits and skilled personnel. Therefore, there is an urgent need for a fast, accurate, and contact-less diagnosis technique for COVID-19. In this work, both CXI and CCTI databases were used to diagnose COVID-19 using the FBSED method and pre-trained CNN with a classifier.
FBSED is used to decompose the CXI and CCTI into SBIs. From each SBI, deep features are extracted from different CNN models: ResNet-50, AlexNet, Inception-ResNet-v2, NASNet, and EfficientNet. The deep features are ensembled using fusion operations and finally fed to a different classifier: RF, J48, Naive Bayes, AdaBoost, and Softmax classifier. The study is done to find the best level of decomposition of FBSED, CNN model, classifier, optimizer method, and combination of channels for classification of COVID-19. It was observed that level-2 is the best level of decomposition, ResNet-50 is the best CNN model, SGDM is the best optimizer, and Softmax is the best classifier for both CXI and CCTI databases. Any channel out of channel-2 to channel-9 gives the best performance for CXI database and combination of channel-9, -1, -2, and -8 for CCTI database. With the proposed method, classification performance for the CXI database is 100%. For the CCTI database, PRE, SEN, SPE, F1-score, ACC, and AUC are 98.40%, 97.60%, 98.36%, 0.98, 97.6%, and 0.98, respectively. The 5-fold and 10-fold cross-validation were also used to evaluate our proposed framework. For CCTI database, 5-fold cross-validation process offered the best results. For CXI database, both 5-fold and 10-fold cross-validation processes provide the same results with 100% classification performance. For the CCTI database, 5-fold cross-validation process provided the best results with average standard deviation of PRE, SEN, SPE, F1-score, ACC, and AUC are 97.40 1.2%, 97 0.5%, 96.5 2%, 0.97 0.01%, 97.6 0.4%, and 0.98 0%, respectively. Performance of proposed model was also computed for two-class classification of CXI database: pneumonia class and pneumonia due to COVID-19 class. The 100% classification performance is obtained for both 5-fold and 10-fold cross-validation approaches. Thus, the proposed model can be a useful COVID-19 diagnostic technique for doctors. Despite having several advantages, FBSED based approach suffers from the problem of higher computational complexity due to the extra time taken in the image decomposition step. Although the computational time is high, the proposed method provides significant improvement in classification performance. Moreover, in this application, level of decomposition was restricted to level-2 so proposed method is feasible.
This work can be further extended to perform diagnosis for different stages of COVID-19. Additionally, instead of considering whole CXI and CCTI directly, segmented CXI and CCTI lesions can be used for diagnostic purposes. Segmentation may increase the performance of the proposed model but it will add an extra step which may affect the computational complexity. This methodology can also be modified to diagnose other diseases caused by infections, such as tuberculosis and influenza, from X-rays, CT scans and other imaging modalities.
CRediT authorship contribution statement
Pradeep Kumar Chaudhary: Resources, formal analysis, software, data curation, conceptualization, methodology, original draft preparation, reviewing and editing. Ram Bilas Pachori: Supervision, conceptualization, methodology, validation, visualization, Resources, reviewing and editing.
Declaration of competing interest
The authors, Pradeep Kumar Chaudhary and Ram Bilas Pachori, declare no conflict of interest.
Biographies

Pradeep Kumar Chaudhary received B.E. degree in Electrical and Electronics Engineering from Rajiv Gandhi Technological University, Bhopal, India in 2016 and M.Tech degree in Electrical Engineering from National Institute of Technology Hamirpur, India in 2018. Currently he is pursuing Ph.D in Electrical Engineering from Indian Institute of Technology Indore, Indore, India. His current research interests include medical signal processing, image processing, and machine learning. He has published several research papers for reputed international journals and conference papers. He served as a reviewer in Biomedical Signal Processing and Control and IEEE Sensor Journal.

Ram Bilas Pachori received the B.E. degree with honours in Electronics and Communication Engineering from Rajiv Gandhi Technological University, Bhopal, India in 2001, the M.Tech. and Ph.D. degrees in Electrical Engineering from Indian Institute of Technology (IIT) Kanpur, Kanpur, India in 2003 and 2008, respectively. He worked as a Postdoctoral Fellow at Charles Delaunay Institute, University of Technology of Troyes, France during 2007–2008. He served as an Assistant Professor at Communication Research Center, International Institute of Information Technology, Hyderabad, India during 2008–2009. He served as an Assistant Professor at Department of Electrical Engineering, IIT Indore, Indore, India during 2009–2013. He worked as an Associate Professor at Department of Electrical Engineering, IIT Indore, Indore, India during 2013–2017 where presently he has been working as a Professor since 2017. He is also an Associated Faculty with Department of Biosciences and Biomedical Engineering and Center for Advanced Electronics at IIT Indore. He was a Visiting Professor at School of Medicine, Faculty of Health and Medical Sciences, Taylor's University, Subang Jaya, Malaysia during 2018–2019. He worked as a Visiting Scholar at Intelligent Systems Research Center, Ulster University, Northern Ireland, UK during December 2014. He is an Associate Editor of Electronics Letters, Biomedical Signal Processing and Control journal and an Editor of IETE Technical Review journal. He is a senior member of IEEE and a Fellow of IETE and IET. He has supervised 12 Ph.D., 20 M.Tech., and 37 B.Tech. students for their theses and projects. He has 220 publications to his credit which include journal papers (132), conference papers (66), books (04), and book chapters (18). His publications have around 8000 citations with h index of 47 (Google Scholar, April 2021). He has been listed in the top h index scientists in the area of Computer Science and Electronics by Guide2Research website. He has been listed in the world's top 2% scientists in the study carried out at Stanford University, USA. He has served on review boards for more than 100 scientific journals and served for scientific committees of various national and international conferences. He has delivered more than 140 lectures in various conferences, workshops, short term courses, and institutes. His research interests are in the areas of Signal and Image Processing, Biomedical Signal Processing, Non-stationary Signal Processing, Speech Signal Processing, Brain-Computer Interfacing, Machine Learning, and Artificial Intelligence in Healthcare.
References
- 1.World Health Organization Coronavirus disease (COVID-19) (12 october 2020) https://www.who.int/emergencies/diseases/novel-coronavirus-2019/question-and-answers-hub/q-a-detail/coronavirus-disease-covid-19
- 2.WHO updates on COVID-19 Coronavirus disease (COVID-19) (3 April 2020) https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen/q-a-detail/coronavirus-disease-covid-19
- 3.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Experts explain the different COVID-19 tests: rapid antigen vs RT-PCR test, which is better? 07 August 2020. https://swachhindia.ndtv.com/experts-explain-the-different-covid-19-tests-rapid-antigen-vs-rt-pcr-test-which-is-better-48040/
- 5.S.-H. Wang, D. R. Nayak, D. S. Guttery, X. Zhang, Y.-D. Zhang, nCOVID-19 Classification by CCSHNet with Deep Fusion Using Transfer Learning and Discriminant Correlation Analysis, Information Fusion. [DOI] [PMC free article] [PubMed]
- 6.Kong W., Agarwal P.P. Chest imaging appearance of COVID-19 infection. Radiology: Cardiothoracic Imaging. 2020;2(1) doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee E.Y., Ng M.-Y., Khong P.-L. COVID-19 pneumonia: what has CT taught us? Lancet Infect. Dis. 2020;20(4):384–385. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., Diao K., Lin B., Zhu X., Li K., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020:200463. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., Zeng B., Li Z., Li X., Li H. Diagnosis of the coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.E. E.-D. Hemdan, M. A. Shouman, M. E. Karar, Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images, arXiv preprint arXiv:2003.11055.
- 11.Sethy P.K., Behera S.K. Detection of Coronavirus Disease (COVID-19) Based on Deep Features. Preprints. 2020 doi: 10.20944/preprints202003.0300.v1. [DOI] [Google Scholar]
- 12.Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: a comprehensive study. Biomed. Signal Process Contr. 2020;64:102365. doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., Lin Z.Q., Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest X-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.İmre A. A typical chest CT appearance of a case with Coronavirus Disease 2019 (COVID-19) Radiology. 2020:200490. [Google Scholar]
- 15.Ni Q., Sun Z.Y., Qi L., Chen W., Yang Y., Wang L., Zhang X., Yang L., Fang Y., Xing Z., et al. A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest CT images. Eur. Radiol. 2020;30(12):6517–6527. doi: 10.1007/s00330-020-07044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.L. Li, L. Qin, Z. Xu, Y. Yin, X. Wang, B. Kong, J. Bai, Y. Lu, Z. Fang, Q. Song, et al., Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy, Radiology 296 (2). [DOI] [PMC free article] [PubMed]
- 17.Chaudhary P.K., Pachori R.B. 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE; 2020. Automatic diagnosis of COVID-19 and pneumonia using FBD method; pp. 2257–2263. [Google Scholar]
- 18.L. S. V. R. Challenge, Imagenet http://www.image-net.org/challenges,LSVRC/2012/results.html.
- 19.J. P. Cohen, P. Morrison, L. Dao, COVID-19 image data collection, arXiv preprint arXiv:2003.11597.
- 20.Kaggle chest X-ray images (pneumonia) dataset. 2018. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
- 21.E. Soares, P. Angelov, S. Biaso, M. H. Froes, D. K. Abe, SARS-CoV-2 CT-scan Dataset: A Large Dataset of Real Patients CT Scans for SARS-CoV-2 Identification.
- 22.Shensa M.J. The discrete wavelet transform: wedding the a trous and Mallat algorithms. IEEE Trans. Signal Process. 1992;40(10):2464–2482. [Google Scholar]
- 23.Mallat S., Zhong S. Characterization of signals from multiscale edges. IEEE Trans. Pattern Anal. Mach. Intell. 1992;14(7):710–732. doi: 10.1109/34.142909. [DOI] [Google Scholar]
- 24.P. K. Chaudhary, R. B. Pachori, Automatic diagnosis of glaucoma using two-dimensional Fourier-Bessel series expansion based empirical wavelet transform, Biomed. Signal Process Contr. 64 102237.
- 25.Coifman R., Meyer Y., Wickerhauser M. 1992. Wavelet analysis and signal processing in Wavelets and Their Applications (ed. ruskai et al.)(1992), 153–178; and Size Properties of Wavelet Packets (1992) pp. 453–470. [Google Scholar]
- 26.Porter R., Canagarajah N. Robust rotation-invariant texture classification: wavelet, Gabor filter and GMRF based schemes. IEE Proc. Vis. Image Signal Process. 1997;144(3):180–188. [Google Scholar]
- 27.Manthalkar R., Biswas P.K., Chatterji B.N. Rotation and scale invariant texture features using discrete wavelet packet transform. Pattern Recogn. Lett. 2003;24(14):2455–2462. [Google Scholar]
- 28.Pachori R.B., Sircar P. EEG signal analysis using FB expansion and second-order linear TVAR process. Signal Process. 2008;88(2):415–420. [Google Scholar]
- 29.Schroeder J. Signal processing via Fourier-Bessel series expansion. Digit. Signal Process. 1993;3(2):112–124. [Google Scholar]
- 30.Gupta V., Pachori R.B. Epileptic seizure identification using entropy of FBSE based EEG rhythms. Biomed. Signal Process Contr. 2019;53:101569. [Google Scholar]
- 31.Pachori R.B., Sircar P. Analysis of multicomponent AM-FM signals using FB-DESA method. Digit. Signal Process. 2010;20(1):42–62. [Google Scholar]
- 32.Pisano E.D., Zong S., Hemminger B.M., DeLuca M., Johnston R.E., Muller K., Braeuning M.P., Pizer S.M. Contrast limited adaptive histogram equalization image processing to improve the detection of simulated spiculations in dense mammograms. J. Digit. Imag. 1998;11(4):193. doi: 10.1007/BF03178082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 34.Krizhevsky A., Sutskever I., Hinton G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM. 2017;60(6):84–90. [Google Scholar]
- 35.Christian S., Sergey I., Vincent V., Alexander A. Inception-v4 inception-resnet and the impact of residual connections on learning. AAAI. 2017;4 [Google Scholar]
- 36.Zoph B., Vasudevan V., Shlens J., Le Q.V. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. Learning transferable architectures for scalable image recognition; pp. 8697–8710. [Google Scholar]
- 37.M. Tan, Q. V. Le, Efficientnet: Rethinking model scaling for convolutional neural networks, arXiv preprint arXiv:1905.11946.
- 38.Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009;22(10):1345–1359. [Google Scholar]
- 39.Pal M. Random forest classifier for remote sensing classification. Int. J. Rem. Sens. 2005;26(1):217–222. [Google Scholar]
- 40.G. Kaur, A. Chhabra, Improved J48 classification algorithm for the prediction of diabetes, Int. J. Comput. Appl. 98 (22).
- 41.Rish I., et al. vol. 3. 2001. An empirical study of the naive Bayes classifier; pp. 41–46. (IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence). [Google Scholar]
- 42.Hastie T., Rosset S., Zhu J., Zou H. Multi-class adaboost. Stat. Interface. 2009;2(3):349–360. [Google Scholar]
- 43.Liu W., Wen Y., Yu Z., Yang M. Large-margin softmax loss for convolutional neural networks. ICML. 2016;2:7. [Google Scholar]
- 44.Buckland M., Gey F. The relationship between recall and precision. J. Am. Soc. Inf. Sci. 1994;45(1):12–19. [Google Scholar]
- 45.Huang J., Ling C.X. Using AUC and accuracy in evaluating learning algorithms. IEEE Trans. Knowl. Data Eng. 2005;17(3):299–310. [Google Scholar]
- 46.Y. Pathak, P. K. Shukla, K. Arya, Deep bidirectional classification model for COVID-19 disease infected patients, IEEE ACM Trans. Comput. Biol. Bioinf. [DOI] [PubMed]
- 47.Khan A.I., Shah J.L., Bhat M.M. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput. Methods Progr. Biomed. 2020:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutskever I., Martens J., Dahl G., Hinton G. International Conference on Machine Learning. 2013. On the importance of initialization and momentum in deep learning; pp. 1139–1147. [Google Scholar]
- 49.G. Hinton, N. Srivastava, K. Swersky, Neural networks for machine learning lecture 6a overview of mini-batch gradient descent, Cited on 14 (8).
- 50.D. P. Kingma, J. Ba, Adam: A method for stochastic optimization, arXiv preprint arXiv:1412.6980.
- 51.Silva P., Luz E., Silva G., Moreira G., Silva R., Lucio D., Menotti D. COVID-19 detection in CT images with deep learning: a voting-based scheme and cross-datasets analysis. Informatics in Medicine Unlocked. 2020;20:100427. doi: 10.1016/j.imu.2020.100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z., Liu Q., Dou Q. Contrastive cross-site learning with redesigned net for COVID-19 CT classification. IEEE Journal of Biomedical and Health Informatics. 2020;24(10):2806–2813. doi: 10.1109/JBHI.2020.3023246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Singh V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-scan images, Chaos. Solitons & Fractals. 2020;140:110190. doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]