Dung beetles are a very important part of an ecosystem because of their role in the removal and decomposition of vertebrate dung. It has been suspected that symbiotic gut bacteria facilitate this role, a hypothesis that we have explored with high-throughput barcoding.
KEYWORDS: symbiosis, Australian endemic genera, gut morphology, detritivore, Desulfovibrio, coevolution
ABSTRACT
Gut microbes play an important role in the biology and evolution of insects. Australian native dung beetles (Scarabaeinae) present an opportunity to study gut microbiota in an evolutionary context as they come from two distinct phylogenetic lineages and some species in each lineage have secondarily adapted to alternative or broader diets. In this study, we characterized the hindgut bacterial communities found in 21 species of dung beetles across two lineages, using 16S rRNA sequencing. We found that gut microbial diversity was more dependent on host phylogeny and gut morphology than specific dietary preferences or environment. In particular, gut microbial diversity was highest in the endemic, flightless genus Cephalodesmius, which feeds on a broad range of composted organic matter. The hindgut of Cephalodesmius beetles harbors a highly conserved core set of bacteria, suggesting that the bacteria are symbiotic. Symbiosis is supported by the persistence of the core microbiota across isolated beetle populations and between species in the genus. A coevolutionary relationship is supported by the expansion of the hindgut to form a fermentation chamber and the fermentative nature of the core microbes. In contrast, Australian species of the widespread dung beetle genus Onthophagus that specialize on a single food resource, such as dung or fungus, exhibit minimal food processing behavior and have a short, narrow hindgut and a variable gut microbiota with relatively few core bacterial taxa. A conserved, complex gut microbiota is hypothesized to be unnecessary for this highly mobile genus.
IMPORTANCE Dung beetles are a very important part of an ecosystem because of their role in the removal and decomposition of vertebrate dung. It has been suspected that symbiotic gut bacteria facilitate this role, a hypothesis that we have explored with high-throughput barcoding. We found that differences in hindgut morphology had the greatest effect on the bacterial community composition. Species with a hindgut fermentation chamber harbored a distinctly different hindgut community compared to those species with a narrow, undifferentiated hindgut. Diet and phylogeny were also associated with differences in gut community. Further understanding of the relationships between dung beetles and their gut microbes will provide insights into the evolution of their behaviors and how gut communities contribute to their fitness.
INTRODUCTION
The insect gut can be colonized by various microorganisms, but the composition, abundance, and stability of microbial taxa vary considerably across the diverse orders of insects (1). The observed differences in gut bacterial communities can be attributed to several factors, including host diet, phylogeny, environment, gut morphology, and behavior (2–6). Microbes are known to be of functional significance, especially in insects with nutritionally limited diets or difficult-to-digest diets (1). Often, insects with specialized diets, such as honeybees (Apis mellifera) have a small number of specialized core gut taxa (4, 7). In contrast, insects with broad diets, such as omnivores and detritivores, have gut communities that are diverse, and some insects can have hundreds of taxa (4, 8–10). Detritivores, in particular, share a distinct and diverse microbial gut community, even though they occur in divergent taxonomic groups, which suggests that a specialized microbiota is required to consume decaying organic material (2).
While many insects have an undifferentiated gut morphology, many of the detritivores have enlarged regions in the hindgut (11–16). Insects such as termites (16), detritus-feeding fly larvae (15), and scarab beetle larvae (11, 17) all have a dilated hindgut region that forms an anoxic fermentation chamber. This provides a suitable microhabitat for anaerobic microbes to establish residence and in turn aid with the digestion of plant polysaccharides and other lignocellulosic matter. In several species of soil-dwelling scarab beetle larvae (Melolonthinae and Cetoniinae), the hindgut microbial community has been found to be highly diverse and many of the gut microbes are consistently present, suggesting a level of symbiosis (11, 17, 18). Most of these studies have focused on the gut communities in the larval stages (19), while comparative studies of adult scarab beetles are limited.
Among the scarabs, the true dung beetles (Scarabaeidae: Scarabaeinae) are an ecologically important group because of their association with vertebrate dung. Dung feeding is a specialized form of detritivory, and it is suspected that dung beetles rely on gut microbiota to aid with digestion as dung is considered to be a nutritionally limited food source (20–23). The dung beetle larvae, in particular, have a hindgut fermentation chamber, as is seen in other subfamilies of scarab beetle larvae (23), yet only a few studies have investigated gut microbiota in dung beetles. Many have focused on transmission of microbes between adults and larvae in a few selected species (24, 25). A comparative study of adult beetles from five different families included four dung beetle species from the genus Onthophagus (3). These dung beetles had a more diverse gut microbial community than all other beetle families, but the microbiota were highly variable (3). Another study, also investigating Onthophagus spp., found that their gut microbiota shared some core elements yet was significantly influenced by the local environment when the insects were introduced to new locations (5). In the dung beetle genus Euoniticellus, adult male and female gut communities were significantly different, and the composition of the female gut was more similar to that of the larval gut (26). This difference may reflect the fact that in this species, only the female is engaged in preparing the brood material. Two African congeneric dung beetle species (Pachysoma), both with atypical diets—one a dry-dung feeder and the other a plant detritus feeder—had a small core microbiota; however, the gut bacterial compositions differed between the two species. The detritivorous species had the greater bacterial diversity overall (27).
Australian native dung beetles (Scarabaeinae) are ideal for a comparative study of gut microbiota in an evolutionary context. From the phylogenetic perspective, the Australian fauna is composed of two distinct groups: the Onthophagini, which contains the cosmopolitan genus Onthophagus (∼250 Australian spp. and over 2,000 spp. worldwide) that dispersed into Australia from Asia around 20 to 24 million years ago (Mya) (28), and the Australian endemic genera (AuEG) (∼250 spp.), which are a relictual Gondwanan lineage, with mid-Cretaceous origins (∼80 Mya) (29). The Australian dung beetles present a useful test case for dietary specialization and gut microbiota because a number of species in both groups have broadened their diet (30–33). In addition, there are distinct differences in behaviors and feeding strategies. The Onthophagus species tunnel directly beneath their food source (paracoprids), where they mass provision for their offspring, lay eggs, and then leave, engaging in little parental care (34). In contrast, the AuEG are telecoprids: i.e., they transport their food away from the sources to avoid competition, lay fewer eggs, and exhibit higher levels of parental care.
The genus Cephalodesmius stands out among the AuEG as having undergone the most extreme dietary shift, together with associated food processing behaviors and a high level of parental care. Males and females pair bond and work together to gather a range of organic materials, including leaves, flowers, fruit, and fungi, which are worked into a composting brood mass, thus creating a dung substitute with which to feed their larvae. The adults continue to progressively provision their larvae throughout their development to the pupal stage, exhibiting a level of subsocial behavior (33). In addition, the anterior hindgut of adult Cephalodesmius beetles is dilated into a “large, thin-walled, sac-like structure” that is proposed to be a fermentation chamber, a novel gut structure for an adult dung beetle, which may be capable of housing symbiotic bacteria (35). Given their subsocial behavior and the presence of a putative fermentation chamber, we hypothesized that members of the genus Cephalodesmius would possess a diverse and stable gut microbiota that would support their unusual food processing and brood care behavior. This specialized gut structure has not been noted in any other adult dung beetle.
Here, we examined the hindgut microbiota found in 21 Australian dung beetle species from the genera Onthophagus and Cephalodesmius and seven other Australian endemic genera (AuEG) that show different dietary adaptations. We focused on the microbial communities found in the anterior hindgut as this region of the gut provides the most suitable habitat for microbes and often has the largest microbial populations (36). Dung beetles that utilize a single food resource had a simple core gut microbial community, but in dung beetle species that pursue a greater range of food resources, we found an increasingly more complex microbial community. The gut community was strikingly different in the genus Cephalodesmius, where we discovered a persistent, distinct, and diverse community of gut microbes.
RESULTS
Bacterial diversity in the anterior hindgut.
We determined the microbiota composition from the hindgut of 18 individuals across seven species of Onthophagus, 32 individuals across three species of Cephalodesmius, and 30 individuals across seven genera (11 species) from the AuEG (Table 1; see Table S1 in the supplemental material). Although Cephalodesmius is part of the AuEG, it was considered separately because of the unusual feeding and nesting behavior of the beetles.
TABLE 1.
Species list from a study of dung beetle gut microbiotaa
| Species | Taxonomic group | Presumed diet |
Flightless | |||
|---|---|---|---|---|---|---|
| Mainly dung | Mainly mushroom | Mixed | Unknown | |||
| Amphistomus NSW1 | AuEG | x | No | |||
| Cephalodesmius armiger | AuEG | x | Yes | |||
| Cephalodesmius laticollis | AuEG | x | Yes | |||
| Cephalodesmius quadridens | AuEG | x | Yes | |||
| Demarziella intermedius | AuEG | x | No | |||
| Demarziella scarpensis | AuEG | x | No | |||
| Diorygopyx simpliciclunis | AuEG | x | Yes | |||
| Diorygopyx tibialis | AuEG | x | Yes | |||
| Labroma umbratilis | AuEG | x | Yes | |||
| Lepanus australis | AuEG | x | No | |||
| Lepanus NSW2 | AuEG | x | No | |||
| Lepanus ustulatus | AuEG | x | No | |||
| Mentophilus hollandiae | AuEG | x | Yes | |||
| Onthophagus arrilla | Onthophagini | x | No | |||
| Onthophagus CQ2 | Onthophagini | x | No | |||
| Onthophagus dunningi | Onthophagini | x | No | |||
| Onthophagus fuliginosus | Onthophagini | x | No | |||
| Onthophagus granulatus | Onthophagini | x | No | |||
| Onthophagus kumbaingeri | Onthophagini | x | No | |||
| Onthophagus pugnax | Onthophagini | x | No | |||
| Tesserodon pilicrepus | AuEG | x | Yes | |||
Species are listed alphabetically with their taxonomic group, known dietary information, and whether they are flightless or not. Dietary information was compiled from Matthews (31, 32) and Ebert et al. (30), as well as unpublished data from G. B. Monteith. Undescribed species were recorded according to the nomenclature coding system devised by Geoff Monteith (Queensland Museum) and Tom Weir (Australian National Insect Collection) for Australian museum collections.
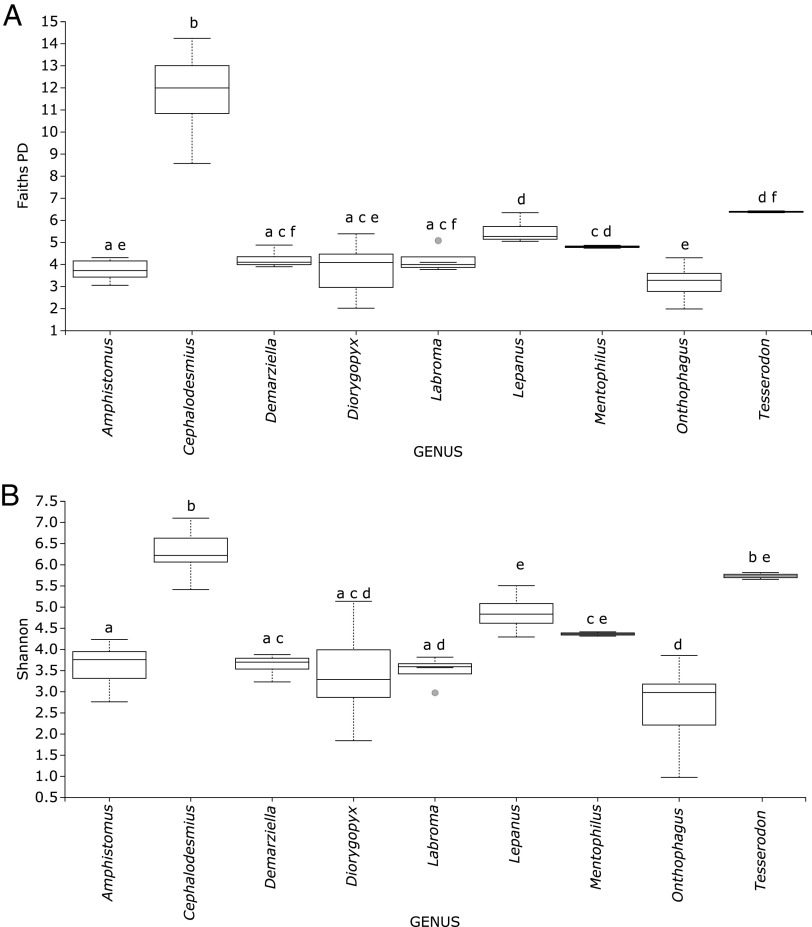
The diversity of the hindgut bacteria within each of the 9 dung beetle genera was compared using three indices: (i) Shannon’s diversity, H, which measures species richness (numbers of distinct taxa) and evenness (similarity of abundance); (ii) Faith’s phylogenetic diversity (PD), which incorporates phylogenetic relationships to provide an evolutionary measure of biodiversity; and (iii) evenness (pielou_e), which measures the similarity of abundance of the different taxa (distinct sequences) within the gut community (Table 2). The Cephalodesmius gut community consistently had the highest measures of diversity using all three indices, with more than 600 different bacterial taxa present. In contrast, the gut community of each of the other genera had fewer than 200 bacterial taxa present. The lowest-diversity measures using all indices were seen in members of the Onthophagus, which had fewer than 100 different bacterial taxa in the gut. A Kruskal-Wallis test showed that the mean values for both Shannon’s diversity and Faith’s PD were significantly different between the genera (Fig. 1) (Shannon’s H = 65.4, P < 0.0001; Faith’s PD H = 65.9, P < 0.0001). Evenness values ranged from 0.5 to 0.8, with the mean evenness value for Onthophagus being 0.59 ± 0.11: for Cephalodesmius, it was 0.8 ± 0, and for the remaining AuEG, it was 0.71 ± 0.08 (Table 2). A higher value of evenness (values between 0 and 1) indicates that the bacterial community is more evenly distributed; a lower value of evenness, such as was seen in Onthophagus, indicates that some bacterial taxa are more dominant.
TABLE 2.
Diversity of hindgut bacterial communities in 21 Australian native dung beetle speciesa
| Species | n | Total no. of sequences | Median no. of sequences/indvidual | Total no. of bacterial taxa | Diversity index value |
||
|---|---|---|---|---|---|---|---|
| Shannon’s H | Faith’s PD H | Evenness (pielou_e) | |||||
| Cephalodesmius armiger | 19 | 636,465 | 30,187 | 967 | 6.2 | 11.9 | 0.8 |
| Cephalodesmius laticollis | 4 | 132,798 | 34,882 | 640 | 6.5 | 11.8 | 0.8 |
| Cephalodesmius quadridens | 9 | 219,842 | 24,578 | 700 | 6.4 | 10.9 | 0.8 |
| Tesserodon pilicripus | 2 | 73,390 | 36,695 | 127 | 5.7 | 6.1 | 0.8 |
| Mentophilus hollandiae | 2 | 29,378 | 14,689 | 77 | 4.4 | 4.6 | 0.8 |
| Labroma umbratilis | 4 | 35,919 | 8,627 | 110 | 3.4 | 4.0 | 0.6 |
| Lepanus australis | 1 | 13,135 | 13,135 | 64 | 4.6 | 5.6 | 0.7 |
| Lepanus NSW2 | 2 | 23,407 | 11,703 | 93 | 4.9 | 5.2 | 0.8 |
| Lepanus ustulatus | 2 | 19,851 | 9,925 | 91 | 4.9 | 5.5 | 0.8 |
| Amphistomus NSW1 | 6 | 74,748 | 12,124 | 123 | 3.6 | 3.6 | 0.7 |
| Demarziella interrupta | 2 | 29,511 | 14,755 | 85 | 3.5 | 4.5 | 0.6 |
| Demarziella scarpensis | 2 | 22,643 | 11,321 | 74 | 3.8 | 4.2 | 0.7 |
| Diorygopyx tibialis | 3 | 19,756 | 6,112 | 91 | 3.7 | 3.8 | 0.7 |
| Diorygopyx simpliciclunis | 4 | 15,275 | 3,636 | 66 | 3.1 | 3.7 | 0.6 |
| Onthophagus arrilla | 1 | 15,839 | 15,839 | 23 | 3.8 | 3.0 | 0.8 |
| Onthophagus CQ2 | 1 | 11,870 | 11,870 | 32 | 3.1 | 3.1 | 0.6 |
| Onthophagus pugnax | 4 | 21,832 | 5,513 | 84 | 2.4 | 3.1 | 0.5 |
| Onthophagus fuliginosus | 2 | 25,856 | 12,928 | 71 | 2.6 | 3.0 | 0.5 |
| Onthophagus granulatus | 4 | 32,547 | 8,097 | 78 | 2.2 | 2.9 | 0.5 |
| Onthophagus dunningib | 5 | 73,517 | 18,743 | 92 | 3.1 | 3.6 | 0.6 |
| Onthophagus kumbaingerib | 1 | 16,304 | 16,304 | 49 | 3.3 | 3.4 | 0.6 |
n represents the number of samples. The total number of sequences found in each species is followed by the median number of sequences per individual. The number of taxa represents the different bacterial taxa (OTUs) found in each beetle species. Larger values of diversity indicate a greater measure of richness and evenness (Shannon’s diversity [H]) or phylogenetic richness (Faith’s PD). Kruskal-Wallis tests showed a significant difference in means between genera for both measures of diversity (Shannon’s H = 65.4, P < 0.0001; Faith’s PD H = 65.9, P < 0.0001). Cephalodesmius shows the highest gut community diversity, and Onthophagus shows the lowest community diversity. A higher value of evenness (values between 0 and 1) indicates that the bacterial community is more evenly distributed; a lower value of evenness indicates that some species are more dominant.
Mushroom specialist species.
FIG 1.
Comparisons of gut microbial community diversity within nine dung beetle genera. Box plots show diversity measured by (A) Faith’s phylogenetic diversity (PD) and (B) Shannon’s diversity index. Kruskal-Wallis tests showed a significant difference in means between genera for both measures of diversity (Shannon’s H = 65.4, P < 0.0001; Faith’s PD H = 65.9, P < 0.0001). Cephalodesmius shows the highest gut community diversity, and Onthophagus shows the lowest community diversity.
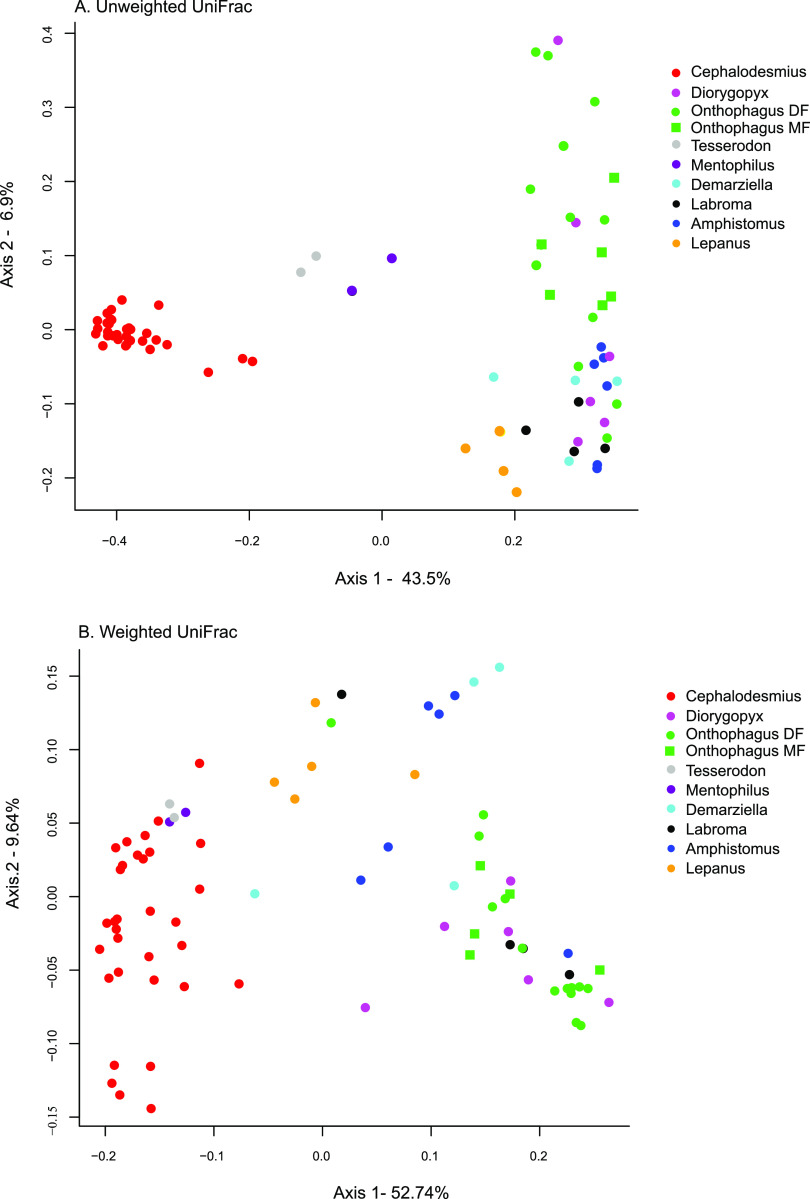
Comparisons of gut bacterial communities between beetle samples were made using unweighted and weighted UniFrac measures of diversity: the unweighted analysis takes into account the presence or absence of a particular bacterial sequence (operational taxonomic unit [OTU]) and its phylogenetic relationship to other bacterial sequences, while the weighted analysis adds the abundance of the bacterial sequences (OTUs) to the analysis. A principal-coordinate analysis (PCoA) based on unweighted and weighted UniFrac distances (Fig. 2A and B, respectively) was used to visualize comparisons of bacterial community composition between beetle genera. Bacterial communities of all individuals across three species of Cephalodesmius formed a distinct cluster, indicating that they shared similar gut microbiota that differed from all the other genera (Fig. 2). In contrast, Onthophagus gut microbiota did not cluster closely together, indicating that few taxa were shared between individuals within the genus (Fig. 2). Four genera from the AuEG had a gut community composition that overlapped Onthophagus, indicating some shared taxa. Two of the genera from the AuEG (Mentophilus and Tesserodon) clustered closest to Cephalodesmius, suggesting a similar gut composition that was distinct from the rest of AuEG and Onthophagus. One of the AuEG (Lepanus) was loosely clustered and overlapped with some of the AuEG but did not overlap Onthophagus in the unweighted analysis (Fig. 2A). Overall, the gut community composition of most of the AuEG appears to have more in common with Onthophagus than Cephalodesmius, with the exception of Mentophilus and Tesserodon.
FIG 2.
Comparisons of gut microbial community diversity between nine genera of dung beetles. β diversity measures were compared using a principal-coordinate analysis of unweighted UniFrac (A) and weighted UniFrac (B) distances. Each point represents an individual beetle sample. Genera are designated by different colors. Species in the genus Onthophagus are designated separately as mushroom feeders (MF) or dung feeders (DF). Proportion of variance for each axis is denoted by the corresponding axis label.
Comparisons of gut bacterial community composition between dung beetle genera.
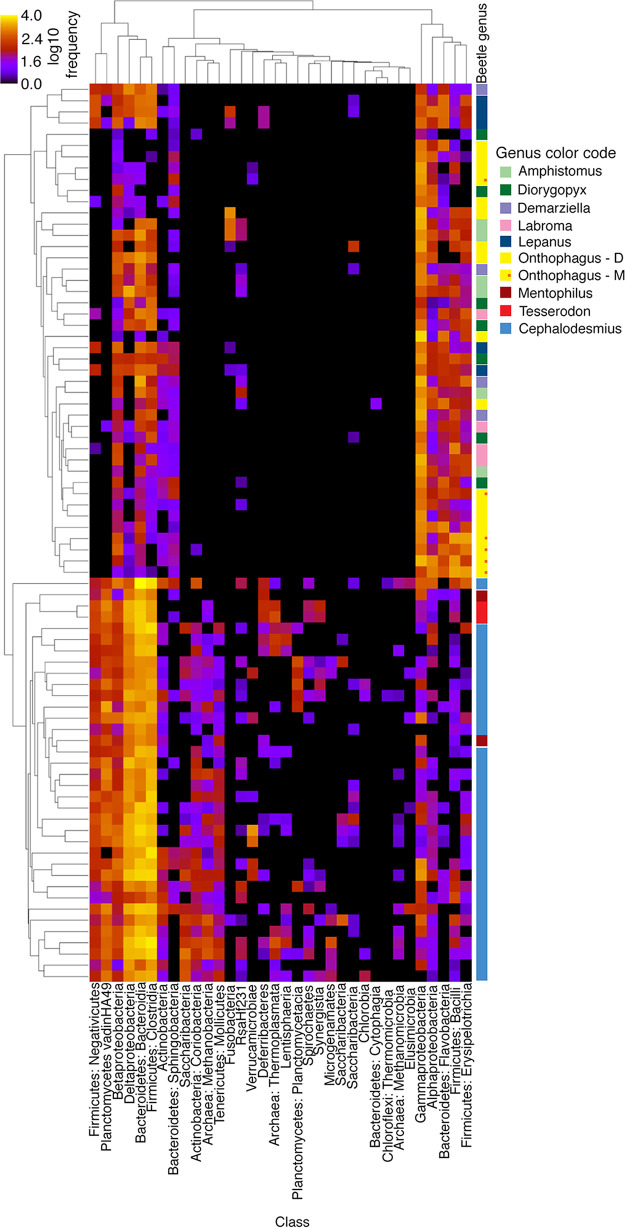
Three main bacterial phyla, Proteobacteria, Firmicutes, and Bacteroidetes, are commonly found in various proportions in the digestive tracts of other insects and animals and are also present in all nine beetle genera considered here (Fig. 3; see Fig. S12 in the supplemental material). At the taxonomic level of bacterial class, differences between dung beetle groups become much more apparent (Fig. 3). The Onthophagus gut community is dominated by the class Gammaproteobacteria within the phylum Proteobacteria. The mushroom-feeding Onthophagus beetles also contain Gammaproteobacteria but have a large proportion of the classes Bacilli and Erysipelotrichia from the phylum Firmicutes (Fig. 3; Fig. S12). In contrast, the Cephalodesmius gut community is dominated by three different classes of bacteria in roughly the same proportions: Clostridia (in the Firmicutes), Bacteroidia (in the Bacteroidetes), and Deltaproteobacteria (in the Proteobacteria) (Fig. 3; Fig. S12). The remaining AuEG have various gut community compositions. Some of the gut communities seen in the AuEG, such as Labroma and Diorygopyx, are dominated by Gammaproteobacteria, similar to Onthophagus, but gut communities in other AuEG, such as Mentophilus and Tesserodon, share the bacterial classes of Deltaproteobacteria, Clostridia, and Bacteroidia with Cephalodesmius (Fig. 3). Further comparisons of the gut communities using a heat map representation of the gut taxa from each sample shows a clear relationship between the gut communities of Cephalodesmius, Mentophilus, and Tesserodon distinct from the other genera (Fig. 4). Traces of Archaea were present in only a few of the samples (Fig. 3).
FIG 3.
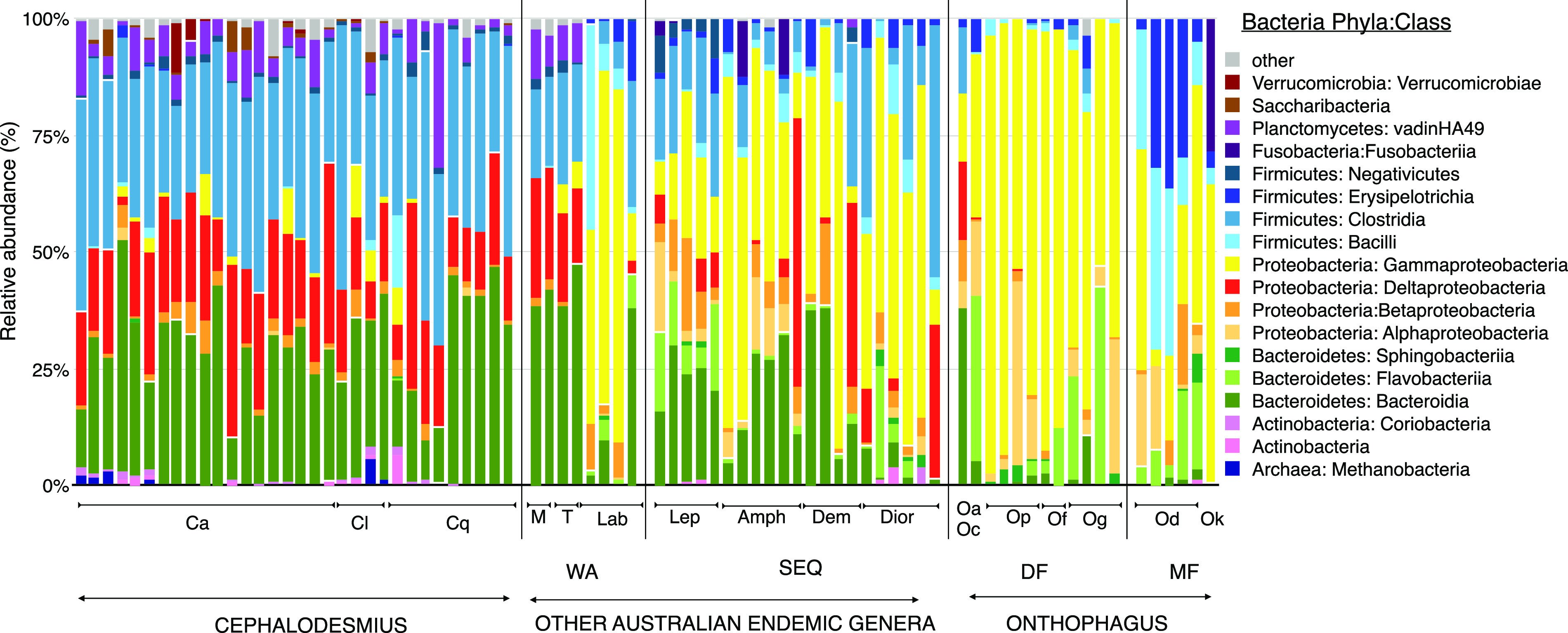
Relative abundance of bacteria (shown as phylum: class) and archaea in dung beetle gut samples. The bacterial classes are color coded by phyla, such that shades of green represent Bacteroidetes, yellow-orange-red are Proteobacteria classes, and shades of blue represent Firmicutes. Each bar represents a beetle sample. Members of the beetle genus Cephalodesmius are on the left (Ca, Cephalodesmius armiger; Cl, C. laticollis; Cq, C. quadridens). The other Australian endemic genera are separated into genera collected from Western Australia (WA) (M, Mentophilus; T, Tesserodon; Lab, Labroma) and southeast Queensland (SEQ) (Lep, Lepanus; Amph, Amphistomus; Dem, Demarziella; Dior, Diorygopyx). The Onthophagus gut community is shown on the right. DF indicates the dung-feeding Onthophagus group, and MF indicates the mushroom-feeding Onthophagus group (Oa, O. arrilla; Oc, Onthophagus CQ2; Op, O. pugnax; Of, O. fuliginosus; Og, O. granulatus; Od, O. dunningi; Ok, O. kumbaingeri).
FIG 4.
Heat map showing the composition of the gut bacterial community present in each host beetle sample. Each row represents a different beetle specimen. Beetle genera for each row are indicated by color on the right. Each column shows the presence and abundance of bacteria at the taxonomic level of class. Heat map colors indicate abundance normalized to a log scale.
Comparisons of core bacteria of the gut microbial community at the family level.
Because bacteria make up a large part of the dung beetle diet, it is important to distinguish between bacteria that are stably associated with a taxon of beetle rather than being transient and presumably environmentally acquired. To do this, we identified core families of bacteria for each of the beetle genera (i.e., bacterial families found in over 92% of individuals in the genus). The focus on core bacterial families allowed us to make generalized comparisons between the gut microbiota of different dung beetle genera at the lowest reliable taxonomic level.
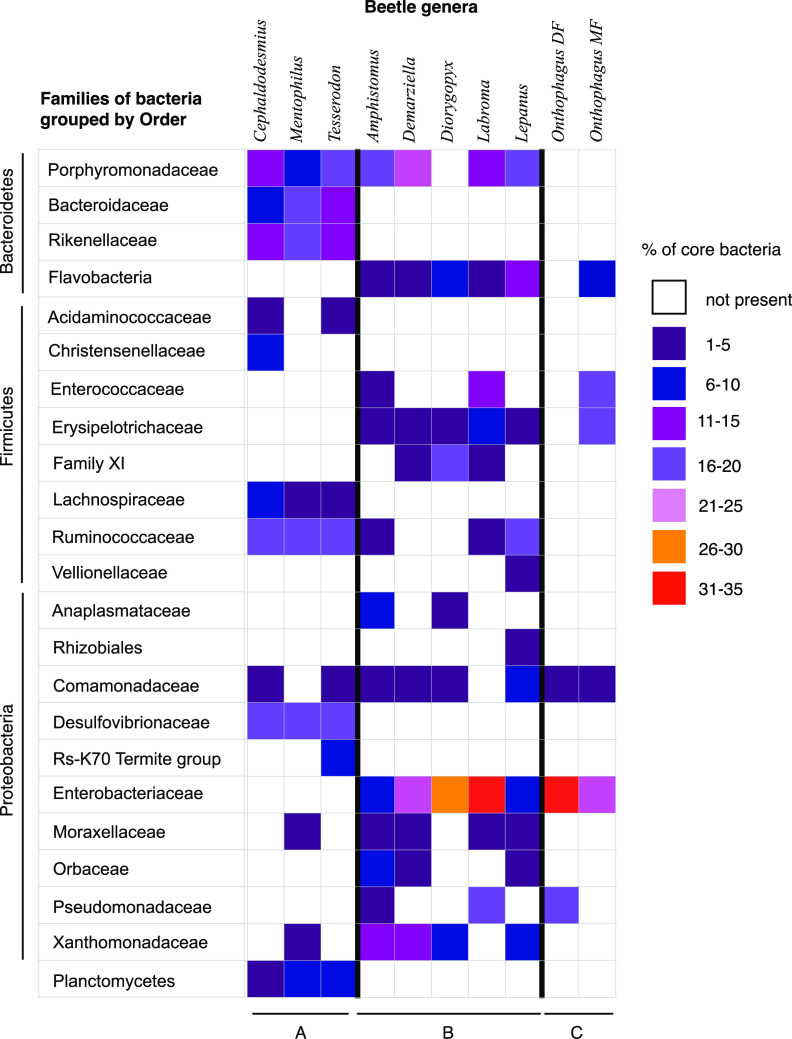
Thirty-two individuals from three species of Cephalodesmius had a consistent core gut community of 10 bacterial families regardless of the geographical location from which they were collected (Fig. 5; group A; see Fig. S3 in the supplemental material). These 10 core families made up an average of 84% of the total gut bacteria for all individuals. A large proportion of these families were anaerobic, fermentative bacteria, from the phyla Firmicutes and Bacteroidetes, including the families Ruminococcaceae and Lachnospiraceae, known to degrade complex plant material (37), and Rikenellaceae, which is common in fungus-cultivating termites and cockroaches and aids with the digestion of proteins (38–40). Fifteen percent of the gut bacteria were anaerobic, sulfate-reducing bacteria from the family Desulfovibrionaceae (Proteobacteria-Deltaproteobacteria), a family also found in the digestive tracts of fungus-cultivating termites, cockroaches, and humus-feeding scarab larvae (11, 38, 39). Five percent of the gut community of Cephalodesmius were from the phylum Planctomycetes, a phylum also found in detritivorous species (39, 41, 42), but not recorded in dung beetles, with the exception of a single detritivorous species that eats decaying leaves (27). Archaea were present in small amounts in some specimens but do not appear to be a consistent component of the hindgut fauna.
FIG 5.
Comparisons of core bacterial families found in the hindgut of nine genera of dung beetles. Colors represent different relative proportions of the bacterial families present in each genus. Group A contains the AuEG: Cephalodesmius, Mentophilus, and Tesserodon. Group B contains the remaining AuEG. Group C contains the two Onthophagus groups: dung feeding (DF) and mushroom feeding (MF).
Within the genus Onthophagus, the composition of the gut community varied considerably. Only three bacterial families could be identified as core in the 12 individuals from five species of dung-feeding Onthophagus beetles: Enterobacteriaceae and Pseudomonadaceae (Gammaproteobacteria) and Comamondaceae (Betaproteobacteria) make up 55% of the total gut bacteria (Fig. 5; group C; see Fig. S10 in the supplemental material). The gut community composition of the six individuals of two species of mushroom-feeding Onthophagus contained five core families, including two, the Enterobacteriaceae and Comamonadaceae, that were found in the dung-feeding Onthophagus beetles. Additionally, two families, Enterococcaceae and Erysipelotrichaceae, from the phylum Firmicutes were major components of the core gut microbiota of mushroom feeders (Fig. 5; group C; see Fig. S11 in the supplemental material). Only one minor core bacterial family (Comamonadaceae) was shared between the core microbiota of Onthophagus and Cephalodesmius (Fig. 5).
The core gut communities of the remaining endemic genera had bacterial families that were shared with either Onthophagus or Cephalodesmius (Fig. 5; group B; see Fig. S2 to S9 in the supplemental material). The core gut communities of Mentophilus and Tesserodon were most similar to those of Cephalodesmius, while the remaining five endemic genera had more families in common with Onthophagus, especially the mushroom-feeding Onthophagus (Fig. 5).
Further analysis of the gut community of Cephalodesmius collected from widely separated geographical regions revealed that even at the level of the OTU, several taxa were abundant and present in at least 80% of all samples from the three species (Table 3). Most notable were the sulfate-reducing anaerobic Desulfovibrio (Deltaproteobacteria) and the fermentative, anaerobic Clostridiales “Candidatus Soleaferrea” (Ruminococcaceae) and Tyzzerella (Lachnospiraceae). Similar analysis of Onthophagus samples revealed that none of the OTUs were abundant in more than 30% of samples in the dung feeders (Table 4). However, the gut communities from two mushroom-feeding species of Onthophagus all shared OTUs from the Firmicutes family Erysipelotrichaceae (Table 4).
TABLE 3.
Top five most abundant OTUs found in the hindgut of species of the dung beetle genus Cephalodesmiusa
| Taxonomy of top 5 OTUs found in 80% of samples | No. of: |
|
|---|---|---|
| Samples | Reads | |
| Deltaproteobacteria | ||
| Desulfovibrio sp. 1 | 32 | 59,177 |
| Desulfovibrio sp. 2 | 31 | 52,519 |
| Bacteroidetes | ||
| Rikenellaceae: Alistipes | 27 | 27,217 |
| Firmicutes: Clostridiales | ||
| Ruminococcaceae: “Candidatus Soleaferrea” | 32 | 10,727 |
| Lachnospiraceae: Tyzzerella | 27 | 10,034 |
There were three species of Cephalodesmius (n = 32 beetles). The OTUs are identified to the lowest taxonomic level available. The total number of reads was 989,105, and the total number of OTUs was 1,157.
TABLE 4.
Top five most abundant OTUs found in the hindgut of the dung beetle genus Onthophagusa
| Onthophagus group | Taxonomy of top 5 OTUs found in 30% of samples | No. of: |
|
|---|---|---|---|
| Samples | Reads | ||
| Dung feeding | Gammaproteobacteria | ||
| Enterobacteriaceae: Providencia | 4 | 12,238 | |
| Enterobacteriaceae: Escherichia-Shigella | 5 | 1,463 | |
| Bacteroidetes | |||
| Flavobacteriaceae | 5 | 9,124 | |
| Alphaproteobacteria | |||
| Wolbachia | 5 | 3,071 | |
| Bacteroidetes: Chitinophagaceae | 9 | 415 | |
| Mushroom feeding | Firmicutes | ||
| Erysipelotrichaceae: Erysipelothrix | 6 | 9,390 | |
| Erysipelotrichaceae | 6 | 6,237 | |
| Enterococcaceae: Vagococcus | 6 | 3,238 | |
| Gammaproteobacteria | |||
| Enterobacteriaceae: Morganella | 6 | 1,672 | |
| Bacteroidetes: Chitinophagaceae | 6 | 433 | |
In the dung-feeding Onthophagus beetles (n = 13), there were no abundant OTUs common to more than 9 of the samples: most OTUs occurred in 30% of samples. There were 118,046 reads and 230 OTUs total. In the mushroom-feeding Onthophagus beetles (n = 6), the five most abundant OTUs were found in all six samples. There were 89,892 reads and 128 OTUs. OTUs are identified to the lowest taxonomic level available.
Hindgut morphology comparisons.
The observed differences in microbial communities might be anticipated to coincide with differences in gut structure if features of the gut were associated with unique functions of the microbiota. Two primary and two intermediate hindgut morphologies were observed in the 21 dung beetle species in this study (Fig. 6; Fig. S2 to S11), and these differences did indeed coincide with composition of the microbiota. The first was characteristic of the genus Onthophagus. It consisted of a short hindgut configured in a simple U-shape bend starting from the pylorus (valve between midgut and hindgut), with no dilation of the anterior region (Fig. 6A and B). The second form was characteristic of the genus Cephalodesmius. In the second form, the hindgut was lengthened about 2-fold to form two loops, with a dilation of the anterior region of the hindgut just after the pylorus (Fig. 6E). Intermediate forms exhibited lengthening but no dilation of the anterior region (Fig. 6F) or dilation of the anterior hindgut, but less lengthening (Fig. 6C and D). Differences in hindgut morphology coincided with differences in gut microbiota. The beetle genera with dilated anterior hindguts (Cephalodesmius, Mentophilus, and Tesserodon) shared a distinctly different microbiota containing Deltaproteobacteria and Planctomycetes, while those beetle genera with short, nondilated hindguts were dominated by Gammaproteobacteria.
FIG 6.
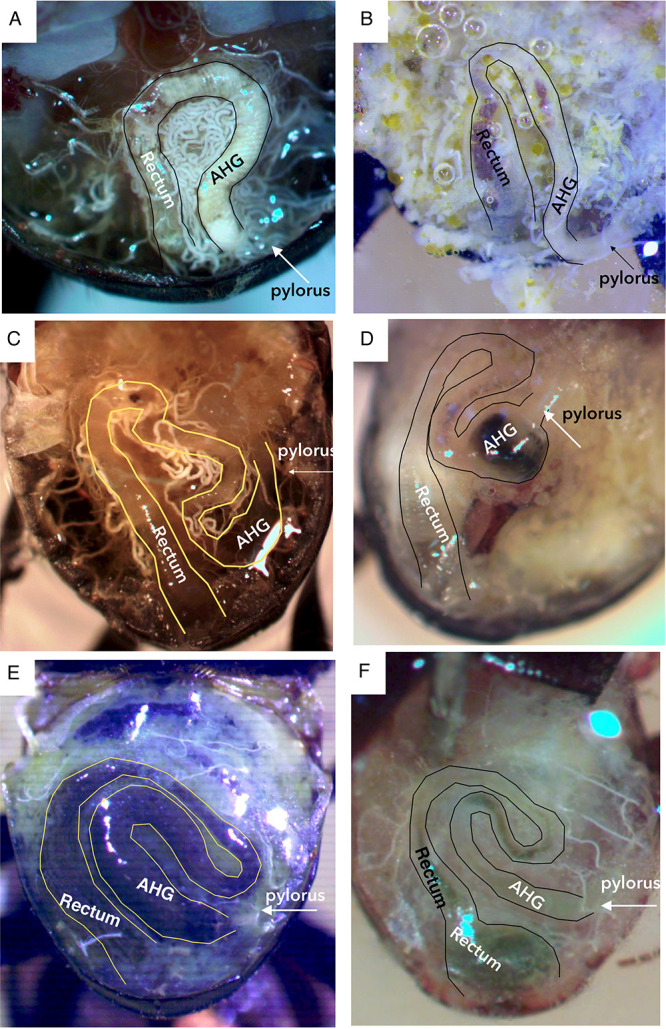
A dorsal view of in situ gross hindgut morphologies seen in six representative adult dung beetle species. (A) Onthophagus pugnax. The hindgut is short, with only a single U-shaped bend, and it does not have an enlarged anterior hindgut (AHG). (B) Amphistomus NSW1 also has a single U-shaped bend and a narrow anterior hindgut region. (C) Mentophilus hollandiae has a short, dilated region just after the pylorus, followed by a slight clockwise loop, which then loops anticlockwise. (D) Tesserodon pilicrepus also shows a short, dilated region in the anterior hindgut. (E) Cephalodesmius quadridens. From the pylorus, the anterior hindgut dilates, then it loops in a clockwise direction before contracting and reversing to an anticlockwise loop, which dilates again and terminates with the rectum. (F) Lepanus NSW2 hindgut shows an anterior hindgut looped clockwise first, then looped anticlockwise, without dilation in the anterior hindgut. The pylorus marks the beginning of the hindgut.
DISCUSSION
Australian dung beetles provide an opportunity to explore the adaptive potential of gut microbiota due to their phylogenetic diversification and different feeding strategies, which include dietary diversification and elaborate food processing behavior, as well as distinctive hindgut morphologies. Each of these factors has the potential to significantly influence the gut microbial community and, in turn, be influenced by the resident gut microbes, providing an opportunity to identify potential associations.
An important indication of whether or not the microbiota contributes to the evolution of their host is whether the composition of the microbiota follows the evolutionary trajectory of the host. It is clear from our results that the microbiota of three genera of the AuEG, most notably Cephalodesmius, are distinct from the microbiota of phylogenetically distinct Australian Onthophagus species, while the remaining members of AuEG exhibit some commonalities. The differences in microbiota within the AuEG may be related to their phylogenetic relationships. Tarasov and Genier (43) have proposed that the Australian endemic genera should be divided into two separate clades. Cephalodemius and Mentophilus are considered to be in a separate clade from several of the other AuEG, suggesting a phylogenetic component to the differences in microbiota.
An important caveat is that an apparent difference in microbiota between distinct taxa could simply relate to the location from which the insects were sampled. For Onthophagus, this appeared to be the case, as the composition of the microbial gut community was quite variable even between beetles of the same species. Environmental acquisition of microbial gut fauna has been noted in Onthophagus by others and may contribute to its adaptability (5). Onthophagus is a cosmopolitan genus that readily disperses to new environments (28, 44). It has been suggested that animals that are not dependent upon gut symbionts may be more able to switch to new habitats or food resources (45). The cosmopolitan distribution of Onthophagus species provides evidence for their adaptability, as does the fact that some have shifted from coprophagy to diets of carrion, fungi, or fruits in some habitats (29, 31, 46–50).
The situation within the genus Cephalodesmius, however, is quite different. Rather than having the variability of the microbiota seen within Onthophagus, Cephalodesmius species share a conserved gut microbiota of 10 core bacterial families, whereas Onthophagus has only two. Even at the level of the OTU, several bacterial taxa are conserved across all three species of Cephalodesmius. The conservation of the core microbiota in Cephalodesmius species is particularly remarkable in that the gut community composition is conserved across isolated populations in disjunct patches of remnant rainforest. The fact that the core microbiota is stable across geographic regions despite Cephalodesmius beetles being flightless suggests that the core microbiota has been stable for a long time. This stability of the association between the insect host and its microbiota is indicative of a mutually beneficial relationship that is under stabilizing selection.
An obvious possibility is that the microbiota might simply correlate with diet. Superficially, it would seem that food choice and microbiota are related as Onthophagus and Cephalodesmius exhibit the most extreme dietary preferences and also have the most divergent microbiota, with the remaining AuEG having intermediate food preferences and microbiota. We find, however, that specific food preferences are not reflected in the overall composition of the gut microbial community or in specific gut microbe taxa. Almost all of the dung beetles sampled are attracted to dung, yet some are attracted to a wider variety of non-dung food sources. Those species that feed mainly on dung did not share a consistent gut microbiota. Similarly, dung beetles with broader, varied diets did not share a consistent gut microbiota either, although there are some common taxa. For example, both Lepanus and Cephalodesmius species have been collected at a variety of different types of bait (30), yet the two genera had different gut communities. This indicates that other factors, in addition to diet, influence the gut community composition.
There is another intriguing possibility: that a relationship between diet and gut microbiota of dung beetles does exist, but that it relates to the way food is processed and the brood are fed. Onthophagus beetles are opportunistic, strong flyers that locate and exploit ephemeral, fresh dung resources. They dig tunnels beneath a dung source, where the female deposits dung provisions for her brood, lays eggs, then moves on to other fresh dung resources, thus producing a large number of offspring but engaging in minimal parental care (34). In contrast, Cephalodesmius beetles are flightless and inhabit remnant rainforests. Consistent with their isolation and restricted mobility, members of the genus Cephalodesmius rely on dependable, local food resources, which may, however, require processing to improve the nutritional value. The food processing activity of Cephalodesmius species involves a pair-bonded male and female, working together to gather dung, carrion, fungi, leaves, fruits, or flowers into a permanent nest burrow, where it is manipulated into a ball of composting material called a brood mass (33). Adult feces are added to this material, in essence inoculating the brood mass with hindgut microbiota. Both parents maintain the brood mass throughout the development of their offspring in order to provide continuous provisions. Not as much is known about the nesting behavior of the other AuEG, but they, like Cephalodesmius species, have low fecundity and transfer food resources to a new location away from the source (telecoprids), in contrast to Onthophagus species, which have high fecundity and bury the food resource at its source (paracoprids).
This study revealed that the gut community of Cephalodesmius beetles has more in common with gut communities of insect detritivores than with that of other coprophagic dung beetles. In a meta-analysis of insect gut communities, the most basal split in community composition separated detritivores and xylophages (dead wood feeding) from all other dietary guilds (2). The detritivore gut microbiota is distinctive, dominated by Clostridiales, Bacteroidales, and Deltaproteobacteria—the same classes of bacteria we see in Cephalodesmius. These groups form only minor components in the gut community of nondetritivorous insect diet guilds (2). The higher termites comprise much of the detritivore guild (51); however, convergences to the same gut community structure are seen in other detritivores, such as humus-feeding scarab larvae (11) and detritus-feeding fly larvae (15). Subsequently, gut microbiota studies of a number of omnivorous insects have revealed a picture of different degrees of convergence to this detritivore-type microbiota: they include field crickets (10), the New Zealand weta (Orthoptera) (42), and cockroaches (39). An emerging picture is that detritivores tend to converge on a similar gut community, but each major taxonomic group has unique components.
A common anatomical feature shared between Cephalodesmius and other detritivores is the expanded hindgut, which appears to have a significant impact on the composition and function of the gut microbiota. Similar gut alterations have never been observed in Onthophagus, providing support for the notion that the structural changes may be closely tied to the retention of the distinctive Cephalodesmius microbiota across evolutionary time and geographical distance. It is interesting to note that members of two other genera in the AuEG, Mentophilus and Tesserodon, have microbiota very similar to that of Cephalodesmius species, as well as a degree of hindgut dilation. The detritivore-type microbial community, the diverse and stable core microbiota, and corresponding gut dilation in the three beetle genera Cephalodesmius, Mentophilus, and Tesserodon support the hypothesis that the expanded anterior hindgut is functioning as a fermentation chamber that houses symbiotic bacteria to assist with digestion. The distinctive digestive system may have provided an essential microhabitat for the establishment of environmental bacteria in the gut, therefore allowing Cephalodesmius to exploit a new behavioral niche.
The most abundant OTUs common to all the Cephalodesmius samples were Desulfovibrio in the Deltaproteobacteria. Desulfovibrio has also been found to be abundant in the hindgut of larvae in other scarab subfamilies (17, 18). In Melolontha larvae, Desulfovibrio species specifically colonize the hindgut wall, while many of other gut bacteria are restricted to the gut lumen, suggesting that the sulfate-reducing Desulfovibrio species are adapted to colonize this microhabitat (18). It would seem plausible that environmentally acquired Desulfovibrio could be preferentially selected and adapted to live in this particular gut microhabitat in adult Cephalodesmius beetles, similar to what has been documented in cockroaches (52).
Why do Cephalodesmius, Mentophilus, and Tesserodon beetles share a core community that more closely resembles that of detritivores such as cockroaches than it does that of pure dung feeders such as members of the Onthophagus? The community likeness does not appear to be habitat based because while Cephalodesmius beetles live in rainforest areas in eastern Australia, Mentophilus and Tesserodon beetles are found in arid, open regions along the dry central coastal regions of Western Australia. The only behavioral observations of Mentophilus and Tesserodon beetles indicate that they bury old, dried fecal pellets deep in the ground below the moisture line (32), where the moisture may revive bacterial and fungal activity in the fecal pellets, thus providing a microbial food source for the beetles or their larvae. It is possible that feeding on once-dried fecal pellets necessitates a core gut microbiota more typical of detritus feeders, and Cephalodesmius beetles’ feeding on composting organic matter places the same demands. It has also been noted that Mentophilus beetles have been found frequently under mushrooms and once feeding on dead beetle larvae (32). A closer examination of Mentophilus and Tesserodon beetle behavior is warranted to establish whether their diet includes additional detritus components or even possibly fungus cultivation.
In beetles from the Cephalodesmius genus, we suspect that the detritus-associated microbiota is associated with the fact that the brood ball materials they consume are made of composted plant and organic matter, fed upon and added to over time. This is evidenced by the abundance of plant-degrading bacteria in their gut community. Also, the filtering mandibles used by adult dung feeders to obtain nutrients from fresh dung might not be as effective when compost is the food source, leading to a requirement for detritivore-style bacterial community and a gut fermentation chamber. A point to be borne in mind is that the diet and, consequently, the gut microbiota of the adult gut might also be related to larval nutrition. Larvae are reliant on nutrients in the brood ball, and consequently, the adults might carry a microbiota whose substantial role is to facilitate larval nutrition, as reported for members of the dung beetle genus Euoniticellus (26).
The question arises that since dung beetles arose from detritivorous ancestors (53), are the diet and gut microbiota found in Cephalodesmius species an ancestral condition? Monteith and Storey (33) suggest that the advanced food processing and complex subsocial behaviors seen in Cephalodesmius beetles are highly specialized rather than ancestral. To understand the evolution of these gut microbial communities, it would be useful to examine the gut microbiota in other dung beetles that are closely related to members of the Cephalodesmius, in addition to more primitive beetle relatives, such as those in Geotrupidae. Examination of the gut microbiota of other dung beetles from the two different clades of AuEG may provide further evidence of phylogenetic associations in gut community composition.
Conclusions.
Overall, this study has revealed an unexpected diversity in the gut microbiota of dung beetles, which may have facilitated the adaptation and evolution required to expand into new habitats or new behaviors. This is most apparent in the dichotomy between Onthophagus and Cephalodesmius species. Our findings suggest that a stable evolutionary partnership between members of the genus Cephalodesmius and its highly conserved gut microbiota may have allowed the exploitation of abundant, but low-quality, non-dung food resources. The strongly conserved microbiota across evolutionary time and geographical isolation indicates a coevolved, mutually beneficial association. The distinctive morphology of the gut of Cephalodesmius beetles may provide a fermentation chamber that facilitates the function of the microbiota or that ensures that essential microbes are retained. It seems likely that the microbiota has helped members of the Cephalodesmius to adapt to a niche where dung is less abundant, leading to the coevolution of behaviors required for the processing of alternative food resources. The time and effort required to process the food items that are collected necessitates the continuous provisioning of the brood, resulting in extended biparental care. Clearly, multiple factors affect the composition of gut microbiota, and further examination of gut microbial communities will add to the overall understanding of the evolutionary influence of gut microbiota.
MATERIALS AND METHODS
Beetle collection.
A total of 81 dung beetle specimens from nine genera and 21 species were collected from southeast Queensland and Western Australia between 2016 and 2018 (see Fig. S1 and Table S1 in the supplemental material). Beetle species were selected to represent different phylogenetic groups (Onthophagini and Australian endemic genera [AuEG]) as well as different diet groups. Beetles were collected mainly from southeast Queensland in areas where sampling has been done extensively in the past so that the dung beetles could be readily identified. Western Australian species were included in order to have additional species from a different biogeographical region. Of the 21 species, seven were from Onthophagus, three were from Cephalodesmius, and 11 were from other AuEG (Table S1). Sampling was particularly focused on areas where Cephalodesmius beetles occur, since this was a genus of particular interest.
Beetles were collected alive: either from pitfall traps baited with kangaroo (Macropus giganteus) dung or rotting mushroom or directly excavated from nest burrows. Specimens were identified to the species level. Undescribed species were recorded according to the nomenclature coding system devised by Geoff Monteith (Queensland Museum, Brisbane, QLD) and Tom Weir (Australian National Insect Collection, Canberra, ACT) for Australian museum collections. Voucher specimens are stored at the Queensland Museum.
Dissections.
After collection, beetles were housed for 4 to 5 days in plastic containers without food prior to dissections. Beetles were euthanized by being placed in a freezer (–20°C) for at least 1 h and then were dissected immediately upon removal. All dissection equipment was sterilized in 30% sodium hypochlorite solution and then rinsed in distilled water. Beetles were surface sterilized by immersion in 75% ethanol for 1 min prior to dissection. The dissections took place in three stages: (i) removal of elytra, (ii) removal of dorsal abdominal cuticle, with the hindgut photographed in situ, and then (iii) removal of anterior hindgut (starting from the end of the narrow pyloric valve to the first bend of the hindgut). Tools were sterilized between each stage to limit contamination of the final sample.
Prior to the removal of the anterior hindgut from the abdominal cavity, the entire hindgut was photographed in situ for each species using a Touptek USB microscope camera and Touplite photographic software for MacOS.
DNA extraction.
DNA was extracted from each sample using the HotSHOT DNA extraction procedure (54). Hindgut samples were immersed in alkaline lysis reagent (25 μM NaOH, 0.2 μM EDTA [pH 12]), mashed with a sterile pipette tip, heated to 95°C for 30 min, and then neutralized with Tris-HCl (40 μM). Samples were stored at –20°C until use.
DNA sequencing and taxon classification.
Amplicon sequencing of 16S rRNA genes was performed by the Australian Centre for Ecogenomics (ACE) (University of Queensland, St. Lucia, QLD, Australia) using the 803F forward primer (5′-TTAGAKACCCBNGTAGTC-3′) and the 1392wR reverse primer (5′-ACGGGCGGTGWGTRC-3′) for the V6 to V8 region on the MiSeq sequencing system (Illumina, Inc.). All sequence data were initially processed and quality filtered through the ACE pipeline with fastQC. Sequences were trimmed to 250 bases to remove primer sequences and poor-quality sequences using Trimmomatic (55) and then processed using the DADA2 denoising algorithm (56). Taxonomic assignments for sequences were based on 97% identity and obtained from the Silva and UNITE reference databases using BLAST+ (57).
Sequence filtering and data analysis.
All processing and analyses of microbial data were conducted using QIIME 2-2018.2 Microbiome analysis software (58). All unassigned (failed to classify) and Eukaryota sequences and any sequences not identifiable beyond domain were removed. Sequences that only occurred in one individual or had a frequency of less than 100 reads were also removed. For diversity analyses, data were rarefied to a sampling depth of 2,500 (98.8%) reads to minimize effects of uneven sequence counts between samples.
Data were analyzed using the QIIME 2 Core Metrics Phylogeny, which aligns sequences phylogenetically to produce diversity data. Bacterial community diversity within samples (α diversity) was assessed using Shannon’s and Faith’s phylogenetic diversity (PD) indices (59, 60). Effects of beetle genus on alpha diversity were assessed using Kruskal-Wallis pairwise comparisons. Bacterial community diversity between beetle samples (β diversity) was calculated using unweighted and weighted UniFrac distances to assess phylogenetic diversity. UniFrac, an abbreviation of “unique fraction,” is a phylogenetic technique developed specifically for microbial communities that measures community similarity based on the bacterial lineages they contain (61). Principal-coordinate analysis (PCoA) was used to visualize the community similarity between beetle genera. Heat maps of the gut microbial communities in each beetle specimen were also generated using QIIME2.
Defining and analyzing core microbiota.
In order to make detailed comparisons, a core group of bacteria was identified for each genus of beetle. Since many of the bacterial sequences could not be reliably identified below the family level, we chose to look at core families, as this taxonomic level would achieve the most detail, yet still be reliable. Core families were determined to be those which were present in 100% of individuals within a given genus, with the exception of Onthophagus and Cephalodesmius. As these two genera had larger sample sizes, we defined core families as those being present in all but one individual (31 out of 32 for Cephalodesmius and 11 out of 12 for Onthophagus). The proportions of taxa comprising the core were calculated by dividing the number of core families by the total number of families. The quantitative contribution of each core bacterial family taxon was calculated as the proportion of reads assigned to each core relative to total reads for each beetle genus.
Data availability.
All 16S rRNA sequences are accessible in the NCBI Sequence Read Archive (SRA) under accession no. PRJNA638479.
Supplementary Material
ACKNOWLEDGMENTS
We are extremely grateful to Geoff Monteith from the Queensland Museum, who provided invaluable assistance as mentor for both fieldwork and species identification. We also thank Cara Conradsen for assistance with R coding.
K.M.E., P.R.E., and D.J.M. designed the project. K.M.E. and W.G.A. collected beetles. K.M.E. and W.G.A. performed the experiments, with guidance from P.R.E. K.M.E. and W.G.A. analyzed the data. K.M.E. and D.J.M. wrote the paper, with revisions from P.R.E. All authors have approved the manuscript.
Support for this study was provided by University of Queensland—Australian Postgraduate Award (APA) to K.M.E.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Engel P, Moran NA. 2013. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 2.Colman DR, Toolson EC, Takacs-Vesbach CD. 2012. Do diet and taxonomy influence insect gut bacterial communities? Mol Ecol 21:5124–5137. doi: 10.1111/j.1365-294X.2012.05752.x. [DOI] [PubMed] [Google Scholar]
- 3.Kolasa M, Ścibior R, Mazur MA, Kubisz D, Dudek K, Kajtoch Ł. 2019. How hosts taxonomy, trophy and endosymbionts shape microbiome diversity in beetles. Microb Ecol 78:995–1013. doi: 10.1007/s00248-019-01358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yun J-H, Roh SW, Whon TW, Jung M-J, Kim M-S, Park D-S, Yoon C, Nam Y-D, Kim Y-J, Choi J-H, Kim J-Y, Shin N-R, Kim S-H, Lee W-J, Bae J-W. 2014. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage and phylogeny of host. Appl Environ Microbiol 80:5254–5264. doi: 10.1128/AEM.01226-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker ES, Newton ILG, Moczek AP. 2020. (My microbiome) would walk 10,000 miles: maintenance and turnover of microbial communities in introduced dung beetles. Microb Ecol 80:435–446. doi: 10.1007/s00248-020-01514-9. [DOI] [PubMed] [Google Scholar]
- 6.Jones RT, Sanchez LG, Fierer N. 2013. A cross-taxon analysis of insect-associated bacterial diversity. PLoS One 8:e61218. doi: 10.1371/journal.pone.0061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raymann K, Moran NA. 2018. The role of the gut microbiome in health and disease of adult honey bee workers. Curr Opin Insect Sci 26:97–104. doi: 10.1016/j.cois.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gontang EA, Aylward FO, Carlos C, Glavina del Rio T, Chovatia M, Fern A, Lo C-C, Malfatti SA, Tringe SG, Currie CR, Kolter R. 2017. Major changes in microbial diversity and community composition across gut sections of a juvenile Panchlora cockroach. PLoS One 12:e0177189. doi: 10.1371/journal.pone.0177189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng SH, Stat M, Bunce M, Simmons LW. 2018. The influence of diet and environment on the gut microbial community of field crickets. Ecol Evol 8:4704–4720. doi: 10.1002/ece3.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tinker KA, Ottesen EA. 2016. The core gut microbiome of the American cockroach, Periplaneta americana, is stable and resilient to dietary shifts. Appl Environ Microbiol 82:6603–6610. doi: 10.1128/AEM.01837-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andert J, Marten A, Brandl R, Brune A. 2010. Inter- and intraspecific comparison of the bacterial assemblages in the hindgut of humivorous scarab beetle larvae (Pachnoda spp.). FEMS Microbiol Ecol 74:439–449. doi: 10.1111/j.1574-6941.2010.00950.x. [DOI] [PubMed] [Google Scholar]
- 12.Arias-Cordero E, Ping L, Reichwald K, Delb H, Platzer M, Boland W. 2012. Comparative evaluation of the gut microbiota associated with the below- and above-ground life stages (larvae and beetles) of the forest cockchafer, Melolontha hippocastani. PLoS One 7:e51557. doi: 10.1371/journal.pone.0051557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brune A, Dietrich C. 2015. Termites: digesting the diversity in the light of ecology and evolution. Annu Rev Microbiol 69:145–166. doi: 10.1146/annurev-micro-092412-155715. [DOI] [PubMed] [Google Scholar]
- 14.Canhoto C, Graca MAS. 2006. Digestive tract and leaf processing capacity of the stream invertebrate Tipula lateralis. Can J Zool 84:1087–1095. doi: 10.1139/z06-092. [DOI] [Google Scholar]
- 15.Cook DM, DeCrescenzo Henriksen E, Upchurch R, Peterson JBD. 2007. Isolation of polymer-degrading bacteria and characterization of the hindgut bacterial community from the detritus-feeding larvae of Tipula abdominalis (Diptera: Tipulidae). Appl Environ Microbiol 73:5683–5686. doi: 10.1128/AEM.00213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hongoh Y. 2011. Toward the functional analysis of uncultivable, symbiotic microorganisms in the termite gut. Cell Mol Life Sci 68:1311–1325. doi: 10.1007/s00018-011-0648-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang S, Zhang H. 2013. The impact of environmental heterogeneity and life stage on the hindgut microbiota of Holotrichia parallela larvae (Coleoptera: Scarabaeidae). PLoS One 8:e57169. doi: 10.1371/journal.pone.0057169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egert M, Stingl U, Bruun LD, Pommerenke B, Brune A, Friedrich MW. 2005. Structure and topology of microbial communities in the major gut compartments of Melolontha melolontha larvae (Coleoptera: Scarabaeidae). Appl Environ Microbiol 71:4556–4566. doi: 10.1128/AEM.71.8.4556-4566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alonso-Pernas P, Arias-Cordero E, Novoselov A, Ebert C, Rybak J, Kaltenpoth M, Westermann M, Neugebauer U, Boland W. 2017. Bacterial community and PHB-accumulating bacteria associated with the wall and specialized niches of the hindgut of the forest cockchafer (Melolontha hippocastani). Front Microbiol 8:291. doi: 10.3389/fmicb.2017.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank K, Brückner A, Hilpert A, Heethoff M, Blüthgen N. 2017. Nutrient quality of vertebrate dung as a diet for dung beetles. Sci Rep 7:12141. doi: 10.1038/s41598-017-12265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holter P. 2016. Herbivore dung as food for dung beetles: elementary coprology for entomologists. Ecol Entomol 41:367–377. doi: 10.1111/een.12316. [DOI] [Google Scholar]
- 22.Rougon D, Rougon C, Levieux J, Trichet J. 1990. Variations in the amino-acid content in zebu dung in the Sahel during nesting by dung beetles (Coleoptera, Scarabaeideae). Soil Biol Biochem 22:217–223. doi: 10.1016/0038-0717(90)90090-M. [DOI] [Google Scholar]
- 23.Halffter G, Matthews EG. 1971. The natural history of dung beetles. A supplement on associated biota. Rev Latinoam Microbiol 13:147–164. [PubMed] [Google Scholar]
- 24.Estes AM, Hearn DJ, Snell-Rood EC, Feindler M, Feeser K, Abebe T, Dunning Hotopp JC, Moczek AP. 2013. Brood ball-mediated transmission of microbiome members in the dung beetle, Onthophagus taurus (Coleoptera: Scarabaeidae). PLoS One 8:e79061. doi: 10.1371/journal.pone.0079061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwab DB, Riggs HE, Newton ILG, Moczek AP. 2016. Developmental and ecological benefits of the maternally transmitted microbiota in a dung beetle. Am Nat 188:679–692. doi: 10.1086/688926. [DOI] [PubMed] [Google Scholar]
- 26.Shukla SP, Sanders JG, Byrne MJ, Pierce NE. 2016. Gut microbiota of dung beetles correspond to dietary specializations of adults and larvae. Mol Ecol 25:6092–6106. doi: 10.1111/mec.13901. [DOI] [PubMed] [Google Scholar]
- 27.Franzini PZN, Ramond JB, Scholtz CH, Sole CL, Ronca S, Cowan DA. 2016. The gut microbiomes of two Pachysoma Macleay desert dung beetle species (Coleoptera: Scarabaeidae: Scarabaeinae) feeding on different diets. PLoS One 11:e0161118-19. doi: 10.1371/journal.pone.0161118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breeschoten T, Doorenweerd C, Tarasov S, Vogler AP. 2016. Phylogenetics and biogeography of the dung beetle genus Onthophagus inferred from mitochondrial genomes. Mol Phylogenet Evol 105:86–95. doi: 10.1016/j.ympev.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Gunter NL, Monteith GB, Cameron SL, Weir TA. 2019. Evidence from Australian mesic zone dung beetles support Gondwanan vicariance and Mesozoic origin of the Scarabaeinae (Coleoptera: Scarabaeidae). Insect Syst Evol 50:162–188. doi: 10.1163/1876312X-00002171. [DOI] [Google Scholar]
- 30.Ebert KM, Monteith GB, Menéndez R, Merritt DJ. 2019. Bait preferences of Australian dung beetles (Coleoptera: Scarabaeidae: Scarabaeinae) in tropical and subtropical Queensland forests. Aust Entomol 58:772–782. doi: 10.1111/aen.12396. [DOI] [Google Scholar]
- 31.Matthews EG. 1972. A revision of the scarabaeine dung beetles of Australia. I. Tribe Onthophagini. Aust J Zool Suppl Ser 9:1–330. [Google Scholar]
- 32.Matthews EG. 1976. A revision of the scarabaeine dung beetles of Australia. II. Tribe Scarabaeini. Aust J Zoo Suppl 24:1–211. doi: 10.1071/AJZS038. [DOI] [Google Scholar]
- 33.Monteith GB, Storey RI. 1981. The biology of Cephalodesmius, a genus of dung beetles which synthesizes “dung” from plant material (Coleoptera: Scarabaeidae: Scarabaeinae). Memoirs Queensland Museum 20:253–277. [Google Scholar]
- 34.Halffter G, Edmonds WD. 1982. The nesting behavior of dung beetles (Scarabaeinae)—an ecological and evolutive approach. Instituto de Ecologia, Mexico D.F., Mexico. [Google Scholar]
- 35.Lopez-Guerrero Y. 2002. Anatomy and histology of the digestive system of Cephalodesmius armiger Westwood (Coleoptera, Scarabaeidae, Scarabaeinae). Coleopterists Bull 56:97–106. doi: 10.1649/0010-065X(2002)056[0097:AAHOTD]2.0.CO;2. [DOI] [Google Scholar]
- 36.Douglas AE. 2015. Multiorganismal insects: diversity and function of resident microorganisms. Annu Rev Entomol 60:17–34. doi: 10.1146/annurev-ento-010814-020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biddle A, Stewart L, Blanchard J, Leschine S. 2013. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 5:627–640. doi: 10.3390/d5030627. [DOI] [Google Scholar]
- 38.Otani S, Mikaelyan A, Nobre T, Hansen LH, Kone NA, Sorensen SJ, Aanen DK, Boomsma JJ, Brune A, Poulsen M. 2014. Identifying the core microbial community in the gut of fungus-growing termites. Mol Ecol 23:4631–4644. doi: 10.1111/mec.12874. [DOI] [PubMed] [Google Scholar]
- 39.Schauer C, Thompson CL, Brune A. 2012. The bacterial community in the gut of the cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites. Appl Environ Microbiol 78:2758–2767. doi: 10.1128/AEM.07788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauer E, Lampert N, Mikaelyan A, Köhler T, Maekawa K, Brune A. 2015. Physicochemical conditions, metabolites and community structure of the bacterial microbiota in the gut of wood-feeding cockroaches (Blaberidae: Panesthiinae). FEMS Microbiol Ecol 91:1–14. doi: 10.1093/femsec/fiu028. [DOI] [PubMed] [Google Scholar]
- 41.Makonde HM, Boga HI, Osiemo Z, Mwirichia R, Mackenzie LM, Goker M, Klenk H-P. 2013. 16S-rRNA-based analysis of bacterial diversity in the gut of fungus-cultivating termites (Microtermes and Odontotermes species). Antonie Van Leeuwenhoek 104:869–883. doi: 10.1007/s10482-013-0001-7. [DOI] [PubMed] [Google Scholar]
- 42.Waite DW, Dsouza M, Biswas K, Ward DF, Deines P, Taylor MW. 2015. Microbial community structure in the gut of the New Zealand insect Auckland tree weta (Hemideina thoracica). Arch Microbiol 197:603–612. doi: 10.1007/s00203-015-1094-3. [DOI] [PubMed] [Google Scholar]
- 43.Tarasov S, Genier F. 2015. Innovative Bayesian and parsimony phylogeny of dung beetles (Coleoptera, Scarabaeidae, Scarabaeinae) enhanced by ontology-based partitioning of morphological characters. PLoS One 10:e0116671. doi: 10.1371/journal.pone.0116671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silva DP, Vilela B, Buzatto BA, Moczek AP, Hortal J. 2016. Contextualized niche shifts upon independent invasions by the dung beetle Onthophagus taurus. Biol Invasions 18:3137–3148. doi: 10.1007/s10530-016-1204-4. [DOI] [Google Scholar]
- 45.Hammer TJ, Janzen DH, Hallwachs W, Jaffe SP, Fierer N. 2017. Caterpillars lack a resident gut microbiome. Proc Natl Acad Sci U S A 114:9641–9646. doi: 10.1073/pnas.1707186114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bornemissza GF. 1971. Mycetophagous breeding in the Australian dung beetle, Onthophagus dunningi. Pedobiologia 11:133–142. [Google Scholar]
- 47.Storey RI, Weir TA. 1988. New localities and biological notes for the genus Onthophagus Latreille (Coleoptera: Scarabaeidae) in Australia. Aust Entomologist Mag 15:17–24. [Google Scholar]
- 48.Halffter G, Halffter V. 2009. Why and where coprophagous beetles (Coleoptera: Scarabaeinae) eat seeds, fruits or vegetable detritus. Bol Soc Entomologica Aragonesa 45:1–22. [Google Scholar]
- 49.Halffter G, Matthews EG. 1966. The natural history of dung beetles of the subfamily Scarabaeinae (Coleoptera, Scarabaeidae). Folia Entomol Mexicana 12–14:1–312. [Google Scholar]
- 50.Howden HF, Cartwright OL. 1963. Scarab beetles of the genus Onthophagus Latreille north of Mexico (Coleoptera: Scarabaeidae). Proc U S Natl Museum 114:1–135. doi: 10.5479/si.00963801.114-3467.1. [DOI] [Google Scholar]
- 51.Mikaelyan A, Meuser K, Brune A. 2017. Microenvironmental heterogeneity of gut compartments drives bacterial community structure in wood- and humus-feeding higher termites. FEMS Microbiol Ecol 93:fiw210. doi: 10.1093/femsec/fiw210. [DOI] [PubMed] [Google Scholar]
- 52.Mikaelyan A, Thompson CL, Hofer MJ, Brune A. 2016. Deterministic assembly of complex bacterial communities in guts of germ-free cockroaches. Appl Environ Microbiol 82:1256–1263. doi: 10.1128/AEM.03700-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scholtz CH, Davis ALV, Kryger U. 2009. Evolutionary biology and conservation of dung beetles. Pensoft Publishers, Sofia, Bulgaria. [Google Scholar]
- 54.Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT. Biotechniques 29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 55.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol Conservation 61:1–10. doi: 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- 60.Shannon CE. 2001. A mathematical theory of communication. SIGMOBILE Mob Comput Commun Rev 5:3–55. doi: 10.1145/584091.584093. [DOI] [Google Scholar]
- 61.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All 16S rRNA sequences are accessible in the NCBI Sequence Read Archive (SRA) under accession no. PRJNA638479.