Besides the infection properties in human and animals, S. aureus can produce different enterotoxins in food. The enterotoxins can cause vomiting and diarrhea, often involving many people.
KEYWORDS: Staphylococcus aureus, egc, enterotoxin
ABSTRACT
Currently, only 5 (SEA to SEE) out of 27 known staphylococcal enterotoxins can be analyzed using commercially available kits. Six genes (seg, sei, sem, sen, seo, and seu), encoding putative and undetectable enterotoxins, are located on the enterotoxin gene cluster (egc), which is part of the Staphylococcus aureus genomic island vSaβ. These enterotoxins have been described as likely being involved in staphylococcal food-poisoning outbreaks. The aim of the present study was to determine if whole-genome data can be used for the prediction of staphylococcal egc enterotoxin production, particularly enterotoxin G (SEG) and enterotoxin I (SEI). For this purpose, whole-genome sequences of 75 Staphylococcus aureus strains from different origins (food-poisoning outbreaks, human, and animal) were investigated by applying bioinformatics methods (phylogenetic analysis using the core genome and different alignments). SEG and SEI expression was tested in vitro using a sandwich enzyme-linked immunosorbent assay method. Strains could be allocated to 14 different vSaβ types, each type being associated with a single clonal complex (CC). In addition, the vSaβ type and CC were associated with the origin of the strain (human or cattle derived). The amount of SEG and SEI produced also correlated with the vSaβ type and the CC of a strain. The present results show promising indications that the in vitro production of SEG and SEI can be predicted based on the vSaβ type or CC of a strain.
IMPORTANCE Besides having infectious properties in human and animals, S. aureus can produce different enterotoxins in food. The enterotoxins can cause vomiting and diarrhea, often involving many people. Most of these outbreaks remain undiscovered, as detection methods for enterotoxins are only available for a few enterotoxins but not for the more recently discovered enterotoxins G (SEG) and I (SEI). In this study, we show promising results that in vitro production of SEG and SEI can be predicted based on the whole-genome sequencing data of a strain. In addition, these data could be used to find the source (human or cattle derived) of an outbreak strain, which is the key for a better understanding of the role SEG and SEI play in foodborne outbreaks caused by S. aureus.
INTRODUCTION
Staphylococcus aureus can produce a variety of heat-stable enterotoxins, which, when they are secreted in food, can cause staphylococcal food-poisoning outbreaks (SFPO). According to the European Food Safety Authority (EFSA), staphylococcal enterotoxins (SE) in mixed foods and meat products are among the top 10 pathogen/food vehicle pairs, causing the highest number of hospitalizations in strong-evidence outbreaks. By looking at the number of cases, this trend seems to be on the rise (1). In addition, most of the SFPO are classified as weak-evidence outbreaks, since only the so-called classical enterotoxins (SEA, SEB, SEC, SED, and SEE) can be detected and quantified by commercially available kits (2). Besides these five well-known SE, another 20 have been described recently, and some of them were shown to have an emetic activity (SE) and, hence, could be involved in SFPO (3–9). Enterotoxins for which emetic activity has not yet been proved are considered staphylococcal enterotoxin-like (SEl) proteins. As not all SE can be detected directly in food, different methods have been applied in the past to better characterize the S. aureus strains involved in food-poisoning outbreaks, such as pulsed-field gel electrophoresis typing, PCR for detection of the enterotoxin genes, and other methods (10–14). These methods allowed us to evaluate the toxigenic profile of strains or to establish the link between strains and secreted toxins. With the recent advance of whole-genome sequencing (WGS), often each strain involved in an outbreak can be sequenced and characterized genetically, opening new doors to the understanding of the role different SE play in SFPO as well as prediction of antimicrobial resistance and infectivity (15–20).
Twenty years ago, a novel cluster of SE genes, the enterotoxin gene cluster (egc), was described containing the so-called new enterotoxins seg, sei, sem, sen, seo, and seu (21, 22). The egc is located on the genomic island vSaβ and is incorporated in the chromosome as a prophage (16). Literature suggests that about 50% of S. aureus strains harbor an egc (21, 23, 24).
For SEG, SEI, SEM, SEN, and SEO, emetic activity has been demonstrated, and it appears that some SFPO might be caused by these enterotoxins (3, 5). A lot is known on the expression of the classical SE (25–27), yet studies on the expression of the new SE are still very limited (28). Genetic backbones and regulatory systems of SE genes vary among S. aureus strains, causing diverse SE expression patterns. Hence, quantities of toxin production vary between strains (25–27).
Due to the lack of information, new methods and tools need to be developed to better understand and predict the expression and regulation mechanisms of the new enterotoxins, including those of the egc (29). For this reason, the aim of the present study was to determine whether WGS data can be used to predict staphylococcal enterotoxin production of the egc in vitro, particularly of SEG and SEI. These enterotoxins (SEG and SEI) were chosen because they are the only ones (of egc enterotoxins) for which a quantitative method for detection is currently available, allowing a direct link for the corresponding WGS data.
RESULTS
Strain characterization.
Multilocus sequence typing (MLST) of the 75 S. aureus strains isolated from different sources, like food, humans, animals, and the environment, showed that the most frequently found clonal complexes (CC) are CC5 (n = 17), CC20 (n = 15), CC30 (n = 13), and CC705 (n = 11), followed by CC45, CC22, CC50, and CC9 (6, 3, 2, and 2 strains, respectively). In contrast, CC10, CC72, and CC121, as well as an unknown CC, were detected only once (Table 1).
TABLE 1.
Genotypic characteristics (i.e., clonal complex, enterotoxin genes present on the genome, vSaβ type, and spa type) and origins of the 75 studied strainsa
| Strain | Country | Origin | Source of isolation | CC | Enterotoxin genes | vSaβ type | spa type |
|---|---|---|---|---|---|---|---|
| 07CEB94STA | Belgium | Food (SFPO) | Ready to eat | 5 | a, g, i, m, n, o, x | I | t704 |
| 11CEB145STA | Japan | Human | Infection | 5 | a, c, g, i, m, n, o, x | I | * |
| 13CEB178STA | Ireland | SFPO | NA | 5 | d, j, g, i, m, n, o, r, x | I | t463 |
| 13CEB188STA | Ireland | Food (SFPO) | Milk product | 5 | g, i, m, n, o, x | I | t5829 |
| 13CEB191STA | Ireland | Food (SFPO) | Milk product | 5 | d, g, i, j, m, n, o, r, x | I | t837 |
| 13CEB329STA | Belgium | Human (SFPO) | Nose and throat | 5 | g, i, m, n, o, x | I | t7506 |
| 15SBCL1507STA | Ireland | Food (SFPO) | Meat | 5 | g, i, m, n, o, x | I | * |
| 15SBCL1550STA | Ireland | Food (SFPO) | Ready to eat | 5 | g, i, m, n, o, x | I | t450 |
| 17SBCL08STA | France | Food (SFPO) | Meat | 5 | g, i, m, n, o, x | I | t111 |
| 17SBCL09STA | France | Food (SFPO) | Meat | 5 | g, i, m, n, o, x | I | t586 |
| 17SBCL580STA | Bulgaria | Food (SFPO) | Ready to eat | 5 | a, d, g, i, j, m, n, o, r, x | I | t535 |
| 17SBCL585STA | Bulgaria | Food (SFPO) | Ready to eat | 5 | a, d, g, i, j, m, n, o, r, x | I | t535 |
| 502A | USA | Human | Infection | 5 | g, i, m, n, o | I | t010 |
| Mu50 | Japan | Human | Infection | 5 | a, c, g, i, l, m, n, o, tst, x | I | t002 |
| N315 | Japan | Human | Human faeces | 5 | c, g, i, l, m, n, o, p, tst, x | I | t002 |
| NZAK3 | New Zealand | Human | Skin | 5 | c, g, i, l, m, n, o, p, x | I | t002 |
| ST288 | England | Human | Urine | 5 | g, i, m, n, o | I | t1003 |
| 18SBCL679 | Switzerland | Food (SFPO) | Milk product | 9 | g, i, m, n, o, u, x, y, 27 | XIII | t899 |
| G19F | Italy | Animal | Mastitis (cow) | 9 | g, i, m, n, o, u | XIII | t100 |
| 13CEB177STA | Ireland | NA | FPO | 10 | c, g, i, m, n, o, u, x | XVII | t148 |
| 11CEB277STA | Italy | Food | Milk product | 20 | g, i, m, n, o, u, x | XII | t3929 |
| 11CEB279STA | Italy | environment | NA | 20 | g, i, m, n, o, u, x, y | XII | t325 |
| 15SBCL1292STA | France | Food (SFPO) | Milk product | 20 | g, i, m, n, o, u, x | XII | * |
| 15SBCL1299STA | France | Food (SFPO) | Ready to eat | 20 | g, i, m, n, o, u. x. y | XII | t164 |
| 15SBCL1397STA | France | Food (SFPO) | Milk product | 20 | g, i, m, n, o, tst, u, x | XII | t164 |
| 15SBCL1409STA | France | Food (SFPO) | Milk product | 20 | g, i, m, n, o, u, x, y | XII | * |
| 15SBCL1428STA | France | Food (SFPO) | Milk product | 20 | g, i, m, n, o, u, x, y | XII | * |
| 17SBCL202STA | France | Food (SFPO) | Milk product | 20 | g, i, m, n, o, u, x, y | XII | t164 |
| 17SBCL208STA | France | Food (SFPO) | Milk product | 20 | g, i, m, n, o, u, x, y | XII | t458 |
| 17SBCL214STA | France | Food (SFPO) | Milk product | 20 | g, i, m, n, o, u, x, y | XII | * |
| 17SBCL220STA | France | Food (SFPO) | Milk product | 20 | g, i, m, n, o, u, x, y | XII | t10134 |
| 17SBCL225STA | France | Food (SFPO) | Milk product | 20 | g, i, m, n, o, u, x, y | XII | * |
| 18 SBCL 680 | Switzerland | Food | Milk product | 20 | g, i, m, n, o, u, x, y | XII | t1544 |
| 18 SBCL667 | Switzerland | Food | Milk product | 20 | g, i, m, n, o, u, x, y | XII | * |
| G11F | Switzerland | Animal | Mastitis (cow) | 20 | g, i, m, n, o, u | XII | t2736 |
| 13CEB179STA | Ireland | NA | FPO | 22 | c, g, i, m, n, o, u, x | XVI | * |
| 15SBCL1517STA | Ireland | Food (SFPO) | Meat | 22 | c, g, i, l, m, n, o, u, x | XVI | t645 |
| 15SBCL1527STA | Ireland | Food (SFPO) | Ready to eat | 22 | g, i, m, n, o, u, x | XVI | * |
| 13CEB181STA | Ireland | Food (SFPO) | Ready to eat | 30 | a, g, i, m, n, o, u | III | t3018 |
| 13CEB312STA | Belgium | Food (SFPO) | Ready to eat | 30 | a, g, i, m, n, o, u | III | t022 |
| 13CEB313STA | Belgium | Human (SFPO) | Human faeces | 30 | a, g, i, m, n, o, u | III | * |
| 13CEB317STA | Belgium | Human (SFPO) | Nose and throat | 30 | a, g, i, m, n, o, u | III | * |
| 13CEB318STA | Belgium | Human (SFPO) | Nose and throat | 30 | a, g, i, m, n, o, u | III | * |
| 13CEB327STA | Belgium | Human (SFPO) | Nose and throat | 30 | g, i, m, n, o, u | III | * |
| 13CEB328STA | Belgium | Human (SFPO) | Nose and throat | 30 | g, i, m, n, o, u | III | * |
| 18 SBCL671 | Switzerland | Food (SFPO) | Milk product | 30 | g, i, m, n, o, tst, u | III | t021 |
| 18SBCL675 | Switzerland | Food | Ready to eat | 30 | g, i, m, n, o, tst, u | III | t021 |
| 18SBCL678 | Switzerland | Food | Ready to eat | 30 | g, i, m, n, o, u | III | t166 |
| ATCC 25923 | USA | Human | Skin | 30 | g, i, m, n, o, u | III | t021 |
| KS90 | Switzerland | Food (SFPO) | Ready to eat | 30 | g, i, m, n, o, u | III | t021 |
| MRSA252 | USA | Human | Infection | 30 | g, i, m, n, o, u | III | t018 |
| 07CEB90STA | Belgium | Food (SFPO) | Ready to eat | 45 | c, g, i, m, n, o, u | XXII | t1040 |
| 18 SBCL 676 | Switzerland | Food | Ready to eat | 45 | g, i, l, m, n, o, u | XXII | t505 |
| 18SBCL673 | Switzerland | Food (SFPO) | Milk product | 45 | g, i, m, n, o, u | XXII | t015 |
| 18SBCL674 | Switzerland | Food (SFPO) | Milk product | 45 | g, i, m, n, o, u | XXII | t015 |
| 18SBCL677 | Switzerland | Food | Ready to eat | 45 | g, i, l, m, n, o, u | XXII | t505 |
| USA600 | USA | Human | Infection | 45 | g, i, m, n, o, u | XXII | t004 |
| 18SBCL672 | Switzerland | Food | Milk product | 50 | i, m, n, o, u, x, z | XXI | t246 |
| GN3 | Japan | Human | NA | 50 | i, m, n, o, u | XXI | t185 |
| 13CEB323STA | Belgium | Human (SFPO) | Nose and throat | 72 | c, x, g, i, m, n, o, u | XX | t022 |
| 05CEB52STA | NA | Human | FPO | 121 | b, g, i, m, n, o, u, y, x | XIX | * |
| 18SBCL669 | Switzerland | Food | Milk product | 479 | d, g, i, m, n, o, u, x | XI | t7013 |
| G68P | Switzerland | Animal | Mastitis (cow) | 479 | g, i, m, n, o, u | XI | t7013 |
| 13CEB182STA | Ireland | Food (SFPO) | Milk product | 705 | c, i, m, n, o, tst, u, x | IV | t529 |
| 13CEB190STA | Ireland | Food (SFPO) | Milk product | 705 | c, i, m n, o, tst, u, x | IV | t529 |
| 15SBCL1438STA | France | Food (SFPO) | Milk product | 705 | c, i, m n, o, tst, u, y, x | IV | t529 |
| 18SBCL670 | Switzerland | Food | Milk product | 705 | c, i, m n, o, tst, u, y, x | IV | t529 |
| M1280 | Switzerland | Animal | Mastitis (cow) | 705 | c, i, m, n, o, u | IV | t529 |
| M1655 | Switzerland | Animal | Mastitis (cow) | 705 | c, i, m, n, o, u | IV | t529 |
| M2323 | Switzerland | Animal | Mastitis (cow) | 705 | c, l, i, m n, o, tst, u | IV | t529 |
| M2682 | Switzerland | Animal | Mastitis (cow) | 705 | c, i, m, n, o, u | IV | t529 |
| M2839 | Switzerland | Animal | Mastitis (cow) | 705 | c, l, i, n, o, tst, u | IV | t529 |
| M3783 | Switzerland | Animal | Mastitis (cow) | 705 | i, m, n, o, u | IV | t529 |
| RF122 | Ireland | Animal | Mastitis (cow) | 705 | c, i, l, m, n, o, u, tst, x, y, z | IV | t529 |
| 17SBCL13STA | France | Food (SFPO) | Meat | ** | a, g, i, m, n, o, x | XVIII | t13785 |
NA, data not available; SFPO, food poisoning outbreak; *, unknown spa type; **, unknown clonal complex (CC).
The strains from the most frequently found CCs (CC5 and CC30) originated from a vast geographical range and were isolated from either human or food. In contrast, the CC20 and CC705 strains, always originating from France, Italy, and Switzerland, were isolated either from dairy products or bovine mastitis (Table 1).
spa typing of the 75 strains revealed that in most cases the strains belonging to a single CC were allocated to different spa types. Perfect agreement between CC and spa type was found only for CC705 (n = 11), where all strains were allocated to t529. For 15 strains, spa typing resulted in an unknown type, of which the majority belonged to CC30 and CC20 (5 and 6 unknown spa types, respectively).
Besides egc, the 75 strains also harbored other non-egc SE genes (Table 1). Indeed, from genome assembly, all 27 SE genes were detected in one of the strains at least once, yet it is noteworthy that the five strains belonging to CC5 often carried additional SE genes, such as selx (in 4 strains), sea (in 3 strains), and a plasmid containing sed, sej, and ser (in 2 strains). Furthermore, CC30 (n = 13) harbored sea in 6 strains and tst (toxic-shock toxin) in 2 strains.
CC705 was comprised of sec, tst, selx, and sel, whereas CC20 often carried selx and sey (in 14 and 11 out of 15 strains, respectively).
Allocation of the strains to their vSaβ types and diversity of SEG and SEI.
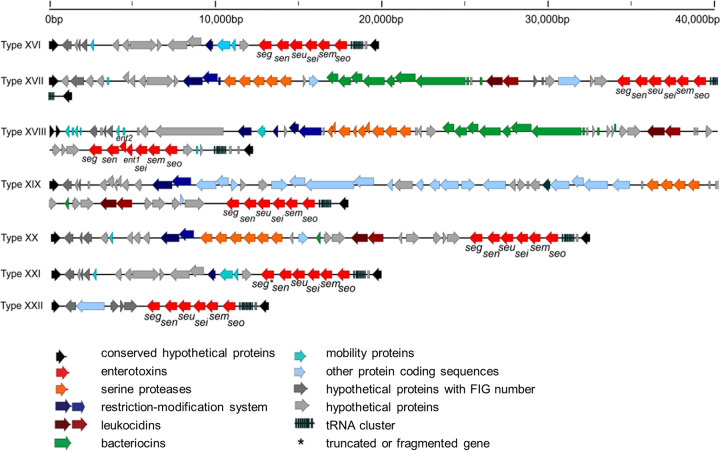
In 59 of 75 strains (79%), the vSaβ type could be allocated to an existing one with overall similarities of >90%. For the remaining 16 strains, new vSaβ types were defined by numbering continuously from XVI onward (Fig. 1), resulting in seven new vSaβ types (XVI to XXII). Three strains were allocated to vSaβ type XVI, two strains to vSaβ type XXI, and six strains to vSaβ type XXII, respectively (Table 1). For the remaining vSaβ types (XVII, XVIII, XIX, and XX), only one strain of each was found.
FIG 1.
Representation of the newly defined S. aureus genomic island vSaβ types XVI to XXII. The virulence-associated genes, and other hypothetical genes located on vSaβ, are also presented. For each vSaβ type, one reference strain is shown. Arrows show the orientation of open reading frames. FIG numbers are hp genes that were assigned to a FIG number by the RAST (Rapid Annotations using Subsystem Technology) pipeline. ent1 and ent2 of vSaβ type XVIII are genes that were already described by Collery and Smyth (78). *, truncated or fragmented gene.
The seven newly defined vSaβ types (Fig. 1) all contained, in addition to the egc genes, virulence-associated and hypothetical genes. vSaβ types XVII and XVIII carry bacteriocins and serine proteases, whereas vSaβ type XIX was notably (approximately 20,000 bp) longer than the other vSaβ types and carried numerous genes coding for hypothetical proteins. vSaβ type XXII was shorter than all other vSaβ types (approximately 13,000 bp) and did not carry any additional virulence-associated genes besides the egc genes.
Within each vSaβ type, an amino acid identity of 100% for each SE was observed. However, SE differences were observed among different vSaβ types (Table 2). Among all strains included in the study, the SEG amino acid similarity varied between 96% and 100%, with a maximum of 9 amino acids of difference, compared to strain Mu50 (reference). For SEI, the similarity varied between 93% and 100%, with a maximum difference of 19 amino acids.
TABLE 2.
Amino acid similarity of SEG and SEI compared to the reference strainsa (Mu50 and vSaβ type I)
| vSaβ type | Amino acid similarity (%) |
|
|---|---|---|
| SEG | SEI | |
| I | 100R | 100R |
| III | 97 | 95 |
| IV | * | 95 |
| XI | 97 | 93 |
| XII | 100 | 100 |
| XIII | 99 | 100 |
| XVI | 100 | 99 |
| XVII | 96 | 97 |
| XVIII | 100 | 100 |
| XIX | 97 | 93 |
| XX | 99 | 100 |
| XXI | * | 97 |
| XXII | 100 | 99 |
Each vSaβ type sequence is represented based on 100% intergroup similarity. Superscript R, reference; *, gene absent.
Phylogenetic analysis of the core genome.
To evaluate the evolutionary relationship of S. aureus strains included in the present study, their phylogeny was evaluated based on their core genomes. The tree shows a perfect concordance between the phylogenetic clades, CCs, and vSaβ type of the strain (Fig. 2). For vSaβ type IV, XI, XII, and XIII, a perfect concordance was observed between strains isolated from milk products, and animal mastitis can be observed (no human strains harbored these vSaβ types). On the other side, strains harboring vSaβ type I, III, and XXII were only found in humans (including infections) and food isolates. No animal strains harbored these vSaβ types. SFPO strains were found in every vSaβ type.
FIG 2.
Maximum likelihood phylogenetic tree based on the core genome (nucleotidic sequences) showing the evolutionary relationship among 75 isolates of Staphylococcus aureus (all strains positive for the enterotoxin gene cluster) recovered from human, animal, environment, and food samples (left). At the right, for each strain its clonal complex (CC), origin of the strain, source of the strain, and involvement in staphylococcal food poisoning outbreak (SFPO) is given. Bootstrap values of >80 are shown. Production of enterotoxin G (SEG) and I (SEI) for the 32 analyzed strains is also given (last two columns). These are shown as L for low enterotoxin production and H for high enterotoxin production. *, statistical outliers; **, unknown CC.
Enterotoxin production.
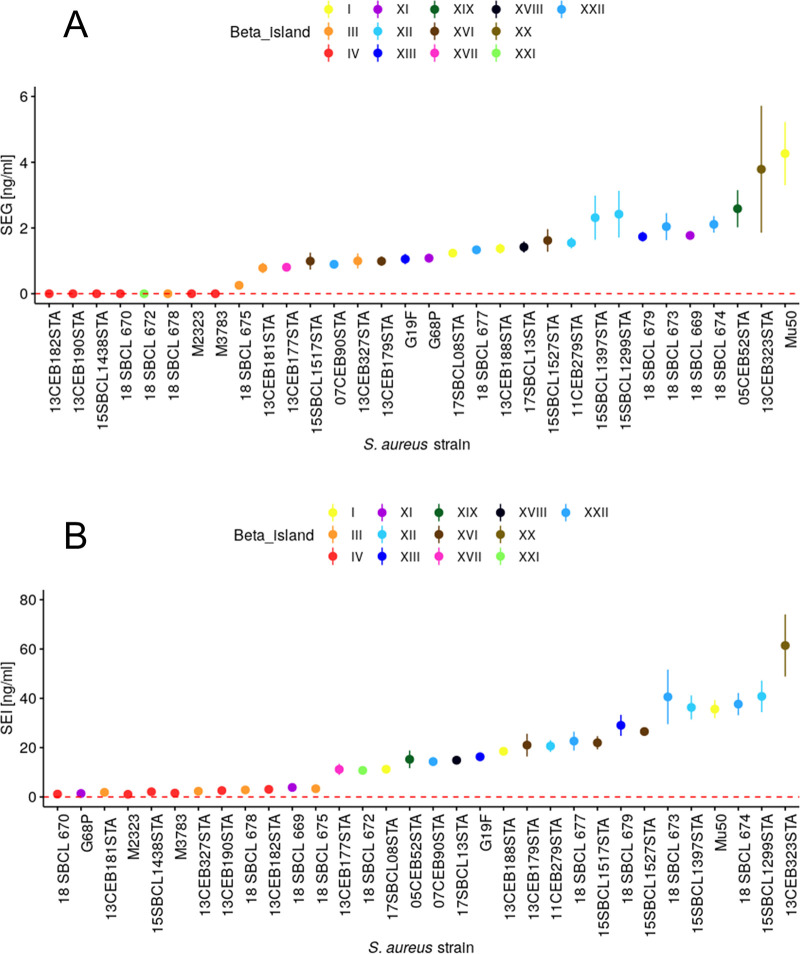
SEG production ranged from below the limit of detection (LOD; 0.001 ng/ml) to 4.26 ± 0.78 ng/ml, with a median of 1.17 ng/ml. SEG production below the LOD (0.001 ng/ml) was observed for vSaβ IV and XXI. One strain carrying vSaβ III (18SBCL675) showed nondetectable quantities of SEG, whereas the other two strains harboring vSaβ III had values between 0.26 ± 0.01 and 0.78 ± 0.13 ng/ml. All the other strains showed values between 0.80 ± 0.11 and 4.26 ± 0.78 g/ml. By visual data inspection (Fig. 3A), two levels of SEG production can be distinguished: 9 strains that generated low (L) and 23 strains that produced high (H) concentrations of SEG. The median concentration for the L producer was 0 ng/ml (minimum [min], 0 ng/ml; maximum [max], 0.26 ± 0.01 ng/ml) and for the H producer was 1.42 ± 0.14 ng/ml (min, 0.783 ± 0.13 ng/ml; max, 4.26 ± 0.78 ng/ml). The difference between medians was highly significantly (P < 0.001).
FIG 3.
Amount (nanograms per milliliter) of staphylococcal enterotoxins G and I (A and B, respectively), measured with enzyme-linked immunosorbent assay. Each point is the average measurement from three biological replicates, and the corresponding bars represent standard deviations. Strains were incubated in brain heart infusion (BHI) for 24 h at 37°C. The limit of detection of the corresponding enterotoxin is presented by red dashed line (LOD SEG, 0.001 ng/ml; LOD SEI, 0.037 ng/ml).
The amount of SEI produced (LOD, 0.037 ng/ml) by the strains ranged from 1.06 ± 0.17 ng/ml to 61.43 ± 10.29 ng/ml (median, 14.31 ng/ml) (Table 1 and Fig. 3B). According to their SEI production, strains could again be visually allocated to two different levels, L producers (producing 1.06 ± 0.17 to 3.85 ± 0.99 ng/ml; median, 2.22 ng/ml) and H producers (10.77 ± 1.22 to 61.43 ± 10.29 ng/ml; median, 21.51 ng/ml). The L strains belonged to the vSaβ types III, IV, and XI, whereas the H strains belonged to vSaβ types I, XII, XIII, XVI, XVII, XVIII, XIX, XX, XXI, and XXII (P < 0.001 between L and H).
To assess a possible relationship between SEG and SEI production, first a robust linear regression (see Fig. S1 and S2 in the supplemental material) was performed, identifying four outliers (G68P, 18SBCL669, Mu50, and 05CEB52). These outliers were not taken into consideration for a second, ordinary least-square linear regression analysis (Fig. S3). This regression was modeled to [SEI] = 15.49 × [SEG] + 0.63, with R = 0.940 (P < 0.001), where brackets indicate the SE concentrations in nanograms per milliliter.
DISCUSSION
In the present study, we demonstrate that SEG and SEI production in vitro can be predicted using genomic data. In fact, there are strong indications that the amount these SE produced depends on the vSaβ type. Furthermore, with the analysis and findings described here, it is now possible to infer the origin of an egc-containing S. aureus strain (human derived, cattle derived) that is involved in an SFPO. As the vSaβ type is perfectly linked to the CC of a strain, as shown in the present study and in a previous report from Kläui et al. (30), the SE production and the origin of the SFPO also can be predicted based on the CC of the strain obtained by MLST, a typing method that is well established.
Previous studies already demonstrated that different strains can produce different amounts of SE, but in most cases the link to the genome was missing (31, 32).
In this study, the focus was on the egc enterotoxins that, according to previous studies (3, 12, 33, 34), are harbored by about 50% of S. aureus strains. The importance of the egc enterotoxins regarding food safety has been shown by Johler et al. (3), who described the probable egc enterotoxins’ involvement in foodborne outbreaks. However, strong evidence could not be confirmed, as the enterotoxin measurement in the food and from the bacteria could not be performed due to lack of appropriate methods. This could also be the reason why a lot of egc-caused SFPO remain undiscovered. In this study, for two enterotoxins (SEG and SEI) out of the five egc enterotoxins, an enzyme-linked immunosorbent assay (ELISA) method was available, whereas for the other egc enterotoxins this still is not the case. seu was not considered at all, as there is no literature demonstrating its emetic activity. Due to this lack of information about egc enterotoxins, new methods and tools need to be developed to better understand and predict their expression and regulation mechanisms (29). As a consequence, the aim of the present study was to determine whether WGS data can be used to predict staphylococcal enterotoxin production of the egc in vitro, particularly of SEG and SEI.
Prediction of SEG and SEI production in vitro.
For the present study, 75 strains were chosen, originating from both human hosts and animal (cattle) as well as from environmental and food sources, with special attention on SFPO strains (35). Out of the 75 strains, 60 were allocated to the 15 previously defined vSaβ types (30). The remaining 15 strains could be grouped into 7 newly defined vSaβ types (Fig. 1). According to these new insights, using the vSaβ types seems to be a very precise tool to characterize the different egc present in S. aureus strains instead of using egc types I to VI, as has been described previously (14, 21, 22, 36, 37).
The present study shows that there are two groups of SE producers, strains that produce low levels of SEG and SEI and strains with increased SE production (for both, SE P < 0.001). A special case is the absence of SEG production for vSaβ IV and XXI. This is explained by the fact that both had a truncated seg gene, resulting in an incomplete, nondetectable protein.
A very high linear dependency was observed between the production of SEG and SEI (R = 0.98, P < 0.001), while the amount of SEI measured was approximately 16 times higher than that of SEG. The high correlation between SEG and SEI production suggests that both SE are regulated primarily by the same transcription factor as that proposed by Kusch et al. (38). This hypothesis, however, neglects the fact that the SEG production is 16× lower than that for SEI, accounting for a fine tuning by additional transcription factors, as observed for other SE (38, 39).
During the first robust linear regression analysis, outliers were observed (G68P, 18SBCL669, and 05CEB52). For these strains, all members of vSaβ types XI and XIX, the production of SEI was always lower than SEG production (see Fig. S2 in the supplemental material). As demonstrated in Table 2, SEI of both vSaβ types showed the lowest similarity (93%) compared to the reference (Mu50). These findings indicate that the monoclonal antibody used for the present study matches incompletely with the SEI epitopes produced by vSaβ type XI- and XIX-producing strains, resulting in a reduced detection of SEI quantities. Besides the technical aspect, it cannot be ruled out; however, regulation of SEI production is special for these vSaβ types. To clarify these ambiguities, additional studies are required.
The results of the present European study were not in agreement with the results published by Omoe et al. (40), who detected SEI in only 40% of the strains and SEG was not detected at all. In our study, SEG was produced by 96% of the strains and SEI for 100% of the strains, being positive for the two enterotoxin genes detected by NAuRa (35). Only for one strain (18SBCL678) was seg predicted, but SEG enterotoxin was not detected. As our results were generated from a large variety of strains, the involvement of the egc enterotoxins in SFPO should be reconsidered.
Inferring the origin of an SFPO-involved strain.
Looking at the major vSaβ types found in this study (I, III, IV, XII, and XXII), it was observed that in each group there are SFPO-associated strains (isolated from food) and strains that are human (infection) or cattle (mastitis) derived but never both for the same vSaβ type.
In addition to our previous study with 15 allocated vSaβ type observed (30), we found 9 new types. Again, a perfect concordance between vSaβ type and CC was found, confirming this observation as a general principle in S. aureus. This principle can now be applied for evaluation of egc-containing strains involved in SFPO. In fact, instead of inferring the vSaβ type involved in the SFPO, the common and simpler method of CC assessment can be performed. This is particularly easy for WGS data, as the reads can be directly uploaded to an Internet app, such as cge.cbs.dtu.dk, for inferring of the sequence type (ST), which is then used together with the pubMLST database program (41) to obtain the corresponding CC. If WGS data are not available, the standard MLST procedure can be performed using standard PCR and Sanger sequencing for the seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpiA, and yqiL) (42). Instead of the original primers (35), the newly designed primer by Boss et al. (43) can be applied. They enable unidirectional Sanger sequencing, which considerably saves cost, work, and time.
The suspected reason for this strong link between CC and vSaβ type is that vSaβ acquisition by horizontal transfer in the S. aureus genome happened immediately before or simultaneously to clonal diversification of S. aureus (30). This hypothesis is also supported by the phylogenetic analysis of the core genomes of the present study (Fig. 2), showing a perfect agreement between the phylogenetic clade, CC, and vSaβ type.
The CC can be used to perform an association of an egc-carrying SFPO strain to a specific origin (human or cattle). As can be seen in Fig. 2, CC705 and CC20 are strains strictly associated with bovine mastitis and dairy products. In fact, CC705-positive strains are classical pathogens of bovine mastitis observed in- and outside of Europe (43–45). In addition, they are also frequently present in delivered milk (43) and cheese (46). CC705 strains are uniquely positive for spa type t529 (Table 1) and are typical colonizers of bovine skin as well as infections of bovine wounds (47). Similar findings are also true for CC20-positive strains. These can also cause bovine mastitis and are present in delivered milk, but they are less abundant than CC705 (43).
On the other hand, strains allocated to CC5, CC30, and CC45 were exclusively isolated from human samples (infection, skin, feces, nose, and throat) and from food (Fig. 2), where human handling was very likely (ready-to-eat products). Furthermore, these CCs are widely described in the literature as being found in human infections (48–51). This is a further advantage of CC nomenclature as literature about them is broad (23, 52, 53), enabling us to extend the scope beyond an egc enterotoxin-caused SFPO.
Application of new insights in evaluation of egc-caused SFPO.
The involvement of egc enterotoxins in foodborne outbreaks is highlighted by the fact that vSaβ types (and the corresponding CC) from S. aureus strains producing high levels of SEG and SEI are also described to be involved in foodborne outbreaks, especially CC5, CC20, and CC45 (23, 34, 53). Furthermore, certain strains of CC45 (harboring egc) do not harbor any classical enterotoxin (34, 54), yet these strains could have been involved in foodborne outbreaks.
As an example, we deal with strain 18SBCL673, which was involved in a foodborne outbreak related to artisanal goat cheese in southern Switzerland (54) and was included in the present study. It is characterized by the presence of just egc enterotoxins, as shown by NAuRa, and produces a substantial amount of SEG (2.04 ± 0.33 ng/ml) and SEI (40.58 ± 9.03 ng/ml). It is positive to vSaβ type XXII and CC45. As the strain had been isolated from goat cheese, it could be hypothesized that goat milk was the probable source. However, according to the present study (Fig. 2), it is clear that the origin of the involved strain is, with a high probability, human. As a consequence, the SFPO caused by this strain was a highly human contamination during cheese manufacturing. This conclusion is supported by the fact that CC45 is never found in goats and goat milk (55, 56).
Conclusions.
The presented study demonstrates that the in vitro production of SEG and SEI can be predicted based on the vSaβ type and the CC of a strain. Furthermore, the vSaβ type/CC enables us to predict the source of an egc-positive SFPO strain (animal or human derived). Due to the perfect correlation between CC and vSaβ type, the use of common CC typing is an easy and quick way to characterize a strain involved in an SFPO. Therefore, it is a good alternative to the proposed egc typing (I to IV), a method that results in only four biologically irrelevant types.
This information will enhance the ability to better understand the involvement of the egc enterotoxins in SFPO. The fact that the egc is found in more than 50% of the S. aureus strains and, according to our study, exactly 75% expressed SEG and 100% expressed SEI are further indications that these and other egc enterotoxins are involved in SFPO.
MATERIALS AND METHODS
Bacterial strain and genome collection.
The general aim was to use egc-harboring S. aureus strains representing a large diversity in their genomes and origins. To achieve this, 75 strains and genomes from different sources (food, environment, animal, and human) as well as different geographical origins were chosen (Table 1). SFPO genome sequences and strains (42 genomes and strains) were obtained from the collection of the European National Reference Laboratory for Coagulase-Positive Staphylococci (EURL CPS; Maisons-Alfort, France). Nine Swiss bovine mastitis strains were used from the Agroscope strain and genome collection; these strains were sampled previously by Fournier et al. (57) and their genome sequenced by Kläui et al. (30). For genomic and phylogenetic analysis, seven strains of human and animal origins were obtained from NCBI (reference sequence database; https://www.ncbi.nlm.nih.gov) to increase the sample size and variation of the strains. Two strains (Mu50 and N315) were obtained from P. Moreillon (University of Lausanne). Thirteen egc-containing strains were isolated from food in Switzerland (details are described below). An overview of the bacterial strain collection used in study is listed in Table 1.
Characterization of Swiss food strains.
Forty-five Swiss S. aureus strains originating from food were obtained from the Federal Food Safety and Veterinary Office (kindly provided by A. Baumgartner). The presence of egc genes in these strains was determined by applying a real-time PCR assay with melting curve analysis (mPCR) for detection of seg, sei, sem, sen, and seo. For detection of seg and sei, primers and PCR conditions were applied as described by Cosandey et al. (58). For detection of sem, sen, and seo, new primers were designed (Table 3). The detection of seu was omitted, as its emetic activity has not been shown so far. After being cultured at 37°C for 24 h on blood agar (bioMérieux Suisse s.a., Geneva, Switzerland), DNA was extracted from single colonies of S. aureus. A colony was picked and resuspended in 100 μl of 10 mM Tris-HCl and 10 mM EDTA (pH 8.5), incubated at 95°C for 10 min, and immediately stored on ice. The lysates were diluted 1:100 in H2O to be used as templates for the different mPCRs (43). For all mPCRs, the total volume was 20 μl, containing 300 nM corresponding forward and reverse primer (Table 3), 1× Kapa Sybr Fast (Kapa Biosystems Inc., Woburn, MA), and 2.5 μl of 1:100 diluted DNA template. The mPCR run began with an initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 3 s, annealing and extension in a single step at 60°C for 50 s, and a final extension step at 60°C for 5 min. Melting of the amplicons was performed between 60°C and 94°C, with increments of 1°C and a 5-s waiting time at each step. The mPCRs were performed using a Rotor-Gene 6000 real-time thermal cycler (Corbett Life Science, Mortlake, Australia).
TABLE 3.
Primers for detection of enterotoxin genes developed and used in this study
| Gene | Primera | Sequence 5′–3′ | Amplicon size (bp) |
|---|---|---|---|
| sem | Gsem_S | GATGTCGGAGTTTTGAATCTTA | 584 |
| Gsem_AS | ACTTTCAGCTTGCCCTGTT | ||
| sen | Gsen_S | TTCTTCCAGTTAAGCCTACACA | 218 |
| Gsen_AS | CTGATATAACGTGGCAATTAG | ||
| seo | Gseo_S | TAAAGCGCATTGTCATGGTGAG | 348 |
| Gseo_AS | ACATCACTAGTCATTCGGTCATA |
S, sense primers; AS, antisense primers.
Primer specificity (Table 3) was tested with S. aureus strains that were previously sequenced, namely, G11F, G19P, M1280, M1655, M2323, M2682, M3783, Mu50, and N315 (Table 1).
Applying the mPCR for detection of the egc genes showed that only 40% of the strains were egc positive. Based on the diversity of their origins, 14 egc-positive strains were selected.
These Swiss strains, isolated from food, were sequenced as follows. Strains were cultured at 37°C for 24 h on blood agar, 3 to 4 colonies were suspended in 4.5 ml tryptic soy broth (TSB; Becton, Dickinson), and incubated 18 h (37°C, with shaking). From this overnight culture (ONC), 1 ml was suspended in 500 ml TSB and incubated under the same conditions. The resulting ONC was centrifuged for 23 min (7°C, 6,000 × g) (Cellsep 6/720R; Henderson Biomedical Ltd., Lower Sydenham, UK). The supernatant was discarded and the pellet resuspended in 15 ml 10 mM Tris-HCl, pH 7.8, and transferred to a falcon tube, which again was centrifuged for 5 min (4°C, 18,000 × g). After centrifugation, the pellet was treated using the NucleoBond Xtra Maxi kit (Machery Nagel, Düren, Germany) according to the manufacturer’s protocol, with the following modifications: instead of resuspending the pellet directly in 24 ml RES (from the kit), the pellet was resuspended in 2 ml RES containing 350 mg glass beads (425 to 600 μm; Merck, Darmstadt, Germany) and shaken on a Bead Ruptor at level 6 for 45 s (Bead Ruptor Elite; Omni International, Kennesaw, GA, USA). After centrifugation for 5 min (4°C, 13,500 × g), 22 ml was added to the supernatant, and DNA was then extracted according to the protocol of the manufacturer of the kit. The pellet was resuspended in 200 μl ddH2O (double-distilled water) and further purified by applying the High Pure PCR template preparation kit protocol (Roche, Basel, Switzerland). DNA quality was considered sufficient if the optical density at 260 nm (OD260)/OD280 was ≥1.8 and OD260/OD230 was ≥1.9 (measured with a QuickDrop spectrophotometer; Molecular Devices, San Jose, CA). The extracted DNA (representing the whole genome) was sequenced by an Illumina HiSeq at Eurofins GATC (Constance, Germany), generating more than 1.5 Gb of reads.
Bioinformatics.
The reads from the strains from EURL CPS were obtained from the European Nucleotide Archive database (https://www.ebi.ac.uk/ena). For these reads and the reads from the Swiss food strains, the method for assembly and annotation was applied according to Merda et al. (35). Before the assembly, reads were normalized using BBnorm (https://jgi.doe.gov/data-and-tools/bbtools/) to have a maximum coverage of 100×. Normalized reads were trimmed using Trimmomatic (59). Quality filtering then was performed, removing reads shorter than 50 bp as well as excluding bases having a Phred quality score lower than 30. With these filtered reads, assembly was performed in three steps: (i) a de novo assembly was generated using SPAdes (v.3.9.1) (60) applying the default parameters, (ii) scaffolding was performed in MeDuSa (61), using the nearest complete public genome of S. aureus estimated by Mash (62), and (iii) gaps were closed using GMcloser (63). The quality of each assembled genome was assessed with QUAST (v.4.3) (64). The assemblies were annotated using Prokka (v.1.11) (65) and RAST (66) for the prediction of coding sequences (CDSs).
MLST, spa type, and vSaβ type allocation.
For all 75 genomes used in this study, three typing methods were applied to further characterize the strains genomically: (i) multilocus sequence typing (MLST), (ii) spa typing, and (iii) vSaβ typing. The MLST of the seven housekeeping genes (67) and spa typing (68) were done by using the Center for Genomic Epidemiology online platform (http://www.genomicepidemiology.org/). In the pubMLST database program (41), the sequence types (STs) from MLST were used to allocate each strain to a CC. For ST504 in the actual pubMLST database, no corresponding CC is available; as a consequence, this ST was allocated to CC705, as also described in the literature (43). vSaβ islands were identified in the genome by applying the method described by Kläui et al. (30). Briefly, if the vSaβ island of a strain had a sequence similarity of ≥90% to the reference strain of any existing vSaβ type, it was considered of the same type (30). If the sequence similarity was <90%, the vSaβ island was defined as a new type. All alignments were performed by using the Needleman-Wunsch algorithm of Clone Manager Professional 9 software (Scientific & Educational Software, Denver, CO).
Enterotoxin gene profiles.
The enterotoxin gene profiles of the S. aureus strains, based on the WGS, were determined using the NAuRA tool (https://github.com/afelten-Anses/NAuRA). The screening of the enterotoxins was performed using the gene sequence and their relative protein sequence of the already described 27 SE and the estimated parameters of BLAST by Merda et al. (35).
Phylogenetic analysis.
The core genome of each of the 75 strains was determined by the Roary pipeline (69). For this, the previously obtained GFF3 file from Prokka was used as an input containing all of the strains’ genes as detected by the software. All genes of a strains’ core genome were then concatenated. A multiple-sequence alignment (MSA) (using MAFFT [70]) was performed using the concatenated core genomes of all the strains. The MSA then was imported into the Gblock program (71) for quality checking using the default setup and removing any misaligned regions. A phylogenetic tree was constructed using the maximum-likelihood method in IQtree (72). This program estimated the evolutionary model of sequences, and the best model, according to Akaike criterion, was GTR + I + gamma. The branch support was calculated by the bootstrap method, using 1,000 replicates. The graphic representation of the phylogeny was obtained by using iTOL web viewer (https://itol.embl.de/) (73).
Staphylococcal enterotoxin measurement.
The 32 S. aureus strains used for the enterotoxin measurements are shown in Fig. 3A and B. These were selected based on their allocation to the different vSaβ islands (Table 1). If available, three strains per vSaβ type were used. The selected strains were cultivated on plate count agar (PCA; Becton, Dickinson, Franklin Lakes, NJ) for 24 h at 37°C, and then 3 single colonies were taken and suspended in 45 ml brain heart infusion broth (BHI; Becton, Dickinson). The inoculated broth was then incubated at 37°C for 24 h in a flask with shaking. After 24 h, the optical density of the culture was measured to check the growing of the cells (OD480 > 1.8). The culture was transferred to a falcon tube and centrifuged at 8,000 × g for 15 min at room temperature. The supernatant was then filtered through a 0.2-μm syringe filter, and the resulting filtrate was used for the downstream analysis. Quantitative analysis of SEG and SEI was performed by using an in-house sandwich quantitative ELISA. seg (GenBank accession no. CP001781.1) and sei (GenBank accession no. CP001781.1) genes from S. aureus were synthesized (Genecust) and inserted into a bacterial plasmid [isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible pET22b(+) vector; Novagen, Merck] for inducible expression of recombinant SEG and SEI toxins (here used as immunogens and standards). Specific laboratory-made monoclonal antibodies were used as coating and probing biotinylated antibodies. Briefly, Biozzi mice were immunized 4 times at 3-week intervals with 10 μg of recombinant SEG or SEI toxin in alum adjuvant (intraperitoneal injection). After intravenous boost injections, hybridomas were produced by fusing spleen cells with NS1 myeloma cells, as previously described by Köhler and Milstein (74). Monoclonal antibodies were produced from hybridoma culture supernatants and further purified by protein A or protein G affinity chromatography using the AKTAxpress system (GE Healthcare, Chicago, USA).
Two separate 96-well polystyrene microtiter plates (MaxiSorp; Nunc, Roskilde, Denmark) were coated with 100 μl of monoclonal anti-SEG IgG or anti-SEI IgG (SEG41 and SEI27) at 10 μg/ml in 50 mM phosphate-buffered saline (PBS), pH 7.4, overnight at room temperature (RT), and blocked with 300 μl/well of enzyme immunoassay buffer (0.1 M PBS, pH 7.4, 1 g/liter bovine serum albumin, 0.1 g/liter sodium azide) for at least 4 h at RT. Saturated microplates were washed by 300 μl of phosphate-Tween 20 before use. A calibration curve was prepared with dilutions of SEG- and SEI-purified recombinant toxins with five concentrations between 0 and 0.3 ng SEG/ml and 0 and 2.0 ng SEI/ml, respectively (duplicate calibration points per level). Samples and recombinant standard toxins (100 μl/well) were distributed and incubated at RT for 60 min and washed three times with PBS-Tween 20, followed by addition of 100 ng/ml of biotinylated monoclonal anti-SEG or anti-SEI antibody (SEG27 and SEI26) at RT for 60 min. After extensive washing, 100 μl/well of poly-horseradish peroxidase-labeled streptavidin (dilution 1/50,000; Thermo Fisher Scientific) was used for detection at RT for 30 min and washed 5 times again. Substrate solution (100 μl/well) containing tetramethylbenzidine (TMB; Thermo Fisher Scientific, Waltham, MA) then was added for 30 min. Finally, the reaction was stopped by addition of 100 μl of H2SO4 2 N. Absorbances were read at 450 nm on a microplate reader (SAFAS; Monaco). Quantification was performed by using a calibration curve based on the quadratic fit model. Validation data (sensitivity, specificity, and repeatability) of the above-described method are unpublished (Cécile Féraudet-Tarisse, Céline Goulard-Huet, Yacine Nia, Karine Devilliers, Dominique Marcé, Chloé Dambrune, Donatien Lefebvre, Jacques-Antoine Hennekinne, and Stéphanie Simon, unpublished data).
Statistical analysis.
For analysis of potential correlation between production of SEG and SEI, a regression analysis was performed. First, the robust method was applied to verify the regression model and to identify outliers. Four outliers were identified and eliminated from the data set before calculating an ordinary least-square regression model.
To proof the two different levels of SEG and SEI production, a Kruskal-Wallis test was performed. For all statistical analyses, measured values under the limit of detection were taken as the value 0.
All statistical analyses were performed in Systat (version 13; Systat, Chicago, IL).
The graphical presentation of the enterotoxin data was performed using R (version 3.4.4) with the packages ggplot (75), ggsignif (76), and ggpubr (77). With these packages, the data of the enterotoxin production of the single strains were plotted in increasing order of production (means ± standard deviations) and a color given according to their relative vSaβ type.
Data availability.
Sequencing data for all isolates analyzed in this study have been deposited in the NCBI GenBank database under BioProject accession number PRJNA633807. Accession numbers for individual genomes and assembly statistics are listed in Tables S2 and S3.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the Agroscope research program. The work is a collaboration project between Anses and Agroscope. The development of the ELISA method for G and I was funded in part by the French joint ministerial program of R&D against CBRNE threats.
In addition, a particular thanks goes to L. Fritsch for support in using R as well as to N. Vingadassalon and I. Ivanovic for the help in their laboratories.
REFERENCES
- 1.European Food Safety Authority, European Centre for Disease Prevention and Control. 2018. The European Union One Health zoonoses report. EFSA J 17:e05926. doi: 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hennekinne JA, Guillier F, Perelle S, De Buyser ML, Dragacci S, Krys S, Lombard B. 2007. Intralaboratory validation according to the EN ISO 16 140 standard of the Vidas SET2 detection kit for use in official controls of staphylococcal enterotoxins in milk products. J Appl Microbiol 102:1261–1272. doi: 10.1111/j.1365-2672.2006.03183.x. [DOI] [PubMed] [Google Scholar]
- 3.Johler S, Giannini P, Jermini M, Hummerjohann J, Baumgartner A, Stephan R. 2015. Further evidence for staphylococcal food poisoning outbreaks caused by egc-encoded enterotoxins. Toxins (Basel) 7:997–1004. doi: 10.3390/toxins7030997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ono HK, Omoe K, Imanishi K, Iwakabe Y, Hu DL, Kato H, Saito N, Nakane A, Uchiyama T, Shinagawa K. 2008. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect Immun 76:4999–5005. doi: 10.1128/IAI.00045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ono HK, Hirose S, Naito I, Sato'o Y, Asano K, Hu DL, Omoe K, Nakane A. 2017. The emetic activity of staphylococcal enterotoxins, SEK, SEL, SEM, SEN and SEO in a small emetic animal model, the house musk shrew. Microbiol Immunol 61:12–16. doi: 10.1111/1348-0421.12460. [DOI] [PubMed] [Google Scholar]
- 6.Ono HK, Sato'o Y, Narita K, Naito I, Hirose S, Hisatsune J, Asano K, Hu DL, Omoe K, Sugai M, Nakane A. 2015. Identification and characterization of a novel staphylococcal emetic toxin. Appl Environ Microbiol 81:7034–7040. doi: 10.1128/AEM.01873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langley RJ, Ting YT, Clow F, Young PG, Radcliff FJ, Choi JM, Sequeira RP, Holtfreter S, Baker H, Fraser JD. 2017. Staphylococcal enterotoxin-like X (SElX) is a unique superantigen with functional features of two major families of staphylococcal virulence factors. PLoS Pathog 13:e1006549. doi: 10.1371/journal.ppat.1006549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benkerroum N. 2018. Staphylococcal enterotoxins and enterotoxin-like toxins with special reference to dairy products: an overview. Crit Rev Food Sci Nutr 58:1943–1970. doi: 10.1080/10408398.2017.1289149. [DOI] [PubMed] [Google Scholar]
- 9.Zhang DF, Yang XY, Zhang J, Qin X, Huang X, Cui Y, Zhou M, Shi C, French NP, Shi X. 2018. Identification and characterization of two novel superantigens among Staphylococcus aureus complex. Int J Med Microbiol 308:438–446. doi: 10.1016/j.ijmm.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Ciupescu LM, Auvray F, Nicorescu IM, Meheut T, Ciupescu V, Lardeux AL, Tanasuica R, Hennekinne JA. 2018. Characterization of Staphylococcus aureus strains and evidence for the involvement of non-classical enterotoxin genes in food poisoning outbreaks. FEMS Microbiol Lett 365:fny139. doi: 10.1093/femsle/fny139. [DOI] [PubMed] [Google Scholar]
- 11.Kerouanton A, Hennekinne JA, Letertre C, Petit L, Chesneau O, Brisabois A, De Buyser ML. 2007. Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int J Food Microbiol 115:369–375. doi: 10.1016/j.ijfoodmicro.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 12.Yan X, Wang B, Tao X, Hu Q, Cui Z, Zhang J, Lin Y, You Y, Shi X, Grundmann H. 2012. Characterization of Staphylococcus aureus strains associated with food poisoning in Shenzhen, China. Appl Environ Microbiol 78:6637–6642. doi: 10.1128/AEM.01165-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fusco V, Quero GM, Morea M, Blaiotta G, Visconti A. 2011. Rapid and reliable identification of Staphylococcus aureus harbouring the enterotoxin gene cluster (egc) and quantitative detection in raw milk by real time PCR. Int J Food Microbiol 144:528–537. doi: 10.1016/j.ijfoodmicro.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Chieffi D, Fanelli F, Cho GS, Schubert J, Blaiotta G, Franz C, Bania J, Fusco V. 2020. Novel insights into the enterotoxigenic potential and genomic background of Staphylococcus aureus isolated from raw milk. Food Microbiol 90:103482. doi: 10.1016/j.fm.2020.103482. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald JR, Monday SR, Foster TJ, Bohach GA, Hartigan PJ, Meaney WJ, Smyth CJ. 2001. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J Bacteriol 183:63–70. doi: 10.1128/JB.183.1.63-70.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol 190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macori G, Bellio A, Bianchi DM, Chiesa F, Gallina S, Romano A, Zuccon F, Cabrera-Rubio R, Cauquil A, Merda D, Auvray F, Decastelli L. 2019. Genome-wide profiling of enterotoxigenic Staphylococcus aureus strains used for the production of naturally contaminated cheeses. Genes 11:33. doi: 10.3390/genes11010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manara S, Pasolli E, Dolce D, Ravenni N, Campana S, Armanini F, Asnicar F, Mengoni A, Galli L, Montagnani C, Venturini E, Rota-Stabelli O, Grandi G, Taccetti G, Segata N. 2018. Whole-genome epidemiology, characterisation, and phylogenetic reconstruction of Staphylococcus aureus strains in a paediatric hospital. Genome Med 10:82. doi: 10.1186/s13073-018-0593-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, Kearns AM, Pichon B, Young B, Wilson DJ, Llewelyn MJ, Paul J, Peto TE, Crook DW, Walker AS, Golubchik T. 2014. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52:1182–1191. doi: 10.1128/JCM.03117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price J, Gordon NC, Crook D, Llewelyn M, Paul J. 2013. The usefulness of whole genome sequencing in the management of Staphylococcus aureus infections. Clin Microbiol Infect 19:784–789. doi: 10.1111/1469-0691.12109. [DOI] [PubMed] [Google Scholar]
- 21.Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, Etienne J, Vandenesch F, Bonneville M, Lina G. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J Immunol 166:669–677. doi: 10.4049/jimmunol.166.1.669. [DOI] [PubMed] [Google Scholar]
- 22.Thomas DY, Jarraud S, Lemercier B, Cozon G, Echasserieau K, Etienne J, Gougeon ML, Lina G, Vandenesch F. 2006. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect Immun 74:4724–4734. doi: 10.1128/IAI.00132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Argudin MA, Mendoza MC, Gonzalez-Hevia MA, Bances M, Guerra B, Rodicio MR. 2012. Genotypes, exotoxin gene content, and antimicrobial resistance of Staphylococcus aureus strains recovered from foods and food handlers. Appl Environ Microbiol 78:2930–2935. doi: 10.1128/AEM.07487-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth DS, Hartigan PJ, Meaney WJ, Fitzgerald JR, Deobald CF, Bohach GA, Smyth CJ. 2005. Superantigen genes encoded by the egc cluster and SaPIbov are predominant among Staphylococcus aureus isolates from cows, goats, sheep, rabbits and poultry. J Med Microbiol 54:401–411. doi: 10.1099/jmm.0.45863-0. [DOI] [PubMed] [Google Scholar]
- 25.Sihto HM, Tasara T, Stephan R, Johler S. 2015. Temporal expression of the staphylococcal enterotoxin D gene under NaCl stress conditions. FEMS Microbiol Lett 362:fnv024. doi: 10.1093/femsle/fnv024. [DOI] [PubMed] [Google Scholar]
- 26.Valihrach L, Alibayov B, Zdenkova K, Demnerova K. 2014. Expression and production of staphylococcal enterotoxin C is substantially reduced in milk. Food Microbiol 44:54–59. doi: 10.1016/j.fm.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 27.Zeaki N, Radstrom P, Schelin J. 2015. Evaluation of potential effects of NaCl and sorbic acid on staphylococcal enterotoxin A formation. Microorganisms 3:551–566. doi: 10.3390/microorganisms3030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Zhu A, Tang J, Tang C, Chen J. 2017. Identification and measurement of staphylococcal enterotoxin M from Staphylococcus aureus isolate associated with staphylococcal food poisoning. Lett Appl Microbiol 65:27–34. doi: 10.1111/lam.12751. [DOI] [PubMed] [Google Scholar]
- 29.Zeaki N, Johler S, Skandamis PN, Schelin J. 2019. The role of regulatory mechanisms and environmental parameters in staphylococcal food poisoning and resulting challenges to risk assessment. Front Microbiol 10:1307. doi: 10.3389/fmicb.2019.01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kläui AJ, Boss R, Graber HU. 2019. Characterization and comparative analysis of the Staphylococcus aureus genomic island vSabeta: an in silico approach. J Bacteriol 201:e00777-18. doi: 10.1128/JB.00777-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derzelle S, Dilasser F, Duquenne M, Deperrois V. 2009. Differential temporal expression of the staphylococcal enterotoxins genes during cell growth. Food Microbiol 26:896–904. doi: 10.1016/j.fm.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Borst DW, Betley MJ. 1994. Phage-associated differences in staphylococcal enterotoxin A gene (sea) expression correlate with sea allele class. Infect Immun 62:113–118. doi: 10.1128/IAI.62.1.113-118.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song M, Shi C, Xu X, Shi X. 2016. Molecular typing and virulence gene profiles of enterotoxin gene cluster (egc)-positive Staphylococcus aureus isolates obtained from various food and clinical specimens. Foodborne Pathog Dis 13:592–601. doi: 10.1089/fpd.2016.2162. [DOI] [PubMed] [Google Scholar]
- 34.Umeda K, Nakamura H, Yamamoto K, Nishina N, Yasufuku K, Hirai Y, Hirayama T, Goto K, Hase A, Ogasawara J. 2017. Molecular and epidemiological characterization of staphylococcal foodborne outbreak of Staphylococcus aureus harboring seg, sei, sem, sen, seo, and selu genes without production of classical enterotoxins. Int J Food Microbiol 256:30–35. doi: 10.1016/j.ijfoodmicro.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 35.Merda D, Felten A, Vingadassalon N, Denayer S, Titouche Y, Decastelli L, Hickey B, Kourtis C, Daskalov H, Mistou MY, Hennekinne JA. 2020. NAuRA: genomic tool to identify staphylococcal enterotoxins in Staphylococcus aureus Strains responsible for foodborne outbreaks. Front Microbiol 11:1483. doi: 10.3389/fmicb.2020.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letertre C, Perelle S, Dilasser F, Fach P. 2003. Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J Appl Microbiol 95:38–43. doi: 10.1046/j.1365-2672.2003.01957.x. [DOI] [PubMed] [Google Scholar]
- 37.Collery MM, Smyth DS, Tumilty JJG, Twohig JM, Smyth CJ. 2009. Associations between enterotoxin gene cluster types egc1, egc2 and egc3, agr types, enterotoxin and enterotoxin-like gene profiles, and molecular typing characteristics of human nasal carriage and animal isolates of Staphylococcus aureus. J Med Microbiol 58:13–25. doi: 10.1099/jmm.0.005215-0. [DOI] [PubMed] [Google Scholar]
- 38.Kusch K, Hanke K, Holtfreter S, Schmudde M, Kohler C, Erck C, Wehland J, Hecker M, Ohlsen K, Broker B, Engelmann S. 2011. The influence of SaeRS and sigma(B) on the expression of superantigens in different Staphylococcus aureus isolates. Int J Med Microbiol 301:488–499. doi: 10.1016/j.ijmm.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Fisher EL, Otto M, Cheung GYC. 2018. Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Front Microbiol 9:436. doi: 10.3389/fmicb.2018.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omoe K, Ishikawa M, Shimoda Y, Hu DL, Ueda S, Shinagawa K. 2002. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates Harboring seg, seh, or sei genes. J Clin Microbiol 40:857–862. doi: 10.1128/jcm.40.3.857-862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boss R, Cosandey A, Luini M, Artursson K, Bardiau M, Breitenwieser F, Hehenberger E, Lam T, Mansfeld M, Michel A, Mosslacher G, Naskova J, Nelson S, Podpecan O, Raemy A, Ryan E, Salat O, Zangerl P, Steiner A, Graber HU. 2016. Bovine Staphylococcus aureus: subtyping, evolution, and zoonotic transfer. J Dairy Sci 99:515–528. doi: 10.3168/jds.2015-9589. [DOI] [PubMed] [Google Scholar]
- 44.Kappeli N, Morach M, Corti S, Eicher C, Stephan R, Johler S. 2019. Staphylococcus aureus related to bovine mastitis in Switzerland: clonal diversity, virulence gene profiles, and antimicrobial resistance of isolates collected throughout 2017. J Dairy Sci 102:3274–3281. doi: 10.3168/jds.2018-15317. [DOI] [PubMed] [Google Scholar]
- 45.Cremonesi P, Pozzi F, Raschetti M, Bignoli G, Capra E, Graber HU, Vezzoli F, Piccinini R, Bertasi B, Biffani S, Castiglioni B, Luini M. 2015. Genomic characteristics of Staphylococcus aureus strains associated with high within-herd prevalence of intramammary infections in dairy cows. J Dairy Sci 98:6828–6838. doi: 10.3168/jds.2014-9074. [DOI] [PubMed] [Google Scholar]
- 46.Hummerjohann J, Naskova J, Baumgartner A, Graber HU. 2014. Enterotoxin-producing Staphylococcus aureus genotype B as a major contaminant in Swiss raw milk cheese. J Dairy Sci 97:1305–1312. doi: 10.3168/jds.2013-7643. [DOI] [PubMed] [Google Scholar]
- 47.Leuenberger A, Sartori C, Boss R, Resch G, Oechslin F, Steiner A, Moreillon P, Graber HU. 2019. Genotypes of Staphylococcus aureus: on-farm epidemiology and the consequences for prevention of intramammary infections. J Dairy Sci 102:3295–3309. doi: 10.3168/jds.2018-15181. [DOI] [PubMed] [Google Scholar]
- 48.Elie-Turenne MC, Fernandes H, Mediavilla JR, Rosenthal M, Mathema B, Singh A, Cohen TR, Pawar KA, Shahidi H, Kreiswirth BN, Deitch EA. 2010. Prevalence and characteristics of Staphylococcus aureus colonization among healthcare professionals in an urban teaching hospital. Infect Control Hosp Epidemiol 31:574–580. doi: 10.1086/652525. [DOI] [PubMed] [Google Scholar]
- 49.Espadinha D, Faria NA, Miragaia M, Lito LM, Melo-Cristino J, de Lencastre H, Medicos Sentinela N, Médicos Sentinela Network. 2013. Extensive dissemination of methicillin-resistant Staphylococcus aureus (MRSA) between the hospital and the community in a country with a high prevalence of nosocomial MRSA. PLoS One 8:e59960. doi: 10.1371/journal.pone.0059960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Donker GA, Deurenberg RH, Driessen C, Sebastian S, Nys S, Stobberingh EE. 2009. The population structure of Staphylococcus aureus among general practice patients from The Netherlands. Clin Microbiol Infect 15:137–143. doi: 10.1111/j.1469-0691.2008.02662.x. [DOI] [PubMed] [Google Scholar]
- 51.Resman F, Thegerstrom J, Mansson F, Ahl J, Tham J, Riesbeck K. 2016. The prevalence, population structure and screening test specificity of penicillin-susceptible Staphylococcus aureus bacteremia isolates in Malmo, Sweden. J Infect 73:129–135. doi: 10.1016/j.jinf.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 52.Baumgartner A, Niederhauser I, Johler S. 2014. Virulence and resistance gene profiles of staphylococcus aureus strains isolated from ready-to-eat foods. J Food Prot 77:1232–1236. doi: 10.4315/0362-028X.JFP-14-027. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki Y, Omoe K, Hu DL, Sato'o Y, Ono HK, Monma C, Arai T, Konishi N, Kato R, Hirai A, Nakama A, Kai A, Kamata Y. 2014. Molecular epidemiological characterization of Staphylococcus aureus isolates originating from food poisoning outbreaks that occurred in Tokyo, Japan. Microbiol Immunol 58:570–580. doi: 10.1111/1348-0421.12188. [DOI] [PubMed] [Google Scholar]
- 54.Johler S, Weder D, Bridy C, Huguenin MC, Robert L, Hummerjohann J, Stephan R. 2015. Outbreak of staphylococcal food poisoning among children and staff at a Swiss boarding school due to soft cheese made from raw milk. J Dairy Sci 98:2944–2948. doi: 10.3168/jds.2014-9123. [DOI] [PubMed] [Google Scholar]
- 55.Porrero MC, Hasman H, Vela AI, Fernandez-Garayzabal JF, Dominguez L, Aarestrup FM. 2012. Clonal diversity of Staphylococcus aureus originating from the small ruminants goats and sheep. Vet Microbiol 156:157–161. doi: 10.1016/j.vetmic.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 56.Merz A, Stephan R, Johler S. 2016. Staphylococcus aureus isolates from goat and sheep milk seem to be closely related and differ from isolates detected from bovine milk. Front Microbiol 7:319. doi: 10.3389/fmicb.2016.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fournier C, Kuhnert P, Frey J, Miserez R, Kirchhofer M, Kaufmann T, Steiner A, Graber HU. 2008. Bovine Staphylococcus aureus: association of virulence genes, genotypes and clinical outcome. Res Vet Sci 85:439–448. doi: 10.1016/j.rvsc.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 58.Cosandey A, Boss R, Luini M, Artursson K, Bardiau M, Breitenwieser F, Hehenberger E, Lam T, Mansfeld M, Michel A, Mosslacher G, Naskova J, Nelson S, Podpecan O, Raemy A, Ryan E, Salat O, Zangerl P, Steiner A, Graber HU. 2016. Staphylococcus aureus genotype B and other genotypes isolated from cow milk in European countries. J Dairy Sci 99:529–540. doi: 10.3168/jds.2015-9587. [DOI] [PubMed] [Google Scholar]
- 59.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bosi E, Donati B, Galardini M, Brunetti S, Sagot MF, Lio P, Crescenzi P, Fani R, Fondi M. 2015. MeDuSa: a multi-draft based scaffolder. Bioinformatics 31:2443–2451. doi: 10.1093/bioinformatics/btv171. [DOI] [PubMed] [Google Scholar]
- 62.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kosugi S, Hirakawa H, Tabata S. 2015. GMcloser: closing gaps in assemblies accurately with a likelihood-based selection of contig or long-read alignments. Bioinformatics 31:3733–3741. doi: 10.1093/bioinformatics/btv465. [DOI] [PubMed] [Google Scholar]
- 64.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 66.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bartels MD, Petersen A, Worning P, Nielsen JB, Larner-Svensson H, Johansen HK, Andersen LP, Jarlov JO, Boye K, Larsen AR, Westh H. 2014. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 52:4305–4308. doi: 10.1128/JCM.01979-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 72.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Köhler G, Milstein C. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 75.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 76.Ahlmann-Eltze C. 2019. ggsignif: significance brackets for ggplot2, R package version 0.5. https://cran.r-project.org/web/packages/ggsignif/index.html.
- 77.Kassambara A. 2018. ggpubr: ggplot2 based publication ready plots, R package version 0.1.7. https://cran.r-project.org/web/packages/ggpubr/index.html.
- 78.Collery MM, Smyth CJ. 2007. Rapid differentiation of Staphylococcus aureus isolates harbouring egc loci with pseudogenes psient1 and psient2 and the selu or seluv gene using PCR-RFLP. J Med Microbiol 56:208–216. doi: 10.1099/jmm.0.46948-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data for all isolates analyzed in this study have been deposited in the NCBI GenBank database under BioProject accession number PRJNA633807. Accession numbers for individual genomes and assembly statistics are listed in Tables S2 and S3.