DMSP is present in the marine environment, produced in large quantities by marine phytoplankton as an osmoprotectant, and is an important component of the global geochemical sulfur cycle. This algal osmolyte has not been previously investigated for its role in marine heterotrophic bacterial osmotic stress response. Vibrionaceae species are marine species, many of which are halophiles exemplified by V. parahaemolyticus, a species that possesses at least six transporters for the uptake of osmolytes.
KEYWORDS: BCCT transporters, dimethylsulfoniopropionate, osmolyte, osmotic stress
ABSTRACT
Dimethylsulfoniopropionate (DMSP), a key component of the global geochemical sulfur cycle, is a secondary metabolite produced in large quantities by marine phytoplankton and utilized as an osmoprotectant, thermoprotectant, and antioxidant. Marine bacteria can use two pathways to degrade and catabolize DMSP, a demethylation pathway and a cleavage pathway that produces the climate-active gas dimethylsulfide (DMS). Whether marine bacteria can also accumulate DMSP as an osmoprotectant to maintain the turgor pressure of the cell in response to changes in external osmolarity has received little attention. The marine halophile Vibrio parahaemolyticus contains at least six osmolyte transporters, namely four betaine carnitine choline transport (BCCT) carriers (BccT1 to BccT4) and two ATP-binding cassette (ABC) family ProU transporters. In this study, we showed that DMSP is used as an osmoprotectant by V. parahaemolyticus and by several other Vibrio species, including Vibrio cholerae and Vibrio vulnificus. Using a V. parahaemolyticus proU double mutant, we demonstrated that these ABC transporters are not required for DMSP uptake. However, a bccT null mutant lacking all four BCCTs had a growth defect compared to the wild type (WT) in high-salinity medium supplemented with DMSP. Using mutants possessing only one functional BCCT in growth pattern assays, we identified two BCCT family transporters, BccT1 and BccT2, that are carriers of DMSP. The only V. parahaemolyticus BccT homolog that V. cholerae and V. vulnificus possess is BccT3, and functional complementation in Escherichia coli MKH13 showed that V. cholerae VcBccT3 could transport DMSP. In V. vulnificus strains, we identified and characterized an additional BCCT family transporter, which we named BccT5, that was also a carrier for DMSP.
IMPORTANCE DMSP is present in the marine environment, produced in large quantities by marine phytoplankton as an osmoprotectant, and is an important component of the global geochemical sulfur cycle. This algal osmolyte has not been previously investigated for its role in marine heterotrophic bacterial osmotic stress response. Vibrionaceae species are marine species, many of which are halophiles exemplified by V. parahaemolyticus, a species that possesses at least six transporters for the uptake of osmolytes. Here, we demonstrated that V. parahaemolyticus and other Vibrio species can accumulate DMSP as an osmoprotectant and show that several BCCT family transporters uptake DMSP. These studies suggest that DMSP is a significant bacterial osmoprotectant that may be important for understanding the fate of DMSP in the environment. DMSP is produced and present in coral mucus, and Vibrio species form part of the microbial communities associated with corals. The function of DMSP in these interactions is unclear, but it could be an important driver for these associations, allowing Vibrio proliferation. This work suggests that DMSP likely has a more important role in heterotrophic bacteria ecology than previously appreciated.
INTRODUCTION
Compatible solutes (osmolytes) are accumulated by bacteria to maintain the turgor pressure of the cell in response to high external osmolarity (1–3). Known compatible solutes of bacteria include glycine betaine, ectoine, proline, glutamate, glycerol, and trehalose (2, 4–9). Compatible solutes, as the name suggests, are compounds that can be accumulated to high levels and are compatible with the molecular machinery and processes of the cell. These osmolytes allow organisms to continue to grow and divide in unfavorable environments that may be characterized by changes in osmolarity and temperature shifts (10–19). The uptake and biosynthesis of compatible solutes in response to osmotic stress has been studied extensively in Escherichia coli and Bacillus subtilis; both species can biosynthesize glycine betaine from exogenous choline and can uptake this and glycine betaine, among other osmolytes (10, 20–32).
Organisms that live in marine environments have adapted to grow optimally in high salinity and have also evolved mechanisms to maintain cellular homeostasis to cope with fluctuations in salinity that they encounter. Members of the family Vibrionaceae have the ability to grow optimally at 0.5 to 1.0 M NaCl, but regularly encounter fluctuations in osmolarity ranging from 0.1 to 1.5 M NaCl (33). One of the key responses of Vibrio species to fluctuations in osmolarity is the accumulation of compatible solutes, either through biosynthesis of the osmolytes ectoine and glycine betaine or the rapid uptake of these osmolytes from the environment using as many as six osmolyte transporters (33–35). Vibrio parahaemolyticus, a halophile, biosynthesizes glycine betaine from exogenously supplied choline and ectoine from aspartic acid de novo, and it can uptake at least 14 different compatible solutes (33–36). This species contains two osmolyte ATP-binding cassette (ABC) family transporters, ProU1 (VP1726 to VP1728) and ProU2 (VPA1112 to VPA1114), and four betaine carnitine choline transporter (BCCT) family transporters, BccT1 (VP1456), BccT2 (VP1723), BccT3 (VP1905), and BccT4 (VPA0356) (33). All BCCTs described have 12 predicted transmembrane (TM) α-helical domains, TM1 to TM12, which is a defining feature in their classification (21, 37–41). In addition to the 12 TM domains, these proteins contain hydrophilic N- and C-terminal tails of various lengths (21, 39–41). Some species, such as Vibrio cholerae, Vibrio vulnificus, and Aliivibrio fischeri, contain only a single BccT, a homolog of BccT3 (33). We have previously demonstrated that in Vibrio parahaemolyticus, BCCTs can transport glycine betaine, choline, dimethylglycine (DMG), ectoine, and proline, with some redundancy in the substrates transported by each BCCT (35, 36).
Dimethylsulfoniopropionate (DMSP) is an organosulfur compound abundant in marine surface waters that is produced by phytoplankton and some halophytic vascular plants in large quantities and used by these primary producers as an osmoprotectant, thermoprotectant, and antioxidant (42–47). Particulate DMSP levels can range from nanomolar amounts in surface water to micromolar amounts during phytoplankton blooms, and intracellular concentrations of DMSP in marine phytoplankton can range from 120 mM to 1 M (48–50). It was reported previously that DMSP is also produced in significant quantities by bacteria present in marine and estuarine sediments, as well as in seawater (51, 52). DMSP is an important component of the global geochemical sulfur cycle as a precursor for dimethylsulfide (DMS). DMS is a climate-active gas that is produced from the degradation of oceanic DMSP, releasing sulfur-containing aerosols into the atmosphere (44, 45, 53–57). Many bacteria and phytoplankton catabolize DMSP as a source of reduced sulfur and carbon using two pathways, a demethylation pathway and a cleavage pathway that produces DMS (43, 58–62). DMSP-catabolizing bacteria are mainly confined to marine Alphaproteobacteria taxa such as SAR11, SAR116, and Roseobacter, and some Gammaproteobacteria taxa (63–72). Given that species of Vibrionaceae interact and associate with DMSP producers, it is surprising that there are no studies on the use of DMSP as an osmoprotectant in these bacteria or in marine bacteria in general (3, 73–75), although studies have shown that DMSP is assimilated by cyanobacteria Prochlorococcus and Synechococcus (45, 76). The first direct evidence that DMSP can be used as an osmoprotectant for bacteria came from a study in E. coli, which does not encounter this compound naturally. Osmotically stressed E. coli bacteria responded to DMSP, and the ABC family ProU transporter was shown to take up DMSP to relieve NaCl stress (73). More recently, the Gram-positive bacterium Bacillus was shown to utilize DMSP as an osmoprotectant by uptake via the ABC family transporters OpuC and OpuF (74, 77). The BCCT family transporter DddT, which is linked to DMSP cleavage pathways in Halomonas sp. HTNK1, was demonstrated to take up DMSP in a heterologous E. coli background (70, 75). Outside of these studies, little is known about bacterial utilization of DMSP as an osmolyte or the bacterial response to this abundant marine compound.
In this study, we examined Vibrio species for their ability to utilize DMSP as an osmoprotectant. A V. parahaemolyticus ectB mutant, which cannot grow in high NaCl in the absence of an exogenous osmolyte, was rescued in M9 minimal medium supplemented with glucose (M9G) 6% NaCl supplemented with DMSP. The effectiveness of DMSP as an osmoprotectant was determined in several Vibrio species, including V. cholerae, which has only a single known osmolyte transporter, and V. vulnificus. Vibrio parahaemolyticus has six osmolyte transporters, two ProUs, and four BCCTs. We examined a proU double mutant and a bccT null mutant for defects when grown under high-salt conditions with DMSP as an osmolyte. The ProU transporters did not play a significant role in DMSP uptake. The role of each of the BCCTs in DMSP uptake was investigated using four triple mutants that each contained a single bccT gene, showing that two BccTs were responsible for DMSP uptake. Vibrio cholerae contained only a single BCCT family transporter, a BccT3 homolog, and we determined its ability to take up DMSP. In V. vulnificus, an additional BCCT family transporter that could uptake DMSP, named BccT5, was identified.
RESULTS
Vibrio parahaemolyticus can utilize dimethylsulfoniopropionate as a compatible solute.
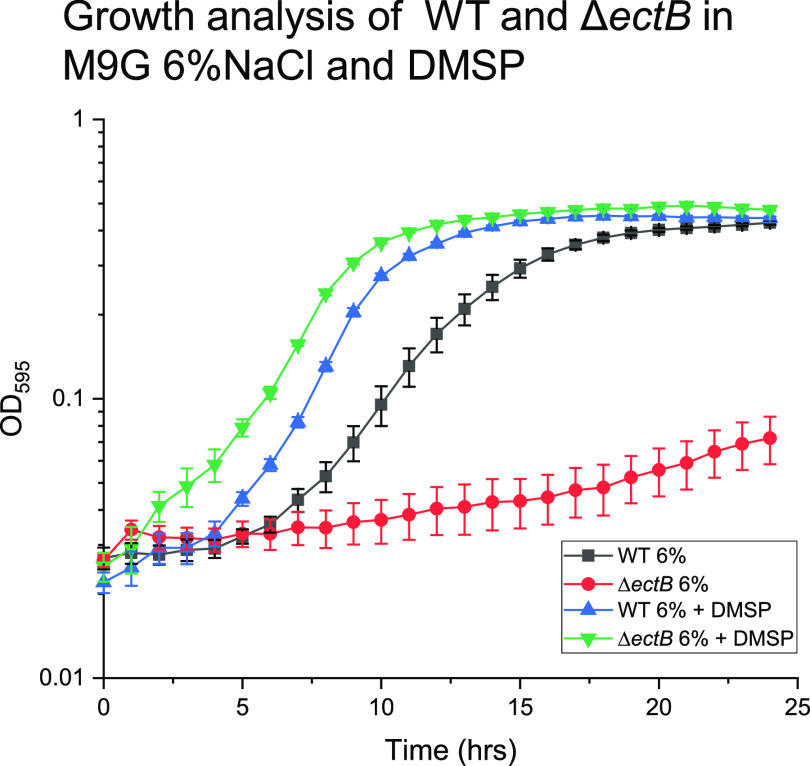
As a halophile, V. parahaemolyticus grows optimally in 0.5 M NaCl (∼3% NaCl) and can grow in salinities up to 1.5 M (∼9%) (34). We examined the ability of V. parahaemolyticus to grow in high-salinity conditions using DMSP as an osmolyte. The V. parahaemolyticus ectB deletion mutant (ΔectB) cannot grow under high-NaCl growth conditions in the absence of exogenous osmolytes (34, 36). The wild type (WT) and the ectB mutant were grown in M9 minimal medium supplemented with glucose and 6% NaCl (M9G 6%NaCl), in the presence and absence of 500 µM DMSP at 37°C for 24 h. Growth pattern analysis showed that the lag phase of the wild-type strain without exogenous compatible solutes is ∼6 h with a growth rate of 0.042 h−1, while the ΔectB mutant did not grow (Fig. 1). However, in M9G 6% NaCl supplemented with DMSP, the ΔectB mutant grew similarly to the wild type, with growth rates of 0.082 h−1 and 0.074 h−1, respectively. Lag phases were reduced to ∼3 h for the wild-type strain and to <1 h for the ΔectB mutant (Fig. 1). It is likely that the ΔectB mutant has a shorter lag phase than that of the wild type in the presence of DMSP because it does not produce ectoine, which is energetically costly (78–80). The data demonstrate that V. parahaemolyticus can uptake and utilize DMSP as an osmolyte.
FIG 1.
Growth analysis of wild-type (WT) Vibrio parahaemolyticus RIMD2210633 and an ectB mutant was conducted in M9 minimal medium supplemented with glucose (M9G) supplemented with 6% NaCl and 500 µM DMSP. Optical density at 595 nm (OD595) was measured every hour for 24 h; means and standard errors of at least two biological replicates are displayed.
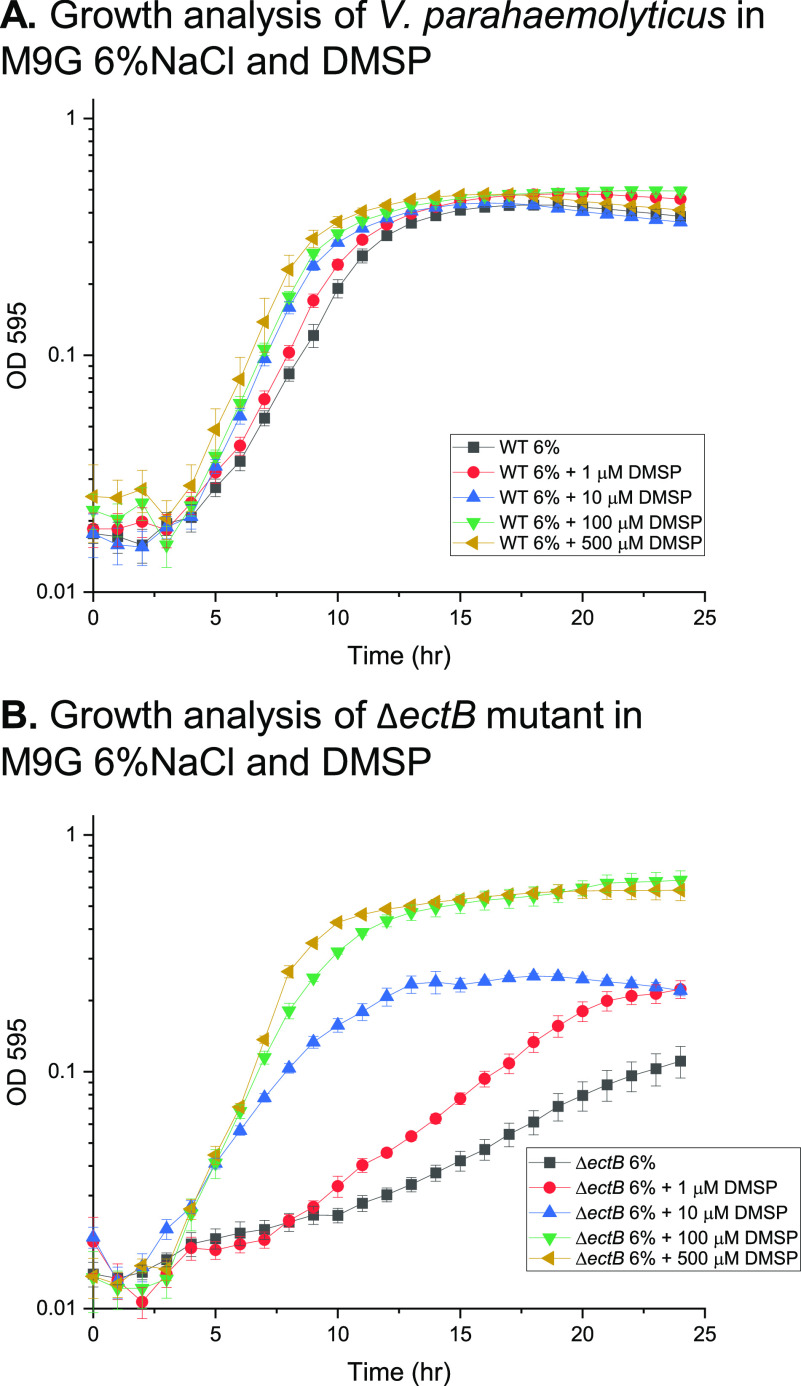
Since DMSP levels in the environment can vary from nanomolar to micromolar amounts in algal blooms, we examined the growth of V. parahaemolyticus in a range of DMSP concentrations (Fig. 2). At DMSP micromolar concentrations (1, 10, 100, and 500 µM), in the wild type a shift in the growth curves was observed for all four concentrations, indicating that DMSP is taken up and utilized as an osmolyte (Fig. 2A). Similarly, we examined the rescue of the ΔectB mutant and found that all four DMSP concentrations rescued the mutant, with the higher concentrations being more effective (Fig. 2B). At DMSP nanomolar concentrations (10 nM to 500 nM), a shift in the lag phase in the wild type is observed, with the greatest shift at 500 nM DMSP (see Fig. S1 in the supplemental material). The ΔectB mutant was not rescued under nanomolar concentrations (data not shown). These data demonstrate that V. parahaemolyticus responded to physiologically relevant DMSP concentrations.
FIG 2.
(A) Growth analysis of wild-type (WT) V. parahaemolyticus RIMD2210633 in M9G supplemented with 6% NaCl (M9G6%) and 1 µM to 500 µM DMSP. (B) Growth analysis of the ΔectB mutant in M9G6% supplemented with 1 µM to 500 µM DMSP. Optical density (OD595) was measured every hour for 24 h; means and standard errors of at least three biological replicates are displayed.
Next, we compared the effectiveness of DMSP and glycine betaine as osmoprotectants in the wild-type and ΔectB mutant strains. We compared the growth patterns of both strains in M9G 6% NaCl in 100 µM DMSP or 100 µM glycine betaine (see Fig. S2A and B in the supplemental material). Both strains responded to glycine betaine more effectively than to DMSP, exhibiting shorter lag phases and higher growth rates in glycine betaine (Fig. S2A and B).
DMSP is utilized as an osmolyte by other Vibrio species.
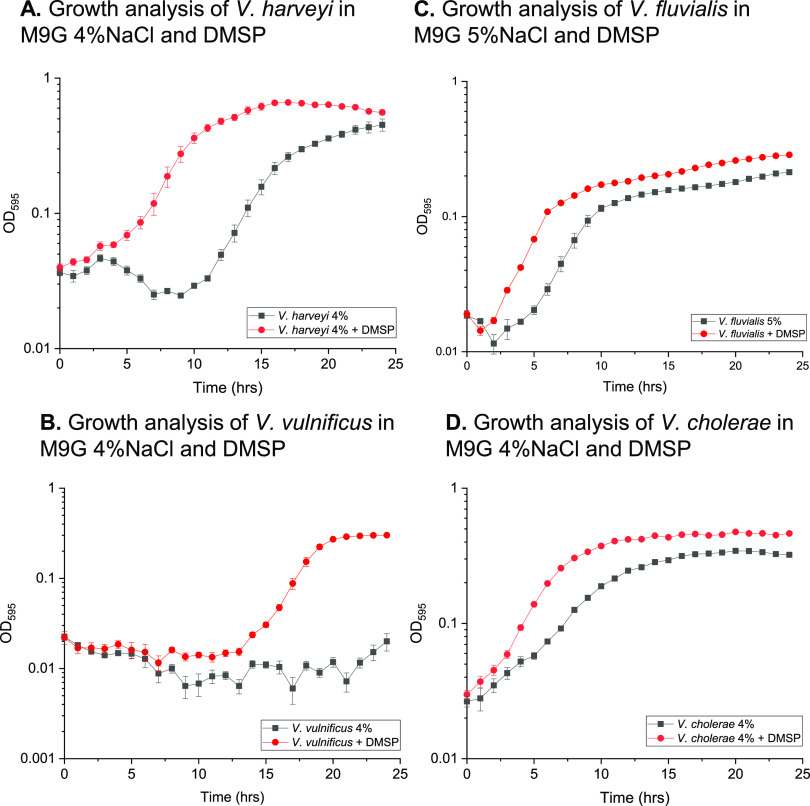
To determine whether other Vibrio species can utilize DMSP as an osmolyte, we examined Vibrio harveyi 393, V. vulnificus YJ016, V. cholerae N16961, and Vibrio fluvialis ATCC 33809. These species are members of divergent Vibrionaceae clades, and each contains a different complement of osmolyte transporters. The growth of V. harveyi 393, V. vulnificus YJ016, and V. cholerae N16961 was examined in M9G 4% NaCl and that of V. fluvialis ATCC 33809 in M9G 5% NaCl, with 500 µM DMSP (Fig. 3A to D). The growth of V. harveyi 393 showed a 10-h lag phase with a growth rate of 0.06 h−1 in the absence of DMSP; however, the addition of DMSP resulted in a lag phase of less than 2 h, with a maximum growth rate of 0.088 h−1. Vibrio vulnificus did not grow in M9G 4% NaCl, but the addition of DMSP rescued growth with a 13-h lag phase and a growth rate of 0.065 h−1 (Fig. 3B). V. fluvialis had a growth rate of 0.026 h−1 with a lag phase of ∼4 h when grown without DMSP, which was reduced to ∼2 h and the growth rate increased to 0.041 h−1 in the presence of DMSP (Fig. 3C). Vibrio cholerae cells had a reduced lag phase, from 2 h to less than 1 h, and an increased growth rate, from 0.034 h−1 to 0.059 h−1, in the presence of DMSP (Fig. 3D). These data demonstrated that all Vibrio species tested can utilize DMSP as an osmolyte.
FIG 3.
Growth analyses of (A) Vibrio harveyi 393, (B) Vibrio vulnificus YJ016, (C) Vibrio fluvialis ATCC 33809, and (D) Vibrio cholerae N16961 was conducted in M9G 4% NaCl (5% NaCl for V. fluvialis) with and without 500 µM DMSP. Optical density (OD595) was measured every hour for 24 h. Means and standard errors of two biological replicates are shown.
Some marine bacteria are able to catabolize DMSP, although this has never been shown in any Vibrio species (71, 72, 81). To ensure that the reduced lag phase in each Vibrio species tested was not due to the use of DMSP as a carbon source, we grew each strain in M9 minimal medium with DMSP as the sole carbon source, utilizing M9G (glucose as the sole carbon source) as a control. None of the Vibrio species tested grew with DMSP as the sole carbon source, and all grew on M9G, which indicated that they cannot utilize DMSP as a carbon source (see Fig. S3 in the supplemental material). In addition, in silico analysis of the published genomes of these species did not identify any known DMSP catabolism genes. These data demonstrated that DMSP appears to be a bona fide compatible solute for the Vibrio species tested.
BCCT transporters required for DMSP uptake.
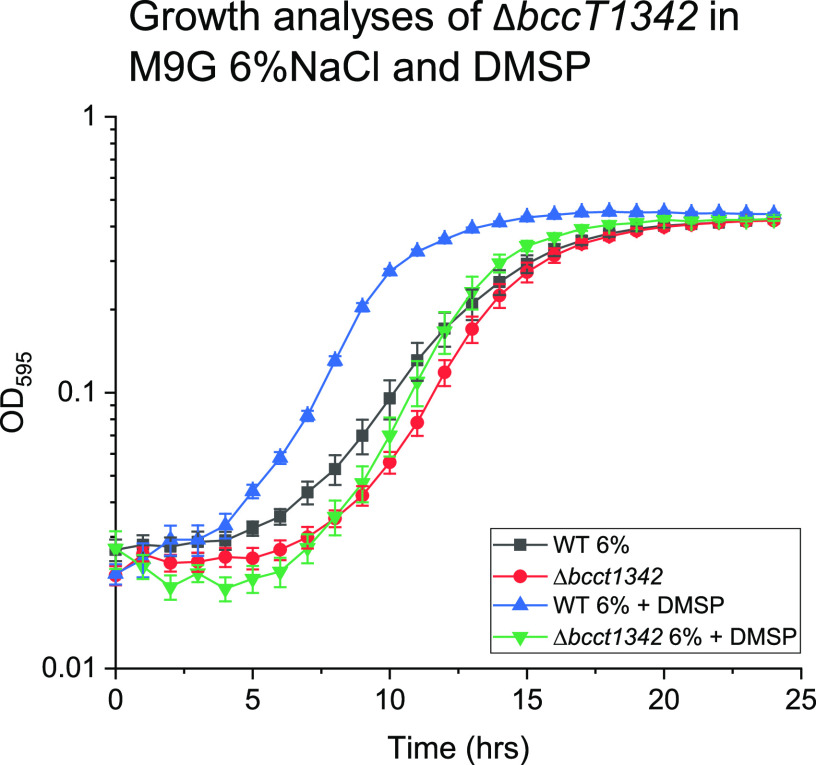
Previous studies in E. coli and B. subtilis showed that DMSP was transported into cells by an ABC family osmolyte transporter (73, 74). Vibrio parahaemolyticus contains two ABC-type osmolyte transporters, ProU1 (VP1726 to VP1728) and ProU2 (VPA1112 to VPA1114). Growth analyses with a double proU mutant (ΔproU1 ΔproU2) were performed to determine whether either is required for DMSP uptake. In these assays, no difference between the mutant and the wild type in the absence or presence of DMSP was seen (see Fig. S4 in the supplemental material). These data demonstrated that ProU transporters were not required for uptake of DMSP in V. parahaemolyticus. Next, we examined whether any of the four BccTs in V. parahaemolyticus were responsible for the uptake of DMSP into the cell. To accomplish this, the wild type and a bccT null strain were grown in the presence or absence of DMSP in M9G 6% NaCl. The lag phase of the wild-type strain was reduced from ∼6 h to ∼3 h when grown in the presence of DMSP, which indicates that DMSP is an osmoprotectant for V. parahaemolyticus (Fig. 4). However, the bccT null mutant did not exhibit a reduced lag phase in the presence of DMSP (Fig. 4). This indicated that at least one of the BCCTs is responsible for transport of DMSP into the cell and confirms that neither ProU plays a significant role in uptake of DMSP.
FIG 4.
Growth analysis of wild-type (WT) V. parahaemolyticus RIMD2210633 and a ΔbccT1 ΔbccT3 ΔbccT4 ΔbccT2 mutant was conducted in M9G6% and 500 µM DMSP. Optical density (OD595) was measured every hour for 24 h; means and standard errors of at least two biological replicates are displayed.
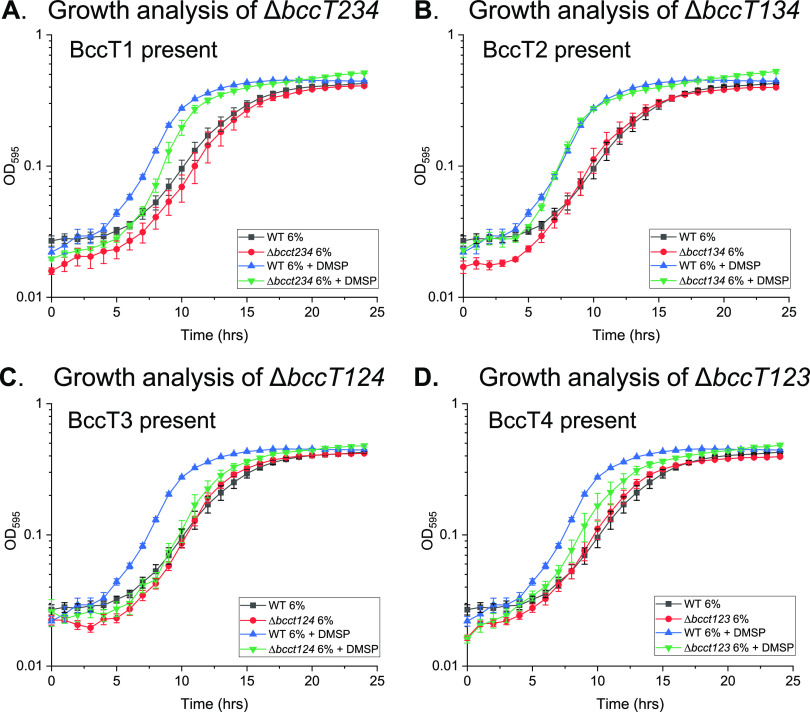
To determine which of the BCCTs were responsible for uptake of DMSP, four triple bccT deletion mutants were utilized, each containing only one functional bccT gene. The ΔbccT2 ΔbccT3 ΔbccT4 mutant, which expresses only bccT1 (VP1456), had a slight reduction in the lag phase and had a higher growth rate through the exponential phase when grown in the presence of DMSP. This suggests that BccT1 can transport DMSP with low efficiency (Fig. 5A). The ΔbccT1 ΔbccT3 ΔbccT4 mutant, which expresses only bccT2 (VP1723), showed a nearly identical reduction in the lag phase as that in the wild-type strain, which indicated that it also transports DMSP into the cell (Fig. 5B). The ΔbccT1 ΔbccT2 ΔbccT4 mutant, which expresses only bccT3 (VP1905), and ΔbccT1 ΔbccT2 ΔbccT3, which expresses only bccT4 (VPA0356), had slight reductions in the lag phase in the presence of DMSP (Fig. 5C and D), which indicated that BccT3 and BccT4 do not play a significant role in DMSP transport in V. parahaemolyticus.
FIG 5.
DMSP uptake analysis of wild-type (WT) V. parahaemolyticus RIMD2210633 and (A) ΔbccT2 ΔbccT3 ΔbccT4, (B) ΔbccT1 ΔbccT3 ΔbccT4, (C) ΔbccT1 ΔbccT2 ΔbccT4, or (D) ΔbccT1 ΔbccT2 ΔbccT3 in M9G6% with and without the addition of 500 µM DMSP. Optical density (OD595) was measured every hour for 24 h; means and standard errors of at least two biological replicates are displayed.
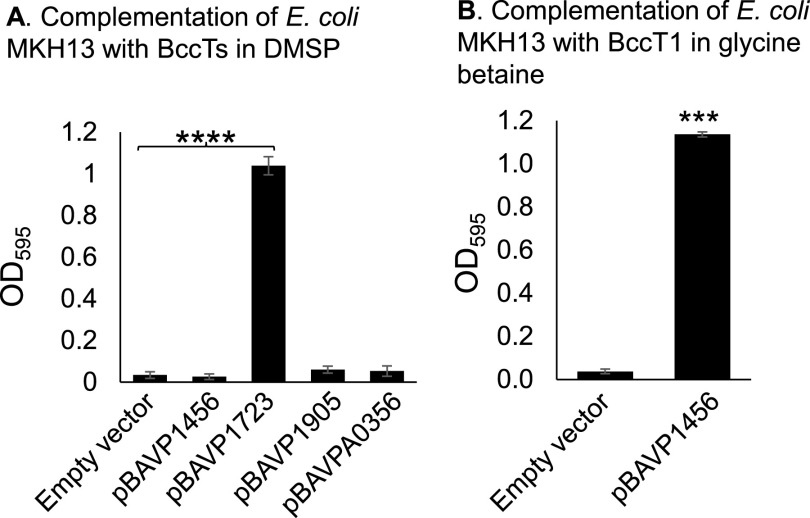
Next, we examined the DMSP transport capabilities of each BccT using functional complementation in E. coli MKH13, a mutant strain that has deletions in betIBA-betT, proU, proP, and putP and cannot grow in M9G 4% NaCl (82). Strains were grown in M9G 4% NaCl in the presence of 500 µM DMSP, with an arabinose-inducible expression plasmid, pBAD33, harboring a full-length copy of a single bccT. We measured the optical density at 595 nm (OD595) after 24 h for each complemented strain and compared the values to that of the empty vector strain. Only the BccT2-complemented strain (pBAVP1723) could grow in the presence of DMSP (Fig. 6A). The E. coli BccT1-complemented strain (pBAVP1456) was unable to grow in the presence of DMSP (Fig. 6A). In V. parahaemolyticus expressing only BccT1, DMSP was taken up, though the shift in growth was not as significant as that when BccT2 was expressed, suggesting that it is not an efficient DMSP transporter (Fig. 5A). To confirm that bccT1 is expressed and produces a functional protein in E. coli, we examined the ability of BccT1 to take up glycine betaine, a known substrate for this transporter. In this assay, E. coli pBAVP1456 grew with the addition of glycine betaine, which indicated that BccT1 is functional (Fig. 6B). It is important to note that functional complementation assays do not provide information about efficiency of transport of a particular substrate, only that the substrate in question is able to be imported into the cell. Here, we have shown that only BccT2 is capable of transporting DMSP in the E. coli background.
FIG 6.
E. coli strain MKH13, which has deletions in all compatible solute transporters, was grown in M9G supplemented with 4% NaCl and functionally complemented with V. parahaemolyticus RIMD2210633 (A) bccT1 (pBAVP1456), bccT2 (pBAVP1723), bccT3 (pBAVP1905), or bccT4 (pBAVPA0356). Strains were grown for 24 h in the presence of 500 µM DMSP and the final optical density (OD595) was compared to that of a strain harboring empty pBAD33. (B) E. coli MKH13 complemented with bccT1 was grown in the presence of 500 µM glycine betaine and compared to a strain harboring empty pBAD33. Means and standard errors of at three biological replicates are shown. Statistics were calculated using Student’s t test (***, P < 0.001; ****, P < 0.0001).
BccT3 from V. cholerae is a DMSP transporter.
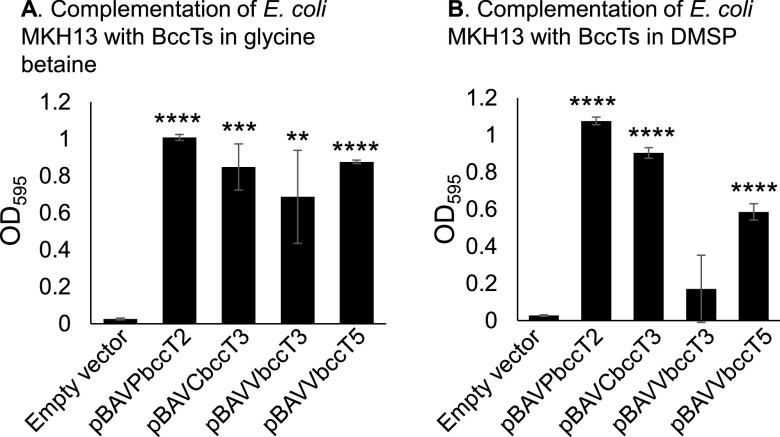
Of the four additional Vibrio species that used DMSP as an osmolyte, V. harveyi and V. fluvialis contain homologs of BccT1 and BccT2, which transport DMSP in V. parahaemolyticus. However, V. vulnificus contains homologs of only BccT3 and ProU2, while V. cholerae possesses only a BccT3 homolog and no ProU transporter. Given that V. vulnificus and V. cholerae can utilize DMSP as an osmolyte and contained only a BccT3 homolog, we cloned V. vulnificus bccT3 (VV2103 and VVbccT3) and V. cholerae bccT3 (FY484_RS06475 and VCbccT3) homologs into the expression plasmid pBAD33 and performed functional complementation assays in E. coli strain MKH13. We tested the growth of MKH13 in the presence of glycine betaine to determine whether these transporters were functional and found that both were glycine betaine carriers, as evidenced by growth of the MKH13 strain in M9G4% (Fig. 7A). Next, we examined DMSP uptake by these transporters and showed that VCBccT3 functionally complemented MKH13, while VVBccT3 did not (Fig. 7B). Thus, the V. cholerae VCBccT3 is functional and can transport DMSP into the cell.
FIG 7.
E. coli strain MKH13 was grown in M9G supplemented with 4% NaCl and functionally complemented with VPbccT2 (pBAVP1723), VCbccT3, VVbccT5, or VVbccT5. Strains were grown for 24 h in the presence of (A) 500 µM glycine betaine or (B) 500 µM DMSP, and the final optical density (OD595) was compared to that of a strain harboring empty pBAD33 expression vector. Means and standard errors of three biological replicates are shown. Statistics were calculated using Student’s t test (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
BccT5, an additional BCCT transporter present in V. vulnificus.
Since VVBccT3 did not transport DMSP, we reexamined the V. vulnificus genome and identified an additional BCCT family transporter that shared <32% homology with BCCTs from V. parahaemolyticus. This BCCT, which we named BccT5 (VV0783), was annotated as a 637-amino acid protein. It was demonstrated via hydropathy profile analysis to have 12 TM domains, a feature of all BCCT transporters (see Fig. S5 in the supplemental material). E. coli MKH13 complemented with VVBccT5 was unable to grow in the presence of 500 µM DMSP and grew poorly in the presence of 500 µM glycine betaine (data not shown) in M9G 4% NaCl. Further analysis identified an alternative start codon in VV0783, which resulted in a larger 672-amino-acid protein, which was used in functional complementation of E. coli MKH13 and resulted in growth in the presence of either glycine betaine or DMSP (Fig. 7). It is interesting to note that the N-terminal tail of BccT5 appears to be required for full functionality of the transporter. Overall, the data show that Vibrio species have at least four BCCT family transporters that are capable of DMSP uptake.
DISCUSSION
Bacteria have adapted to hyperosmotic conditions by changing the level of osmolytes in their cells to maintain turgor pressure. The most prevalent and energetically effective method to maintain turgor pressure is the uptake of osmolytes from the surrounding environment. DMSP is produced in vast quantities by marine phytoplankton and utilized as an osmoprotectant for these primary producers. The concentrations of DMSP in the environment reach micromolar amounts during algal blooms, and many marine bacteria associate with phytoplankton during these blooms. The contribution of DMSP to the bacterial osmotic stress response is largely unknown. Our studies demonstrate that DMSP can play a significant role in adaptation to osmotic stress by the uptake of DMSP as an osmolyte. Additionally, we demonstrated that the BCCT family of transporters are important components of DMSP uptake for use as an osmolyte, and BccT transporters are widespread among Vibrionaceae.
Here, we have shown that marine bacteria in the genus Vibrio can use DMSP as an osmolyte demonstrating the ecological significance of this compound to marine bacteria. Our studies demonstrate a more widespread use of DMSP as an osmolyte among marine bacteria that was previously unrecognized. BCCT family transporters were solely responsible for the uptake of DMSP by V. parahaemolyticus, and among Vibrio species, four different BCCTs could uptake DMSP. The ABC family transporters ProU1 and ProU2 did not contribute significantly to DMSP uptake, suggesting that BCCTs likely play a major role in DMSP uptake in Vibrio species. BCCT family proteins were highly prevalent among Vibrio with most species containing multiple BccT proteins.
Recently, an increased incidence of algal blooms has led to increased V. parahaemolyticus and V. vulnificus proliferation (83–85). Our data show that V. parahaemolyticus does not utilize DMSP as a carbon source, but it can grow and divide rapidly in high salinity when exogenous DMSP is present. We speculate that the ability of V. parahaemolyticus and other Vibrio species to utilize DMSP as a compatible solute increases their ability to proliferate in algal blooms that occur both under high salinity conditions and in warmer months. Dinoflagellates are some of the main producers of DMSP, and V. parahaemolyticus proliferation has been found in conjunction with dinoflagellate algal blooms (84, 85). Likewise, V. vulnificus can grow and divide rapidly in higher salinity when exogenous DMSP is present, likely contributing to its fitness. Coral reefs are often described as DMSP hot spots, and DMSP production has been linked to the coral stress response (86). Several Vibrio species are associated with coral disease such as Vibrio coralliilyticus that infects reef-building corals and Vibrio mediterranei (Vibrio shilonii) associated with coral deaths worldwide (87–90). Our in silico analysis of the V. coralliilyticus genome sequences identified five BccT proteins, which included a copy present on a plasmid prevalent among strains. This suggests a mechanism of horizontal transfer of BccT among Vibrio. In addition, in V. coralliilyticus CN52H-1, a strain recovered from the surface mucus layer of the coral Colpophyllia natans, we identified a BccT protein that clustered with a putative DMSP lyase gene, suggesting a potential for this species to metabolize DMSP. In the V. mediterranei genome sequences, we identified up to eight different BCCTs, which included homologs of BccT1 to BccT5, suggesting that these species have the ability to take up and utilize DMSP as an osmolyte that could enhance their association with corals. Indeed, it has been proposed that the concentrations of DMSP and DMS in corals drives the structure of the microbial community associated with them (91). In fact, it was reported that DMSP alone was sufficient to elicit a chemotactic response from V. coralliilyticus, and this response was increased at higher temperatures, when heat-stressed corals emit five times as much DMSP (92). This highlights the importance of DMSP in marine communities, and our work suggests that the BCCT family transporters play a significant role in these interactions. It will be interesting to learn whether Vibrio species associated with coral, such as V. coralliilyticus, can also use DMSP as an osmolyte and/or for thermoprotection.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
All strains and plasmids used in this study are listed in Table 1. A streptomycin-resistant V. parahaemolyticus RIMD2210633 was used as the wild-type (WT) strain. Vibrio parahaemolyticus were grown either in lysogeny broth (LB) (Fisher Scientific, Fair Lawn, NJ) with 3% (wt/vol) NaCl (LB3%) or M9 minimal medium (47.8 mM Na2HPO4, 22 mM KH2PO4, 18.7 mM NH4Cl, and 8.6 mM NaCl; Sigma-Aldrich) supplemented with 2 mM MgSO4, 0.1 mM CaCl2, and 20 mM glucose as the sole carbon source (M9G) and NaCl (wt/vol), as indicated. Dimethylsulfoniopropionate (DMSP) was used at a final concentration of 20 mM when supplied as a carbon source. E. coli strains were grown in either LB supplemented with 1% NaCl (LB1%) or M9G supplemented with 1% NaCl (M9G1%). All strains were grown at 37°C with aeration. Chloramphenicol (Cm) was used at a final concentration of 25 μg/ml as necessary.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Reference(s) or source |
|---|---|---|
| Strain | ||
| Vibrio parahaemolyticus | ||
| RIMD2210633 | O3:K6 clinical isolate, Strr | 15, 98 |
| ΔectB | RIMD2210633 ΔectB (VP1721), Strr | 33 |
| SOYBCCT124 | RIMD2210633 ΔVP1456 ΔVP1723 ΔVPA0356, Strr | 35 |
| SOYBCCT123 | RIMD2210633 ΔVP1456 ΔVP1723 ΔVP1905, Strr | 35 |
| SOYBCCT134 | RIMD2210633 ΔVP1456 ΔVP1905 ΔVPA0356, Strr | 35 |
| SOYBCCT234 | RIMD2210633 ΔVP1723 ΔVP1905 ΔVPA0356, Strr | 35 |
| SOYBCCT1342 | RIMD2210633 ΔVP1456 ΔVP1723 ΔVP1905 ΔVPA0356, Strr | 35 |
| ΔproU1 | RIMD2210633 ΔproU1, Strr | 33 |
| ΔproU1ΔproU2 | RIMD2210633 ΔproU1 ΔproU2, Strr | This study |
| Vibrio harveyi 393 | Isolated from barramundi in Australia | 99 |
| Vibrio fluvialis ATCC 33809 | [NCTC 11327][606] Clinical isolate | 100 |
| Vibrio vulnificus YJ016 | Clinical isolate | 101 |
| Vibrio cholerae N16961 | O1, El Tor strain, Bangladesh, clinical, 1975 | 102 |
| Escherichia coli | ||
| DH5α λpir | Δlac pir | |
| β2155 λpir | ΔdapA::erm pir for bacterial conjugation | 103 |
| MKH13 | MC4100 (ΔbetTIBA) Δ(putPA)101 Δ(proP)2 Δ(proU); Spr | 82 |
| Plasmids | ||
| pBAD33 | Expression vector; araBAD promoter; Cmr; p15a origin | 94 |
| pBAVP1456 | pBAD33 harboring full-length VP1456 (bccT1) | 36 |
| pBAVP1723 | pBAD33 harboring full-length VP1723 (bccT2) | 35 |
| pBAVP1905 | pBAD33 harboring full-length VP1905 (bccT3) | 35 |
| pBAVPA0356 | pBAD33 harboring full-length VPA0356 (bccT4) | 36 |
| pBAVCBccT3 | pBAD33 harboring full-length BccT3 from V. cholerae | This study |
| pBAVVBccT3 | pBAD33 harboring full-length BccT3 from V. vulnificus | This study |
| pBAVVBccT5 | pBAD33 harboring 637-amino-acid BccT5 from V. vulnificus | This study |
| pBAVVBccT5long | pBAD33 harboring 672-amino-acid BccT5 from V. vulnificus | This study |
Str, streptomycin; Sp, spectinomycin; Cm, chloramphenicol; r, resistance.
Mutant strain construction.
The ΔproU1 ΔproU2 double mutant was constructed by creating an in-frame deletion of proU2 in a proU1 background (33). Splicing by overlap extension PCR and allelic exchange were performed as previously described (93). V. parahaemolyticus RIMD2210633 genomic DNA template and splicing by overlap extension (SOE) primers listed in Table 2 were used in the SOE PCR amplification to create a truncated proU2 PCR fragment of 750 bp in size by removing ∼1,150 bp from the wild-type proU2 gene (1,900 bp). The truncated proU2 fragment was subsequently cloned into a pDS132 suicide vector, transformed in E. coli β2155 λpir DAP auxotroph, and mobilized into a recipient wild-type V. parahaemolyticus RIMD2210633 strain by conjugation (33). The generated V. parahaemolyticus mutant strain harboring a truncated version of the proU2 gene was designated ΔproU2. To create the double mutant ΔproU1 ΔproU2 strain, a V. parahaemolyticus ΔproU2 strain was used as a background, and a previously created ΔproU1 inserted into the ΔproU2 background by conjugation and homologous recombination. All deletions were confirmed by PCR and sequencing.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′–3′)a | Length (bp)b |
|---|---|---|
| VCBCCT3F | tgggctagcgaattcgagctTGATTAAAAAACCGAAAATGCTTTG | 1,675 |
| VCBCCT3R | ggatccccgggtaccgagctTTAAGATTGGTGAAGACGCTTG | |
| VVBCCT3F | tgggctagcgaattcgagctGGCTGTATTTTCAAAATTAGTTAG | 1,630 |
| VVBCCT3R | ggatccccgggtaccgagctTTATTGCTTCTCGGTGCG | |
| VVBCCT5F | tgggctagcgaattcgagctTATCGAACAATCGGTATTTCATAC | 1,966 |
| VVBCCT5R | ggatccccgggtaccgagctTTAACTGAGTGATTCTGACTGAAG | |
| VVBCCT5Flong | tgggctagcgaattcgagctAAGCAATTACTCACCAACCAATTTAG | 2,069 |
| VVBCCT5Rlong | ggatccccgggtaccgagctTTAACTGAGTGATTCTGACTGAAG | |
| VPA1109A | GCATGCTGCCATTGCCGCACCAATC | 355 |
| VPA1109B | CAGCTGAGATCTGGTACCGTCCATGATTAAGCCTCCT | |
| VPA1109C | GGTACCAGATCTCAGCTGGCTTAGTTGAGATAAACGT | 380 |
| VPA1109D | GAGCTCCTTCAACGGTCATTGGAC | |
| VPA1109FL-F | CATTGTACCGGGACT | 2,008 |
| VPA1109FL-R | AGCAGATATGTACAACACC |
Lowercase letters indicate complementary regions for Gibson assembly.
Length (bp) indicates length of PCR product for each primer pair.
Growth analysis of mutant strains in DMSP.
V. parahaemolyticus RIMD2210633 or an in-frame deletion mutant of ectB were grown overnight in M9G1%. Cultures were subsequently diluted 1:50 into fresh medium, grown for 5 h, and used as previously described for analysis of DMG as an osmolyte (36). These strains, the proU double mutant, and a quadruple ΔbccT1 ΔbccT3 ΔbccT4 ΔbccT2 mutant (bccT null) strain exponential culture were then diluted 1:40 into 200 µl of M9G6% medium with and without exogenous compatible solutes in a 96-well microplate and grown at 37°C with intermittent shaking for 24 h. Dimethylsulfoniopropionate (DMSP) was added to a final concentration of 500 µM. Growth analysis was repeated following the above procedure with each of four triple bccT deletion mutants, ΔbccT2 ΔbccT3 ΔbccT4, ΔbccT1 ΔbccT3 ΔbccT4, ΔbccT1 ΔbccT2 ΔbccT4, and ΔbccT1 ΔbccT2 ΔbccT3, to determine which BCCT was responsible for transport of DMSP.
Functional complementation of E. coli strain MKH13.
The 1,623-bp opuD gene (bccT3 homolog) was amplified from the V. cholerae N16961 genome using the primers listed in Table 2. The 1,572-bp bccT3 homolog, the 1,914-bp bccT5, and the 2,016-bp bccT5 gene were amplified from the V. vulnificus YJ016 genome using primers listed in Table 2. All primers were purchased from Integrated DNA Technologies (Coralville, IA). Gibson assembly protocol using NEBuilder HiFi DNA assembly mastermix (New England Biolabs, Ipswich, MA) was followed to ligate the amplified fragments with the expression vector pBAD33 (94), which had been linearized with SacI. Regions of complementarity for Gibson assembly are indicated by lowercase letters in the primer sequences in Table 2. The resulting expression plasmids, pBAVCbccT3, pBAVVbccT3, pBAVVbccT5, and pBAVVbccT5long, were transformed into E. coli Dh5α cells for propagation. Plasmids were then purified, sequenced, and subsequently transformed into E. coli strain MKH13, which has large deletions that include compatible solute transporters (putP, proP, and proU) and the choline uptake and glycine betaine biosynthesis loci (betT-betIBA) (82).
E. coli MKH13 strains containing pBAD expressing a single bccT gene were grown overnight in minimal medium supplemented with 1% NaCl and 20 mM glucose (M9G1%) with chloramphenicol and subsequently diluted 1:100 into M9G supplemented with 4% NaCl (M9G4%) and 500 µM the indicated compatible solute and chloramphenicol. Expression of each BCCT was induced with 0.01% arabinose, and functional complementation was determined by measuring OD595 after 24 h of growth at 37°C with aeration. Growth was compared to that of an MKH13 strain harboring empty pBAD33, which cannot grow in M9G4% without exogenous compatible solutes. Statistics were calculated using Student’s t test.
Bioinformatics analyses.
Transmembrane helix probabilities of V. parahaemolyticus BccT1 (GenPept accession number Q87PP5.1) and V. vulnificus BccT5 (GenPept accession number BAC93547.1) were generated using OCTOPUS and aligned via the AlignMe program (http://www.bioinfo.mpg.de/AlignMe) (95–97).
Supplementary Material
ACKNOWLEDGMENTS
We are especially appreciative to three anonymous reviewers for their constructive feedback, suggestions, and comments. We thank members of the Boyd Group for reviewing the manuscript.
This research was supported by a National Science Foundation grant (award IOS-1656688) to E.F.B. G.J.G. was funded in part by a University of Delaware graduate fellowship award.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Csonka LN. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev 53:121–147. doi: 10.1128/MR.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galinski EA. 1995. Osmoadaptation in bacteria. Adv Microb Physiol 37:272–328. [PubMed] [Google Scholar]
- 3.Wood JM. 2011. Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu Rev Microbiol 65:215–238. doi: 10.1146/annurev-micro-090110-102815. [DOI] [PubMed] [Google Scholar]
- 4.da Costa MS, Santos H, Galinski EA. 1998. An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv Biochem Eng Biotechnol 61:117–153. doi: 10.1007/BFb0102291. [DOI] [PubMed] [Google Scholar]
- 5.Galinski EA, Oren A. 1991. Isolation and structure determination of a novel compatible solute from the moderately halophilic purple sulfur bacterium Ectothiorhodospira marismortui. Eur J Biochem 198:593–598. doi: 10.1111/j.1432-1033.1991.tb16055.x. [DOI] [PubMed] [Google Scholar]
- 6.Sleator RD, Hill C. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev 26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 7.Roberts MF. 2004. Osmoadaptation and osmoregulation in archaea: update 2004. Front Biosci 9:1999–2019. doi: 10.2741/1366. [DOI] [PubMed] [Google Scholar]
- 8.Roberts MF. 2005. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst 1:5. doi: 10.1186/1746-1448-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kempf B, Bremer E. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol 170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- 10.Wood JM, Bremer E, Csonka LN, Kraemer R, Poolman B, van der Heide T, Smith LT. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp Biochem Physiol A Mol Integr Physiol 130:437–460. doi: 10.1016/s1095-6433(01)00442-1. [DOI] [PubMed] [Google Scholar]
- 11.Browne N, Dowds B. 2001. Heat and salt stress in the food pathogen Bacillus cereus. J Appl Microbiol 91:1085–1094. doi: 10.1046/j.1365-2672.2001.01478.x. [DOI] [PubMed] [Google Scholar]
- 12.Holtmann G, Bremer E. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J Bacteriol 186:1683–1693. doi: 10.1128/jb.186.6.1683-1693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budde I, Steil L, Scharf C, Völker U, Bremer E. 2006. Adaptation of Bacillus subtilis to growth at low temperature: a combined transcriptomic and proteomic appraisal. Microbiology (Reading) 152:831–853. doi: 10.1099/mic.0.28530-0. [DOI] [PubMed] [Google Scholar]
- 14.Kuhlmann AU, Bursy J, Gimpel S, Hoffmann T, Bremer E. 2008. Synthesis of the compatible solute ectoine in Virgibacillus pantothenticus is triggered by high salinity and low growth temperature. Appl Environ Microbiol 74:4560–4563. doi: 10.1128/AEM.00492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitaker WB, Parent MA, Naughton LM, Richards GP, Blumerman SL, Boyd EF. 2010. Modulation of responses of Vibrio parahaemolyticus O3:K6 to pH and temperature stresses by growth at different salt concentrations. Appl Environ Microbiol 76:4720–4729. doi: 10.1128/AEM.00474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bashir A, Hoffmann T, Smits SH, Bremer E. 2014. Dimethylglycine provides salt and temperature stress protection to Bacillus subtilis. Appl Environ Microbiol 80:2773–2785. doi: 10.1128/AEM.00078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bashir A, Hoffmann T, Kempf B, Xie X, Smits S, Bremer E. 2014. Plant-derived compatible solutes proline betaine and betonicine confer enhanced osmotic and temperature stress tolerance to Bacillus subtilis. Microbiology (Reading) 160:2283–2294. doi: 10.1099/mic.0.079665-0. [DOI] [PubMed] [Google Scholar]
- 18.Kalburge SS, Whitaker WB, Boyd EF. 2014. High-salt preadaptation of Vibrio parahaemolyticus enhances survival in response to lethal environmental stresses. J Food Prot 77:246–253. doi: 10.4315/0362-028X.JFP-13-241. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann T, Bremer E. 2011. Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J Bacteriol 193:1552–1562. doi: 10.1128/JB.01319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.May G, Faatz E, Lucht JM, Haardt M, Bolliger M, Bremer E. 1989. Characterization of the osmoregulated Escherichia coli proU promoter and identification of ProV as a membrane-associated protein. Mol Microbiol 3:1521–1531. doi: 10.1111/j.1365-2958.1989.tb00138.x. [DOI] [PubMed] [Google Scholar]
- 21.Lamark T, Kaasen I, Eshoo MW, Falkenberg P, McDougall J, Strom AR. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol Microbiol 5:1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 22.Lucht J, Bremer E. 1991. Characterization of mutations affecting the osmoregulated proU promoter of Escherichia coli and identification of 5′ sequences required for high-level expression. J Bacteriol 173:801–809. doi: 10.1128/jb.173.2.801-809.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hengge-Aronis R. 1996. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol 21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 24.Gunasekera T, Csonka L, Paliy O. 2008. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J Bacteriol 190:3712–3720. doi: 10.1128/JB.01990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belitsky BR, Brill J, Bremer E, Sonenshein AL. 2001. Multiple genes for the last step of proline biosynthesis in Bacillus subtilis. J Bacteriol 183:4389–4392. doi: 10.1128/JB.183.14.4389-4392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boch J, Kempf B, Bremer E. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J Bacteriol 176:5364–5371. doi: 10.1128/jb.176.17.5364-5371.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boch J, Kempf B, Schmid R, Bremer E. 1996. Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J Bacteriol 178:5121–5129. doi: 10.1128/jb.178.17.5121-5129.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boch J, Nau-Wagner G, Kneip S, Bremer E. 1997. Glycine betaine aldehyde dehydrogenase from Bacillus subtilis: characterization of an enzyme required for the synthesis of the osmoprotectant glycine betaine. Arch Microbiol 168:282–289. doi: 10.1007/s002030050500. [DOI] [PubMed] [Google Scholar]
- 29.Brill J, Hoffmann T, Bleisteiner M, Bremer E. 2011. Osmotically controlled synthesis of the compatible solute proline is critical for cellular defense of Bacillus subtilis against high osmolarity. J Bacteriol 193:5335–5346. doi: 10.1128/JB.05490-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahne H, Mäder U, Otto A, Bonn F, Steil L, Bremer E, Hecker M, Becher D. 2010. A Comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J Bacteriol 192:870–882. doi: 10.1128/JB.01106-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steil L, Hoffmann T, Budde I, Völker U, Bremer E. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J Bacteriol 185:6358–6370. doi: 10.1128/jb.185.21.6358-6370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeter R, Hoffmann T, Voigt B, Meyer H, Bleisteiner M, Muntel J, Jürgen B, Albrecht D, Becher D, Lalk M, Evers S, Bongaerts J, Maurer K, Putzer H, Hecker M, Schweder T, Bremer E. 2013. Stress responses of the industrial workhorse Bacillus licheniformis to osmotic challenges. PLoS One 8:e80956. doi: 10.1371/journal.pone.0080956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naughton LM, Blumerman SL, Carlberg M, Boyd EF. 2009. Osmoadaptation among Vibrio species and unique genomic features and physiological responses of Vibrio parahaemolyticus. Appl Environ Microbiol 75:2802–2810. doi: 10.1128/AEM.01698-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ongagna-Yhombi SY, Boyd EF. 2013. Biosynthesis of the osmoprotectant ectoine, but not glycine betaine, is critical for survival of osmotically stressed Vibrio parahaemolyticus cells. Appl Environ Microbiol 79:5038–5049. doi: 10.1128/AEM.01008-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ongagna-Yhombi SY, McDonald ND, Boyd EF. 2015. Deciphering the role of multiple betaine-carnitine-choline transporters in the halophile Vibrio parahaemolyticus. Appl Environ Microbiol 81:351–363. doi: 10.1128/AEM.02402-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregory GJ, Dutta A, Parashar V, Boyd EF. 2020. Investigations of dimethylglycine (DMG), glycine betaine and ectoine uptake by a BCCT family transporter with broad substrate specificity in Vibrio species. J Bacteriol 202:e00314-20. doi: 10.1128/JB.00314-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saier MH, Jr. 2001. Evolution of transport proteins. Genet Eng (N Y) 23:1–10. [DOI] [PubMed] [Google Scholar]
- 38.Saier MH, Jr, Paulsen IT. 2001. Phylogeny of multidrug transporters. Semin Cell Dev Biol 12:205–213. doi: 10.1006/scdb.2000.0246. [DOI] [PubMed] [Google Scholar]
- 39.Ziegler C, Bremer E, Kramer R. 2010. The BCCT family of carriers: from physiology to crystal structure. Mol Microbiol 78:13–34. doi: 10.1111/j.1365-2958.2010.07332.x. [DOI] [PubMed] [Google Scholar]
- 40.Farwick M, Siewe RM, Kramer R. 1995. Glycine betaine uptake after hyperosmotic shift in Corynebacterium glutamicum. J Bacteriol 177:4690–4695. doi: 10.1128/jb.177.16.4690-4695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peter H, Burkovski A, Kramer R. 1996. Isolation, characterization, and expression of the Corynebacterium glutamicum betP gene, encoding the transport system for the compatible solute glycine betaine. J Bacteriol 178:5229–5234. doi: 10.1128/jb.178.17.5229-5234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner SM, Nightingale PD, Broadgate W, Liss PS. 1995. The distribution of dimethyl sulphide and dimethylsulphoniopropionate in Antarctic waters and sea ice. Deep Sea Res Part II Top Stud Oceanogr 42:1059–1080. doi: 10.1016/0967-0645(95)00066-Y. [DOI] [Google Scholar]
- 43.Keller MD, Bellows WK, Guillard RRL. 1989. Dimethyl sulfide production in marine phytoplankton, p 167–182. In Biogenic sulfur in the environment. ACS Publications, Washington, DC. [Google Scholar]
- 44.Burgermeister S, Zimmerman RL, Georgii HW, Bingemer HG, Kirst GO, Janssen M, Ernst W. 1990. On the biogenic origin of dimethylsulfide: relation between chlorophyll, ATP, organismic DMSP, phytoplankton species, and DMS distribution in Atlantic surface water and atmosphere. J Geophys Res 95:607–615. [Google Scholar]
- 45.Kiene RP, Linn LJ, Bruton JA. 2000. New and important roles for DMSP in marine microbial communities. J Sea Res 43:209–224. doi: 10.1016/S1385-1101(00)00023-X. [DOI] [Google Scholar]
- 46.Malin G, Erst GO. 1997. Algal production of dimethyl sulfide and its atmospheric role. J Phycol 33:889–896. doi: 10.1111/j.0022-3646.1997.00889.x. [DOI] [Google Scholar]
- 47.Sunda W, Kieber D, Kiene R, Huntsman S. 2002. An antioxidant function for DMSP and DMS in marine algae. Nature 418:317–320. doi: 10.1038/nature00851. [DOI] [PubMed] [Google Scholar]
- 48.Speeckaert G, Borges AV, Champenois W, Royer C, Gypens N. 2018. Annual cycle of dimethylsulfoniopropionate (DMSP) and dimethylsulfoxide (DMSO) related to phytoplankton succession in the Southern North Sea. Sci Total Environ 622–623:362–372. doi: 10.1016/j.scitotenv.2017.11.359. [DOI] [PubMed] [Google Scholar]
- 49.Kiene RP, Nowinski B, Esson K, Preston C, Marin R, III, Birch J, Scholin C, Ryan J, Moran MA. 2019. Unprecedented DMSP concentrations in a massive dinoflagellate bloom in Monterey Bay, CA. Geophys Res Lett 46:12279–12288. doi: 10.1029/2019GL085496. [DOI] [Google Scholar]
- 50.Yoch DC. 2002. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl Environ Microbiol 68:5804–5815. doi: 10.1128/aem.68.12.5804-5815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams BT, Cowles K, Bermejo Martínez A, Curson ARJ, Zheng Y, Liu J, Newton-Payne S, Hind AJ, Li C-Y, Rivera PPL, Carrión O, Liu J, Spurgin LG, Brearley CA, Mackenzie BW, Pinchbeck BJ, Peng M, Pratscher J, Zhang X-H, Zhang Y-Z, Murrell JC, Todd JD. 2019. Bacteria are important dimethylsulfoniopropionate producers in coastal sediments. Nat Microbiol 4:1815–1825. doi: 10.1038/s41564-019-0527-1. [DOI] [PubMed] [Google Scholar]
- 52.Curson A, Liu J, Bermejo MA, Green R, Chan Y, Carrión O, Williams B, Zhang S, Yang G, Bulman Page P, Zhang X, Todd J. 2017. Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat Microbiol 2:17009. doi: 10.1038/nmicrobiol.2017.9. [DOI] [PubMed] [Google Scholar]
- 53.Lovelock J, Maggs R, Rasmussen R. 1972. Atmospheric dimethyl sulphide and the natural sulphur cycle. Nature 237:452–453. doi: 10.1038/237452a0. [DOI] [Google Scholar]
- 54.Kiene RP. 1990. Dimethyl sulfide production from dimethylsulfoniopropionate in coastal seawater samples and bacterial cultures. Appl Environ Microbiol 56:3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X, Liu J, Liu J, Yang G, Xue C, Curson A, Todd J. 2019. Biogenic production of DMSP and its degradation to DMS—their roles in the global sulfur cycle. Sci China Life Sci 62:1296–1319. doi: 10.1007/s11427-018-9524-y. [DOI] [PubMed] [Google Scholar]
- 56.Carrión O, Curson A, Kumaresan D, Fu Y, Lang A, Mercadé E, Todd J. 2015. A novel pathway producing dimethylsulphide in bacteria is widespread in soil environments. Nat Commun 6:6579. doi: 10.1038/ncomms7579. [DOI] [PubMed] [Google Scholar]
- 57.Carrión O, Pratscher J, Curson A, Williams B, Rostant W, Murrell J, Todd J. 2017. Methanethiol-dependent dimethylsulfide production in soil environments. ISME J 11:2379–2390. doi: 10.1038/ismej.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonzalez JM, Simo R, Massana R, Covert JS, Casamayor EO, Pedros-Alio C, Moran MA. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol 66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buchan A, LeCleir GR, Gulvik CA, Gonzalez JM. 2014. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol 12:686–698. doi: 10.1038/nrmicro3326. [DOI] [PubMed] [Google Scholar]
- 60.Archer SD, Widdicombe CE, Tarran GA, Rees AP, Burkill PH. 2001. Production and turnover of particulate dimethylsulphoniopropionate during a coccolithophore bloom in the northern North Sea. Aquat Microb Ecol 24:225–241. doi: 10.3354/ame024225. [DOI] [Google Scholar]
- 61.Howard EC, Henriksen JR, Buchan A, Reisch CR, Burgmann H, Welsh R, Ye W, Gonzalez JM, Mace K, Joye SB, Kiene RP, Whitman WB, Moran MA. 2006. Bacterial taxa that limit sulfur flux from the ocean. Science 314:649–652. doi: 10.1126/science.1130657. [DOI] [PubMed] [Google Scholar]
- 62.Stefels J. 2000. Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J Sea Res 43:183–197. doi: 10.1016/S1385-1101(00)00030-7. [DOI] [Google Scholar]
- 63.Ansede J, Pellechia P, Yoch D. 2001. Nuclear magnetic resonance analysis of [1-13C]dimethylsulfoniopropionate (DMSP) and [1-13C]acrylate metabolism by a DMSP lyase-producing marine isolate of the α-subclass of Proteobacteria. Appl Environ Microbiol 67:3134–3139. doi: 10.1128/AEM.67.7.3134-3139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ansede J, Friedman R, Yoch D. 2001. Phylogenetic analysis of culturable dimethyl sulfide-producing bacteria from a Spartina-dominated salt marsh and estuarine water. Appl Environ Microbiol 67:1210–1217. doi: 10.1128/AEM.67.3.1210-1217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Todd J, Curson A, Kirkwood M, Sullivan M, Green R, Johnston A. 2011. DddQ, a novel, cupin-containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria. Environ Microbiol 13:427–438. doi: 10.1111/j.1462-2920.2010.02348.x. [DOI] [PubMed] [Google Scholar]
- 66.Todd J, Kirkwood M, Newton-Payne S, Johnston A. 2012. DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J 6:223–226. doi: 10.1038/ismej.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hehemann J, Law A, Redecke L, Boraston A. 2014. The structure of RdDddP from Roseobacter denitrificans reveals that DMSP lyases in the DddP-family are metalloenzymes. PLoS One 9:e103128. doi: 10.1371/journal.pone.0103128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burkhardt I, Lauterbach L, Brock N, Dickschat J. 2017. Chemical differentiation of three DMSP lyases from the marine Roseobacter group. Org Biomol Chem 15:4432–4439. doi: 10.1039/c7ob00913e. [DOI] [PubMed] [Google Scholar]
- 69.Onda D, Azanza R, Lluisma A. 2015. Potential DMSP-degrading Roseobacter clade dominates endosymbiotic microflora of Pyrodinium bahamense var. compressum (Dinophyceae) in vitro. Arch Microbiol 197:965–971. doi: 10.1007/s00203-015-1133-0. [DOI] [PubMed] [Google Scholar]
- 70.Todd JD, Curson AR, Nikolaidou-Katsaraidou N, Brearley CA, Watmough NJ, Chan Y, Page PC, Sun L, Johnston AW. 2010. Molecular dissection of bacterial acrylate catabolism—unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environ Microbiol 12:327–343. doi: 10.1111/j.1462-2920.2009.02071.x. [DOI] [PubMed] [Google Scholar]
- 71.Curson AR, Todd JD, Sullivan MJ, Johnston AW. 2011. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat Rev Microbiol 9:849–859. doi: 10.1038/nrmicro2653. [DOI] [PubMed] [Google Scholar]
- 72.Moran MA, Reisch CR, Kiene RP, Whitman WB. 2012. Genomic insights into bacterial DMSP transformations. Annu Rev Mar Sci 4:523–542. doi: 10.1146/annurev-marine-120710-100827. [DOI] [PubMed] [Google Scholar]
- 73.Cosquer A, Pichereau V, Pocard JA, Minet J, Cormier M, Bernard T. 1999. Nanomolar levels of dimethylsulfoniopropionate, dimethylsulfonioacetate, and glycine betaine are sufficient to confer osmoprotection to Escherichia coli. Appl Environ Microbiol 65:3304–3311. doi: 10.1128/AEM.65.8.3304-3311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Broy S, Chen C, Hoffmann T, Brock NL, Nau-Wagner G, Jebbar M, Smits SH, Dickschat JS, Bremer E. 2015. Abiotic stress protection by ecologically abundant dimethylsulfoniopropionate and its natural and synthetic derivatives: insights from Bacillus subtilis. Environ Microbiol 17:2362–2378. doi: 10.1111/1462-2920.12698. [DOI] [PubMed] [Google Scholar]
- 75.Sun L, Curson ARJ, Todd JD, Johnston AWB. 2012. Diversity of DMSP transport in marine bacteria, revealed by genetic analyses. Biogeochemistry 110:121–130. doi: 10.1007/s10533-011-9666-z. [DOI] [Google Scholar]
- 76.Vila-Costa M, Simo R, Harada H, Gasol JM, Slezak D, Kiene RP. 2006. Dimethylsulfoniopropionate uptake by marine phytoplankton. Science 314:652–654. doi: 10.1126/science.1131043. [DOI] [PubMed] [Google Scholar]
- 77.Teichmann L, Kummel H, Warmbold B, Bremer E. 2018. OpuF, a new Bacillus compatible solute ABC transporter with a substrate-binding protein fused to the transmembrane domain. Appl Environ Microbiol 84 doi: 10.1128/AEM.01728-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oren A. 1999. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev 63:334–348. doi: 10.1128/MMBR.63.2.334-348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shao Z, Deng W, Li S, He J, Ren S, Huang W, Lu Y, Zhao G, Cai Z, Wang J. 2015. GlnR-mediated regulation of ectABCD transcription expands the role of the GlnR regulon to osmotic stress management. J Bacteriol 197:3041–3047. doi: 10.1128/JB.00185-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pastor JM, Bernal V, Salvador M, Argandona M, Vargas C, Csonka L, Sevilla A, Iborra JL, Nieto JJ, Canovas M. 2013. Role of central metabolism in the osmoadaptation of the halophilic bacterium Chromohalobacter salexigens. J Biol Chem 288:17769–17781. doi: 10.1074/jbc.M113.470567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reisch CR, Moran MA, Whitman WB. 2011. Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front Microbiol 2:172. doi: 10.3389/fmicb.2011.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haardt M, Kempf B, Faatz E, Bremer E. 1995. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol Gen Genet 246:783–786. doi: 10.1007/BF00290728. [DOI] [PubMed] [Google Scholar]
- 83.Paranjpye R, Hamel OS, Stojanovski A, Liermann M. 2012. Genetic diversity of clinical and environmental Vibrio parahaemolyticus strains from the Pacific Northwest. Appl Environ Microbiol 78:8631–8638. doi: 10.1128/AEM.01531-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Greenfield DI, Gooch Moore J, Stewart JR, Hilborn ED, George BJ, Li Q, Dickerson J, Keppler CK, Sandifer PA. 2017. Temporal and environmental factors driving Vibrio vulnificus and V. parahaemolyticus populations and their associations with harmful algal blooms in South Carolina detention ponds and receiving tidal creeks. GeoHealth 1:306–317. doi: 10.1002/2017GH000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thickman JD, Gobler CJ. 2017. The ability of algal organic matter and surface runoff to promote the abundance of pathogenic and non-pathogenic strains of Vibrio parahaemolyticus in Long Island Sound, USA. PLoS One 12:e0185994. doi: 10.1371/journal.pone.0185994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guibert I, Bourdreux F, Bonnard I, Pochon X, Dubousquet V, Raharivelomanana P, Berteaux-Lecellier V, Lecellier G. 2020. Dimethylsulfoniopropionate concentration in coral reef invertebrates varies according to species assemblages. Sci Rep 10:9922. doi: 10.1038/s41598-020-66290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ben-Haim Y, Zicherman-Keren M, Rosenberg E. 2003. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl Environ Microbiol 69:4236–4242. doi: 10.1128/aem.69.7.4236-4242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rubio-Portillo E, Yarza P, Peñalver C, Ramos-Esplá AA, Antón J. 2014. New insights into Oculina patagonica coral diseases and their associated Vibrio spp. communities. ISME J 8:1794–1807. doi: 10.1038/ismej.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ushijima B, Meyer JL, Thompson S, Pitts K, Marusich MF, Tittl J, Weatherup E, Reu J, Wetzell R, Aeby GS, Häse CC, Paul VJ. 2020. Disease diagnostics and potential coinfections by Vibrio coralliilyticus during an ongoing coral disease outbreak in Florida. Front Microbiol 11:569354. doi: 10.3389/fmicb.2020.569354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ushijima B, Videau P, Burger AH, Shore-Maggio A, Runyon CM, Sudek M, Aeby GS, Callahan SM. 2014. Vibrio coralliilyticus strain OCN008 is an etiological agent of acute Montipora white syndrome. Appl Environ Microbiol 80:2102–2109. doi: 10.1128/AEM.03463-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raina JB, Dinsdale EA, Willis BL, Bourne DG. 2010. Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol 18:101–108. doi: 10.1016/j.tim.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 92.Garren M, Son K, Raina JB, Rusconi R, Menolascina F, Shapiro OH, Tout J, Bourne DG, Seymour JR, Stocker R. 2014. A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J 8:999–1007. doi: 10.1038/ismej.2013.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 94.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khafizov K, Staritzbichler R, Stamm M, Forrest L. 2010. A study of the evolution of inverted-topology repeats from LeuT-fold transporters using AlignMe. Biochemistry 49:10702–10713. doi: 10.1021/bi101256x. [DOI] [PubMed] [Google Scholar]
- 96.Stamm M, Staritzbichler R, Khafizov K, Forrest L. 2013. Alignment of helical membrane protein sequences using AlignMe. PLoS One 8:e57731. doi: 10.1371/journal.pone.0057731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stamm M, Staritzbichler R, Khafizov K, Forrest L. 2014. AlignMe—a membrane protein sequence alignment web server. Nucleic Acids Res 42:W246–W251. doi: 10.1093/nar/gku291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 99.Pedersen K, Verdonck L, Austin B, Austin DA, Blanch AR, Grimont PAD, Jofre J, Koblavi S, Larsen JL, Tiainen T, Vigneulle M, Swings J. 1998. Taxonomic evidence that Vibrio carchariae Grimes et al, 1985 is a junior synonym of Vibrio harveyi (Johnson and Shunk 1936) Baumann et al. 1981. Intern J Syst Bacteriol 48:749–748. doi: 10.1099/00207713-48-3-749. [DOI] [Google Scholar]
- 100.Lee J, Shread P, Furniss A, Bryant T. 1981. Taxonomy and description of Vibrio fluvialis sp. nov. (synonym group F vibrios, group EF6). J Appl Bacteriol 50:73–94. doi: 10.1111/j.1365-2672.1981.tb00873.x. [DOI] [PubMed] [Google Scholar]
- 101.Chen C, Wu K, Chang Y, Chang C, Tsai H, Liao T, Liu Y, Chen H, Shen A, Li J, Su T, Shao C, Lee C, Hor L, Tsai S. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res 13:2577–2587. doi: 10.1101/gr.1295503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kaper J, Lockman H, Baldini M, Levine M. 1984. Recombinant nontoxinogenic Vibrio cholerae strains as attenuated cholera vaccine candidates. Nature 308:655–658. doi: 10.1038/308655a0. [DOI] [PubMed] [Google Scholar]
- 103.Dehio C, Meyer M. 1997. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J Bacteriol 179:538–540. doi: 10.1128/jb.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.