Pseudomonas aeruginosa causes a variety of acute and chronic infections in humans. The type III secretion system (T3SS) plays an important role in acute infection, and the type VI secretion system (T6SS) and biofilm formation are associated with chronic infections.
KEYWORDS: biofilm, Pseudomonas aeruginosa, RetS, type III secretion system, YbeY, sRNA
ABSTRACT
YbeY is a highly conserved RNase in bacteria and plays essential roles in the maturation of 16S rRNA, regulation of small RNAs (sRNAs), and bacterial responses to environmental stresses. Previously, we verified the role of YbeY in rRNA processing and ribosome maturation in Pseudomonas aeruginosa and demonstrated YbeY-mediated regulation of rpoS through an sRNA, ReaL. In this study, we demonstrate that mutation of the ybeY gene results in upregulation of the type III secretion system (T3SS) genes as well as downregulation of the type VI secretion system (T6SS) genes and reduction of biofilm formation. By examining the expression of the known sRNAs in P. aeruginosa, we found that mutation of the ybeY gene leads to downregulation of the small RNAs RsmY/Z, which control the T3SS, T6SS, and biofilm formation. Further studies revealed that the reduced levels of RsmY/Z are due to upregulation of retS. Taken together, our results reveal the pleiotropic functions of YbeY and provide detailed mechanisms of YbeY-mediated regulation in P. aeruginosa.
IMPORTANCE Pseudomonas aeruginosa causes a variety of acute and chronic infections in humans. The type III secretion system (T3SS) plays an important role in acute infection, and the type VI secretion system (T6SS) and biofilm formation are associated with chronic infections. Understanding of the mechanisms that control the virulence determinants involved in acute and chronic infections will provide clues for the development of effective treatment strategies. Our results reveal a novel RNase-mediated regulation of T3SS, T6SS, and biofilm formation in P. aeruginosa.
INTRODUCTION
YbeY is a highly conserved bacterial RNase that is involved in the maturation of 16S rRNA, ribosome quality control, regulation of sRNAs, and stress responses (1–6). Previous studies in Escherichia coli identified YbeY as a UPF0054 family metal-dependent hydrolase, and the three-dimensional crystal structure of YbeY revealed a conserved metal ion-binding region (7). The YbeY protein purified from Sinorhizobium meliloti displays metal-dependent endoribonuclease activity that cleaves both single-stranded (ssRNA) and double-stranded (dsRNA) RNA substrates (6). Deletion of ybeY in E. coli reduces protein translation efficiency by affecting the 30S ribosome subunits (8). Jacob et al. demonstrated that YbeY is a single-strand-specific endoribonuclease that plays key roles in ribosome quality control and 16S rRNA maturation together with RNase R in E. coli (1). The structural model of YbeY revealed a positively charged cavity similar to the middle domain of Argonaute (AGO) proteins involved in RNA silencing in eukaryotes (9). Recent studies in Vibrio cholerae, S. meliloti, and E. coli demonstrated that a defect in YbeY results in aberrant expression of small RNAs (sRNAs) and the corresponding target mRNAs (2, 9, 10).
In pathogenic bacteria, YbeY has been found to play important roles in bacterial virulence. In V. cholerae, the absence of YbeY reduces the production of the cholera toxin and intestinal colonization in mice (2). In Yersinia enterocolitica, YbeY is required for intestinal adhesion and bacterial virulence (11). A defect in ybeY severely impairs the ability of Brucella to infect macrophages (12). However, the mechanisms by which YbeY affects bacterial virulence and stress response remain largely unknown.
Pseudomonas aeruginosa is an opportunistic Gram-negative pathogen that causes acute and chronic infections in humans (13). The bacterium possesses a variety of virulence determinants that contribute to pathogenesis. The type III secretion system (T3SS) is one of the major virulence factors that play critical roles in acute infections (14). It is a syringe-like machinery that directly injects effector proteins into mammalian cells, interfering with cell physiological functions or leading to cell death (14). The chronic infection caused by P. aeruginosa is usually accompanied by the formation of biofilm in which bacteria are protected by an extracellular matrix against host immune cells and antibacterial substances (15).
The type VI secretion system (T6SS) is a weapon for bacterial warfare and interfering with the functions of host cells (16). A number of T6SSs have been demonstrated to target competing bacteria and efficiently kill the competitors (17–20), which may play a key role in the survival and proliferation of the producer cells in a multimicrobial environment (21). P. aeruginosa harbors three T6SS clusters, namely, H1-, H2-, and H3-T6SS. The H1-T6SS is related to the adaptability of this bacterium to chronic infection (22, 23). A recent study in reference strain PA14 revealed that all the three T6SSs are under the control of the RetS-GacS/GacA-RsmA pathway and the transcriptional regulator AmrZ (24).
The RetS/LadS-GacS/GacA-RsmY/RsmZ-RsmA regulatory pathway plays a key role in the transition between acute and chronic infections. RetS inhibits the GacS-mediated phosphorylation of GacA through directly binding to GacS, whereas LadS promotes the phosphorylation of GacA. The two-component system GacS/GacA directly activates the expression of RsmY/RsmZ sRNAs that antagonize the function of RsmA through direct interaction. RsmA is an RNA binding protein that represses expression of T6SS genes and biofilm formation and activates the expression of T3SS genes (25–29). AmrZ is a DNA binding protein that controls gene expression at the transcriptional level. Unlike RsmA, which represses the expression of all three T6SS genes, AmrZ represses the expression of the H2-T6SS genes but activates the expression of the H1- and H3-T6SS genes (24).
Previously, we demonstrated that the P. aeruginosa endoribonuclease YbeY is involved in 16S rRNA maturation and ribosome assembly. In addition, we found that YbeY controls bacterial resistance to oxidative stresses through an sRNA, ReaL (30). In this study, we demonstrate that YbeY regulates the expression of T3SS and T6SS genes and biofilm formation through the RetS-GacS/GacA-RsmY/RsmZ-RsmA pathway, further revealing the pleiotropic function of YbeY in P. aeruginosa.
RESULTS
Mutation of ybeY enhances the expression of the T3SS genes and bacterial cytotoxicity.
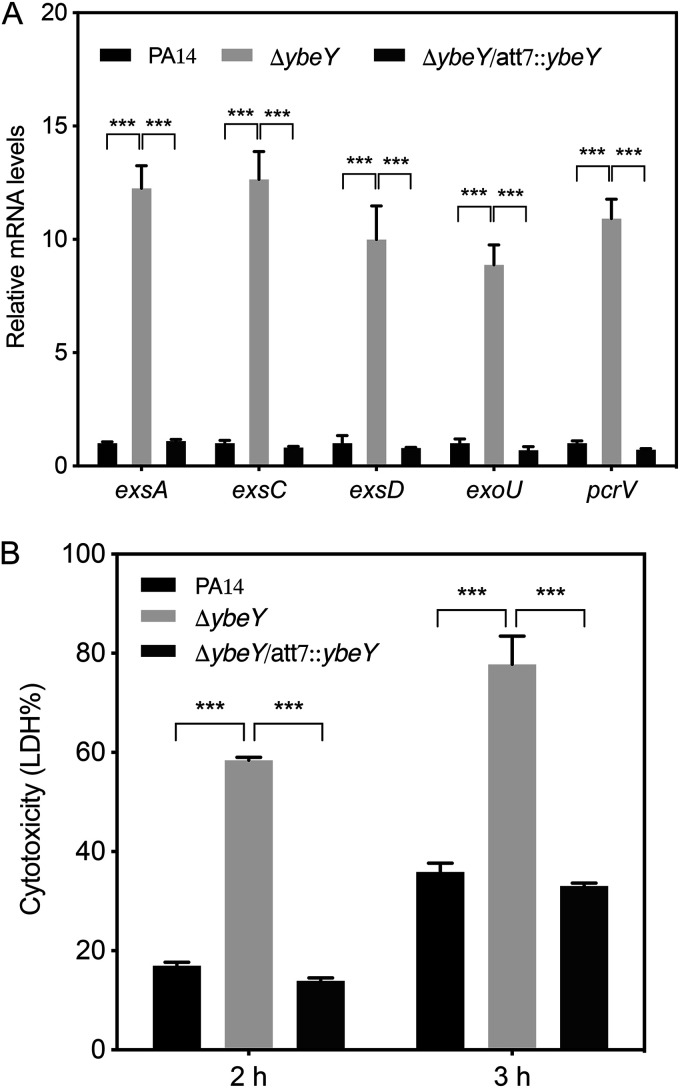
Our previous transcriptomic analyses revealed an upregulation of the T3SS genes in a PA14 ΔybeY mutant (30). To understand the relationship between YbeY and the T3SS genes, we utilized reverse transcription-quantitative PCR (RT-qPCR) to verify the expression levels of the T3SS regulatory genes exsA, exsC, and exsD, the structural gene pcrV, and the effector gene exoU. All of the tested genes were upregulated about 9- to 13-fold in the ΔybeY mutant and returned to wild-type levels by the complementation of the ybeY gene (Fig. 1A). Since T3SS plays a major role in the bacterial cytotoxicity, we performed an LDH release assay with the A549 human lung carcinoma cell line. Compared to the wild-type strain, the ΔybeY mutant displayed enhanced cytotoxicity (Fig. 1B).
FIG 1.
YbeY is involved in the regulation of the T3SS. (A) Wild-type PA14, the ΔybeY mutant, and the complemented strain were grown in LB to an OD600 of 1. The relative mRNA levels of the T3SS genes were determined by RT-qPCR. Results represent means ± standard deviations (SD). (B) A549 cells were infected with the indicated strains at an MOI of 50 for 2 or 3 h. The relative cytotoxicity was determined by the LDH release assay. Results represent means ± SD. ***, P < 0.001 by Student's t test.
YbeY influences the expression of the T3SS and T6SS genes and biofilm formation through the RsmY/RsmZ-RsmA pathway.
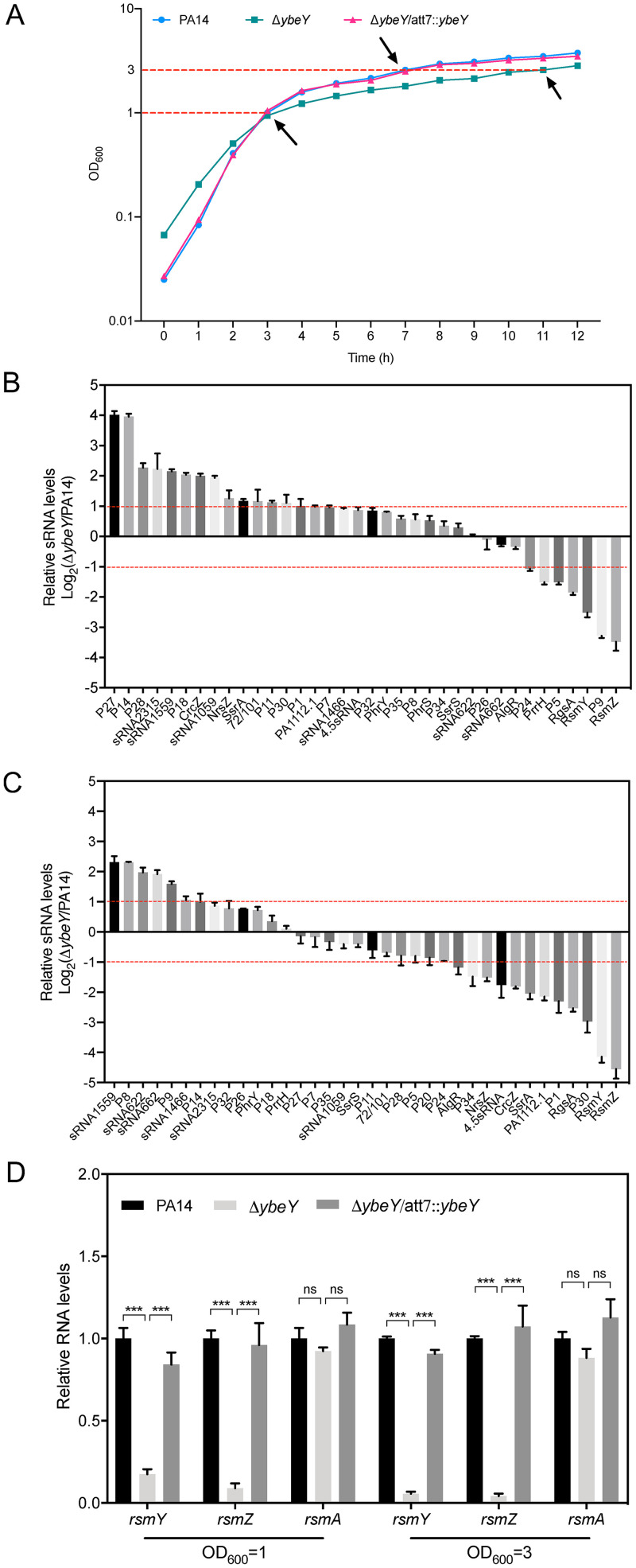
YbeY is an endoribonuclease that has been shown to control the expression of rpoS through the sRNA ReaL (30). We hypothesized that YbeY affects the expression of the T3SS genes through sRNAs. Thus, we examined the levels of 36 known P. aeruginosa sRNAs by RT-qPCR. Previously, we found that mutation of ybeY reduces the bacterial growth rate (30). Therefore, we increased the inoculum of the ΔybeY mutant to achieve an optical density at 600 nm (OD600) of 1, the same as the wild-type strain at the same time before RNA isolation. However, it took longer for the ΔybeY mutant to achieve an OD600 of 3.0. The growth curves and sample collection points are shown in Fig. 2A. The expression of 21 and 18 sRNAs was altered (fold change, >2) by the mutation of ybeY in exponential and stationary growth phases, respectively (Fig. 2B and C). Of note, the sRNAs RsmY and RsmZ were two of the most downregulated sRNAs in the exponential and stationary growth phases in the ΔybeY mutant. Complementation of the ybeY gene in the ΔybeY mutant restored the levels of RsmY and RsmZ (Fig. 2D). The mRNA level of rsmA was not affected by the mutation of ybeY (Fig. 2D).
FIG 2.
YbeY controls the expression of sRNAs but not the expression of rsmA. (A) The growth curves of wild-type PA14, the ΔybeY mutant, and the complemented strain. The bacteria were grown in LB overnight. Aliquots of 0.3 ml of the cultures of the wild-type PA14 and the complemented strain or 0.9 ml of the culture of the ΔybeY mutant were subcultured into 30 ml fresh LB medium and grown at 37°C with agitation at 200 rpm. The OD600 was monitored every hour for 12 h. The sample collection points are indicated by arrows. Bacteria were grown to an OD600 of 1 (B) or 3 (C). Total RNA was purified, and the relative sRNA levels were determined by RT-qPCR. The relative levels of the small RNAs in the ΔybeY mutant compared to those in wild-type PA14 are shown. Results represent means ± SD. The red lines represent a fold change of 2. (D) The bacteria were grown in LB to an OD600 of 1 or 3. The relative RNA levels of rsmY-rsmZ and rsmA were determined by RT-qPCR. Results represent means ± SD. ***, P < 0.001 by Student's t test. ns, not significant.
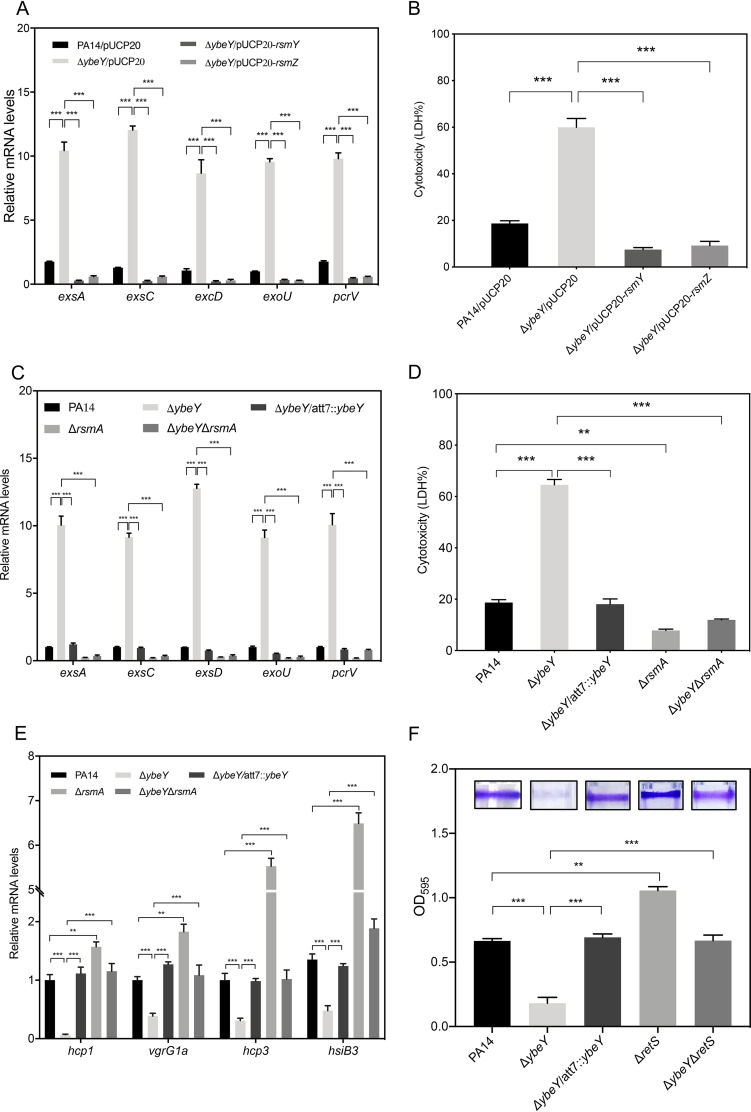
Previous studies revealed that upregulation of RsmY/Z leads to downregulation of the T3SS genes (31, 32). To investigate whether RsmY/Z is involved in the regulation of the T3SS genes by YbeY, we overexpressed RsmY or RsmZ in the ΔybeY mutant, which reduced the expression levels of the T3SS genes and the bacterial cytotoxicity (Fig. 3A and B). Deletion of the rsmA gene in the ΔybeY mutant reduced the expression of the T3SS genes and the cytotoxicity (Fig. 3C and D).
FIG 3.
YbeY controls the expression of the T3SS and T6SS genes and biofilm formation through RsmY/Z-RsmA. (A) The indicated strains were grown in LB to an OD600 of 1. The relative mRNA levels of the T3SS genes were determined by RT-qPCR. Results represent means ± SD. (B) A549 cells were infected with the indicated strains at an MOI of 50 for 2 h. The relative cytotoxicity was determined by the LDH release assay. (C) The relative mRNA levels of the T3SS genes were determined by RT-qPCR. Results represent means ± SD. (D) The relative bacterial cytotoxicity was determined by the LDH release assay. (E) The indicated strains were grown in LB to an OD600 of 1. The relative mRNA levels of the T6SS genes were determined by RT-qPCR. (F) The indicated strains were grown in 96-well plates for 20 h. The wells were washed with PBS and stained with 1% crystal violet. The crystal violet was dissolved in ethanol and measured at a wavelength of 595 nm. Results represent means ± SD. **, P < 0.01; ***, P < 0.001; both by Student's t test.
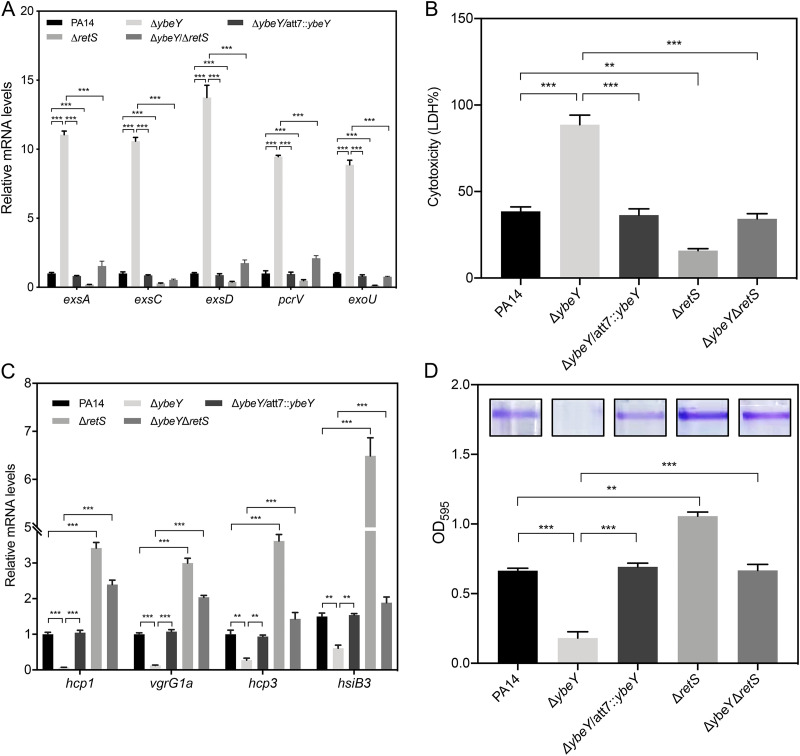
Since the GacS/GacA-RsmY/Z-RsmA pathway reciprocally regulates the T3SS, T6SS, and biofilm formation (24, 25), we suspected that YbeY is involved in the regulation of the T6SS and biofilm formation. We then examined the expression levels of H1- and H3-T6SS genes that are regulated in the same patterns by GacS/GacA and AmrZ. Indeed, RT-qPCR results revealed downregulation of hcp-1, vgrG1a, hcp3, and hsiB3 in the ΔybeY mutant (Fig. 3E). In addition, the ΔybeY mutant displayed reduced biofilm formation (Fig. 3F). Deletion of rsmA in the ΔybeY mutant restored the expression of the T6SS genes and biofilm formation (Fig. 3E and F). These results demonstrate that YbeY plays an important role in the transition between acute and chronic infections through the RsmY/RsmZ-RsmA regulatory pathway.
YbeY regulates the expression of RsmY/Z through RetS.
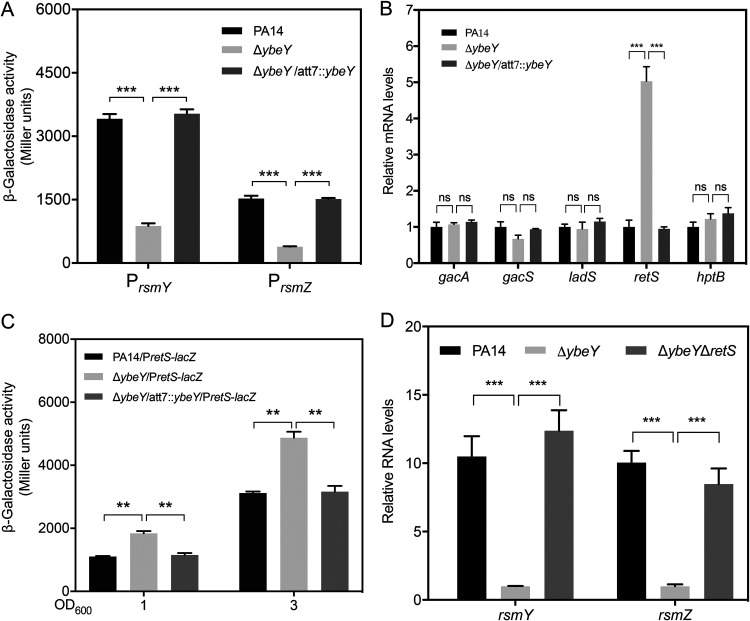
The expression of rsmY and rsmZ is directly activated by the GacS/GacA two-component system. RetS inhibits the GacS-mediated phosphorylation of GacA through directly binding to GacS, whereas LadS promotes the phosphorylation of GacA (25–29). To understand the mechanism of the downregulation of rsmY and rsmZ in the ΔybeY mutant, we monitored the promoter activities of the two genes by lacZ transcriptional fusions (PrsmY-lacZ and PrsmZ-lacZ). The LacZ levels of both of the constructs were lower in the ΔybeY mutant and returned to wild-type levels by the complementation of the ybeY gene (Fig. 4A), indicating a reduction at the transcriptional level. The transcription of rsmY and rsmZ is directly activated by the GacS/GacA two-component regulatory system (28). However, the mRNA levels of gacS and gacA were not affected by the mutation of ybeY (Fig. 4B). We then examined the genes regulating the activity of the GacS/GacA system. Mutation of ybeY resulted in upregulation of retS, whereas the expression of ladS and hptB was not affected (Fig. 4B). By utilizing a transcriptional fusion between the retS promoter and a lacZ gene (PretS-lacZ), we found the promoter activity of retS was increased in the ybeY mutant and returned to wild-type levels by the complementation of the ybeY gene (Fig. 4C). These results led us to speculate that the upregulation of retS represses the expression of rsmY and rsmZ and subsequently leads to the activation of the T3SS genes and suppression of the T6SS genes and biofilm formation. To test our hypothesis, we knocked out retS in the ΔybeY mutant, which resulted in increased levels of RsmY/Z (Fig. 4D). In addition, deletion of retS in the ΔybeY mutant reduced expression of the T3SS genes and cytotoxicity (Fig. 5A and B) and increased the expression of the H1- and H3-T6SS genes as well as biofilm formation (Fig. 5C and D). These results demonstrate that YbeY plays an important role in the regulation of T3SS, T6SS, and biofilm formation through RetS.
FIG 4.
YbeY controls the expression of rsmY and rsmZ through RetS. (A) PA14, the ΔybeY mutant, and the complemented strain containing the PrsmY-lacZ or PrsmZ-lacZ transcriptional fusion were cultured in LB to an OD600 of 1. The bacteria were collected and subjected to the β-galactosidase activity assay. (B) Wild-type PA14, the ΔybeY mutant, and the complemented strain were grown in LB to an OD600 of 1. The relative mRNA levels of gacA, gacS, ladS, retS, and hptB were determined by RT-qPCR. Results represent means ± SD. (C) PA14, the ΔybeY mutant, and the complemented strain containing the PretS-lacZ transcriptional fusion were cultured in LB to an OD600 of 1 or 3. The bacteria were collected and subjected to the β-galactosidase activity assay. (D) Wild-type PA14, the ΔybeY mutant, and the ΔybeY ΔretS mutant were grown in LB to an OD600 of 1. The relative RNA levels of rsmY and rsmZ were determined by RT-qPCR. Results represent means ± SD. ns, not significant; **, P < 0.01; ***, P < 0.001; by Student's t test.
FIG 5.
YbeY controls biofilm formation and the expression of T3SS and T6SS genes through RetS. (A) The indicated strains were grown in LB to an OD600 of 1. The relative mRNA levels of the T3SS genes were determined by RT-qPCR. Results represent means ± SD. (B) A549 cells were infected with the indicated strains at an MOI of 50 for 3 h. The relative cytotoxicity was determined by the LDH release assay. (C) The indicated strains were grown in LB to an OD600 of 1. The relative mRNA levels of the T6SS genes were determined by RT-qPCR. Results represent means ± SD. (D) The indicated strains were grown in 96-well plates for 20 h. The wells were washed with PBS and stained with 1% crystal violet. The crystal violet was dissolved in ethanol and measured at a wavelength of 595 nm. Results represent means ± SD. **, P < 0.01; ***, P < 0.001; both by Student's t test.
Mutation of ybeZ results in phenotypes similar to those of the ΔybeY mutant.
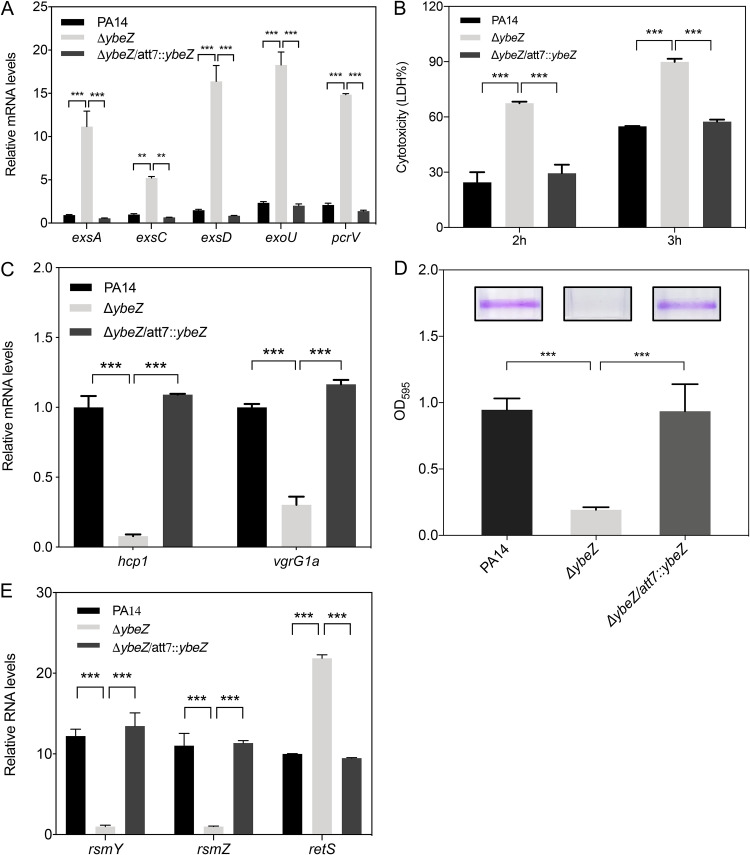
In our previous research, we found that YbeZ binds to YbeY and is involved in the maturation of 16S rRNA and the response to oxidative stress (30). Therefore, we speculated that YbeZ plays a role in the regulation of the T3SS, T6SS, and biofilm formation. Indeed, mutation of ybeZ resulted in upregulation of T3SS genes and enhanced cytotoxicity (Fig. 6A and B). In addition, the ΔybeZ mutant displayed downregulation of T6SS genes and reduced biofilm formation (Fig. 6C and D). Consistent with this, the ΔybeZ mutant displayed similar expression levels of the genes encoding RsmY, RsmZ, and RetS (Fig. 6E). In combination, these results demonstrate that YbeZ is involved in the regulation of transition between acute and chronic infections through RetS.
FIG 6.
YbeZ influences the expression of the T3SS and T6SS genes and biofilm formation. (A) Wild-type PA14, the ΔybeZ mutant, and the complemented strain were grown in LB to an OD600 of 1. The relative mRNA levels of the T3SS genes were determined by RT-qPCR. Results represent means ± SD. (B) Cytotoxicity of wild-type PA14, the ΔybeZ mutant, and the complemented strain. A549 cells were infected with the indicated strains at an MOI of 50 for 2 or 3 h. The relative cytotoxicity was determined by the LDH release assay. (C) The indicated strains were grown in LB to an OD600 of 1. The relative mRNA levels of the T6SS genes were determined by RT-qPCR. Results represent means ± SD. (D) The indicated strains were grown in 96-well plates for 20 h. The wells were washed with PBS and stained with 1% crystal violet. The crystal violet was dissolved in ethanol and measured at a wavelength of 595 nm. (E) The bacteria were grown in LB to an OD600 of 1. The relative RNA levels of rsmY, rsmZ, and retS were determined by RT-qPCR. Results represent means ± SD. **, P < 0.01; ***, P < 0.001; both by Student's t test.
DISCUSSION
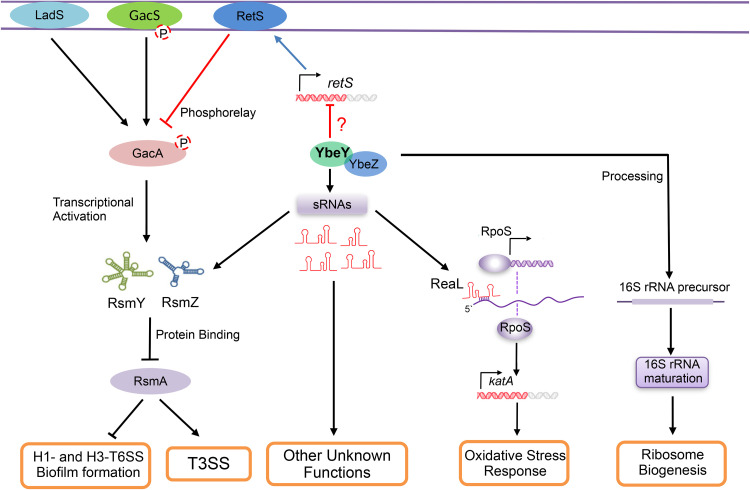
Ribonucleases play important roles in bacterial stress responses and regulation of virulence factors. YbeY is a conserved endoribonuclease that plays pleiotropic roles in bacterial physiology and virulence (1–6). In V. cholera, mutation in the ybeY gene resulted in complete loss of mouse colonization and biofilm formation (2). In E. coli, YbeY has been shown to play important roles in bacterial resistance to heat shock, oxidative stresses, and a variety of antibiotics (33). Deletion of the ybeY gene in the plant pathogen Agrobacterium tumefaciens reduced the bacterial growth rate, motility, and stress tolerance (34). In Yersinia enterocolitica serotype O:3, YbeY is involved in the regulation of the genes of the Yersinia virulence plasmid (pYV) and multiple regulatory small RNAs (11). In enterohemorrhagic E. coli (EHEC), YbeY is required for the expression of the T3SS genes. Further studies revealed that mutation of ybeY reduces the amount of initiating ribosomes, leading to destabilization of the T3SS gene mRNA (35). Previously, we found that YbeY controls bacterial resistance to oxidative stresses through a small RNA (sRNA), ReaL, and participates in the maturation of 16S rRNA in P. aeruginosa. Here, we demonstrated that YbeY is involved in the regulation of serval sRNAs in P. aeruginosa. In addition, we found that mutation of ybeY results in the upregulation of the T3SS genes. Further studies revealed that YbeY regulates the expression of the T3SS genes through the GacA/S-RsmY/Z-RsmA pathway by regulating the expression of retS (Fig. 7).
FIG 7.
Roles of YbeY/YbeZ in P. aeruginosa. YbeY/YbeZ are involved in the maturation of 16S rRNA and ribosome assembly (30). Meanwhile, YbeY influences the levels of multiple sRNAs. One of the direct regulatory targets of YbeY is the sRNA ReaL. ReaL binds to the 5′-untranslated region of the rpoS mRNA, inhibiting its translation (58). RpoS is an alternative sigma factor that has been demonstrated to contribute to bacterial responses to oxidative stresses by activating the expression of the major catalase KatA in P. aeruginosa (59). YbeY directly degrades ReaL, thereby positively regulating the expression of RpoS (30). In addition, YbeY and YbeZ control the expression of the T3SS and T6SS genes as well as biofilm formation through the RetS-GacS/GacA-RsmY/Z-RsmA pathway. YbeY and YbeZ repress the transcription of retS through an unknown mechanism. RetS directly binds to GacS, which inhibits the phosphorelay-dependent activation of GacA. GacA activates the expression of two sRNAs, RsmY and RsmZ, that antagonize the function of RsmA. RsmA is a posttranscriptional regulator that activates the expression of the T3SS genes and represses the expression of the T6SS genes as well as the extracellular polysaccharide biosynthesis pel and psl genes, which are important for biofilm formation (25–29).
Previous studies revealed that the expression of retS is repressed by the two-component system PhoP/PhoQ and activated by the transcriptional regulator CysB (36, 37). Our results revealed the downregulation of phoP and upregulation of cysB in the ΔybeY mutant (data not shown). However, overexpression of phoP or a knockout of cysB in the ΔybeY mutant did not reduce the expression of retS and the T3SS genes (data not shown). Thus, the mechanism of the upregulation of retS in the ybeY mutant remains elusive and requires further studies.
The T6SS is a weapon that targets competing bacteria and efficiently kills the competitors (17–20). We found that the ybeY mutation resulted in downregulation of all three T6SS genes and a reduction of the ability to kill other bacteria (data not shown). A recent study revealed that all the three T6SSs are under the control of the RetS-GacS/GacA-RsmA pathway, and the H2-T6SS plays a major role in bacterial killing in the reference strain PA14 (24). In our study, we found that knocking out rsmA or retS in the context of ybeY mutation could not restore the expression of the H2-T6SS genes and the ability to kill other bacteria (data not shown), indicating that additional factors control the expression of the H2-T6SS genes. Previous study has shown that the transcriptional regulator AmrZ directly represses the expression of the H2-T6SS genes but activates the expression of the H1- and H3-T6SS genes (24). Our preliminary results demonstrated an upregulation of amrZ in the ybeY mutant (data not shown). Currently, we are making efforts to understand the mechanism of YbeY-mediated regulation on amrZ.
sRNAs affect the stabilities and translation efficiencies of mRNAs through complementary base pairing, which is a key regulatory mechanism of bacterial gene expression (31, 38, 39). Although bacterial sRNAs affect a wide range of biological processes, including energy utilization and metabolism, pathogenicity, and antibiotic resistance, our understanding of the regulation of sRNAs is still limited (40–43). Ribonucleases play an important role in cellular RNA metabolism processes, such as mRNA degradation and rRNA/tRNA maturation, and have emerged as the main posttranscriptional regulators of sRNAs (44–46). RNase E and PNPase have been shown to be involved in the degradation of the free pool of sRNAs (47, 48). In addition, RNases are also involved in the maturation process of sRNAs (45). Recent studies in V. cholerae, S. meliloti, and E. coli have shown that YbeY is involved in the regulation of sRNAs (2, 9, 10).
In this study, we found that mutation of the ybeY gene influenced the expression of multiple sRNAs. For example, crcZ, which is related to carbon metabolism, is upregulated in the exponential phase but downregulated in the stationary phase, indicating that YbeY is involved in the growth phase-dependent metabolism regulation. The production of sRNA P27, PrrH, and NrsZ, involved in quorum sensing, was altered by the ybeY mutation (49–51). The RpoS‐dependent sRNA RgsA, which regulates Fis and AcpP, is downregulated, which might be due to the defective expression of rpoS in the ybeY mutant (52). These results imply that YbeY plays a role in the regulation of quorum-sensing genes. SsrA is a critical component of the trans-translation system that is involved in the release of ribosomes stalled on mRNAs (53). In the ybeY mutant, SsrA is upregulated in the exponential phase but downregulated in the stationary phase, indicating that YbeY affects mRNA translation in a growth phase-dependent manner. However, the functions of the remaining sRNAs are not known. Nevertheless, these results indicate that YbeY participates in multiple sRNA-mediated regulation processes in physiological functions in P. aeruginosa. Further studies are warranted to understand the functions of these sRNAs and the mechanisms of YbeY-mediated regulation of them.
In many bacterial species, including P. aeruginosa, Staphylococcus aureus, and E. coli, the ybeZ gene is in the same operon as the ybeY gene (30, 33, 54). We previously demonstrated the interaction between YbeY and YbeZ in P. aeruginosa and found that mutation of ybeZ resulted in a defective response to oxidative stresses similar to that of the ybeY mutant. In this study, we found that mutation of ybeZ resulted in the increased expression of T3SS and cytotoxicity, as seen in the ybeY mutant. These results suggest that YbeY and YbeZ function together in the transition between acute and chronic infections through RetS. YbeZ contains a nucleoside triphosphate hydrolase and an ATP binding domain. However, the exact function of YbeZ remains elusive and warrants further studies.
Overall, our results reveal pleiotropic roles of YbeY in the regulation of T3SS, T6SS, biofilm formation, and oxidative stress response in P. aeruginosa. Analyses of the global gene and sRNA expression profiles under various environmental stresses might reveal additional roles of YbeY and the regulatory pathways mediated by this endonuclease.
MATERIALS AND METHODS
Bacteria strains and plasmids.
The bacterial strains, primers, and plasmids used in this study are listed in Table 1. Bacteria were cultured in L-broth medium (LB; 10 g/liter tryptone, 5 g/liter yeast, 5 g/liter NaCl) at 37°C with agitation at 200 rpm (27). Antibiotics were used at the following concentrations: for E. coli, 100 μg/ml ampicillin, 50 μg/ml kanamycin, 10 μg/ml gentamicin, and 10 μg/ml tetracycline; for P. aeruginosa, 50 μg/ml tetracycline, 50 μg/ml gentamicin, and 150 μg/ml carbenicillin. Chromosomal gene mutations were generated as described previously (55).
TABLE 1.
Bacterial strains, plasmids and primers used in this studya
| Strain, plasmid, or primer | Description or sequence (5′–3′) | Source (reference) or function |
|---|---|---|
| E. coli | ||
| DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 φ80dlacZΔM15 | 57 |
| S17-1 | Δ(lacZYA-argF)U169 hsdR17 (rK−mK+) λ-thi pro hsdR recA traC+ | 57 |
| P. aeruginosa | ||
| PA14 | Wild type | 60 |
| △ybeY | PA14 deleted of ybeY | 30 |
| △ybeY/att7::ybeY | △ybeY with ybeY inserted on chromosome with mini-Tn7T insertion; GENr | 30 |
| △ybeZ | PA14 deleted of ybeZ | 30 |
| △ybeZ/att7::ybeZ | △ybeZ with ybeZ inserted on chromosome with mini-Tn7T insertion; GENr | 30 |
| △rsmA | PA14 deleted of rsmA | This study |
| △ybeY△rsmA | PA14 deleted of ybeY and rsmA | This study |
| △retS | PA14 deleted of retS | This study |
| △ybeY△retS | PA14 deleted of ybeY and retS | This study |
| PA14/pUCP20 | PA14 with empty plasmid pUCP20; CARr | This study |
| △ybeY/pUCP20 | △ybeY with empty plasmid pUCP20; CARr | This study |
| △ybeY/pUCP20-rsmY | △ybeY with plasmid pUCP20-rsmY; CARr | This study |
| △ybeY/pUCP20-rsmZ | △ybeY with plasmid pUCP20-rsmZ; CARr | This study |
| Plasmids | ||
| pUCP20 | Escherichia-Pseudomonas shuttle vector without lac promoter; AMPr | 61 |
| pEX18Tc | Gene replacement vector; TETr, oriT+, sacB+ | 61 |
| pUC18T-mini-Tn7T-Gm | Mini-Tn7 base vector from insertion into chromosome attTn7 site; GENr | 61 |
| pDN19lacΩ | Promoterless lacZ fusion vector; SPTr, STRr, TETr | 57 |
| pEX18Tc-△rsmA | rsmA gene of PA14 deletion on pEX18Tc; TETr | This study |
| pEX18Tc-△retS | retS gene of PA14 deletion on pEX18Tc; TETr | This study |
| pUCP20-rsmY | Overpression of rsmY on pUCP20; CARr | This study |
| pUCP20-rsmZ | Overpression of rsmZ on pUCP20; CARr | This study |
| Primers | ||
| RsmA-L-F | CCGGAATTCGCACATCGACGACACCCAC | rsmA deletion |
| RsmA-L-R | TGCTCTAGACCCGACGAGTCAGAATCAGC | rsmA deletion |
| RsmA-R-F | TGCTCTAGAAGAAAGATCAAGAGCCAAACCA | rsmA deletion |
| RsmA-R-R | CCCAAGCTTCTTAGTCTTGCCCCCTATGGA | rsmA deletion |
| RsmA-T-F | AGGGTGAGTGACGCTGGCA | △rsmA screen |
| RsmA-T-R | GCCGCCTGAATCAACCTCTA | △rsmA screen |
| RetS-L-F | CCCAAGCTTGAAGCCAAGTGCGAGAACGT | retS deletion |
| RetS-L-R | TGCTCTAGAGAGCAGAAGCAGCAGGAAGC | retS deletion |
| RetS-R-F | TGCTCTAGAGGTGCTGATGGACTGCGAGA | retS deletion |
| RetS-T-F | CGGCCACTTGGCTATAATCC | △retS screen |
| RetS-T-R | CAGGACAGCACGAAGAAGGG | △retS screen |
| PA1805-RT-F | ATCAGTCTCAATGAAGTC | RT-PCR |
| PA1805-RT-R | CATGGATGGATCGAAATC | RT-PCR |
| RpsL-RT-F | GTAAGGTATGCCGTGTACG | RT-PCR |
| RpsL-RT-R | CACTACGCTGTGCTCTTG | RT-PCR |
| ExsA-RT-F | GCTATGTCGTAAGTACCA | RT-PCR |
| ExsA-RT-R | GAAGCCTTGTAGAAACTG | RT-PCR |
| ExsC-RT-F | CAGCTTCAACCGCCATTG | RT-PCR |
| ExsC-RT-R | CGCATACAACTGGACCTTG | RT-PCR |
| ExsD-RT-F | AGAGGTGCGGCAGATTCTCC | RT-PCR |
| ExsD-RT-R | ATCATCGACTGCGGCACG | RT-PCR |
| ExoU-RT-F | AACACATTAGCAGCGAGAT | RT-PCR |
| ExoU-RT-R | AGCAGCAACTCAGAGAAG | RT-PCR |
| PcrV-RT-F | CACGCTCTATGGCTATGC | RT-PCR |
| PcrV-RT-R | AAGGTATCCAGATTGCTCAG | RT-PCR |
| RsmA-RT-F | GAAGGAAGTCGCCGTACA | RT-PCR |
| RsmA-RT-R | TAATGGTTTGGCTCTTGATCTTT | RT-PCR |
| RsmY-RT-F | CCAAAGACAATACGGAAA | RT-PCR |
| RsmY-RT-R | GTTTTGCAGACCTCTATC | RT-PCR |
| RsmZ-RT-F | CAACCCCGAAGGTTC | RT-PCR |
| RsmZ-RT-R | CAGTCCCTCGTCATC | RT-PCR |
| GacA-RT-F | CCTGATGATCGCCAACTG | RT-PCR |
| GacA-RT-R | ATAGGTATTCACGGTCTTCG | RT-PCR |
| GacS-RT-F | GAGGAAATGCAGCACAAC | RT-PCR |
| GacS-RT-R | GTTCTGGATCTCGATGGT | RT-PCR |
| RetS-RT-F | GACTACGTGCAGACCATC | RT-PCR |
| RetS-RT-R | CTTGGAGATGTCGAGGAT | RT-PCR |
| LadS-RT-F | GATGCTGATCTACAACCT | RT-PCR |
| LadS-RT-R | GAAGCGATATAGAGGATGT | RT-PCR |
| HptB-RT-F | CATCTCGATGATCGTGTTC | RT-PCR |
| HptB-RT-R | GAAGGTATCCAGCAGGAC | RT-PCR |
| Hcp1-RT-F | AGGACCTGTCGTTCACCAA | RT-PCR |
| Hcp1-RT-R | ATAGTGCTTGCCGCTGGA | RT-PCR |
| VgrG1a-RT-F | GAGACCAGCTTCGACTTCATC | RT-PCR |
| VgrG1a-RT-R | CTTCTGCTCATGGCGGAAC | RT-PCR |
| Hcp3-RT-F | ACATCAAAGGCGACAGCC | RT-PCR |
| Hcp3-RT-R | GTTGCTGACGTCGTTGGT | RT-PCR |
| HsiB3-RT-F | ATCACCTACGACGTCGAGAT | RT-PCR |
| HsiB3-RT-R | GTCGATGTCGACGAAACGC | RT-PCR |
GENr, gentamicin resistance; AMPr, ampicillin resistance; TETr, tetracycline resistance; CARr, carbenicilin resistance; STRr, streptomycin resistance; SPTr, spectinomycin resistance; KANr, kanamycin resistance. Enzyme cleavage sites are underlined.
RNA isolation and RT-qPCR.
Bacteria cultured overnight were diluted 1:100 into fresh LB and cultured at 37°C to the late log phase (OD600 of 1). Aliquots of 1.5 ml bacteria were collected by centrifugation and resuspended in 0.5 ml TRIzol reagent (Thermo Fisher Scientific, USA). Total RNA was extracted by chloroform extraction and isopropanol precipitation. Residual DNA was digested with RNase-free recombinant DNase I (TaKaRa, Dalian, China). RNA was dissolved in RNase-free water. cDNAs were synthesized using random primers and reverse transcriptase (TaKaRa, Dalian, China). RT-qPCR was performed with the SYBR II green supermix (TaKaRa, Dalian, China). The ribosomal gene rpsL or PA1805 was used as the internal control (56).
Cytotoxicity assays.
Bacterial cytotoxicity was determined by the lactate dehydrogenase (LDH) release assay. The A549 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) with 10% (vol/vol) thermally inactivated fetal bovine serum, streptomycin (100 mg/ml), and penicillin G (100 U/ml) at 37°C with 5% CO2. A total of 2 × 105 cells were inoculated into each well of a 24-well plate and cultured overnight. Bacteria were cultured at 37°C in LB to the late log phase (OD600 of 1), and then the bacterial cells were washed twice in phosphate-buffered saline (PBS). Before infection, the cell culture medium was replaced by DMEM with 2.5% bovine serum albumin (BSA). The cells were infected with the indicated strains of bacteria at a multiplicity of infection (MOI) of 50. After adding bacteria to the cells, the plate was centrifuged at 700 × g for 10 min to synchronize the infection. The LDH level in the medium was determined with the LDH cytotoxicity assay kit (Beyotime, Shanghai, China) at 2 or 3 h postinfection. Treatment with the LDH release reagent provided by the kit was used as a control for total LDH release. The percentage of cytotoxicity was calculated according to the manufacturer's instructions.
Biofilm formation assays.
The bacteria were cultured at 37°C to an OD600 of 1 and then diluted 1:40 into fresh LB to an OD600 of 0.025. A volume of 150 μl of the bacterial suspension was added into each well of a 96-well plate and cultured at 37°C for 20 h. The culture medium was discarded, and the wells were washed three times with fresh PBS and dried at 65°C for 15 min. The wells were then stained with 1% crystal violet for 20 min, washed with PBS, and dried at 65°C. Aliquots of 200 μl ethanol were added into each well and incubated with gentle shaking at room temperature. The crystal violet solution was measured at a wavelength of 595 nm.
β-Galactosidase assay.
The bacteria were cultured at 37°C to an OD600 of 1. A volume of 0.5 ml of the bacterial culture was collected by centrifuging and resuspended in 1.5 ml Z buffer (60 mM NaH2PO4, 60 mM Na2HPO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0). The β-galactosidase activity was determined as previously described (57).
Data availability.
The transcriptome data that support the findings of this study have been deposited in the NCBI Sequence Read Archive (SRA) with the accession code PRJNA574019. The plasmids constructed in this study are available from Weihui Wu (wuweihui@nankai.edu.cn).
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Project of China (2017YFE0125600), the National Science Foundation of China (31670130, 31970680, and 31870130), the Tianjin Municipal Science and Technology Commission (19JCYBJC24700), and the program of China Scholarships Council (201906200035). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Jacob AI, Kohrer C, Davies BW, RajBhandary UL, Walker GC. 2013. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol Cell 49:427–438. doi: 10.1016/j.molcel.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vercruysse M, Kohrer C, Davies BW, Arnold MF, Mekalanos JJ, RajBhandary UL, Walker GC. 2014. The highly conserved bacterial RNase YbeY is essential in Vibrio cholerae, playing a critical role in virulence, stress regulation, and RNA processing. PLoS Pathog 10:e1004175. doi: 10.1371/journal.ppat.1004175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies BW, Kohrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, Rajbhandary UL, Walker GC. 2010. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol 78:506–518. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Zhou W, Liu G, Yang C, Sun Y, Wu W, Cao S, Wang C, Hai G, Wang Z, Bock R, Huang J, Cheng Y. 2015. The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis. Plant Physiol 168:205–221. doi: 10.1104/pp.114.255000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies BW, Walker GC. 2008. A highly conserved protein of unknown function is required by Sinorhizobium meliloti for symbiosis and environmental stress protection. J Bacteriol 190:1118–1123. doi: 10.1128/JB.01521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saramago M, Peregrina A, Robledo M, Matos RG, Hilker R, Serrania J, Becker A, Arraiano CM, Jimenez-Zurdo JI. 2017. Sinorhizobium meliloti YbeY is an endoribonuclease with unprecedented catalytic features, acting as silencing enzyme in riboregulation. Nucleic Acids Res 45:1371–1391. doi: 10.1093/nar/gkw1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhan C, Fedorov EV, Shi W, Ramagopal UA, Thirumuruhan R, Manjasetty BA, Almo SC, Fiser A, Chance MR, Fedorov AA. 2005. The ybeY protein from Escherichia coli is a metalloprotein. Acta Crystallogr Sect F Struct Biol Cryst Commun 61:959–963. doi: 10.1107/S1744309105031131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasouly A, Davidovich C, Ron EZ. 2010. The heat shock protein YbeY is required for optimal activity of the 30S ribosomal subunit. J Bacteriol 192:4592–4596. doi: 10.1128/JB.00448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey SP, Minesinger BK, Kumar J, Walker GC. 2011. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res 39:4691–4708. doi: 10.1093/nar/gkr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandey SP, Winkler JA, Li H, Camacho DM, Collins JJ, Walker GC. 2014. Central role for RNase YbeY in Hfq-dependent and Hfq-independent small-RNA regulation in bacteria. BMC Genomics 15:121. doi: 10.1186/1471-2164-15-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leskinen K, Varjosalo M, Skurnik M. 2015. Absence of YbeY RNase compromises the growth and enhances the virulence plasmid gene expression of Yersinia enterocolitica O:3. Microbiology (Reading) 161:285–299. doi: 10.1099/mic.0.083097-0. [DOI] [PubMed] [Google Scholar]
- 12.Budnick JA, Sheehan LM, Colquhoun JM, Dunman PM, Walker GC, Roop RM, Jr, Caswell CC. 2018. Endoribonuclease YbeY is linked to proper cellular morphology and virulence in Brucella abortus. J Bacteriol 200:e00105-18. doi: 10.1128/JB.00105-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gellatly SL, Hancock RE. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 14.Sawa T. 2014. The molecular mechanism of acute lung injury caused by Pseudomonas aeruginosa: from bacterial pathogenesis to host response. J Intensive Care 2:10. doi: 10.1186/2052-0492-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall CW, Mah TF. 2017. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev 41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 16.Cianfanelli FR, Monlezun L, Coulthurst SJ. 2016. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol 24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. 2011. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol 193:6057–6069. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. 2010. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A 107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durand E, Cambillau C, Cascales E, Journet L. 2014. VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol 22:498–507. doi: 10.1016/j.tim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potvin E, Lehoux DE, Kukavica-Ibrulj I, Richard KL, Sanschagrin F, Lau GW, Levesque RC. 2003. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol 5:1294–1308. doi: 10.1046/j.1462-2920.2003.00542.x. [DOI] [PubMed] [Google Scholar]
- 24.Allsopp LP, Wood TE, Howard SA, Maggiorelli F, Nolan LM, Wettstadt S, Filloux A. 2017. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 114:7707–7712. doi: 10.1073/pnas.1700286114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhagirath AY, Pydi SP, Li Y, Lin C, Kong W, Chelikani P, Duan K. 2017. Characterization of the direct interaction between hybrid sensor kinases PA1611 and RetS that controls biofilm formation and the type III secretion system in Pseudomonas aeruginosa. ACS Infect Dis 3:162–175. doi: 10.1021/acsinfecdis.6b00153. [DOI] [PubMed] [Google Scholar]
- 26.Francis VI, Waters EM, Finton-James SE, Gori A, Kadioglu A, Brown AR, Porter SL. 2018. Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa. Nat Commun 9:2219. doi: 10.1038/s41467-018-04640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chambonnier G, Roux L, Redelberger D, Fadel F, Filloux A, Sivaneson M, de Bentzmann S, Bordi C. 2016. The hybrid histidine kinase LadS forms a multicomponent signal transduction system with the GacS/GacA two-component system in Pseudomonas aeruginosa. PLoS Genet 12:e1006032. doi: 10.1371/journal.pgen.1006032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A 103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 30.Xia Y, Weng Y, Xu C, Wang D, Pan X, Tian Z, Xia B, Li H, Chen R, Liu C, Jin Y, Bai F, Cheng Z, Kuipers OP, Wu W. 2020. Endoribonuclease YbeY is essential for RNA processing and virulence in Pseudomonas aeruginosa. mBio 11:e00659-20. doi: 10.1128/mBio.00659-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonnleitner E, Romeo A, Bläsi U. 2012. Small regulatory RNAs in Pseudomonas aeruginosa. RNA Biol 9:364–371. doi: 10.4161/rna.19231. [DOI] [PubMed] [Google Scholar]
- 32.Intile PJ, Diaz MR, Urbanowski ML, Wolfgang MC, Yahr TL. 2014. The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 196:357–366. doi: 10.1128/JB.01199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vercruysse M, Kohrer C, Shen Y, Proulx S, Ghosal A, Davies BW, RajBhandary UL, Walker GC. 2016. Identification of YbeY-protein interactions involved in 16S rRNA maturation and stress regulation in Escherichia coli. mBio 7:e01785-16. doi: 10.1128/mBio.01785-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Möller P, Busch P, Sauerbrei B, Kraus A, Förstner KU, Wen TN, Overlöper A, Lai EM, Narberhaus F. 2019. The RNase YbeY is vital for ribosome maturation, stress resistance, and virulence of the natural genetic engineer Agrobacterium tumefaciens. J Bacteriol 201:e00730-18. doi: 10.1128/JB.00730-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAteer SP, Sy BM, Wong JL, Tollervey D, Gally DL, Tree JJ. 2018. Ribosome maturation by the endoribonuclease YbeY stabilizes a type 3 secretion system transcript required for virulence of enterohemorrhagic Escherichia coli. J Biol Chem 293:9006–9016. doi: 10.1074/jbc.RA117.000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulcahy H, Lewenza S. 2011. Magnesium limitation is an environmental trigger of the Pseudomonas aeruginosa biofilm lifestyle. PLoS One 6:e23307. doi: 10.1371/journal.pone.0023307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song Y, Yang C, Chen G, Zhang Y, Seng Z, Cai Z, Zhang C, Yang L, Gan J, Liang H. 2019. Molecular insights into the master regulator CysB-mediated bacterial virulence in Pseudomonas aeruginosa. Mol Microbiol 111:1195–1210. doi: 10.1111/mmi.14200. [DOI] [PubMed] [Google Scholar]
- 38.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Law COK, Huang C, Pan Q, Lee J, Hao Q, Chan TF, Lo NWS, Ang IL, Koon A, Ip M, Chan E, Lau TCK. 2019. A small RNA transforms the multidrug resistance of Pseudomonas aeruginosa to drug susceptibility. Mol Ther Nucleic Acids 16:218–228. doi: 10.1016/j.omtn.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parmeciano Di Noto G, Molina MC, Quiroga C. 2019. Insights into non-coding RNAs as novel antimicrobial drugs. Front Genet 10:57. doi: 10.3389/fgene.2019.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonnleitner E, Haas D. 2011. Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl Microbiol Biotechnol 91:63–79. doi: 10.1007/s00253-011-3332-1. [DOI] [PubMed] [Google Scholar]
- 43.Arrieta-Ortiz ML, Hafemeister C, Shuster B, Baliga NS, Bonneau R, Eichenberger P. 2020. Inference of bacterial small RNA regulatory networks and integration with transcription factor-driven regulatory networks. mSystems 5:e00057-20. doi: 10.1128/mSystems.00057-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpousis AJ, Luisi BF, McDowall KJ. 2009. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci 85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- 45.Saramago M, Bárria C, Dos Santos RF, Silva IJ, Pobre V, Domingues S, Andrade JM, Viegas SC, Arraiano CM. 2014. The role of RNases in the regulation of small RNAs. Curr Opin Microbiol 18:105–115. doi: 10.1016/j.mib.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 46.Anderson KL, Dunman PM. 2009. Messenger RNA turnover processes in Escherichia coli, Bacillus subtilis, and emerging studies in Staphylococcus aureus. Int J Microbiol 2009:525491. doi: 10.1155/2009/525491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrade JM, Arraiano CM. 2008. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA 14:543–551. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, Silva IJ, Viegas SC. 2010. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev 34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 49.Lu Y, Li H, Pu J, Xiao Q, Zhao C, Cai Y, Liu Y, Wang L, Li Y, Huang B, Zeng J, Chen C. 2019. Identification of a novel RhlI/R-PrrH-LasI/Phzc/PhzD signalling cascade and its implication in P aeruginosa virulence. Emerg Microbes Infect 8:1658–1667. doi: 10.1080/22221751.2019.1687262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen R, Wei X, Li Z, Weng Y, Xia Y, Ren W, Wang X, Jin Y, Bai F, Cheng Z, Jin S, Wu W. 2019. Identification of a small RNA that directly controls the translation of the quorum sensing signal synthase gene rhlI in Pseudomonas aeruginosa. Environ Microbiol 21:2933–2947. doi: 10.1111/1462-2920.14686. [DOI] [PubMed] [Google Scholar]
- 51.Wenner N, Maes A, Cotado-Sampayo M, Lapouge K. 2014. NrsZ: a novel, processed, nitrogen-dependent, small non-coding RNA that regulates Pseudomonas aeruginosa PAO1 virulence. Environ Microbiol 16:1053–1068. doi: 10.1111/1462-2920.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu P, Wang Y, Zhang Y, Hu Y, Thompson KM, Chen S. 2016. RpoS-dependent sRNA RgsA regulates Fis and AcpP in Pseudomonas aeruginosa. Mol Microbiol 102:244–259. doi: 10.1111/mmi.13458. [DOI] [PubMed] [Google Scholar]
- 53.Feaga HA, Viollier PH, Keiler KC. 2014. Release of nonstop ribosomes is essential. mBio 5:e01916. doi: 10.1128/mBio.01916-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood A, Irving SE, Bennison DJ, Corrigan RM. 2019. The (p)ppGpp-binding GTPase Era promotes rRNA processing and cold adaptation in Staphylococcus aureus. PLoS Genet 15:e1008346. doi: 10.1371/journal.pgen.1008346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 56.Son MS, Matthews WJ, Jr, Kang Y, Nguyen DT, Hoang TT. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun 75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weng Y, Chen F, Liu Y, Zhao Q, Chen R, Pan X, Liu C, Cheng Z, Jin S, Jin Y, Wu W. 2016. Pseudomonas aeruginosa enolase influences bacterial tolerance to oxidative stresses and virulence. Front Microbiol 7:1999. doi: 10.3389/fmicb.2016.01999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thi Bach Nguyen H, Romero AD, Amman F, Sorger-Domenigg T, Tata M, Sonnleitner E, Bläsi U. 2018. Negative control of RpoS synthesis by the sRNA ReaL in Pseudomonas aeruginosa. Front Microbiol 9:2488. doi: 10.3389/fmicb.2018.02488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. 2013. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol 195:2011–2020. doi: 10.1128/JB.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi K-H, Schweizer HP. 2006. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The transcriptome data that support the findings of this study have been deposited in the NCBI Sequence Read Archive (SRA) with the accession code PRJNA574019. The plasmids constructed in this study are available from Weihui Wu (wuweihui@nankai.edu.cn).