Abstract
Background
The gut microbiota are reported to be altered in critical illness. The pattern and impact of dysbiosis on prognosis has not been thoroughly investigated in the ICU setting.
Aims
We aimed to evaluate changes in the gut microbiota of ICU patients via 16S rRNA gene deep sequencing, assess the association of the changes with antibiotics use or disease severity, and explore the association of gut microbiota changes with ICU patient prognosis.
Methods
Seventy-one mechanically ventilated patients were included. Fecal samples were collected serially on days 1–2, 3–4, 5–7, 8–14, and thereafter when suitable. Microorganisms of the fecal samples were profiled by 16S rRNA gene deep sequencing.
Results
Proportions of the five major phyla in the feces were diverse in each patient at admission. Those of Bacteroidetes and Firmicutes especially converged and stabilized within the first week from admission with a reduction in α-diversity (p < 0.001). Significant differences occurred in the proportional change of Actinobacteria between the carbapenem and non-carbapenem groups (p = 0.030) and that of Actinobacteria according to initial SOFA score and changes in the SOFA score (p < 0.001). An imbalance in the ratio of Bacteroidetes to Firmicutes within seven days from admission was associated with higher mortality when the ratio was > 8 or < 1/8 (odds ratio: 5.54, 95% CI: 1.39–22.18, p = 0.015).
Conclusions
Broad-spectrum antibiotics and disease severity may be associated with gut dysbiosis in the ICU. A progression of dysbiosis occurring in the gut of ICU patients might be associated with mortality.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10620-021-07000-7.
Keywords: Gut microbiota, Critical care, 16S rRNA, Dysbiosis, Antibiotics, Sequential organ failure assessment score
Introduction
Human beings live together with numerous different bacteria. The majority of these symbiotic bacteria, estimated to be 1014 microbes [1] comprising over 1000 different bacterial species, reside in the host’s colon [2]. These bacteria have a close connection with human metabolism and homeostasis of the immune system that is more complex and widespread than we ever imagined [3, 4]. Dysbiosis, defined as an imbalance in the microbial communities living in or on the body [5], leads to various diseases such as obesity, metabolic syndrome, cardiovascular diseases, cancer, and autoimmune diseases [6]. Dysbiosis has been confirmed to occur not only in chronic diseases but also in critical illnesses such as sepsis [7], major burn [8], acute care surgery [9], and acute respiratory distress syndrome [10]. Recently, 16S rRNA gene sequencing has been developed as a method to perform sequence-based studies of organisms previously thought to be inaccessible, including obligate anaerobes and other microorganisms that cannot alive outside their hosts without symbionts [11].
Several studies of dysbiosis in critical illness using 16S rRNA gene sequencing [9, 12, 13] reported similar findings: The gut microbiota change rapidly and dramatically during the intensive care unit (ICU) stay. However, the patients were evaluated only at certain points in their ICU stay or by cross-sectional analysis, and thus, it was unclear how the gut microbiota of each patient change at multiple time points during the acute phase of critical care. From this viewpoint, we reported a pilot study that prospectively monitored 12 ICU patients and observed serial changes in their gut microbiota [14]. It revealed that dynamic changes in the gut microbiota occurred at the phylum level of each ICU patient during the acute phase and suggested that an extreme imbalance of the gut microbiota may be associated with prognosis in critically ill patients. However, because of the small population studied, we could not confirm whether this new finding would be generalizable. The study also raised a further question about the factors that can cause these changes in the human gut microbiota during the acute phase of critical illness.
The present study had three objectives: to investigate via 16S rRNA gene deep sequencing how the gut microbiota change at phylum level during the acute phase in ICU patients, to evaluate the associations of antibiotics and disease severity with the change of the host gut microbiota during the acute phase in ICU patients, and to explore the association of changes in the gut microbiota with the prognosis of ICU patients.
Materials and Methods
Enrollment
This study was conducted in the ICU of the Emergency Department of Osaka University Hospital, an academic urban tertiary referral hospital in Osaka, Japan, from February 2014 to January 2015. Patients who were intubated and required mechanical ventilation at admission or during their ICU stay were consecutively enrolled in this study. Exclusion criteria included postoperative patients with surgery of the rectum, patients with perianal infections, and patients expected to be on mechanical ventilation for less than 3 days. We included 71 patients in the study group, of whom 12 patients were enrolled in our previous study [14]. Informed consent for participation in this study was obtained from each patient or their next of kin. Nine healthy adult subjects were also included as a control group. They had never taken antibiotics and had not been admitted to hospitals within 3 months before participating in the study. This study was approved by the institutional review board of Osaka University Hospital (approval number: 12035).
Study Protocol
In this observational prospective cohort study, fecal samples were collected serially from patients during hospitalization on days 1–2, 3–4, 5–7, 8–14, and thereafter when the researchers thought suitable. Fecal samples in the control group were collected on an arbitrary day without any symptoms. All samples were collected by inserting a sterile cotton-tipped swab 1–2 cm beyond the anus and rotating the swab for several seconds. Swabs were placed in sterile centrifuge tubes and immediately stored in a freezer at − 78 °C until use. DNA was extracted from these fecal samples using a DNeasy PowerSoil Kit (QIAGEN). To monitor changes in gut microbiota of the study population, 16S rRNA gene (V1–V2 region) deep sequencing was performed on a MiSeq system using a MiSeq v2 500 cycle kit (both, Illumina). The samples from each patient were sequenced in the same batch. The paired-end sequences obtained were merged, filtered, and denoised using DADA2. Taxonomic assignment was performed using the QIIME2 feature-classifier plugin with the Greengenes 13_8 database. The QIIME2 pipeline, version 2020.2, was used as the bioinformatics environment for the processing of all relevant raw sequencing data [15]. We collected and analyzed 238 samples of feces. Antibiotics class and duration were determined by the attending physician. Enteral nutrition with Glucerna®-Ex (Abbott Japan Co., Ltd.) was started via nasogastric tube within 48 h of admission if there were no contraindications. If infections occurred, patients were initially treated empirically for the underlying clinical syndrome using Gram staining and then according to the results of antibiotic susceptibility testing of the bacterial isolate causing the infection [16, 17]. Antibiotics were administered under the same policy during the entire study period. In trauma cases, antibiotics were administered prophylactically to prevent wound infections for about one week from admission.
Patient Data Collection
Patient age, sex, Acute Physiology and Chronic Health Evaluation (APACHE II) score [18], and diagnosis were recorded on admission. The Sequential Organ Failure Assessment (SOFA) score [19] and the class of administered antibiotics were also recorded every day during hospitalization. Each patient’s outcome and date of death were also archived.
Statistical Analyses
Patient baseline demographic and clinical variables are presented as median and interquartile range (IQR, 25th–75th percentile). The composition of the five major phyla (Bacteroidetes, Firmicutes, Proteobacteria, Actinobacteria and Fusobacteria) and each genus of gut microbiota are presented as percentages. The proportion of the five major phyla at the first sample was compared according to disease types (trauma, cardiac arrest, and sepsis) by Kruskal–Wallis test. To estimate the change in gut microbiota, we focused on two variables. One was the absolute proportional change in each phylum from the first sample to that on the sampling day to observe the overall changes from admission. The other was the relative proportional change of each phylum between the sampling day and the preceding sample to observe the daily changes (Additional file 1: Fig. S1). The composition rate of each phylum and the proportional changes at phylum level as well as the number of OTUs (operational taxonomic units: clusters of similar sequence variants of the 16S rRNA marker gene sequence, considered one of the milestones of biological diversity [α-diversity] [20]) of the healthy controls and the patient group on days 1–3, 4–7, and 8–14 were evaluated by Kruskal–Wallis test with the Dunn’s test used as a post hoc test. We also performed UniFrac distance analysis to compare the microbial flora structures [β-diversity] [21] between the healthy controls and the patient group on days 1–3, 4–7, and 8–14, and the distances were evaluated in the same way.
Carbapenems are thought to have one of the broadest spectrums of activity among all antibiotic classes; hence, we thought they would have the strongest influence on the gut microbiota. Samples were classified as belonging to the carbapenem group or the non-carbapenem group according to the antibiotic used on the day of sampling for a priori analyses. To account for dependencies in repeatedly measured observations within a subject, a nonlinear mixed-effect model was used with group, day, and interaction of group and day. Nonlinear associations between the difference in percentage in the phylum and day were assessed by including restricted cubic splines in the model. To establish the normality of the residuals of the mixed model and the nonlinear model, we performed a log transformation on the outcome. Because log conversion was not possible when the outcome was zero, we added a small enough value not to affect the results of the outcome before log conversion. Multivariable regression analyses were adjusted for age, sex, APACHE II score on admission, and SOFA scores. The absolute proportional changes of each phylum between the carbapenem group, penicillin group, and cephalosporin group were also estimated in the same way.
We also used a nonlinear mixed-effect model with initial SOFA score, SOFA score changes from the initial score on the sampling day, and interaction of initial SOFA score and SOFA score changes. Nonlinear associations between the difference in percentage in the phylum and SOFA score changes were assessed by including restricted cubic splines in the model. Multivariable regression analyses were adjusted for age, sex, and APACHE II score on admission. The absolute proportional changes of microbiota at each hospital day were tested using contrasts of the estimated regression coefficients in each nonlinear mixed effect model.
The changes in gut microbiota were also evaluated by the ratio of Bacteroidetes to Firmicutes (B/F ratio). Bacteroidetes and Firmicutes are the most predominant phyla in humans, and the B/F ratio is thought to be one of the indicators of change in the gut microbiota [22]. The maximum and minimum B/F ratios calculated during the study period were taken as representative values of the changes and of dysbiosis. We previously reported in a preliminary study that a B/F ratio > 10 or < 0.10 may be associated with poor prognosis [14]. The maximum and minimum B/F ratios within seven days from admission were analyzed by classification and regression tree (CART) analysis to explore the association with mortality. CART analysis is a binary recursive partitioning method using nonparametric approaches to identify high-risk and poor prognosis groups. The overall study group is split into two subgroups based on the most powerful predictor of the outcome [23]. Multivariate logistic regression analyses adjusted by sex and APACHE II score on admission were performed to evaluate the association of B/F ratio suggested by CART analysis. A receiver operating characteristic (ROC) curve was created to evaluate classification performance of B/F ratio suggested by CART analysis for the outcome of mortality, and the area under the curve (AUC) was calculated. Kaplan–Meier survival curves were constructed to display the probability of survival over the 56-day period in the patients with B/F ratios suggested by CART analysis and in those without them. The difference between them was evaluated by log-rank test.
All statistical inferences were made with a two-sided significance level of 5%. All statistical analyses were performed using SPSS (version 22, SPSS, Chicago, IL), R software version 3.3.2 using the “rms” package, and JMP Pro (version 11.2.0, SAS Institute, Cary, NC), and the survival curve graph was created using GraphPad Prism (version 6.04, GraphPad Software, La Jolla, CA).
Results
Patient Characteristics
Patient characteristics are shown in Table 1. During the study period, all patients required mechanical ventilation and received antibiotics intravenously. The antibiotic types are summarized in Table 1. Except for two patients, none of the patients were administered antibiotics within 3 months before study enrollment. None of the patients developed Clostridioides (Clostridium) difficile infection. Enteral nutrition was administered within 48 h from admission in 52 patients (73.2%), and 15 patients (21.1%) did not receive enteral feeding during the study period. The median duration of ICU stay was 17 (IQR: 9–37) days. In this study, 24 patients (34%) died from causes of death that included multiple organ failure in 10 patients, cardiac failure in 8 patients, and brain death in 6 patients.
Table 1.
Patient characteristics (N = 71)
| Characteristic | Value |
|---|---|
| Age (years) | 66.0 [51.0–78.5] |
| Sex (male) | 48 (68%) |
| APACHE II score on admission | 23.0 [16.0–31.0] |
| SOFA score on admission | 6.0 [3.0–9.0] |
| Diagnosis | |
| Trauma | 18 (25%) |
| Cardiac arrest | 18 (25%) |
| Sepsis | 12 (17%) |
| Central nervous system disorder | 7 (10%) |
| Acute cardiac failure | 6 (8%) |
| Acute respiratory failure | 6 (8%) |
| Others | 4 (6%) |
| Antibiotics when feces were collected (no. of feces samples = 238) | |
| Penicillins | 108 (45%) |
| Carbapenems | 58 (24%) |
| Cephalosporins | 37 (16%) |
| Vancomycin or linezolid or daptomycin | 30 (13%) |
| Quinolones | 9 (4%) |
| Others | 38 (16%) |
| Mortality | 24 (34%) |
| Cause of death | |
| Multiple organ failure | 10 (14%) |
| Cardiac failure | 8 (11%) |
| Brain death | 6 (8%) |
Data are presented as median [interquartile range] or number (percentage)
APACHE Acute Physiology and Chronic Health Evaluation, SOFA Sequential Organ Failure Assessment
Gut Microbiota at Admission and Progression of Dysbiosis During the Acute-Phase ICU stay
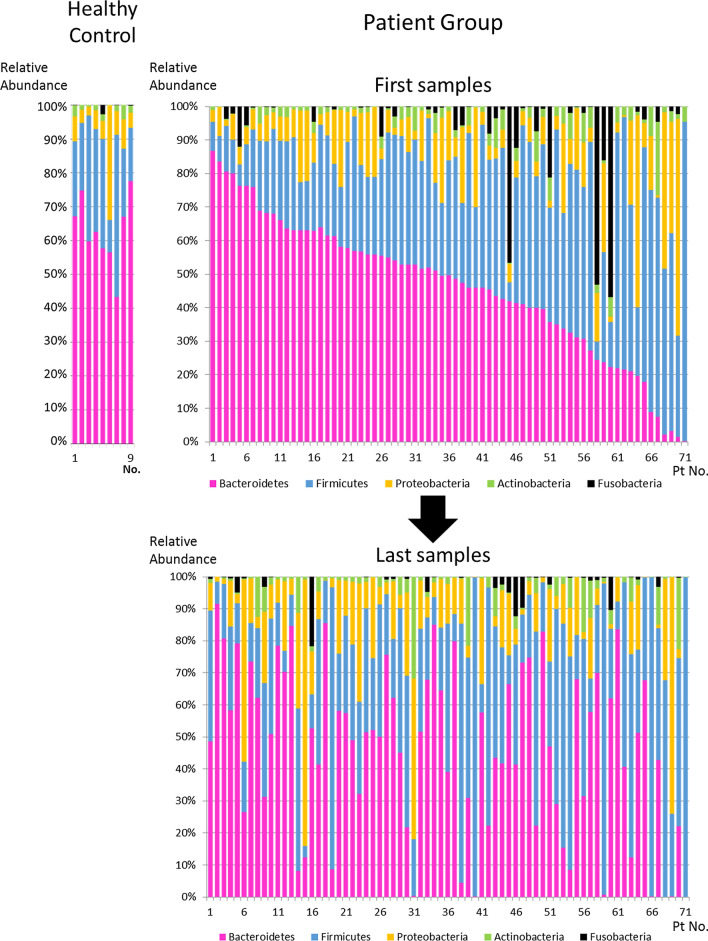
All sampled data are summarized in Additional file 2: Table S1. The taxonomic composition of the gut microbiota at phylum level of the first sample and the last sample in each patient and the healthy controls is shown in Fig. 1. Bacteroidetes and Firmicutes were the predominant phyla in most patients, and their proportions varied from patient to patient at the first sample. The composition of the gut microbiota dramatically changed during the ICU stay. The occupancy rates of the top 10 taxonomic compositions at phylum, class, order, family, and genus level of all patient samples are shown in Additional file 1: Fig. S2. The five major phyla in the patients were Bacteroidetes (44.1%), Firmicutes (34.1%), Proteobacteria (12.7%), Actinobacteria (4.2%), and Fusobacteria (3.1%). The proportion of each phylum at the first sample between patients with trauma, cardiac arrest, and sepsis is shown in Additional file 1: Table S2. There were no significant differences between diseases types except for Proteobacteria (p = 0.032).
Fig. 1.
Taxonomic composition of the gut microbiota at the phylum level in the first and the last samples of the 71 patients and healthy controls (n = 9). Color coding: red = Bacteroidetes, blue = Firmicutes, yellow = Proteobacteria, green = Actinobacteria, black = Fusobacteria
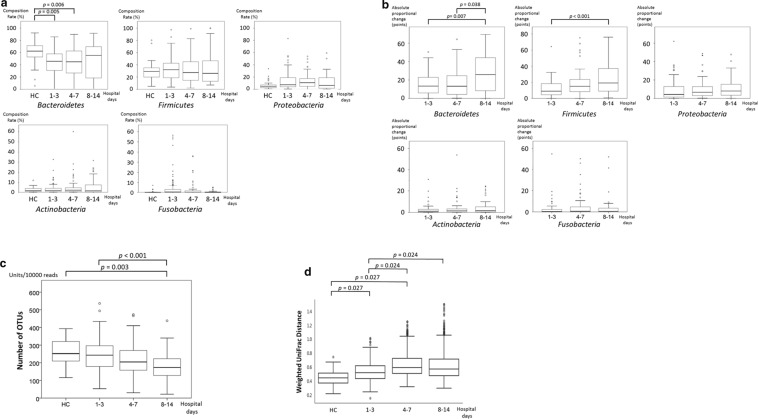
The composition rate and absolute proportional changes of each phylum as well as changes in the number of OTUs in the healthy controls and the patient group on days 1–3, 4–7, and 8–14 from admission are shown in Fig. 2. The composition rate of each phylum was not significantly different between each sampling day. However, the absolute proportional differences of Bacteroidetes and Firmicutes significantly increased over time (p < 0.001; Fig. 2a, b), and the number of OTUs significantly decreased over time (p < 0.001; Fig. 2c). To compare the microbial community between the healthy controls and the patient group on each sampling day, we also performed UniFrac distance analysis of our 16S rRNA gene sequencing data. The distance between the healthy controls and the patient group on each sampling day significantly increased until day 7 (p < 0.05; Fig. 2d), which meant that the microbial flora structures gradually changed until day 7. The taxonomic composition of the gut microbiota at genus level during the ICU stay and that of the healthy controls is shown in Additional file 1: Fig. S3. There were significant changes in the proportions of Blautia, Clostridium, Coprococcus, Enterococcus, Faecalibacterium, Lacnospiraceae, Roseburia, Ruminococcus, and Streptococcus between the healthy controls and each sampling day.
Fig. 2.
a Serial changes of composition rate at the phylum level in the patients and healthy controls (HC). Data are presented as median (interquartile range). b Serial absolute proportional changes at the phylum level from admission in the patient group. Data are presented as median (interquartile range). c Serial changes in the number of OTUs (operational taxonomic units) from admission. Data are presented as median (interquartile range). The number of units per 10,000 reads is indicated on the y axis. HC healthy controls. (d) Serial changes of weighted UniFrac distances from the healthy controls (HC). Data are presented as median (interquartile range)
These results suggested that each ICU patient had their own unique gut microbiota at admission, and the dysbiosis was gradually proceeding with the loss of diversity of gut microbiota during the acute phase of the ICU stay.
Convergence of Gut Microbiota at the Phylum Level by Administered Antibiotics
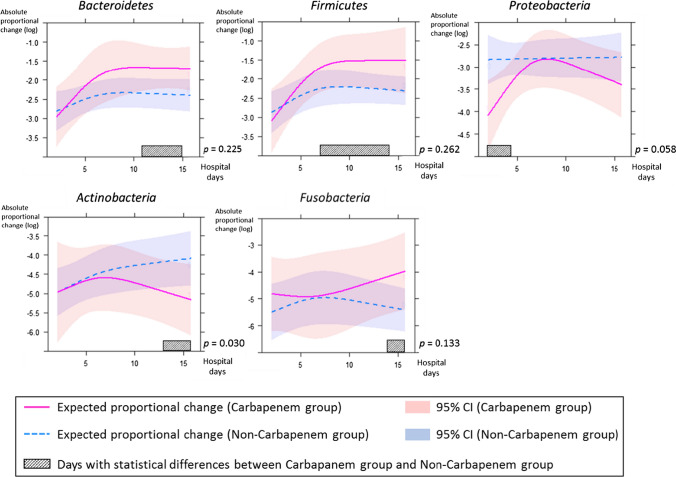
To assess the association between the use of broad-spectrum antibiotics and the gut microbiota of the ICU patients, the absolute proportional change from the first sample and the relative proportional change from the preceding sample were estimated between the carbapenem and non-carbapenem groups by use of a mixed-effect model.
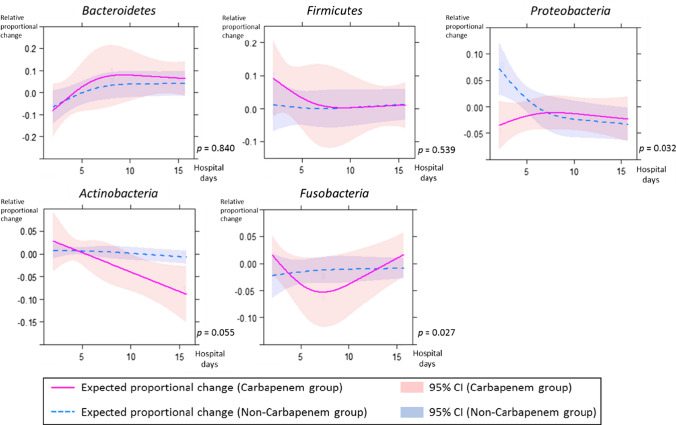
The absolute proportional changes of Bacteroidetes and Firmicutes increased and stabilized approximately on the 7th hospital day in both groups (Fig. 3). Statistically significant differences were found between the carbapenem and non-carbapenem groups in Bacteroidetes from the 11th to 15th hospital day and in Firmicutes from the 7th to 14th hospital day (p < 0.05). The relative proportional changes of Bacteroidetes and Firmicutes converged almost to zero approximately on the 7th hospital day in both groups (Fig. 4).
Fig. 3.
The absolute proportional changes from the first sample of each phylum between the carbapenem group and non-carbapenem group are shown. The amount of variation is indicated as a natural logarithmic value on the y axis. Each line and zone indicate an expected proportional change and the 95% confidence interval (CI) (carbapenem group = red, non-carbapenem group = blue). Shaded bars indicate days with a statistically significant difference between two groups. P indicates the p-value for interaction between hospital days and groups
Fig. 4.
The relative proportional changes from the preceding sample of each phylum between the carbapenem group and non-carbapenem group are shown. The amount of variation is indicated in fraction form on the y axis. Each line and zone indicate an expected proportional change and the 95% confidence interval (CI) (carbapenem group = red, non-carbapenem group = blue). P indicates the p-value for interaction between hospital days and groups
Among the other phyla, Actinobacteria showed a significant difference in the absolute proportional change between the carbapenem and non-carbapenem groups (p = 0.030; Fig. 3). Significant differences in the absolute proportional changes in Proteobacteria from the 1st to 4th hospital day, in Actinobacteria from the 13th to 17th hospital day, and in Fusobacteria from the 14th to 17th hospital day (p < 0.05) were also found by testing for contrasts in interaction (Fig. 3). The relative proportional change of Proteobacteria converged almost to zero approximately on the 7th hospital day in both groups (Fig. 4).
To identify differences between other antibiotics, the absolute proportional changes of each phylum between the carbapenem group, penicillin group, and cephalosporin group were also estimated (Additional file 1: Fig. S4). Each phylum reached inflection points approximately on the 7th hospital day in each antibiotics group, but the differences did not reach statistical significance between the groups.
These results suggested that the major changes of the gut microbiota, especially of Bacteroidetes and Firmicutes, may be completed within the first week after admission to ICU, and broad-spectrum antibiotics may be associated with the change of the composition of gut microbiota from the early phase after ICU admission. Actinobacteria may be more sensitive to carbapenems than other phyla.
Association Between Disease Severity and the Transition of Gut Microbiota in the Acute-Phase ICU Patients
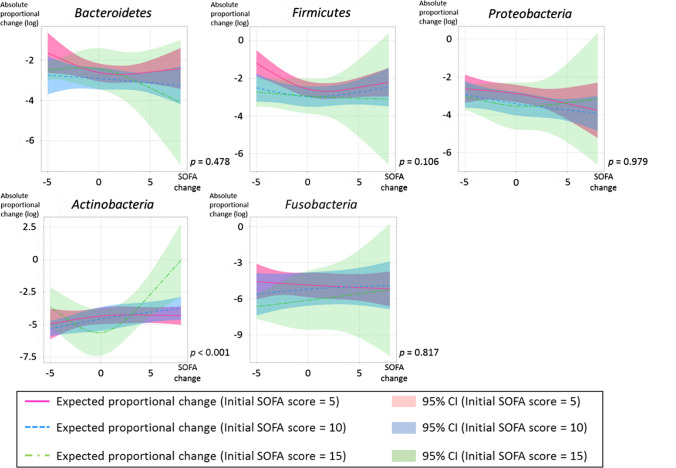
To estimate the association between disease severity and the gut microbiota of the ICU patients, the absolute proportional changes from the first sample were estimated by a linear mixed-effect model with interaction of initial SOFA score and changes in SOFA score. There was a statistically significant difference in the absolute proportional change in Actinobacteria with interaction of the initial SOFA score and changes in SOFA score during the patients’ ICU stay (p < 0.001; Fig. 5). This result suggested that disease severity may be associated with the change in the proportion of Actinobacteria during the acute phase of ICU stay.
Fig. 5.
The absolute proportional changes from the first sample of each phylum, with interaction of initial SOFA score and changes in SOFA score, are shown. The amount of variation is indicated as a natural logarithmic value on the y axis. Each line and zone indicate an expected proportional change and the 95% confidence interval (CI) (initial SOFA score is indicated by color: 5 = red, 10 = blue, 15 = green). P indicates the p-value for interaction between initial SOFA score and changes in SOFA score
Association Between Gut Dysbiosis and Mortality During the Acute-Phase ICU Stay
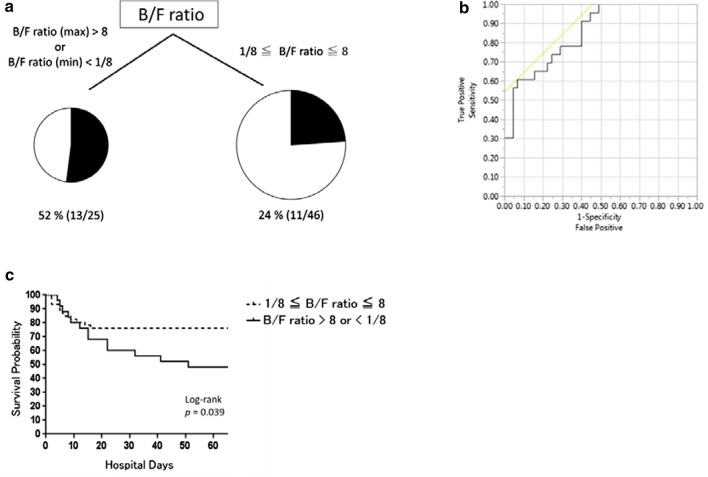
We explored whether gut dysbiosis during the acute-phase ICU stay could be associated with mortality by using CART and multivariate logistic regression analyses. The cutoff values of the maximum and minimum B/F ratios within seven days of admission were determined to be 8 and 1/8, respectively. The diagnostic characteristics of this tree have a sensitivity of 54.2% and specificity of 74.5% (Fig. 6a). ROC curve analysis using these B/F ratios for the outcome of ICU mortality was performed, and the AUC was 0.853 (Fig. 6b). Differences in the rates of ICU mortality were calculated by Kaplan–Meier analysis using these B/F ratios, and the hazard ratio was 2.41 (95% confidence interval [CI]: 1.04–5.51, p = 0.039) (Fig. 6c). Multivariate logistic regression analysis showed an association between an imbalance in B/F ratio and higher mortality when the B/F ratio was > 8 or < 1/8 (odds ratio: 5.54, 95% CI: 1.39–22.18, p = 0.015; Table 2). We also performed multivariate logistic regression analysis adjusted for APACHE II score on admission, sex, and B/F ratio (> 8 or < 1/8) of the fecal samples on the first three days from admission or at seven to nine days from admission to estimate ICU mortality (Additional file 1: Table S3). The B/F ratio on the first three days from admission was not associated with ICU mortality, whereas the B/F ratio one week after admission was associated with ICU mortality.
Fig. 6.
a Mortality partitioned by B/F ratios using classification and regression tree analysis. B/F ratio (max), maximum B/F ratio; B/F ratio (min), minimum B/F ratio. b Receiver operating characteristic curve analysis using B/F ratios (maximum B/F ratio > 8 or minimum B/F ratio < 1/8) for the outcome of ICU mortality. The area under the curve was 0.853. c The 56-day overall survival rate of patients with B/F ratios (maximum B/F ratio > 8 or minimum B/F ratio < 1/8) and those without them. Solid line, overall survival of patients with B/F ratios (maximum B/F ratio > 8 or minimum B/F ratio < 1/8). Dotted line, overall survival of patients without B/F ratios (maximum B/F ratio ≦ 8 and minimum B/F ratio ≧ 1/8)
Table 2.
Results of multivariate logistic regression analysis
| Covariates | Coeff (β) | SE (β) | p value | Odds ratio | 95% CI | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| APACHE II score on admission | 0.162 | 0.045 | < 0.001 | 1.176 | 1.077 | 1.283 |
| B/F ratio | 1.713 | 0.707 | 0.015 | 5.543 | 1.386 | 22.176 |
| Sex | − 0.681 | 0.711 | 0.338 | 0.506 | 0.125 | 2.040 |
Coeff coefficient, SE standard error, CI confidence interval, APACHE Acute Physiology and Chronic Health Evaluation, B/F ratio ratio of Bacteroidetes to Firmicutes
These results suggested that the B/F ratio within seven days from admission might be useful as an indicator of poor prognosis and that an extreme change in the composition of the gut microbiota may be associated with mortality in the ICU. Although it may be difficult to predict prognosis based on the value of the B/F ratio on admission, changes in the B/F ratio during the ICU stay may be more useful for the prediction of prognosis.
Discussion
The main findings of this study are that (1) each ICU patient had his/her own unique gut microbiota at admission, (2) the gut microbiota of the ICU patients converged and stabilized with loss of diversity within the first week after admission, (3) both antibiotics and the severity of illness may be associated with migration of the gut microbiota and Actinobacteria might be more sensitive to these factors than other phyla, and (4) an extreme change in the composition of the gut microbiota as represented by the B/F ratio may be associated with higher mortality in the ICU.
Healthy individuals have been shown to have widely different gut microbiota and microbial diversity [24]. However, the composition of gut microbiota in ICU patients at admission is uncertain, and in the present study, it differed widely between the patients. McDonald et al. reported that patients had developed their own unique gut microbiota within 48 h of ICU admission [12]. These findings suggest that ICU patients also have their own unique gut microbiota at admission.
The uniqueness of each healthy individual’s gut microbiota remains stable over time [14, 24], whereas a dynamic change in gut microbiota has been confirmed in critically ill patients [9, 12, 13, 25, 26]. However, only a few reports followed the serial changes of gut microbiota in the same patients in critical care situations [9, 25]. These reports suggested dynamic variations of gut microbiota along with the loss of diversity and disruption of inherent commensal microbes with the growth of pathogens within one week after ICU admission both inter- and intra-individually. We newly found that the rates of occupancy of Bacteroidetes and Firmicutes, predominant phyla in the human gut microbiome, continued to change until one week after ICU admission along with a reduction in diversity, and after that, no further convergence could be observed. Proteobacteria, which includes major pathogens such as Enterobacteria and Pseudomonas, was also subject to similar effects. It also appeared that convergence occurred independently of the antibiotics groups. These findings suggested that in ICU patients, convergence of the gut microbiota occurred during the period until one week after admission, and thus, it may be effective to administer enteric therapy such as probiotics or synbiotics in the early ICU period to prevent the progression of dysbiosis. Although there have been positive reports of the efficacy of probiotics/synbiotics therapy for ICU patients, this therapy remains controversial. Shimizu et al. recently reported that prophylactic synbiotics administered from the early period could modulate the gut microbiota and may reduce enteritis and ventilator-associated pneumonia in sepsis patients [27]. A cystamatic review by Manzanares et al. also reported that probiotics show promise in reducing infections in the ICU [28]. Further study is needed to clarify the connection between gut microbiota and patient outcome in critical illness.
Antibiotics-induced dysbiosis in the ICU has been reported in a few studies. Two small observational studies reported the reduction of microbial diversity with overgrowth of pathogenic microorganisms such as Enterococcus in relation to prolonged use of antibiotics [29, 30]. However, these reports dealt with feces collected later than one week after ICU admission, so the impact of antibiotics on gut microbiota during the acute ICU phase was unknown. We found in the present study that antibiotics were associated with the changes in gut microbiota in the acute-phase ICU patients, and the change of Actinobacteria was significantly associated with carbapenems. The differences in response to antibiotics by each phylum were probably attributable to the antibiotics spectrum or antibiotic excretion [31]. We also found that the change of Actinobacteria was significantly associated with disease severity. Shimizu et al. previously showed that Bifidobacterium, the major genus belonging to Actinobacteria, was decreased in patients with severe systemic inflammatory syndrome [7]. The decrease in Bifidobacterium was also reported in various diseases such as necrotizing enterocolitis, colorectal cancer, and inflammatory bowel disease [32]. Actinobacteria might be more vulnerable to external stimuli such as exposure to antibiotics, inflammation, ischemia, or hypoxia. Bifidobacterium is also thought to have an important role in preventing growth of pathogenic microorganisms in the gut, maintaining the intestinal environment, and modulating the host immunity system [33]. The change of Bifidobacterium related to antibiotics and disease severity might affect these functions. Further study is needed to clarify these points.
The present study suggested that extreme changes in the composition of gut microbiota represented by the change of B/F ratio may be associated with mortality in the ICU. Bacteroidetes and Firmicutes contain a number of obligate anaerobes occupying the majority of the gut microbiota in healthy adults. They are involved in the regulation of lipids and bile acids metabolism to maintain energy homeostasis in the host [34]. However, lipids and bile acids can modulate the gut microbiota via their direct or indirect antibacterial effects and regulate host immune homeostasis [35, 36]. Accumulating evidence suggests that the disruption of this crosstalk relates to various diseases such as cancer, obesity, inflammatory bowel disease, or Clostridium difficile infection [37]. It was also reported that butyrate, which is a major short fatty acid in feces, is mainly produced by Firmicutes along with some Bacteroidetes [38]. Butyrate has an essential role in the maintenance of gut homeostasis and immunity by controlling gut inflammatory response and inducing regulatory T cells [39]. The decrease in butyrate-producing microbiomes was reported in various diseases such as inflammatory bowel diseases and in patients with allogenic hematopoietic cell transplantation [40, 41]. A decrease in the proportion of butyrate-producing microorganisms (such as Blautia, Clostridium, Coprococcus, Faecalibacterium, Lacnospiraceae, Roseburia, or Ruminococcus) was also found in our study (Additional file 1: Fig. S3).
In the ICU setting, Shimizu et al. also reported patients with severe systemic inflammatory response syndrome had lower concentrations of short-chain fatty acids in feces that contained lower total anaerobic bacterial counts including Bacteroidetes and Firmicutes species, and a decrease in anaerobic bacteria was also associated with septic complications and death [7, 42]. Osuka et al. also reported that fecal pH extending beyond the normal range, meaning that the feces contained lower numbers of bacteria producing short-chain fatty acids including Bacteroidetes and Firmicutes species, was associated with the prognosis of critically ill patients [43]. The rationale for the association between the changes of gut microbiota or gut environment and the outcome of ICU patients is unclear. There are also controversies regarding the relation of dysbiosis to patient prognosis in the ICU. Lankelma et al. reported no association of B/F ratio with patient prognosis in an ICU setting by means of 16S rRNA gene sequencing [13]. We focused on the maximum and minimum B/F ratios within seven days from admission as indicators of a change in gut microbiota, which may make a difference in the interpretation of the results as they relate to prognosis. We suppose the imbalance of gut microbiota represented by the change in B/F ratio might be associated with the disruption of lipids or bile acids metabolism or the host immune system in ICU patients. Further study is needed to elucidate these points.
The extreme change of gut microbiota during the ICU stay as represented by the B/F ratio might be a predictor of ICU mortality. Recently, some observational studies reported that disruption of the gut microbiota was associated with higher mortality in patients undergoing allogenic hematopoietic cell transplantation or in neurocritically ill patients [44–46]. The finding in our study was consistent with these reports, and we newly found that the B/F ratio might be a marker indicating disruption of the gut microbiota. We suppose that maintenance of the gut microbiota as represented by a certain range of B/F ratio might be a therapeutic target to ameliorate ICU mortality. Enteric therapy such as probiotics/synbiotics therapy or fecal transplantation might be a promising treatment. Further study is needed from this viewpoint in the ICU setting.
There are several limitations in this study. First, we assessed the data of each sample in terms of the antibiotics used on the sampling days. However, the gut microbiota could also be impacted by prior antibiotics exposure. We did not consider this factor in the statistical analyses because of concern of overfitting of the model due to the small sample size. Further study is needed to evaluate the effects of the duration of antibiotics exposure on gut microbiota. Second, all of the patients required and received antibiotics, and so we could not evaluate the effects of critical illness on gut microbiota in patients not receiving antibiotics. Third, the composition of gut microbiota is influenced by ethnicity or living environments [47–49]. Thus, it is unclear whether the findings in this study would be valid in other countries.
Conclusions
In conclusion, each patient had a unique gut microbiota even on admission to the ICU. Each patient’s gut microbiota dramatically changed with a reduction in diversity during the ICU stay and converged at new levels within the first week after admission. The transition of gut microbiota was associated with broad-spectrum antibiotics and disease severity. The extreme change of gut microbiota during the acute-phase ICU stay might be associated with mortality.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- APACHE
Acute Physiology and Chronic Health Evaluation
- B/F ratio
Ratio of Bacteroidetes to Firmicutes
- CART
Classification and regression tree
- CI
Confidence interval
- DNA
Deoxyribonucleic acid
- ICU
Intensive care unit
- IQR
Interquartile range
- OTUs
Operational taxonomic units
- rRNA
Ribosomal ribonucleic acid
- SOFA
Sequential Organ Failure Assessment
Author’s contribution
OM and SK participated in the conception and design of the study, acquisition of data, interpretation of data analysis, all statistical analyses, and drafting of the manuscript. MD and NS participated in the analysis of feces by 16S rRNA gene deep sequencing, interpretation of data analysis, statistical analysis (Fig. 2 and Fig. S3), and drafting of the manuscript. IT and SA participated in the interpretation of data analysis, statistical analysis (Figs. 3, 4, 5 and Fig S4), and drafting of the manuscript. OH and YK participated in the conception and design of the study, interpretation of data analysis, and drafting and review of the manuscript. TI and TS reviewed the manuscript. All authors read and approved the final manuscript.
Funding
The study was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant Nos. 17K17049, 19H03760, 19H03761). The funding source had no involvement in study design, analysis, or interpretation of the results.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This study was approved by the Institutional Review Board of Osaka University Hospital (approval number: 12035).
Informed consent
Informed consent was obtained from all individual participants included in the study or their next of kin.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Masahiro Ojima, Email: ojimarionet999@hp-emerg.med.osaka-u.ac.jp.
Kentaro Shimizu, Email: shimiken@hp-emerg.med.osaka-u.ac.jp.
Daisuke Motooka, Email: daisukem@gen-info.osaka-u.ac.jp.
Takuma Ishihara, Email: t.ishihara.1989.2013@gmail.com.
Shota Nakamura, Email: nshota@gen-info.osaka-u.ac.jp.
Ayumi Shintani, Email: ayumi.shintani@gmail.com.
Hiroshi Ogura, Email: ogura@hp-emerg.med.osaka-u.ac.jp.
Tetsuya Iida, Email: iida@biken.osaka-u.ac.jp.
Kazuhisa Yoshiya, Email: yoshiya@hp-emerg.med.osaka-u.ac.jp.
Takeshi Shimazu, Email: shimazu@hp-emerg.med.osaka-u.ac.jp.
References
- 1.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 2.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 4.Sommer F, Backhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 5.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu K, Ogura H, Hamasaki T, et al. Altered gut flora are associated with septic complications and death in critically ill patients with systemic inflammatory response syndrome. Dig Dis Sci. 2011;56:1171–1177. doi: 10.1007/s10620-010-1418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimizu K, Ogura H, Asahara T, et al. Gut microbiota and environment in patients with major burns—a preliminary report. Burns. 2015;41:e28–33. doi: 10.1016/j.burns.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 9.Yeh A, Rogers MB, Firek B, Neal MD, Zuckerbraun BS, Morowitz MJ. Dysbiosis across multiple body sites in critically ill adult surgical patients. Shock. 2016;46:649–654. doi: 10.1097/SHK.0000000000000691. [DOI] [PubMed] [Google Scholar]
- 10.Dickson RP, Singer BH, Newstead MW, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tringe SG, Rubin EM. Metagenomics: DNA sequencing of environmental samples. Nat Rev Genet. 2005;6:805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 12.McDonald D, Ackermann G, Khailova L, et al. Extreme Dysbiosis of the Microbiome in Critical Illness. mSphere. 2016;1. [DOI] [PMC free article] [PubMed]
- 13.Lankelma JM, van Vught LA, Belzer C, et al. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: a pilot study. Intensive Care Med. 2017;43:59–68. doi: 10.1007/s00134-016-4613-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ojima M, Motooka D, Shimizu K, et al. Metagenomic analysis reveals dynamic changes of whole gut microbiota in the acute phase of intensive care unit patients. Dig Dis Sci. 2016;61:1628–1634. doi: 10.1007/s10620-015-4011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikasa K, Aoki N, Aoki Y, et al. JAID/jsc guidelines for the treatment of respiratory infectious diseases: the Japanese association for infectious diseases/Japanese society of chemotherapy—the JAID/JSC guide to clinical management of infectious disease/guideline-preparing committee respiratory infectious disease WG. J Infect Chemother. 2016;22:S1–S65. doi: 10.1016/j.jiac.2015.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshimura J, Kinoshita T, Yamakawa K, et al. Impact of Gram stain results on initial treatment selection in patients with ventilator-associated pneumonia: a retrospective analysis of two treatment algorithms. Crit Care. 2017;21:156. doi: 10.1186/s13054-017-1747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13:818–829. doi: 10.1097/00003246-198510000-00009. [DOI] [PubMed] [Google Scholar]
- 19.Vincent JL, Moreno R, Takala J, et al. The SOFA (sepsis-related organ failure assessment) score to describe organ dysfunction/failure. On behalf of the working group on sepsis-related problems of the european society of intensive care medicine. Intensive Care Med. 1996;22:707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 20.Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev. 2011;35:343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 21.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 23.Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and regression trees. New York: Chapman Hall CRC; 1984. [Google Scholar]
- 24.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada T, Shimizu K, Ogura H, et al. Rapid and sustained long-term decrease of fecal short-chain fatty acids in critically ill patients with systemic inflammatory response syndrome. JPEN J Parenter Enteral Nutr. 2015;39:569–577. doi: 10.1177/0148607114529596. [DOI] [PubMed] [Google Scholar]
- 26.Hayakawa M, Asahara T, Henzan N, et al. Dramatic changes of the gut flora immediately after severe and sudden insults. Dig Dis Sci. 2011;56:2361–2365. doi: 10.1007/s10620-011-1649-3. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu K, Yamada T, Ogura H, et al. Synbiotics modulate gut microbiota and reduce enteritis and ventilator-associated pneumonia in patients with sepsis: a randomized controlled trial. Crit Care. 2018;22:239. doi: 10.1186/s13054-018-2167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manzanares W, Lemieux M, Langlois PL, Wischmeyer PE. Probiotic and synbiotic therapy in critical illness: a cystamatic review and meta-analysis. Crit Care. 2016;19:262. doi: 10.1186/s13054-016-1434-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iapichino G, Callegari ML, Marzorati S, et al. Impact of antibiotics on the gut microbiota of critically ill patients. J Med Microbiol. 2008;57:1007–1014. doi: 10.1099/jmm.0.47387-0. [DOI] [PubMed] [Google Scholar]
- 30.Zaborin A, Smith D, Garfield K, et al. Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. mBio. 2014;5:e01361–14. [DOI] [PMC free article] [PubMed]
- 31.Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;65:1906–1915. doi: 10.1136/gutjnl-2016-312297. [DOI] [PubMed] [Google Scholar]
- 32.Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sánchez B, Margolles A. Bifidobacteria and their health-promoting effects. Microbiol Spectr. 2017;5. [DOI] [PMC free article] [PubMed]
- 33.Wieërs G, Belkhir L, Enaud R, et al. How probiotics affect the microbiota. Front Cell Infect Microbiol. 2019;9:454. doi: 10.3389/fcimb.2019.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krajmalnik-Brown R, Ilhan ZE, Kang DW, DiBaise JK. Effects of gut microbes on nutrient absorption and energy regulation. Nutr Clin Pract. 2012;27:201–214. doi: 10.1177/0884533611436116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15:111–128. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staley C, Weingarden AR, Khoruts A, Sadowsky MJ. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol. 2017;101:47–64. doi: 10.1007/s00253-016-8006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vital M, Karch A, Pieper DH. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems. [DOI] [PMC free article] [PubMed]
- 39.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 40.Devaux CA, Million M, Raoult D. The butyrogenic and lactic bacteria of the gut microbiota determine the outcome of allogenic hematopoietic cell transplant. Front Microbiol. 2020;11:1642. doi: 10.3389/fmicb.2020.01642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu K, Ogura H, Goto M, et al. Altered gut flora and environment in patients with severe SIRS. J Trauma. 2006;60:126–133. doi: 10.1097/01.ta.0000197374.99755.fe. [DOI] [PubMed] [Google Scholar]
- 43.Osuka A, Shimizu K, Ogura H, et al. Prognostic impact of fecal pH in critically ill patients. Crit Care. 2012;16:R119. doi: 10.1186/cc11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peled JU, Gomes ALC, Devlin SM, et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. New Engl J Med. 2020;382:822–834. doi: 10.1056/NEJMoa1900623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kusakabe S, Fukushima K, Maeda T, et al. Pre- and post-serial metagenomic analysis of gut microbiota as a prognostic factor in patients undergoing haematopoietic stem cell transplantation. Br J Haematol. 2020;188:438–449. doi: 10.1111/bjh.16205. [DOI] [PubMed] [Google Scholar]
- 46.Xu R, Tan C, Zhu J, et al. Dysbiosis of the intestinal microbiota in neurocritically ill patients and the risk for death. Crit Care. 2019;23. [DOI] [PMC free article] [PubMed]
- 47.Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017;8:1162. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen L, Zhang YH, Huang T, Cai YD. Gene expression profiling gut microbiota in different races of humans. Sci Rep. 2016;6:23075. doi: 10.1038/srep23075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishijima S, Suda W, Oshima K, et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016;23:125–133. doi: 10.1093/dnares/dsw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.