The term “microbiota” invokes images of mucosal surfaces densely populated with bacteria. These surfaces and the luminal compartments they form indeed predominantly harbor bacteria.
KEYWORDS: mycobiome, mycobiota, fungi, interkingdom interactions, fungal-bacterial interactions, Candida, bacterial-fungal interactions, commensal fungi, host-pathogen interactions, microbiome, microbiota
ABSTRACT
The term “microbiota” invokes images of mucosal surfaces densely populated with bacteria. These surfaces and the luminal compartments they form indeed predominantly harbor bacteria. However, research from this past decade has started to complete the picture by focusing on important but largely neglected constituents of the microbiota: fungi, viruses, and archaea. The community of commensal fungi, also called the mycobiota, interacts with commensal bacteria and the host. It is thus not surprising that changes in the mycobiota have significant impact on host health and are associated with pathological conditions such as inflammatory bowel disease (IBD). In this review we will give an overview of why the mycobiota is an important research area and different mycobiota research tools. We will specifically focus on distinguishing transient and actively colonizing fungi of the oral and gut mycobiota and their roles in health and disease. In addition to correlative and observational studies, we will discuss mechanistic studies on specific cross-kingdom interactions of fungi, bacteria, and the host.
INTRODUCTION
WHAT IS THE MYCOBIOTA?
Fungi are microeukaryotes that can be found on various mammalian mucosal surfaces, such as the lungs (1–3), the vaginal tract (4, 5), the urinary tract (6, 7), the oral cavity (8–10), and the intestines (9, 11, 12) as well as on the skin (5, 9, 13–15), breast, and in breastmilk (16) (Fig. 1). Historically, research on fungi focused on pathological conditions and fungi as pathogens or pathobionts. For example, expansion of the commensal yeast Malassezia on the skin is associated with a disease known as pityriasis versicolor (17). Inhaled spores of the mold Aspergillus fumigatus can germinate and cause invasive lung disease in immunocompromised patients (18, 19). The yeast Candida albicans is arguably the most-studied pathogenic fungus and is responsible for a variety of disease conditions, which include vulvovaginal candidiasis in women, oropharyngeal candidiasis in infants and immunocompromised patients, and invasive candidiasis with systemic dissemination of Candida to peripheral organs (20–23).
FIG 1.
Fungal populations in and on different anatomical sites. Fungal populations have been identified in and on almost all human body sites. This figure is a schematic representation of the most commonly identified fungal genera under nonpathological conditions in the oral cavity (53, 54, 56, 57, 184), skin (92, 103), urinary tract (6, 7), vagina (4, 185), breast milk (93), lungs (186, 187), and intestine (44, 96, 104, 188–190).
Fungi not only are the causative agents of disease but also can be isolated from mammals in the absence of disease (11, 15, 24–26). C. albicans, for example, can frequently be isolated as a commensal of the oral cavity, vagina, or gut of healthy individuals and only causes infections if the host immune system is compromised or the local microbiota is disturbed (22, 23). However, a culture-dependent approach has a high likelihood to yield an incomplete picture of the total fungal diversity (11, 12). The advent of sequencing technology allowed us to answer important questions about fungi: do complex commensal communities of fungi exist in or on different mammalian anatomical sites? Which fungi do they comprise? Are they transiently present or do they stably colonize? What are their functions? The last decade has seen a stark increase in publications addressing these questions. We now know that diverse commensal fungal communities exist in and on mammals. These fungal communities are commonly referred to as the fungal microbiota or mycobiota and are the subject of exciting new research.
METHODS TO ANALYZE THE MYCOBIOTA
A variety of different methods have been used to detect live fungal cells or fungal genomes. Some of the techniques include direct culturing, enriched culturing, microscopy with fluorescence in situ hybridization (FISH) or immunofluorescence, flow cytometry, amplicon sequencing (e.g., internal transcribed spacer [ITS]), and whole-genome shotgun sequencing. However, there are distinct advantages and disadvantages to each approach. Therefore, a combination of different methods described in the following paragraphs will help solve important current questions regarding what constitutes a core mycobiota and which fungi are transient or resident.
Culturing of fungi.
Many environmental, commensal, and pathogenic fungi can be cultured on standard media (11, 25). However, some fungi found in mammals require specific medium conditions. Malassezia species, for example, fail to grow in the absence of fatty acids (17), and anaerobic fungi from ruminants require an anaerobic environment and specific additions such as wheat straw for culture (27). A broad “culturomics” approach identified the highest number of fungi from human gut samples on Dixon medium, a complex medium that includes malt extract, ox bile, and different fatty acids (11). In the gut, bacteria greatly outnumber fungi, which comprise approximately 0.1% of the gut microbiota (28). Isolation of fungi from gut samples therefore usually requires the addition of antibiotics (11, 25). Nevertheless, fungal species with a low abundance might not be recovered. Colonies can be identified to the species level by species-specific PCR or amplification and sequencing of the ITS regions (11, 25). Species can also be identified via matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis, but identification is limited by the available databases, which are focused on pathogenic rather than commensal species (29, 30). One of the drawbacks of microbial culture is that it will identify all viable fungi, including fungal spores or transiently present fungi that might not be metabolically active in the gastrointestinal tract.
Visualization of fungi.
Culturing identifies viable organisms in a given sample. However, unless sampling is performed in specific sections (e.g., mucosa versus lumen), it gives no spatial information. Staining for fungi in fixed or frozen tissue samples can provide such spatial information. Fungi can be visualized in sectioned samples via immunostaining with fungal specific antibodies (12, 31), soluble conjugated receptors (12), or fluorescence in situ hybridization (32, 33). These approaches are limited by the specificity of antibody or probe used. However, they will identify fungal cells in a given sample and omit relic DNA.
Metagenomic analyses.
The specific technical and bioinformatic demands of mycobiome sequencing are expertly outlined elsewhere (34–36). Sequencing detects DNA of fungi present in a given sample regardless of whether they are culturable or not. Amplicon sequencing uses fungal specific primers to amplify ITS or 18S regions of the rRNA gene locus, which contain hypervariable domains and allow for species discrimination (12, 37–39), analogous to 16S bacterial sequencing. This method illuminated a diverse mycobiome in humans and virtually all other species analyzed, for example, mice (12, 32, 40–44), pigs (45), dogs (46), bees (47), and lizards (48). We will discuss the most frequently identified fungi of the human and mouse oral and gut mycobiomes in the following sections. Shortcomings of amplicon sequencing include amplification bias and the lack of comprehensive and fully annotated reference databases that take the complex fungal taxonomy into account (35, 39). Whole-genome shotgun sequencing does not require amplification and can be used to analyze bacterial and fungal metagenomes simultaneously (28). An advantage is that publicly available data sets generated for bacteriome analysis can be reanalyzed for fungi (49). However, the presence of fungi might be underestimated, since fungal sequences are vastly outnumbered by bacterial sequences and some are not accurately identified as fungal due to the scarcity of available fungal genomes (36). A general drawback of DNA sequencing-based methods is the inability to discern between metabolically active organisms and relic DNA (36). The amount of relic DNA content in human feces seems currently unknown, but relic DNA accounts for >40% of recovered sequences from soil samples (50). In the future, amplicon sequencing of the fungal ITS region derived from total RNA (36) or metatranscriptomic approaches might help to address this important question (51).
THE ORAL MYCOBIOTA
The gateway to the gastrointestinal tract is the oral cavity. Interactions of oral mycobiome members influence the local environment but can also have more distant effects. For example, C. albicans abundance in fecal matter is diminished when oral C. albicans abundance is reduced by more frequent brushing of teeth (52). Here, we will address which members of the oral mycobiota might constitute a core mycobiome, identify which are transient and which are colonizing, and highlight examples of interactions with fungi occurring in the oral cavity.
Members of the oral mycobiota.
In their seminal study, Ghannoum and colleagues identified a total of 101 species in the oral cavities of 20 healthy individuals. However, 39 genera were present in only one subject and just 15 genera, including Candida and Cladosporium, were present in more than four individuals (53) (Fig. 1). Another study aimed to define the oral mycobiome found Malassezia to be the most prevalent genus in the oral mycobiomes of six different healthy individuals (54). The oral mycobiome thus appears to be more subject specific than the oral bacteriome, where 47% of bacterial operational taxonomic units (OTUs) were shared between three analyzed samples (55). Despite the high interindividual variability, the core oral mycobiome within individuals seems largely stable, as it maintained a similar composition and relative abundance in human subjects over the course of a 30-week-long study (56). A recently published review article summarized the fungal species identified in different studies with both culturing and sequencing techniques (57). Interestingly, many of these fungi are known to be associated with plants, such as Aureobasidium, Fusarium, and Alternaria, and food commonly consumed, i.e., Saccharomyces and Penicillium, or found as mold both indoors and outdoors, such as Aspergillus species and Cladosporium (Fig. 1). This poses the question if all identified fungi are actively colonizing or if some are transiently present.
Active and transient colonizers.
Among the most commonly found fungi in the oral cavity are Aspergillus species, e.g., A. niger, and Candida species, such as C. albicans, C. tropicalis, and C. parapsilosis (58). A recent study identified them as the most abundant species in both healthy individuals and patients with periodontal disease (59). Aspergillus species are spore-forming filamentous fungi ubiquitously present in the environment (60). Even though Aspergillus species have been found as members of the oral mycobiome, pathological conditions due to Aspergillus are rare in the oral cavity and are usually restricted to the lungs and respiratory tract (61). On the other hand, Candida infections of the oral cavity are very common and affect up to 7% of infants, 30% of HIV patients, and 20% of cancer patients (58). This discrepancy might be due to the possibility that Aspergillus is a transient member of the oral mycobiome, acquired via diet intake or inhalation. A report from 1966 showed that Aspergillus flavus was cultured from 60% of analyzed wheat flours and comprised 5.8% of the total fungal load (62). Candida species actively colonize the oral cavity and erupt in infections when conditions allow it. Both culture-dependent and culture-independent studies have identified Candida species as components of the oral mycobiome. C. albicans was present in 90% (63–65) or even 100% of analyzed subjects (66). Older age, poor oral hygiene, and a fewer number of teeth are some of the associations found with an increased level of colonization of Candida species (53). C. albicans has also been shown to take an active part in biofilm formation and plaque virulence in combination with Streptococcus mutans (67) (Fig. 2). The oral mycobiome thus consists of both resident fungi and fungi that might only be transiently present. Mechanistic research on fungi in the oral cavity has been focused on the resident yeast C. albicans, which will be outlined in the next section. Other fungi of the oral mycobiota, such as Saccharomyces, Aspergillus, Penicillium, and Malassezia, might also represent active members of the oral microbiota, but their function has yet to be identified (53, 56).
FIG 2.
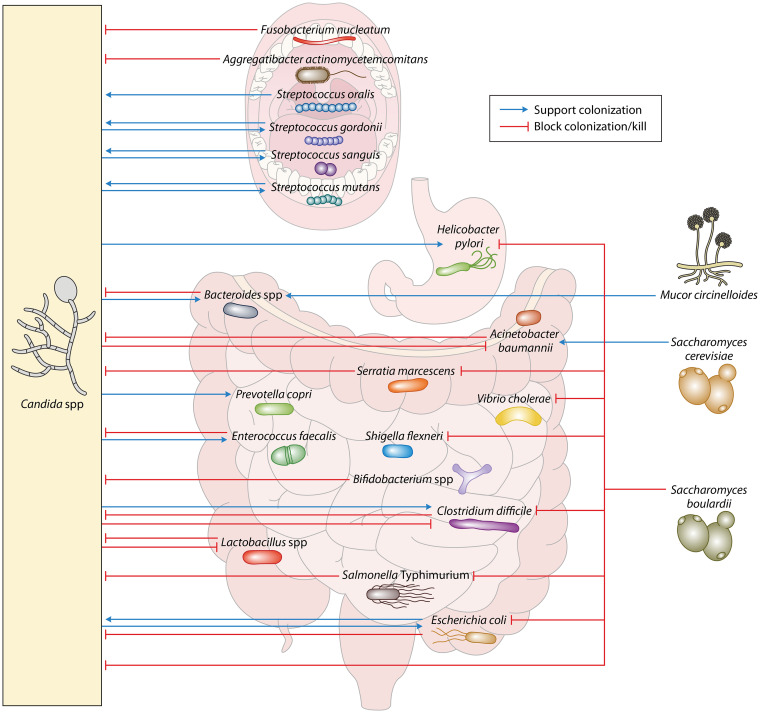
Specific fungal-bacterial interactions in the gastrointestinal tract. Schematic representation of the bacteria-fungi interactions discussed in this review. Localization in the cartoon is not a representation of where the interactions occur within a specific organ. Fungi are depicted outside the organs for schematic purposes.
Examples of fungal-bacterial interactions in the oral cavity.
(i) Protective interactions. Most of the studies investigating the cross-kingdom interactions of fungi and bacteria in the oral cavity have focused on models involving C. albicans. Usually existing in its commensal yeast form, C. albicans can be stimulated to form invasive hyphae (68). The oral commensal Fusobacterium nucleatum has been shown to adhere to both the yeast and hyphal forms of C. albicans (69, 70). This interaction limits C. albicans hyphal formation, thus reducing its ability to kill macrophages in vitro (71). The commensal Aggregatibacter actinomycetemcomitans inhibits biofilm production by C. albicans through the secretion of the quorum-sensing molecule autoinducer-2 (72) and limits polymicrobial biofilm formation by C. albicans and Streptococcus mutans in vitro (73) (Fig. 2). The oral bacterial microbiota thus has an active role in limiting the conversion of C. albicans yeast cells to invasive hyphae.
(ii) Pathogenic interactions. Analysis of the salivary microbiomes of older adults revealed that Candida species abundance is associated with decreased bacterial diversity and increased abundance of Streptococcus species (74). Various studies have investigated mutualistic interactions between C. albicans and species of Streptococcus that promote infection. Glucose-starved C. albicans has been shown to coaggregate with multiple Streptococcus species, including S. sanguis, S. gordonii, and S. oralis (75) (Fig. 2). Further studies revealed that this coaggregation is mediated by cell wall polysaccharides, salivary proteins, and adhesins on the surface of S. gordonii (76–78). Adhesion is also mediated by the receptors Als1p, Als3p, and Als5p on the surface of C. albicans (79, 80). An in vitro study demonstrated that S. gordonii can enhance C. albicans biofilm formation (81, 82) (Fig. 2). S. mutans also enhanced biofilm production in the oral cavity of infected mice through interactions between a glucosyltransferase secreted by S. mutans and surface mannan expressed by C. albicans (67, 83). Another study used an oral infection model of immunosuppressed mice to show that coinfection with S. oralis enhances mucosal invasion of C. albicans by synergistically promoting E-cadherin degradation (84). These studies demonstrate that some members of the oral bacterial microbiota, specifically the genus Streptococcus, facilitate fungal overgrowth. More interactions between C. albicans and Streptococcus species have been reviewed in detail in reference 85.
THE GUT MYCOBIOTA
The vast majority of microorganisms in mammals can be found in the gut, and fungi have emerged as an important component of the gut environment. Much research has therefore focused on understanding how the gut mycobiota is shaped and its interaction with human health.
Development of the gut mycobiota.
The formation of the human mycobiota begins very early in life. Willis and colleagues recently suggested that fungal species might be present prior to birth and that C. albicans specifically could be associated with preterm delivery (86). Additional studies have shown that vaginal delivery allows vertical transmission of Candida species from mother to infant (87, 88). Infants born by C-section harbor a bacterial microbiome similar to the mother’s skin microbiome (89). They could therefore also harbor higher Malassezia species in their gastrointestinal tract, as this genus is the major colonizer of the human skin (90–92) (Fig. 1). After delivery, the gastrointestinal mycobiota is modulated by diet intake. For many infants, the main food source during the first months of age is breast milk. Boix-Amorós and colleagues found a core breast milk mycobiome, composed of Malassezia, Davidiella, Sistotrema, and Penicillium, that was shared by study participants despite a varied geographical origin (93). Accordingly, the infant gut mycobiome is initially dominated by Malasseziales, most likely taken up through lactation. After the first 6 months of age, the infant gut mycobiome undergoes a dramatic change and is no longer dominated by Malasseziales but by Saccharomycetales instead (94). This change in mycobiome coincides with a change from breast milk to solid food. The gut microbiota further changes and matures during the development from childhood to adulthood (95). These changes are most likely driven by the development of the immune system and by the microorganisms that humans are exposed to through their diet and environment. Similarly to oral mycobiome research, recent gut mycobiome research has focused on determining which fungal species are transient colonizers and which species are residents of the gastrointestinal tract.
Active and transient colonizers.
Analysis of the Human Microbiome Project data determined Saccharomyces, Malassezia, Candida, and Cyberlindnera as the four most abundant genera present in the human gut (Fig. 1). However, researchers found very high mycobiome variability between individuals and within individuals over time (96–98). Compared to the bacterial gut microbiome, the gut mycobiome thus seems to be less consistent and stable over time (96). The high fluctuation might in part be explained by fungi being introduced in the gastrointestinal lumen via diet intake and environment. Indeed, a standard human diet contains high levels of live fungi and fungal DNA (62, 99–102), and some fungal genera identified in the human gastrointestinal (GI) tract are thought to lack the ability to grow at the temperature, pH, and low oxygen present in the gut environment (100). Saccharomyces is ubiquitously present in the human diet, while Malassezia is the most abundant fungus colonizing the human skin (92, 103). Cyberlindnera is a food additive and most likely acquired through the diet (96), while Candida is the most identified fungus in the oral cavity (104). Indeed, a small-scale study showed that presence of Saccharomyces in human feces was directly dependent on diet intake, while, as previously mentioned, C. albicans was associated with oral hygiene (52).
Current data thus suggest that some fungi in the human gastrointestinal tract can be classified as transient. However, evidence for true colonizers can also be found. C. albicans, Malassezia restricta, Cryptococcus neoformans, and others have been shown to bloom in the gut under inflammatory conditions. This is particularly highlighted in patients affected with inflammatory bowel disease (IBD), where C. albicans increases in abundance during inflammation. However, it is not yet clear if C. albicans creates the inflammatory environment or if its increase in abundance is a consequence of inflammation (12, 105, 106). A unique phylum of resident fungi can be found in some herbivores but is not detected humans or mice. Neocallimastigomycota are strictly anaerobic fungi that are present in all foregut fermenters (e.g., cows) and some hindgut fermenters (e.g., elephants), where they aid in the digestion of lignocellulose (107). Despite being an anaerobic environment, the human gut does not seem to support the colonization of strictly anaerobic fungi.
So, what proportion of the mycobiota is transiently present and what represents true residents? The bulk of research beginning to address this question has been performed in the mouse model, which we will focus on in the following section. Fungi in mice have been predominantly identified with culture-independent techniques (12, 32, 33, 40–44, 104). Interestingly, fungal sequences can be detected not only in feces but also in mouse chow (12, 32, 41). Some of the identified genera, such as Aspergillus, Cladosporium, and Alternaria, can be found in both feces and chow. However, other fungi, e.g., Candida, Fusarium, and Saccharomyces, can be found only in feces and not in chow (12, 32) or their abundance is expanded in feces compared to that in chow (41). Similarly, a more recent study found that 80% of the fungal taxa identified in mice fed a standard diet were not present in the diet and that 90% of fungal taxa identified in mice fed a high-fat diet were absent in the respective diet (40). These studies thus suggest that the composition of the mycobiota in the gut distinctly differs from the fungi present in the diet.
Even though culture-independent techniques identified fungal species unique to feces, reports showing fungi cultured from mouse feces are rare and restricted to non-specific-pathogen-free (non-SPF) conditions (43, 108). Possible reasons include (i) the relatively low abundance of fungi compared to that of bacteria or (ii) the sampling site, as feces might harbor a different composition of live fungi compared to that in sites in the upper gastrointestinal tract (109). Interestingly, laboratory mice released in an outdoor environment show an increased alpha-diversity of the mycobiome, and researchers were able to culture several fungal species from the mice’s feces. Most of the fungi that researchers were able to culture were Aspergillus species (43). This increase of fungi found in rewilded mice could be due to the presence of spores passing through the GI tract and/or an indication of live Aspergilli that have colonized the gut. The SPF environment of most laboratory mice might therefore be “too clean” to allow for the acquisition of living fungi to colonize their gut. This is supported by the discovery that 22% of laboratory mice do not survive cohousing with pet store mice (110). Pet store mice harbor bacteria that are absent in laboratory mice, and this might also be true for the fungal component of the microbiota. Collectively, studies in mice and humans support the idea that two mycobiomes are present in the mammalian gut: a transient mycobiome originating from diet intake and a resident mycobiome with persistently colonizing fungi. However, discriminating between transient fungi and active colonizers is still challenging. A combination of the different techniques outlined in Methods to Analyze the Mycobiota, as well as other parameters, i.e., activation of the host immune response and interaction between bacteria and fungi, as postulated by Fiers and colleagues (102), will be essential to characterize the role of transient and resident fungi in the gut.
Bacterial-fungal interactions in the gastrointestinal tract.
Recent studies have suggested that the mycobiota plays roles in maintaining homeostasis of the bacterial microbiota and influencing overall gut health. One study found that the administration of antifungal drugs to dextran sulfate sodium (DSS)-treated mice exacerbates colitis and induces changes in the microbiome. Here, the microbiome undergoes an expansion of the bacterial genera Hallella, Barnesiella, Bacteroides, Alistipes, and Lactobacillus and a reduction of Clostridium XIVa and Anaerostipes (41). Another group found that the ingestion of the pathogenic fungus Mucor circinelloides by mice induced changes in the microbiota, notably with an increase in the genus Bacteroides and a decrease in Akkermansia (111). Another study demonstrated that C. albicans affects the recolonization of the cecum by the microbiota in mice treated with antibiotics. The presence of the fungus increased the recovery of bacterial diversity, specifically the return of Bacteroides species. However, it also allowed colonization by the pathobiont Enterococcus faecalis and reduced colonization of probiotic Lactobacillus strains (112) (Fig. 2). The mechanism of how C. albicans influences bacterial colonization is still unclear. A follow-up study revealed that antibiotic-treated C. albicans-colonized mice showed reduced expression of specific immune genes but no visible changes in inflammation. These changes in expression could be limiting the host’s ability to maintain microbial homeostasis, but there is still a possibility that C. albicans directly interacts with bacteria (113). A study that investigated differences in the microbiome between Japanese and Indian individuals proposed an interesting diet-fungal-bacterial interaction. The microbiome of the Indian participants showed a higher abundance of Candida and Prevotella. Since plants make up a major part of Indian diets, Pareek and colleagues (114) went on to show that arabinoxylan, a plant polysaccharide, can be used as a growth factor by various Candida species. Finally, they showed that Candida supernatant enhances the growth of Prevotella copri and that prior colonization by C. albicans is required for the colonization of germ-free mice by P. copri (114). These studies indicate that interactions between fungi and bacterial species influence gut homeostasis and are relevant to human health.
Protective interactions.
Specific cross-kingdom interactions between fungi and bacteria are currently being explored as a tool to maintain intestinal homeostasis. The yeast Saccharomyces boulardii has been extensively studied as a potential probiotic due to its protective effect against various bacterial gastrointestinal pathogens, including Clostridium difficile, Helicobacter pylori, Vibrio cholerae, Salmonella enterica serovar Typhimurium, Shigella flexneri, and Escherichia coli (115–122) (Fig. 2). Protection against C. difficile is at least partially due to the production of a protease by S. boulardii that degrades toxins A and B of C. difficile (123, 124). Protection against V. cholerae seems to involve the recognition of cholera toxin and subsequent activation of cyclic AMP signaling by S. boulardii (125). Even though S. boulardii has shown efficacy in a rat model of V. cholerae infection (119), this has yet to show clinical significance for humans (126). Both E. coli and S. Typhimurium bind to the surface of S. boulardii, potentially preventing adhesion to intestinal epithelial cells and thus allowing quicker excretion through fecal matter (127, 128). This interaction is inhibited by the addition of exogenous mannose, indicating that E. coli and S. Typhimurium are adhering to surface mannose residues present on S. boulardii (129). S. boulardii may also interact with commensal Enterobacteriaceae to alleviate DSS-induced colitis, as this protective effect is lost in mice treated with Enterobacteriaceae-depleting antibiotics (130). The depicted interactions underline the antipathogenic potential for commensal fungi, but the inverse also occurs, where commensal bacteria can protect against pathogenic fungi.
The most intensely studied examples of pathogenic fungi being antagonized by commensal bacteria involve C. albicans. Four probiotic strains, Lactobacillus acidophilus, Lactobacillus reuteri, Lactobacillus casei GG, and Bifidobacterium animalis, have shown efficacy in limiting the severity of C. albicans infection in both immunocompromised and germ-free mice (131) (Fig. 2). Another probiotic mixture, consisting of S. boulardii, L. acidophilus, Lactobacillus rhamnosus, and Bifidobacterium breve, successfully inhibited the in vitro formation of polymicrobial biofilms containing E. coli, Serratia marcescens, and either C. albicans or C. tropicalis (132). These polymicrobial biofilms may be relevant to intestinal disease, as E. coli, S. marcescens, and C. albicans have shown higher abundance in fecal samples from Crohn’s disease patients (133). Various bacterial species have been shown to inhibit the transition of C. albicans to its invasive hyphal form. The widely studied probiotic L. rhamnosus GG produces an exopolysaccharide that limits hyphal formation and blocks C. albicans binding to intestinal epithelial cells in vitro (134). L. rhamnosus GG also inhibits C. albicans hyphal formation in liquid medium via the peptidoglycan hydrolase MspI, which degrades chitin present in the cell wall (135). Enterococcus faecalis produces the bacteriocin EntV to inhibit C. albicans hyphal formation, reducing pathogenicity in a murine oropharyngeal candidiasis infection model (136). Studies have also reported that soluble factors produced by E. coli show antifungal activity against C. albicans. A soluble factor from the E. coli K-12 strain induced the death of C. albicans in vitro (137), and supernatant from an E. coli biofilm inhibited biofilm formation on polystyrene plates for a variety of Candida species (138). Furthermore, metabolites produced by a consortium of bacterial species derived from healthy human fecal samples effectively inhibited the growth of C. albicans in liquid culture. Species of Roseburia and Bacteroides ovatus were directly responsible for these antifungal effects (139). Interestingly, C. albicans also demonstrates probiotic properties by enhancing the growth of two strictly anaerobic commensal bacteria, Bacteroides fragilis and Bacteroides vulgatus, in liquid media. Possible mechanisms of this interaction are utilization of surface mannan as a carbon source or reduction of culture oxygen levels by C. albicans (140). Beneficial interactions between bacteria and fungi are continuously being explored as potential probiotic interventions for intestinal disease.
Pathogenic interactions.
Alternatively, interactions between fungal and bacterial commensals and pathogens have the potential to enhance pathogenesis. For example, mice treated with DSS to induce colitis showed increased disease when C. albicans was present. However, when mice were administered colistin to eliminate resident Enterobacteriaceae, the presence of C. albicans did not exacerbate colitis severity. Supplementation with colistin-resistant E. coli restored the C. albicans effect on DSS-induced colitis, suggesting that Enterobacteriaceae are required for C. albicans-mediated enhancement of colitis (130). Other studies have found that enterohemorrhagic E. coli enhances C. albicans invasion of intestinal epithelial cells in vitro (141). There is also evidence that E. coli strain O111:B4 enhances C. albicans infection of mice (142). E. coli 07KL was also found to enhance C. albicans attachment to epithelial cells in vitro, with a mechanism that likely involves bacterial pili (143). As mentioned in the previous section, the guts of Crohn’s disease patients can harbor an expansion of E. coli, C. tropicalis, and S. marcescens, which together have the ability to form polymicrobial biofilms in vitro (133). These studies thus underline that the interactions between E. coli and Candida species and their effect on pathogenesis are complex and strain dependent. C. albicans allows the growth of the strict anaerobe C. difficile under aerobic culture conditions (144). The ability of C. albicans to protect anaerobic bacteria under aerobic conditions is due to the rapid reduction of dissolved oxygen in the vicinity of the yeast (145). When examined in a mouse model of infection, C. albicans enhanced C. difficile pathogenicity when delivered orally 1 day prior to C. difficile infection (146). Another study found that the colonization of mice with C. albicans 3 weeks before C. difficile infection protected mice from infection (147). These two different experimental setups and outcomes indicate that the effect C. albicans has on C. difficile infection is dependent on the colonization state of C. albicans. Studies have also shown an interaction between C. albicans and H. pylori in gastric biopsy samples, where H. pylori was found within vacuoles in C. albicans cells (148, 149). It has been suggested that this behavior provides an environment that H. pylori can use to survive the low pH of the stomach (150) (Fig. 2). By analyzing whole stomachs of mice, Mason and colleagues show that antibiotic treatment allows for C. albicans colonization, triggering inflammation and inhibiting recolonization by commensal Lactobacillus strains (151). Alternatively, the commensal yeast S. cerevisiae enhances the growth of the opportunistic pathogen Acinetobacter baumannii by producing ethanol. Furthermore, ethanol-stimulated A. baumannii shows enhanced pathogenicity in a Caenorhabditis elegans model of infection (152). It is important to keep these potentially detrimental interactions between pathogens, opportunistic pathogens, commensal bacteria, and fungi in mind when designing therapeutics involving probiotics.
Antagonistic interactions between pathogens.
There are also several antagonistic interactions between intestinal pathogens that do not have a clear benefit for intestinal health. One example is observed with S. marcescens, which employs a type VI secretion system to deliver antifungal toxins that kill both the yeast and hyphal form of C. albicans in liquid culture (153) (Fig. 2). S. Typhimurium also demonstrates a similar antifungal behavior by injecting type III secretion system effectors into C. albicans, blocking hyphal formation during C. elegans infection (154, 155). A. baumannii also demonstrates antifungal activity by binding to C. albicans filaments via OmpA and inducing apoptosis, preventing biofilm formation on polystyrene plates and limiting infection of C. elegans (156, 157). Conversely, C. albicans seems to express a mechanism to limit A. baumannii growth in vitro by producing the quorum-sensing molecule farnesol during late-stage biofilm formation (157). The previously mentioned symbiotic interaction provided by C. albicans to C. difficile is not reciprocated. The same study that found that C. albicans provides C. difficile with the means to grow under aerobic conditions also found that C. difficile inhibits C. albicans hyphal growth through the secretion of the small molecule p-cresol (144) (Fig. 2). These studies highlight bidirectional antagonistic interactions between pathogenic species of bacteria and fungi that are relevant to human health.
MYCOBIOTA AND IMMUNE SYSTEM INTERACTION
All bacterial-fungal interactions within the host occur in an environment that is ultimately regulated by the host immune response. The immunological changes stimulated by a specific microbial colonizer can have a profound effect on the intestinal environment, affecting a wide variety of microbial species already present. This is illustrated, for example, in a study that found that Bacteroides thetaiotaomicron stimulates expression of the innate immune genes encoding hypoxia-inducible factor 1 alpha (HIF-1α) and the antimicrobial peptide LL-37-CRAMP, and this differential expression provides colonization resistance against C. albicans in mice (158). Numerous studies have been performed focusing on the impact of both bacterial and fungal species on the host immune system and vice versa, as summarized previously (104, 159–165). We will focus on how the immune system recognizes fungi and some of the most recent studies on mycobiota and immune system interactions.
Recognition of fungi by the immune system.
The prerequisite for the host to respond to fungi is the ability of cells, in particular, immune cells, to identify and respond to different molecular patterns present on fungi. Among the pattern recognition receptors (PRRs) that can identify fungi are Toll-like receptors (TLRs), C-type lectin receptors (CLRs), and NOD-like receptors (NLRs). Fungal structures that are recognized by PRRs include surface polysaccharides, such as mannans or mannoproteins (TLR2, TLR4, Dectin-2, Mincle, and DC-SIGN), β-glucans (TLR2, Dectin-1, and NKp30), and unmethylated DNA (TLR9). Phagocytosed fungi can also activate NLRs, which leads to inflammasome formation and the production of the inflammatory protein interleukin 1 beta (IL-1β) (109, 166–169). Mutations in the receptors highlight the importance of proper recognition of fungi by the immune system. Mutations in the gene encoding Dectin-1 have been associated with increased C. tropicalis invasion in mice and exacerbated colitis in both mice and humans (12). Mutations in Dectin-2 were found to be associated with increased Candida glabrata infections due to a deficient immune response to the fungus (170). Lack of TLR4 and TLR2 responses were shown to affect disseminated candidiasis in mice: lack of TLR4 caused an increased C. albicans kidney burden, while blocking of TLR2 inhibited the production of inflammatory cytokines such as tumor necrosis factor alpha (TNF-α) and IL-1β (171). Several other receptors are involved in the recognition of fungi. These include the recently identified MelLec, a CLR able to bind melanin on A. fumigatus conidia (172), soluble receptors such as pentraxins and mannose-binding lectin (MBL), involved in the recognition of galactomannan and mannan, respectively, and the intracellular RIG-I-like receptor (RLR) MDA5, which was found to be involved in the immune response to systemic C. albicans infection (167, 173, 174).
Immune system-mycobiota interaction.
Much emphasis has been placed on understanding the roles of the mycobiota in shaping the immune system. A recent study highlighted the important contribution of fungi in the maturation of the immune system. The authors showed that fungi colonizing the guts of mice kept in a natural outdoor environment were sufficient to induce an increase in circulating granulocytes to a level more similar to that in humans than in laboratory mice (43). This finding expanded on results showing that mice colonized with a “wild-mouse microbiota” would respond to immunotherapy in a manner more similar to that in humans (175). Fungi are not only important during homeostatic conditions but also necessary for the development of a healthy immune system. Most of the research performed tries to understand the involvement of the mycobiota in the development and origin of inflammatory and pathogenic conditions. Candida and Malassezia are among the fungal genera that have been most studied in this context. Candida species, particularly C. albicans, are known to be able to exacerbate gut inflammation (161). Fungal dysbiosis and increased C. albicans colonization were identified in association with IBD in human patients (176). In a mouse model of DSS-induced colitis, the presence of C. albicans also worsened local and systemic inflammation (177). Like C. albicans, M. restricta was shown to increase disease severity in DSS-treated mice. Increased relative abundance of M. restricta in the colons of Crohn’s disease patients was linked to a mutation in the CARD9 gene, CARDS12N, previously associated with the onset of IBD (105). This ability of Malassezia to elicit inflammation was later connected to the activation of the NLRP3 inflammasome (178). Dysbiosis of the gut mycobiota can also affect distal organs. A recent study found that gut dysbiosis, specifically, increased abundance of Malassezia species, promoted pancreatic ductal adenocarcinoma development through the activation of the complement cascade via the engagement of the mannose-binding lectin (33). Gut dysbiosis has also been linked to lung inflammation. During gut inflammation, C. albicans was shown to induce the generation of Th17 cells cross-reactive against the airborne pathogen A. fumigatus, thus contributing to the exacerbation of allergic bronchopulmonary aspergillosis (179). Similarly, CX3CR1+ mononuclear phagocytes, present in the lamina propria and involved in trafficking bacteria from the gut to mesenteric lymph nodes (180), were identified to play an important role in the immunity against fungi in the gut (181) and are able to create a gut-lung axis that exacerbates allergic airway disease following gut fungal dysbiosis (182). Another example of interconnection between the gut and lungs is the expansion of Wallemia mellicola in the gut following antibiotic treatment. This intestinal expansion exacerbates lung inflammation by increasing eosinophil recruitment in a mouse model of allergic airway disease (44). A recently published study finally highlighted the importance of a balanced immune response to control fungal infections. Th17 immune responses are known to be important during mucocutaneous fungal infections, especially C. albicans infections (162). However, Break and colleagues showed that in mice and human patients with mutation in the gene Aire that had an intact Th17 response, excessive gamma interferon (IFN-γ) production by T cells at the mucosal level was the cause of increased susceptibility to chronic mucocutaneous candidiasis (183). Future studies will continue to expand our knowledge on the role of fungi during homeostatic and pathological conditions and dissect their role in the development of a balanced immune system.
CONCLUSION AND OUTLOOK
Mycobiome research is a rapidly expanding scientific field, but many questions are currently still unanswered. Due to the high inter- and intraindividual variability, it is unclear if core mycobiomes can be defined. Future research will expand our knowledge on which fungi are resident and which are transiently present in the gastrointestinal tract. However, it is indisputable that the mycobiome fulfills crucial roles. Irrespective of their ability to colonize, fungi interact with and train the immune system and contribute to gastrointestinal homeostasis. Analogous to bacteriome research, mycobiome research is also moving from describing composition to ascribing function. Highly interesting mechanisms of how specific gut commensal fungi modulate immune responses and interact with bacteria are beginning to emerge. Future research directions include the characterization of the fungal metabolome in the gastrointestinal tract to identify which products are produced by fungi and how they influence the microbiome and the host. A recent analysis of the metabolome of differently colonized gnotobiotic mice found that fungi significantly contributed to microbial ecology and host immune functionality but contributed only a small extent to the overall gut metabolome (175). More research with an extended spectrum of commensal gut fungi and additional models will be needed to define the fungal metabolome and the role of fungi in the gut ecosystem.
ACKNOWLEDGMENTS
Work in J.B.’s lab was funded by the NIH (AI143641-01 and DK098170-05). The funders had no role in decision to publish or preparation of the manuscript.
We thank Dara Kiani and Amisha Rana for providing helpful comments on the manuscript. We declare no conflict of interest.
Biographies

William Santus, Ph.D., is a postdoctoral research associate in the laboratory of Dr. Judith Behnsen in the Department of Microbiology and Immunology at the University of Illinois at Chicago. He received his Ph.D. from the University of Milano-Bicocca in Milan, Italy. In his Ph.D. research, he studied the interaction between the innate immune system and the fibrinolytic system with the main focus on the role of NFATc2 and IFN-γ during skin infection of Candida albicans and Staphylococcus aureus. In 2018, he joined the Behnsen lab and started focusing on the role of the mycobiota during Salmonella Typhimurium pathogenesis.

Jason R. Devlin received his B.S. degree in biology from Benedictine University in 2017. He is currently a graduate research assistant in the laboratory of Dr. Judith Behnsen at the University of Illinois at Chicago, where he is working towards a Ph.D. in microbiology and immunology. His research interests involve understanding the interactions between pathogens, the microbiota, and the host and their contributions to infection. His current research investigates the roles of Salmonella Typhimurium chitinases during gastrointestinal infection.

Judith Behnsen, Ph.D., is an Assistant Professor in the Department of Microbiology and Immunology at the University of Illinois at Chicago. She received her Ph.D. from the Friedrich Schiller University in Jena, Germany. In her Ph.D. research, performed at the Leibniz Institute for Natural Product Research and Infection Biology, she studied complement system evasion strategies of the opportunistic fungal pathogen Aspergillus fumigatus. Her postdoctoral research at the University of California Irvine focused on the role of interleukin-22 and the bacterial microbiota during Salmonella Typhimurium pathogenesis. Her lab combines the two areas of expertise and studies the role of the intestinal fungal microbiota during Salmonella Typhimurium pathogenesis.
REFERENCES
- 1.Hamm PS, Taylor JW, Cook JA, Natvig DO. 2020. Decades-old studies of fungi associated with mammalian lungs and modern DNA sequencing approaches help define the nature of the lung mycobiome. PLoS Pathog 16:e1008684. 10.1371/journal.ppat.1008684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kramer R, Sauer-Heilborn A, Welte T, Guzman CA, Abraham WR, Hofle MG. 2015. Cohort study of airway mycobiome in adult cystic fibrosis patients: differences in community structure between fungi and bacteria reveal predominance of transient fungal elements. J Clin Microbiol 53:2900–2907. 10.1128/JCM.01094-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rick EM, Woolnough KF, Seear PJ, Fairs A, Satchwell J, Richardson M, Monteiro WR, Craner M, Bourne M, Wardlaw AJ, Pashley CH. 2020. The airway fungal microbiome in asthma. Clin Exp Allergy 50:1325–1341. 10.1111/cea.13722. [DOI] [PubMed] [Google Scholar]
- 4.Bradford LL, Ravel J. 2017. The vaginal mycobiome: a contemporary perspective on fungi in women's health and diseases. Virulence 8:342–351. 10.1080/21505594.2016.1237332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward TL, Dominguez-Bello MG, Heisel T, Al-Ghalith G, Knights D, Gale CA. 2018. Development of the human mycobiome over the first month of life and across body sites. mSystems 3:e00140-17. 10.1128/mSystems.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ackerman AL, Underhill DM. 2017. The mycobiome of the human urinary tract: potential roles for fungi in urology. Ann Transl Med 5:31. 10.21037/atm.2016.12.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ackerman AL, Chai TC. 2019. The bladder is not sterile: an update on the urinary microbiome. Curr Bladder Dysfunct Rep 14:331–341. 10.1007/s11884-019-00543-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumgardner DJ. 2019. Oral fungal microbiota: to thrush and beyond. J Patient Cent Res Rev 6:252–261. 10.17294/2330-0698.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L, Zeng T, Deligios M, Milanesi L, Langille MGI, Zinellu A, Rubino S, Carru C, Kelvin DJ. 2020. Age-related variation of bacterial and fungal communities in different body habitats across the young, elderly, and centenarians in Sardinia. mSphere 5:e00558-18. 10.1128/mSphere.00558-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandara H, Panduwawala CP, Samaranayake LP. 2019. Biodiversity of the human oral mycobiome in health and disease. Oral Dis 25:363–371. 10.1111/odi.12899. [DOI] [PubMed] [Google Scholar]
- 11.Hamad I, Ranque S, Azhar EI, Yasir M, Jiman-Fatani AA, Tissot-Dupont H, Raoult D, Bittar F. 2017. Culturomics and amplicon-based metagenomic approaches for the study of fungal population in human gut microbiota. Sci Rep 7:16788. 10.1038/s41598-017-17132-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, Wang HL, McGovern DP, Brown GD, Underhill DM. 2012. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336:1314–1317. 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keum HL, Kim H, Kim HJ, Park T, Kim S, An S, Sul WJ. 2020. Structures of the skin microbiome and mycobiome depending on skin sensitivity. Microorganisms 8:1032. 10.3390/microorganisms8071032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu T, Duan YY, Kong FQ, Galzote C, Quan ZX. 2020. Dynamics of skin mycobiome in infants. Front Microbiol 11:1790. 10.3389/fmicb.2020.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leong C, Schmid B, Toi MJ, Wang J, Irudayaswamy AS, Goh JPZ, Bosshard PP, Glatz M, Dawson TL, Jr.. 2019. Geographical and ethnic differences influence culturable commensal yeast diversity on healthy skin. Front Microbiol 10:1891. 10.3389/fmicb.2019.01891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heisel T, Nyaribo L, Sadowsky MJ, Gale CA. 2019. Breastmilk and NICU surfaces are potential sources of fungi for infant mycobiomes. Fungal Genet Biol 128:29–35. 10.1016/j.fgb.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A. 2012. The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev 25:106–141. 10.1128/CMR.00021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinekamp T, Schmidt H, Lapp K, Pahtz V, Shopova I, Koster-Eiserfunke N, Kruger T, Kniemeyer O, Brakhage AA. 2015. Interference of Aspergillus fumigatus with the immune response. Semin Immunopathol 37:141–152. 10.1007/s00281-014-0465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baltussen TJH, Zoll J, Verweij PE, Melchers WJG. 2019. Molecular mechanisms of conidial germination in Aspergillus spp. Microbiol Mol Biol Rev 84:e00049-19. 10.1128/MMBR.00049-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mba IE, Nweze EI. 2020. Mechanism of Candida pathogenesis: revisiting the vital drivers. Eur J Clin Microbiol Infect Dis 39:1797–1819. 10.1007/s10096-020-03912-w. [DOI] [PubMed] [Google Scholar]
- 21.Köhler JR, Hube B, Puccia R, Casadevall A, Perfect JR. 2017. Fungi that Infect Humans. Microbiol Spectr 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.d'Enfert C, Kaune AK, Alaban LR, Chakraborty S, Cole N, Delavy M, Kosmala D, Marsaux B, Frois-Martins R, Morelli M, Rosati D, Valentine M, Xie Z, Emritloll Y, Warn PA, Bequet F, Bougnoux ME, Bornes S, Gresnigt MS, Hube B, Jacobsen ID, Legrand M, Leibundgut-Landmann S, Manichanh C, Munro CA, Netea MG, Queiroz K, Roget K, Thomas V, Thoral C, Van den Abbeele P, Walker AW, Brown AJP. 24 November 2020. The impact of the fungus-host-microbiota interplay upon Candida albicans infections: current knowledge and new perspectives. FEMS Microbiol Rev 10.1093/femsre/fuaa060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jabra-Rizk MA, Kong EF, Tsui C, Nguyen MH, Clancy CJ, Fidel PL, Jr, Noverr M. 2016. Candida albicans pathogenesis: fitting within the host-microbe damage response framework. Infect Immun 84:2724–2739. 10.1128/IAI.00469-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Chen Z, Guo R, Chen N, Lu H, Huang S, Wang J, Li L. 2011. Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn Microbiol Infect Dis 70:492–498. 10.1016/j.diagmicrobio.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Scanlan PD, Marchesi JR. 2008. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J 2:1183–1193. 10.1038/ismej.2008.76. [DOI] [PubMed] [Google Scholar]
- 26.Prohic A, Jovovic Sadikovic T, Krupalija-Fazlic M, Kuskunovic-Vlahovljak S. 2016. Malassezia species in healthy skin and in dermatological conditions. Int J Dermatol 55:494–504. 10.1111/ijd.13116. [DOI] [PubMed] [Google Scholar]
- 27.Griffith GW, Ozkose MK, Theodorou MK, Davies DR. 2009. Diversity of anaerobic fungal populations in cattle revealed by selective enrichment culture using different carbon sources. Fungal Ecol 2:87–97. 10.1016/j.funeco.2009.01.005. [DOI] [Google Scholar]
- 28.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu JJ, Lo HJ, Lee CH, Chen MJ, Lin CC, Chen YZ, Tsai MH, Wang SH. 2021. The use of MALDI-TOF mass spectrometry to analyze commensal oral yeasts in nursing home residents. Microorganisms 9:142. 10.3390/microorganisms9010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel R. 2019. A moldy application of MALDI: MALDI-ToF mass spectrometry for fungal identification. J Fungi (Basel) 5:4. 10.3390/jof5010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bohm L, Torsin S, Tint SH, Eckstein MT, Ludwig T, Perez JC. 2017. The yeast form of the fungus Candida albicans promotes persistence in the gut of gnotobiotic mice. PLoS Pathog 13:e1006699. 10.1371/journal.ppat.1006699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scupham AJ, Presley LL, Wei B, Bent E, Griffith N, McPherson M, Zhu F, Oluwadara O, Rao N, Braun J, Borneman J. 2006. Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol 72:793–801. 10.1128/AEM.72.1.793-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, Shadaloey SA, Wu D, Preiss P, Verma N, Guo Y, Saxena A, Vardhan M, Diskin B, Wang W, Leinwand J, Kurz E, Kochen Rossi JA, Hundeyin M, Zambrinis C, Li X, Saxena D, Miller G. 2019. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574:264–267. 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halwachs B, Madhusudhan N, Krause R, Nilsson RH, Moissl-Eichinger C, Hogenauer C, Thallinger GG, Gorkiewicz G. 2017. Critical issues in mycobiota analysis. Front Microbiol 8:180. 10.3389/fmicb.2017.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang J, Iliev ID, Brown J, Underhill DM, Funari VA. 2015. Mycobiome: approaches to analysis of intestinal fungi. J Immunol Methods 421:112–121. 10.1016/j.jim.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilsson RH, Anslan S, Bahram M, Wurzbacher C, Baldrian P, Tedersoo L. 2019. Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat Rev Microbiol 17:95–109. 10.1038/s41579-018-0116-y. [DOI] [PubMed] [Google Scholar]
- 37.Yang RH, Su JH, Shang JJ, Wu YY, Li Y, Bao DP, Yao YJ. 2018. Evaluation of the ribosomal DNA internal transcribed spacer (ITS), specifically ITS1 and ITS2, for the analysis of fungal diversity by deep sequencing. PLoS One 13:e0206428. 10.1371/journal.pone.0206428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium . 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246. 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frau A, Kenny JG, Lenzi L, Campbell BJ, Ijaz UZ, Duckworth CA, Burkitt MD, Hall N, Anson J, Darby AC, Probert CSJ. 2019. DNA extraction and amplicon production strategies deeply influence the outcome of gut mycobiome studies. Sci Rep 9:9328. 10.1038/s41598-019-44974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heisel T, Montassier E, Johnson A, Al-Ghalith G, Lin YW, Wei LN, Knights D, Gale CA. 2017. High-fat diet changes fungal microbiomes and interkingdom relationships in the murine gut. mSphere 2:e00351-17. 10.1128/mSphere.00351-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu X, Zhang F, Yang X, Wu N, Jiang W, Li X, Li X, Liu Y. 2015. Changes in the composition of intestinal fungi and their role in mice with dextran sulfate sodium-induced colitis. Sci Rep 5:10416. 10.1038/srep10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, McCulloch JA, Anastasakis DG, Sarshad AA, Leonardi I, Collins N, Blatter JA, Han SJ, Tamoutounour S, Potapova S, Foster St Claire MB, Yuan W, Sen SK, Dreier MS, Hild B, Hafner M, Wang D, Iliev ID, Belkaid Y, Trinchieri G, Rehermann B. 2019. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365:eaaw4361. 10.1126/science.aaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeung F, Chen YH, Lin JD, Leung JM, McCauley C, Devlin JC, Hansen C, Cronkite A, Stephens Z, Drake-Dunn C, Fulmer Y, Shopsin B, Ruggles KV, Round JL, Loke P, Graham AL, Cadwell K. 2020. Altered immunity of laboratory mice in the natural environment is associated with fungal colonization. Cell Host Microbe 27:809.e6–822.e6. 10.1016/j.chom.2020.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skalski JH, Limon JJ, Sharma P, Gargus MD, Nguyen C, Tang J, Coelho AL, Hogaboam CM, Crother TR, Underhill DM. 2018. Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog 14:e1007260. 10.1371/journal.ppat.1007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arfken AM, Frey JF, Summers KL. 2020. Temporal dynamics of the gut bacteriome and mycobiome in the weanling pig. Microorganisms 8:868. 10.3390/microorganisms8060868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foster ML, Dowd SE, Stephenson C, Steiner JM, Suchodolski JS. 2013. Characterization of the fungal microbiome (mycobiome) in fecal samples from dogs. Vet Med Int 2013:658373. 10.1155/2013/658373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yun JH, Jung MJ, Kim PS, Bae JW. 2018. Social status shapes the bacterial and fungal gut communities of the honey bee. Sci Rep 8:2019. 10.1038/s41598-018-19860-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montoya-Ciriaco N, Gomez-Acata S, Munoz-Arenas LC, Dendooven L, Estrada-Torres A, Diaz de la Vega-Perez AH, Navarro-Noya YE. 2020. Dietary effects on gut microbiota of the mesquite lizard Sceloporus grammicus (Wiegmann, 1828) across different altitudes. Microbiome 8:6. 10.1186/s40168-020-0783-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donovan PD, Gonzalez G, Higgins DG, Butler G, Ito K. 2018. Identification of fungi in shotgun metagenomics datasets. PLoS One 13:e0192898. 10.1371/journal.pone.0192898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carini P, Marsden PJ, Leff JW, Morgan EE, Strickland MS, Fierer N. 2016. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat Microbiol 2:16242. 10.1038/nmicrobiol.2016.242. [DOI] [PubMed] [Google Scholar]
- 51.Marcelino VR, Irinyi L, Eden JS, Meyer W, Holmes EC, Sorrell TC. 2019. Metatranscriptomics as a tool to identify fungal species and subspecies in mixed communities - a proof of concept under laboratory conditions. IMA Fungus 10:12. 10.1186/s43008-019-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Auchtung TA, Fofanova TY, Stewart CJ, Nash AK, Wong MC, Gesell JR, Auchtung JM, Ajami NJ, Petrosino JF. 2018. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere 3:e00092-18. 10.1128/mSphere.00092-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. 2010. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6:e1000713. 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, Dongari-Bagtzoglou A, Diaz PI, Strausbaugh LD. 2014. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One 9:e90899. 10.1371/journal.pone.0090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaura E, Keijser BJ, Huse SM, Crielaard W. 2009. Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol 9:259. 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monteiro-da-Silva F, Araujo R, Sampaio-Maia B. 2014. Interindividual variability and intraindividual stability of oral fungal microbiota over time. Med Mycol 52:498–505. 10.1093/mmy/myu027. [DOI] [PubMed] [Google Scholar]
- 57.Diaz PI, Hong BY, Dupuy AK, Strausbaugh LD. 2017. Mining the oral mycobiome: methods, components, and meaning. Virulence 8:313–323. 10.1080/21505594.2016.1252015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patil S, Rao RS, Majumdar B, Anil S. 2015. Clinical appearance of oral Candida infection and therapeutic strategies. Front Microbiol 6:1391. 10.3389/fmicb.2015.01391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peters BA, Wu J, Hayes RB, Ahn J. 2017. The oral fungal mycobiome: characteristics and relation to periodontitis in a pilot study. BMC Microbiol 17:157. 10.1186/s12866-017-1064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mousavi B, Hedayati MT, Hedayati N, Ilkit M, Syedmousavi S. 2016. Aspergillus species in indoor environments and their possible occupational and public health hazards. Curr Med Mycol 2:36–42. 10.18869/acadpub.cmm.2.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho H, Lee KH, Colquhoun AN, Evans SA. 2010. Invasive oral aspergillosis in a patient with acute myeloid leukaemia. Aust Dent J 55:214–218. 10.1111/j.1834-7819.2010.01219.x. [DOI] [PubMed] [Google Scholar]
- 62.Graves RR, Hesseltine CW. 1966. Fungi in flour and refrigerated dough products. Mycopathol Mycol Appl 29:277–290. 10.1007/BF02128456. [DOI] [PubMed] [Google Scholar]
- 63.O’Connell LM, Santos R, Springer G, Burne RA, Nascimento MM, Richards VP. 2020. Site-specific profiling of the dental mycobiome reveals strong taxonomic shifts during progression of early-childhood caries. Appl Environ Microbiol 86:e02825-19. 10.1128/AEM.02825-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, Wang K, Zhang B, Tu Q, Yao Y, Cui B, Ren B, He J, Shen X, Van Nostrand JD, Zhou J, Shi W, Xiao L, Lu C, Zhou X. 2019. Salivary mycobiome dysbiosis and its potential impact on bacteriome shifts and host immunity in oral lichen planus. Int J Oral Sci 11:13. 10.1038/s41368-019-0045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Persoon IF, Buijs MJ, Özok AR, Crielaard W, Krom BP, Zaura E, Brandt BW. 2017. The mycobiome of root canal infections is correlated to the bacteriome. Clin Oral Invest 21:1871–1881. 10.1007/s00784-016-1980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fechney JM, Browne GV, Prabhu N, Irinyi L, Meyer W, Hughes T, Bockmann M, Townsend G, Salehi H, Adler CJ. 2019. Preliminary study of the oral mycobiome of children with and without dental caries. J Oral Microbiol 11:1536182. 10.1080/20002297.2018.1536182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai C-H, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, Koo H. 2014. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun 82:1968–1981. 10.1128/IAI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gow NAR, van de Veerdonk FL, Brown AJP, Netea MG. 2011. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol 10:112–122. 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grimaudo NJ, Nesbitt WE. 1997. Coaggregation of Candida albicans with oral Fusobacterium species. Oral Microbiol Immunol 12:168–173. 10.1111/j.1399-302x.1997.tb00374.x. [DOI] [PubMed] [Google Scholar]
- 70.Wu T, Cen L, Kaplan C, Zhou X, Lux R, Shi W, He X. 2015. Cellular components mediating coadherence of Candida albicans and Fusobacterium nucleatum. J Dent Res 94:1432–1438. 10.1177/0022034515593706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bor B, Cen L, Agnello M, Shi W, He X. 2016. Morphological and physiological changes induced by contact-dependent interaction between Candida albicans and Fusobacterium nucleatum. Sci Rep 6:27956. 10.1038/srep27956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bachtiar EW, Bachtiar BM, Jarosz LM, Amir LR, Sunarto H, Ganin H, Meijler MM, Krom BP. 2014. AI-2 of Aggregatibacter actinomycetemcomitans inhibits Candida albicans biofilm formation. Front Cell Infect Microbiol 4:94. 10.3389/fcimb.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bachtiar EW, Bachtiar BM. 2020. Effect of cell-free spent media prepared from Aggregatibacter actinomycetemcomitans on the growth of Candida albicans and Streptococcus mutans in co-species biofilms. Eur J Oral Sci 128:395–404. 10.1111/eos.12725. [DOI] [PubMed] [Google Scholar]
- 74.Kraneveld EA, Buijs MJ, Bonder MJ, Visser M, Keijser BJF, Crielaard W, Zaura E. 2012. The relation between oral Candida load and bacterial microbiome profiles in Dutch older adults. PLoS One 7:e42770. 10.1371/journal.pone.0042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jenkinson HF, Lala HC, Shepherd MG. 1990. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect Immun 58:1429–1436. 10.1128/IAI.58.5.1429-1436.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holmes AR, Gopal PK, Jenkinson HF. 1995. Adherence of Candida albicans to a cell surface polysaccharide receptor on Streptococcus gordonii. Infect Immun 63:1827–1834. 10.1128/IAI.63.5.1827-1834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O’Sullivan JM, Jenkinson HF, Cannon RD. 2000. Adhesion of Candida albicans to oral streptococci is promoted by selective adsorption of salivary proteins to the streptococcal cell surface. Microbiology 146:41–48. 10.1099/00221287-146-1-41. [DOI] [PubMed] [Google Scholar]
- 78.Holmes AR, McNab R, Jenkinson HF. 1996. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect Immun 64:4680–4685. 10.1128/IAI.64.11.4680-4685.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klotz SA, Gaur NK, De Armond R, Sheppard D, Khardori N, Edwards JE, Jr, Lipke PN, El-Azizi M. 2007. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med Mycol 45:363–370. 10.1080/13693780701299333. [DOI] [PubMed] [Google Scholar]
- 80.Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. 2010. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun 78:4644–4652. 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bamford CV, d'Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. 2009. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun 77:3696–3704. 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Montelongo-Jauregui D, Saville SP, Lopez-Ribot JL. 2019. Contributions of Candida albicans dimorphism, adhesive interactions, and extracellular matrix to the formation of dual-species biofilms with Streptococcus gordonii. mBio 10:e01179-19. 10.1128/mBio.01179-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hwang G, Liu Y, Kim D, Li Y, Krysan DJ, Koo H. 2017. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog 13:e1006407. 10.1371/journal.ppat.1006407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu H, Sobue T, Bertolini M, Thompson A, Dongari-Bagtzoglou A. 2016. Streptococcus oralis and Candida albicans synergistically activate μ-calpain to degrade E-cadherin from oral epithelial junctions. J Infect Dis 214:925–934. 10.1093/infdis/jiw201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Förster TM, Mogavero S, Dräger A, Graf K, Polke M, Jacobsen ID, Hube B. 2016. Enemies and brothers in arms: Candida albicans and Gram-positive bacteria. Cell Microbiol 18:1709–1715. 10.1111/cmi.12657. [DOI] [PubMed] [Google Scholar]
- 86.Willis KA, Purvis JH, Myers ED, Aziz MM, Karabayir I, Gomes CK, Peters BM, Akbilgic O, Talati AJ, Pierre JF. 2019. Fungi form interkingdom microbial communities in the primordial human gut that develop with gestational age. FASEB J 33:12825–12837. 10.1096/fj.201901436RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Al-Rusan RM, Darwazeh AM, Lataifeh IM. 2017. The relationship of Candida colonization of the oral and vaginal mucosae of mothers and oral mucosae of their newborns at birth. Oral Surg Oral Med Oral Pathol Oral Radiol 123:459–463. 10.1016/j.oooo.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 88.Waggoner-Fountain LA, Walker MW, Hollis RJ, Pfaller MA, Ferguson JE, II, Wenzel RP, Donowitz LG. 1996. Vertical and horizontal transmission of unique Candida species to premature newborns. Clin Infect Dis 22:803–808. 10.1093/clinids/22.5.803. [DOI] [PubMed] [Google Scholar]
- 89.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107:11971–11975. 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nagata R, Nagano H, Ogishima D, Nakamura Y, Hiruma M, Sugita T. 2012. Transmission of the major skin microbiota, Malassezia, from mother to neonate. Pediatr Int 54:350–355. 10.1111/j.1442-200X.2012.03563.x. [DOI] [PubMed] [Google Scholar]
- 91.Ward TL, Knights D, Gale CA. 2017. Infant fungal communities: current knowledge and research opportunities. BMC Med 15:30. 10.1186/s12916-017-0802-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, NIH Intramural Sequencing Center Comparative Sequencing Program, Kong HH, Segre JA. 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boix-Amorós A, Puente-Sánchez F, Du Toit E, Linderborg KM, Zhang Y, Yang B, Salminen S, Isolauri E, Tamames J, Mira A, Collado MC. 2019. Mycobiome profiles in breast milk from healthy women depend on mode of delivery, geographic location, and interaction with bacteria. Appl Environ Microbiol 85:e02994-18. 10.1128/AEM.02994-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, Wegienka G, Boushey HA, Ownby DR, Zoratti EM, Levin AM, Johnson CC, Lynch SV. 2016. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 22:1187–1191. 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nagpal R, Mainali R, Ahmadi S, Wang S, Singh R, Kavanagh K, Kitzman DW, Kushugulova A, Marotta F, Yadav H. 2018. Gut microbiome and aging: physiological and mechanistic insights. Nutr Healthy Aging 4:267–285. 10.3233/NHA-170030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, Ajami NJ, Petrosino JF. 2017. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5:153. 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486:215–221. 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tournas VH, Niazi NS. 2018. Potentially toxigenic fungi from selected grains and grain products. J Food Saf 38:e12422. 10.1111/jfs.12422. [DOI] [Google Scholar]
- 100.Suhr MJ, Hallen-Adams HE. 2015. The human gut mycobiome: pitfalls and potentials–a mycologist's perspective. Mycologia 107:1057–1073. 10.3852/15-147. [DOI] [PubMed] [Google Scholar]
- 101.Raimondi S, Amaretti A, Gozzoli C, Simone M, Righini L, Candeliere F, Brun P, Ardizzoni A, Colombari B, Paulone S, Castagliuolo I, Cavalieri D, Blasi E, Rossi M, Peppoloni S. 2019. Longitudinal survey of fungi in the human gut: ITS profiling, phenotyping, and colonization. Front Microbiol 10:1575. 10.3389/fmicb.2019.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fiers WD, Gao IH, Iliev ID. 2019. Gut mycobiota under scrutiny: fungal symbionts or environmental transients? Curr Opin Microbiol 50:79–86. 10.1016/j.mib.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oh J, Freeman AF, NISC Comparative Sequencing Program, Park M, Sokolic R, Candotti F, Holland SM, Segre JA, Kong HH. 2013. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res 23:2103–2114. 10.1101/gr.159467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Underhill DM, Iliev ID. 2014. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 14:405–416. 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, Iliev ID, Skalski JH, Brown J, Landers C, Borneman J, Braun J, Targan SR, McGovern DPB, Underhill DM. 2019. Malassezia is associated with Crohn's disease and exacerbates colitis in mouse models. Cell Host Microbe 25:377.e6–388.e6. 10.1016/j.chom.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Q, Wang C, Tang C, He Q, Li N, Li J. 2014. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn's disease. J Clin Gastroenterol 48:513–523. 10.1097/MCG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gruninger RJ, Puniya AK, Callaghan TM, Edwards JE, Youssef N, Dagar SS, Fliegerova K, Griffith GW, Forster R, Tsang A, McAllister T, Elshahed MS. 2014. Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. FEMS Microbiol Ecol 90:1–17. 10.1111/1574-6941.12383. [DOI] [PubMed] [Google Scholar]
- 108.Stejskal V, Hubert J, Kubatova A, Vanova M. 2005. Fungi associated with rodent feces in stored grain environment in the Czech Republic. Z Pflanzenkr Pflanzenschutz 112:98–102. [Google Scholar]
- 109.Richard ML, Sokol H. 2019. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 16:331–345. 10.1038/s41575-019-0121-2. [DOI] [PubMed] [Google Scholar]
- 110.Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, Sekaly RP, Jenkins MK, Vezys V, Haining WN, Jameson SC, Masopust D. 2016. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532:512–516. 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mueller KD, Zhang H, Serrano CR, Billmyre RB, Huh EY, Wiemann P, Keller NP, Wang Y, Heitman J, Lee SC. 2019. Gastrointestinal microbiota alteration induced by Mucor circinelloides in a murine model. J Microbiol 57:509–520. 10.1007/s12275-019-8682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mason KL, Erb Downward JR, Mason KD, Falkowski NR, Eaton KA, Kao JY, Young VB, Huffnagle GB. 2012. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun 80:3371–3380. 10.1128/IAI.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Erb Downward JR, Falkowski NR, Mason KL, Muraglia R, Huffnagle GB. 2013. Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci Rep 3:2191–2191. 10.1038/srep02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pareek S, Kurakawa T, Das B, Motooka D, Nakaya S, Rongsen-Chandola T, Goyal N, Kayama H, Dodd D, Okumura R, Maeda Y, Fujimoto K, Nii T, Ogawa T, Iida T, Bhandari N, Kida T, Nakamura S, Nair GB, Takeda K. 2019. Comparison of Japanese and Indian intestinal microbiota shows diet-dependent interaction between bacteria and fungi. NPJ Biofilms Microbiomes 5:37. 10.1038/s41522-019-0110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kelesidis T, Pothoulakis C. 2012. Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Therap Adv Gastroenterol 5:111–125. 10.1177/1756283X11428502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Surawicz CM, McFarland LV, Greenberg RN, Rubin M, Fekety R, Mulligan ME, Garcia RJ, Brandmarker S, Bowen K, Borjal D, Elmer GW. 2000. The search for a better treatment for recurrent Clostridium difficile disease: use of high-dose vancomycin combined with Saccharomyces boulardii. Clin Infect Dis 31:1012–1017. 10.1086/318130. [DOI] [PubMed] [Google Scholar]
- 117.Szajewska H, Horvath A, Piwowarczyk A. 2010. Meta-analysis: the effects of Saccharomyces boulardii supplementation on Helicobacter pylori eradication rates and side effects during treatment. Aliment Pharmacol Ther 32:1069–1079. 10.1111/j.1365-2036.2010.04457.x. [DOI] [PubMed] [Google Scholar]
- 118.Zhou BG, Chen LX, Li B, Wan LY, Ai YW. 2019. Saccharomyces boulardii as an adjuvant therapy for Helicobacter pylori eradication: a systematic review and meta-analysis with trial sequential analysis. Helicobacter 24:e12651. 10.1111/hel.12651. [DOI] [PubMed] [Google Scholar]
- 119.Czerucka D, Rampal P. 2002. Experimental effects of Saccharomyces boulardii on diarrheal pathogens. Microbes Infect 4:733–739. 10.1016/s1286-4579(02)01592-7. [DOI] [PubMed] [Google Scholar]
- 120.Rodrigues AC, Nardi RM, Bambirra EA, Vieira EC, Nicoli JR. 1996. Effect of Saccharomyces boulardii against experimental oral infection with Salmonella Typhimurium and Shigella flexneri in conventional and gnotobiotic mice. J Appl Bacteriol 81:251–256. 10.1111/j.1365-2672.1996.tb04325.x. [DOI] [PubMed] [Google Scholar]
- 121.Martins FS, Dalmasso G, Arantes RME, Doye A, Lemichez E, Lagadec P, Imbert V, Peyron J-F, Rampal P, Nicoli JR, Czerucka D. 2010. Interaction of Saccharomyces boulardii with Salmonella enterica serovar Typhimurium protects mice and modifies T84 cell response to the infection. PLoS One 5:e8925. 10.1371/journal.pone.0008925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Czerucka D, Dahan S, Mograbi B, Rossi B, Rampal P. 2000. Saccharomyces boulardii preserves the barrier function and modulates the signal transduction pathway induced in enteropathogenic Escherichia coli-infected T84 cells. Infect Immun 68:5998–6004. 10.1128/iai.68.10.5998-6004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Castagliuolo I, LaMont JT, Nikulasson ST, Pothoulakis C. 1996. Saccharomyces boulardii protease inhibits Clostridium difficile toxin A effects in the rat ileum. Infect Immun 64:5225–5232. 10.1128/IAI.64.12.5225-5232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Castagliuolo I, Riegler MF, Valenick L, LaMont JT, Pothoulakis C. 1999. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect Immun 67:302–307. 10.1128/IAI.67.1.302-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brandão RL, Castro IM, Bambirra EA, Amaral SC, Fietto LG, Tropia MJ, Neves MJ, Dos Santos RG, Gomes NC, Nicoli JR. 1998. Intracellular signal triggered by cholera toxin in Saccharomyces boulardii and Saccharomyces cerevisiae. Appl Environ Microbiol 64:564–568. 10.1128/AEM.64.2.564-568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sheele J, Cartowski J, Dart A, Poddar A, Gupta S, Stashko E, Ravi BS, Nelson C, Gupta A. 2015. Saccharomyces boulardii and bismuth subsalicylate as low-cost interventions to reduce the duration and severity of cholera. Pathog Glob Health 109:275–282. 10.1179/2047773215Y.0000000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gedek BR. 1999. Adherence of Escherichia coli serogroup O 157 and the Salmonella Typhimurium mutant DT 104 to the surface of Saccharomyces boulardii. Mycoses 42:261–264. 10.1046/j.1439-0507.1999.00449.x. [DOI] [PubMed] [Google Scholar]
- 128.Pontier-Bres R, Munro P, Boyer L, Anty R, Imbert V, Terciolo C, André F, Rampal P, Lemichez E, Peyron J-F, Czerucka D. 2014. Saccharomyces boulardii modifies Salmonella Typhimurium traffic and host immune responses along the intestinal tract. PLoS One 9:e103069. 10.1371/journal.pone.0103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tiago FCP, Martins FS, Souza ELS, Pimenta PFP, Araujo HRC, Castro IM, Brandão RL, Nicoli JR. 2012. Adhesion to the yeast cell surface as a mechanism for trapping pathogenic bacteria by Saccharomyces probiotics. J Med Microbiol 61:1194–1207. 10.1099/jmm.0.042283-0. [DOI] [PubMed] [Google Scholar]
- 130.Sovran B, Planchais J, Jegou S, Straube M, Lamas B, Natividad JM, Agus A, Dupraz L, Glodt J, Da Costa G, Michel M-L, Langella P, Richard ML, Sokol H. 2018. Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome 6:152. 10.1186/s40168-018-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wagner RD, Pierson C, Warner T, Dohnalek M, Farmer J, Roberts L, Hilty M, Balish E. 1997. Biotherapeutic effects of probiotic bacteria on candidiasis in immunodeficient mice. Infect Immun 65:4165–4172. 10.1128/IAI.65.10.4165-4172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hager CL, Isham N, Schrom KP, Chandra J, McCormick T, Miyagi M, Ghannoum MA. 2019. Effects of a novel probiotic combination on pathogenic bacterial-fungal polymicrobial biofilms. mBio 10:e00338-19. 10.1128/mBio.00338-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, Poulain D, Sendid B, Ghannoum MA. 2016. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio 7:e01250-16. 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Allonsius CN, van den Broek MFL, De Boeck I, Kiekens S, Oerlemans EFM, Kiekens F, Foubert K, Vandenheuvel D, Cos P, Delputte P, Lebeer S. 2017. Interplay between Lactobacillus rhamnosus GG and Candida and the involvement of exopolysaccharides. Microb Biotechnol 10:1753–1763. 10.1111/1751-7915.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Allonsius CN, Vandenheuvel D, Oerlemans EFM, Petrova MI, Donders GGG, Cos P, Delputte P, Lebeer S. 2019. Inhibition of Candida albicans morphogenesis by chitinase from Lactobacillus rhamnosus GG. Sci Rep 9:2900. 10.1038/s41598-019-39625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Graham CE, Cruz MR, Garsin DA, Lorenz MC. 2017. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc Natl Acad Sci U S A 114:4507–4512. 10.1073/pnas.1620432114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cabral DJ, Penumutchu S, Norris C, Morones-Ramirez JR, Belenky P. 2018. Microbial competition between Escherichia coli and Candida albicans reveals a soluble fungicidal factor. Microb Cell 5:249–255. 10.15698/mic2018.05.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bandara HMHN, Cheung BPK, Watt RM, Jin LJ, Samaranayake LP. 2013. Secretory products of Escherichia coli biofilm modulate Candida biofilm formation and hyphal development. J Invest Clin Dent 4:186–199. 10.1111/jicd.12048. [DOI] [PubMed] [Google Scholar]
- 139.García C, Tebbji F, Daigneault M, Liu N-N, Köhler JR, Allen-Vercoe E, Sellam A. 2017. The human gut microbial metabolome modulates fungal growth via the TOR signaling pathway. mSphere 2:e00555-17. 10.1128/mSphere.00555-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Valentine M, Benadé E, Mouton M, Khan W, Botha A. 2019. Binary interactions between the yeast Candida albicans and two gut-associated Bacteroides species. Microb Pathog 135:103619. 10.1016/j.micpath.2019.103619. [DOI] [PubMed] [Google Scholar]
- 141.Yang W, Zhou Y, Wu C, Tang J. 2016. Enterohemorrhagic Escherichia coli promotes the invasion and tissue damage of enterocytes infected with Candida albicans in vitro. Sci Rep 6:37485. 10.1038/srep37485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Klaerner HG, Uknis ME, Acton RD, Dahlberg PS, Carlone-Jambor C, Dunn DL. 1997. Candida albicans and Escherichia coli are synergistic pathogens during experimental microbial peritonitis. J Surg Res 70:161–165. 10.1006/jsre.1997.5110. [DOI] [PubMed] [Google Scholar]
- 143.Centeno A, Davis CP, Cohen MS, Warren MM. 1983. Modulation of Candida albicans attachment to human epithelial cells by bacteria and carbohydrates. Infect Immun 39:1354–1360. 10.1128/IAI.39.3.1354-1360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van Leeuwen PT, van der Peet JM, Bikker FJ, Hoogenkamp MA, Oliveira Paiva AM, Kostidis S, Mayboroda OA, Smits WK, Krom BP. 2016. Interspecies interactions between Clostridium difficile and Candida albicans. mSphere 1:e00187-16. 10.1128/mSphere.00187-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lambooij JM, Hoogenkamp MA, Brandt BW, Janus MM, Krom BP. 2017. Fungal mitochondrial oxygen consumption induces the growth of strict anaerobic bacteria. Fungal Genet Biol 109:1–6. 10.1016/j.fgb.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 146.Panpetch W, Somboonna N, Palasuk M, Hiengrach P, Finkelman M, Tumwasorn S, Leelahavanichkul A. 2019. Oral Candida administration in a Clostridium difficile mouse model worsens disease severity but is attenuated by Bifidobacterium. PLoS One 14:e0210798. 10.1371/journal.pone.0210798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Markey L, Shaban L, Green ER, Lemon KP, Mecsas J, Kumamoto CA. 2018. Pre-colonization with the commensal fungus Candida albicans reduces murine susceptibility to Clostridium difficile infection. Gut Microbes 9:497–509. 10.1080/19490976.2018.1465158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Siavoshi F, Heydari S, Shafiee M, Ahmadi S, Saniee P, Sarrafnejad A, Kolahdoozan S. 2019. Sequestration inside the yeast vacuole may enhance Helicobacter pylori survival against stressful condition. Infect Genet Evol 69:127–133. 10.1016/j.meegid.2019.01.029. [DOI] [PubMed] [Google Scholar]
- 149.Siavoshi F, Saniee P. 2014. Vacuoles of Candida yeast as a specialized niche for Helicobacter pylori. World J Gastroenterol 20:5263–5273. 10.3748/wjg.v20.i18.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sánchez-Alonzo K, Parra-Sepúlveda C, Vega S, Bernasconi H, Campos VL, Smith CT, Sáez K, García-Cancino A. 2020. In vitro incorporation of Helicobacter pylori into Candida albicans caused by acidic pH stress. Pathogens 9:489. 10.3390/pathogens9060489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mason KL, Erb Downward JR, Falkowski NR, Young VB, Kao JY, Huffnagle GB. 2012. Interplay between the gastric bacterial microbiota and Candida albicans during postantibiotic recolonization and gastritis. Infect Immun 80:150–158. 10.1128/IAI.05162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Smith MG, Des Etages SG, Snyder M. 2004. Microbial synergy via an ethanol-triggered pathway. Mol Cell Biol 24:3874–3884. 10.1128/mcb.24.9.3874-3884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Trunk K, Peltier J, Liu Y-C, Dill BD, Walker L, Gow NAR, Stark MJR, Quinn J, Strahl H, Trost M, Coulthurst SJ. 2018. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol 3:920–931. 10.1038/s41564-018-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tampakakis E, Peleg AY, Mylonakis E. 2009. Interaction of Candida albicans with an intestinal pathogen, Salmonella enterica serovar Typhimurium. Eukaryot Cell 8:732–737. 10.1128/EC.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kim Y, Mylonakis E. 2011. Killing of Candida albicans filaments by Salmonella enterica serovar Typhimurium is mediated by sopB effectors, parts of a type III secretion system. Eukaryot Cell 10:782–790. 10.1128/EC.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77:3150–3160. 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Peleg AY, Tampakakis E, Fuchs BB, Eliopoulos GM, Moellering RC, Mylonakis E. 2008. Prokaryote–eukaryote interactions identified by using Caenorhabditis elegans. Proc Natl Acad Sci U S A 105:14585–14590. 10.1073/pnas.0805048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, Simms-Waldrip TR, Xie Y, Hooper LV, Koh AY. 2015. Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med 21:808–814. 10.1038/nm.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Forbes JD, Bernstein CN, Tremlett H, Van Domselaar G, Knox NC. 2018. A fungal world: could the gut mycobiome be involved in neurological disease? Front Microbiol 9:3249. 10.3389/fmicb.2018.03249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Iliev ID, Leonardi I. 2017. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol 17:635–646. 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li XV, Leonardi I, Iliev ID. 2019. Gut mycobiota in immunity and inflammatory disease. Immunity 50:1365–1379. 10.1016/j.immuni.2019.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lionakis MS, Iliev ID, Hohl TM. 2017. Immunity against fungi. JCI Insight 2:1365. 10.1172/jci.insight.93156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Richard ML, Lamas B, Liguori G, Hoffmann TW, Sokol H. 2015. Gut fungal microbiota: the yin and yang of inflammatory bowel disease. Inflamm Bowel Dis 21:656–665. 10.1097/MIB.0000000000000261. [DOI] [PubMed] [Google Scholar]
- 164.Rizzetto L, De Filippo C, Cavalieri D. 2014. Richness and diversity of mammalian fungal communities shape innate and adaptive immunity in health and disease. Eur J Immunol 44:3166–3181. 10.1002/eji.201344403. [DOI] [PubMed] [Google Scholar]
- 165.Lai GC, Tan TG, Pavelka N. 2019. The mammalian mycobiome: a complex system in a dynamic relationship with the host. Wiley Interdiscip Rev Syst Biol Med 11:e1438. 10.1002/wsbm.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Romani L. 2011. Immunity to fungal infections. Nat Rev Immunol 11:275–288. 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 167.Brown GD. 2011. Innate antifungal immunity: the key role of phagocytes. Annu Rev Immunol 29:1–21. 10.1146/annurev-immunol-030409-101229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hoving JC, Wilson GJ, Brown GD. 2014. Signalling C-type lectin receptors, microbial recognition and immunity. Cell Microbiol 16:185–194. 10.1111/cmi.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Patin EC, Thompson A, Orr SJ. 2019. Pattern recognition receptors in fungal immunity. Semin Cell Dev Biol 89:24–33. 10.1016/j.semcdb.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ifrim DC, Bain JM, Reid DM, Oosting M, Verschueren I, Gow NA, van Krieken JH, Brown GD, Kullberg BJ, Joosten LA, van der Meer JW, Koentgen F, Erwig LP, Quintin J, Netea MG. 2014. Role of Dectin-2 for host defense against systemic infection with Candida glabrata. Infect Immun 82:1064–1073. 10.1128/IAI.01189-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Netea MG, Van Der Graaf CA, Vonk AG, Verschueren I, Van Der Meer JW, Kullberg BJ. 2002. The role of toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J Infect Dis 185:1483–1489. 10.1086/340511. [DOI] [PubMed] [Google Scholar]
- 172.Stappers MHT, Clark AE, Aimanianda V, Bidula S, Reid DM, Asamaphan P, Hardison SE, Dambuza IM, Valsecchi I, Kerscher B, Plato A, Wallace CA, Yuecel R, Hebecker B, da Glória Teixeira Sousa M, Cunha C, Liu Y, Feizi T, Brakhage AA, Kwon-Chung KJ, Gow NAR, Zanda M, Piras M, Zanato C, Jaeger M, Netea MG, van de Veerdonk FL, Lacerda JF, Campos A, Carvalho A, Willment JA, Latgé JP, Brown GD. 2018. Recognition of DHN-melanin by a C-type lectin receptor is required for immunity to Aspergillus. Nature 555:382–386. 10.1038/nature25974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jaeger M, van der Lee R, Cheng SC, Johnson MD, Kumar V, Ng A, Plantinga TS, Smeekens SP, Oosting M, Wang X, Barchet W, Fitzgerald K, Joosten LAB, Perfect JR, Wijmenga C, van de Veerdonk FL, Huynen MA, Xavier RJ, Kullberg BJ, Netea MG. 2015. The RIG-I-like helicase receptor MDA5 (IFIH1) is involved in the host defense against Candida infections. Eur J Clin Microbiol Infect Dis 34:963–974. 10.1007/s10096-014-2309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Salazar F, Brown GD. 2018. Antifungal innate immunity: a perspective from the last 10 years. J Innate Immun 10:373–397. 10.1159/000488539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.van Tilburg Bernardes E, Pettersen VK, Gutierrez MW, Laforest-Lapointe I, Jendzjowsky NG, Cavin JB, Vicentini FA, Keenan CM, Ramay HR, Samara J, MacNaughton WK, Wilson RJA, Kelly MM, McCoy KD, Sharkey KA, Arrieta MC. 2020. Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat Commun 11:2577. 10.1038/s41467-020-16431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L. 2017. Fungal microbiota dysbiosis in IBD. Gut 66:1039–1048. 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Panpetch W, Hiengrach P, Nilgate S, Tumwasorn S, Somboonna N, Wilantho A, Chatthanathon P, Prueksapanich P, Leelahavanichkul A. 2020. Additional Candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by Lactobacillus rhamnosus L34. Gut Microbes 11:465–480. 10.1080/19490976.2019.1662712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wolf AJ, Limon JJ, Nguyen C, Prince A, Castro A, Underhill DM. 2021. Malassezia spp. induce inflammatory cytokines and activate NLRP3 inflammasomes in phagocytes. J Leukoc Biol 109:161–172. 10.1002/JLB.2MA0820-259R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann S, Röhmel J, Eschenhagen P, Grehn C, Seidel K, Rickerts V, Lozza L, Stervbo U, Nienen M, Babel N, Milleck J, Assenmacher M, Cornely OA, Ziegler M, Wisplinghoff H, Heine G, Worm M, Siegmund B, Maul J, Creutz P, Tabeling C, Ruwwe-Glösenkamp C, Sander LE, Knosalla C, Brunke S, Hube B, Kniemeyer O, Brakhage AA, Schwarz C, Scheffold A. 2019. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell 176:1340.e15–1355.e15. 10.1016/j.cell.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 180.Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. 2013. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature 494:116–120. 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, Pla J, Iliev ID. 2018. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science 359:232–236. 10.1126/science.aao1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li X, Leonardi I, Semon A, Doron I, Gao IH, Putzel GG, Kim Y, Kabata H, Artis D, Fiers WD, Ramer-Tait AE, Iliev ID. 2018. Response to fungal dysbiosis by gut-resident CX3CR1+ mononuclear phagocytes aggravates allergic airway disease. Cell Host Microbe 24:847.e4–856.e4. 10.1016/j.chom.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Break TJ, Oikonomou V, Dutzan N, Desai JV, Swidergall M, Freiwald T, Chauss D, Harrison OJ, Alejo J, Williams DW, Pittaluga S, Lee CR, Bouladoux N, Swamydas M, Hoffman KW, Greenwell-Wild T, Bruno VM, Rosen LB, Lwin W, Renteria A, Pontejo SM, Shannon JP, Myles IA, Olbrich P, Ferré EMN, Schmitt M, Martin D, Genomics and Computational Biology Core, Barber DL, Solis NV, Notarangelo LD, Serreze DV, Matsumoto M, Hickman HD, Murphy PM, Anderson MS, Lim JK, Holland SM, Filler SG, Afzali B, Belkaid Y, Moutsopoulos NM, Lionakis MS. 2021. Aberrant type 1 immunity drives susceptibility to mucosal fungal infections. Science 371:eaay5731. 10.1126/science.aay5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mukherjee PK, Chandra J, Retuerto M, Sikaroodi M, Brown RE, Jurevic R, Salata RA, Lederman MM, Gillevet PM, Ghannoum MA. 2014. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog 10:e1003996. 10.1371/journal.ppat.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Drell T, Lillsaar T, Tummeleht L, Simm J, Aaspõllu A, Väin E, Saarma I, Salumets A, Donders GG, Metsis M. 2013. Characterization of the vaginal micro- and mycobiome in asymptomatic reproductive-age Estonian women. PLoS One 8:e54379. 10.1371/journal.pone.0054379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nguyen LD, Viscogliosi E, Delhaes L. 2015. The lung mycobiome: an emerging field of the human respiratory microbiome. Front Microbiol 6:89. 10.3389/fmicb.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.van Woerden HC, Gregory C, Brown R, Marchesi JR, Hoogendoorn B, Matthews IP. 2013. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC Infect Dis 13:69. 10.1186/1471-2334-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. 2013. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 8:e66019. 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Strati F, Di Paola M, Stefanini I, Albanese D, Rizzetto L, Lionetti P, Calabrò A, Jousson O, Donati C, Cavalieri D, De Filippo C. 2016. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front Microbiol 7:1227. 10.3389/fmicb.2016.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Goren I, Godny L, Reshef L, Yanai H, Gophna U, Tulchinsky H, Dotan I. 2019. Starch consumption may modify antiglycan antibodies and fecal fungal composition in patients with ileo-anal pouch. Inflamm Bowel Dis 25:742–749. 10.1093/ibd/izy370. [DOI] [PubMed] [Google Scholar]