Abstract
Proteomic analysis of urinary extracellular vesicles (EVs) is a powerful approach to discover potential bladder cancer (BCa) biomarkers, however urine contains numerous EVs derived from the kidney and normal urothelial epithelium, which can obfuscate information related to BCa cell‐derived EVs. In this study, we combined proteomic analysis of urinary EVs and tissue‐exudative EVs (Te‐EVs), which were isolated from culture medium of freshly resected viable BCa tissues. Urinary EVs were isolated from urine samples of 11 individuals (7 BCa patients and 4 healthy individuals), and Te‐EVs were isolated from 7 BCa tissues. We performed tandem mass tag (TMT)‐labeling liquid chromatography (LC‐MS/MS) analysis for both urinary EVs and Te‐EVs and identified 1960 proteins in urinary EVs and 1538 proteins in Te‐EVs. Most of the proteins identified in Te‐EVs were also present in urinary EVs (82.4%), with 55 of these proteins showing upregulated levels in the urine of BCa patients (fold change > 2.0; P < .1). Among them, we selected 22 membrane proteins as BCa biomarker candidates for validation using selected reaction monitoring/multiple reaction monitoring (SRM/MRM) analysis on urine samples from 70 individuals (40 BCa patients and 30 healthy individuals). Six urinary EV proteins (heat‐shock protein 90, syndecan‐1, myristoylated alanine‐rich C‐kinase substrate (MARCKS), MARCKS‐related protein, tight junction protein ZO‐2, and complement decay‐accelerating factor) were quantified using SRM/MRM analysis and validated as significantly upregulated in BCa patients (P < .05). In conclusion, the novel strategy that combined proteomic analysis of urinary EVs and Te‐EVs enabled selective detection of urinary BCa biomarkers.
Keywords: bladder cancer, exosome, extracellular vesicle, proteomics, urinary biomarker
Proteomic analysis of urinary extracellular vesicles (EVs) is a powerful approach to discovering potential BCa biomarkers, however urine contains numerous EVs derived from kidney and normal urothelial epithelium that could dilute the information of cancer BCa cell‐derived EVs. In this study, we performed combined proteomic analysis of both urinary EVs and tissue‐extracted EVs (Te‐EVs) to identify reliable BCa biomarkers. This novel strategy presented here identified reliable urinary EV biomarker proteins exhibiting high levels of specificity and sensitivity for non‐invasive BCa detection.
Abbreviations
- AUC
area under curve
- BCa
bladder cancer
- CD55
complement decay‐accelerating factor
- CE
collision energy
- CI
confidence interval
- D2O
deuterium oxide
- eHSP90
extracellular HSP90
- EV
extracellular vesicle
- GO
Gene Ontology
- HSP90
heat‐shock protein 90
- IHC
immunohistochemistry
- LC‐MS/MS
liquid chromatography‐tandem mass spectrometry
- MARCKS
myristoylated alanine‐rich C‐kinase substrate
- MARCKSL
MARCKS‐related protein
- MIBCa
muscle‐invasive bladder cancers
- NMIBCa
non‐muscle‐invasive bladder cancers
- NTA
NanoSight particle‐tracking analysis
- PTS
phase transfer surfactant
- ROC
receiver operating characteristic
- SDC1
syndecan‐1
- SI‐peptide
stable isotope‐labeled peptide
- SRM/MRM
selected reaction monitoring/multiple reaction monitoring
- TCGA
the Cancer Genome Atlas
- Te‐EV
tissue‐exudative extracellular vesicle
- TEM
transmission electron microscopy
- THP
Tamm‐Horsfall protein
- TJP2
tight junction protein ZO‐2
- TMT
tandem mass tag
- TURBT
transurethral resection of bladder tumor
1. INTRODUCTION
Bladder cancer (BCa) is one of the most common malignant epithelial tumors, with an estimated 24 300 new cases and 9500 deaths in Japan in 2020. 1 At initial diagnosis, approximately 30% of the cases are diagnosed as muscle‐invasive BCa (MIBCa), which are clinically aggressive, progress and metastasize rapidly, and are usually fatal, therefore screening for early detection of BCa is important. The remaining 70% cases are diagnosed as non‐muscle‐invasive BCa (NMIBCa), which do not typically pose a threat to survival, although ~30% to 50% of cases show recurrence, necessitating lifelong surveillance, which is a burden on patients. 2 , 3 Cystoscopy remains the gold standard for diagnosing this malignancy, but it is invasive and uncomfortable and has limited ability to detect flat lesions, such as carcinoma in situ. Moreover, urine cytology exhibits poor sensitivity for detecting low‐grade BCa and depends on the level of expertise of the pathologist for accurate interpretation. 4 To date, urinary protein biomarkers, such as NMP22, BTA, and BFP, have been approved for diagnostic purposes by the Japanese Ministry of Health, Labor, and Welfare. However, these markers are not widely adopted because of their limited sensitivity and/or specificity. Therefore, the development of accurate biomarkers for this disease is urgently needed.
EVs are lipid bilayer particles secreted by almost all cells into various bodily fluids; they play an important role in intercellular communication. 5 , 6 EVs harbor various bioactive molecules, including nucleic acids (miRNAs, RNAs, and DNA), proteins, and lipids that are characteristic of the host cells. 6 , 7 In particular, EVs derived from cancer cells reportedly contain cancer‐specific proteins capable of promoting tumor progression, survival, invasion, and angiogenesis. 8 The proteins in EVs are encapsulated in membrane vesicles, which protect them from proteases and are stable in biological fluids. 5 , 9 , 10 Urine is among the most accessible bodily fluids used for clinical diagnosis; it is consistently in contact with the bladder epithelial layer and can be collected noninvasively, making it an ideal specimen for detecting molecules associated with BCa. 11 , 12 , 13 Recent developments in quantitative proteomic technology have enabled large‐scale quantitation and validation of biomarker candidates. 14 , 15 Therefore, proteomic analysis of urinary EVs is a powerful approach to discover potential BCa biomarkers, with several reports focusing on urinary EV proteins. 16 , 17 , 18 , 19
However, urine contains a large number of EVs from the renal epithelium and normal urothelial epithelium, which can obfuscate information on cancer‐specific EVs, making proteomic analysis of urinary EVs cumbersome. To address this, we focused on tissue‐exudative extracellular vesicles (Te‐EVs), secreted and isolated from freshly resected tissue following a brief incubation in serum‐free medium. In contrast with bodily fluid samples, Te‐EVs harbor minimal contaminants, such as urine‐abundant proteins and whole‐body‐derived EVs. 20
We performed combined proteomic analysis for urinary EVs and Te‐EVs to identify potential urinary biomarker candidates for the diagnosis of BCa and then verified the candidate proteins.
2. MATERIALS AND METHODS
2.1. Patients and biological sample collection
In the discovery phase for TMT‐labeling liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) analysis, voided urine samples (>50 mL each) were collected from 7 BCa patients (NMIBCa [n = 3], MIBCa [n = 4]) who were TURBT at Osaka University Hospital (Osaka, Japan) and 4 healthy individuals. Healthy individuals were defined as those without a current malignant disease and a medical history of urinary cancer. All BCa patients were histologically diagnosed. Collected urine samples were kept at 4°C for up to 6 h before processing and then centrifuged at 2000 g for 30 min. After removing the pellets, the supernatants were stored at −80°C until subsequent processing. In addition to the urine samples, BCa tissue samples were also obtained following TURBT from the 7 BCa patients who provided urine samples.
In the validation phase for selected reaction monitoring/multiple reaction monitoring (SRM/MRM) analysis, urine samples were collected from 30 healthy individuals and 40 BCa patients (NMIBCa [n = 20], MIBCa [n = 20]) at Osaka University Hospital and Osaka General Medical Center, Japan. Approval was obtained from the Institutional Review Board before initiating the study, and all patients provided written informed consent. All investigations were performed following relevant guidelines and regulations.
Histological diagnosis was determined based on standard hematoxylin and eosin‐stained sections by experienced senior pathologists. Patients were staged according to the 7th AJCC TNM staging system, and tumors were graded according to the 2016 World Health Organization criteria. The urine cytology was also evaluated by specialists according to our strict institutional criteria, in which negative urine cytology is defined to be no more than class III, and positive urine cytology is defined to be classes IV and V.
2.2. Urinary EV isolation using ultracentrifugation on a 30% sucrose/deuterium oxide (D2O) cushion
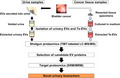
Ultracentrifugation on a 30% sucrose/D2O cushion was performed for urinary EV isolation as previously described. 21 Details are provided in Materials and Methods (Supporting Information). The procedure for urinary EV isolation is shown in Figure 1A. The protein concentration of urinary EVs was measured using a Micro BCA protein assay kit (Thermo Fisher Scientific).
FIGURE 1.
A, Isolation of urinary EVs using ultracentrifugation on a 30% sucrose/D2O cushion. B, Secretion and isolation of Te‐EVs. C, Experimental design of the discovery and validation of bladder cancer biomarkers. D2O, deuterium oxide; EVs, extracellular vesicles; Te‐EVs, tissue‐exudative extracellular vesicles
2.3. Te‐EV isolation
Following excision, BCa tissue samples were immediately immersed in 4 mL Dulbecco modified Eagle medium (Wako Pure Chemical Co.) without fetal bovine serum and stored at 4°C for 2 h. The tissue‐immersed medium was then centrifuged at 2000 g for 30 min at 4°C, and the collected supernatant was centrifuged at 17 000 g for 30 min at 4°C to remove cell debris and large EVs. The supernatant was filtered through a 0.22‐μm filter and transferred to a 5‐mL Ultra‐Clear Tube (Beckman Coulter), followed using ultracentrifugation at 100 000 g for 90 min at 4°C using an SW 55Ti rotor (ravg = 84.6 mm and adjusted k‐factor = 139.1; Beckman Coulter). The pellet was then washed with PBS and ultracentrifuged at 100 000 g for 90 min at 4°C using an SW 55Ti rotor (ravg = 84.6 mm and adjusted k‐factor = 139.1; Beckman Coulter), and the final pellet was resuspended in 100 μL PBS and frozen at −80°C. The procedure for Te‐EV secretion and isolation is shown in Figure 1B. The protein concentration of Te‐EVs was also measured using a Micro BCA protein assay kit (Thermo Fisher Scientific).
2.4. Sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and western blotting
The identity of EVs was established by the presence of specific surface proteins (CD9 and CD63), and the ability to remove the urinary non‐EV‐associated proteins using ultracentrifugation with a 30% sucrose/D2O cushion, evaluated by the absence of THP. Details are provided in Materials and Methods (Supporting Information).
2.5. NanoSight particle‐tracking analysis
The size and concentration of the isolated urinary EVs and Te‐EVs were analyzed using the NTA system (NanoSight). Details are provided in Materials and Methods (Supporting Information).
2.6. Transmission electron microscopy
TEM was performed as previously reported. 13 , 22 Details are provided in Materials and Methods (Supporting Information).
2.7. Solubilization and digestion of proteins in EVs
EV proteins (20 µg) were solubilized and digested using a PTS protocol. 23 Details are provided in Materials and Methods (Supporting Information).
2.8. TMT‐labeling
We used the TMT 10‐plex system (Thermo Fisher Scientific) for TMT‐labeling. Dried peptide samples were resuspended in 10 μL 100 mmol/L tetraethylammonium bicarbonate buffer and labeled with 4 μL 10‐plex TMT reagents for 1 h at room temperature. The reaction was quenched by adding 0.8 μL 5% hydroxylamine, and labeled samples were pooled and subjected to LC‐MS/MS analysis.
2.9. Shotgun proteomics
Digested and TMT‐labeled peptides were separated into 7 fractions using a C18‐SCX StageTip chromatography column according to a previously reported method, 24 , 25 and analyzed using a Q‐Exactive Plus mass spectrometer (Thermo Fisher Scientific) with an UltiMate 3000 Nano‐flow high‐performance LC system (Dionex) and an HTC‐PAL autosampler (CTC Analytics). Details are provided in Materials and Methods (Supporting Information). The values of each urinary EV sample were normalized according to the values of CD9 for deviations in EV collection from urine.
2.10. Selection of biomarker candidate proteins and target peptides for SRM/MRM analysis
We used the following criteria to select biomarker candidate proteins: (a) highly expressed proteins in urinary EVs from BCa patients (fold change > 2.0; P < .1); (b) proteins identified in Te‐EVs; and (c) membrane proteins reported in the UniProt Knowledgebase (UniProtKB). To verify candidate proteins as BCa biomarkers, we performed SRM/MRM analysis. Target peptides of biomarker candidate proteins were selected according to the following criteria: (a) peptides with sequences not shared among multiple genes; (b) peptides that were completely cleaved and had no methionine; (c) peptides less than 20 amino acids in length for higher sensitivities of SRM/MRM analysis. Stable isotope‐labeled internal standard peptides (SI‐peptide) with the same sequence as the selected peptide and a C‐terminal 15N‐ and 13C‐labeled arginine or lysine residue (isotopic purity > 99%) were purchased from JPT Peptide Technologies GmbH.
2.11. Target proteomics
SRM/MRM analyses were performed as previously described. 22 , 23 The digested peptides from 20 µg of EV proteins were dissolved in a 2% acetonitrile solution containing 0.1% trifluoroacetic acid and analyzed using a TSQ‐Vantage triple quadrupole mass spectrometer (Thermo Fisher Scientific) with a nano‐LC interface (AMR Alliance), Paradigm MS2 (Michrom BioResources), and an HTC‐PAL autosampler (CTC Analytics). The analytical column was packed with a reversed‐phase material (ReproSil‐Pur C18‐AQ; 1.9‐μm resin; Dr Maisch HPLC GmbH) into a self‐pulled needle (100‐mm length × 75‐μm i.d.). The mobile phases comprised buffers A (0.1% formic acid and 2% acetonitrile) and B (0.1% formic acid and 90% acetonitrile). Digested peptides were dissolved in buffer A with the SI‐peptide internal standard mixture before LC‐MS measurements and loaded onto a trap column (Acclaim PepMap RSLC Nano‐Trap column; 0.075 × 20 mm; Thermo Fisher Scientific). The nano‐LC gradient was delivered at 200 nL/min and comprised a linear gradient of buffer B developed from 5% to 35% over 75 min. The Instrument parameters were set as follows: 0.002 m/z scan width, 0.7 fwhm Q1 resolution, 2.5 s cycle time, and 1.8 mTorr gas pressure. Transition settings (pairs of precursor m/z and product m/z) to monitor a target peptide in SRM/MRM analysis were optimized for target peptides by performing a test run using the SI‐peptide mixture, as previously described. 26 The CE for each peptide was obtained using the following equations: CE = 0.034 × precursor m/z − 0.848 for double‐charged precursor ions; and CE = 0.022 × precursor m/z + 5.953 for triple‐charged precursor ions. Data acquisition was performed in scheduled SRM mode (time window: 10 min). The raw files acquired from SRM/MRM analyses were evaluated using Skyline software. 27 SRM signal peaks corresponding to each target peptide were assigned using comparison with an SI‐peptide internal standard for each counterpart. The quantitative values of the target peptides were obtained as ratios of the endogenous target peptides to the isotope‐labeled‐peptide internal standard using 1 transition per peptide with the highest signal. The obtained quantification values of each urinary EV sample were normalized according to the values of CD9 for deviations in EV collection from urine. If a protein had multiple detectable peptides, the target sequence with the highest intensity was adopted.
2.12. Immunohistochemistry (IHC)
Immunohistochemical staining was performed as previously reported. 28 Details are provided in Materials and Methods (Supporting Information). IHC score was evaluated by the percentage of stained tumor cells and staining intensity and was scored from 0 to 3+ (0: no staining, 1+: weak, 2+: moderate, 3+: strong). The average IHC score of the 3 randomly selected fields (400×) was evaluated as the final result. Twenty BCa tissues (NMIBCa [n = 10], MIBCa [n = 10]) in the validation cohort of this study were stained with anti‐HSP90, anti‐SDC1, and anti‐MARCKS, and the IHC score of each protein was compared between NMIBCa and MIBCa samples.
2.13. Survival analysis
We evaluated the association between the expression level of HSP 90, SDC1, and MARCKS, and overall survival using BCa patients in TCGA cohort. Patients were divided into 2 groups (high‐expression and low‐expression groups) according to the expression level of each protein, and survival curves were compared.
2.14. Statistical analysis
Identified EV proteins were analyzed using DAVID tools (https://david.ncifcrf.gov/) for GO annotation. 29 Statistical analyses were performed using JMP Pro software (v.14.0.0; SAS Institute), and visualization quantification was performed using GraphPad Prism software (v.7.05; GraphPad Software). Patient characteristics were compared using Fisher exact test and Mann‐Whitney U test. Univariate analysis was performed using Welch t test and Mann‐Whitney U test. The survival rates were determined using the Kaplan‐Meier method, and the log‐rank test was used for comparison. Differences were considered statistically significant at P < .05. The optimal cut‐off value for each EV protein was determined from the ROC curve using Youden index, and the sensitivity and specificity for detecting BCa using each EV protein were calculated according to each optical cut‐off value.
3. RESULTS
3.1. Isolation and characterization of urinary EVs and Te‐EVs
Figure 1C shows the study design. To explore the urinary EV proteome for detecting BCa biomarkers, we recruited a discovery cohort of 11 participants (4 healthy individuals and 7 BCa patients). Clinical and pathological information of patients is shown in Table 1.
TABLE 1.
Clinical and pathological information of patients in the discovery cohort
| HC (n = 4) | NMIBCa (n = 3) | MIBCa (n = 4) | |||
|---|---|---|---|---|---|
| Age (y), median (range) | 67.5 (42‐79) | 75 (67‐80) | 73 (66‐83) | ||
| Gender, n (%) | |||||
| Male | 3 (75.0) | 2 (66.7) | 4 (100) | ||
| Female | 1 (25.0) | 1 (33.3) | 0 (0) | ||
| Pathological T stage | – | pTa | pT2 | ||
| Pathological grade, n (%) | |||||
| Low grade | – | 2 (66.7) | 0 (0) | ||
| High grade | – | 1 (33.3) | 4 (100) | ||
| Te‐EVs | – | + | + | ||
Abbreviations: HC, healthy control; MIBCa, muscle‐invasive bladder cancer; NMIBCa, non‐muscle‐invasive bladder cancer; Te‐EVs, tissue‐exudative extracellular vesicles.
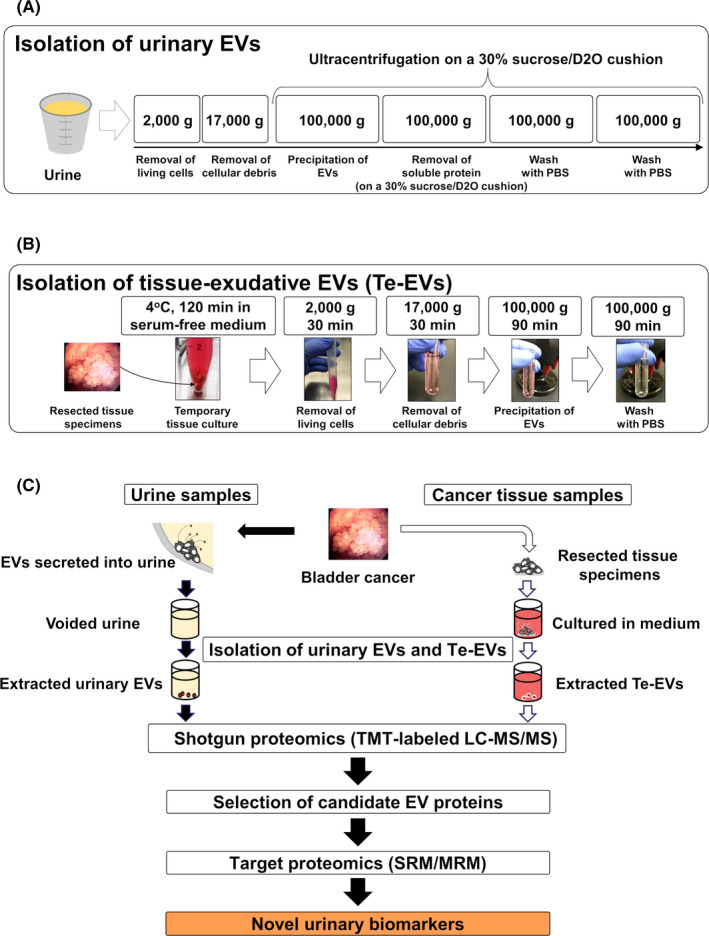
The quality of the isolated EVs was evaluated using western blot (Figure 2A,B), NTA (Figure 2C), and TEM (Figure 2D). Figure 2A shows that urinary THP can be removed using a 30% sucrose/D2O cushion. Our results showed that EVs were successfully isolated from urine using ultracentrifugation on a 30% sucrose/D2O cushion and from viable BCa tissues by short‐term culture, followed by ultracentrifugation.
FIGURE 2.
Verification of the quality of isolated urinary EVs and Te‐EVs. A, Western blot showing levels of EV‐specific proteins and THP in urinary EVs isolated using ultracentrifugation with or without a 30% sucrose/D2O cushion. B, Western blot showing levels of EV‐specific proteins in Te‐EVs. C, NTA revealing that almost all the extracted particles were <200 nm in size. D, Immunoelectron microscopy of urinary EVs and Te‐EVs immunolabeled with the anti‐CD9 antibody and secondary antibody conjugated to 20 nm of colloidal gold. BCa, bladder cancer; D2O, deuterium oxide; EVs, extracellular vesicles; NTA, NanoSight particle‐tracking analysis; Te‐EVs, tissue‐exudative extracellular vesicles; THP, Tamm‐Horsfall protein
3.2. TMT‐labeling LC‐MS/MS analysis of urinary EVs and Te‐EVs
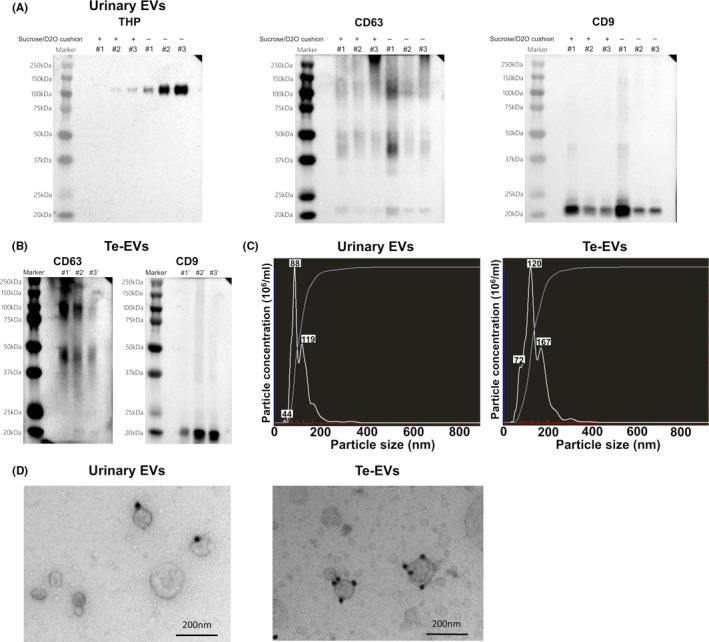
For BCa biomarker discovery, we performed TMT‐labeling LC‐MS/MS analysis of urinary EV and Te‐EV samples. We identified 1960 proteins in urinary EVs and 1538 proteins in Te‐EVs. GO analysis of cellular components revealed that the identified proteins in both urinary EVs and Te‐EVs were enriched in the “extracellular exosome” term (Figure 3A,B). Additionally, 82.4% (1268/1538) of the proteins identified in Te‐EVs were also detected in urinary EVs, and 64.7% (1268/1960) of those identified in urinary EVs were also detected in Te‐EVs (Figure 3C).
FIGURE 3.
GO annotation (cellular component) of identified proteins using shotgun proteomic analysis in (A) urinary EVs and (B) Te‐EVs. C, Venn diagram showing the distribution of EV proteins identified using shotgun proteomic analysis. EVs, extracellular vesicles; GO, Gene Ontology; Te‐EVs, tissue‐exudative extracellular vesicles
3.3. Verification of biomarker candidate proteins using SRM/MRM analysis
According to the criteria for biomarker candidate‐protein selection, we focused on highly expressed proteins in urinary EVs from BCa patients (fold change > 2.0; P < .1). Among them, 55 proteins, which were also identified in Te‐EVs, were selected as candidate biomarker proteins (Table S1). Considering the development of EV‐based clinical assays in the future, we focused on membrane proteins, with 22 of the 55 EV proteins identified as membrane proteins according to UniProtKB and subsequently selected as candidate biomarker proteins.
We then performed SRM/MRM analysis on the urinary EVs isolated from 70 individuals (30 healthy individuals and 40 BCa patients). Clinical and pathological information of patients is shown in Table 2. Target peptides of the 22 biomarker candidate proteins were then selected for SRM/MRM analysis, resulting in 21 target peptide sequences for 14 of the candidate proteins (Table S2). They were quantified in SRM/MRM analysis, and 13 target peptide sequences for 8 proteins were quantifiable. SRM/MRM transitions and quantified values of 13 peptides are shown in Tables S3 and S4.
TABLE 2.
Clinical and pathological information of patients in the validation cohort
| HC (n = 30) | BCa patients (n = 40) | P | |
|---|---|---|---|
| Age (y), median (range) | 57 (41‐74) | 71 (31‐87) | <.001 |
| Gender, n (%) | |||
| Male | 17 (56.7) | 29 (72.5) | .167 |
| Female | 13 (43.3) | 11 (27.5) | |
| Urine cytology, n (%) | |||
| Negative | 30 (100) | 20 (50.0) | <.001 |
| Positive | 0 (0) | 19 (47.5) | |
| Unknown | 1 (2.5) | ||
| Pathological T stage n, (%) | |||
| pTa | – | 20 (50.0) | |
| pT2 ≤ | – | 20 (50.0) | |
| Pathological grade, n (%) | |||
| Low grade | – | 12 (30.0) | |
| High grade | – | 28 (70.0) | |
Abbreviations: BCa, bladder cancer; HC, healthy controls.
SRM/MRM analysis revealed levels of 6 proteins (HSP90, SDC1, MARCKS, MARCKS‐related protein [MARCKSL], tight junction protein ZO‐2 [TJP2], and complement decay‐accelerating factor [CD55]) were significantly raised in urinary EVs from BCa patients compared with those from healthy individuals (P < .05; Mann‐Whitney U test) (Figure 4). ROC curve analysis indicated AUC values of these 6 urinary EV proteins for BCa diagnosis ranging from 0.706 to 0.813 (Figure 5). Among the 6 proteins, HSP90 had the highest AUC value (0.813, 95% confidence interval [CI]: 0.707‐0.918), with a sensitivity of 82.5% and specificity of 70.0%. The second highest AUC value was for SDC1 (0.785, 95% CI: 0.678‐0.892), with a sensitivity of 82.5% and specificity of 63.3%, followed by MARCKS (0.772, 95% CI: 0.663‐0.881), with a sensitivity of 65.0% and specificity of 80.0%. These urinary EV proteins showed better AUC values for BCa diagnosis than urine cytology (AUC: 0.744, 95% CI: 0.628‐0.859, with a sensitivity of 48.7% and specificity of 100%). The AUCs of HSP90, SDC1, and MARCKS were better for high‐grade BCa diagnosis (n = 28) than for low‐grade BCa diagnosis (n = 12) in SRM/MRM analysis (Figure S1). In addition, the levels of HSP90, SDC1, and MARCKS were significantly higher in urinary EVs from MIBCa patients than in those from NMIBCa patients (P < .05; Mann‐Whitney U test) (Figure S2). However, there was no association between overall survival and the expression levels of these proteins in BCa patients from the TCGA cohort (Figure S3).
FIGURE 4.
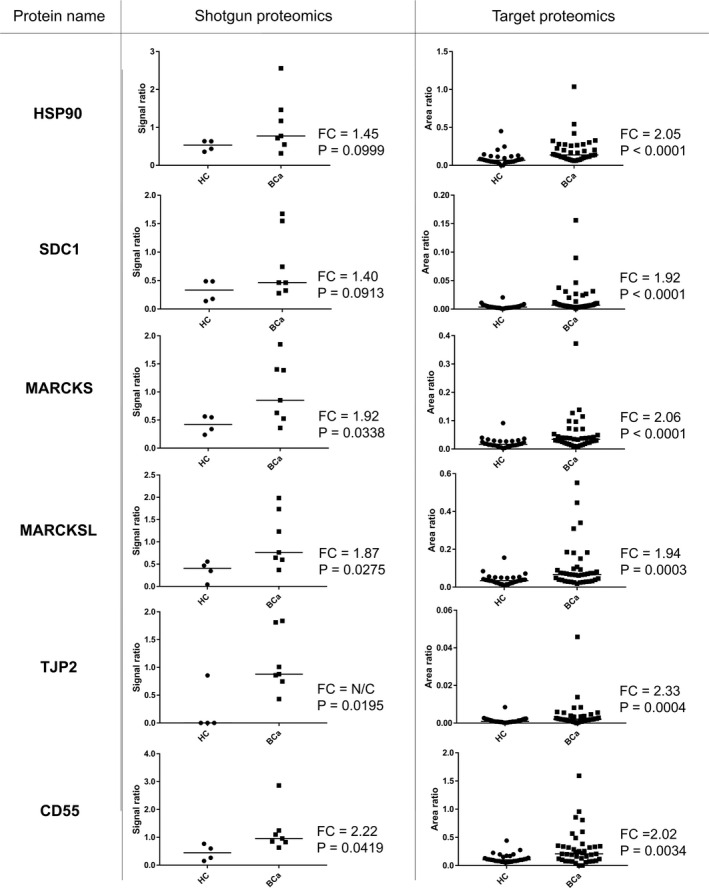
Relative quantitation data of identified BCa biomarker proteins in urinary EVs from HCs vs BCa patients using TMT‐labeling LC‐MS/MS and SRM/MRM analyses. FC is defined [median BCa]/[median HC]. BCa, bladder cancer; EVs, extracellular vesicles; FC, fold change; HC, healthy control; LC‐MS/MS, liquid chromatography‐tandem mass spectrometry; SRM/MRM, selected reaction monitoring/multiple reaction monitoring; Te‐EVs, tissue‐exudative extracellular vesicles; TMT, tandem mass tag
FIGURE 5.
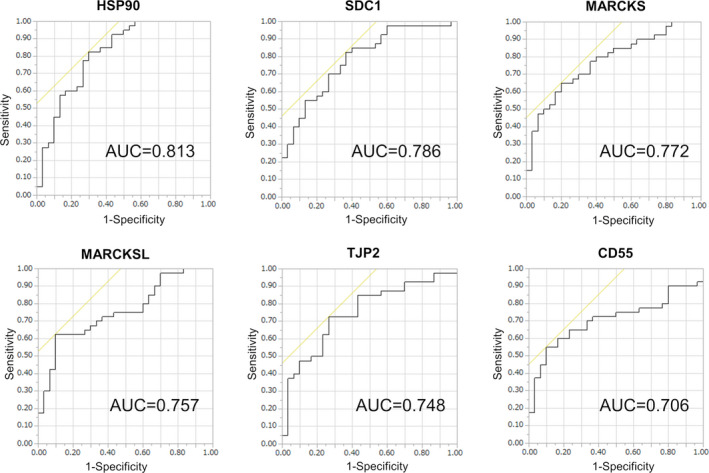
ROC curves for the 6 urinary EV proteins identified as potential BCa biomarkers according to SRM/MRM analysis. AUC value is shown on each graph. AUC, area under the curve; BCa, bladder cancer; EV, extracellular vesicle; ROC, receiver operating characteristic; SRM/MRM, selected reaction monitoring/multiple reaction monitoring
3.4. IHC analysis
Examples of strong (+3), moderate (+2), and weak (+1) staining for HSP90, SDC1, and MARCKS are shown in Figure 6A. Clinical and pathological information of patients is shown in Table S5. We evaluated the IHC scores and found no significant difference between NMIBCa and MIBCa samples in the IHC scores of HSP90, SDC1, and MARCKS (P = .222, P = .396, and P = .132, respectively; Mann‐Whitney U test) (Figure 6B). IHC analysis of BCa samples established that HSP90, SDC1, and MARCKS were present in BCa tissues regardless of NMIBCa or MIBCa, suggesting that they could be the source of these proteins in urinary EVs from BCa patients.
FIGURE 6.
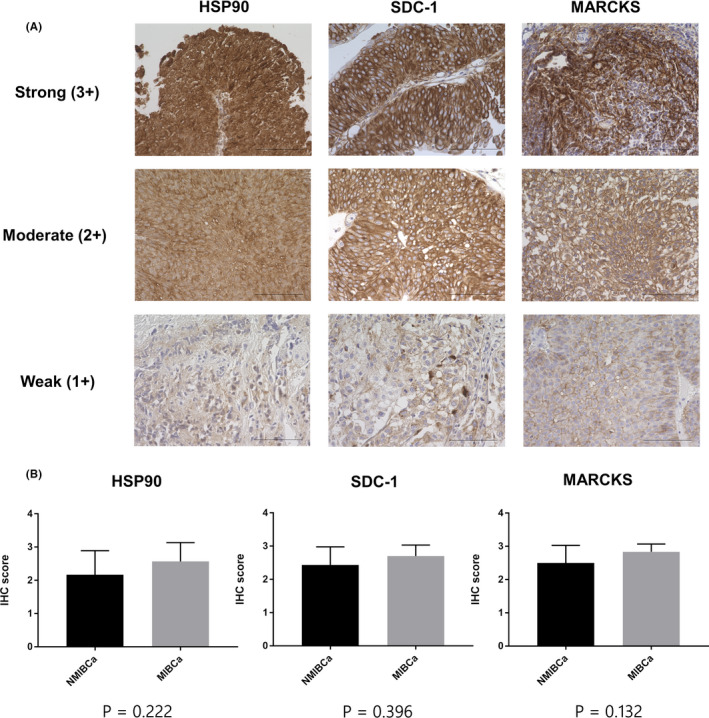
Immunohistochemical analysis of BCa. A, Typical patterns of strong (+3), moderate (+2), and weak (+1) staining of HSP90, SDC1, and MARCKS (scale bars, 100 μm). B, Comparison of IHC scores of HSP90, SDC1, and MARCKS between NMIBCa and MIBCa tissues. Data are presented as mean with SD. BCa, bladder cancer; HSP90, heat‐shock protein 90; MIBCa, muscle‐invasive bladder cancer; MARCKS, myristoylated alanine‐rich C‐kinase substrate; NMIBCa, non‐muscle‐invasive bladder cancer; SDC1, syndecan‐1
4. DISCUSSION
The presence of EVs in human urine was first demonstrated in 2004. 30 EVs are not only stable in urine but also harbor a potential wealth of diagnostic and prognostic information, therefore urinary EVs have recently garnered increased interest as a potential source of non‐invasive biomarkers for urological malignancies. 11 , 13 , 24 , 31
In this study, we performed an MS‐based quantitative proteomic analysis of both urinary EVs and Te‐EVs and identified reliable urinary EV biomarker proteins for BCa detection. To date, several studies have performed comparative proteomic analysis of urinary EVs in search of BCa biomarkers. 16 , 17 , 19 , 32 A study identified mucin‐1, carcinoembryonic antigen, epidermal growth factor receptor kinase substrate 8‐like protein 2, and moesin as BCa biomarkers according to comparative proteomic analysis results of urinary EVs from BCa patients and healthy individuals. 16 Using the same approach, other studies identified potential protein biomarkers, however urinary EVs acquired in these reports were isolated using ultracentrifugation alone, which tends to co‐isolate high levels of non‐EV‐associated proteins, such as THP. 33 , 34 , 35 Contaminating proteins can compete with EV protein identification by LC‐MS/MS analysis, thereby limiting the detection of low‐content EV proteins in urine and leading to false‐negative identification of cancer‐specific biomarkers. 31 , 35 Notably, it was reported that the proteomic profile differed significantly between urinary EVs isolated using ultracentrifugation alone and urinary EVs isolated using density fractionation in prostate cancer patients. 35 Therefore, in this study, we introduced an additional purification step using a sucrose/D2O cushion to isolate urinary EVs with minimal THP and other soluble protein contamination, 8 , 36 , 37 thereby increasing the reliability and integrity of the acquired proteomic data for biomarker discovery. We quantified 1960 proteins using deep proteomic analysis, representing the largest number of EV proteins detected in urine from BCa patients reported to date.
It should be noted that EVs are also secreted by the renal epithelium and normal urothelial epithelium, 30 , 38 and that not all urinary EVs necessarily reflect cancer‐specific EVs. We previously reported that the number of urinary EVs is strongly associated with urinary creatinine levels and that no difference was seen in particle counts in urinary EVs between BCa patients and healthy individuals. 13 These findings indicated that most urinary EVs were derived from the kidney and normal urothelial epithelium and that BCa‐derived EVs represented a very small population. Therefore, in this study, we performed an MS‐based quantitative proteomic analysis of both urinary EVs and Te‐EVs, finding that the majority of proteins identified in Te‐EVs was also present in urinary EVs (1268/1538; 82.4%), indicating urine as a suitable biofluid for detecting BCa‐specific EV proteins. In contrast, of the 1960 proteins identified in urinary EVs, only 1268 (64.7%) proteins were also present in Te‐EVs, suggesting that urinary EVs contain proteins unrelated to cancer. Therefore, exploring cancer‐specific EV proteins from multiple sources appeared to be an excellent strategy for identifying reliable biomarkers. To date, several studies have reported the results of proteomic analysis of EVs derived from BCa cell lines for the detection of cancer‐specific EVs, as well as their validation in urine samples. 39 , 40 , 41 However, the biosynthesis of cancer‐specific EVs is perhaps strongly influenced by the complex interactions between cancer cells and their surroundings, suggesting that Te‐EVs more accurately reflect the physiological characteristics and behavior of EVs in the human body compared with EVs derived from cultured cancer cells. 18 Therefore, the analysis of Te‐EVs might be more advantageous for discovering EV biomarkers. Presumably, this is the first comprehensive proteomic analysis of both urinary EVs and Te‐EVs for the discovery of BCa biomarkers.
During the verification phase, it is difficult to verify multiple biomarkers using antibody‐based immunoassays (eg, western blot or ELISA) as a result of the limited amount of extractable EV proteins from clinical urine samples. Recently, SRM/MRM analysis emerged as a powerful method for targeted protein quantification, enabling measurement of multiple proteins with high sensitivity and throughput without antibodies. Here, we identified the 6 urinary EV proteins as significantly upregulated in BCa patients and further evaluated the 3 most upregulated proteins (HSP90, SDC1, and MARCKS).
HSP90 is among the most abundantly expressed proteins in almost all nucleated cells and historically characterized as a cytoplasmic chaperone protein. 42 , 43 Intracellular HSP90 assists in the conformational activation of >200 client proteins under physiological and stress conditions. 44 , 45 Additionally, cancer cells use HSP90 to protect an array of mutated and overexpressed oncoproteins from misfolding and degradation, thereby characterizing HSP90 as a crucial facilitator of oncogene addiction and cancer cell survival. 43 Recent studies indicated that HSP90 is not confined to the intracellular environment but expressed on the surface of cancer cells 46 and secreted into the extracellular space as extracellular HSP90 (eHSP90), which acts differently from its intracellular counterpart. 47 Recently, it has been shown that the majority of eHSP90 protein is associated with tumor‐cell‐secreted EVs. 48 Moreover, highly metastatic oral‐cancer‐derived EVs are enriched with HSP90 compared with those from low metastatic oral cancers, 49 and they play an essential role in cancer‐malignancy properties, such as tumor invasion and metastasis. 50 HSP90 in EVs derived from BCa cells can show cancer‐promoting functions, thereby making it a potential therapeutic target.
SDC1 (CD138) is a cell‐surface proteoglycan expressed on various epithelial and vascular endothelial cells and involved in cell proliferation, adhesion, migration, epithelial morphogenesis, and angiogenesis. 51 , 52 SDC1 reportedly plays a key role in survival and progression of urothelial carcinoma cells. 53 Additionally, SDC1 is shed in a soluble form from cell surfaces through degradation of its extracellular domain by heparanases 54 and is a marker of systemic inflammation. 55 Moreover, soluble SDC1 reportedly exhibits tumor‐growth‐promoting effects and correlates with poor prognosis in lung cancer. 56 Because SDC1 was previously identified using proteomic analysis of EVs purified from cultured BCa cells, 57 it is reliable to assume that SDC1 is an EV protein derived from BCa cells.
MARCKS, a major substrate of protein kinase C, is a ubiquitously expressed protein that plays a critical role in cancer development and progression. 58 MARCKS shuttles between the membrane and the cytoplasm according to its phosphorylation status and affects the dynamics of membrane‐actin interactions. 59 , 60 Increases in MARCKS phosphorylation reportedly contribute to tumor‐cell survival, 58 with MARCKS identified as essential for tumor‐cell survival and differentially expressed between healthy and tumor tissues. These findings suggest that MARCKS is a potential target for cancer therapy. Furthermore, MARCKS was previously identified in EVs purified from cultured BCa cells, 56 supporting that MARCKS is a BCa‐specific EV protein.
This study has several limitations. First, ultracentrifugation is time consuming and SRM/MRM analysis requires expensive equipment, limiting its use beyond a scientific research setting. Therefore, it is necessary to develop a simple assay system with clinical value, such as ELISA or microfluidic chips capable of measuring potential EV protein biomarkers in a high‐throughput manner. Second, our cohort of non‐BCa patients did not include hematuria and cystitis patients. It is necessary to validate the biomarkers in these patients, as they are expected in clinical practice. Third, BCa is a heterogeneous disease, and selection bias could exist in the discovery phase. Patients with large tumor volume were selected for the discovery cohort, and patients with low‐grade BCa were not included, which might have excluded candidate biomarkers that were raised in low‐grade tumors. Fourth, the biological significance of high amounts of EV proteins as BCa biomarkers in urine was not investigated.
In conclusion, proteomic analysis of urinary EVs and Te‐EVs revealed that most of the proteins identified in Te‐EVs also present in urinary EVs. The novel strategy that combined proteomic analysis of urinary EVs and Te‐EVs enabled the identification of reliable urinary EV biomarker proteins (HSP90, SDC1, and MARCKS) for BCa detection. These BCa‐specific EV proteins represent both potential biomarkers and therapeutic targets. Our future work will focus on the development of a high‐throughput detection system of BCa biomarker EV proteins for clinical applications and the analysis of the function of these EV proteins.
DISCLOSURE
The authors have no conflict of interest.
Supporting information
Fig S1
Fig S2
Fig S3
Supplementary Material
Table S1‐S5
ACKNOWLEDGMENTS
This study was supported by the Japan Society for the Promotion of Science under KAKENHI (grant number JP17K16788) and the Japan Agency for Medical Research and Development under Translational Research (grant number A102).
Tomiyama E, Matsuzaki K, Fujita K, et al. Proteomic analysis of urinary and tissue‐exudative extracellular vesicles to discover novel bladder cancer biomarkers. Cancer Sci. 2021;112:2033–2045. 10.1111/cas.14881
Tomiyama and Matsuzaki contributed equally to this work.
REFERENCES
- 1. Projected Cancer Statistics, 2020 . Center for Cancer Control and Information Services, National Cancer Center. https://ganjoho.jp/reg_stat/statistics/stat/short_pred.html. Accessed February 5, 2021
- 2. Yoshida T, Kates M, Fujita K, Bivalacqua TJ, McConkey DJ. Predictive biomarkers for drug response in bladder cancer. Int J Urol. 2019;26(11):1044‐1053. 10.1111/iju.14082 [DOI] [PubMed] [Google Scholar]
- 3. Hayashi T, Fujita K, Hayashi Y, et al. Mutational landscape and environmental effects in bladder cancer. Int J Mol Sci. 2020;21(17):1‐14. 10.3390/ijms21176072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lotan Y, Roehrborn CG. Sensitivity and specificity of commonly available bladder tumor markers versus cytology: results of a comprehensive literature review and meta‐analyses. Urology. 2003;61(1):109‐118. 10.1016/S0090-4295(02)02136-2 [DOI] [PubMed] [Google Scholar]
- 5. Yáñez‐Mó M, Siljander P‐M, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;2015(4):1‐60. 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jia S, Zocco D, Samuels ML, et al. Emerging technologies in extracellular vesicle‐based molecular diagnostics. Expert Rev Mol Diagn. 2014;14(3):307‐321. 10.1586/14737159.2014.893828 [DOI] [PubMed] [Google Scholar]
- 8. Junker K, Heinzelmann J, Beckham C, Ochiya T, Jenster G. Extracellular vesicles and their role in urologic malignancies. Eur Urol. 2016;70(2):323‐331. 10.1016/j.eururo.2016.02.046 [DOI] [PubMed] [Google Scholar]
- 9. Nawaz M, Camussi G, Valadi H, et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol. 2014;11(12):688‐701. 10.1038/nrurol.2014.301 [DOI] [PubMed] [Google Scholar]
- 10. Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non‐exosomal miRNA in human urine. Kidney Int. 2014;86(2):433‐444. 10.1038/ki.2013.502 [DOI] [PubMed] [Google Scholar]
- 11. Linxweiler J, Junker K. Extracellular vesicles in urological malignancies: an update. Nat Rev Urol. 2020;17(1):11‐27. 10.1038/s41585-019-0261-8 [DOI] [PubMed] [Google Scholar]
- 12. Hayashi Y, Fujita K. A new era in the detection of urothelial carcinoma by sequencing cell‐free DNA. Transl Androl Urol. 2019;8(Suppl 5):S497‐S501. 10.21037/tau.2019.08.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsuzaki K, Fujita K, Jingushi K, et al. MiR‐21‐5p in urinary extracellular vesicles is a novel biomarker of urothelial carcinoma. Oncotarget. 2017;8(15):24668‐24678. 10.18632/oncotarget.14969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine‐reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3(12):1154‐1169. 10.1074/mcp.M400129-MCP200 [DOI] [PubMed] [Google Scholar]
- 15. Kume H, Muraoka S, Kuga T, et al. Discovery of colorectal cancer biomarker candidates by membrane proteomic analysis and subsequent verification using Selected Reaction Monitoring (SRM) and Tissue Microarray (TMA) Analysis. Mol Cell Proteomics. 2014;13(6):1471‐1484. 10.1074/mcp.M113.037093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee J, McKinney KQ, Pavlopoulos AJ, et al. Altered proteome of extracellular vesicles derived from bladder cancer patients urine. Mol Cells. 2018;41(3):179‐187. 10.14348/molcells.2018.2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smalley DM, Sheman NE, Nelson K, Theodorescu D. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J Proteome Res. 2008;7(5):2088‐2096. 10.1021/pr700775x [DOI] [PubMed] [Google Scholar]
- 18. Liu YR, Ortiz‐Bonilla CJ, Lee YF. Extracellular vesicles in bladder cancer: biomarkers and beyond. Int J Mol Sci. 2018;19(9):2822. 10.3390/ijms19092822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen C‐L, Lai Y‐F, Tang P, et al. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J Proteome Res. 2012;11(12):5611‐5629. 10.1021/pr3008732 [DOI] [PubMed] [Google Scholar]
- 20. Jingushi K, Uemura M, Ohnishi N, et al. Extracellular vesicles isolated from human renal cell carcinoma tissues disrupt vascular endothelial cell morphology via azurocidin. Int J Cancer. 2018;142(3):607‐617. 10.1002/ijc.31080 [DOI] [PubMed] [Google Scholar]
- 21. Tomiyama E, Fujita K, Nonomura N. Urinary extracellular vesicles. In: Salvi S, Casadio V, eds. Urinary Biomarkers: Methods and Protocols, Methods in Molecular Biology, vol. 2292. New York, NY: Humana Press; 2021:173‐181. [DOI] [PubMed] [Google Scholar]
- 22. Fujita K, Kume H, Matsuzaki K, et al. Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci Rep. 2017;7(February):1‐9. 10.1038/srep42961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shiromizu T, Kume H, Ishida M, et al. Quantitation of putative colorectal cancer biomarker candidates in serum extracellular vesicles by targeted proteomics. Sci Rep. 2017;7(1):1‐13. 10.1038/s41598-017-13092-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rappsilber J, Mann M, Ishihama Y. Protocol for micro‐purification, enrichment, pre‐fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2(8):1896‐1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- 25. Adachi J, Hashiguchi K, Nagano M, et al. Improved proteome and phosphoproteome analysis on a cation exchanger by a combined acid and salt gradient. Anal Chem. 2016;88(16):7899‐7903. 10.1021/acs.analchem.6b01232 [DOI] [PubMed] [Google Scholar]
- 26. Narumi R, Shimizu Y, Ukai‐Tadenuma M, et al. Mass spectrometry‐based absolute quantification reveals rhythmic variation of mouse circadian clock proteins. Proc Natl Acad Sci USA. 2016;113(24):E3461‐E3467. 10.1073/pnas.1603799113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maclean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966‐968. 10.1093/bioinformatics/btq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tomiyama E, Fujita K, Rodriguez Pena MDC, et al. Expression of nectin‐4 and pd‐l1 in upper tract urothelial carcinoma. Int J Mol Sci. 2020;21(15):1‐13. 10.3390/ijms21155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44‐57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 30. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101(36):13368‐13373. 10.1073/pnas.0403453101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fujita K, Nonomura N. Urinary biomarkers of prostate cancer. Int J Urol. 2018;25(9):770‐779. 10.1111/iju.13734IJU [DOI] [PubMed] [Google Scholar]
- 32. Lin S‐Y, Chang C‐H, Wu H‐C, et al. Proteome profiling of urinary exosomes identifies alpha 1‐antitrypsin and H2B1K as diagnostic and prognostic biomarkers for urothelial carcinoma. Sci Rep. 2016;6:34446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gonzales P, Pisitkun T, Knepper MA. Urinary exosomes: is there a future? Nephrol Dial Transplant. 2008;23(6):1799‐1801. 10.1093/ndt/gfn058 [DOI] [PubMed] [Google Scholar]
- 34. Rood IM, Deegens JKJ, Merchant ML, et al. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010;78 (8):810–816. 10.1038/ki.2010.262 [DOI] [PubMed] [Google Scholar]
- 35. Dhondt B, Geeurickx E, Tulkens J, et al. Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density‐based fractionation of urine. J Extracell Vesicles. 2020;9(1):1736935. 10.1080/20013078.2020.1736935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Raj DAA, Fiume I, Capasso G, Pocsfalvi G. A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int. 2012;81(12):1263‐1272. 10.1038/ki.2012.25 [DOI] [PubMed] [Google Scholar]
- 37. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30(1):3.22.1‐3.22.29. 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- 38. Turco AE, Lam W, Rule AD, et al. Specific renal parenchymal‐derived urinary extracellular vesicles identify age‐associated structural changes in living donor kidneys. J Extracell Vesicles. 2016;5(1):29642. 10.3402/jev.v5.29642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Welton JL, Khanna S, Giles PJ, et al. Proteomics analysis of bladder. Cancer. 2010;9:1324‐1338. 10.1074/mcp.M000063-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beckham CJ, Olsen J, Yin P‐N, et al. Bladder cancer exosomes contain EDIL‐3/Del1 and facilitate cancer progression. J Urol. 2014;192(2):583‐592. 10.1016/j.juro.2014.02.035 [DOI] [PubMed] [Google Scholar]
- 41. Silvers CR, Liu YR, Wu CH, Miyamoto H, Messing EM, Lee YF. Identification of extracellular vesicle‐borne periostin as a feature of muscle‐invasive bladder cancer. Oncotarget. 2016;7(17):23335‐23345. 10.18632/oncotarget.8024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein‐folding tool. J Cell Biol. 2001;154(2):267‐274. 10.1083/jcb.200104079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whitesell L, Lindquist SL. HSP90 and the chaperoning. Nat Rev Cancer. 2005;5:761‐772. 10.1038/nrc1716 [DOI] [PubMed] [Google Scholar]
- 44. Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10(8):537‐549. 10.1038/nrc2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li W, Tsen F, Sahu D, et al. Extracellular Hsp90 (EHsp90) as the Actual Target in Clinical Trials: Intentionally or Unintentionally. Vol 303. Oxford, UK: Elsevier; 2013. 10.1016/B978-0-12-407697-6.00005-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Becker B, Multhoff G, Farkas B, et al. Induction of Hsp90 protein expression in malignant melanomas and melanoma metastases. Exp Dermatol. 2004;13(1):27‐32. 10.1111/j.0906-6705.2004.00114.x [DOI] [PubMed] [Google Scholar]
- 47. Eustace BK, Sakurai T, Stewart JK, et al. Functional proteomic screens reveal an essential extracellular role for hsp90α in cancer cell invasiveness. Nat Cell Biol. 2004;6(6):507‐514. 10.1038/ncb1131 [DOI] [PubMed] [Google Scholar]
- 48. Tang X, Chang C, Guo J, et al. Tumour‐secreted Hsp90α on external surface of exosomes mediates tumour – stromal cell communication via autocrine and paracrine mechanisms. Sci Rep. 2019;9(1):1‐13. 10.1038/s41598-019-51704-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Taha EA, Ono K, Eguchi T. Roles of extracellular HSPs as biomarkers in immune surveillance and immune evasion. Int J Mol Sci. 2019;20(18):4588. 10.3390/ijms20184588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ono K, Sogawa C, Kawai H, et al. Triple knockdown of CDC37, HSP90‐alpha and HSP90‐beta diminishes extracellular vesicles‐driven malignancy events and macrophage M2 polarization in oral cancer. J Extracell Vesicles. 2020;9(1):1769373. 10.1080/20013078.2020.1769373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fujii T, Shimada K, Tatsumi Y, et al. microRNA‐145 promotes differentiation in human urothelial carcinoma through down‐regulation of syndecan‐1. BMC Cancer. 2015;15(1):1‐8. 10.1186/s12885-015-1846-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mennerich D, Vogel A, Klaman I, et al. Shift of syndecan‐1 expression from epithelial to stromal cells during progression of solid tumours. Eur J Cancer. 2004;40(9):1373‐1382. 10.1016/j.ejca.2004.01.038 [DOI] [PubMed] [Google Scholar]
- 53. Shimada K, Nakamura M, De Velasco MA, et al. Human urothelial carcinoma. Cancer Sci. 2010;101(1):155‐160. 10.1111/j.1349-7006.2009.01379.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ramani VC, Yang Y, Ren Y, Nan L, Sanderson RD. Heparanase plays a dual role in driving hepatocyte growth factor (HGF) signaling by enhancing HGF expression and activity. J Biol Chem. 2011;286(8):6490‐6499. 10.1074/jbc.M110.183277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Johansson PI, Stensballe J, Rasmussen LS, Ostrowski SR. A high admission syndecan‐1 Level, a marker of endothelial glycocalyx degradation, is associated with inflammation, protein C depletion, fibrinolysis, and increased mortality in trauma patients. Ann Surg. 2011;254(2):194‐200. 10.1097/SLA.0b013e318226113d [DOI] [PubMed] [Google Scholar]
- 56. Joensuu H, Anttonen A, Eriksson M, et al. Soluble syndecan‐1 and serum basic fibroblast growth factor are new prognostic factors in lung cancer. Cancer Res. 2002;62(18):5210‐5217. [PubMed] [Google Scholar]
- 57. Welton JL, Khanna S, Giles PJ, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9(6):1324‐1338. 10.1074/mcp.M000063-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fong LWR, Yang DC, Chen CH. Myristoylated alanine‐rich C kinase substrate (MARCKS): a multirole signaling protein in cancers. Cancer Metastasis Rev. 2017;36(4):737‐747. 10.1007/s10555-017-9709-6 [DOI] [PubMed] [Google Scholar]
- 59. Arbuzova A, Schmitz AAP, Vergères G. Cross‐talk unfolded: MARCKS proteins. Biochem J. 2002;362(1):1‐12. 10.1042/0264-6021:3620001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Laux T, Fukami K, Thelen M, Golub T, Frey D, Caroni P. GAP43, MARCKS, and CAP23 modulate PI(4,5)P2 at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J Cell Biol. 2000;149(7):1455‐1471. 10.1083/jcb.149.7.1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Supplementary Material
Table S1‐S5


