Due to the worldwide emergence of A. fumigatus azole resistance, this opportunistic pathogen poses a serious health threat, and therefore, it has been included in the watch list in the CDC publication Antibiotic Resistance Threats in the United States, 2019 (CDC, 2019). Azoles play a critical role in the control and management of fungal diseases, not only in the clinical setting but also in agriculture.
KEYWORDS: Aspergillus fumigatus, azole resistance, azole drugs, DMIs, plant pathogens
ABSTRACT
Drug resistance poses a serious threat to human health and agricultural production. Azole drugs are the largest group of 14-α sterol demethylation inhibitor fungicides that are used both in agriculture and in clinical practice. As plant-pathogenic molds share their natural environment with fungi that cause opportunistic infections in humans, both are exposed to a strong and persistent pressure of demethylase inhibitor (DMI) fungicides, including imidazole and triazole drugs. As a result, a loss of efficacy has occurred for this drug class in several species. In the clinical setting, Aspergillus fumigatus azole resistance is a growing public health problem, and finding the source of this resistance has gained much attention. It is urgent to determine if there is a direct link between the agricultural use of azole compounds and the different A. fumigatus resistance mechanisms described for clinical triazoles. In this study, we performed A. fumigatus susceptibility testing against clinical triazoles and crop protection DMIs using a collection of azole-susceptible and -resistant strains which harbor most of the described azole resistance mechanisms. Various DMI susceptibility profiles have been found in the different A. fumigatus population groups based on their azole resistance mechanism and previous whole-genome sequencing (WGS) analysis, which suggests that the different resistance mechanisms have different origins and are specifically associated with the local use of a particular DMI.
IMPORTANCE Due to the worldwide emergence of A. fumigatus azole resistance, this opportunistic pathogen poses a serious health threat, and therefore, it has been included in the watch list in the CDC publication Antibiotic Resistance Threats in the United States, 2019 (CDC, 2019). Azoles play a critical role in the control and management of fungal diseases, not only in the clinical setting but also in agriculture. Thus, azole resistance leads to a limited therapeutic arsenal which reduces the treatment options for aspergillosis patients, increasing their mortality risk. Evidence is needed to understand whether A. fumigatus azole resistance is emerging from an agricultural source due to the extended use of demethylase inhibitors as fungicides or whether it is coming from somewhere else, such as the clinical setting. If the environmental route is demonstrated, the current use and management of azole antifungal compounds might be forced to change in the coming years.
INTRODUCTION
Aspergillus fumigatus is responsible for the increased incidence of invasive aspergillosis, with high mortality rates in some immunocompromised hosts (1). In this context, azole drugs play a major role in the prevention and treatment of these infections (2). Generally, these drugs are called demethylation inhibitors (DMIs) and are widely used because of their high efficiency and broad-spectrum activity; in fact, azoles are the only class of compounds that are used in both agriculture and clinical management (3, 4).
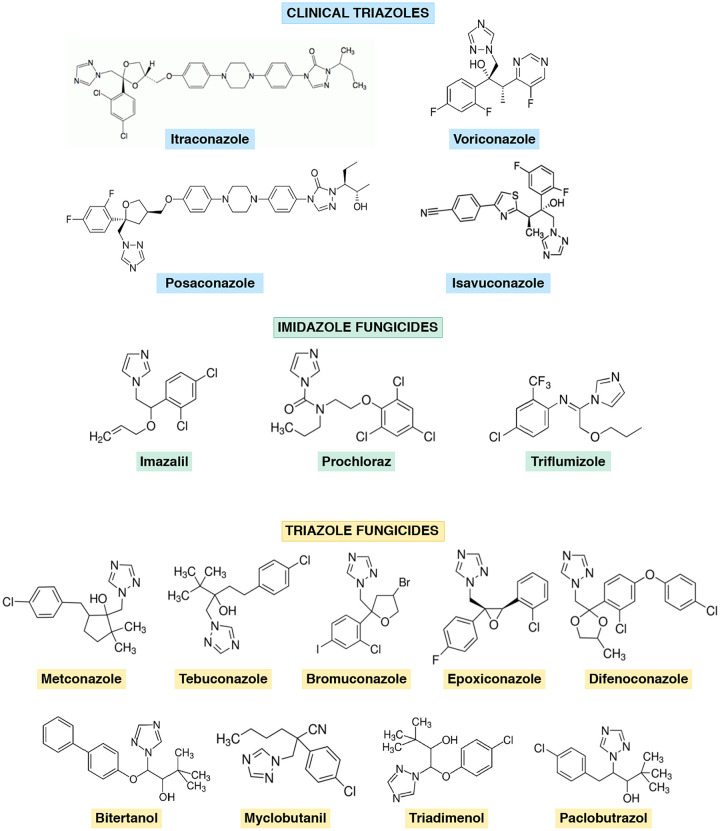
Azole drugs have dominated the agricultural fungicide market since they were approved in the 1970s; however, their capacity to induce resistance in the target pathogens is weaker than that of other agricultural fungicides. Chemically, azoles are divided into imidazoles and triazoles (5). Several azole drugs used in crop protection have a molecular structure similar to that of medical triazoles (Fig. 1), and cross-resistance between them has been demonstrated through lab evolution under selective pressure of agricultural azoles (6, 7). In the clinical setting, the introduction of azole drugs initiated a new era in therapy for systemic fungal diseases. Nowadays, the treatment of invasive aspergillosis mainly relies on triazole drugs approved in the late 1990s to 2000s, such as itraconazole (ITZ), voriconazole (VRZ), posaconazole (PSZ), and, more recently, isavuconazole (ISZ) (8).
FIG 1.
Chemical structures of clinical triazoles and demethylation inhibitor compounds used in this study, grouped as imidazole or triazole fungicides based on the number of nitrogen atoms in the azole aromatic ring.
Along with the increased use of DMI fungicides globally, a rise in the number of A.fumigatus azole-resistant isolates has been reported (2). This is especially worrisome due to the critical role that these drugs play in the control and management of fungal diseases. Azole resistance is directly associated with treatment failure; in fact, there is a subset of patients on azole prophylaxis who develop breakthrough aspergillosis that are theoretically untreatable because the use of azole is precluded, which leads to high mortality rates (9). Due to the worldwide emergence of azole resistance, A. fumigatus has been included in the watch list in the CDC publication Antibiotic Resistance Threats in the United States, 2019 (10).
Azole drugs act inhibiting the activity of Cyp51 enzymes, the azole target. Many filamentous fungi, particularly ascomycetes, harbor one, two, or even three cyp51 paralogous genes encoding these enzymes (11). In A. fumigatus, the azole target 14-α sterol demethylase is encoded by two paralogous genes (cyp51A and cyp51B) (12). In general, cyp51 mutations resulting in acquired azole resistance are usually restricted to just one paralog, most often cyp51A; thus, any cost associated with a change in the protein might be eluded by the other wild-type paralogs with an unchanged enzyme activity (13).
Multiple studies of human and plant pathogens have identified two main mechanisms of azole resistance, which are quite common in both scenarios: (i) mutations in the Cyp51 target resulting in decreased enzyme affinity for inhibitors and (ii) overexpression of the cyp51 target gene caused by insertions in the predicted promoter regions. Both azole resistance mechanisms can also appear in different Cyp51 combinations resulting in various azole susceptibility profiles (2, 14).
In plant pathogens, the variety of DMIs used for crop protection is high and sometimes the use of various compounds is the rule, which makes it more difficult to link a particular Cyp51 mutation to the specific use of a DMI. In addition, the number of resistance mechanisms and plant pathogens under investigation is quite diverse too (Table 1). However, some Cyp51 point mutations and promoter modifications are consistently found, independently or in combination, in several species of fungi (2, 15–22).
TABLE 1.
Main Cyp51 resistance mechanisms to DMIs found in plant pathogens from 2000 to 2020a
| Plant pathogen | DMI resistance | Cyp51 modification(s) | Promoter alteration | Overexpression | Cyp51 gene | Reference |
|---|---|---|---|---|---|---|
| Penicillium digitatum | TFZ, FNM, BTN | Absent | 126-bp TR | Yes | Cyp51 | 2 |
| IMZ | Absent | 199-bp TR | Yes | Cyp51B | 2 | |
| Blumeriella jaapii | FBZ | Absent | Truncated retrotransposon | Yes | Cyp51 | 2 |
| Venturia inaequalis | MCB | Absent | 553-bp insertion | Yes | Cyp51A | 2 |
| DFZ | Absent | EL3,1,2 repeated element | Yes | Cyp51A | 2 | |
| Monilinia fructicola | PPZ | Absent | Mona genetic element | Yes | Cyp51B | 2 |
| Ustilaginoidea virens | PPZ | Absent | CC insertion | Yes | Cyp51 | 2 |
| Pyrenopeziza brassicae | TBZ, MTZ, FSZ, PTZ, PRZ | G460S, S508T | 151-bp insertion | Yes | Cyp51 | 2 |
| Erysiphe necator | MCB, TBZ, FNM | Y136F | ND | Yes | Cyp51B | 2 |
| MCB | Y136F | ND | Yes | Cyp51 | 2 | |
| Puccinia triticina | EPZ | Y134F | ND | Yes | Cyp51B | 2 |
| Villosiclava virens | TBZ | Y137H | ND | Yes | Cyp51B | 15 |
| Pyrenophora teres | TBZ, MTZ, TRZ, DFZ, PRZ | F489L | ND | Yes | Cyp51A | 2 |
| Uncinula necator | TDM | Y136F | ND | No | Cyp51B | 2 |
| Erysiphe graminis f. sp. hordei (Blumeria graminis f. sp. hordei) | TDM, TBZ | Y136F, S509T | ND | ND | Cyp51B | 2 |
| BZZ | Y136F | ND | ND | Cyp51B | 2 | |
| TDM | Y136F, K147Q | ND | ND | Cyp51 | 2 | |
| Mycosphaerella graminicola (Zymoseptoria tritici) | TDM, TBZ, PRZ, TBZ, EPZ | Y137F, I381V, V136A, ΔY459, ΔG460 | ND | No | Cyp51 | 2 |
| TBZ, DFZ | I381V | ND | ND | Cyp51 | 2 | |
| TBZ, EPZ | Y461S, Y137F | ND | ND | Cyp51 | 16 | |
| PTZ, EPZ | S524T | ND | ND | Cyp51 | 16 | |
| Fusarium graminearum | TBZ | Y137H | ND | ND | Cyp51B | 17 |
| Penicillium digitatum | PRZ | Y136H, Q309H, G459S, F506I | ND | ND | Cyp51B | 18 |
| Ustilago maydis | PPZ | G464S | ND | ND | Cyp51 | 2 |
| Mycosphaerella fijiensis | TDM, FSZ, PPZ | Y136F, A313G, Y461D, Y463D/N/H | ND | No | Cyp51A | 19 |
| Cercospora beticola | TTZ | Absent | Absent | Yes | Cyp51B | 20 |
| EPZ | Absent | Absent | Yes | Cyp51 | 21 | |
| Sclerotinia homoeocarpa | PPZ | Absent | Absent | Yes | Cyp51 | 22 |
ND, not determined or not described; IMZ, imazalil; PRZ, prochloraz; TFZ, triflumizole; MTZ, metconazole; TBZ, tebuconazole; EPZ, epoxiconazole; BRZ, bromuconazole; DFZ, difenoconazole; BTN, bitertanol; MCB, myclobutanil; TDM, triadimenol; PPZ, propiconazole; FNM, fenarimol; FBZ, fenbuconazole; FSZ, flusilazole; PTZ, prothioconazole; BZZ, benzimidazole; TTZ, tetraconazole; TRZ, triticonazole.
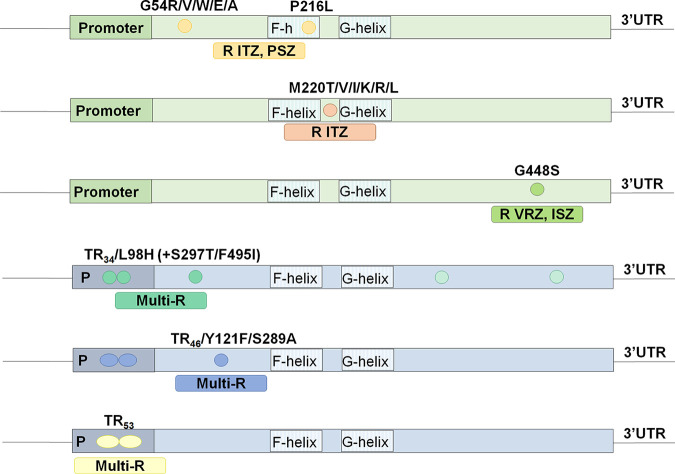
In A. fumigatus, the different susceptibility profiles depend on the specific Cyp51A amino acid substitution (Fig. 2). Such is the case of G54 and P216 mutations in the A. fumigatus Cyp51A enzyme, responsible for cross-resistance to the long-tailed azole drugs ITZ and PSZ but with unaffected MICs to short-tailed azoles such as VRZ and ISZ (23, 24). Mutation M220 leads to ITZ resistance and variable MIC values to VRZ, PSZ, and ISZ (25), while point mutation G448S yields resistance to VRZ and ISZ and variable MIC values to ITZ and PSZ (26, 27). On the other hand, A. fumigatus strains with promoter integrations (tandem repeat [TR]) and cyp51A point mutations (TR34/L98H, TR34/L98H/S297T/F495I, TR46/Y121F/T289A, and TR53) normally show a multiazole resistance phenotype (28–30).
FIG 2.
The most common azole resistance mechanisms in A. fumigatus and susceptibility profiles to clinical azoles associated with each Cyp51A modification. UTR, untranslated region.
Given the similarity among clinical azoles and those used in crop protection, cross-resistance among DMIs and clinical azoles is common. This suggests an association between the azole susceptibility phenotypes and the resistance mechanism shown by both class of fungal pathogens. Moreover, some Cyp51 alterations at equivalent positions in both human and plant pathogens have been found (2).
In this study, a collection of azole-resistant and -susceptible A. fumigatus strains were tested against the most commonly used DMIs to analyze whether the susceptibility phenotypes provide enough evidence to ultimately point toward the pathway involved in the A. fumigatus environmental source of azole resistance. Different patterns of azole cross-resistance were observed depending on the azole resistance mechanism.
RESULTS AND DISCUSSION
The worldwide emergence of A. fumigatus azole-resistant isolates poses a significant threat to the management of these infections (2, 31). The environmental use of azole drugs as agricultural fungicides is believed to be one of the driving forces of the A. fumigatus azole resistance emergence, although solid evidence is still lacking (32).
The A. fumigatus strain collection represents a heterogeneous population.
Several authors have demonstrated the huge genetic diversity among A. fumigatus strains using data from various typing techniques and whole-genome sequencing (WGS) (33–36). All the strains used in this study were identified as A. fumigatus sensu stricto. Their azole resistance mechanism was analyzed by PCR amplification and sequencing of the cyp51A gene, including its promoter. Since both genetic background and phenotypic features, such as antifungal resistance, may influence susceptibility testing results, the isolates included in this study were distributed in different groups according to their Cyp51A modifications, susceptibility to clinical azole drugs, and WGS cluster based on a previous A. fumigatus study performed in our group (33).
A description of each group, resistance mechanism, and number of strains within it is provided in Table 2. The strains used in this work belonged to what we called cluster I, i.e., azole-susceptible cyp51A wild-type (WT) strains together with azole-resistant cyp51A single-point mutation strains, cluster II, i.e., azole-susceptible and -resistant strains with both cyp51A single point mutations combined with TR promoter integrations mechanisms, cluster III, i.e., strains with five particular cyp51A modifications (F46Y, M172V, N248T, D255E, and E427K), and cluster IV, i.e., strains with three particular cyp51A modifications (F46Y, M172V, and E427K) (33).
TABLE 2.
Ranges of MICs to clinical and agricultural azole antifungalsa
| Cyp51A modification (no. of isolates) |
WGS cluster |
Range of MICs to clinical azoles (mg/liter) |
Range of MICs to agricultural DMIs (mg/liter) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITZ | VRZ | PSZ | ISZ | IMZ | PRZ | TFZ | MTZ | TBZ | EPZ | BRZ | DFZ | ||
| AZL-S | |||||||||||||
| WT (20) | I-II | 0.25 to 0.5 | 0.125 to 0.5 | 0.06 to 0.125 | 0.25 to 1 | 0.125 to 0.5 | 0.125 to 0.5 | 8 to 16 | 0.125 to 0.5 | 1 to 4 | 2 to 4 | 1 to 4 | 0.5 to 2 |
| 5SNPsb (6) | III | 0.5 to 1 | 1 to 2 | 0.125 to 0.5 | 1 | 0.25 to 0.5 | 0.25 to 0.5 | 16 to >32 | 0.25 to 1 | 2 to 8 | 8 to 16 | 4 to 16 | 2 to 8 |
| 3SNPsc (11) | IV | 0.25 to 1 | 0.5 to 1 | 0.06 to 0.125 | 1 to 2 | 0.125 to 0.25 | 0.125 to 0.25 | 8 to 16 | 0.25 to 1 | 2 to 4 | 2 to 4 | 2 to 8 | 2 to 4 |
| AZL-R point mutations | |||||||||||||
| G54 (12) | I-II | >8 | 0.25 to 0.5 | 1 to >8 | 0.25 to 1 | 0.06 to 0.125 | 0.125 to 0.25 | 2 to 4 | 0.06 to 0.125 | 0.5 to 2 | 0.5 to 2 | 0.5 to 1 | 0.06 to 0.25 |
| M220 (7) | I-II | >8 | 0.25 to 1 | 0.25 to 2 | 1 to 4 | 0.25 to 2 | 0.25 to 1 | 8 to 32 | 0.25 to 2 | 2 to 16 | 4 to 16 | 1 to 4 | 2 to 16 |
| G448S (5) | I-II | 1 to 2 | >8 | 0.25 to 1 | 4 to 8 | 0.5 to 2 | 1 to 2 | 32 to >32 | 4 to 8 | 8 to >32 | 8 to >32 | 4 to >32 | 4 to >32 |
| AZL-R TR integrationsd | |||||||||||||
| TR34/L98H (12) | II | >8 | 4 to 8 | 0.5 to 1 | 8 | 1 to 8 | 2 to 8 | >32 | 1 to 2 | 16 to 32 | >32 | 8 to 32 | 16 to >32 |
| TR34/L98H/S297T/F495I (3) | II | >8 | 4 to 8 | 0.5 to 1 | >8 | 8 | >32 | >32 | 4 to 16 | 16 to 32 | >32 | >32 | >32 |
| TR46/Y121F/T289A (4) | II | 2 to 4 | 4 to >8 | 0.5 | >8 | 32 to >32 | 16 to >32 | >32 | 8 to 16 | >32 | >32 | >32 | >32 |
| TR53 (3) | II | > 8 | 2 to 4 | 0.5 to 1 | 8 | 2 to 8 | 2 to 8 | >32 | 2 | 16 to 32 | >32 | 32 | 16 to 32 |
A. fumigatus isolates are grouped based on their azole susceptibility profiles and their Cyp51A modifications. AZL-S, azole susceptible; AZL-R, azole resistant; ITZ, itraconazole; VRZ, voriconazole; PSZ, posaconazole; ISZ, isavuconazole; IMZ, imazalil; PRZ, prochloraz; TFZ, triflumizole; MTZ, metconazole; TBZ, tebuconazole; EPZ, epoxiconazole; BRZ, bromuconazole; DFZ, difenoconazole.
5SNPs, F46Y/M172V/N248T/D255E/E427K.
3SNPs, F46Y/M172V/E427K.
Tandem repeat (TR) integration in the cyp51A promoter in combination, or not, with single point mutations.
Antifungal susceptibility testing (AFST).
(i) Clinical azole drugs. Following the European Committee on Antifungal Susceptibility Testing (EUCAST) guidelines (see Materials and Methods), the analyzed strains showed a wide range of MIC values to all four clinical antifungals tested—itraconazole (ITZ), voriconazole (VRZ), posaconazole (PSZ), and isavuconazole (ISZ). These differences were based on the specific genetic background (WGS cluster) and azole resistance mechanism. In vitro susceptibility testing showed ranges within one or two 2-fold MICs for each strain, which suggests stable and reliable results. However, MIC ranges per group may be broader since several isolates are included in a group. MIC ranges for each clinical azole and group of strains are shown in Table 2. There was no relevant difference in MIC values among the Cyp51A WT strains (from cluster I or II) to the clinical azoles tested. All the A. fumigatus azole-resistant strains with G54 mutation were resistant to ITZ and PSZ, while the strains with M220 were resistant to ITZ but variable to VRZ, ISZ, and PSZ. Strains harboring the G448S mutation were resistant to VRZ and ISZ but variable to ITZ and PSZ. Finally, the isolates with the combined resistance mechanism which includes a TR insertion in the cyp51A promoter showed a multiazole resistance profile to all clinical azoles tested. No differences in susceptibility to amphotericin B or echinocandin drugs were seen among all the strains tested (see Table S1 in the supplemental material).
(ii) DMIs. Susceptibility testing to eight DMI fungicides used for crop protection, consisting of three imidazole drugs (imazalil [IMZ], prochloraz [PRZ], and triflumizole [TFZ]) and five triazole drugs (metconazole [MTZ], tebuconazole [TBZ], epoxiconazole [EPZ], bromuconazole [BRZ], and difenoconazole [DFZ]), was performed using the A. fumigatus strain collection. Again, in vitro susceptibility testing showed ranges within one or two 2-fold MICs for each strain. MIC ranges for each DMI and group of strains are shown in Table 2.
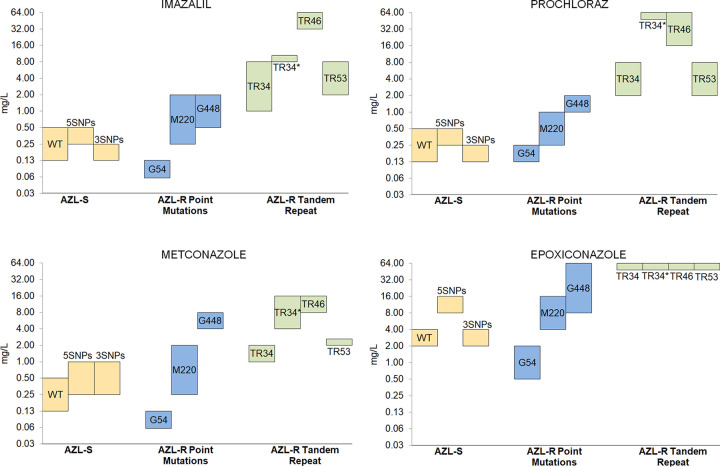
There were no remarkable differences in the MIC values to DMI drugs among the isolates that formed the azole-susceptible group (Cyp51A-WT, Cyp51A-3SNPS, and Cyp-5SNPs from clusters I, II, III, and IV), showing that their different genomic backgrounds do not influence their DMI susceptibility profiles (Table 2). However, there were several relevant differences depending on the azole resistance mechanism groups (Table 2 and Fig. 3). In general, most A. fumigatus azole-resistant strains showed high MICs to all DMIs tested except for the strains with the Cyp51A-G54 mutation, which exhibited a hypersusceptible phenotype to all the agricultural fungicides tested. Moreover, strains that harbored the resistance mechanisms TR46/Y121F/T289A and TR34/L98H/S297T/F495I were highly resistant to imidazoles, to both IMZ and PRZ and just PRZ, respectively. Strains with the G448S mutation showed a pattern of high resistance to triazole DMIs but not that much to imidazole drugs.
FIG 3.
Ranges of MICs to four agricultural azole antifungals. A. fumigatus isolates are grouped based on their azole susceptibility profile and their Cyp51A modifications.
Role of clinical azoles and agriculture DMIs in the emergence and development of A. fumigatus azole resistance.
It is presumed, but still currently debated, that the development of azole resistance in A. fumigatus may be linked either to a medical or patient-acquired route or to an environmental route (9, 14). Although azole resistance is acquired by selection pressure in both cases, it is proposed that as a result, different resistance mechanisms and susceptibility patterns are developed.
Most A. fumigatus isolates with cyp51A single point mutations—G54, P216, M220, and G448—were isolated from patients who had received long-term azole treatment (26, 37). However, mutations at position G54, and occasionally M220 and P216, have also been reported for strains from an environmental origin (38–40).
It is well known that G54 mutation may emerge after long-term ITZ therapy in patients with chronic aspergillosis or cystic fibrosis (41). However, the fact that it has also been isolated from the environment in very different geographical locations (several European countries, India, China, Tanzania, and Thailand) points to a possible agricultural origin (38–40, 42). The results obtained in this study do not point toward the environmental route to explain this resistance mechanism, as all G54 strains tested are resistant to long-tailed clinical azoles but highly susceptible to agricultural DMIs and short-tailed clinical azoles, such as VRZ and ISZ (Fig. 3 and Table 2). A. fumigatus Cyp51A homology model studies have showed that the G54R mutation can prevent long-tailed azoles from entering the channel but not the more compact molecule VRZ (43). In addition, the equivalent Cyp51 mutation has never been identified in plant pathogens related to DMI resistance (Table 1). These strains showed even lower MIC values to the new triazole DMIs tested than the cyp51A-WT strains (Table S2). Alternatively, the possibility that G54 A. fumigatus azole-resistant isolates may develop during azole therapy within an infected or colonized patient and then spread into the environment has been proposed (44). The G448S mutation has been shown to confer resistance to VRZ and ISZ, together with elevated MICs to ITZ and PSZ (26). Although to date this mutation has mainly been reported in the clinical setting, the associated high triazole DMI resistance (Table 2) and the recent finding of A. fumigatus isolates with environmental origin, which harbor this resistance mechanism (45, 46), would suggest that this mutation could emerge under VRZ selective pressure in the clinical setting or under selective pressure from other DMI triazoles, such as MTZ, in the environment (Fig. 3).
Currently, the more frequent A. fumigatus mechanism of azole resistance involves the overexpression of the cyp51A gene, sometimes together with point mutations (TR34/L98H, TR46/Y121F/T289A, and TR53) (28–30), and is associated with the environmental route and the extended use of DMI fungicides in crop protection (14). Moreover, strains with these resistance mechanisms have been found in azole-naive patients but also in the environment throughout multiple worldwide locations (32, 47). Since azole fungicides are used on a global scale, several resistance mechanisms have been described to be common between plant pathogens and A. fumigatus azole-resistant isolates (Table 1).
In this context, the most common cyp51 mutation in plant pathogens associated with DMI resistance is the 134/136/137 tyrosine (Y) substitution to phenylalanine (F) or to histidine (H) (Cyp51 amino acid position varies depending on the fungal species) without known alterations in the Cyp51 promoter (Table 1). This mutation would correspond to the Y121F modification commonly found in A. fumigatus together with other modifications in the cyp51A gene, e.g., TR46/Y121F/T289A (26, 30). Interestingly, the Y121F mutation without TR integration in A. fumigatus has been found only in one clinical isolate, but the patient was never exposed to azole drugs. This strongly suggests a resistance of environmental origin and could represent the missing link between the wild-type gene and the TR46/Y121F/T289A resistance mechanism (48). The sole Y121F mutation confers resistance only to VRZ and not to ITZ or PSZ, whereas the TR46/Y121F/T289A mutation is associated with multiazole resistance. High-resolution X-ray crystal structure analysis demonstrated that the Y140F/H mutation in Saccharomyces cerevisiae Erg11 disrupted the binding of short-tailed triazoles but not long-tailed ones (49).
The A. fumigatus strains which harbor the TR46/Y121F/T289A mutation combination have a pattern of resistance to all DMIs tested but particularly high resistance to imidazole drugs. Apart from A. fumigatus, other fungal human pathogens present the equivalent Cyp51/ERG11 mutations (Cryptococcus neoformans, Histoplasma capsulatum, Candida albicans, and Candida auris) (50–53), which lead to resistance to only short-tailed triazoles. Similarly, the sole Y121F mutation in A. fumigatus leads just to VRZ resistance (48). This mechanism of resistance commonly found in both plant pathogens and A. fumigatus leads to similar activity and therefore might be developed from azole selection pressure in both cases. In Erysiphe necator, a strong association between cyp51 gene copy number variation, which influenced expression in a gene-dose-dependent manner and was correlated with fungal growth in the presence of a DMI fungicide, has been found (54).
Several authors have observed elevated MIC values to the imidazole PRZ among A. fumigatus isolates harboring the TR34/L98H/S297T/F495I mutation (55–57). Our results are in agreement with them, as these strains showed a substantially stronger increase in the MIC value to PRZ (range, 8 to 32 mg/liter) than did the strains harboring the TR34/L98H mutation (1 to 8 mg/liter).
It has been described that most of the A. fumigatus strains with the TR34/L98H/S297T/F495I mutation are more genetically related than strains with the TR34/L98H mutation, which might be due to an extremely adaptive recombinant event under the selection pressure of imidazole fungicides in some countries (55–58). In one of our previous studies using WGS, the strains with the TR34/L98H/S297T/F495I mutation grouped together in a small subcluster even when their geographical origins were nonrelated, such as in the case of strains from Spain, Denmark, or the Netherlands (data not shown). Moreover, if we compare the agricultural pathogen Cyp51 proteins to the Cyp51A protein of A. fumigatus, the role of these mutations in PRZ resistance has been demonstrated even with structural in silico modeling (18). For instance in Penicillium digitatum, the F506I mutation arose in combination with a 199-bp insertion in the cyp51 promoter, showing even higher resemblance to the A. fumigatus TR resistance mechanism therefore suggesting a common and environmental evolutionary route (18, 55). Moreover, in this plant pathogen the single F495I mutation is not responsible for the whole increase in the imidazole MIC values, as L98H on its own does not lead to the same MIC values as its combination with the promoter insertion (18, 28). The possibility that the S297T mutation might be required to compensate for the deleterious effect of F495I on the protein function, as T289A does in the case of the TR46/Y121F/T289A mutation, has been previously proposed (59).
In general, resistant strains with TR insertions in the cyp51A promoter are grouped together into one cluster based on our previous WGS phylogenetic analysis (33), which indicates genetic closeness independently of the geographic origin. This common genetic background may help them to adapt to the environment or may confer on them improved fitness that favors their selection and spread. Moreover, different TR mutations are emerging in different geographic locations (32), which suggests that the local use of DMIs may affect the development of a specific resistance mechanism (41, 58, 60).
In conclusion, this study suggests that the environmental use of imidazole fungicides might confer selection pressure for the emergence of TR34/L98H/S297T/F495I and TR46/Y121F/T289A A. fumigatus azole-resistant isolates. In any case, cross-resistance to all of them is the rule. Therefore, the use of DMIs should be further controlled and contained in order to minimize the development and spread of azole-resistant A. fumigatus strains. Finally, it is very unlikely that the G54 mutation is being selected from the most common DMIs used in crop protection, and thus, the fact that it has been isolated from the environment should be investigated further.
MATERIALS AND METHODS
Aspergillus fumigatus strain collection.
A total of 83 unrelated strains of A. fumigatus from different countries with clinical origin were included in this study. Fungal genomic DNA was extracted as described previously (12). All isolates were identified at the species level by PCR amplification and sequencing of ITS1-5.8S-ITS2 regions and a portion of the β-tubulin gene (61).
Characterization of azole resistance molecular mechanisms in A. fumigatus strains.
Azole resistance mechanisms were studied by sequencing the main azole target gene cyp51A in the A. fumigatus collection. Conidia from each strain were cultured in 3 ml of GYEP broth (2% glucose, 0.3% yeast extract, 1% peptone) and grown overnight at 37°C, after which mycelium mats were harvested and DNA was extracted (62). The full coding sequence of the cyp51A gene, including its promoter sequence, was amplified and sequenced using the PCR conditions described before (28). Each isolate was independently analyzed twice. DNA cyp51A sequences were compared against the cyp51A sequence of the A. fumigatus reference strain CBS 144.89 (GenBank accession number AF338659). A total of 46 independent A. fumigatus strains with known azole resistance mechanisms were included in this work, as well as 37 azole-susceptible strains.
TRESPERG genotyping and whole-genome sequence analysis.
All A. fumigatus isolates included in this study were genotyped following the previously described TRESPERG typing assay (36). Whole-genome sequencing previously performed in a collection of 101 A. fumigatus genomes, including azole-susceptible and azole-resistant strains, was used to divide the A. fumigatus collection into four different clusters (33).
Antifungal susceptibility testing.
(i) Clinical azoles. Antifungal susceptibility testing (AFST) was performed using a broth microdilution method following the European Committee on Antifungal Susceptibility Testing (EUCAST) reference method 9.3.1 (63). The antifungal clinical azoles used were itraconazole (Janssen Pharmaceutica, Madrid, Spain), voriconazole (Pfizer SA, Madrid, Spain), posaconazole (Schering-Plough Research Institute, Kenilworth, NJ), and isavuconazole (Basilea Pharmaceutica, Basel, Switzerland; tested from January 2017). In addition, we performed AFST to amphotericin B (Sigma-Aldrich Química, Madrid, Spain) as well as the echinocandins caspofungin (Merck & Co., Inc., Rahway, NJ) and anidulafungin (Pfizer SA, Madrid, Spain). The final concentrations tested ranged from 0.015 to 8 mg/liter for azoles, 0.03 to 16 mg/liter for amphotericin B and caspofungin, and 0.008 to 4 mg/liter for anidulafungin. Aspergillus flavus ATCC 204304 and A. fumigatus ATCC 204305 were used as quality control strains in all tests performed. MICs were visually read after 24 and 48 h of incubation at 37°C in a humid atmosphere. MIC determinations were performed at least three independent times for each isolate (biological triplicates). A. fumigatus clinical breakpoints for interpreting AFST results established by EUCAST were used to classify each isolate as susceptible (S) or resistant (R) against a specific antifungal, in this case ITZ (S ≤ 1; R > 1), VCZ (S ≤ 1; R > 1), PSZ (S ≤ 0.125; R > 0.25), or ISZ (S ≤ 1; R > 2) (64).
(ii) Agricultural azoles (DMIs). AFST was also performed against 14-α demethylation-inhibiting fungicides (DMIs) following the EUCAST methodology as described before. The antifungal DMIs tested were three imidazole drugs (prochloraz, imazalil, and triflumizole) and five triazole compounds (tebuconazole, bromuconazole, metconazole, epoxiconazole, and difenoconazole), all purchased at Sigma-Aldrich, Química (Madrid, Spain). All DMIs were dissolved in dimethyl sulfoxide (DMSO) and autosterilized for 30 min at room temperature, as stated in the EUCAST protocol for clinical azoles. The final concentrations tested ranged from 0.064 to 32 mg/liter. Clinical breakpoints for interpreting AFST results have not been established, so isolates were considered susceptible or resistant based on the MIC shown by the group of clinical azole-susceptible strains. MIC determinations were performed at least three independent times for each isolate (biological triplicates). In addition, four new DMIs that have been recently introduced into the market—bitertanol, myclobutanil, triadimenol and paclobutrazol (all purchased at Sigma-Aldrich, Química)—were also tested against our A. fumigatus strain collection following the same methodology.
Supplementary Material
ACKNOWLEDGMENTS
E.M. conceived and designed the experiments. R.G.-R., I.G.-J., and J.L. performed the experiments. R.G.-R., I.G.-J., and E.M. analyzed the data. R.G.-R., I.G.-J., and E.M. drafted the manuscript. All authors read and approved the final manuscript.
This research was funded by Fondo de Investigacion Sanitaria (FIS) (PI18CIII/00045) and also by Plan Nacional de I+D+i 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/CIII/0004/0003), cofinanced by the European Regional Development Fund (ERDF) A Way To Achieve Europe Operative program Intelligent Growth 2014–2020.
We declare no conflict of interest.
The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the result.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Latgé JP, Chamilos G. 2019. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev 33:e00140-18. doi: 10.1128/CMR.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Rubio R, Cuenca-Estrella M, Mellado E. 2017. Triazole resistance in Aspergillus species: an emerging problem. Drugs 77:599–613. doi: 10.1007/s40265-017-0714-4. [DOI] [PubMed] [Google Scholar]
- 3.Meneau I, Sanglard D. 2005. Azole and fungicide resistance in clinical and environmental Aspergillus fumigatus isolates. Med Mycol 43:307–311. doi: 10.1080/13693780500090826. [DOI] [PubMed] [Google Scholar]
- 4.Buil JB, Hare RK, Zwaan BJ, Arendrup MC, Melchers WJG, Verweij PE. 2019. The fading boundaries between patient and environmental routes of triazole resistance selection in Aspergillus fumigatus. PLoS Pathog 15:e1007858. doi: 10.1371/journal.ppat.1007858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andes DR, Dismukes WE. 2011. Azoles, p 61–93. In Kauffman C, Pappas P, Sobel J, Dismukes W (ed), Essentials of clinical mycology, 2nd ed. Springer, New York, NY. [Google Scholar]
- 6.Snelders E, Camps SM, Karawajczyk A, Schaftenaar G, Kema GH, van der Lee HA, Klaassen CH, Melchers WJ, Verweij PE. 2012. Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PLoS One 7:e31801. doi: 10.1371/journal.pone.0031801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, van den Heuvel J, Debets AJM, Verweij PE, Melchers WJG, Zwaan BJ, Schoustra SE. 2017. Evolution of cross-resistance to medical triazoles in Aspergillus fumigatus through selection pressure of environmental fungicides. Proc R Soc B 284:20170635. doi: 10.1098/rspb.2017.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nett JE, Andes DR. 2016. Antifungal agents: spectrum of activity, pharmacology, and clinical indications. Infect Dis Clin North Am 30:51–83. doi: 10.1016/j.idc.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Verweij PE, Chowdhary A, Melchers WJ, Meis JF. 2016. Azole resistance in Aspergillus fumigatus: can we retain the clinical use of mold-active antifungal azoles? Clin Infect Dis 62:362–368. doi: 10.1093/cid/civ885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CDC. 2019. Antibiotic resistance threats in the United States, 2019. US Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 11.Becher R, Wirsel SG. 2012. Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl Microbiol Biotechnol 95:825–840. doi: 10.1007/s00253-012-4195-9. [DOI] [PubMed] [Google Scholar]
- 12.Mellado E, Diaz-Guerra TM, Cuenca-Estrella M, Rodriguez-Tudela JL. 2001. Identification of two different 14-alpha sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol 39:2431–2438. doi: 10.1128/JCM.39.7.2431-2438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawkins NJ, Cools HJ, Sierotzki H, Shaw MW, Knogge W, Kelly SL, Kelly DE, Fraaije BA. 2014. Paralog re-emergence: a novel, historically contingent mechanism in the evolution of antimicrobial resistance. Mol Biol Evol 31:1793–1802. doi: 10.1093/molbev/msu134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger S, El Chazli Y, Babu AF, Coste AT. 2017. Azole resistance in Aspergillus fumigatus: a consequence of antifungal use in agriculture? Front Microbiol 8:1024. doi: 10.3389/fmicb.2017.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Lin Y, Yin WX, Peng YL, Schnabel G, Huang JB, Luo CX. 2015. The Y137H mutation of VvCYP51 gene confers the reduced sensitivity to tebuconazole in Villosiclava virens. Sci Rep 5:17575. doi: 10.1038/srep17575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald MC, Renkin M, Spackman M, Orchard B, Croll D, Solomon PS, Milgate A. 2018. Rapid parallel evolution of azole fungicide resistance in Australian populations of the wheat pathogen Zymoseptoria tritici. Appl Environ Microbiol 85:e01908-18. doi: 10.1128/AEM.01908-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian H, Du J, Chi M, Sun X, Liang W, Huang J, Li B. 2018. The Y137H mutation in the cytochrome P450 FgCYP51B protein confers reduced sensitivity to tebuconazole in Fusariumgraminearum. Pest Manag Sci 74:1472–1477. doi: 10.1002/ps.4837. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Yu J, Liu J, Yuan Y, Li N, He M, Qi T, Hui G, Xiong L, Liu D. 2014. Novel mutations in CYP51B from Penicillium digitatum involved in prochloraz resistance. J Microbiol 52:762–770. doi: 10.1007/s12275-014-4112-2. [DOI] [PubMed] [Google Scholar]
- 19.Cañas-Gutiérrez GP, Angarita-Velásquez MJ, Restrepo-Flórez JM, Rodríguez P, Moreno CX, Arango R. 2009. Analysis of the CYP51 gene and encoded protein in propiconazole-resistant isolates of Mycosphaerella fijiensis. Pest Manag Sci 65:892–899. doi: 10.1002/ps.1770. [DOI] [PubMed] [Google Scholar]
- 20.Bolton MD, Birla K, Rivera-Varas V, Rudolph KD, Secor GA. 2012. Characterization of CbCyp51 from field isolates of Cercospora beticola. Phytopathology 102:298–305. doi: 10.1094/PHYTO-07-11-0212. [DOI] [PubMed] [Google Scholar]
- 21.Nikou D, Malandrakis A, Konstantakaki M, Vontas J, Markoglou A, Ziogas B. 2009. Molecular characterization and detection of overexpressed C-14 alpha-demethylase-based DMI resistance in Cercospora beticola field isolates. PesticBiochemPhysiol 95:18–27. doi: 10.1016/j.pestbp.2009.04.014. [DOI] [Google Scholar]
- 22.Ma B, Tredway LP. 2013. Induced overexpression of cytochrome P450 sterol 14α‐demethylase gene (CYP51) correlates with sensitivity to demethylation inhibitors (DMIs) in Sclerotinia homoeocarpa. Pest Manag Sci 69:1369–1378. doi: 10.1002/ps.3513. [DOI] [PubMed] [Google Scholar]
- 23.Diaz-Guerra TM, Mellado E, Cuenca-Estrella M, Rodriguez-Tudela JL. 2003. A point mutation in the 14α-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother 47:1120–1124. doi: 10.1128/aac.47.3.1120-1124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camps SM, van der Linden JW, Li Y, Kuijper EJ, van Dissel JT, Verweij PE, Melchers WJ. 2012. Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature. Antimicrob Agents Chemother 56:10–16. doi: 10.1128/AAC.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellado E, Garcia-Effron G, Alcazar-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL. 2004. Substitutions at methionine 220 in the 14-alpha sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob Agents Chemother 48:2747–2750. doi: 10.1128/AAC.48.7.2747-2750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelaez T, Gijon P, Bunsow E, Bouza E, Sanchez-Yebra W, Valerio M, Gama B, Cuenca-Estrella M, Mellado E. 2012. Resistance to voriconazole due to a G448S substitution in Aspergillus fumigatus in a patient with cerebral aspergillosis. J Clin Microbiol 50:2531–2534. doi: 10.1128/JCM.00329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregson L, Goodwin J, Johnson A, McEntee L, Moore CB, Richardson M, Hope WW, Howard SJ. 2013. In vitro susceptibility of Aspergillus fumigatus to isavuconazole: correlation with itraconazole, voriconazole, and posaconazole. Antimicrob Agents Chemother 57:5778–5780. doi: 10.1128/AAC.01141-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellado E, Garcia-Effron G, Alcazar-Fuoli L, Melchers WJ, Verweij PE, Cuenca-Estrella M, Rodriguez-Tudela JL. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob Agents Chemother 51:1897–1904. doi: 10.1128/AAC.01092-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodiamont CJ, Dolman KM, Ten Berge IJ, Melchers WJ, Verweij PE, Pajkrt D. 2009. Multiple-azole-resistant Aspergillus fumigatus osteomyelitis in a patient with chronic granulomatous disease successfully treated with long-term oral posaconazole and surgery. Med Mycol 47:217–220. doi: 10.1080/13693780802545600. [DOI] [PubMed] [Google Scholar]
- 30.van der Linden JW, Camps SM, Kampinga GA, Arends JP, Debets-Ossenkopp YJ, Haas PJ, Rijnders BJ, Kuijper EJ, van Tiel FH, Varga J, Karawajczyk A, Zoll J, Melchers WJ, Verweij PE. 2013. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clin Infect Dis 57:513–520. doi: 10.1093/cid/cit320. [DOI] [PubMed] [Google Scholar]
- 31.van der Linden JW, Snelders E, Kampinga GA, Rijnders BJ, Mattsson E, Debets-Ossenkopp YJ, Kuijper EJ, van Tiel FH, Melchers WJG, Verweij PE. 2011. Clinical implications of azole resistance in Aspergillus fumigatus, The Netherlands, 2007–2009. Emerg Infect Dis 17:1846–1854. doi: 10.3201/eid1710.110226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeanvoine A, Rocchi S, Bellanger AP, Reboux G, Millon L. 2020. Azole-resistant Aspergillus fumigatus: a global phenomenon originating in the environment? Med Mal Infect 50:389–395. doi: 10.1016/j.medmal.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Rubio R, Monzon S, Alcazar-Fuoli L, Cuesta I, Mellado E. 2018. Genome-wide comparative analysis of Aspergillus fumigatus strains: the reference genome as a matter of concern. Genes (Basel) 9:363. doi: 10.3390/genes9070363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdolrasouli A, Rhodes J, Beale MA, Hagen F, Rogers TR, Chowdhary A, Meis JF, Armstrong-James D, Fisher MC. 2015. Genomic context of azole resistance mutations in A. fumigatus determined using whole-genome sequencing. mBio 6:e00536-15. doi: 10.1128/mBio.00536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashu EE, Hagen F, Chowdhary A, Meis JF, Xu J. 2017. Global population genetic analysis of Aspergillus fumigatus. mSphere 2:e00019-17. doi: 10.1128/mSphere.00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Rubio R, Escribano P, Gomez A, Guinea J, Mellado E. 2018. Comparison of two highly discriminatory typing methods to analyze Aspergillus fumigatus azole resistance. Front Microbiol 9:1626. doi: 10.3389/fmicb.2018.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meis JF, Chowdhary A, Rhodes JL, Fisher MC, Verweij PE. 2016. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos Trans R Soc B 371:20150460. doi: 10.1098/rstb.2015.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tangwattanachuleeporn M, Minarin N, Saichan S, Sermsri P, Mitkornburee R, Groß U, Chindamporn A, Bader O. 2017. Prevalence of azole-resistant Aspergillus fumigatus in the environment of Thailand. Med Mycol 55:429–435. doi: 10.1093/mmy/myw090. [DOI] [PubMed] [Google Scholar]
- 39.Sharma C, Hagen F, Moroti R, Meis JF, Chowdhary A. 2015. Triazole-resistant Aspergillus fumigatus harbouring G54 mutation: is it de novo or environmentally acquired? J Glob Antimicrob Resist 3:69–74. doi: 10.1016/j.jgar.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Bader O, Tünnermann J, Dudakova A, Tangwattanachuleeporn M, Weig M, Groß U. 2015. Environmental isolates of azole-resistant Aspergillus fumigatus in Germany. Antimicrob Agents Chemother 59:4356–4359. doi: 10.1128/AAC.00100-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh A, Sharma B, Mahto KK, Meis JF, Chowdhary A. 2020. High-frequency direct detection of triazole resistance in Aspergillus fumigatus from patients with chronic pulmonary fungal diseases in India. J Fungi (Basel) 6:67. doi: 10.3390/jof6020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng S, Zhang L, Ji Y, Verweij PE, Tsui KM, Hagen F, Houbraken J, Meis JF, Abliz P, Wang X, Zhao J, Liao W. 2017. Triazole phenotypes and genotypic characterization of clinical Aspergillus fumigatus isolates in China. Emerg Microbes Infect 6:e109. doi: 10.1038/emi.2017.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talbot JJ, Subedi S, Halliday CL, Hibbs DE, Lai F, Lopez-Ruiz FJ, Harper L, Park RF, Cuddy WS, Biswas C, Cooley L, Carter D, Sorrell TC, Barrs VR, Chen SC. 2018. Surveillance for azole resistance in clinical and environmental isolates of Aspergillus fumigatus in Australia and cyp51A homology modelling of azole-resistant isolates. J Antimicrob Chemother 73:2347–2351. doi: 10.1093/jac/dky187. [DOI] [PubMed] [Google Scholar]
- 44.Engel TGP, Erren E, van den Driessche KSJ, Melchers WJG, Reijers MH, Merkus P, Verweij PE. 2019. Aerosol transmission of Aspergillus fumigatus in cystic fibrosis patients in the Netherlands. Emerg Infect Dis 25:797–799. doi: 10.3201/eid2504.181110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoustra SE, Debets AJM, Rijs AJMM, Zhang J, Snelders E, Leendertse PC, Melchers WJG, Rietveld AG, Zwaan BJ, Verweij PE. 2019. Environmental hotspots for azole resistance selection of Aspergillus fumigatus, the Netherlands. Emerg Infect Dis 25:1347–1353. doi: 10.3201/eid2507.181625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao D, Wu R, Dong S, Wang F, Ju C, Yu S, Xu S, Fang H, Yu Y. 2020. Five-year survey (2014 to 2018) of azole resistance in environmental Aspergillus fumigatus isolates from China. Antimicrob Agents Chemother 64:e00904-20. doi: 10.1128/AAC.00904-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Astvad KM, Jensen RH, Hassan TM, Mathiasen EG, Thomsen GM, Pedersen UG, Christensen M, Hilberg O, Arendrup MC. 2014. First detection of TR46/Y121F/T289A and TR34/L98H alterations in Aspergillus fumigatus isolates from azole-naive patients in Denmark despite negative findings in the environment. Antimicrob Agents Chemother 58:5096–5101. doi: 10.1128/AAC.02855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lescar J, Meyer I, Akshita K, Srinivasaraghavan K, Verma C, Palous M, Mazier D, Datry A, Fekkar A. 2014. Aspergillus fumigatus harbouring the sole Y121F mutation shows decreased susceptibility to voriconazole but maintained susceptibility to itraconazole and posaconazole. J Antimicrob Chemother 69:3244–3247. doi: 10.1093/jac/dku316. [DOI] [PubMed] [Google Scholar]
- 49.Sagatova AA, Keniya MV, Wilson RK, Sabherwal M, Tyndall JD, Monk BC. 2016. Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14α-demethylase. Sci Rep 6:26213. doi: 10.1038/srep26213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sionov E, Chang YC, Garraffo HM, Dolan MA, Ghannoum MA, Kwon-Chung KJ. 2012. Identification of a Cryptococcus neoformans cytochrome P450 lanosterol 14α-demethylase (Erg11) residue critical for differential susceptibility between fluconazole/voriconazole and itraconazole/posaconazole. Antimicrob Agents Chemother 56:1162–1169. doi: 10.1128/AAC.05502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wheat LJ, Connolly P, Smedema M, Durkin M, Brizendine E, Mann P, Patel R, McNicholas PM, Goldman M. 2006. Activity of newer triazoles against Histoplasma capsulatum from patients with AIDS who failed fluconazole. J Antimicrob Chemother 57:1235–1239. doi: 10.1093/jac/dkl133. [DOI] [PubMed] [Google Scholar]
- 52.Flowers SA, Colon B, Whaley SG, Schuler MA, Rogers PD. 2015. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother 59:450–460. doi: 10.1128/AAC.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones L, Riaz S, Morales-Cruz A, Amrine KC, McGuire B, Gubler WD, Walker MA, Cantu D. 2014. Adaptive genomic structural variation in the grape powdery mildew pathogen, Erysiphe necator. BMC Genomics 15:1081. doi: 10.1186/1471-2164-15-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Li Z, Han X, Tian S, Zhao J, Chen F, Su X, Zhao J, Zou Z, Gong Y, Qu F, Qiu G, Wang S, Jia X, Lu Z, Hu M, Huang L, Verweij PE, Han L. 2018. Elevated MIC values of imidazole drugs against Aspergillus fumigatus isolates with TR34/L98H/S297T/F495I mutation. Antimicrob Agents Chemother 62:e01549-17. doi: 10.1128/AAC.01549-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang HC, Huang JC, Lin YH, Chen YH, Hsieh MI, Choi PC, Lo HJ, Liu WL, Hsu CS, Shih HI, Wu CJ, Chen YC. 2018. Prevalence, mechanisms and genetic relatedness of the human pathogenic fungus Aspergillus fumigatus exhibiting resistance to medical azoles in the environment of Taiwan. Environ Microbiol 20:270–280. doi: 10.1111/1462-2920.13988. [DOI] [PubMed] [Google Scholar]
- 57.Pontes L, Beraquet CAG, Arai T, Pigolli GL, Lyra L, Watanabe A, Moretti ML, Schreiber AZ. 2019. Aspergillus fumigatus clinical isolates carrying CYP51A with TR34/L98H/S297T/F495I substitutions detected after four-year retrospective azole resistance screening in Brazil. Antimicrob Agents Chemother 64:e02059-19. doi: 10.1128/AAC.02059-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren J, Jin X, Zhang Q, Zheng Y, Lin D, Yu Y. 2017. Fungicides induced triazole-resistance in A. fumigatus associated with mutations of TR46/Y121F/T289A and its appearance in agricultural fields. J Hazard Mater 326:54–60. doi: 10.1016/j.jhazmat.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 59.Snelders E, Camps SM, Karawajczyk A, Rijs AJ, Zoll J, Verweij PE, Melchers WJ. 2015. Genotype-phenotype complexity of the TR46/Y121F/T289A cyp51A azole resistance mechanism in Aspergillus fumigatus. Fungal Genet Biol 82:129–135. doi: 10.1016/j.fgb.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 60.Siopi M, Rivero-Menendez O, Gkotsis G, Panara A, Thomaidis NS, Alastruey-Izquierdo A, Pournaras S, Meletiadis J. 2020. Nationwide surveillance of azole-resistant Aspergillus fumigatus environmental isolates in Greece: detection of pan-azole resistance associated with the TR46/Y121F/T289A cyp51A mutation. J Antimicrob Chemother 75:3181–3188. doi: 10.1093/jac/dkaa316. [DOI] [PubMed] [Google Scholar]
- 61.Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, Cuenca-Estrella M, Rodriguez-Tudela JL. 2008. Aspergillus section Fumigati: antifungal susceptibility patterns and sequence-based identification. Antimicrob Agents Chemother 52:1244–1251. doi: 10.1128/AAC.00942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang CM, Cohen J, Holden DW. 1992. An Aspergillus fumigatus alkaline protease mutant constructed by gene disruption is deficient in extracellular elastase activity. Mol Microbiol 6:1663–1671. doi: 10.1111/j.1365-2958.1992.tb00891.x. [DOI] [PubMed] [Google Scholar]
- 63.Arendrup MC, Meletiadis J, Mouton JW, Lagrou K, Howard SJ. 2017. European Committee for Antimicrobial Susceptibility Testing Subcommittee on Antifungal Susceptibility Testing (EUCAST-AFST). Definitive Document E.Def 9.3.1: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia forming moulds. Clin Microbiol Infect 14:982–984. [DOI] [PubMed] [Google Scholar]
- 64.European Committee on Antimicrobial Susceptibility Testing. 2018. Antifungal agents. Breakpoint tables for interpretation of MICs. Version 9.0. www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Clinical_breakpoints/Antifungal_breakpoints_v_9.0_180212.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.