Abstract
The Ton complex is a molecular motor that uses the proton gradient at the inner membrane of Gram-negative bacteria to apply forces on outer membrane proteins, allowing active transport of nutrients into the periplasmic space. For decades, contradictory data has been reported on the structure and stoichiometry of the Ton complex. However, recent reports strongly support a subunit stoichiometry of 5:2 for the ExbB-ExbD subcomplex. In this review, we summarize the recent discoveries of the structures and proposed mechanisms of the Ton system, as well as similar protein motor complexes in Gram-negative bacteria.
Keywords: Ton motor complex, ExbB, ExbD, proton motive force, flagellar motor, MotA, MotB
Introduction
Gram-negative bacteria are characterized by two cellular membranes: an inner cytoplasmic membrane and protective outer membrane. The outer membrane acts as a barrier against antimicrobials, toxic compounds, and other environmental hazards [1]. Specialized porins in the outer membrane facilitate the diffusion of small molecules (under ~600 Da) into the periplasm [1, 2], while scarce nutrients are actively transported into the periplasmic space by the Ton system [3]. Ton is a multi-subunit membrane protein complex (TonB-ExbB-ExbD) that uses the proton gradient across the inner membrane as its energy source. The elongated TonB subunit physically interacts with nutrient bound TonB Dependent Transporters (TBDTs) at the outer membrane. ExbB and ExbD harness the proton motive force (pmf), and transfer it to TonB which then opens a gate in the TBDT, allowing the nutrient to enter the periplasmic space [4]. The type of nutrient actively transported by the Ton complex is determined by the specificity of the TBDT, ranging from iron-siderophores, divalent metals, carbohydrates, vitamins or peptides [3, 5–8]. The Ton system can also be hijacked by bacteriocins that bind to a specific TBDT, gaining access to the interior of the cell and killing the infected bacterium with high efficiency [9–12].
The structures of numerous TBDTs have been solved and all show the same architecture: a 22-stranded ß-barrel C-terminal domain, occluded with an N-terminal plug domain inserted into the interior of the barrel [3]. Binding of the ligand on the extracellular face of the TBDT has been shown to induce conformational changes leading to the exposure of the TonB box, a short, conserved N-terminal sequence, to the periplasm. The C-terminal periplasmic domain of the TonB subunit then interacts with the TBDT TonB box, forming a stable complex that physically connects the TBDT to the inner membrane (Fig. 1) [13, 14]. What happens after the formation of the TBDT/TonB complex is largely unknown, but it is believed that the energy derived from the pmf in the inner membrane is transmitted to TonB and used to alter the conformation of the TBDT. In the pulling model, the TonB C-terminal domain bound to the TBDT TonB box is pulled into the periplasm and gradually unfolds the TBDT plug domain, eventually opening a channel in the TBDT [15, 16]. The pulling model is supported by in vitro experiments that show that the TBDT plug domain can reversibly unfold, and that the interaction between the TonB C-terminal domain and the TBDT TonB box is strong enough to sustain a pulling force that unfolds a significant part of the plug domain [17, 18].
Figure 1 –
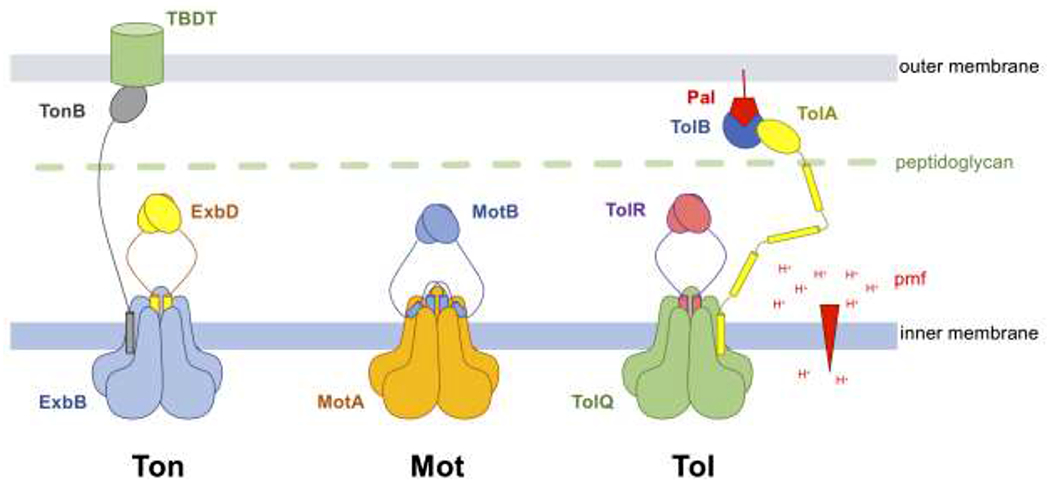
Schematic representation of the Ton, Mot and Tol complexes.
The Ton complex is composed of five ExbB (blue), two ExbD (yellow) and at least one TonB (grey). The elongated TonB subunit interacts with nutrient-loaded TBDTs at the outer membrane.
The Mot complex contains five MotA (orange) and two MotB (blue). The MotA-MotB is represented here in its plugged state. The plug domain of MotB, a short helix C-terminal of the TM domain, binds at the interface between two MotA subunits and blocks the movement of protons (the positions of the plug domains are shown according to [24]). Upon incorporation with the flagellar apparatus, the plug domains dissociate from MotA, allowing the MotB C-terminal domains to interact with the peptidoglycan layer (green).
The stoichiometry of the Tol complex is currently unknown. Because of extensive homologies with the Ton and Mot complexes, the complex is shown here to contain five TolQ (green), two TolR (purple), and at least one TolA (yellow) [21]. The elongated TolA periplasmic domain interacts with Tol-Pal (blue-red) complexes at the outer membrane.
The proton (H+) gradient across the inner membrane that forms the pmf is shown as a red arrowhead.
The Ton complex is considered a stator complex, a motor protein complex which converts potential energy from the ion motor force into torque. Other stator complexes evolutionarily related to the ExbB-ExbD-TonB system, with analogous components, include the TolQ-TolR-TolA and MotA-MotB complexes (Fig. 1) [19]. Although the complexes fulfill different functions including cell membrane integrity (Tol) and flagellar rotation (Mot), they are characterized in the same class of ion-driven, membrane associated mechanical motors as the Ton complex [20, 21]. Recently, the cryo-EM structures of ExbB-ExbD and MotA-MotB from various bacterial species have been reported [22–24]. In this review, we will compare and discuss the stoichiometry and structures of the Ton and Mot complexes and their predicted mechanisms for harnessing energy.
Main
Stoichiometry of the ExbB-ExbD complex
Since the discovery of the Ton system in the 1970s, contradictory results have been reported on the stoichiometry of the Ton complex [4]. However, recent reports strongly support that the ExbB-ExbD subcomplex has a 5:2 stoichiometry. The crystallographic structure of a truncated ExbB-ExbD complex first showed the pentameric structure of ExbB [25]. Mass spectrometry experiments performed on native membranes also identified a pentamer of ExbB, suggesting that the pentamer was not an artefact [26]. Finally cryo-EM high resolution structures of ExbB-ExbD complexes from Escherichia coli and Pseudomonas savastanoi clearly show a 5:2 ExbB-ExbD arrangement, with ExbB forming a pentameric hydrophobic central pore encompassing a dimer of ExbD transmembrane helices (TMHs) (Fig. 1, 2a) [22, 23].
Figure 2 –
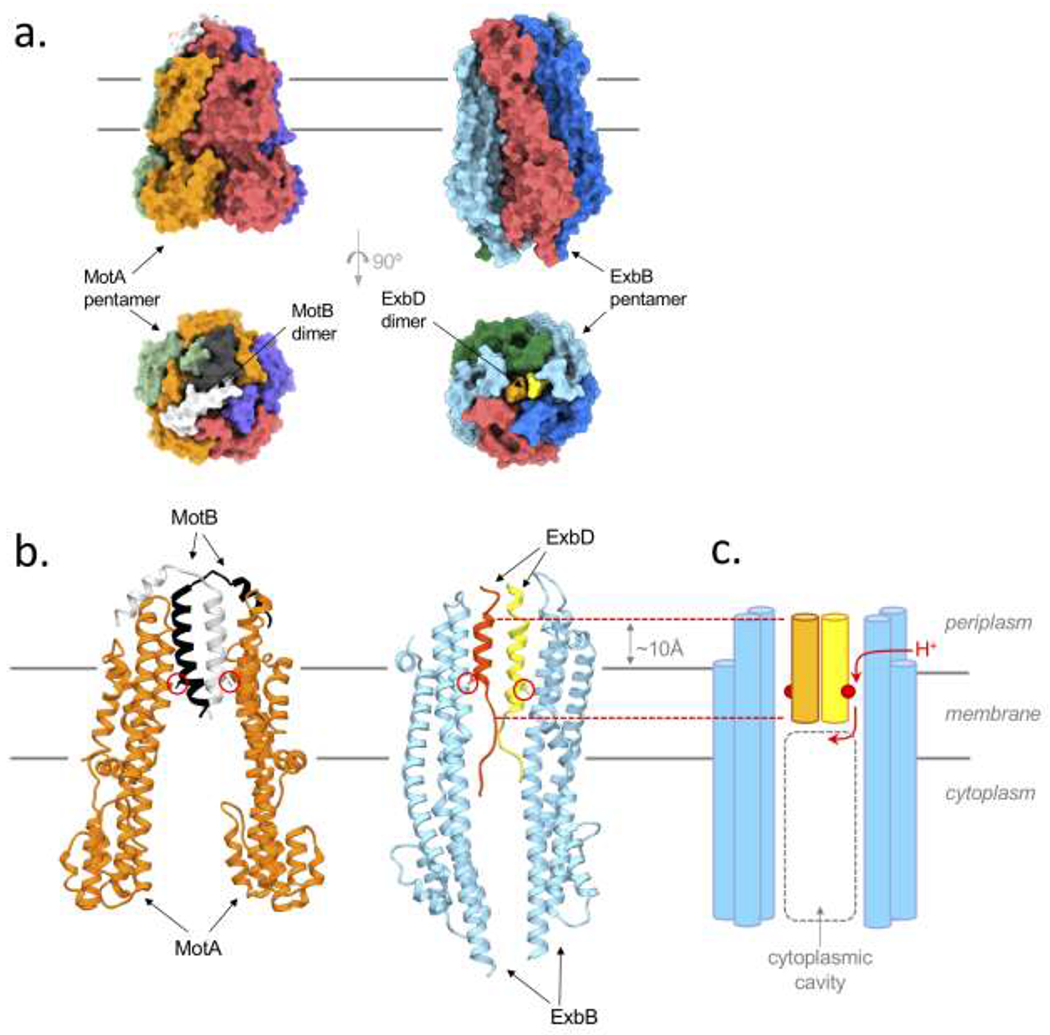
Structure of the ExbB-ExbD complex
a. Surface representation of the MotA-MotB (left, pdb id 6YKM) and ExbB-ExbD structures (right, pdb id6TYI) [22,24]. Top view perpendicular to the inner membrane plane, shown with black solid lines, bottom view from the periplasmic space. The elongated MotA subunits (purple, orange, red, green) and ExbB subunits (blue, cyan, red and green) associate as a pentamer, defining a central pore in which are located the TMHs of the MotB (white and black) and ExbD (orange and yellow) dimers. The soluble periplasmic domains of MotB and ExbD were not visible in the cryo-EM structure.
b. Ribbon representation of the MotA-MotB (left) and ExbB-ExbD (right) structures. For clarity, only two MotA subunits (orange) and two ExbB subunits (cyan) are shown. The inner membrane is illustrated by solid black lines. The sidechains of the essential aspartate residue on the MotB and ExbD TMHs are shown as sticks and are circled in red. The position of the central hydrophobic pore of the ExbB pentamer is shifted ~10Å toward the periplasm (shown with dashed red lines).
c. Schematic representation of ExbB-ExbD (b), the conserved Asp are shown as red circles. The path of the proton (as described for MotA-MotB [24]) is shown with red arrows: a lateral opening between two ExbB subunits allows the proton to travel to the Asp on ExbD and is subsequently delivered into the cytoplasmic cavity (dashed grey lines).
The stoichiometry of the MotA-MotB stator complex has been postulated to be 4:2 for decades [27]. Nevertheless, the reported cryo-EM structures of several MotA-MotB complexes, as well as PomA-PomB (equivalent to MotA-MotB, but utilizing a sodium gradient in Gram positive bacteria) show the same 5:2 arrangement (Fig. 1) [23, 24]. Comparison of the ExbB-ExbD and MotA-MotB structures show a high level of conservation for the transmembrane helices that form the central hydrophobic pore (see extended data fig. 9 in [23]).
Because of the high homology between the Ton, Mot and Tol complexes, it is likely that the TolQ-TolR complex shares the same 5:2 ratio (Fig. 1, 3) [21]. While the dimerization of the ExbD, MotB, and TolR subunits has been well characterized (see [4]), the stoichiometry of the TonB or TolA subunit in the Ton or Tol complexes has not been clearly established. Only for the Ton complex has the co-purification of all three subunits been reported [23, 25, 28]. An attempt to solve the cryo-EM structure of the P. savastanoi TonB-ExbB-ExbD purified complex has been reported and suggests that one TonB TMH binds at the periphery of the ExbB-ExbD TM region (see extended data fig. 10 in [23]). However, the observed densities identified as TonB were only seen at low contour levels and therefore need to be further investigated [23].
Figure 3 –
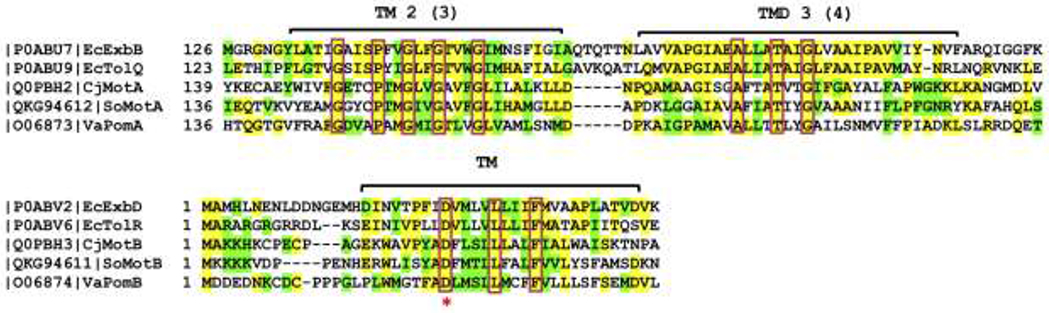
Sequence alignment of Ton, Tol, Mot, and Pom systems at conserved TM regions that construct the central pore.
All gene sequences represented are from recent cryo-EM structures [21–23]. Yellow depicts similar residues, while green represents identical residues. The red boxes indicate conserved residues between all five proteins and species, including the conserved ExbD Asp residue depicted in Figure 2b, c. This conserved Asp residue is indicated with a red star. Only TMs 2 and 3 of ExbB and TolQ are shown, and their equivalent TMs 3 and 4 of MotA and PomA. Accession numbers and species are indicated at the beginning of the sequence: Ec, Escherichia coli; Cj, Campylobacter jejuni; So, Shewanella oneidensis; Va, Vibrio alginolyticus. The alignment was created with T-coffee multiple sequence alignment [33].
Structural homology among stators
Sequence alignments between Ton, Tol and Mot from different species reveal a high degree of similarity in the TM domains that form the central pore (TMs 2 and 3 of ExbB/TolQ, TMs 3 and 4 of MotA, and the TM of ExbD/TolR/MotB, Fig. 3).
A strikingly similar feature of the ExbB-ExbD and MotA-MotB structures is that the central TM pore domain is not embedded in the cytoplasmic membrane plane, but shifted about 10A toward the periplasmic space. As shown in Fig. 2b–c, the ExbB TMHs 2 and 3 extend toward the periplasm, bringing the dimer of ExbD TMHs into a position above the membrane plane [22]. A similar arrangement is observed for the reported structures of the MotA-MotB complexes (Fig. 2b) [23, 24]. As a consequence, the central hydrophilic cavity defined by the ExbB/MotA pentamer expands almost halfway into the membrane plane, and the conserved essential aspartate residue on the ExbD/MotB TMH is in close proximity to the inner membrane periplasmic leaflet.
A rotary mechanism has been postulated for the Mot complexes based on the MotA-MotB structures [23, 24]. In these models, the proton travels from the periplasm to the MotB conserved Asp residue. The protonation of Asp induces a structural rearrangement resulting in the rotation of the MotA pentamer around the MotB TM dimer. When the protonated Asp finally delivers the proton into the central cytoplasmic cavity, the second Asp on the other TMH becomes primed for protonation. The cryo-EM structure of the un-plugged Campylobacter jejuni MotA-MotB (CjMotA-MotB) complex reveals a channel that connects the periplasm to the conserved MotB Asp, which opens through the rearrangement of a conserved hydrophobic residue on TM4 of MotA [24]. Interestingly, the entry of the channel on the periplasmic side is not located at the top of the hydrophobic pore, but instead through a lateral opening between two MotA subunits. This configuration greatly reduces the distance traveled by the translocated proton, as it minimizes the distance between the conserved Asp on MotB with both the periplasmic lateral opening and the cytoplasmic cavity (Fig. 2c).
The cryo-EM structure of GldL-GldM, another stator complex involved in bacterial gliding motility in Bacteroides bacteria, was recently reported [29]. GldL-GldM uses the proton motive force to generate stroke and the GldL subunit forms a pentamer with a central pore that encompasses the GldM dimer. Again, a rotary mechanism is postulated in which proton translocation induces the rotation of the GldM dimer that is transmitted to components in the outer membrane via the elongated periplasmic domains of GldM [29]. GldL-GldM is not closely related to Ton, Mot and Tol, as they do not share a high degree of sequence similarity. In the Ton, Mot and Tol complexes, the only charged residue in the central TM pore is the essential Asp on the ExbD/MotB/TolR TMH, and the remainder of the pore is lined with hydrophobic residues. In contrast, the hydrophobic pore of GldL-GldM contains several charged residues (GldL-Glu49 and GldM-Arg9) that form ion-pairs with tyrosine residues (GldL-Tyr13 and GldM-Tyr17). Furthermore, the TM pore of GldL-GldM sits in register with the membrane plane and is not shifted toward the periplasm as observed for ExbB-ExbD and MotA-MotB. These differences suggest that the mechanism of proton translocation is not precisely the same for GldL-GldM and the other stators. However, the 5:2 arrangement shared by these two families of pmf driven stators suggest that they are derived from a common ancestor.
Ton mechanism of action
As mentioned previously, a rotary mechanism has been proposed for the MotA-MotB complexes [23, 24]. A piston like mechanism could also be envisioned in which the TMHs of MotB (or ExbD/TolR) would travel up and down the hydrophobic pore such that the conserved Asp residue would grab the proton on the periplasmic side and deliver it in the cytoplasmic cavity. However, the piston mechanism is energetically unfavorable, as it would need to expose a stretch of hydrophobic residues of the MotB/ExbD/TolR TMH to the periplasm aqueous environment. Santiveri et al. solved the cryo-EM structure of the CjMotA-MotB unplugged construct with the MotB conserved Asp residue replaced with Asn, so as to mimic the protonated state of Asp [24]. Comparison of the two unplugged structures shows a reorientation of the Asn sidechain, pointing toward the cytoplasmic cavity, suggesting that upon protonation of the essential Asp, the MotB TMH does not shift its position up or down the pore to deliver the proton into the cytoplasm. Therefore, while there is no direct evidence for rotation of MotA around MotB, the reported structures are in good agreement with a rotary mechanism.
Thus, a rotary mechanism is also likely to take place in the Ton and Tol complexes. Within the Mot complex, MotB has a conserved plug domain that folds between two MotA subunits, preventing proton leakage (Fig. 1, 2a–b). The incorporation into the flagellum apparatus activates MotA-MotB through dissociation of the MotB plug domain from MotA, opening the proton channel, and allowing the C-terminal MotB domain to reach and bind the peptidoglycan layer [30]. The ExbB-ExbD cryo-EM structure most likely corresponds to the plugged state of MotA-MotB, as there is no evidence of a proton channel that would connect the conserved ExbD-Asp25 to the periplasm [22]. There is no equivalent of the MotB plug domain in the Ton complex. The activation mechanism of ExbB-ExbD is currently unknown, but probably relies on its association with the TonB subunit and/or the signaling of an interaction between the TonB C-terminal domain and a ligand loaded TBDT.
How a rotary motion of either ExbD or ExbB is transmitted to the C-terminal domain of TonB, and how a gate opens inside the TBDT, have yet to be determined. The interaction between the TonB C-terminal domain of the TBDT TonB box is strong enough to sustain pulling forces that would partially unfold the TBDT N-terminal plug domain [15, 16, 18]. Once bound to the TBDT TonB box, the TonB C-terminal domain becomes mobile and can adopt different orientations compared to the TBDT [13, 14]. In vivo studies have shown that the TonB C-terminal domain interacts with the ExbD C-terminal domain, and that this interaction is dependent on the pmf [31, 32]. Studies of the translocation process of the Ton dependent bacteriocin S2 through the Pseudomonas aeruginosa TBDT FpvA provide in vivo evidence that the TBDT plug domain partially unfolds during the cycle [9, 10]. Using a GFP fused S2 construct and incorporation of the photoreactive amino acid analogue pBpa (p-benzoyl-L-phenylalanine), it was shown for the first time that a bacteriocin can physically translocate through a TBDT, but also that forces exerted on TonB lead to the unfolding of a labile domain of the TBDT plug, opening a channel wide enough for the unfolded bacteriocin to pass through [10].
Further work is needed to elucidate at the molecular level how ExbB, ExbD and TonB work together to promote transport of nutrients through the TBDTs at the outer membrane. The structure determination of a full TonB-ExbB-ExbD complex may give insight on how TonB associates with both ExbB in the membrane, and ExbD in the periplasm. Ultimately, we will need structures of the full Ton complex bound to a TBDT with and without trapped ligand.
Conclusion
In this review, we discuss the similar features of the recently reported ExbB-ExbD and MotA-MotB structures. These complexes share a common 5:2 subunit ratio, and show a central hydrophobic pore shifted above the membrane plane such that the essential Asp involved in proton translocation is in close proximity to the periplasmic space.
A great deal of further research is needed in the field of Ton motor complexes. Many questions still exist including: How is the proton gradient regulated at the inner membrane? How do protons enter the inner membrane in order to interact with ExbB and ExbD? How does TonB interact with ExbB-ExbD and at what stoichiometric ratio? Similar questions can also be asked about TolA and the Tol complex due to an almost complete lack of structural data for this complex.
An additional barrier to this area of research is the lack of an in vitro assay to assess proper pmf function of the Ton motor complex. The difficulty is purifying two lipid membranes to mimic the outer membrane encompassing a TDBT and the inner membrane to house TonB-ExbB-ExbD. In addition, the pmf must stay intact, while also monitoring the structural changes of each protein involved. A similar issue arises when assessing the activity levels of the Tol-Pal complex.
Acknowledgements
We thank Dr. Nicholas M. I. Taylor for generously providing atomic coordinates of the MotA-MotB complexes prior to publication. A.C.R, H.C., and S.K.B. are supported by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing declared.
References
- 1.Nikaido H, Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev, 2003. 67(4): p. 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vergalli J, et al. , Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat Rev Microbiol, 2019. [DOI] [PubMed] [Google Scholar]
- 3.Noinaj N, et al. , TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol, 2010. 64: p. 43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.••.Celia H, Noinaj N, and Buchanan SK, Structure and Stoichiometry of the Ton Molecular Motor. Int J Mol Sci, 2020. 21(2). [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive review on the stoichiometry, mechanism of action, and structures of the Ton complex and its subunits.
- 5.Madej M, et al. , Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis. Nat Microbiol, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita M, et al. , A TonB-dependent receptor constitutes the outer membrane transport system for a lignin-derived aromatic compound. Commun Biol, 2019. 2: p. 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolam DN and van den Berg B, TonB-dependent transport by the gut microbiota: novel aspects of an old problem. Curr Opin Struct Biol, 2018. 51: p. 35–43. [DOI] [PubMed] [Google Scholar]
- 8.Calmettes C, et al. , The molecular mechanism of Zinc acquisition by the neisserial outer-membrane transporter ZnuD. Nat Commun, 2015. 6: p. 7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atanaskovic I and Kleanthous C, Tools and Approaches for Dissecting Protein Bacteriocin Import in Gram-Negative Bacteria. Front Microbiol, 2019. 10: p. 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White P, et al. , Exploitation of an iron transporter for bacterial protein antibiotic import. Proc Natl Acad Sci USA, 2017. 114(45): p. 12051–12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behrens HM, et al. , Pyocin S5 Import into Pseudomonas aeruginosa Reveals a Generic Mode of Bacteriocin Transport. mBio, 2020. 11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruhe ZC, Low DA, and Hayes CS, Polymorphic Toxins and Their Immunity Proteins: Diversity, Evolution, and Mechanisms of Delivery. Annual Review of Microbiology, 2020. 74(1): p. null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Josts I, Veith K, and Tidow H, Ternary structure of the outer membrane transporter FoxA with resolved signalling domain provides insights into TonB-mediated siderophore uptake. Elife, 2019. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarver JL, et al. , A Dynamic Protein-Protein Coupling between the TonB-Dependent Transporter FhuA and TonB. Biochemistry, 2018. 57(6): p. 1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gumbart J, Wiener MC, and Tajkhorshid E, Mechanics of force propagation in TonB-dependent outer membrane transport. Biophys J, 2007. 93(2): p. 496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noinaj N, et al. , Structural basis for iron piracy by pathogenic Neisseria. Nature, 2012. 483(7387): p. 53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Udho E, et al. , Reconstitution of bacterial outer membrane TonB-dependent transporters in planar lipid bilayer membranes. Proc Natl Acad Sci U S A, 2009. 106(51): p. 21990–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hickman SJ, et al. , Gating of TonB-dependent transporters by substrate-specific forced remodelling. Nat Commun, 2017. 8: p. 14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai YW, et al. , Evolution of the Stator Elements of Rotary Prokaryote Motors. J Bacteriol, 2020. 202(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cascales E, Lloubès R, and Sturgis JN, The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Molecular Microbiology, 2008. 42(3): p. 795–807. [DOI] [PubMed] [Google Scholar]
- 21.•.Szczepaniak J, Press C, and Kleanthous C, The multifarious roles of Tol-Palin Gram-negative bacteria. FEMS Microbiol Rev, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Review on the structures of the Tol complex and its mechanism of action.
- 22.••.Celia H, et al. , Cryo-EM structure of the bacterial Ton motor subcomplex ExbB-ExbD provides information on structure and stoichiometry. Commun Biol, 2019. 2: p. 358. [DOI] [PMC free article] [PubMed] [Google Scholar]; First evidence for the ExbB-ExbD 5:2 stoichiometry with a dimer of ExbD TM in the central hydrophobic pore. The cryo-EM structure altered the predicted mechanism for motor function.
- 23.••.Deme JC, et al. , Structures of the stator complex that drives rotation of the bacterial flagellum. Nat Microbiol, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Confirmation of the 5:2 stoichiometric ratio of MotAB, PomAB and ExbB-ExbD in several species of Gram-negative bacteria. The authors hypothesize that MotAB uses a bi-directional rotary mechanism for flagella motion. First structural evidence suggesting that TonB associates at the outside of the ExbB-ExbD subcomplex.
- 24.••.Santiveri M, et al. , Structure and function of stator units of the bacterial flagellar motor. Cell, 2020.183: p1–14. [DOI] [PubMed] [Google Scholar]; In addition to solving the cryo-EM structure of MotAB, these authors solved the structure of the unplugged MotAB construct, with and without the substitution of the MotB conserved Asp residue with Asn, as to mimic the protonated state of Asp. Comparison of the cryo-EM structures showed a path for the proton, and reorientation of the Asn sidechain.
- 25.Celia H, et al. , Structural insight into the role of the Ton complex in energy transduction. Nature, 2016. 538(7623): p. 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.•.Chorev DS, et al. , Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science, 2018. 362(6416): p. 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mass spectrometry experiments performed on E. coli native membrane showed evidence of the physiological pentameric nature of ExbB.
- 27.Morimoto YV and Minamino T, Structure and function of the bi-directional bacterial flagellar motor. Biomolecules, 2014. 4(1): p. 217–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sverzhinsky A, et al. , Membrane Protein Complex ExbB4-ExbD1-TonB1 from Escherichia coli Demonstrates Conformational Plasticity. J Bacteriol, 2015. 197(11): p. 1873–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.•.James RH, et al. , Structure of a proton-powered molecular motor that drives protein transport and gliding motility. bioRxiv, 2020: p. 2020.05.11.089193. [Google Scholar]; Structure of the GldLM complex in F. johnsoniae via cryo-EM at a 5:2 stoichometric ratio. The authors identified amino acids within the transmembrane domains that are protonatable and propose a rotary mechanism of the dimer of GldM.
- 30.Hosking ER, et al. , The Escherichia coli MotAB proton channel unplugged. J Mol Biol, 2006. 364(5): p. 921–37. [DOI] [PubMed] [Google Scholar]
- 31.Ollis AA and Postle K, Identification of functionally important TonB-ExbD periplasmic domain interactions in vivo. J Bacteriol, 2012. 194(12): p. 3078–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ollis AA and Postle K, The same periplasmic ExbD residues mediate in vivo interactions between ExbD homodimers and ExbD-TonB heterodimers. J Bacteriol, 2011. 193(24): p. 6852–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Notredame C, Higgins DG, and Heringa J, T-coffee: a novel method for fast and accurate multiple sequence alignment 1 1Edited by J. Thornton. Journal of Molecular Biology, 2000. 302(1): p. 205–217. [DOI] [PubMed] [Google Scholar]


