Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped virus initially detected in Wuhan in December 2019, responsible for coronavirus disease 2019 (COVID-19), a respiratory syndrome currently affecting >220 countries around the world, with >80 million cases registered and >1.8 million deaths.
Objective
As several vaccines are still being developed and 2 have been approved, it is particularly important to perform evolutionary surveillance to identify mutations potentially affecting vaccine efficacy.
Methods
DynaMut server has been used to evaluate the impact of the mutation found on SARS-CoV-2 isolates available on GISAID.
Results
In this article, we analyze whole genomes sequenced from Italian patients, and we report the characterization of 3 mutations, one of which presents in the spike protein.
Conclusion
The mutations analyzed in this article can be useful to evaluate the evolution of SARS-CoV-2.
Keywords: Bioinformatic, Coronavirus disease 2019, Evolutionary analysis, Protein modeling, Syndrome coronavirus 2
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) initially detected in Wuhan in December 2019 [1 ] is responsible for the coronavirus disease 2019 (COVID-19) [2 ], a respiratory syndrome, and currently, it is affecting >220 countries around the world, with >80 million cases registered and more than 1.8 million deaths. SARS-CoV-2 is an enveloped virus, belonging to the beta-CoV genus, with a positive-sense single-stranded RNA genome of approximately 30,000 bases, with 5′-cap structure and 3′-poly-A tail [3 ]. SARS-CoV-2 has 16 nonstructural proteins and, like other coronaviruses, 4 structural proteins, known as the spike (S), envelope (E), membrane, and nucleocapsid proteins [4 , 5 ]. The main responsibility of the nucleocapsid protein is to hold the RNA genome, while the S, envelope, and membrane proteins together create the viral envelope. In particular, the S protein is responsible for allowing the virus to attach to and fuse with the membrane of a host cell by recognizing and interacting with a specific receptor, the angiotensin-converting enzyme 2 (ACE2) [6 ]. Italy is now experiencing the second epidemic wave of COVID-19, and as several vaccines are still being developed and 2 have been approved [7 ], it is particularly important to perform evolutionary surveillance to identify mutations potentially affecting vaccine efficacy [8 ]. Several reports have demonstrated how a single amino acidic substitution, first observed in Europe emerging from China [9 ], can affect the viral infectious phenotype [10 , 11 , 12 , 13 ], and several new mutations have been reported that have the possibility to influence the spreading of the SARS-CoV-2 [14 , 15 , 16 , 17 , 18 , 19 ]. In this article, we report 3 mutations possibly affecting its phenotypical characteristics.
Materials and Methods
A total of 870 COVID-19 whole genome sequences isolated from Italian patients from January 29 to November 10, 2020 have been downloaded from GISAID (https://www.gisaid.org/) database. The dataset has been aligned using multiple sequence alignment (MAFFT) online tool [20 ] and manually edited using BioEdit program v7.0.5 [21 ]. Sequence alignments and analyses were obtained through the Jalview editor [22 ], and structural models have been built relying on the website I-TASSER [23 ], protein data bank HHPred [24 ], and CUPSAT [25 ], and DynaMut [26 ] online server has been used to estimate the stability of potential mutations found using the selective pressure analysis. Three-dimensional structures have been analyzed and displayed using PyMOL.
Results
The analysis of the alignment has revealed the presence of 3 mutations on helicase, S, and papain-like protease (NSP3) proteins in COVID-19 sequences isolated from September to November 2020 from Italian patients for a total of 11 isolates. The helicase protein has shown nonsynonymous mutation from a histidine to a tyrosine in the 39th amino acid position. They are both polar amino acid but with different side chain structures; the first one has an imidazole side chain, while the other has a phenol side chain. Using the 3-dimensional structure available on I-TASSER server, the implication of this mutation has been analyzed by DynaMut server. The results point out both mutations reduce the stability of the protein (ΔΔG [kcal/mol] 0.377), while the Δ vibrational entropy energy between wild-type and mutant papain-like protease protein has been calculated to be ΔΔSVib ENCoM: −0.472 kcal mol-1 K−1. In the S glycoprotein, the transition from an asparagine to a lysine occurs on the 439th amino acid position. Asparagine is a polar aliphatic amino acid which frequently occurs at the beginning of alpha helices, while lysine is a basic, chiral, charged, and aliphatic amino acid, and its ε-amino group often participates in hydrogen bonding, salt bridges, and covalent interactions. This mutation is located in the SD2 region of the RBD-containing S1 subunit and particularly in the most exposed region of the protein that interacts with the ACE2 receptor on human cells [13 ]. Using the crystallographic 3-dimensional structure of the S protein, the implications of this mutation have been analyzed using CUPSAT and DynaMut servers. The results of these analyses have shown that the mutation from asparagine to lysine reduces the stability of the protein (ΔΔG [kcal/mol] −0.209) favoring the torsion potential. The Δ vibrational entropy energy between wild-type and mutant S protein has been calculated to be ΔΔSVib ENCoM: 0.262 kcal mol−1 K−1 (Fig. 1 ). This mutation falls in one of the 2 N-terminal zinc-binding domain (1–99 a.a) [27 ]. Regarding the papain-like protease protein, the transition from an isoleucine to a threonine occurs on the 1,683th amino acid position. Isoleucine is a nonpolar, uncharged, branched chain, aliphatic amino acid, while threonine is a polar, uncharged amino acid and is susceptible to numerous posttranslational modifications. The homology modeling analysis performed using HHpred server has shown structural similarity of the subdomains where the mutation is with the ubiquitin-like protein domain of papain-like protease protein of MERS-CoV (Fig. 2 ). Using the crystallographic 3-dimensional structure of the helicase protein, the implications of this mutation have been analyzed using CUPSAT and DynaMut servers. The results of these analyses have shown that the mutation from isoleucine to a threonine reduces the stability of the protein (ΔΔG [kcal/mol] −1.963) favoring the torsion potential. The Δ vibrational entropy energy between wild-type and mutant helicase protein has been calculated to be ΔΔSVib ENCoM: 0.781 kcal mol−1 K−1 (Fig. 3 ).
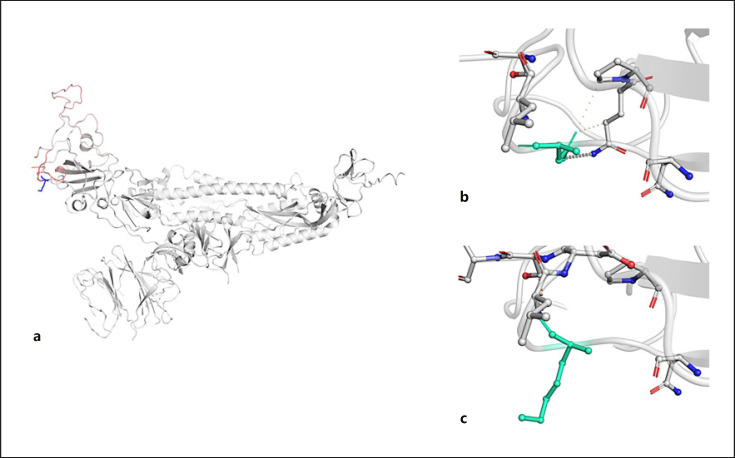
Fig. 1.
a Model of S glycoprotein monomer displaying the amino acids colored according to the vibrational entropy change upon mutation. Red regions are those gaining in flexibility, whereas blue regions are those gaining stability; the top image shows the molecular interaction between the side chain of the wild-type amino acid and the side chains of the surrounding amino acid (b); the bottom image shows the molecular interaction between the side chain of the mutated amino acid and the side chains of the surrounding amino acid (c). S, spike.
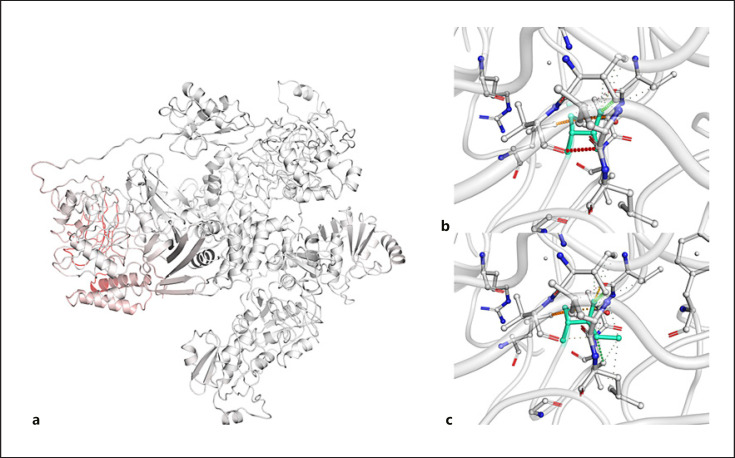
Fig. 2.
a Model of papain-like protease protein displaying the amino acids colored according to the vibrational entropy change upon mutation. Red regions are those gaining in flexibility, whereas blue regions are those gaining stability; the top image shows the molecular interaction between the side chain of the wild-type amino acid and the side chains of the surrounding amino acid (b); the bottom image shows the molecular interaction between the side chain of the mutated amino acid and the side chains of the surrounding amino acid (c).
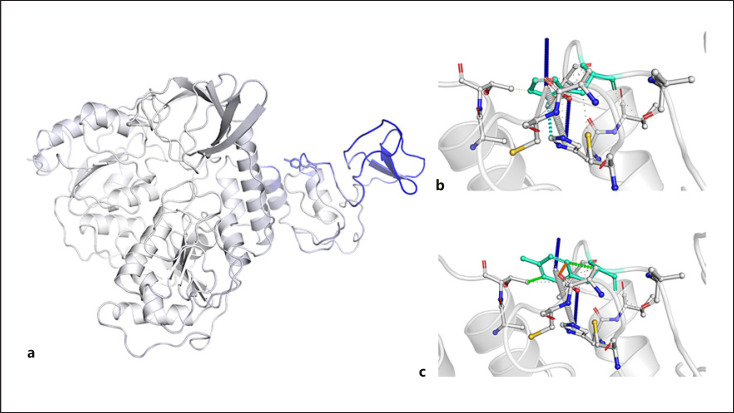
Fig. 3.
a Model of helicase protein displaying the amino acids colored according to the vibrational entropy change upon mutation (a). Red regions are those gaining flexibility, whereas blue regions are those gaining stability; the top image shows the molecular interaction between the side chain of the wild-type amino acid and the side chains of the surrounding amino acid (b); the bottom image shows the molecular interaction between the side chain of the mutated amino acid and the side chains of the surrounding amino acid (c).
Discussion
The presence of new mutations in Italian sequences isolated in October 2020 could partially clarify the higher spread of the virus reached in that month; moreover, nowadays a new strain from the UK probably responsible for a higher infection has been observed. The mutation analysis tool GISAID has also shown that the mutations evaluated in this article are highly prevalent in the UK. We have identified 3 nonsynonymous mutations, H39Y on helicase protein, N439K on S glycoprotein, and I1683T on papain-like protease. Previous studies have shown how single amino acidic substitutions could scientifically influence the COVID-19 characteristics [10 , 11 , 12 , 13 ] and how bioinformatics approach could be useful to predict the impact of amino acidic mutation on the clinical features of the virus [28 ]. The structural analysis of the mutated helicase protein revealed an increase in stability of zinc-binding domain; moreover, the mutations affect the stability of the whole ZBD region. This could possibly make the structure more rigid and less capable of performing its normal activity and partially explain the presence of other mutations in these isolates. The N439K substitution on the S glycoprotein resides on the amino acidic region responsible for binding ACE2 on human cells. It is well known that lysine increases the ability of making new bindings, and our results suggests that this substitution has an effect on the structure of S glycoprotein, increasing its flexibility and potentially increasing the ability of the virus to bind cellular receptors, as already seen for the previous D614G S mutations. The last mutation on papain-like protease falls on a ZBD region. This specific region is involved in multiple molecular fundamental processes including autophagy and host cell regulation. The autophagy pathway seems to be crucial for SARS-CoV-2 since other mutations on this pathway have been found [14 ].
Statement of Ethics
The paper is exempt from ethical committee approval because the dataset used for this analysis has been downloaded from a public open online repository and no human or animal has been involved in the research.
Conflict of Interest Statement
The authors declare no conflict of interest.
Funding Sources
The authors have no funding to declare.
Author Contributions
All authors wrote sections of this manuscript and approved the submitted version.
Acknowledgement
We thank the GISAID website and all the colleagues who have immediately made available the SARS-CoV-2 sequences. Their laborious and important work is highly appreciated.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382((8)):727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya AE, Baker S, Baric R, de Groot RJ, Drosten C, Gulyaeva AA, et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5((4)):536–44. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579((7798)):265–9. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giovanetti M, Benedetti F, Campisi G, Ciccozzi A, Fabris S, Ceccarelli G, et al. Evolution patterns of SARS-CoV-2: snapshot on its genome variants. Biochem Biophys Res Commun. 2021 Jan 29;538:88–91. doi: 10.1016/j.bbrc.2020.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene reports. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181((2)):281–e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020 Dec 31;383((27)):2603–15. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koyama T, Weeraratne D, Snowdon JL, Parida L. Emergence of drift variants that may affect COVID-19 vaccine development and antibody treatment. Pathogens. 2020;9((5)):324. doi: 10.3390/pathogens9050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pachetti M, Marini B, Benedetti F, Giudici F, Mauro E, Storici P, et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18((1)):179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182((4)):812–e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH, et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370((6523)):1464. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butowt R, Bilinska K, Von Bartheld CS. Chemosensory dysfunction in COVID-19: integration of genetic and epidemiological data points to D614G spike protein variant as a contributing factor. ACS Chem Neurosci. 2020;11((20)):3180–4. doi: 10.1021/acschemneuro.0c00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benvenuto D, Angeletti S, Giovanetti M, Bianchi M, Pascarella S, Cauda R, et al. Evolutionary analysis of SARS-CoV-2: how mutation of non-structural protein 6 (NSP6) could affect viral autophagy. J Infect. 2020;81((1)):e24–7. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benedetti F, Snyder GA, Giovanetti M, Angeletti S, Gallo RC, Ciccozzi M, et al. Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1. J Transl Med. 2020;18((1)):329. doi: 10.1186/s12967-020-02507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holland LA, Kaelin EA, Maqsood R, Estifanos B, Wu LI, Varsani A, et al. An 81-nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (January–March 2020) J Virol. 2020;94((14)):e00711–20. doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bal A, Destras G, Gaymard A, Bouscambert-Duchamp M, Valette M, Escuret V, et al. Molecular characterization of SARS-CoV-2 in the first COVID-19 cluster in France reveals an amino acid deletion in nsp2 (Asp268del) Clin Microbiol Infect. 2020;26((7)):960–2. doi: 10.1016/j.cmi.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young BE, Fong SW, Chan YH, Mak TM, Ang LW, Anderson DE, et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396((10251)):603–11. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20((4)):1160–6. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/ NT. 1999.
- 22.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25((9)):1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12((1)):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmermann L, Stephens A, Nam SZ, Rau D, Kübler J, Lozajic M, et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol. 2018;430((15)):2237–43. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34((Web Server issue)):W239–42. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigues CH, Pires DE, Ascher DB. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46((W1)):W350–355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirza MU, Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J Pharm Anal. 2020;10((4)):320–8. doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benvenuto D, Demir AB, Giovanetti M, Bianchi M, Angeletti S, Pascarella S, et al. Evidence for mutations in SARS-CoV-2 Italian isolates potentially affecting virus transmission. J Med Virol. 2020;92((10)):2232–7. doi: 10.1002/jmv.26104. [DOI] [PMC free article] [PubMed] [Google Scholar]