Abstract
Background:
The aim of this study was to quantify the association between subgingival microbiota and periodontal disease progression in older women, for which limited published data exist.
Methods:
A total of 1,016 postmenopausal women, aged 53–81 years, completed baseline (1997–2001) and 5-year (2002–2006) dental exams that included probing depth, clinical attachment level, gingival bleeding, and radiographic alveolar crestal height (ACH). Baseline microbiota were measured in subgingival plaque using 16S rRNA sequencing. Associations between 52 microbiota we previously found statistically significantly associated with clinical periodontal disease at baseline, were examined with disease progression. The traditional Socransky microbiota complexes also were evaluated. Side-by-side radiograph comparisons were used to define progression as ≥2 teeth with ≥1mm ACH loss or ≥1 new tooth loss to periodontitis. The association between baseline centered log(2) ratio transformed microbial relative abundances and 5-year periodontal disease progression was measured with generalized linear models.
Results:
Of 36 microbiota we previously showed were elevated in moderate/severe disease at baseline, 24 had statistically significantly higher baseline mean relative abundance in progressing compared with non-progressing women (P<.05, all); which included all Socransky red bacteria (P. gingivalis, T. forsythia, T. denticola). Of 16 microbiota elevated in none/mild disease at baseline, 5 had statistically significantly lower baseline abundance in non-progressing compared with progressing women (P<.05, all), including one Socransky yellow bacteria (S. oralis). When adjusted for baseline age, socioeconomic status, and self-rated general health status, odds ratios for 5-year progression ranged from 1.18–1.51 (per 1-standard deviation increment in relative abundance) for microbiota statistically significantly (P<.05) positively associated with progression, and from 0.77–0.82 for those statistically significantly (P<.05) inversely associated with progression. These associations were similar when stratified on baseline levels of pocket depth, gingival bleeding, ACH, and smoking status.
Conclusions:
These prospective results affirm clearly that subgingival microbiota are measurably elevated several years prior to progression of alveolar bone loss, and include antecedent elevations in previously undocumented taxa additional to known Socransky pathogenic complexes.
Keywords: Oral Microbiome, Women, Periodontal Disease, Alveolar Bone Loss, Longitudinal Study
Summary:
In a prospective cohort of older postmenopausal women, subgingival microbiota measured 5-years prior to alveolar bone loss are elevated and significantly associated with longitudinal bone loss and its severity, and include new taxa additional to known Socransky complexes.
INTRODUCTION
Foundational to periodontal disease etiology is development of a polymicrobial biofilm on the surface of the tooth and tooth root.1 Shifts in biofilm composition and diversity induce altered host immune and inflammatory responses leading to destruction of the periodontium surrounding the tooth.2 Cross-sectional studies have documented a relationship between the subgingival microbiome and periodontal disease presence and severity.3–6 Cross-sectional findings cannot conclusively establish temporality between microbiota and disease, which limits causal arguments inferred from these studies.4, 7
Some studies have published associations between subgingival microbiota and disease progression.8–21 The majority were small studies (e.g., n<150) often in patients selected on periodontal status or undergoing clinical therapy, and subgingival bacteria were measured using targeted approaches at sites after progression had occurred during a preceding time interval. Progression typically was defined using probing measures, which have considerable intra-individual visit-to-visit variability limiting their reliability in quantifying progression.22
We have been prospectively following 1,342 postmenopausal women enrolled in the Buffalo Osteoporosis and Periodontal Disease (OsteoPerio) Study who have serial subgingival plaque sampling and radiographic alveolar crestal height (ACH) measurement.23 In this cohort, 5-year progression of ACH loss and its relationship with baseline periodontal disease and other characteristics has been published.24 Using 16S sequencing, 267 bacterial species have been identified in participant subgingival plaque samples at baseline, of which 56 (20.9%) differed significantly in abundance according to disease severity.25 The aim of this study was to quantify the association of microbiota that differed by baseline disease severity with 5-year disease progression defined by radiographic ACH loss.
MATERIALS AND METHODS
Study Cohort
Participants were postmenopausal women initially recruited from the community setting in 1993–1998 and enrolled into the Women’s Health Initiative Observational Study (WHIOS; N = 2,249; ages 50–79 years) in Buffalo (NY). In 1997–2001, participants further enrolled into the ancillary Buffalo OsteoPerio Study (N = 1,342). Details on the WHIOS and OsteoPerio studies have been published.23, 24, 26 Periodontal disease status was not a criterion for inclusion or exclusion in either study. Of the 1,342 women enrolled at OsteoPerio study baseline, 1,026 were reexamined 5-years later. Baseline microbiome measures, and radiographic ACH measures at both baseline and 5-years to measure progression, were available in 956 women (Figure 1). Written informed consent was obtained from participants. This study was approved by the human subjects ethics board at the University at Buffalo and was conducted in accordance with the Helsinki Declaration of 1975, as revised in 2013. The study conformed to STROBE guidelines for human observational studies.27
Figure 1.
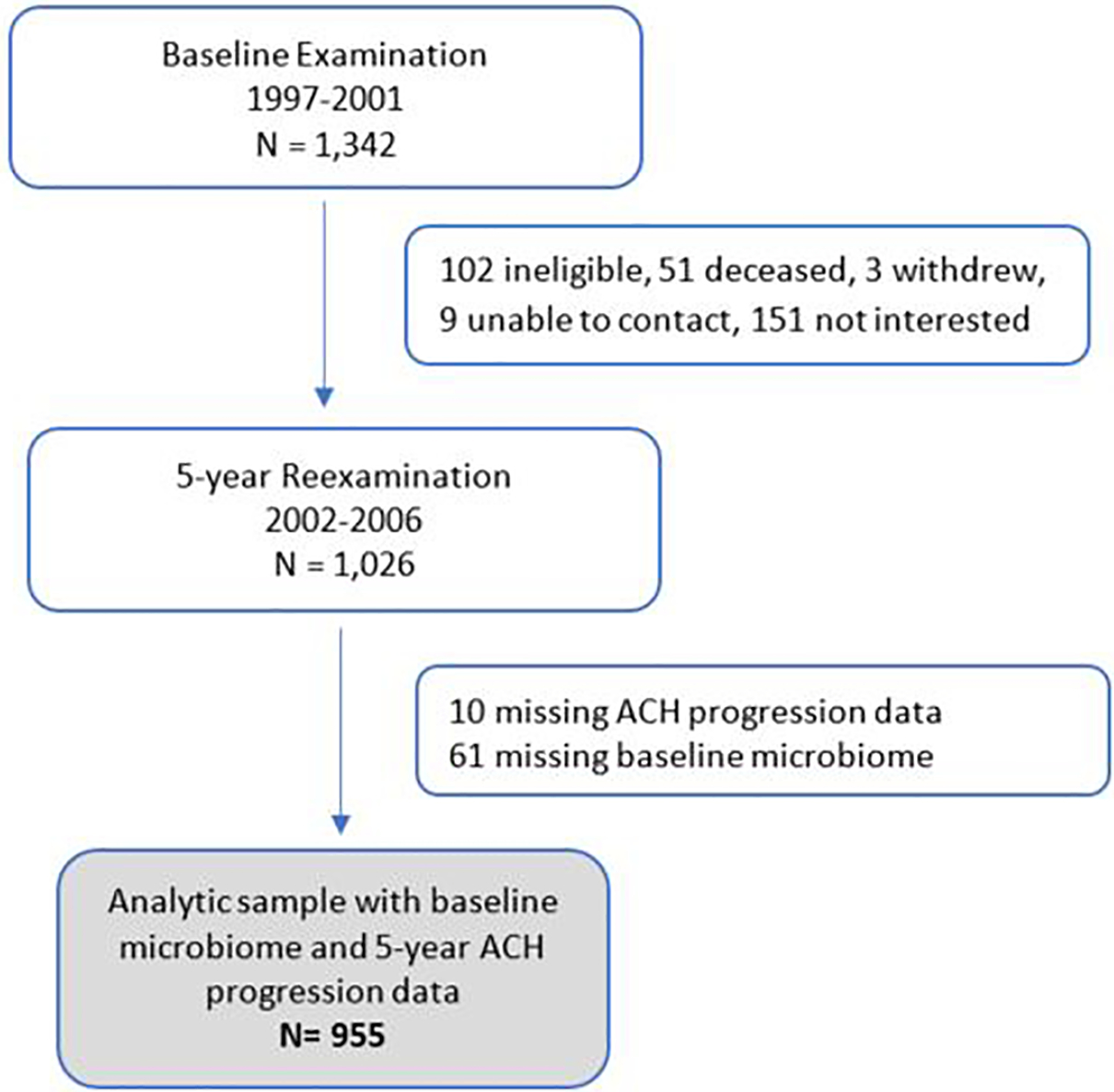
Flow of participants enrolled at OsteoPerio baseline who completed 5-Year reexaminations and are included in the present analysis on baseline microbiome and 5-year periodontal disease progression.
Periodontal Examination and Probing Measures
Participants completed whole mouth dental examinations at both time points conducted by regularly calibrated technicians.23 Reason for missing teeth, including loss due to periodontitis, was documented. Pocket depth (PD) and clinical attachment level (CAL) was measured with an electronic probe# using a standardized probing protocol on six surfaces per tooth except for third molars.24 The within-rater CV was 6% for replicate PD measures in a subset of 724 women. Presence of gingival bleeding was also recorded. Gingival bleeding on probing (absent or present) was assessed at three sites per tooth (buccal, mesiobuccal and lingual) using a manual probe inserted 2 mm into gingival sulcus parallel to long axis of tooth moved in a horizontal direction and is expressed as the percentage of sites bleeding in the whole mouth. Periodontal disease severity was based on PD and CAL classified using CDC/AAP criteria.28
Subgingival Microbiome Measurement
Detailed procedures and quality control steps used to measure the microbiome have been published.23, 25, 29, 30 Subgingival plaque samples were obtained using paper points at pre-specified sites following a published protocol.31 PCR amplification of genomic DNA using the 16S V3 (341F) forward and V4 (805R) reverse primer pairs with added Illumina adapter overhang nucleotide sequences was performed on 96 samples at a time with both positive controls (mock DNA, subgingival plaque pools) and negative controls (PCR grade water, extraction buffer). Samples were multiplexed and 300 bp pair-end sequenced on an Illumina MiSeq**. Sequences were clustered at 97% identity against the Human Oral Microbiome Database (HOMD) version 14.532 with BLAST.33 Batches of 85–88 test samples were processed together, randomly arranged on 96 well plates with negative and positive quality control samples to minimize batch effects. The raw OTU table was filtered at >0.02% abundance of the total read count.
Alveolar Crestal Height Measurement
ACH was measured in seven anterior periapical and four posterior vertical bitewing radiographs using a single radiographic unit††. Projection geometry was controlled by stabilizing participant’s heads with a cephalostat. Radiographs were captured by a digital imaging system.34, 35 ACH was measured as the distance from the CEJ to the most coronal part of the alveolar crest in a plane parallel to the long axis of the tooth. The within-rater CV was 5.1% for replicate ACH measures in a subset of 885 women.
The primary outcome was disease progression defined by ACH change between baseline and 5-years calculated (i.e., “progressing women”) on pairs of digitized images using the “side by side” method.34 For each site, paired radiograph images were displayed on the same monitor and, using a flicker system, the second image was aligned with the first image. This technique allows use of the same landmark to compute changes in ACH over time. If the same CEJ feature is not evident, then an alternative landmark is chosen. The difference between the two sites represents the net change in ACH. The primary case definition for progression was binary, defined as ≥2 teeth with ≥1mm ACH loss or ≥1 new tooth loss to periodontitis. A secondary severity endpoint was defined as moderate (2 or more teeth with ≥1 ACH loss) or severe (2 or more teeth with ≥2 mm ACH loss or ≥1 new tooth loss to periodontitis) progression.
Other Assessments
Body mass index (BMI; kg/m2) was calculated from height (cm) and weight (kg) measured in clinic. Smoking history, hormone therapy use, self-rated general health status, treated diabetes, and frequency of tooth brushing, flossing, dental visits, and history of gum disease/surgery were assessed by questionnaire. Neighborhood socioeconomic status (nSES, score 0–100) was characterized using aggregate census tract information; higher scores indicate greater affluence.36
Statistical Analysis
Prior to analysis, we normalized OTU relative abundance using the centered log(2) ratio (CLR) transformation which accounts for the compositional data structure, reduces the likelihood of spurious correlations, and enhances the meaningfulness of subcomposition comparisons.37 Linear relationships between CLR abundance and baseline ACH measurements were evaluated using Pearson correlations. Comparisons of mean CLR abundance with progression were performed using Student’s t-tests and generalized linear models. Logistic regression was used to estimate odds ratios (OR) and 95% confidence intervals (CI) for 5-year disease progression on a 1-standard deviation increment in baseline CLR abundance. Multivariable analyses controlled for baseline age (years), nSES (continuous), and self-rated general health (Excellent/very good, Good, Fair/poor). To explore the influence that baseline periodontal disease or smoking status might have on associations between microbiota and progression, multivariable models were also stratified according to median values for baseline PD (2.2 mm), percentage of bleeding sites on probing (31%), ACH (2.3 mm), and smoking (never, ever). Progression also was explored in relation to Socransky red, orange, and yellow complex bacteria.6 P-values are two-tailed for hypothesis tests at alpha .05. P-values were not corrected for multiple comparisons; .05/52 tests yields alpha ≈.001. Analyses were performed using SAS software (Carey, NC; v.9.4).
RESULTS
Baseline characteristics according to periodontal disease progression at 5-years are in Table 1. Women who progressed were somewhat older with greater prevalence of current smoking and smoking pack-years compared with non-progressing women. Prevalence of current hormone therapy use was lower among progressing women. Differences in smoking status and pack-years, and prevalence of diabetes and Fair/Poor general health were larger when comparing severe and no progression.
Table 1.
Baseline characteristics according to periodontal disease progression at 5-year examination (N=1,016).
| Characteristic | No Progression (N=791) | Progression (N=225) | Progression Severity | |
|---|---|---|---|---|
| Moderate (N=161) | Severe (N=64) | |||
| Demographics | ||||
| Age, (years) mean (SD) | 65.7 (6.6) | 66.2 (6.9) | 66.0 (6.8) | 66.8 (7.1) |
| < 60 (years), N (%) | 154 (19.5) | 39 (17.3) | 31 (19.3) | 8 (12.5) |
| 60 – 69 | 384 (48.6) | 113 (50.2) | 78 (48.5) | 35 (54.7) |
| 70 – 79 | 240 (30.3) | 68 (30.2) | 51 (31.7) | 17 (26.6) |
| ≥80 | 13 (1.6) | 5 (2.2) | 1 (0.6) | 4 (6.3) |
| Race, N (%) | ||||
| White | 774 (97.9) | 222 (98.7) | 160 (99.4) | 62 (96.9) |
| Black | 9 (1.1) | 2 (0.9) | 1 (0.6) | 1 (1.6) |
| Other | 8 (1.0) | 1 (0.4) | 0 (0) | 1 (1.6) |
| Neighborhood SES, mean (SD) | 76.0 (7.2) | 76.7 (6.0) | 76.9 (5.9) | 76.0 (6.3) |
| BMI (kg/m2), mean (SD) | 26.5 (5.0) | 26.6 (5.6) | 26.6 (5.4) | 26.6 (6.1) |
| Smoking, N (%) | ||||
| Never | 438 (55.4) | 114 (50.7) | 90 (55.9) | 24 (37.5) |
| Former | 336 (42.5) | 101 (44.9) | 65 (40.4) | 36 (56.3) |
| Current | 17 (2.2) | 10 (4.4) | 6 (3.7) | 4 (6.3) |
| Pack-years smoking, mean (SD) | 8.8 (16.6) | 12.4 (19.7) | 10.1 (17.5) | 18.0 (23.5) |
| Treated Diabetes, N (%) | 35 (4.4) | 9 (4.0) | 5 (3.1) | 4 (6.3) |
| Self-rated general health status, N (%) | ||||
| Excellent/Very good | 514(66.9) | 149 (69.0) | 105 (68.6) | 44 (69.8) |
| Good | 219 (28.5) | 60 (27.8) | 46 (30.1) | 14 (22.2) |
| Fair/Poor | 35 (4.5) | 7 (3.3) | 2 (1.3) | 5 (8.0) |
| Hormone Therapy use, N (%) | ||||
| Never | 239 (30.2) | 81 (36.0) | 58 (36.0) | 23 (35.9) |
| Former | 152 (19.2) | 42 (18.7) | 33 (20.5) | 9 (14.1) |
| Current | 400 (50.6) | 102 (45.3) | 70 (43.5) | 32 (50.0) |
| Dental measures | ||||
| Periodontal Disease (CDC/AAP), N (%) | ||||
| None/Mild | 222 (28.3) | 47 (21.2) | 41 (25.8) | 6 (9.6) |
| Moderate | 473 (60.3) | 111 (50.0) | 85 (53.5) | 26 (41.3) |
| Severe | 90 (11.5) | 64 (28.8) | 33 (20.8) | 31 (49.2) |
| Number of teeth present, mean (SD) | 23.6 (5.2) | 23.1 (5.0) | 23.8 (4.4) | 21.2 (6.1) |
| Number of filled/decayed teeth, mean (SD) | 11.2 (4.6) | 10.3 (4.6) | 11.1 (4.6) | 8.2 (4.5) |
| Tooth loss from periodontitis, N (%) | 46 (5.8) | 33 (14.7) | 8 (5.0) | 25 (39.1) |
| Gingival bleeding on probing (%), mean (SD) | 33.2 (21.9) | 34.9 (24.9) | 33.4 (22.9) | 38.4 (29.2) |
| Pocket depth (mm), mean (SD) | ||||
| Whole Mouth Mean | 2.2 (0.4) | 2.3 (0.5) | 2.2 (0.4) | 2.5 (0.5) |
| Worst Site | 4.8 (1.2) | 5.3 (1.6) | 5.0 (1.4) | 6.1 (1.7) |
| Clinical attachment level (mm), mean (SD) | ||||
| Whole Mouth Mean | 2.3 (0.6) | 2.6 (0.8) | 2.4 (0.6) | 3.0 (1.1) |
| Worst Site | 5.4 (1.6) | 6.4 (2.2) | 5.9 (1.9) | 7.7 (2.3) |
| Alveolar crestal height (mm), mean (SD) | ||||
| Whole Mouth Mean | 2.4 (0.7) | 2.7 (0.9) | 2.5 (0.6) | 3.3 (1.2) |
| Worst Site | 4.4 (1.4) | 5.4 (1.9) | 4.9 (1.4) | 6.3 (2.4) |
| Tooth brushing ≥ 2 time/day, N (%) | 605 (76.5) | 180 (80.0) | 130 (80.8) | 50 (78.1) |
| Flossing every day, N (%) | 331 (42.1) | 113 (50.5) | 79 (49.4) | 34 (53.1) |
| Dental visit ≥1 time/year, N (%) | 727 (91.9) | 206 (91.6) | 148 (91.9) | 58 (90.6) |
| History of gum disease/surgery, N (%) | 154 (20.1) | 81 (37.3) | 40 (25.8) | 41 (66.1) |
SD, standard deviation; mm, millimeters; BMI, body mass index; SES, socioeconomic status ranges from 0 to 100, higher score reflects more affluent status; CDC/AAP periodontal disease categories as defined by Eke et al.28
Prevalence of severe periodontal disease at baseline was twice as high in progressing (28.8%) compared with non-progressing (11.5%) women; prevalence was 4-fold higher for women with severe progression (49.2%) (Table 1). On average, the number of teeth present at baseline was similar between non-progressing women and those with any progression, but lower in severe progression. Prevalence of tooth loss to periodontitis at baseline was two-fold higher among progressing women, and nearly 8-fold higher among severe progressors as compared with no progression. Percentage of sites bleeding on probing at baseline was greater in any progression, even more so in severe progression. PD, CAL, and ACH was higher at baseline in progressing compared with non-progressing women; worst site means tended to be larger than whole mouth means. Frequency of dental visits did not differ according to progression, but frequency of tooth brushing and flossing was higher in progression compared with no progression. Baseline history of gum disease/surgery was substantially higher in women with any (37.3%) and severe progression (66.1%) compared with no progression (20.1%).
Pearson correlations between baseline subgingival microbiota and ACH measures are in Table 2. Among 36 microbiota elevated in Moderate/Severe disease at baseline, correlations ranged from r = <0.01 to 0.32 and tended to be stronger for worst site than mean ACH. For the 16 taxa elevated in None/Mild disease at baseline, most correlations were inverse with baseline mean mouth ACH (r = −0.19 to 0.03); all correlations were inverse with worst site ACH (r = −0.06 to −0.19).
Table 2.
Pearson correlations between baseline mean CLR microbial abundance and baseline ACH (mm).
| Subgingival OTU Label | Baseline ACH | |
|---|---|---|
| Mean mouth (Pearson r*) | Worst site (Pearson r*) | |
| 36 Microflora elevated in moderate/severe periodontal categories (CDC/AAP) at baseline† | ||
| Fretibacterium fastidiosum | 0.16 | 0.21 |
| Tannerella forsythia | 0.14 | 0.21 |
| Fretibacterium sp._oral_taxon_360 | 0.14 | 0.19 |
| Fretibacterium sp._oral_taxon_359 | 0.15 | 0.21 |
| Porphyromonas gingivalis | 0.19 | 0.27 |
| Desulfobulbus sp._oral_taxon_041 | 0.17 | 0.19 |
| Anaerolineae_[G-1] sp._oral_taxon_439 | 0.25 | 0.32 |
| Treponema denticola | 0.09 | 0.15 |
| Dialister pneumosintes | 0.06 | 0.10 |
| Peptostreptococcaceae_[XI][G-6] [Eubacterium]_nodatum | 0.19 | 0.27 |
| Treponema maltophilum | 0.16 | 0.21 |
| Selenomonas sp._oral_taxon_134 | 0.03 | 0.08 |
| Treponema socranskii | 0.11 | 0.16 |
| Fretibacterium sp._oral_taxon_362 | 0.19 | 0.24 |
| Pseudoramibacter alactolyticus | 0.23 | 0.24 |
| Streptococcus constellatus | 0.11 | 0.11 |
| Porphyromonas endodontalis | 0.04 | 0.08 |
| Anaeroglobus geminatus | 0.11 | 0.09 |
| Veillonellaceae_[G-1] sp._oral_taxon_150 | 0.09 | 0.09 |
| Prevotella intermedia | 0.05 | 0.09 |
| Bacteroidaceae_[G-1] sp._oral_taxon_272 | 0.21 | 0.27 |
| Lachnospiraceae_[G-8] sp._oral_taxon_500 | 0.12 | 0.17 |
| TM7_[G-1] sp._oral_taxon_349 | 0.06 | 0.09 |
| Filifactor alocis | 0.09 | 0.11 |
| Peptostreptococcaceae_[XI][G-5] [Eubacterium]_saphenum | 0.14 | 0.17 |
| Prevotella dentalis | 0.11 | 0.16 |
| TM7_[G-5] sp._oral_taxon_356 | 0.00 | 0.02 |
| Veillonellaceae_[G-1] sp._oral_taxon_145 | 0.12 | 0.16 |
| Prevotella oralis | 0.11 | 0.09 |
| Prevotella sp._oral_taxon_526 | 0.16 | 0.22 |
| Johnsonella sp._oral_taxon_166 | 0.12 | 0.17 |
| Prevotella baroniae | 0.09 | 0.14 |
| Fusobacterium nucleatum_subsp._vincentii | 0.02 | 0.06 |
| Fretibacterium sp._oral_taxon_361 | 0.14 | 0.19 |
| Parvimonas micra | 0.09 | 0.10 |
| Fretibacterium sp._oral_taxon_358 | 0.16 | 0.16 |
| 16 Microflora elevated in none/mild periodontal categories (CDC/AAP) at baseline† | ||
| Microbacterium flavescens | 0.01 | −0.06 |
| Sphingomonas sp._oral_taxon_006 | 0.03 | −0.07 |
| Porphyrobacter tepidarius | 0.02 | −0.07 |
| Brevundimonas diminuta | 0.00 | −0.08 |
| Actinomyces naeslundii | −0.11 | −0.11 |
| Streptococcus oralis | −0.09 | −0.13 |
| Capnocytophaga sp._oral_taxon_324 | −0.00 | −0.03 |
| Actinomyces massiliensis | −0.17 | −0.14 |
| Haemophilus parahaemolyticus | −0.05 | −0.06 |
| Sphingomonas echinoides | −0.05 | −0.11 |
| Gemella haemolysans | −0.09 | −0.09 |
| Streptococcus sp._oral_taxon_056 | −0.03 | −0.07 |
| Haemophilus parainfluenzae | −0.13 | −0.13 |
| Leptotrichia goodfellowii | −0.14 | −0.14 |
| Rothia aeria | −0.19 | −0.15 |
| Lautropia mirabilis | −0.18 | −0.19 |
Microbiota ordered according to relative abundance (highest to lowest) in overall cohort.
P <0.05 for |r| ≥0.07.
As reported in Genco et al.25
At reexamination 5-years after baseline, 225 (22.2%) women demonstrated disease progression, of whom 161 (71.6%) were classified as moderate and 64 (28.4%) as severe. Table 3 gives mean CLR abundances for baseline microbiota according to 5-year progression. Significantly (P<.05) higher abundance in progressing women was observed for 24 of 36 taxa that were elevated in Moderate/Severe disease at baseline. Socransky/Haffajee6 red bacteria (P. gingivalis, T. forsythia, T. denticola; P<.0001 each) were among microbiota that were of significantly greater abundance among progressors. Of 16 microbiota elevated in None/Mild disease at baseline, 5 had significantly (P<.05) lower baseline abundance in non-progressors, including one Socransky yellow bacteria (S. oralis; P=.002). For most microbiota, differences in abundances were stronger between severe and no progression (see Appendix Table S1 in online Journal of Periodontology).
Table 3.
Relative abundance* of subgingival microbiota at baseline according to periodontal disease progression at 5-year examination.
| Subgingival OTU Label | No Progression Mean (SD)* | Progression Mean (SD)* | P-value† |
|---|---|---|---|
| 36 Microflora elevated in moderate/severe periodontal categories (CDC/AAP) at baseline‡ | |||
| Fretibacterium fastidiosum | 0.75 (3.35) | 1.82 (3.48) | <.0001 |
| Tannerella forsythia (Socransky red complex) | 1.31 (3.38) | 2.40 (3.34) | <.0001 |
| Fretibacterium sp._oral_taxon_360 | 2.26 (3.61) | 3.27 (3.65) | <0.001 |
| Fretibacterium sp._oral_taxon_359 | −0.36 (3.62) | 0.94 (4.01) | <.0001 |
| Porphyromonas gingivalis (Socransky red complex) | −1.08 (3.93) | 0.37 (4.85) | <.0001 |
| Desulfobulbus sp._oral_taxon_041 | −1.14 (3.27) | −0.25 (3.36) | <.001 |
| Anaerolineae_[G-1] sp._oral_taxon_439 | −2.58 (2.47) | −1.98 (2.81) | .003 |
| Treponema denticola (Socransky red complex) | −0.67 (3.46) | 0.45 (3.79) | <.0001 |
| Dialister pneumosintes | 0.11 (3.16) | 0.56 (3.05) | .07 |
| Peptostreptococcaceae_[XI][G-6] [Eubacterium]_nodatum | −2.20 (2.58) | −1.29 (3.11) | <.0001 |
| Treponema maltophilum | −0.92 (2.46) | −0.26 (2.68) | <.001 |
| Selenomonas sp._oral_taxon_134 | 0.36 (3.32) | 0.98 (3.34) | .02 |
| Treponema socranskii | 1.74 (2.58) | 2.29 (2.40) | .006 |
| Fretibacterium sp._oral_taxon_362 | −1.89 (2.76) | −0.98 (3.51) | <.0001 |
| Pseudoramibacter alactolyticus | −2.09 (2.72) | −1.74 (2.86) | .12 |
| Streptococcus constellatus | −0.01 (3.40) | 0.24 (3.44) | .36 |
| Porphyromonas endodontalis | −0.07 (3.97) | 1.61 (4.14) | <.0001 |
| Anaeroglobus geminatus | 1.72 (3.50) | 1.47 (3.10) | .35 |
| Veillonellaceae_[G-1] sp._oral_taxon_150 | 0.90 (3.13) | 0.82 (3.03) | .73 |
| Prevotella intermedia (Socransky orange complex) | −1.74 (3.71) | −1.23 (4.13) | .09 |
| Bacteroidaceae_[G-1] sp._oral_taxon_272 | −2.54 (2.33) | −2.15 (2.53) | .04 |
| Lachnospiraceae_[G-8] sp._oral_taxon_500 | −2.35 (2.37) | −1.32 (2.87) | <.0001 |
| TM7_[G-1] sp._oral_taxon_349 | 2.56 (3.43) | 3.04 (3.18) | .07 |
| Filifactor alocis | −1.48 (3.44) | 0.09 (4.22) | <.0001 |
| Peptostreptococcaceae_[XI][G-5] [Eubacterium]_saphenum | −2.59 (2.58) | −1.71 (3.22) | <.0001 |
| Prevotella dentalis | −2.09 (2.77) | −1.41 (3.12) | 0.003 |
| TM7_[G-5] sp._oral_taxon_356 | 0.19 (3.62) | 0.55 (3.76) | .21 |
| Veillonellaceae_[G-1] sp._oral_taxon_145 | −2.21 (2.69) | −1.64 (2.93) | .009 |
| Prevotella oralis | −0.53 (3.35) | −0.42 (3.11) | .67 |
| Prevotella sp._oral_taxon_526 | −2.88 (2.38) | −2.42 (2.72) | .02 |
| Johnsonella sp._oral_taxon_166 | −3.17 (2.12) | −2.35 (2.77) | <.0001 |
| Prevotella baroniae | −2.35 (2.48) | −2.06 (2.76) | .15 |
| Fusobacterium nucleatum_subsp._vincentii (Socransky orange complex) | 6.29 (2.56) | 6.21 (2.52) | .68 |
| Fretibacterium sp._oral_taxon_361 | −3.29 (1.99) | −2.85 (2.51) | .009 |
| Parvimonas micra (Socransky orange complex) | 4.04 (2.61) | 4.17 (2.55) | .54 |
| Fretibacterium sp._oral_taxon_358 | −3.15 (2.15) | −2.74 (2.94) | .03 |
| 16 Microflora elevated in none/mild periodontal categories (CDC/AAP) at baseline‡ | |||
| Microbacterium flavescens | −3.09 (1.57) | −3.14 (1.59) | .66 |
| Sphingomonas sp._oral_taxon_006 | −3.75 (1.34) | −3.93 (1.25) | .09 |
| Porphyrobacter tepidarius | −3.80 (1.27) | −4.00 (1.25) | .05 |
| Brevundimonas diminuta | −3.48 (1.44) | −3.65 (1.43) | .15 |
| Actinomyces naeslundii | 3.77 (2.10) | 3.33 (2.05) | .009 |
| Streptococcus oralis (Socransky yellow complex) | 7.95 (1.86) | 7.49 (1.81) | .002 |
| Capnocytophaga sp._oral_taxon_324 | −2.46 (2.52) | −2.65 (2.55) | .35 |
| Actinomyces massiliensis | 1.40 (2.37) | 0.78 (2.40) | .001 |
| Haemophilus parahaemolyticus | −2.47 (2.78) | −2.51 (2.66) | .87 |
| Sphingomonas echinoides | −2.03 (2.38) | −2.34 (2.23) | .10 |
| Gemella haemolysans | 1.94 (3.08) | 1.94 (2.90) | .99 |
| Streptococcus sp._oral_taxon_056 | −0.67 (2.96) | −0.86 (2.80) | .41 |
| Haemophilus parainfluenzae | 3.68 (3.08) | 3.10 (2.93) | .02 |
| Leptotrichia goodfellowii | −2.36 (2.50) | −2.36 (2.69) | .99 |
| Rothia aeria | 2.16 (3.13) | 1.71 (3.23) | .07 |
| Lautropia mirabilis | 0.93 (3.04) | 0.31 (3.08) | .01 |
Centered log(base 2) transformed OTU. Microbiota ordered according to relative abundance (highest to lowest) in overall cohort.
P-values are from Student’s t-tests, not corrected for multiple comparisons.
As reported in Genco et al.25
Crude odds ratios for a 1-standard deviation increment in CLR abundance ranged from 1.17 to 1.51 for taxa significantly and positively associated with disease progression (Table 4). Among microbiota inversely and significantly associated with progression, odds ratios ranged from 0.77 to 0.83. Adjustment for baseline age, nSES, and self-rated general health did not materially change these associations. Associations were stronger with severe progression (see Appendix Table S2 in online Journal of Periodontology); statistical significance was more variable due to the small sample size in the severe progression group.
Table 4.
Associations between baseline subgingival microbiota and periodontal disease progression at 5-year examination.
| Subgingival OTU Label | Crude OR (95% CI)* | P-value | Adjusted OR (95% CI)*† | P-value |
|---|---|---|---|---|
| 36 Microflora elevated in moderate/severe periodontal categories (CDC/AAP) at baseline‡ | ||||
| Fretibacterium fastidiosum | 1.37 (1.17–1.60) | <.001 | 1.38 (1.18–1.62) | <.001 |
| Tannerella forsythia (Socransky red complex) | 1.39 (1.18–1.63) | <.001 | 1.40 (1.19–1.65) | <.001 |
| Fretibacterium sp._oral_taxon_360 | 1.33 (1.13–1.56) | <.001 | 1.34 (1.14–1.58) | <.001 |
| Fretibacterium sp._oral_taxon_359 | 1.39 (1.20–1.61) | <.001 | 1.41 (1.21–1.64) | <.001 |
| Porphyromonas gingivalis (Socransky red complex) | 1.38 (1.19–1.60) | <.001 | 1.40 (1.21–1.62) | <.001 |
| Desulfobulbus sp._oral_taxon_041 | 1.30 (1.12–1.51) | <.001 | 1.31 (1.13–1.53) | <.001 |
| Anaerolineae_[G-1] sp._oral_taxon_439 | 1.25 (1.08–1.44) | .003 | 1.27 (1.09–1.47) | .001 |
| Treponema denticola (Socransky red complex) | 1.36 (1.16–1.58) | <.001 | 1.36 (1.17–1.59) | <.001 |
| Dialister pneumosintes | 1.15 (0.99–1.35) | .07 | 1.17 (1.00–1.36) | .09 |
| Peptostreptococcaceae_[XI][G-6] [Eubacterium]_nodatum | 1.37 (1.18–1.58) | <.001 | 1.40 (1.20–1.62) | <.001 |
| Treponema maltophilum | 1.29 (1.11–1.51) | .001 | 1.31 (1.12–1.52) | <.001 |
| Selenomonas sp._oral_taxon_134 | 1.20 (1.03–1.40) | .02 | 1.21 (1.04–1.41) | .03 |
| Treponema socranskii | 1.25 (1.06–1.47) | .007 | 1.27 (1.08–1.50) | .005 |
| Fretibacterium sp._oral_taxon_362 | 1.33 (1.15–1.53) | <.001 | 1.35 (1.17–1.57) | <.001 |
| Pseudoramibacter alactolyticus | 1.13 (0.97–1.31) | .11 | 1.15 (0.98–1.34) | .08 |
| Streptococcus constellatus | 1.07 (0.92–1.25) | .36 | 1.08 (0.93–1.26) | .25 |
| Porphyromonas endodontalis | 1.51 (1.29–1.76) | <.001 | 1.51 (1.29–1.77) | <.001 |
| Anaeroglobus geminatus | 0.93 (0.80–1.09) | .35 | 0.93 (0.79–1.09) | .29 |
| Veillonellaceae_[G-1] sp._oral_taxon_150 | 0.97 (0.83–1.14) | .73 | 0.98 (0.84–1.15) | .89 |
| Prevotella intermedia (Socransky orange complex) | 1.14 (0.98–1.32) | .09 | 1.15 (0.99–1.34) | .06 |
| Bacteroidaceae_[G-1] sp._oral_taxon_272 | 1.17 (1.01–1.36) | .04 | 1.18 (1.02–1.37) | .03 |
| Lachnospiraceae_[G-8] sp._oral_taxon_500 | 1.46 (1.26–1.69) | <.001 | 1.47 (1.27–1.71) | <.001 |
| TM7_[G-1] sp._oral_taxon_349 | 1.16 (0.99–1.35) | .07 | 1.15 (0.99–1.35) | .09 |
| Filifactor alocis | 1.49 (1.29–1.73) | <.001 | 1.50 (1.30–1.74) | <.001 |
| Peptostreptococcaceae_[XI][G-5] [Eubacterium]_saphenum | 1.34 (1.16–1.55) | <.001 | 1.34 (1.16–1.54) | .003 |
| Prevotella dentalis | 1.25 (1.08–1.45) | .003 | 1.26 (1.08–1.46) | .003 |
| TM7_[G-5] sp._oral_taxon_356 | 1.10 (0.95–1.29) | .21 | 1.10 (0.94–1.28) | .29 |
| Veillonellaceae_[G-1] sp._oral_taxon_145 | 1.22 (1.05–1.41) | .009 | 1.23 (1.06–1.42) | .008 |
| Prevotella oralis | 1.03 (0.89–1.21) | .67 | 1.04 (0.89–1.22) | .64 |
| Prevotella sp._oral_taxon_526 | 1.19 (1.03–1.38) | .02 | 1.22 (1.05–1.41) | .009 |
| Johnsonella sp._oral_taxon_166 | 1.37 (1.19–1.58) | <.001 | 1.39 (1.20–1.60) | <.001 |
| Prevotella baroniae | 1.12 (0.96–1.30) | .15 | 1.12 (0.97–1.31) | .17 |
| Fusobacterium nucleatum_subsp._vincentii (Socransky orange complex) | 0.97 (0.83–1.13) | .68 | 0.97 (0.83–1.13) | .62 |
| Fretibacterium sp._oral_taxon_361 | 1.21 (1.05–1.39) | .009 | 1.21 (1.05–1.40) | .005 |
| Parvimonas micra (Socransky orange complex) | 1.05 (0.90–1.23) | .54 | 1.05 (0.90–1.23) | .47 |
| Fretibacterium sp._oral_taxon_358 | 1.17 (1.02–1.36) | .03 | 1.18 (1.02–1.36) | .02 |
| 16 Microflora elevated in none/mild periodontal categories (CDC/AAP) at baseline‡ | ||||
| Microbacterium flavescens | 0.97 (0.83–1.13) | .67 | 0.97 (0.83–1.14) | .62 |
| Sphingomonas sp._oral_taxon_006 | 0.87 (0.74–1.02) | .09 | 0.88 (0.75–1.03) | .09 |
| Porphyrobacter tepidarius | 0.85 (0.73–1.00) | .05 | 0.86 (0.73–1.01) | .05 |
| Brevundimonas diminuta | 0.89 (0.76–1.04) | .15 | 0.90 (0.76–1.05) | .12 |
| Actinomyces naeslundii | 0.82 (0.70–0.95) | .009 | 0.81 (0.70–0.95) | .01 |
| Streptococcus oralis (Socransky yellow complex) | 0.78 (0.66–0.91) | .002 | 0.77 (0.66–0.91) | .002 |
| Capnocytophaga sp._oral_taxon_324 | 0.93 (0.79–1.09) | .35 | 0.92 (0.78–1.08) | .32 |
| Actinomyces massiliensis | 0.77 (0.66–0.90) | .001 | 0.76 (0.65–0.89) | .001 |
| Haemophilus parahaemolyticus | 0.99 (0.84–1.15) | .87 | 0.98 (0.84–1.15) | .95 |
| Sphingomonas echinoides | 0.87 (0.75–1.03) | .10 | 0.87 (0.74–1.03) | .11 |
| Gemella haemolysans | 1.00 (0.86–1.17) | .99 | 1.00 (0.86–1.17) | .72 |
| Streptococcus sp._oral_taxon_056 | 0.94 (0.80–1.10) | .41 | 0.92 (0.79–1.08) | .37 |
| Haemophilus parainfluenzae | 0.83 (0.71–0.96) | .02 | 0.82 (0.70–0.96) | .01 |
| Leptotrichia goodfellowii | 1.00 (0.86–1.17) | .99 | 1.00 (0.86–1.17) | .80 |
| Rothia aeria | 0.87 (0.74–1.01) | .07 | 0.85 (0.73–1.00) | .07 |
| Lautropia mirabilis | 0.82 (0.70–0.95) | .01 | 0.80 (0.69–0.94) | .02 |
OR, odds ratio, and CI, confidence interval, are for a 1-standard deviation increment in CLR transformed OTU.
See Table 3 for standard deviations. Microbiota ordered according to relative abundance (highest to lowest) in overall cohort.
Bold indicates statistical significance, not corrected for multiple comparisons.
Adjusted for age, neighborhood SES, and self-rated health status at baseline.
As reported in Genco et al.25
Analyses were conducted to explore whether baseline periodontal measures (see Appendix Table S3 in online Journal of Periodontology) and smoking status (see Appendix Table S4 in online Journal of Periodontology) might have influenced the associations between microbiota and disease progression. Stratified associations were comparable with the primary results in Table 4. We also explored associations for the microbiota in Socransky’s red, orange, and yellow complexes (see Appendix Table S5 in online Journal of Periodontology). Each red bacteria (adjusted OR 1.36–1.40, P<.001 all) and the summary complex (OR 1.49, P<.001), was positively associated with 5-year progression. Three (S. oralis, S. sanguinis, S. gordonii) of four yellow bacteria and the summary complex (OR 0.75, P<.001) were inversely associated with progression. Orange bacteria were not associated with progression.
Baseline microbiota positively associated with progression, that have been identified less frequently or not at all in previous studies, included F. fastidiosum, F. sp. oral taxa 359, 360, 361, and 362, D. sp oral taxon 041, A. [G-1] sp oral taxon 439, T. maltophilum, P. endodontalis, and J. sp oral taxon 166 (Table 2).
DISCUSSION
In this prospective study on postmenopausal women we observed significant associations with 5-year periodontal disease progression for 29 of 52 subgingival microbiota that differed significantly by disease presence and severity at baseline. The majority (24/29) of these prospective associations were for taxa elevated in moderate/severe disease at baseline, whereas fewer (5/29) were for taxa elevated in none/mild disease at baseline. Associations were not materially different after adjustment for baseline age, nSES, and general health status, or when stratified on baseline periodontal measures or smoking status. Socransky red and several yellow complex microbiota were significantly associated with progression, positively for red, inversely for yellow; the orange complex was not associated with progression in our study. Our prospective findings confirm an association with disease progression for several microbiota previously identified in cross-sectional studies, and for some not yet reported. Three main features deserved further comment.
First, we used a prospective design wherein the subgingival microbiome was measured prior to determination of disease progression. Temporality is a major tenant in establishing causality,38 and has been identified by Teles et al.7 as generally lacking in studies on subgingival microbiota and clinical periodontal measures. Some studies evaluated subgingival microbiota at sites that already had progressed during a preceding time interval (see Appendix Table S6 footnote in online Journal of Periodontology). In one study, 20 adults were examined every 2–4 months for evidence of periodontal breakdown (CAL ≥2 mm at 2–4 sites) and had microbiota cultured in subgingival plaque from progressing sites and non-progressing control sites.14 Sixteen microbiota (including P. gingivalis, F. nucleatum, C. rectus) were significantly elevated at sites where progression already occurred, 14 microbiota (including S. gordonii, S. oralis, S. sanguinis, A. naeslundii) were elevated at control sites that had not progressed. Papapanou et al.15 measured microbiota (targeted checkerboard method) at sites that progressed (≥10 sites with ≥3 mm CAL loss) during the preceding 10 years in 148 adults. Unadjusted ORs for presence of P. gingivalis, T. denticola. C. rectus, T. forsythia, and P. intermedia were 7.01, 5.66, 4.39, 4.02, and 3.62 comparing progressing and non-progressing sites. These ORs exceed those in our study. However, critically important is that different progression case definitions were used and our estimates were for 1-SD difference in CLR abundance measured prior to disease progression. The influence of the microbiota versus the disease itself is challenging to disentangle when measured contemporaneously; that is, the bacteria may be present because of the disease.39 Fundamental to determining pathogenic agents are studies that incorporate established causal criteria; of utmost importance is temporality.7, 40 The prospective findings herein add important new information on periodontal disease etiology.
Second, our progression outcome was defined using radiographic ACH loss. At each examination, the same radiographic instrument and bite-wing procedure was used, care was taken to standardize projection geometry and head position with a cephalostat.24 The Hausmann side-by-side procedure standardizes landmark reference points on the two radiographic images for reliable detection of small ACH differences (e.g., 0.4 mm).35 The Forsyth Longitudinal Study22, 41 showed substantial intra-individual variation in CAL over 12 bimonthly measures, highlighting poor precision when using probing measures to determine disease progression. Few studies have examined longitudinal ACH changes in relation to the subgingival microbiome.9, 17 Slots used immunofluorescence microscopy to identify presence of A. actinomycetemcomitans, P. gingivalis, and P. intermedia in subgingival plaque at sites with radiographic ACH loss (threshold not reported) during the preceding 2–5 years.17 These three bacteria were present in 99.2% of sites with, and only 40% of sites without, ACH loss. The microbiota were measured after ACH loss occurred. Our findings expand on this study by showing several baseline subgingival bacteria are in higher abundance and associated with subsequent occurrence and severity of ACH loss.
Third, we measured the subgingival microbiome using untargeted 16S sequencing, which allows for quantitative characterization of a greater number of bacteria and their association with progression than possible in previous studies that used culture or targeted methods. To enhance statistical efficiency of our progression analysis, we focused on microbiota previously identified in our cohort as having a relationship with periodontal disease presence and severity at baseline.25 Appendix Table S6 (in online Journal of Periodontology) summarizes subgingival microbiota associated with progression in previous studies and in our study. Only nine microbiota (A. naeslundii, F. nucleatum, Haemophilus parainfluenzae, P. gingivalis, P. intermedia, S. oralis, T. forsythia, T. denticola, T. socranskii) that had been associated with a measure of progression in previous studies were also identified in the present study as being significantly associated with progression 5-years after measurement of the microbiome. An additional 20 microbiota were further identified that were significantly associated with progression, which may represent new discovery in periodontal disease etiology. Additional investigation will help refine understanding on how these identified microbiota and those yet to be identified act individually, more likely as an interrelated microbial ecology, to effect periodontal disease presence, severity, and progression.
Strengths of our study include the prospective design, large cohort size, and its community-based enrollment not using periodontal disease or other aspects of oral health as selection criteria. This information provides a benchmark for future studies evaluating the subgingival microbiome and disease progression in clinical and other community populations. The use of untargeted 16S sequencing, with well-documented laboratory procedures and quality control minimizing batch-to-batch variation is another strength of this study. Characterizing progression using radiographic ACH loss minimizes measurement variation common with serial probing assessments. Because progression was documented 5-years after microbiome measurement, results are less likely due to reverse causation bias. Together, these two strengths yield novel study findings. Limitations of the study include sampling subgingival plaque on only a portion of teeth present, although this approach is used in the vast majority of oral microbiome studies. Additionally, the plaque samples were collected and stored frozen for several years prior to 16S metagenomic sequencing. The effect of long-term storage on 16S microbiome results has not been systematically evaluated in the published literature. Available information does suggest that long-term storage at −80°C, or in liquid nitrogen, is not likely to affect DNA-based studies such as used herein.42 Taxonomic OTU annotation was completed using HOMD version 14.5, which could result in an incomplete characterization of microbiota present as additional taxa are added to future database versions. Detailed information on periodontal treatment occurring during the 5-year follow-up interval was not available, which should be considered when generalizing study results. Significance tests were not corrected for multiple comparisons, some results could be due to chance. Associations between individual microbiota and disease progression were quantified. It is likely that bacterial clusters or shifts in relative abundance within such clusters is a key pathogenic factor in progression etiology. Advances in biostatistical and bioinformatics methods are needed to quantify these complex microbial interrelationships. A better test of a causal hypothesis linking the subgingival microbiome with progression would come through evaluation of changes in individual microbial abundance, or composition of clusters, in relation to disease progression thereafter. This approach is being explored in our longitudinal cohort study using additional microbiome measures currently being analyzed. Because our cohort is part of the larger WHI program, men were not included. It is unclear the extent to which our findings extend to men. Enrolling participants without conditioning on periodontal disease reduces potential selection biases, but it also could limit the amount of progression observed.
CONCLUSION
In conclusion, prospectively quantified alveolar bone loss is associated with several subgingival microbiota measured antecedent to ACH loss. Further understanding of both the diversity and functions of microbiota that effect disease progression could lead to etiologic targets for prevention of periodontal disease, tooth loss and impaired oral quality of life in older adults.
Supplementary Material
Acknowledgements
Prior to completion of the work reported in this manuscript, one of the study Co-Principal Investigators and senior scientists, Dr. Robert Genco, passed away. This work would not have been possible without Bob’s sage guidance, unlimited energy, and passion for deeper understanding about periodontal disease microbiology. You are missed dear friend and colleague.
Funding: This study was supported by the following funding sources: National Heart, Lung, and Blood Institute (National Institutes of Health, Bethesda, MD, USA) contract N01WH32122; National Institute for Dental and Craniofacial (National Institutes of Health, Bethesda, MD, USA) Research Grants: DE13505, DE4898, DE022654, and DE024523; National Institute of Allergy and Infectious Diseases (National Institutes of Health, Bethesda, MD, USA) R01Al125982, U.S. Army Reserve Medical Corps (Arlington, VA, USA) Grant: DAMD17-96-1-6319; Feasibility Study Award (AS382) from the Women’s Health Initiative Program (Coordinating Center, Fred Hutchinson Cancer Research Center, Seattle, WA, USA).
Footnotes
Conflictions of Interests: The authors have no conflicts of interest or relevant disclosures.
Availability of data and materials: Data that support the findings of this study are available from the authors upon reasonable request and with permission of the U.S. Women’s Health Initiative program.
The Florida Probe System®, Gainesville, FL, USA.
MiSeq System, Illumina, Inc., San Diego, CA, USA.
Bennet HFQ 300, Bennet X-Ray Corp., Copaigue, NY.
REFERENCES
- 1.Marsh PD. Dental plaque: biological significance of a biofilm and community life-style. J Clin Periodontol 2005;32 Suppl 6:7–15. [DOI] [PubMed] [Google Scholar]
- 2.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 2010;8:481–90. [DOI] [PubMed] [Google Scholar]
- 3.Patini R, Staderini E, Lajolo C, et al. Relationship betwen oral microbiota and periodontal disease: a systematic review. European Review for Medical and Pharmocological Sciences 2018;22:5575–788. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Chaparro PJ, Goncalves C, Figueiredo LC, et al. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res 2014;93:846–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Socransky SS, Haffajee AD. Evidence of bacterial etiology: a historical perspective. Periodontol 2000 1994;5:7–25. [DOI] [PubMed] [Google Scholar]
- 6.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000 2005;38:135–87. [DOI] [PubMed] [Google Scholar]
- 7.Teles R, Teles F, Frias-Lopez J, Paster B, Haffajee A. Lessons learned and unlearned in periodontal microbiology. Periodontol 2000 2013;62:95–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albandar JM, Brown LJ, Loe H. Putative periodontal pathogens in subgingival plaque of young adults with and without early-onset periodontitis. J Periodontol 1997;68:973–81. [DOI] [PubMed] [Google Scholar]
- 9.Bragd L, Dahlen G, Wikstrom M, Slots J. The capability of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius to indicate progressive periodontitis; a retrospective study. J Clin Periodontol 1987;14:95–9. [DOI] [PubMed] [Google Scholar]
- 10.Byrne SJ, Dashper SG, Darby IB, et al. Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol Immunol 2009;24:469–77. [DOI] [PubMed] [Google Scholar]
- 11.Craig RG, Boylan R, Yip J, et al. Serum IgG antibody response to periodontal pathogens in minority populations: relationship to periodontal disease status and progression. J Periodontal Res 2002;37:132–46. [DOI] [PubMed] [Google Scholar]
- 12.Dzink JL, Socransky SS, Haffajee AD. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol 1988;15:316–23. [DOI] [PubMed] [Google Scholar]
- 13.Kinney JS, Morelli T, Braun T, et al. Saliva/pathogen biomarker signatures and periodontal disease progression. J Dent Res 2011;90:752–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore WE, Moore LH, Ranney RR, et al. The microflora of periodontal sites showing active destructive progression. J Clin Periodontol 1991;18:729–39. [DOI] [PubMed] [Google Scholar]
- 15.Papapanou PN, Baelum V, Luan WM, et al. Subgingival microbiota in adult Chinese: prevalence and relation to periodontal disease progression. J Periodontol 1997;68:651–66. [DOI] [PubMed] [Google Scholar]
- 16.Rams TE, Listgarten MA, Slots J. Utility of 5 major putative periodontal pathogens and selected clinical parameters to predict periodontal breakdown in patients on maintenance care. J Clin Periodontol 1996;23:346–54. [DOI] [PubMed] [Google Scholar]
- 17.Slots J, Bragd L, Wikstrom M, Dahlen G. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J Clin Periodontol 1986;13:570–7. [DOI] [PubMed] [Google Scholar]
- 18.Tanner A, Maiden MF, Macuch PJ, Murray LL, Kent RL Jr. Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol 1998;25:85–98. [DOI] [PubMed] [Google Scholar]
- 19.Tanner AC, Dzink JL, Socransky SS, Des Roches CL. Diagnosis of periodontal disease using rapid identification of “activity-related” gram-negative species. J Periodontal Res 1987;22:207–8. [DOI] [PubMed] [Google Scholar]
- 20.Teles RP, Patel M, Socransky SS, Haffajee AD. Disease progression in periodontally healthy and maintenance subjects. J Periodontol 2008;79:784–94. [DOI] [PubMed] [Google Scholar]
- 21.Tran SD, Rudney JD, Sparks BS, Hodges JS. Persistent presence of Bacteroides forsythus as a risk factor for attachment loss in a population with low prevalence and severity of adult periodontitis. J Periodontol 2001;72:1–10. [DOI] [PubMed] [Google Scholar]
- 22.Teles R, Benecha HK, Preisser JS, et al. Modelling changes in clinical attachment loss to classify periodontal disease progression. J Clin Periodontol 2016;43:426–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banack HR, Genco RJ, LaMonte MJ, et al. Cohort profile: the Buffalo OsteoPerio microbiome prospective cohort study. BMJ Open 2018;8:e024263. doi: 10.1136/bmjopen-2018-024263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaMonte MJ, Hovey KM, Genco RJ, et al. Five-year changes in periodontal disease measures among postmenopausal females: the Buffalo OsteoPerio study. J Periodontol 2013;84:572–84. [DOI] [PubMed] [Google Scholar]
- 25.Genco RJ, LaMonte MJ, McSkimming DI, et al. The subgingival microbiome relationship to periodontal disease in older women. J Dent Res 2019;98:975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langer RD, White E, Lewis CE, et al. The Women’s Health Initiative Observational Study: baseline characteristics of participants and reliability of baseline measures. Ann Epidemiol 2003;13 Suppl 9:S107–21. [DOI] [PubMed] [Google Scholar]
- 27.von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet 2007;370:1453–7. [DOI] [PubMed] [Google Scholar]
- 28.Eke PI, Page RC, Wei L, Thornton-Evans G, Genco RJ. Update of the case definitions for population-based surveillance of periodontitis. J Periodontol 2012;83:1449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaMonte MJ, Genco RJ, Buck MJ, et al. Composition and diversity of the subgingival microbiome and its relationship with age in postmenopausal women: an epidemiologic investigation. BMC Oral Health 2019;19:246. 10.1186/s12903-019-0906-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng W, Tsompana M, Ruscitto A, et al. An accurate and efficient experimental approach for characterization of the complex oral microbiota. Microbiome 2015;3:48. doi 10.1186/s40168-015-0110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brennan RM, Genco RJ, Wilding GE, et al. Bacterial species in subgingival plaque and oral bone loss in postmenopausal women. J Periodontol 2007;78:1051–61. [DOI] [PubMed] [Google Scholar]
- 32.Chen T, Yu WH, Izard J, et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- 34.Hausmann E, Allen K, Carpio L, Christersson LA, Clerehugh V. Computerized methodology for detection of alveolar crestal bone loss from serial intraoral radiographs. J Periodontol 1992;63:657–62. [DOI] [PubMed] [Google Scholar]
- 35.Hausmann E, Allen K, Clerehugh V. What alveolar crest level on a bite-wing radiograph represents bone loss? J Periodontol 1991;62:570–2. [DOI] [PubMed] [Google Scholar]
- 36.Dubowitz T, Ghosh-Dastidar M, Eibner C, et al. The Women’s Health Initiative: the food environment, neighborhood socioeconomic status, BMI, and blood pressure. Obesity 2012;20:862–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. Microbiome datasets are compositional: and this is not optional. Front Microbiol 2017;8:2224. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothman KJ, Greenland S. Causation and causal inference in epidemiology. Am J Public Health 2005;95 Suppl 1:S144–50. [DOI] [PubMed] [Google Scholar]
- 39.Tanner AC, Kent R Jr., Kanasi E, et al. Clinical characteristics and microbiota of progressing slight chronic periodontitis in adults. J Clin Periodontol 2007;34:917–30. [DOI] [PubMed] [Google Scholar]
- 40.Salvi GE, Lawrence HP, Offenbacher S, Beck JD. Influence of risk factors on the pathogenesis of periodontitis. Periodontol 2000 1997;14:173–201. [DOI] [PubMed] [Google Scholar]
- 41.Teles R, Moss K, Preisser JS, et al. Patterns of periodontal disease progression based on linear mixed models of clinical attachment loss. J Clin Periodontol 2018;45:15–25. [DOI] [PubMed] [Google Scholar]
- 42.Goodrich JK, Di Rienzi SC, Poole AC, et al. Conducting a microbiome study. Cell 2014;158:250–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


