Abstract
Introduction
SARS-CoV attaches to human angiotensin-converting enzyme 2 receptor with the spike protein and infects cells. It can play a direct role in infection by acting as a receptor and/or co-receptor for blood group antigens, microorganisms, parasites, and viruses.
Objectives
We aimed to compare the blood group distribution of patients with SARS-CoV-2 infection admitted to hospital and that of healthy donors.
Methods
A total of 823 patients with a positive SARS-CoV-2 test and clinical symptoms were included in the study. The results were compared with the normal blood group distribution in the region.
Results
While the prevalence of COVID-19 in the A, B, and AB blood groups was higher than that in the healthy blood donors, it was lower in the O blood group (p = 0.009). The distribution of demographic and clinical characteristics based on blood groups did not differ significantly.
Conclusion
Our results are in agreement with other studies suggesting that blood group O individuals are somewhat more resistant to clinically overt infection with SARS-CoV-2 than other blood groups. However, this tendency is not sufficiently established to allow special prophylaxis recommendations for non-O individuals.
Keywords: SARS-CoV infection, Blood group distribution, Blood group O
Introduction
On March 11, 2020, coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was declared a pandemic by the World Health Organisation (WHO) [1]. Evidence obtained from phylogenetic analysis shows that SARS-CoV-2, which belongs to the Betacoronavirus genus, can infect humans, bats, and wild animals [2]. SARS-CoV attaches to the human angiotensin-converting enzyme 2 (ACE2) receptor with the spike protein and infects cells [3, 4].
COVID-19 spread rapidly around the world. There is a poor prognosis observed in the elderly, which is most commonly associated with hypertension, diabetes mellitus, coronary artery disease, and immunosuppression [1, 5].
The relationship of infectious agents with human blood groups was investigated previously [6, 7, 8, 9, 10, 11, 12, 13, 14]. It has been indicated that the blood group antigens found in erythrocytes and other tissues interact with microorganisms such as bacteria, viruses, parasites, and fungi. Differences in blood group antigen expression can increase or decrease host susceptibility to many infections. This can play a direct role in infection by acting as a receptor and/or coreceptor for blood group antigens, microorganisms, parasites, and viruses [6, 7]. ABO antibodies can also be considered as part of the innate immune system against some bacterial pathogens and enveloped viruses carrying ABO-active antigens [8]. Helicobacter pylori [9], Vibrio cholerae [10, 11], hepatitis C virus [12], human immunodeficiency virus [13], and SARS [8, 14, 15] are some of the infectious agents that have been shown to be associated with human blood groups.
Clinical studies have also been conducted examining the relationship between SARS-CoV-2 and blood groups [16, 17, 18, 19]. However, more studies will be useful for comparing results and to understanding whether findings differ in different populations because the frequency of blood groups is variable between populations [20]. In this study, we aimed to compare blood group distribution in patients with SARS-CoV-2 infection who presented to our hospital with an indication for hospitalization and healthy donors from the same region. A distinctive feature of the Çukurova region is that haemoglobinopathies are endemic in this area [21, 22]. Investigating the relationship between blood groups and SARS-CoV-2 infection is important in this region because the human population living here has been determined by malaria infection throughout history.
Material and Methods
A total of 823 patients followed up at our hospital with a positive PCR test for SARS-CoV-2 RNA between 25 March and 21 December 2020 were included in this study. The Bio-Speedy® (Bioexen, Istanbul, Turkey) nucleic acid isolation kit was used for nucleic acid isolation from respiratory tract (nasopharyngeal swab) samples obtained from the patients. Reverse transcription (RT) and quantitative RT-qPCR procedures were performed with QuantStudio Real-Time PCR Systems (Thermo Fisher Scientific, USA) using the Bio-Speedy® COVID-19 RT-qPCR detection kit, which was developed to detect the Wuhan CoV RdRP-gene region according to the protocol proposed by the WHO [23]. Molecular analysis results were interpreted as SARS-CoV-2-positive if Wuhan RdRP was positive, and SARS-CoV-2-negative if Wuhan RdRP was negative. The medical records of the patients were retrospectively reviewed. Patients' age, sex, white blood cell (WBC), neutrophil, and lymphocyte counts, comorbidities, intubation and intensive care needs, and hospitalization duration were obtained from the medical records. To make the laboratory values of the patient independent from the effects of the drugs, their values at the time of first admission to the hospital were recorded. The clinical results were determined based on the hospitalization duration, the need for intubation and intensive care, and death.
Comorbid diseases came under the following headings. Coronary artery disease, hypertension, cerebrovascular disease, and peripheral artery disease were classified as “cardiovascular disease,” and asthma and chronic obstructive pulmonary disease as “respiratory system diseases.” Cancer patients were not divided according to their types, and they were recorded as a general title. Less common comorbid diseases were classified as “others.”
In the study by Yildiz [24], the blood type distribution in Çukurova region was determined. This study was conducted with healthy blood donors. In this study, the blood groups of 136,038 individuals were determined as A in 38.90%, O in 37.10%, B in 17.00%, and AB in 6.90%. Of the same individuals, 89.90% were Rh(+) and 10.10% were Rh(−). The results of this study were compared with those of our study in terms of blood group distribution.
Statistical Analysis
Statistical evaluation was conducted using SPSS for Windows 20 (IBM SPSS Inc., Chicago, IL, USA). Normality of the distribution of data was evaluated by the Kolmogorov-Smirnov test. Numerical variables with normal distribution are given as mean ± SD and numerical variables without normal distribution as median (interquartile range, 25th–75th percentiles). Categorical variables are indicated as numbers and percentages. The χ2 and Fisher exact tests were used to compare categorical data. The Student t test or Mann-Whitney U test was used for the comparison of numerical variables between 2 groups according to the normality of distribution. Based on the normality of distribution, analysis of variance or the Kruskal-Wallis H test was used for the comparison of numerical variables among blood group ABO patients. The findings associated with the prognosis were analyzed using the Cox regression analysis. The survival chart was evaluated using the Kaplan-Meier analysis. p < 0.05 was considered statistically significant.
Results
The study population consisted of 823 patients (aged 18–97 years; 470 men [57.1%] and 353 women [42.9%]). Of the patients, 351 (42.6%) had blood group A, 153 (18.6%) had group B, 250 (30.4%) had group O, and 69 (8.4%) had group AB; 86 (10.4%) patients were Rh–. The distribution of blood groups in our region is shown in Figure 1. Accordingly, while A, B, and AB blood groups were seen more frequently in COVID-19 cases than in the healthy blood donors, the incidence of COVID-19 was lower in the O blood group than in healthy blood donors (p = 0.009). Among all the patients, 510 (62%) had a comorbidity.
Fig. 1.
ABO blood group prevalence in COVID-19 cases.
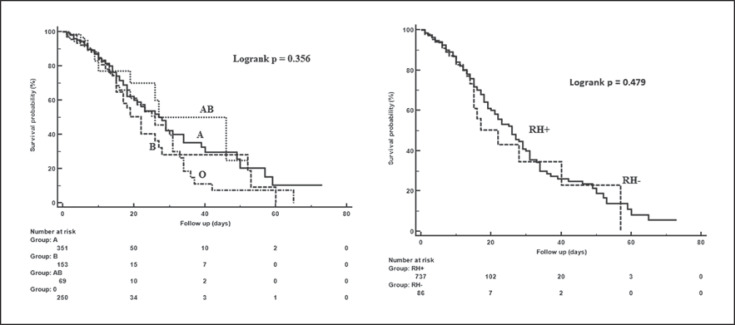
The demographic and clinical characteristics of patients with COVID-19 are shown in Table 1. Distributions of demographic and clinical characteristics did not differ significantly according to blood group. In intubated versus non-intubated patients, male sex ratio (63.4 vs. 45.5%; p = 0.019), mean age (67.0 ± 13.5 vs. 55.2 ± 17.1; p < 0.001), and comorbidity ratio (80.3 vs. 54.5%; p < 0.001) as well as mortality rate (89.9 vs. 1.5%; p < 0.001) were found to be higher. The median WBC (13.2 vs. 7.6; p < 0.001) and neutrophil (11.8 vs. 5.2; p < 0.001) counts were higher and lymphocyte count (0.5 vs. 1.3; p < 0.001) lower in intubated patients than in non-intubated patients (Table 2). The multivariable Cox regression model included age, comorbid diseases, admission to the intensive care unit, intubation status as well as WBC, neutrophil, and lymphocyte counts, all of which were found to be associated with mortality; older age (hazard ratio [HR] 1.02; p = 0.038), T2DM (HR 1.72; p = 0.022), T2DM and cardiovascular disease (HR 1.64; p = 0.029), being intubated (HR 22.84; p < 0.001), and a higher neutrophil count (HR 1.02; p = 0.020) were independent predictors for mortality. There was no significant correlation between blood groups (Fig. 2) and other demographic characteristics and other clinical findings and mortality (Table 3).
Table 1.
Demographic characteristics and clinical findings according to blood groups
| Variables | N = 823 | ABO group |
p value | |||
|---|---|---|---|---|---|---|
| A (n = 351) | B (n = 153) | AB (n = 69) | 0 (n = 250) | |||
| Sex | ||||||
| Female | 353 (42.9) | 137 (39.0) | 66 (43. 1) | 31 (44.9) | 119 (47.6) | 0.211 |
| Male | 470 (57.1) | 214 (61.0) | 87 (56.9) | 38 (55.1) | 131 (52.4) | |
| Age, years (mean ± SD) | 58.6±17.0 | 58.8±16.8 | 60.0±18.3 | 57.3±15.8 | 58.0±16.7 | 0.639 |
| Comorbidity | ||||||
| No | 313 (38.0) | 133 (37.9) | 58 (37.9) | 21 (30.4) | 101 (40.4) | 0.515 |
| Yes | 510 (62.0) | 218 (62.1) | 95 (62.1) | 48 (69.6) | 149 (59.6) | |
| CVD | 179 (21.7) | 82 (23.4) | 37 (24.2) | 13 (18.8) | 47 (18.8) | 0.701 |
| RSD | 28 (3.4) | 10 (2.8) | 6 (3.9) | 5 (7.2) | 7 (2.8) | |
| DM | 81 (9.8) | 35 (10.0) | 14 (9.2) | 6 (8.7) | 26 (10.4) | |
| Psychosis | 7 (0.9) | 1 (0.3) | 1 (0.7) | 2 (2.9) | 3 (1.2) | |
| Cancer | 17 (2.1) | 6 (1.7) | 3 (2.0) | 3 (4.3) | 5 (2.0) | |
| RSD, CVD | 8 (1.0) | 2 (0.6) | 1 (0.7) | − | 5 (2.0) | |
| DM, CVD | 141 (17.1) | 63 (17.9) | 25 (16.3) | 15 (21.7) | 38 (15.2) | |
| DM, CVD, RSD | 12 (1.5) | 5 (1.4) | 2 (1.3) | − | 5 (2.0) | |
| Other | 37 (4.5) | 14 (4.0) | 6 (3.9) | 4 (5.8) | 13 (5.2) | |
| Rh-positive | 737 (89.6) | 308 (87.7) | 138 (90.2) | 68 (98.6) | 223 (89.2) | 0.063 |
| Rh-negative | 86 (10.4) | 43 (12.3) | 15 (9.8) | 1 (1.4) | 27 (10.8) | |
| ICU admission | ||||||
| No | 425 (51.6) | 177 (50.4) | 76 (49.7) | 41 (59.4) | 131 (52.4) | 0.537 |
| Yes | 398 (48.4) | 174 (49.6) | 77 (50.3) | 28 (40.6) | 119 (47.6) | |
| Intubated | ||||||
| No | 585 (71.1) | 251 (71.5) | 105 (68.6) | 53 (76.8) | 176 (70.4) | 0.651 |
| Yes | 238 (28.9) | 100 (28.5) | 48 (31.4) | 16 (23.2) | 74 (29.6) | |
| Prognosis | ||||||
| Alive | 600 (72.9) | 258 (73.5) | 108 (70.6) | 55 (79.7) | 179 (71.6) | 0.510 |
| Exitus | 223 (27.1) | 93 (26.5) | 45 (29.4) | 14 (20.3) | 71 (28.4) | |
| Follow-up period, days WBC, ×103/µL | 9 (6–15) | 9 (6–15) | 9 (6–15) | 8 (5–12) | 9 (6–14) | 0.569 |
| 8.5 (6.2–12) | 8.4 (6.2–11.5) | 8.2 (6.2–11.8) | 8 (5.5–11.2) | 8.8 (6.5–12.5) | 0.380 | |
| NEU, ×103/µL | 6.35 (4.1–10.4) | 6.1 (4.2–10.1) | 5.9 (3.9–9.9) | 5.3 (3.5–9.7) | 7.1 (4.3–11) | 0.144 |
| LY, ×103/µL | 1 (0.6–1.6) | 1.1 (0.6–1.6) | 1.1 (0.5–1.7) | 1.2 (0.6–1.8) | 1 (0.5–1.5) | 0.500 |
Numerical variables without normal distribution are given as median (IQR, 25th–75th percentile). Categorical variables are given as n (%). CVD, cardiovascular disease; RSD, respiratory system diseases; DM, diabetes mellitus; ICU, intensive care unit; WBC, white blood cell; NEU, neutrophil; LY, lymphocyte.
Table 2.
Demographic characteristics and clinical findings according to intubation status
| Variables | Intubated |
p value | |
|---|---|---|---|
| yes (n = 238) | no (n = 585) | ||
| Sex | |||
| Female | 87 (36.6) | 266 (45.5) | 0.019* |
| Male | 151 (63.4) | 319 (54.5) | |
| Age, years (mean ± SD) | 67.0±13.5 | 55.2±17.1 | <0.001* |
| Comorbidity | |||
| No | 47 (19.7) | 266 (45.5) | <0.001* |
| Yes | 191 (80.3) | 319 (54.5) | |
| CVD | 61 (25.6) | 118 (20.2) | <0.001* |
| RSD | 7 (2.9) | 21 (3.6) | |
| DM | 37 (15.5) | 44 (7.5) | |
| Psychosis | 6 (2.5) | 1 (0.2) | |
| Cancer | 10 (4.2) | 7 (1.2) | |
| RSD, CVD | 5 (2.1) | 3 (0.5) | |
| DM, CVD | 45 (18.9) | 96 (16.4) | |
| DM, CVD, RSD | 4 (1.7) | 8 (1.4) | |
| Other | 16 (6.7) | 21 (3.6) | |
| ABO group | |||
| A | 100 (42.0) | 251 (42.9) | 0.651 |
| B | 48 (20.2) | 105 (17.9) | |
| AB | 16 (6.7) | 53 (9.1) | |
| O | 74 (31.1) | 176 (30.1) | |
| Rh-positive | 212 (89.1) | 525 (89.7) | 0.776 |
| Rh-negative | 26 (10.9) | 60 (10.3) | |
| ICU admission | |||
| No | 1 (0.4) | 424 (72.5) | <0.001* |
| Yes | 237 (99.6) | 161 (27.5) | |
| Prognosis | |||
| Alive | 24 (10.1) | 576 (98.5) | <0.001* |
| Exitus | 214 (89.9) | 9 (1.5) | |
| Follow-up period, days | 13 (7–21) | 8 (6–13) | <0.001* |
| WBC, ×103/µL | 13.2 (8.9–18.7) | 7.6 (6–10.2) | <0.001* |
| NEU, ×103/µL | 11.8 (7.3–17.1) | 5.2 (3.7–8.0) | <0.001* |
| LY, ×103/µL | 0.5 (0.2–0.8) | 1.3 (0.8–1.8) | <0.001* |
Numerical variables without normal distribution are given as median (IQR, 25th–75th percentiles). Categorical variables are given as n (%). The asterisks show statistical significance. CVD, cardiovascular disease; RSD, respiratory system diseases; DM, diabetes mellitus; ICU, intensive care unit; WBC, white blood cell; NEU, neutrophil; LY, lymphocyte.
Fig. 2.
Mortality risk by blood group.
Table 3.
Factors associated with prognosis
| Variables | Prognosis |
Univariable |
Multivariable |
|||||
|---|---|---|---|---|---|---|---|---|
| alive (n = 600) | exitus (n = 223) | HR | 95% CI | p value | HR | 95% CI | p value | |
| Sex | ||||||||
| Female | 272 (45.3) | 81 (36.3) | ref. | ref. | ||||
| Male | 328 (54.7) | 142 (63.7) | 1.25 | 0.95–1.65 | 0.105 | − | − | − |
| Age, years (mean ± SD) | 55.2±17 | 67.8±13.2 | 1.03 | 1.02–1.04 | <0.001* | 1.02 | 1.01–1.03 | 0.038* |
| Comorbidity | ||||||||
| No | 272 (45.3) | 41 (18.4) | ref. | ref. | ||||
| CVD | 120 (20.0) | 59 (26.5) | 2.00 | 1.34–2.98 | 0.001* | 1.30 | 0.85–1.99 | 0.225 |
| RSD | 21 (3.5) | 7 (3.1) | 1.45 | 0.65–3.25 | 0.361 | 1.29 | 0.58–2.90 | 0.535 |
| DM | 47 (7.8) | 34 (15.2) | 2.91 | 1.84–4.60 | <0.001* | 1.72 | 1.08–2.75 | 0.022* |
| Psychosis | 1 (.2) | 6 (2.7) | 2.16 | 0.89–5.24 | 0.088 | 0.79 | 0.32–1.93 | 0.605 |
| Cancer | 6 (1.0) | 11 (4.9) | 2.70 | 1.38–5.27 | 0.004* | 1.73 | 0.85–3.48 | 0.130 |
| RSD, CVD | 3 (.5) | 5 (2.2) | 7.17 | 2.82–18.26 | <0.001* | 2.49 | 0.97–6.41 | 0.058 |
| DM, CVD | 98 (16.3) | 43 (19.3) | 2.49 | 1.62–3.82 | <0.001* | 1.64 | 1.05–2.55 | 0.029* |
| DM, CVD, RSD | 10 (1.7) | 2 (0.9) | 1.26 | 0.31–5.24 | 0.747 | 0.57 | 0.14–2.37 | 0.440 |
| Other | 22 (3.7) | 15 (6.7) | 2.55 | 1.41–4.62 | 0.002* | 1.59 | 0.87–2.89 | 0.130 |
| ABO group | ||||||||
| A | 258 (43.0) | 93 (41.7) | ref. | ref. | ||||
| B | 108 (18.0) | 45 (20.2) | 1.22 | 0.85–1.74 | 0.285 | − | − | − |
| AB | 55 (9.2) | 14 (6.3) | 0.90 | 0.51–1.58 | 0.713 | − | − | − |
| O | 179 (29.8) | 71 (31.8) | 1.19 | 0.88–1.63 | 0.264 | − | − | − |
| Rh-positive | 537 (89.5) | 200 (89.7) | ref. | ref. | ||||
| Rh-negative | 63 (10.5) | 23 (10.3) | 1.17 | 0.76–1.80 | 0.479 | − | − | − |
| ICU admission | ||||||||
| No | 425 (70.8) | − | ref. | ref. | ||||
| Yes | 175 (29.2) | 223 (100.0) | 46.65 | 12.13–179.42 | <0.001* | − | − | − |
| Intubated | ||||||||
| No | 576 (96.0) | 9 (4.0) | ref. | ref. | ||||
| Yes | 24 (4.0) | 214 (96.0) | 34.36 | 17.57–67.21 | <0.001* | 22.84 | 11.37–45.86 | <0.001* |
| Follow-up period, days | 9 (6–13) | 12 (6–19) | − | − | − | |||
| WBC, ×103/µL | 7.55 (5.9–10.2) | 13.25 (9.9–19.2) | 1.06 | 1.05–1.08 | <0.001* | − | − | − |
| NEU, ×103/µL | 5.2 (3.7–8) | 12 (8–17.9) | 1.07 | 1.06–1.08 | <0.001* | 1.02 | 1.01–1.04 | 0.020* |
| LY, ×103/µL | 1.3 (0.8–1.8) | 0.4 (0.2–0.8) | 0.29 | 0.21–0.38 | <0.001* | − | − | − |
Numerical variables without normal distribution are given as median (IQR 25th–75th percentiles). Categorical variables are given as n (%). The asterisks show statistical significance. CVD, cardiovascular disease; RSD, respiratory system diseases; DM, diabetes mellitus; ICU, intensive care unit; WBC, white blood cell; NEU, neutrophil; LY, lymphocyte; HR, hazard ratio; CI, confidence interval; ref. reference.
Discussion
In our study, compared with the blood group distribution of healthy blood donors in the region, the prevalence of COVID-19 was higher in blood groups A, B, and AB and lower in blood group O. Our results are in agreement with other studies suggesting that blood group O individuals are somewhat more resistant to clinically overt infection with SARS-CoV-2 than other blood groups [16, 17, 18, 19, 25]. Dzik et al. [26] have stated that the fact that the blood donors were selected particularly from among those with blood group O could be a reason why blood group O was detected less often in the patient group. This is a very reasonable interpretation. However, blood group A was more common than blood group O in the blood group distribution of our donors, even though this was not statistically significant. Nevertheless, blood donors cannot accurately reflect the actual blood type in this area. There are both healthy people and patients in the blood group population that represents the whole society. In addition to the higher O blood group in the donor population, the male gender is also higher. The population mobility of the region may also change the distribution of blood groups over time. Therefore, blood group distribution should be done by screening the whole society and should be up to date. Individuals infected with asymptomatic SARS-CoV-2 should also be identified by screening the whole population. Blood group studies done in this way will give us the most accurate result. Good organization, a long time, a serious workload, and high costs are required to perform this work. It is therefore an ideal but difficult way of working.
In the SARS-CoV-1 epidemic, it was shown that anti-A antibodies in individuals with blood group O prevented the invasion of the S protein into the tissues [14], and it has been claimed that, due to the same mechanism, this will cause less infection in individuals with blood group O in the SARS-CoV-2 pandemic [19, 25]. Although there are anti-A antibodies in blood group B, there are no published studies that show that blood group B is less susceptible to SARS-CoV-2. This situation was explained by Gérard et al. [27], with the anti-A antibodies in blood group O being the IgG type and the anti-A antibodies in blood group B being the IgM type.
The importance of the anti-A antibody titre in SARS-CoV-1 infection has also been emphasized [14]. However, no increase in susceptibility to COVID-19 infection was detected in individuals with blood group AB without any blood group antibodies. In this case, the increased susceptibility of blood group A to infection and lower susceptibility of blood group O to infection must involve other mechanisms. One of these may be that thromboembolic diseases are more common in blood group A, which therefore shows more disease symptoms [28]. In fact, knowing the blood group distribution of SARS-CoV-2-infected asymptomatic cases in the community would help elucidate the relationship between SARS-CoV-2 and the blood group of infected individuals more objectively. However, no such study has yet been published.
There is a symbiotic relationship between blood group expression and maturation of the gastrointestinal microbiome. Bacteria can induce antibodies against blood group antigens, including ABO. Blood group antibodies are stimulated by the intestinal flora, and blood group antibody titres are lower in hyper-hygienic populations and those following Western diets [8, 29, 30]. Guillon et al. [14] showed that SARS-CoV-1 can be blocked with monoclonal anti-A and human anti-A antibodies; this can be accomplished using a high anti-A antibody titre but a lower anti-A antibody titre is ineffective. This finding may indicate that SARS-CoV-2 infection will spread faster in industrialized countries with people who follow Western diets and live in extremely hygienic conditions.
Our results are in agreement with other studies suggesting that blood group O individuals are somewhat more resistant to clinically overt infection with SARS-CoV-2 than others. However, this tendency is not sufficiently established to allow special prophylaxis recommendations for non-O individuals. Of course, blood group is not the only factor that determines contagiousness. For instance, all South American Indians have the O blood type [31]. However, there is no information that the frequency of COVID-19 is lower or the rate of spread is slower in areas where South American natives live.
There are many factors, other than blood groups, that determine the contagiousness and severity of SARS-CoV-2 infection. Many of these are still unexplained. More comprehensive studies are needed to explain the relationship of SARS-CoV-2 infection and blood groups.
Limitations of the Study
Our study has limitations. The blood group distribution of donors was made 11 years ago. The COVID-19 patients were compared with healthy blood donors. Only hospitalized COVID-19 cases were included in the study.
Statement of Ethics
This study was conducted ethically in accordance with the World Medical Association Declaration of Helsinki and was approved by our institutional ethics commission (EC No. 49/2020).
Funding Sources
There was no specific funding for this study.
Conflict of Interest Statement
The authors declare no conflict of interest.
Author Contributions
D.Y.A. designed the study and collected data. M.B. collected and analyzed the data. Both authors read and approved the final version.
References
- 1.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30566-3. published online Mar 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2020 Feb;382((8)):727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss SR, Leibowitz JL. Coronavirus pathogenesis. Adv Virus Res. 2011;81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M, Kleine-Weber H, Krüger N, Müller M, Drosten C, Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. 2020 31 January;:929042. [Google Scholar]
- 5.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020 May;8((5)):475–81. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosseini SM, Feng JJ. How malaria parasites reduce the deformability of infected red blood cells. Biophys J. 2012 Jul;103((1)):1–10. doi: 10.1016/j.bpj.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branch DR. Blood groups and susceptibility to virus infection: new developments. Curr Opin Hematol. 2010 Nov;17((6)):558–64. doi: 10.1097/MOH.0b013e32833ece31. [DOI] [PubMed] [Google Scholar]
- 8.Cooling L. Blood Groups in Infection and Host Susceptibility. Clin Microbiol Rev. 2015 Jul;28((3)):801–70. doi: 10.1128/CMR.00109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martins LC, de Oliveira Corvelo TC, Oti HT, do Socorro Pompeu Loiola R, Aguiar DC, dos Santos Barile KA, et al. ABH and Lewis antigen distributions in blood, saliva and gastric mucosa and H pylori infection in gastric ulcer patients. World J Gastroenterol. 2006 Feb;12((7)):1120–4. doi: 10.3748/wjg.v12.i7.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque AS, Mahalanabis D, Hoque SS, Albert MJ. The relationship between ABO blood groups and susceptibility to diarrhea due to Vibrio cholerae 0139. Clin Infect Dis. 1994 May;18((5)):827–8. doi: 10.1093/clinids/18.5.827. [DOI] [PubMed] [Google Scholar]
- 11.Stowell CP, Stowell SR. Biologic roles of the ABH and Lewis histo-blood group antigens Part I: infection and immunity. Vox Sang. 2019 Jul;114((5)):426–42. doi: 10.1111/vox.12787. [DOI] [PubMed] [Google Scholar]
- 12.Behal R, Jain R, Behal KK, Dhole TN. Variation in the host ABO blood group may be associated with susceptibility to hepatitis C virus infection. Epidemiol Infect. 2010 Aug;138((8)):1096–9. doi: 10.1017/S0950268809991117. [DOI] [PubMed] [Google Scholar]
- 13.Onsten TG, Callegari-Jacques SM, Goldani LZ. The higher frequency of blood group b in a Brazilian population with HIV Infection. Open AIDS J. 2013 Oct;7((1)):47–50. doi: 10.2174/1874613601307010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillon P, Clément M, Sébille V, Rivain JG, Chou CF, Ruvoën-Clouet N, et al. Inhibition of the interaction between the SARS-CoV spike protein and its cellular receptor by anti-histo-blood group antibodies. Glycobiology. 2008 Dec;18((12)):1085–93. doi: 10.1093/glycob/cwn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Y, Cheng G, Chui CH, Lau FY, Chan PK, Ng MH, et al. ABO blood group and susceptibility to severe acute respiratory syndrome. JAMA. 2005 Mar;293((12)):1450–1. doi: 10.1001/jama.293.12.1450-c. [DOI] [PubMed] [Google Scholar]
- 16.Wu BB, Gu DZ, Yu JN, Yang J, Shen WQ. Association between ABO blood groups and COVID-19 infection, severity and demise: A systematic review and meta-analysis. Infect Genet Evol. 2020 Oct;84:104485. doi: 10.1016/j.meegid.2020.104485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Feng Z, Li P, Yu Q. Relationship between ABO blood group distribution and clinical characteristics in patients with COVID-19. Clin Chim Acta. 2020 Oct;509:220–3. doi: 10.1016/j.cca.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Wang X, Chen J, Cai Y, Deng A, Yang M. Association between ABO blood groups and risk of SARS-CoV-2 pneumonia. Br J Haematol. 2020 Jul;190((1)):24–7. doi: 10.1111/bjh.16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J, Yang Y, Huang H, Li D, Gu D, Lu X, et al. Relationship between the ABO Blood Group and the COVID-19 Susceptibility. Clin Infect Dis. 2020 Aug;•••:ciaa1150. doi: 10.1093/cid/ciaa1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPherson R, Pincus M, editors. Henry's Clinical Diagnosis and Management by Laboratory Methods. 21st ed. Philadelphia: Saunders Elsevier; 2007. [Google Scholar]
- 21.Cürük MA, Arpaci A, Attila G, Tuli A, Kilinç Y, Aksoy K, et al. Genetic heterogeneity of beta-thalassemia at Cukurova in southern Turkey. Hemoglobin. 2001 May;25((2)):241–5. doi: 10.1081/hem-100104032. [DOI] [PubMed] [Google Scholar]
- 22.Koçak R, Alparslan ZN, Ağridağ G, Başlamisli F, Aksungur PD, Koltaş S. The frequency of anaemia, iron deficiency, hemoglobin S and beta thalassemia in the south of Turkey. Eur J Epidemiol. 1995 Apr;11((2)):181–4. doi: 10.1007/BF01719485. [DOI] [PubMed] [Google Scholar]
- 23.WHO Protocol 2020 Diagnostic detection of Wuhan coronavirus 2019 by real-time RT-PCR. [Google Scholar]
- 24.Yildiz SM. Distribution of ABO and Rh blood group systems in Cukurova region. Cukurova Med J. 2016;41((4)):658–63. [Google Scholar]
- 25.Göker H, Aladağ Karakulak E, Demiroğlu H, Ayaz Ceylan ÇM, Büyükaşik Y, Inkaya AÇ, et al. The effects of blood group types on the risk of COVID-19 infection and its clinical outcome. Turk J Med Sci. 2020 Jun;50((4)):679–83. doi: 10.3906/sag-2005-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dzik S, Eliason K, Morris EB, Kaufman RM, North CM. COVID-19 and ABO blood groups. Transfusion. 2020 Aug;60((8)):1883–4. doi: 10.1111/trf.15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gérard C, Maggipinto G, Minon JM. COVID-19 and ABO blood group: another viewpoint. Br J Haematol. 2020 Jul;190((2)):e93–4. doi: 10.1111/bjh.16884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sardu C, Marfella R, Maggi P, Messina V, Cirillo P, Codella V, et al. Implications of AB0 blood group in hypertensive patients with covid-19. BMC Cardiovasc Disord. 2020 Aug;20((1)):373. doi: 10.1186/s12872-020-01658-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Springer GF, Horton RE, Forbes M. [Origin of anti-human blood group B agglutinins in white Leghorn chicks] J Exp Med. 1959 Aug;110((2)):221–44. doi: 10.1084/jem.110.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazda T, Yabe R, NaThalang O, Thammavong T, Tadokoro K. Differences in ABO antibody levels among blood donors: a comparison between past and present Japanese, Laotian, and Thai populations. Immunohematology. 2007;23((1)):38–41. [PubMed] [Google Scholar]
- 31.Racial and ethnic distribution of ABO blood types. Available from: http://www.bloodbook.com/world-abo.html.