Abstract
KRAS is the most frequently mutated in ovarian endometriosis. However, it is unclear whether the KRAS mutant allele's mRNA is expressed and plays a biological role in ovarian endometriosis. Here, we performed mutation‐specific RNA in situ hybridization to evaluate mutant allele expression of KRAS p.G12V, the most frequently detected mutation in ovarian endometriosis in our previous study, in formalin‐fixed paraffin‐embedded tissue (FFPE) samples of ovarian endometriosis, cancer cell lines, and ovarian cancers. First, we verified that mutant or wild‐type allele of KRAS were expressed in all 5 cancer cell lines and 9 ovarian cancer cases corresponding to the mutation status. Next, we applied this assay to 26 ovarian endometriosis cases, and observed mutant allele expression of KRAS p.G12V in 10 cases. Mutant or wild‐type allele of KRAS were expressed in line with mutation status in 12 available endometriosis cases for which KRAS gene sequence was determined. Comparison of clinical features between ovarian endometriosis with KRAS p.G12V mutant allele expression and with KRAS wild‐type showed that KRAS p.G12V mutant allele expression was significantly associated with inflammation in ovarian endometriosis. Finally, we assessed the spatial distribution of KRAS mutant allele expression in 5 endometriosis cases by performing multiregional sampling. Intratumor heterogeneity of KRAS mutant allele expression was observed in two endometriosis cases, whereas the spatial distribution of KRAS p.G12V mutation signals were diffuse and homogenous in ovarian cancer. In conclusion, evaluation of oncogene mutant expression will be useful for clarifying the biological significance of oncogene mutations in benign tumors.
Keywords: endometriosis, in situ hybridization, intratumor heterogeneity, KRAS, mutant allele expression
We could visualize mutant allele expression of KRAS p.G12V in ovarian endometriosis epithelial cells harboring KRAS p.G12V mutation by using the in‐situ hybridization assay and revealed biological significance of KRAS mutant allele expression in ovarian endometriosis.
1. INTRODUCTION
Endometriosis affects 10%‐15% of women of reproductive age and causes various symptoms, such as chronic pelvic pain, dysmenorrhea and infertility, leading to a reduction in quality of life. 1 , 2 Another critical issue in the clinical management of women with endometriosis is malignant transformation. For example, there is epidemiological evidence that a personal history of endometriosis increases the risk of ovarian cancer 3 , 4 , 5 and clinical evidence that ovarian clear cell and endometrioid carcinomas are associated with endometriosis. 6 , 7 , 8 , 9 Furthermore, endometriosis itself exhibits potential for invasion or metastasis, including to the colon or lung. 10 , 11 Therefore, endometriosis is considered a precancerous lesion.
Several studies, including by our group, have recently reported that representative oncogenes of human cancers, such as KRAS and PIK3CA, are frequently mutated in ovarian endometriosis, 12 deep infiltrating endometriosis, 13 and iatrogenic endometriosis. 14 Moreover, those oncogenes have also been identified in normal endometrium. 12 , 15 , 16 , 17 , 18 In our study, KRAS was most frequently mutated in ovarian endometriosis, with 23/54 (42.6%) of endometriosis cases harboring somatic KRAS mutations. 12 In general, somatic mutations of oncogenes such as KRAS may be important driver events in the process of endometriosis arising from the normal uterine endometrium as well as in the malignant transformation of endometriosis. Indeed, all KRAS mutations detected to date in ovarian endometriosis are located in so‐called “hot spots”, suggesting that KRAS hotspot mutations have biological significance in endometriosis. 18 However, it has been still uncertain whether the mutated KRAS gene in endometriosis is expressed and affects the tumor phenotype, especially malignant transformation, because it is technically difficult to extract both DNA and RNA from a tiny amount of frozen samples of endometriotic epithelium in ovarian endometriosis. In addition, it is difficult to collect and store endometriotic epithelium samples from ovarian endometriosis in which the existence of endometriotic epithelial cells is extremely limited. Indeed, our previous report described that an average of over 100 endometriosis frozen sections were needed for laser microdissection for exome sequencing. 12 Visualization of oncogene mutations by using RNA‐based in situ hybridization is one solution to overcome this sampling issue because this assay does not require DNA/RNA extraction. Thus, we focused on this RNA‐based in situ hybridization assay to detect mutant allele expression in FFPE tissue samples of ovarian endometriosis. To clarify the biological significance of oncogene mutations in endometriosis, we targeted the KRAS p.G12V (c.G35T) mutation, which was the most frequent mutation in endometriotic epithelium in our previous study. 12 After we confirmed that detected KRAS p.G12V mutant allele expression was consistent with the existence of KRAS p.G12V mutation at the DNA level using ovarian cancer cell lines and ovarian cancer tissues, we applied it to ovarian endometriosis samples. The results revealed mutant allele expression of KRAS p.G12V in ovarian endometriosis and clarified that some ovarian endometriosis specimens exhibit intratumor heterogeneity with regard to oncogene mutations.
2. MATERIALS AND METHODS
2.1. Cancer cell lines
The human cancer cell lines used in this study are listed in Table S1. For each cell line, a cell block was prepared with HistoGelTM and used for subsequent analyses. The mutation status of each cell line was determined by Sanger sequencing (Figure S1).
2.2. Clinical tissue samples
This study was performed in conformity with the Declaration of Helsinki and approved by the institutional ethics review boards of Niigata University, Niigata Chuo General Hospital, and the National Institute of Genetics. All patients provided written informed consent for the collection of samples and subsequent analyses.
We enrolled nine ovarian carcinoma cases in this study to investigate the accuracy of in situ hybridization for clinical cancer tissue samples. For nine cases, we validated the KRAS mutation status of FFPE samples by Sanger sequencing because we had already performed target‐gene sequencing of frozen tissues and detected KRAS mutations in our previous study. 19
We collected 26 ovarian endometriosis FFPE cases with uncertain mutational status (Figure S2). Of them, six were retrospectively selected from our biobank. Although KRAS p.G12V mutation was detected in the frozen tissue samples of all six cases by exome or target‐gene sequencing in our previous study, 12 the KRAS mutation status of the FFPE samples used in this study was not determined because the FFPE and frozen samples were obtained from different endometriosis lesions of the same cases. As the other 20 cases were prospectively collected, the KRAS mutation status in these FFPE samples was unknown. Hematoxylin and eosin‐stained sections of all tissues used in this study were histologically reviewed by an experienced gynecologic pathologist (TM).
2.3. RNA‐based in situ hybridization assay
The BaseScopeTM assay was performed according to the protocol provided by the supplier (Advanced Cell Diagnostics, Newark, CA), with some modifications. FFPE sections (5 μm thick) were prepared on slides. The sections were baked at 60°C for 1 hour before deparaffinizing in xylene (2 × 5 minutes) and ethanol (2 × 4 minutes) and then dried by baking at 60°C for 30 minutes. Pretreat 1 (hydrogen peroxide) was applied for 10 minutes at RT, Pretreat 2 (target retrieval) for 15 minutes at 100°C and Pretreat 3 (protease III, cell block; protease plus, tissue sections) for 30 minutes at 40°C. Each probe was then applied for 2 hours at 40°C in a HybEZ oven before incubation with reagents AMP1 (30 minutes at 40°C), AMP2 (15 minutes at 40°C), AMP3 (15 minutes at 40°C), AMP4 (30 minutes at 40°C), AMP5 (30 minutes at 40°C), AMP6 (15 minutes at 40°C), AMP7 (60 minutes at RT) and AMP8 (15 minutes at RT). The slides were rinsed with wash buffer (2 × 2 minutes) between each AMP incubation. Finally, the slides were incubated with Fast Red for 10 minutes at room temperature in the dark. The slides were counterstained with Gill's hematoxylin before drying for 15 minutes at 60°C and mounting in VectaMount permanent mounting medium (Vector labs).
The RNAscope® assay was also performed according to the protocol provided by the supplier (Advanced Cell Diagnostics).
2.4. Validation of KRAS mutation by Sanger sequencing
To validate the mutation status of cell blocks and ovarian carcinoma FFPE samples, FFPE serial sections following the one used for RNA‐based in situ hybridization assay were prepared for Sanger sequencing. DNA was extracted using a QIAamp DNA FFPE Tissue Kit (QIAGEN Ltd.) according to the manufacturer's instructions. Polymerase chain reaction (PCR) was conducted using a KAPA Taq EXtra HotStart ReadyMix PCR Kit. PCR primers were designed using Primer3 software (http://bioinfo.ut.ee/primer3‐0.4.0/). The forward primer was AGCGTCGATGGAGGAGTTTG and the reverse primer CGTCCACAAAATGATTCTGAATTAGC. The PCR products were purified and sequenced by GENEWIZ (Saitama).
2.5. Laser microdissection and DNA extraction
Laser microdissection was performed to investigate KRAS mutation status in ovarian endometriosis epithelium (Figure S3). Ten‐micrometer‐thick serial FFPE sections that were used for RNA‐based in situ detection assays were cut with a microtome and mounted on RNase‐free foiled slides. The slides were deparaffinized, rehydrated and stained with hematoxylin and eosin before microdissection of mutant and wild‐type regions of the endometriosis epithelium. DNA was extracted using the QiaAMP DNA micro kit (QIAGEN Ltd.).
2.6. Target‐gene sequencing for endometriotic epithelial cells
Target‐gene sequencing of 76 genes in the endometriotic epithelium was performed with the precapture pooling method described in our previous studies. 12 , 20 We selected 12 endometriosis samples to guarantee the quality of results. The average depth was 109.7 reads, with coverage of at least 20 reads for 99.0%. Considering the average depth of each sample, we discuss only KRAS hotspot mutations (codons 12, 13, 61). We confirmed that the number of total reads for KRAS hot spots was sufficient in all 12 endometriosis samples and that there were no alteration reads on the base position of KRAS c.35G in endometriosis samples with wild‐type KRAS.
2.7. Immunohistochemical staining for p‐ERK protein expression
Immunohistochemical analysis of p‐ERK protein expression was performed using FFPE tissue sections. A monoclonal rabbit anti‐phospho‐p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) XP® antibody (#4370, Cell Signaling Technology) was used as the primary antibody. The FFPE tissue sections (4 µm), which were the serial sections used for RNA‐based in situ detection assays, were cut with a microtome. The sections were stained as previously described. 21 Briefly, after deparaffinization, antigen retrieval was carried out with Target Retrieval Solution (10 mM citrate buffer, pH 6.0; Dako) in a microwave for 20 minutes at 98°C. Subsequently, the sections were incubated with the primary antibody (1:700 dilution) overnight and biotinylated secondary antibodies (Vector Laboratories) for 1 hour, followed by incubation with ABC reagent (Dako) and 3,3'‐diaminobenzidine (Sigma‐Aldrich) for 3 minutes. The slides were counterstained with hematoxylin.
2.8. Mapping of KRAS mutant allele expression
For mutational mapping, slides on which we conducted RNA‐based in situ hybridization assays or immunohistochemistry were digitized using a Pannoramic 250 scanner (3D Histech). We manually separated the epithelium by color according to the in situ signals on each section using Adobe Photoshop CS8, with the definition that the clonal regions are sequential epithelial cells from the first positive cell to the last positive cell. 22 For mapping, we defined green, yellow, and red regions as follow: contiguous endometriosis epithelium is considered as one region. If epithelial cell with positive signal is 50 cells apart, the next contiguous region is considered a separate region. The cut‐off for proportion of wild‐type signals or mutation signals that are considered as positive is 2.0%. We defined the region with only wild‐type signals as blue, the region with a mixture of wild‐type signals as yellow and mutation signals and the region with only mutation signals as red.
2.9. Statistical analysis
We conducted all standard statistical tests with the R program (http://www.r‐project.org). We compared categorical variables between two groups by Fisher's exact test and continuous variables between two groups by the Wilcoxon rank‐sum test.
2.10. RNA sequencing
Total RNA was extracted from frozen samples using TRIzol (Invitrogen).
Total RNA was used for the library preparation, which was conducted using VAHTS Stranded mRNA‐seq Library Prep Kit for Illumina (Illumina) according to the manufacturer's protocol. The samples were sequenced on Illumina NovaSeq with the 2 × 150 bp module. The details of the procedure were the same as previously described. 23
3. RESULTS
3.1. RNA‐based in situ hybridization assay in human cell lines
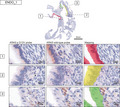
We used an RNA‐based in situ hybridization assay (BaseScopeTM) to detect mutant or wild‐type allele expression of KRAS in ovarian endometriosis. To confirm mutant or wild‐type allele expression of KRAS, we examined five human cancer cell lines (Table S1) with KRAS mutation status determined by Sanger sequencing (Figure S1). In particular, we selected two colorectal cancer cell lines that were analyzed using this assay in a previous report. 22 To detect KRAS mutation status correctly, we utilized four probes: the KRAS p.G12V probe, corresponding KRAS wild‐type, and positive and negative quality control probes (positive control, housekeeping gene PPIB; negative control, bacterial mRNA dapB). Because the in situ hybridization signal was labeled in red within the cell (Figure 1, Figure S4), we calculated the proportion of cells with positive signals in five fields per slide for each probe. In the two cell lines (SW620 and OVCAR5) harboring the KRAS p.G12V homozygous mutation, 37.2% and 30.2% of the total counted cells displayed positive signals when applying the KRAS p.G12V mutation probe but not the KRAS wild‐type probe (Table S1). In one cell line (SW626) carrying the KRAS p.G12V heterozygous mutation, 22.3% and 72.6% of the total counted cells were positive for both KRAS p.G12V mutation and wild‐type probes, respectively. Conversely, when using the KRAS p.G12V mutation probe in KRAS wild‐type cell lines (HT‐29 and SKOV3), no cells showed positive signals. These results demonstrate that mutant or wild‐type allele of KRAS were expressed according to the mutational status in cancer cell lines.
FIGURE 1.
RNA‐based in situ hybridization assay in ovarian cancer cell lines. Representative images of validation of the KRAS p.G12V probe‐set in ovarian cancer cell lines (a wild‐type cell line, a heterozygous mutant cell line, and a homozygous mutant cell line) using a negative control probe (dapB), a positive control probe (PPIB), the wild‐type probe and the mutant probe are shown. The probe signal is visualized as punctate red dots
3.2. RNA‐based in situ hybridization in ovarian cancer tissues
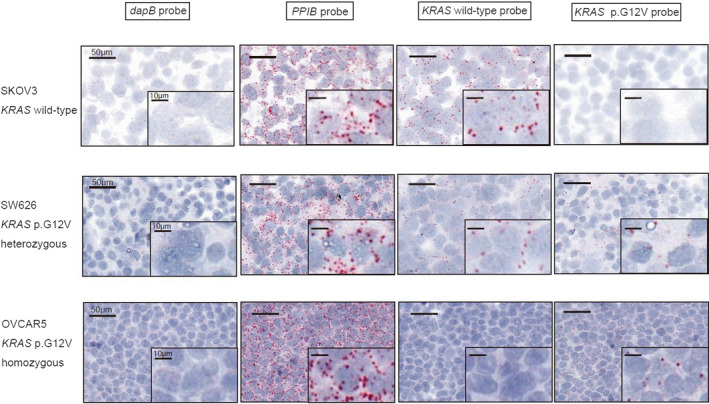
To detect mutant or wild‐type allele expression in clinical samples, we applied this assay to nine ovarian cancer FFPE samples with KRAS mutation status already determined in a previous study (Table S2). 19 A positive signal for the KRAS wild‐type probe was observed in both the epithelial and stromal cells of nine ovarian cancer samples, and the positivity rate was higher in two samples without KRAS mutation. However, KRAS p.G12V mutant allele expression was observed in epithelial cells but not stromal cells among the nine ovarian cancer samples. In ovarian cancer FFPE samples, mutant or wild‐type allele of KRAS were expressed in line with the mutation status. Six of seven samples harboring KRAS p.G12V mutation displayed both wild‐type and mutant KRAS p.G12V signals in the cancer cell area, indicating the presence of a heterozygous mutation. In contrast, one sample (OV9) displayed a predominance of KRAS p.G12V mutant signals in cancer cells, potentially reflecting the KRAS homozygous mutation or mutation with allelic imbalance (Figure 2). Indeed, the positivity rate of cells for the KRAS p.G12V mutation probe in this tissue was 34.6%, similar to the two cancer cell lines with KRAS p.G12V homozygous mutation. In the other samples, the rate of cell positivity for KRAS p.G12V mutation signals varied from 4.0% to 20.7%. These findings indicate that the positivity rate of the KRAS p.G12V mutation probe might be affected by not only the KRAS mutant allele frequency (MAF) at the DNA level but also the quality of the analyzed samples or mutant allele expression of KRAS gene at the RNA level. Most importantly, the spatial distribution of KRAS p.G12V mutation signals was diffuse and homogenous in ovarian cancer samples (Figure S5).
FIGURE 2.
RNA‐based in situ hybridization assay in ovarian cancer cases with known mutational status. Representative images of the validation of the KRAS p.G12V probe‐set using FFPE samples of ovarian cancer (ovarian endometrioid carcinomas with KRAS wild‐type or KRAS p.G12V) using a negative control probe (dapB), a positive control probe (PPIB), the wild‐type probe and the mutant probe are shown. Probe binding is visualized as punctate red dots
To confirm that no cross‐reactivity of the KRAS p.G12V probe with other KRAS mutations occurred, we performed this assay for one ovarian cancer sample harboring KRAS p.G12D (c.G35A) with a high MAF (0.93) but not KRAS p.G12V. As shown in Figure S6, no KRAS p.G12V mutation signal was observed in the KRAS p.G12D‐mutated ovarian cancer sample.
3.3. Mutant allele expression of KRAS p.G12V in retrospectively collected ovarian endometriosis cases
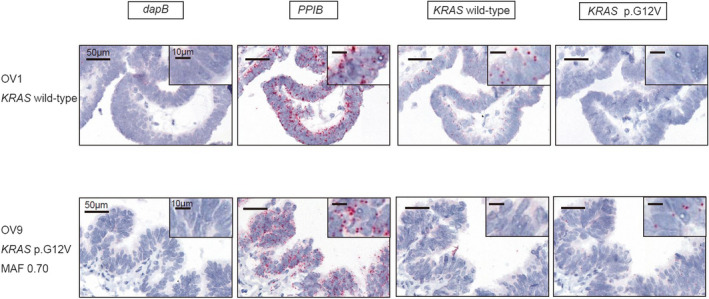
To visualize mutant allele expression of KRAS p.G12V, we selected six retrospectively collected ovarian endometriosis cases (Figure S2). Although we detected KRAS p.G12V mutation in frozen tissue samples obtained from the six cases in our previous study, we could not use FFPE samples adjacent to the frozen tissue samples for the in situ assay. Therefore, FFPE samples of different sites in the same cases were prepared. In four of the six cases, we were able to visualize mutant allele expression of KRAS p.G12V in ovarian endometriotic epithelial cells (Figure 3), whereas only KRAS wild‐type expression in ovarian endometriosis was detected in the other two cases. FFPE and frozen samples were obtained from different endometriosis lesions of the same case, and endometriotic epithelial cells with the KRAS p.G12V mutation might not necessarily be expanded in the entire ovarian endometriotic cyst or expression levels of the KRAS p.G12V mutant allele were low. The proportion of positive cells in ovarian endometriosis cases for each probe is shown in Table S3.
FIGURE 3.
Visualized KRAS p.G12V expression with RNA‐based in situ hybridization assay in ovarian endometriosis epithelial cells. KRAS p.G12V signals and KRAS wild‐type signals are detected in ovarian endometriosis epithelial cells in the upper figure and stromal cells in the lower figure, respectively
3.4. Mutant allele expression of KRAS p.G12V in prospectively collected ovarian endometriosis cases
Next, we assessed expression of KRAS p.G12V in 20 cases collected prospectively. Six of the 20 cases (30.0%) showed KRAS p.G12V expression in ovarian endometriotic epithelial cells (Figure 4, Table S3). After we performed microdissection of endometriotic epithelial cells and target‐gene sequencing for 12 available cases, KRAS p.G12V was detected in all five cases with KRAS p.G12V signals. Of the seven cases without KRAS p.G12V signal, five showed wild‐type KRAS, and the other two harbored KRAS mutations other than KRAS p.G12V. Importantly, mutation status based on target‐gene sequencing corresponded to that based on the in situ hybridization assay (Table 1).
FIGURE 4.
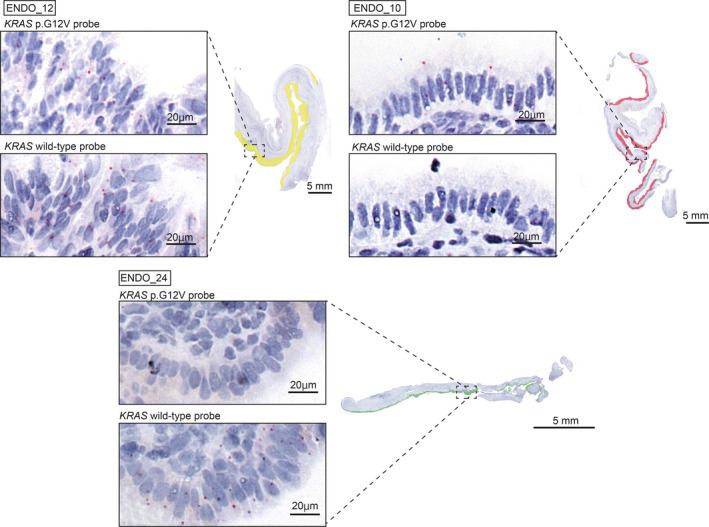
Three patterns of KRAS p.G12V expression in ovarian endometriosis. Whole images of KRAS p.G12V and wild‐type probe signals in three ovarian endometriosis samples are displayed. The areas of endometriotic epithelial cells with only wild‐type signals, both wild‐type and mutational signals, and predominant mutational signals are mapped in green, yellow, and red, respectively
TABLE 1.
Validation of expected mutational status from RNA‐based in situ hybridization analysis with target‐gene sequencing
| Case | RNA‐based in situ hybridization analysis | Target‐gene sequencing | |||||
|---|---|---|---|---|---|---|---|
| dapB | PPIB | KRAS wild‐type | KRAS p.G12V | Expected mutational status of KRAS | Mutation Status of KRAS (MAF) | Mutation depth alt/ref (total read) | |
| ENDO_7 | 0/501 (0.0) | 491/501 (98.0) | 143/492 (29.1) | 30/736 (4.1) | KRAS p.G12V | KRAS p.G12V (0.20) | 23/107 (130) |
| ENDO_8 | 1/981 (0.1) | 495/522 (94.8) | 237/945 (25.1) | 40/822 (4.9) | KRAS p.G12V | KRAS p.G12V (0.29) | 48/117 (165) |
| ENDO_9 | 1/999 (0.1) | 206/210 (98.1) | 101/973 (10.4) | 79/878 (9.0) | KRAS p.G12V | KRAS p.G12V (0.42) | 42/58 (100) |
| ENDO_10 | 0/504 (0.0) | 506/506 (100.0) | 8/520 (1.5) | 76/491 (15.5) | KRAS p.G12V | KRAS p.G12V (0.49) | 61/64 (125) |
| ENDO_12 | 0/1210 (0.0) | 1202/1202 (100.0) | 618/1358 (45.4) | 242/1163 (20.8) | KRAS p.G12V | KRAS p.G12V (0.34) | 53/101 (154) |
| ENDO_15 | 0/690 (0.0) | 715/715 (100.0) | 164/708 (23.2) | 0/669 (0.0) | no KRAS p.G12V | wild‐type | |
| ENDO_20 | 0/440 (0.0) | 610/610 (100.0) | 124/456 (27.2) | 0/407 (0.0) | no KRAS p.G12V | KRAS p.G12A (0.18) | 23/104 (127) |
| ENDO_21 | 1/470 (0.2) | 400/410 (97.6) | 197/619 (31.8) | 0/413 (0.0) | no KRAS p.G12V | wild‐type | |
| ENDO_22 | 0/329 (0.0) | 430/430 (100.0) | 127/393 (32.3) | 0/423 (0.0) | no KRAS p.G12V | wild‐type | |
| ENDO_24 | 0/695 (0.0) | 233/233 (100.0) | 267/741 (36.0) | 0/729 (0.0) | no KRAS p.G12V | KRAS p.G12D (0.36) | 64/114 (178) |
| ENDO_25 | 0/325 (0.0) | 298/310 (96.1) | 154/364 (42.3) | 0/350 (0.0) | no KRAS p.G12V | wild‐type | |
| ENDO_26 | 0/578 (0.0) | 466/466 (100.0) | 265/618 (42.9) | 0/577 (0.0) | no KRAS p.G12V | wild‐type | |
Positive cells / total counted cells (%).
3.5. Comparison of clinical features between ovarian endometriosis with KRAS p.G12V mutant allele expression and KRAS wild‐type
To clarify the significance of KRAS p.G12V mutant allele expression in ovarian endometriosis, we compared the clinical features of ovarian endometriosis cases with KRAS p.G12V mutation expression to those with KRAS wild‐type (Table 2). We defined ten cases with KRAS p.G12V mutation expression as KRAS p.G12V group and five cases which showed both only KRAS wild‐type allele expression and KRAS wild‐type status by target‐gene sequencing as KRAS wild‐type group. Interestingly, preoperative findings representing inflammation tends to be high in KRAS p.G12V group, especially CRP level was significantly higher in KRAS p.G12V group than KRAS wild‐type group (P =.024). Moreover, rASRM (The revised American Society for Reproductive Medicine) score which is a classification of the endometriosis stage scored according to the size and extent region and the degree of adhesion 24 tended to be higher in KRAS p.G12V group than KRAS wild‐type group (P =.053).
TABLE 2.
Comparison of clinical features between ovarian endometriosis with KRAS p.G12V mutant allele expression and KRAS wild‐type
| Characteristics | KRAS p.G12V (10) | KRAS wild‐type (5) | P‐value |
|---|---|---|---|
| Age at operation(SD) a | 43.1 (7.0) | 44.8 (4.3) | .533 |
| Parity b | 4/10 (40.0%) | 1/5 (20.0%) | .600 |
| Null para | 6/10 (60.0%) | 4/5 (80.0%) | |
| Multi para | 4/10 (40.0%) | 1/5 (20.0%) | |
| Preoperative treatment b | 1/10 (10.0%) | 2/5 (40.0%) | .537 |
| DNG | 1/10 (10.0%) | 1/5 (20.0%) | |
| GnRHa | 0/10 (0.0%) | 1/5 (20.0%) | |
| Pre‐operation CA125 (U/mL) a | 119.6 ± 69.0 | 56.0 ± 67.0 | .240 |
| Tumor size (mm) a | 61.8 ± 20.1 | 60.2 ± 30.0 | .972 |
| Laterality b | .231 | ||
| Unilateral | 6/10 (60.0%) | 5/5 (100.0%) | |
| Bilateral | 4/10 (40.0%) | 0/5 (0.0%) | |
| rASRM score a | 78.5 ± 41.3 | 33.6 ± 8.9 | .053 |
| rASRM stage b | .119 | ||
| III | 3/10 (30.0%) | 4/5 (80.0%) | |
| IV | 7/10 (70.0%) | 1/5 (20.0%) | |
| Preoperative laboratory findings | |||
| WBC a | 9596 ± 6160 | 5978 ± 2259 | .254 |
| CRP a | 2.70 ± 5.25 | 0.03 ± 0.04 | .024 |
| Neutrophil to lymphocyte ratio a | 4.5 ± 3.2 | 1.8 ± 0.4 | .099 |
| Fever b | 4/10 (40.0%) | 0/5 (0%) | .231 |
Wilcoxon exact rank test.
Fisher exact test.
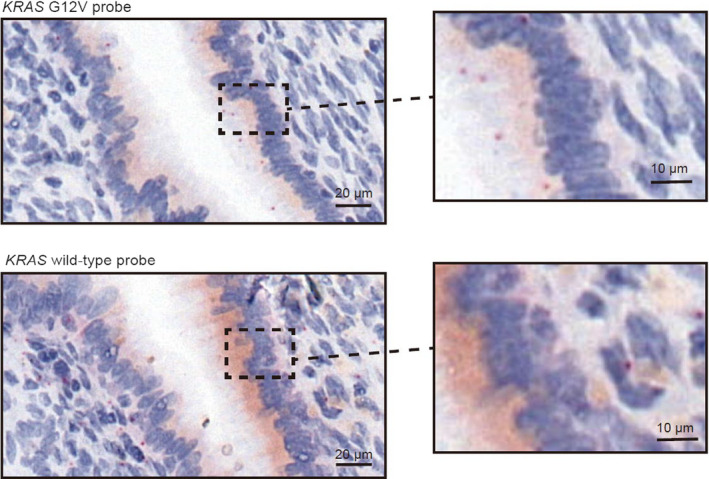
3.6. Spatial assessment of mutant allele expression of KRAS p.G12V in ovarian endometriosis cases
With regard to the spatial assessment of mutant allele expression in the abovementioned ovarian endometriosis cases, eight of 10 cases with KRAS p.G12V mutant allele expression (four and six cases retrospectively and prospectively collected, respectively) displayed a homogenous distribution of KRAS p.G12V signals across continuously arranged epithelial cells (Figure 4). When we applied this assay to five cases for which multisampling for spatially separated epithelial cells was conducted, different expression patterns of KRAS p.G12V mutant allele on the same slide was observed for two (Figure 5, Figure S7). These two cases showed three types of areas: only wild‐type signals (green), both wild‐type and mutational signals (yellow) or predominance of mutational signals (red). We then performed immunohistochemical analysis of phosphorylated ERK (p‐ERK), which is one of the biomarkers of the activated RAS pathway, using serial sections from in situ hybridization assay slides. On the same slide, the staining intensity for p‐ERK was strong in the red region, moderate in the yellow region, and low in the green region (Figure S8).
FIGURE 5.
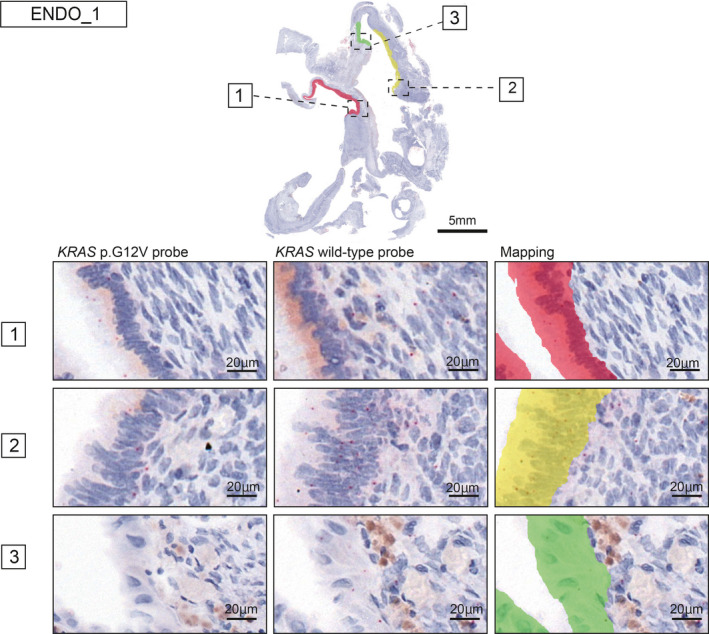
Intratumor heterogeneity of KRAS p.G12V mRNA expression and the topographical map in ENDO_1. Intratumor heterogeneity of KRAS p.G12V mRNA expression was detected in ENDO_1. The areas of endometriotic epithelial cells with only wild‐type signals, both wild‐type and mutational signals, and predominant mutational signals are mapped in green, yellow, and red, respectively. The topographical map contains the mutant subclone (red), mixture mutant and wild‐type subclone (yellow), and wild‐type subclone (green) at the mRNA level
4. DISCUSSION
We demonstrated that KRAS mutant allele's mRNA is expressed in ovarian endometriosis harboring the KRAS p. G12V mutation by using an RNA‐based in situ hybridization assay. In particular, spatial heterogeneity of the KRAS mutation was observed in some ovarian endometriosis cases with the KRAS p.G12V mutation.
The classical approach for detect mutant allele expression is polymerase chain reaction (PCR) of cDNA synthesized from RNA. 25 , 26 Recently, RNA sequencing has been used as a more powerful method to detect mutant allele expression. Nonetheless, as these methods require dissociation of the tissue of interest to obtain sufficient quantity and quality of RNA, crucial information about the spatial distribution of mutant allele expression in the tissue is lost. In contrast, the RNA‐based in situ hybridization assay is an effective tool to scrutinize the distribution of mutant allele expression in a tissue without dissociating the tissue to extract RNA. At present, there are two major technologies used for in situ hybridization. 22 , 27 In the first technology, target transcripts are converted into cDNA molecules and then detected using padlock probes and target primed rolling‐circle amplification (RCA). 27 To date, several studies have applied this in situ padlock probe technology to detect mutations in cancer tissues. 28 , 29 However, this method is strongly dependent on RNA quality because conversion from RNA to cDNA is included in the first step. The key concept of the second technology (BaseScopeTM) is that hybridization of ‘1ZZ’ probes to adjacent regions of RNA target is required to form a signal‐generating “tree” and subsequent signal amplification, allowing detection of short RNA sequences and discrimination between single‐nucleotide alterations. 22 By using this technology, Baker et al. succeeded in both identifying mutant allele expression and mapping the spatial and morphological context of mutant subclones in colon cancer tissue samples. 22 Importantly, this method does not involve conversion of RNA to cDNA and has a high potential of clinical application. Previously, there are two studies that show the potential of this in situ technology for clinical applications in oncology field. The one demonstrated the prognostic value of POLE mutation identified by this assay in uterine cancer, 30 and the other study showed the assay as a tool for discovery of biomarker of EGFR inhibitor in lung cancer. 31 Moreover, this method could be applied as a biomarker for KRAS inhibitor which has been recently clinically applied to cancers with KRAS p.G12C. 32 , 33 Therefore, RNA‐based in situ mutation detection assay can be used for a detection of biomarker and a prediction of prognosis in clinical practice.
Additionally, Baker et al reported that mutant allele expression can be detected in stored FFPE samples (average time since fixation of tissue blocks, 6.4 years; range 0‐20 years). 22 Regardless, optimization of experimental conditions for each type of tissue is required for this in situ hybridization method. To ensure signal intensity and preservation of morphology, we investigated the conditions for RNA quality‐preserving sections, temperature and time of antigen retrieval and protease strength. Accordingly, we first confirmed that detected KRAS p.G12V mutant allele expression was consistent with the existence of KRAS p.G12V mutation at the DNA level using ovarian cancer cell lines and ovarian cancer tissues.
KRAS is one of the most frequently mutated oncogenes in human cancers 34 , 35 and activates several Ras effector pathways, such as the Raf/MEK/ERK (MAPK) kinase cascade and PI3K/AKT‐mediated cascades, and promotes cell proliferation, differentiation, and survival. 36 Mutant KRAS is constitutively in the active GTP‐bound state, resulting in persistent activation of downstream signaling pathways. 37 , 38 Despite numerous studies examining the presence of KRAS mutations at the DNA level, few have investigated mutant allele expression of KRAS, 25 even though RNA sequencing is widely used. In human cancer cell lines submitted to Cancer Cell Line Encyclopedia, 39 the ratio of KRAS mutant allele reads to total reads in RNA sequence data showed a significant positive correlation with KRAS mutation allele frequency at the DNA level (r = 0.91; P <.001; Figure S9). Interestingly, the KRAS mutant allele is dominantly expressed relative to the wild‐type allele in clinical samples of lung cancer with KRAS mutations. 40 Although we detected KRAS mutant signals in all ovarian cancer cell lines and ovarian cancers carrying KRAS mutations, it does not seem appropriate to compare expression levels between mutant and wild‐type alleles based on the proportion of positive cells, because there might be differences in sensitivity between mutant and wild‐type probes in the assay. Furthermore, as well as Baker et al. reported, 22 cell lines had higher percentage of positive rate of mutant and wild‐type signals than clinical samples, which showed only 1‐2 dots per cell. In order to validate mRNA expression of KRAS wild‐type or p.G12V in clinical samples, we performed RNA sequencing to investigate the allele expression in three ovarian cancer cell lines and five ovarian cancer samples with KRAS p.G12V signal. As shown in Figure S10, KRAS mutant allele expression of cell lines and ovarian cancer samples were confirmed by Integrative Genome Viewer (IGV). However, we could not perform RNA sequencing for clinical endometriosis samples because of low quality and quantity of RNA extracted from dissected endometriotic epithelium. Therefore, we conducted RNAscope® in situ assay to assess KRAS mRNA expression in endometriotic epithelium samples. Compared to BaseScopeTM, RNAscope® assay has higher sensitivity for the target because the sequence length of probe for RNAscope® is longer than that for BaseScopeTM. As shown in Figure S11, we could validate that the endometriosis epithelium had KRAS mRNA positive signals, which means KRAS wild‐type or p.G12V actually expressed in endometriosis epithelium. Further improvements are required to achieve quantitative assessments of the expression levels of mutant and wild‐type alleles within the framework of BaseScopeTM.
Next, we discuss the biological significance of KRAS mutant allele expression in ovarian endometriosis. KRAS mutations have been identified in the early stages of tumors, including in cancers of the colon, lung, and pancreas, in which the multistage carcinogenesis theory is elucidated. 41 , 42 , 43 , 44 Mutationally activated KRAS signaling leads to differentiation and proliferation via MAPK signaling in the mouse colon epithelium. 45 , 46 In addition, recent studies, including ours, have demonstrated at the DNA level that KRAS is frequently mutated in the endometriotic epithelium, 12 , 13 , 14 but it remains unclear whether the KRAS mutant allele is expressed and downstream effector proteins are activated in benign tissues. For example, our recent study clarified that ARID1A protein expression is retained in ovarian endometriosis with ARID1A loss‐of‐function mutations. 47 Overall, mutant allele expression is important for assessing the biological significance of cancer‐associated gene mutations. Because the KRAS mutant allele is expressed in ovarian endometriosis with the KRAS p.G12V mutation, it is inferred that by activating Ras effector pathways, KRAS mutation confers a survival advantage to endometriotic epithelial cells. Moreover, in a study of the endometrium of transgenic mice with conditional knock‐in of KRAS p.G12V/+, Cheng et al. reported that KRAS activation promotes the formation of endometriosis and prolongs the survival of endometriotic lesions. 48 Therefore, we conducted immunohistochemical analysis for p‐ERK, which is a representative marker of activated MAPK signaling, in ovarian endometriosis tissue to assess the association between KRAS mutation status and MAPK signaling. Because p‐ERK expression is affected by not only KRAS but also other factors, such as various cytokines and oxidase stress, 49 we did not compare p‐ERK expression among ovarian endometriosis cases. We then focused on one ovarian endometriosis case showing intratumor heterogeneity of KRAS p.G12V mutant allele expression and compared p‐ERK expression among three types of areas with distinct mutant allele expression patterns in the same slide (Figure S8). The intensity of p‐ERK expression varied among these three areas according to the KRAS p.G12V mutant allele expression pattern, suggesting that KRAS activation has an impact on the phenotype of ovarian endometriosis. In addition, comparison of clinical features between ovarian endometriosis with KRAS p.G12V mutant allele expression and KRAS wild‐type suggests that KRAS p.G12V mutant allele expression may be associated with inflammation and progression of the lesion in ovarian endometriosis. Kun et al. demonstrated differences in the impact of KRAS p.G12V and KRAS p.G12D on follicle development using Cre recombination mouse expressing either the KRAS G12V/+ or KRAS G12D/+ mutation. 50 In this study, KRAS p.G12D mice had abnormal follicle structures and developed low‐grade serous ovarian carcinomas, on the other hand KRAS p.G12V mice had normal follicle structures. Remarkably, they found that in KRAS p.G12V mouse the gene networks associated with inflammation, such as IL‐1 and IL‐6, was upregulated using RNA‐seq analysis. Furthermore, several previous literatures showed that KRAS p.G12V or p.G12D in pancreatic cancer cell 51 , 52 or KRAS p.G12D in myeloid leukemia cell 53 induces inflammation. The relationship between inflammation and oncogenic KRAS have been reported in mouse model 53 or human carcinoma cells, 51 , 52 but not in normal human epithelial cells until now. Therefore, we explored the association between KRAS mutation and inflammation related pathways by gene set enrichment analysis (GSEA). 54 From GEO (Gene Expression Omnibus), we obtained RNA expression data from two studies in which human mammary epithelial cells 55 and human pancreatic ductal epithelial cells 56 were transfected with the KRAS p.G12V and KRAS p.G12D mutations. When we compared gene expression profiles between the mutant and control groups by using GSEA, the mutant group was characterized by several inflammation‐related pathways (Figure S12). Additionally, there are several reports that inflammation induced by oncogenic KRAS could be the initiation of carcinogenesis. 57 , 58 , 59 Exploring the relationship between inflammation and KRAS mutations in endometriosis may lead to clarifying molecular mechanisms of malignant transformation in endometriosis. Indeed, our previous report 18 has clarified that RAS oncogenic mutation is an early event in clonal lineage from normal endometrium to endometriosis‐related ovarian cancer via endometriosis. Detection of RAS oncogenic mutations such as KRAS p.G12V in endometriotic epithelium or uterine endometrial epithelium may be a predictive marker for recurrence or malignant transformation of endometriosis. However, mutation profiling of endometriosis or uterine endometrium is extremely difficult 12 because the volume of target cells is very low in surgically resected endometriosis or uterine endometrium tissues. Therefore, this RNA‐based in situ assay could easily detect cancer‐associated gene mutations including KRAS p.G12V and might provide important information about recurrence and malignant transformation of endometriosis in clinical practice.
Recently it has been shown that downstream expression and phenotypes of RAS differ depending on the KRAS mutated amino acids in mouse colon and pancreas. 60 Therefore, spatial analysis of genomic and transcriptomic data with consideration of mutant amino acids is necessary to clarify the biological function of cancer‐associated gene mutations in endometriosis.
Another important issue is the association between KRAS mutation and malignant transformation of ovarian endometriosis. Although we observed expression of the KRAS p.G12V mutant allele in ovarian endometriosis, these samples were histologically non‐malignant tumors. In line with our findings, mouse models of endometriosis with conditional knock‐in KRAS mutations have showed that KRAS mutation alone is insufficient for the development of ovarian carcinoma in mice. 48 In particular, Dinulescu et al. reported that the combination of KRAS activation and conditional deletion of PTEN is needed for malignant transformation of ovarian endometriosis in a mouse model. 61 These findings are similar to the adenoma‐carcinoma sequence in the intestine. 13 , 62 Indeed, we recently identified using whole‐exome sequencing a clonal lineage from the normal uterine endometrium to ovarian endometriosis, atypical endometriosis, and clear cell carcinoma in one ovarian cancer patient. 18 In that case, NRAS p.Q61H was detected in all samples but ARID1A p.E1683fs or LOH at ATM were detected only in atypical endometriosis and clear cell carcinoma samples. In addition, another NRAS mutation (p.G12D) was detected in the normal uterine endometrium and ovarian endometriosis but not in atypical endometriosis and clear cell carcinoma. Most likely, RAS activation alone is not sufficient for the malignant transformation of human ovarian endometriosis. Moreover, we identified a heterogenous spatial distribution of KRAS allele expression in some cases of endometriosis, whereas the spatial distribution of KRAS allele expression was diffuse and homogenous in ovarian cancer. This finding suggests that RAS activation has already expanded in ovarian endometriotic cyst during precancer stage, probably by clonal expansion of the origin cells harboring KRAS mutation or dominant cell growth and expansion of KRAS mutated endometriotic cells. 12
In conclusion, we visualized expression of the KRAS p.G12V mutant allele in ovarian endometriotic epithelial cells through the RNA‐based in situ hybridization technique. In addition, a portion of ovarian endometriosis showed intratumor heterogeneity with regard to KRAS p.G12V mutant allele expression associated with Raf/MEK/ERK signaling activation. Spatial evaluation of oncogene mutant allele expression will be useful for clarifying differences in the biological significance of oncogenes between benign and malignant tumors.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
NY and KY designed and performed experiments, analyzed data and co‐wrote the paper. KS performed experiments and collected samples and the data. HN performed bioinformatics analysis. HU, KS and MY collected samples. YM, KY, RT, TI and HK served as scientific advisors. MT reviewed stained sections histologically. TE critically reviewed the study proposal.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Anna Ishida and Kenji Ohyachi, Naoyuki Yamaguchi for technical assistance. This work was supported in part by JSPS KAKENHI grant number JP16H06279 (Grant‐in‐Aid for Scientific Research on Innovative Areas—Platforms for Advanced Technologies and Research Resources for HN and KY), and JP19K09822 (Grant‐in‐Aid for Scientific Research C for KY) and by "Challenging Exploratory Research Projects for the Future" grant from ROIS (Research Organization of Information and Systems) for HN, research grant from KANAE foundation for the promotion of medical science for KY.
Yachida N, Yoshihara K, Suda K, et al. Biological significance of KRAS mutant allele expression in ovarian endometriosis. Cancer Sci. 2021;112:2020–2032. 10.1111/cas.14871
REFERENCE
- 1. Giudice LC. Clinical practice. Endometriosis. N Engl J Med. 2010;362:2389‐2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hickey M, Ballard K, Farquhar C. Endometriosis. BMJ. 2014;348:g1752. [DOI] [PubMed] [Google Scholar]
- 3. Munksgaard PS, Blaakaer J. The association between endometriosis and gynecological cancers and breast cancer: a review of epidemiological data. Gynecol Oncol. 2011;123:157‐163. [DOI] [PubMed] [Google Scholar]
- 4. Pearce CL, Templeman C, Rossing MA, et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case–control studies. Lancet Oncol. 2012;13:385‐394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heidemann LN, Hartwell D, Heidemann CH, Jochumsen KM. The relation between endometriosis and ovarian cancer ‐ a review. Acta Obstet Gynecol Scand. 2014;93:20‐31. [DOI] [PubMed] [Google Scholar]
- 6. Van Gorp T, Amant F, Neven P, Vergote I, Moerman P. Endometriosis and the development of malignant tumours of the pelvis. A review of literature. Best Pract Res Clin Obstet Gynaecol. 2004;18:349‐371. [DOI] [PubMed] [Google Scholar]
- 7. Prowse AH, Manek S, Varma R, et al. Molecular genetic evidence that endometriosis is a precursor of ovarian cancer. Int J Cancer. 2006;119:556‐562. [DOI] [PubMed] [Google Scholar]
- 8. Yamamoto S, Tsuda H, Takano M, Iwaya K, Tamai S, Matsubara O. PIK3CA mutation is an early event in the development of endometriosis‐associated ovarian clear cell adenocarcinoma. J Pathol. 2011;225:189‐194. [DOI] [PubMed] [Google Scholar]
- 9. Anglesio MS, Bashashati A, Wang YK, et al. Multifocal endometriotic lesions associated with cancer are clonal and carry a high mutation burden. J Pathol. 2015;236:201‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koninckx PR, Ussia A, Adamyan L, Wattiez A, Donnez J. Deep endometriosis: definition, diagnosis, and treatment. Fertil Steril. 2012;98:564‐571. [DOI] [PubMed] [Google Scholar]
- 11. Rousset‐Jablonski C, Alifano M, Plu‐Bureau G, et al. Catamenial pneumothorax and endometriosis‐related pneumothorax: clinical features and risk factors. Hum Reprod. 2011;26:2322‐2329. [DOI] [PubMed] [Google Scholar]
- 12. Suda K, Nakaoka H, Yoshihara K, et al. Clonal expansion and diversification of cancer‐associated mutations in endometriosis and normal endometrium. Cell Rep. 2018;24:1777‐1789. [DOI] [PubMed] [Google Scholar]
- 13. Anglesio MS, Papadopoulos N, Ayhan A, et al. Cancer‐associated mutations in endometriosis without cancer. N Engl J Med. 2017;376:1835‐1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lac V, Verhoef L, Aguirre‐Hernandez R, et al. Iatrogenic endometriosis harbors somatic cancer‐driver mutations. Hum Reprod. 2019;34:69‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moore L, Leongamornlert D, Coorens THH, et al. The mutational landscape of normal human endometrial epithelium. Nature. 2020;580:640‐646. [DOI] [PubMed] [Google Scholar]
- 16. Lac V, Nazeran TM, Tessier‐Cloutier B, et al. Oncogenic mutations in histologically normal endometrium: the new normal? J Pathol. 2019;249:173‐181. [DOI] [PubMed] [Google Scholar]
- 17. Kyo S, Sato S, Nakayama K. Cancer‐associated mutations in normal human endometrium: Surprise or expected? Cancer Sci. 2020;111(10):3458‐3467. 10.1111/cas.14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suda K, Cruz Diaz LA, Yoshihara K, et al. Clonal lineage from normal endometrium to ovarian clear cell carcinoma through ovarian endometriosis. Cancer Sci. 2020;111(8):3000‐3009. 10.1111/cas.14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sugino K, Tamura R, Nakaoka H, et al. Germline and somatic mutations of homologous recombination‐associated genes in Japanese ovarian cancer patients. Sci Rep. 2019;9:17808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahmadloo S, Nakaoka H, Hayano T, et al. Rapid and cost‐effective high‐throughput sequencing for identification of germline mutations of BRCA1 and BRCA2. J Hum Genet. 2017;62:561‐567. [DOI] [PubMed] [Google Scholar]
- 21. Yamawaki K, Ishiguro T, Mori Y, et al. Sox2‐dependent inhibition of p21 is associated with poor prognosis of endometrial cancer. Cancer Sci. 2017;108:632‐640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baker AM, Huang W, Wang XM, et al. Robust RNA‐based in situ mutation detection delineates colorectal cancer subclonal evolution. Nat Commun. 2017;8:1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tamura R, Yoshihara K, Saito T, et al. Novel therapeutic strategy for cervical cancer harboring FGFR3‐TACC3 fusions. Oncogenesis. 2018;7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haas D, Shebl O, Shamiyeh A, Oppelt P. The rASRM score and the Enzian classification for endometriosis: their strengths and weaknesses. Acta Obstet Gynecol Scand. 2013;92:3‐7. [DOI] [PubMed] [Google Scholar]
- 25. Slebos RJ, Habets GG, Evers SG, Mooi WJ, Rodenhuis S. Allele‐specific detection of K‐ras oncogene expression in human non‐small‐cell lung carcinomas. Int J Cancer. 1991;48:51‐56. [DOI] [PubMed] [Google Scholar]
- 26. Hruban RH, van Mansfeld AD, Offerhaus GJ, et al. K‐ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant‐enriched polymerase chain reaction analysis and allele‐specific oligonucleotide hybridization. Am J Pathol. 1993;143:545‐554. [PMC free article] [PubMed] [Google Scholar]
- 27. Larsson C, Koch J, Nygren A, et al. In situ genotyping individual DNA molecules by target‐primed rolling‐circle amplification of padlock probes. Nat Methods. 2004;1:227‐232. [DOI] [PubMed] [Google Scholar]
- 28. Grundberg I, Kiflemariam S, Mignardi M, et al. In situ mutation detection and visualization of intratumor heterogeneity for cancer research and diagnostics. Oncotarget. 2013;4:2407‐2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. El‐Heliebi A, Kashofer K, Fuchs J, et al. Visualization of tumor heterogeneity by in situ padlock probe technology in colorectal cancer. Histochem Cell Biol. 2017;148:105‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu S, Shao H, Ban X, et al. Detection of POLE subtypes in high‐grade endometrioid carcinoma by BaseScope‐ISH assay. Front Oncol. 2019;9:831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu S, Shi X, Si X, et al. EGFR T790M detection in formalin‐fixed paraffin‐embedded tissues of patients with lung cancer using RNA‐based in situ hybridization: a preliminary feasibility study. Thorac Cancer. 2019;10:1936‐1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti‐tumour immunity. Nature. 2019;575:217‐223. [DOI] [PubMed] [Google Scholar]
- 33. Hong DS, Fakih MG, Strickler JH, et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383:1207‐1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457‐2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Timar J, Kashofer K. Molecular epidemiology and diagnostics of KRAS mutations in human cancer. Cancer Metastasis Rev. 2020;39(4):1029‐1038. 10.1007/s10555-020-09915-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322‐327. [DOI] [PubMed] [Google Scholar]
- 37. Stites EC, Ravichandran KS. A systems perspective of ras signaling in cancer. Clin Cancer Res. 2009;15:1510‐1513. [DOI] [PubMed] [Google Scholar]
- 38. Zhou B, Der CJ, Cox AD. The role of wild type RAS isoforms in cancer. Semin Cell Dev Biol. 2016;58:60‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ghandi M, Huang FW, Jane‐Valbuena J, et al. Next‐generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503‐508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Govindan R, Ding L, Griffith M, et al. Genomic landscape of non‐small cell lung cancer in smokers and never‐smokers. Cell. 2012;150:1121‐1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maeshima AM, Maeshima A, Kawashima O, Nakajima T. K‐ras gene point mutation in neogenetic lesions of subpleural fibrotic lesions: either an early genetic event in lung cancer development or a non‐specific genetic change during the inflammatory reparative process. Pathol Int. 1999;49:411‐418. [DOI] [PubMed] [Google Scholar]
- 42. Alsdorf WH, Clauditz TS, Hoenig T, et al. Intratumoral heterogeneity of KRAS mutation is rare in non‐small‐cell lung cancer. Exp Mol Pathol. 2013;94:155‐159. [DOI] [PubMed] [Google Scholar]
- 43. Albury TM, Pandey V, Gitto SB, et al. Constitutively active Akt1 cooperates with KRas(G12D) to accelerate in vivo pancreatic tumor onset and progression. Neoplasia. 2015;17:175‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chang XY, Wu Y, Li Y, Wang J, Chen J. Intraductal papillary mucinous neoplasms of the pancreas: clinical association with KRAS. Mol Med Rep. 2018;17:8061‐8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Feng Y, Bommer GT, Zhao J, et al. Mutant KRAS promotes hyperplasia and alters differentiation in the colon epithelium but does not expand the presumptive stem cell pool. Gastroenterology. 2011;141(1003–1013):e1001‐1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gierut JJ, Lyons J, Shah MS, Genetti C, Breault DT, Haigis KM. Oncogenic K‐Ras promotes proliferation in quiescent intestinal stem cells. Stem Cell Res. 2015;15:165‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yachida N, Yoshihara K, Suda K, et al. ARID1A protein expression is retained in ovarian endometriosis with ARID1A loss‐of‐function mutations: implication for the two‐hit hypothesis. Sci Rep. 2020;10:14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng CW, Licence D, Cook E, et al. Activation of mutated K‐ras in donor endometrial epithelium and stroma promotes lesion growth in an intact immunocompetent murine model of endometriosis. J Pathol. 2011;224:261‐269. [DOI] [PubMed] [Google Scholar]
- 49. McKinnon BD, Kocbek V, Nirgianakis K, Bersinger NA, Mueller MD. Kinase signalling pathways in endometriosis: potential targets for non‐hormonal therapeutics. Hum Reprod Update. 2016;22:382‐403. [DOI] [PubMed] [Google Scholar]
- 50. Kun EHS, Tsang YTM, Lin S, et al. Differences in gynecologic tumor development in Amhr2‐Cre mice with KRAS(G12D) or KRAS(G12V) mutations. Sci Rep. 2020;10:20678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Siddiqui I, Erreni M, Kamal MA, et al. Differential role of Interleukin‐1 and Interleukin‐6 in K‐Ras‐driven pancreatic carcinoma undergoing mesenchymal transition. Oncoimmunology. 2018;7:e1388485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ling J, Kang Y, Zhao R, et al. KrasG12D‐induced IKK2/beta/NF‐kappaB activation by IL‐1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:105‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hamarsheh S, Osswald L, Saller BS, et al. Oncogenic Kras(G12D) causes myeloproliferation via NLRP3 inflammasome activation. Nat Commun. 2020;11:1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rahman M, MacNeil SM, Jenkins DF, et al. Activity of distinct growth factor receptor network components in breast tumors uncovers two biologically relevant subtypes. Genome Med. 2017;9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tsang YH, Dogruluk T, Tedeschi PM, et al. Functional annotation of rare gene aberration drivers of pancreatic cancer. Nat Commun. 2016;7:10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Greten FR, Grivennikov SI. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity. 2019;51:27‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chien Y, Scuoppo C, Wang X, et al. Control of the senescence‐associated secretory phenotype by NF‐kappaB promotes senescence and enhances chemosensitivity. Genes Dev. 2011;25:2125‐2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kuilman T, Michaloglou C, Vredeveld LC, et al. Oncogene‐induced senescence relayed by an interleukin‐dependent inflammatory network. Cell. 2008;133:1019‐1031. [DOI] [PubMed] [Google Scholar]
- 60. Zafra MP, Parsons MJ, Kim J, et al. An in vivo kras allelic series reveals distinct phenotypes of common oncogenic variants. Cancer Discov. 2020;10:1654‐1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dinulescu DM, Ince TA, Quade BJ, Shafer SA, Crowley D, Jacks T. Role of K‐ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med. 2005;11:63‐70. [DOI] [PubMed] [Google Scholar]
- 62. Leslie A, Carey FA, Pratt NR, Steele RJ. The colorectal adenoma‐carcinoma sequence. Br J Surg. 2002;89:845‐860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


