Rickettsia rickettsii, the etiological agent of Rocky Mountain spotted fever (RMSF), a life-threatening tick-borne disease that affects humans and various animal species, has been recognized in medicine and science for more than 100 years. Isolate-dependent differences in virulence of R. rickettsii have been documented for many decades; nonetheless, the specific genetic and phenotypic factors responsible for these differences have not been characterized.
KEYWORDS: RMSF, Rickettsia rickettsii, isolate-dependent, phenotypes, virulence
ABSTRACT
Rickettsia rickettsii, the etiological agent of Rocky Mountain spotted fever (RMSF), a life-threatening tick-borne disease that affects humans and various animal species, has been recognized in medicine and science for more than 100 years. Isolate-dependent differences in virulence of R. rickettsii have been documented for many decades; nonetheless, the specific genetic and phenotypic factors responsible for these differences have not been characterized. Using in vivo and in vitro methods, we identified multiple phenotypic differences among six geographically distinct isolates of R. rickettsii, representing isolates from the United States, Costa Rica, and Brazil. Aggregate phenotypic data, derived from growth in Vero E6 cells and from clinical and pathological characteristics following infection of male guinea pigs (Cavia porcellus), allowed separation of these isolates into three categories: nonvirulent (Iowa), mildly virulent (Sawtooth and Gila), and highly virulent (Sheila SmithT, Costa Rica, and Taiaçu). Transcriptional profiles of 11 recognized or putative virulence factors confirmed the isolate-dependent differences between mildly and highly virulent isolates. These data corroborate previous qualitative assessments of strain virulence and suggest further that a critical and previously underappreciated balance between bacterial growth and host immune response could leverage strain pathogenicity. Also, this work provides insight into isolate-specific microbiological factors that contribute to the outcome of RMSF and confirms the hypothesis that distinct rickettsial isolates also differ phenotypically, which could influence the severity of disease in vertebrate hosts.
INTRODUCTION
Rocky Mountain spotted fever (RMSF) is a life-threatening tick-borne disease that affects humans and various animal species (1). Infected patients demonstrate multiple signs and symptoms that generally include high fever, severe headache, generalized rash, and myalgias. This disease, caused by Rickettsia rickettsii, can evolve rapidly to multiorgan system failure and death from systemic vasculitis (2). In the United States, the estimated case fatality rate of RMSF has ranged from 5% to 10% since the discovery in the 1940s of tetracycline-class antibiotics (3, 4). For reasons not yet understood, case fatality rates of RMSF are considerably higher in many Latin American countries, and contemporary estimates in Mexico and Brazil are approximately 30 to 40% (5–7). Interestingly, previous studies identified genetic differences between various isolates of R. rickettsii that correlate directly with their geographical origin (8–13).
Isolate differences in R. rickettsii virulence have been recognized for more than 60 years (14). Less virulent isolates of R. rickettsii, such as Sawtooth, highly virulent isolates, such as the type strain Sheila Smith, and the avirulent Iowa isolate were described previously, although the phenotypic and molecular basis for their differences have not been fully characterized (8–11, 14–17). Here, we describe an in vitro and in vivo analysis of several phenotypic and transcriptional differences among six geographically distinct isolates of R. rickettsii from the United States, Costa Rica, and Brazil, including three isolated from patients with severe or fatal disease, and three highly pathogenic, minimally pathogenic, or nonpathogenic isolates from ticks (15, 16, 18). These findings provide further insight into isolate-specific microbiological factors and corresponding pathology that contribute to RMSF.
RESULTS
Clinical and pathological characteristics of guinea pigs infected with different R. rickettsii isolates.
To characterize the clinical and pathological differences among isolates of R. rickettsii, 6- to 9-week-old, pathogen-free, male guinea pigs (Cavia porcellus) were infected with standardized amounts of each bacterial isolate (Iowa, Sawtooth, Gila, Sheila SmithT, Costa Rica, and Taiaçu) and the course of infection was documented daily by clinical scoring for 6 days. Sheila SmithT-, Costa Rica-, and Taiaçu-infected animals exhibited consistent and similar signs and lesions of severe disease, as indicated by the clinical parameters, gross abnormalities, and histopathologic findings. In contrast, Sawtooth- and Gila-infected animals showed clinical and pathological findings compatible with mild disease, and animals infected with Iowa remained minimally symptomatic or asymptomatic.
Desquamating dermatitis of the footpads was observed in 20% of Sawtooth-infected, 10% of Gila-infected, 20% of Costa Rica-infected, and 30% of Taiaçu-infected animals, and it was present in the majority (90%) of Sheila SmithT-infected animals (Table 1). In addition, 80 to 100% of Sheila SmithT-, Taiaçu-, and Gila-infected animals exhibited significant scrotal edema, including the presence of necrotic areas in Sheila SmithT- and Taiaçu-infected animals. Only 40% of Sawtooth- and 30% of Costa Rica-infected animals presented these scrotal findings.
TABLE 1.
Clinical signs of infection at 6 days postinoculationa
| Clinical sign | % of animals infected |
|||||
|---|---|---|---|---|---|---|
| Iowa | Sawtooth | Gila | Sheila SmithT | Costa Rica | Taiaçu | |
| Feverb | 50 | 100 | 100 | 100 | 100 | 100 |
| Footpad dermatitis | 0 | 20 | 10 | 90 | 20 | 30 |
| Scrotal edema | 0 | 25 | 80 | 100 | 30 | 90 |
| Wt loss | 0 | 80 | 90 | 100 | 100 | 100 |
| Splenomegalyc | 100 | 100 | 100 | 100 | 100 | 100 |
n = 10 for Iowa, Sawtooth, Gila, Sheila SmithT, and Taiaçu; n = 9 for Costa Rica.
Temperature above 39.5°C during the 6 days of the experiment.
Above 0.13 index of total body weight.
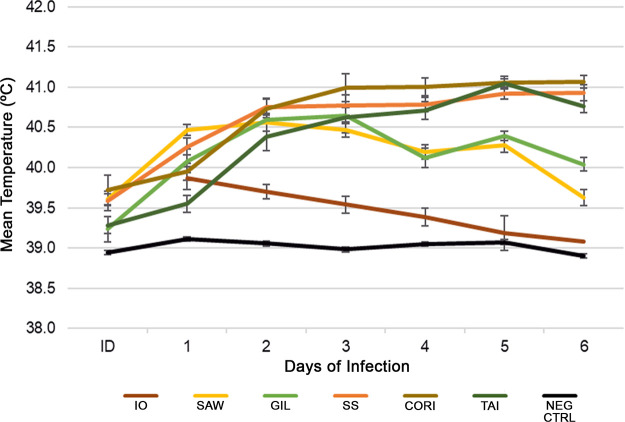
All Sawtooth-, Gila-, Costa Rica-, Taiaçu-, and Sheila SmithT-infected animals presented with fever ≥39.5°C (Fig. 1). Only 50% of the Iowa-infected animals had low-grade fever (40.0°C to 40.5°C) on days 1 and 2 and returned to normal temperature by day 3. Negative-control animals did not develop fever. Each of the Sheila SmithT-, Costa Rica-, and Taiaçu-infected animals presented with fever beginning on day 2 postinfection and continued with high fever between 40.5°C and 41°C until the end of the study. Sawtooth- and Gila-infected animals defervesced after day 4 of infection.
FIG 1.
Body temperature of Rickettsia rickettsii-infected guinea pigs. Temperatures were monitored daily. Each point represents the mean temperature of 10 animals, except for Costa Rica (n = 9), and standard errors of the means (SEM) are shown. Controls received the same amount of noninfected Vero cells. ID, inoculation day; IO, Iowa; SAW, Sawtooth; GIL, Gila; SS, Sheila SmithT; CORI, Costa Rica; TAI, Taiaçu; NEG CTRL, negative control. The ID data point for Iowa was not collected.
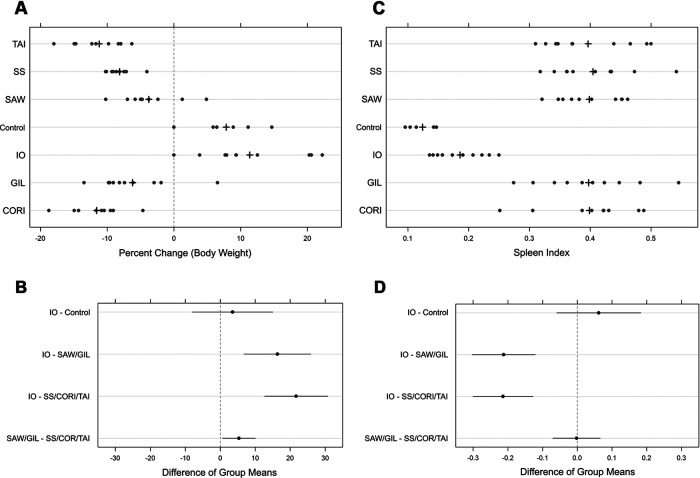
Animal weights were measured at the time of inoculation and at day 6. Both absolute and percent changes in body weight were analyzed. As the results agreed qualitatively, percent change in body weight and confidence intervals were used to provide general interpretation of the comparisons (Fig. 2). By day 6 postinfection, weights were statistically lower among Gila-, Sheila SmithT-, Costa Rica-, and Taiaçu-infected animals than in Iowa-infected animals and the negative-control group, which either maintained weight (n = 1 in each group) or gained weight (n = 9 of each group) over the course of the study (Fig. 2A). Weight loss in the Sawtooth- and Gila-infected animals was also statistically significant, although Sheila SmithT-, Costa Rica-, and Taiaçu-infected-animal losses were about 5% greater than the combined average for Sawtooth- and Gila-infected animals (Fig. 2A and B). We also identified a statistically significant greater day 6 spleen-to-weight index among Sawtooth-, Gila-, Sheila SmithT-, Costa Rica-, and Taiaçu-infected animals than in the Iowa-infected and negative-control groups (Fig. 2C and D).
FIG 2.
Body weight change and spleen size evaluation of all infected and noninfected samples. (A) Percentage of Rickettsia rickettsii-infected guinea pig body weight change. The weight was measured on days 1 and 6 of infection. (C) Spleen index at 6 days postinoculation. The spleen weight index was calculated based on the final body weight. +, mean value. (B and D) Comparative pairwise differences (95% CI) in mean percentage change in body weight by groups of isolates and spleen index, respectively, with the null reference line at 0. Differences in CIs that do not contain 0 are considered statistically significant at 5% significance. IO, Iowa; SAW, Sawtooth; GIL, Gila; SS, Sheila SmithT; CORI, Costa Rica; TAI, Taiaçu.
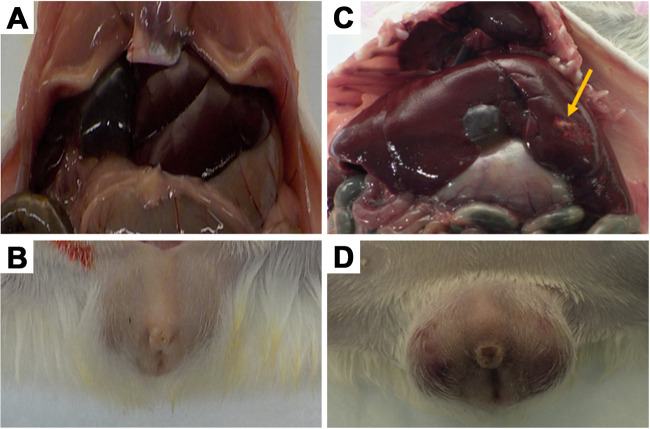
During necropsy, gross abnormalities were observed in the spleens and lungs of the infected animals as well as in livers and testes, as shown in Fig. 3. Pale foci in the liver were identified in 40% to 50% of all Sheila SmithT-, Costa Rica-, and Taiaçu-infected animals, while congested lungs were observed in 30% of these animals (Table 1 and Fig. 3). Prominent congestion and erythema of the testes were identified in almost all individuals infected with the Sheila SmithT (n = 10 of 10) or Taiaçu (n = 9 of 10) isolates (Table 1 and Fig. 3). These lesions were observed in a few Sawtooth-infected (n = 4 of 10) and Costa Rica-infected (n = 3 of 9) animals. No gross abnormalities were observed in Iowa-infected or negative-control animals. All infected animals had enlarged spleens compared to negative-control animals at day 6 of infection.
FIG 3.
Gross pathological findings in one isolate of Rickettsia rickettsii 6 days postinfection. (Top) Internal organs from the abdominal area; (bottom) scrotal area. (A and B) Negative controls; (C and D) Taiaçu infection. The arrow indicates the pale foci in the liver (C). Edema, erythema, and necrotic lesions on infected testes can be seen (D).
Histopathological and immunohistochemical evaluation of spleens, livers, lungs, and testes obtained at necropsy from guinea pigs infected with R. rickettsii revealed a spectrum of pathology that was tissue and isolate dependent (Fig. 4). None of the animals infected with the Iowa isolate showed abnormal histopathologic features or definitive immunohistochemical evidence of infection in any of the tissues examined. For the remaining isolates, inflammatory lesions were identified in the testes and epididymides of all Sheila SmithT-, Costa Rica-, and Taiaçu-infected animals, comprising lymphohistiocytic vasculitis of the tunics and interstitium (Fig. 4A and B). Focal to extensive immunohistochemical staining was identified in inflamed foci of all animals infected with these three isolates but was most abundant with Costa Rica (Fig. 4C). Testes and epididymides were inflamed in only half of Sawtooth- and Gila-infected animals. Infiltrates in these animals were typically less abundant, and rickettsial antigens were identified in only those animals with inflammatory foci. Pulmonary lesions, comprising predominantly mild interstitial mononuclear inflammatory cell infiltrates, were identified in all Sheila SmithT-infected and almost all Costa Rica- and Taiaçu-infected animals, whereas only half of the animals infected with Sawtooth or Gila demonstrated similar lesions. Interestingly, the lungs of most Gila-infected animals also demonstrated multifocal capillaritis, often accompanied by fibrin thrombi (Fig. 4D).
FIG 4.
Histopathological and immunohistochemical findings in tissues from animals infected intraperitoneally with Rickettsia rickettsii. Testes stained with hematoxylin and eosin showing lymphohistiocytic vasculitis of the tunics and interstitium (A and B). Immunohistochemical staining showing focal to extensive rickettsial antigens (red) in Costa Rica-infected testes (C). Capillaritis with fibrin thrombus in the lung of a Gila-infected animal (D). Mild vacuolar degeneration, rare single-cell necrosis, and mild lobular inflammation, comprising the predominant hepatic lesions produced following infection with all isolates except Iowa (E). Spleen histiocytes forming loose granulomas in a Sawtooth-infected animal (F). Magnifications, ×50 (A, B, and E), ×100 (C and D), and ×25 (F).
Antigens of R. rickettsii were detected in inflamed foci of all Sheila SmithT-infected, almost all Costa Rica- and Taiaçu-infected, and only half of Sawtooth- or Gila-infected animals. The predominant hepatic lesion in animals infected with all isolates except Iowa was vacuolar degeneration of hepatocytes, scattered single-cell hepatocyte necrosis, and small lobular foci of mixed inflammatory cell infiltrates (Fig. 4E). Immunohistochemistry revealed generally rare, focal staining of antigens of R. rickettsii in Kupffer cells of all animals infected with Sheila SmithT, Costa Rica, and Taiaçu and half of those infected with Gila but only one animal infected with Sawtooth. Diffuse histiocytic infiltrates were identified in the splenic red pulp of all animals infected with Sheila SmithT and Costa Rica and 40% infected with Taiaçu. In all Sawtooth-infected and 30% of Gila-infected animals, these infiltrates coalesced to form loose granulomas (Fig. 4F). Rare rickettsial antigens were detected by immunohistochemistry (IHC) staining in the spleens of all Costa Rica- and Taiaçu-infected and almost all Sawtooth-, Gila-, and Sheila SmithT-infected animals.
Infection load.
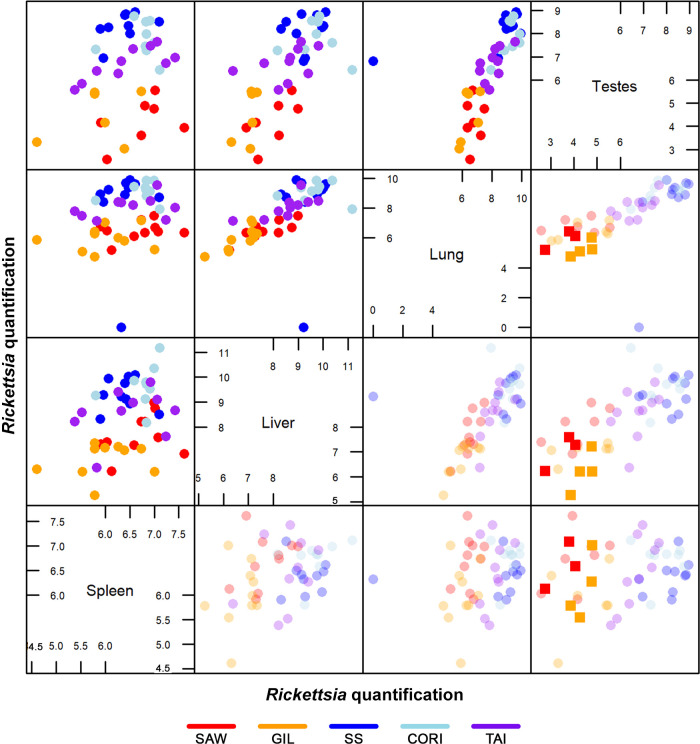
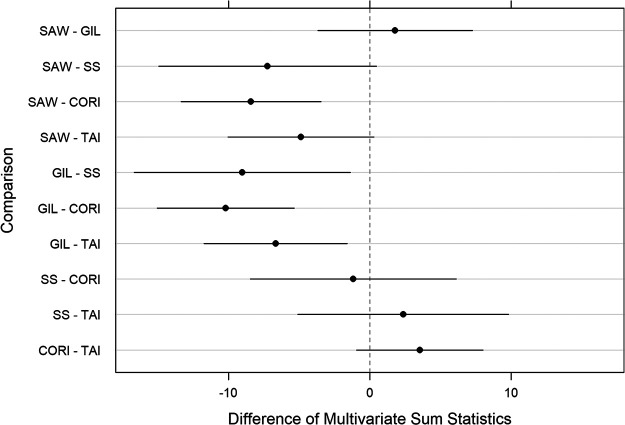
To identify tissue differences in R. rickettsii load among isolates, we evaluated spleens, livers, lungs, and testes from animals infected with all isolates on day 6 postinfection using a multivariate analysis of variance (MANOVA). The Rickettsia-specific real-time quantitative PCR multivariate data are shown in pairwise plots in Fig. 5. Spleen, liver, and lung samples from all animals infected with all isolates except Iowa contained rickettsial DNA, with a positive correlation between bacterial loads in livers and lungs. While testes of all animals infected with Sheila SmithT, Costa Rica, and Taiaçu contained DNA of R. rickettsii, testes from only 70% of Sawtooth- and 60% of Gila-infected animals demonstrated molecular evidence of infection. To evaluate the overall difference in rickettsial load among the isolates, combined tissue data for each isolate were analyzed (Fig. 6). Statistically significant differences were also identified when Sawtooth- and Gila-infected tissues were individually compared to Costa Rica- and Taiaçu-infected tissues (Fig. 6). Interestingly, when tissues were analyzed individually, significant differences in bacterial loads in the testes were seen between Sawtooth- and Gila-infected animals and Sheila SmithT- and Costa Rica-infected animals (see Fig. S1 in the supplemental material). Variation in rickettsial load was less pronounced in the spleen than in other tissues and ranged from 3.63 × 102 to 9.29 × 102 rickettsiae/μg of extracted DNA (Fig. 5 and Fig. S2). Sawtooth- and Gila-infected livers, lungs, and testes had bacterial loads 10- to 100-fold lower than those in Sheila SmithT-, Costa Rica-, and Taiaçu-infected groups (Fig. 5).
FIG 5.
Pairwise plots of a multivariance analysis of Rickettsia rickettsii load in all biological replicates from the five virulent isolates infecting animals 6 days postinfection. Plots represent the original quantification data (top left) and original with mean of multiply imputed quantification data (bottom right) for spleens, livers, lungs, and testes. In the lower half, the means of the multiply imputed values are presented in full color intensity, overlaid on the transparent original data. The imputation of the missing data for Sawtooth- and Gila-infected testes confirmed the patterns already identified in the original data set. Axes represent the log quantity of bacteria obtained by qPCR data. Each dot represents one biological replicate. SAW, Sawtooth; GIL, Gila; SS, Sheila SmithT; CORI, Costa Rica; TAI, Taiaçu.
FIG 6.
Rickettsia rickettsii load differences among isolates, considering all tissues. Pairwise differences (with 95% CI) of multivariate sum mean statistics. The sum of the isolate pairwise differences of the means over the 432 multivariate measurements [log(qPCR)] were used for the four tissues. CIs were adjusted for multiple comparisons and multiple imputation. Estimates were sorted in increasing order of the difference. Differences in CIs that do not contain 0 are considered statistically significant at 5% significance. SAW, Sawtooth; GIL, Gila; SS, Sheila SmithT; CORI Costa Rica; TAI, Taiaçu.
To determine bacterial viability within individual tissues, we inoculated Vero E6 cells with all testicular tissues collected at day 6 of infection and monitored cell cultures for 4 weeks or until the cytopathic effect exceeded 70%. We identified statistically significant differences in isolation rates comparing Sawtooth- to Sheila SmithT-infected (P < 0.01), Sawtooth- to Costa Rica-infected (P < 0.05), and Sawtooth- to Taiaçu-infected (P < 0.05) tissue replicates. Viable R. rickettsii was recovered from 80% of Sheila SmithT-, 77.77% of Costa Rica-, and 80% of Taiaçu-infected samples, while fewer Sawtooth-infected (10%) and Gila-infected (20%) samples contained live Rickettsia. No bacteria were recovered from the Iowa-infected samples or negative-control animals (data not shown).
Based on aggregate phenotypic data, we grouped the 6 studied isolates into three categories: a nonvirulent isolate (Iowa), two mildly virulent isolates (Sawtooth and Gila), and three highly virulent isolates (Sheila SmithT, Costa Rica, and Taiaçu).
Virulence factor gene expression.
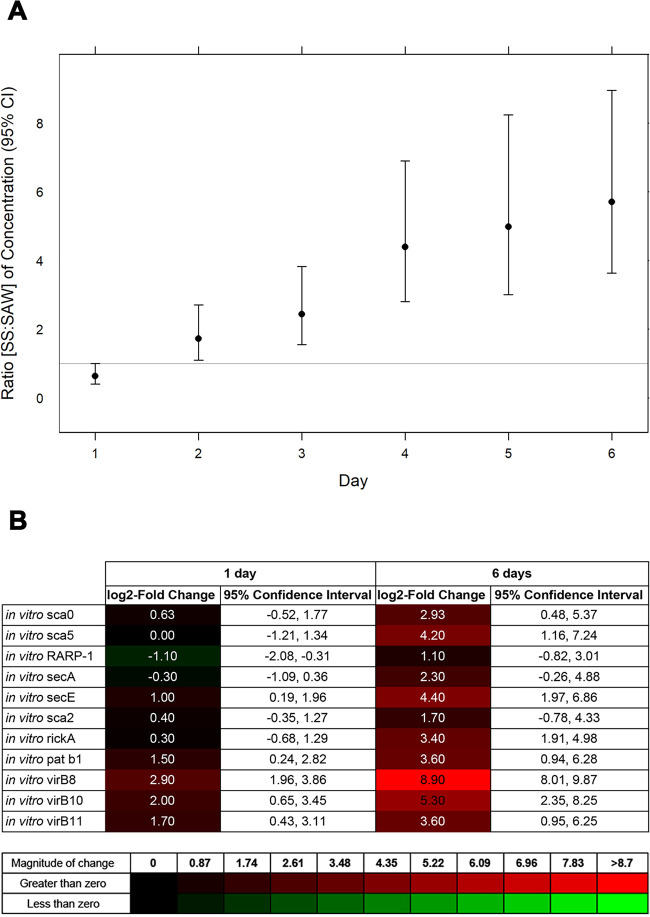
To identify gene expression differences that could account for differential virulence in vitro and in vivo experiments, we compared the transcriptional profiles of one representative of the mildly virulent (Sawtooth) and one representative of the highly virulent (Sheila SmithT) groups. Initially, both isolates were analyzed in vitro, tracking the infected Vero E6 cell cultures for 6 days. Both infections were established successfully, although differences in the number of bacteria per Vero E6 cell were identified over the course of the experiment when the ratio of each isolate per Vero cell (actin) was calculated. Sheila SmithT- and Sawtooth-infected-cell ratios (Rickettsia to actin) were compared. Over time, a statistically significant increase in the number of Sheila SmithT organisms per infected cell relative to the number of Sawtooth organisms per infected cell was identified (Fig. 7A). The ratio of the number of Sheila SmithT organisms per infected cell to the number of Sawtooth organisms per infected cell, a ratio of ratios (Sheila SmithT [Rickettsia to actin] to Sawtooth [Rickettsia to actin]), increased from 0.64 (95% confidence interval [CI], 0.41 to 1.00) at day 1 to 5.70 (95% CI, 3.64 to 8.95) at day 6 of infection, revealing that Sheila SmithT was five times more abundant on a per-cell basis than Sawtooth by day 6. As the rickettsial load per cell was significantly different during the infection, we compared the difference in transcriptional profiles of both isolates at days 1 and 6 of the experiment (Fig. 7B). No transcriptional differences between Sawtooth and Sheila SmithT were identified among most of the 11 potential virulence factors at day 1, including several previously associated with rickettsial virulence following infections in vitro and in vivo (19, 40–44, 50–51). However, a strong induced profile was identified at day 6 in Sawtooth- compared to Sheila SmithT-infected samples, associated mainly with components of secretion systems such as type 4 secretion systems (virB8, virB10, and virB11) as well as the Sec apparatus (secE).
FIG 7.
In vitro infection of Rickettsia rickettsii. (A) Daily comparative quantitative ratios of Sheila SmithT (SS) to Sawtooth (SAW) per Vero E6 cell for 6 days. (B) In vitro relative gene expression (quantified as log2 fold change) of Sawtooth/Sheila SmithT at days 1 and 6 of infection. Expression levels were determined by reverse transcriptase quantitative PCR (RT-qPCR). 95% CIs for the log2 fold change are shown for each time point and target gene.
We then evaluated the transcriptional profiles of these targets in the livers, lungs, and testes of Sawtooth- and Sheila SmithT-infected animals. These tissues were selected because they presented significant pathology and the bacterial load was higher than in the spleen. Similar to the in vitro evaluation, there was a significant difference in the number of bacteria per tissue cell within all tested tissues. The calculated ratio of Rickettsia to actin for Sheila SmithT was significantly higher than that for Sawtooth in all three tissues (Fig. 8A), ranging from 4.79 to 50 times more Sheila SmithT in livers and testes than Sawtooth. However, the absolute number of organisms was lower in the Sawtooth samples. As transcription analyses was carried out in those in vivo samples, transcripts of most of Sawtooth target genes were not detected at day 6 postinoculation, which may be due to the lower number of bacteria in each sample. In contrast, transcripts of 54.55% of the examined genes in liver, 100% in lungs, and 90.91% in testes of animals infected with the Sheila SmithT isolate were detected at day 6 (Fig. 8B).
FIG 8.
Rickettsia rickettsii in vivo infection of livers, lungs, and testes. (A) Comparative quantitative ratios of Sheila SmithT (SS) to Sawtooth (SAW) per liver, lung, and testis cell at day 6 of infection. (B) Detected transcripts (dark gray) and nondetected transcripts (light gray).
DISCUSSION
In this study, we demonstrated distinct and measurable differences in clinical and pathological characteristics of disease in guinea pigs following infection with standardized inocula from six distinct isolates of R. rickettsii. Intraperitoneal (i.p.) inoculation of guinea pigs with R. rickettsii has been used as a convenient and reproducible animal model system for RMSF for almost 100 years (27–30). Although i.p. inoculation does not reflect a natural route of infection, guinea pigs exposed to R. rickettsii in this manner nonetheless develop clinical signs largely identical to those observed in animals exposed to the pathogen via tick bite, including fever, scrotal edema and erythema, footpad dermatitis, and splenomegaly (31, 32). Interestingly, despite being a different animal model, a recent intravenously infected RMSF C3H/HeN mouse model (17) confirmed the rickettsial tissue distribution and load identified herein.
We identified specific differences in bacterial growth and transcriptional profiles of the different isolates following in vitro and in vivo infections. Previous investigators identified distinct genetic differences among various isolates of R. rickettsii (9, 33) that correlate with geographical origin (8–13). However, a recent comparison of four complete genomes of R. rickettsii obtained from eastern and western regions of the United States did not identify any specific genetic factors that could satisfactorily explain geographical differences in virulence (9) among regions reporting fatality rates ranging from 5% to 80% (3, 5–7, 13). To assess the phenotypic differences among isolates that could be associated with differences in disease outcome, we evaluated infections with six isolates originating from widely separated regions of United States, Costa Rica, and Brazil that included isolates obtained from human and tick hosts. The Iowa strain was isolated from a Dermacentor variabilis tick in 1941; it initially exhibited a mildly virulent phenotype in guinea pigs and became avirulent after multiple passages in embryonated chicken eggs (16). The Sawtooth strain was isolated from adult Dermacentor andersoni ticks in western Montana in 1961 (11), is considered virulent (14), and causes a mild cell injury response in vitro (10). Sheila SmithT, isolated in 1952 from a patient with severe RMSF from Montana (15), is perhaps the best-characterized isolate and exhibits a highly virulent phenotype in vitro and in vivo (8–11, 15, 17). The Costa Rica strain also represents a highly virulent phenotype and was isolated in 1980 from the blood of a patient with a fatal case of RMSF from the Limón region of Costa Rica (34). Taiaçu was isolated in 2002 from an Amblyomma aureolatum tick from Taiaçupeba, Brazil (35), where RMSF is endemic (http://www.saude.gov.br/saude-de-a-z/febre-maculosa). The Gila strain was isolated in 2016 from a patient with fatal RMSF in Arizona, USA (this study).
Differences among RMSF phenotypes were initially evaluated in vivo using clinical signs and histopathological findings. Three distinct phenotypes were observed: (i) a nonvirulent phenotype in which no animal was successfully infected (Iowa), (ii) a mildly virulent phenotype that produced mild clinical signs (Sawtooth and Gila), and (iii) a highly virulent phenotype characterized by animals presenting signs of severe disease and extensive tissue inflammation (Sheila SmithT, Costa Rica, and Taiaçu). Iowa-infected animals did not exhibit fever, significant splenomegaly, or weight change by the end of the study. In addition, no bacteria were detected in any organ from any biological replicate, which was confirmed by the unsuccessful reisolation of the organism in vitro. Indeed, the absence of RMSF phenotype in the same animal model was reported previously (8), although antibody titers were identified then but were not investigated in our study. Thus, as the Iowa strain is unable to successfully establish itself in the guinea pig host and cause disease but may promote seroconversion, it could be considered a possible candidate for a R. rickettsii vaccine. However, further studies are needed in order to identify the molecular factors associated with its apparent nonvirulence (36). All animals infected with the other five isolates developed fever and demonstrated significant weight loss and splenomegaly. Sawtooth- and Gila-infected animals exhibited weight loss, only transient fever, and significantly fewer rickettsiae in each tissue than animals infected with each of the highly virulent isolates. Consequently, these isolates were classified as mildly virulent in our animal model. Although bacterial passage history and isolate origin have been associated previously with changes in virulence (37), the passage histories of these isolates did not appear to affect disease outcome in our guinea pig model. Most of the isolates of R. rickettsii used in this evaluation had extensive passage histories (Table 2), while each behaved as described previously in our animal model with respect to having greater virulence (8, 15, 17) or mild or no virulence (10, 16). In comparison, the minimally passaged Gila isolate exhibited relatively mild virulence despite originating from a patient with fatal disease.
TABLE 2.
Rickettsia rickettsii isolate information
| Isolate | CRIRC identification | Host origin (yr of isolation) | Geographical origin | Reference(s)a | Passage historyb |
|---|---|---|---|---|---|
| Iowa | RRI010 | Dermacentor variabilis (1941) | USA | 8, 12 | 271 EP, 1 GP, 9 VR, 1 BC, 16 VR |
| Sawtooth | RRI025 | Dermacentor andersoni (1961) | USA | 7, 13, 14 | 8 VR, 1 MS, 3 VR |
| Gila | RRI017 | Human (2016) | USA | Our unpublished data | 3 VR |
| Sheila SmithT | RRI001T | Human (1952) | USA | 9, 12–15 | 2 VR |
| Costa Rica | RRI020 | Human (1980) | Costa Rica | 16, 17 | 4 VR |
| Taiaçu | RRI011 | Amblyomma aureolatum (2002) | Brazil | 18 | 5 VR, 12 BME26, 2 VR |
Reference where disease severity associated with the host can be found.
Passage history after acquisition of the strain by CDC. EP, egg passage; GP, guinea pig; VR, Vero cells; BC, BALB/c mouse; MS, mouse; BME26, Rhipicephalus microplus cells.
Overall, the variation in rickettsial loads in all tissues among the different virulence groups corroborated the histological differences identified in this study. Fewer animals infected with mildly virulent isolates exhibited inflammatory responses than animals infected with highly virulent isolates. Surprisingly, spleens exhibited low bacterial loads relative to other organs regardless of virulence category, although there was significant splenomegaly associated with histiocytic infiltrates, which could promote rickettsial clearance (38, 39). Indeed, the granulomas detected in all Sawtooth-infected spleen samples could reflect slower growth by this isolate, as identified in vitro. Similarly, testes had lower bacterial loads than lungs and livers, despite prominent vasculitis of the tunics and interstitium in animals infected with highly virulent isolates. The intraperitoneal needle inoculation route used in the study likely affected subsequent bacterial distribution as well as distinct tissue-specific immune responses; nonetheless, this route allowed complete control of the volume of inoculum administered in each biological replicate, which was key in this experimental setting. In contrast to spleens and testes, high and correlative bacterial loads were identified in livers and lungs, suggesting critical roles for these organs during the pathogenesis of this systemic disease.
Differences in rickettsial load among mildly and highly virulent isolates were identified in vivo and in vitro and could reflect differences in the growth rates among individual isolates. When Sawtooth and Sheila SmithT were compared, a lower growth rate and a lower bacterial load per cell in Sawtooth cultures were identified. When the transcription of potential virulence factors previously associated with rickettsial motility (RickA and Sca2) (40, 41), secretion system components or effectors (T4SS, Sec, and RARP-1) (42, 43), and host cell adhesion, attachment, and/or invasion (sca0 and sca5) (19, 44) were investigated, an increase in overall Sawtooth gene expression at the end of the in vitro study was identified, whereas a similar pattern was observed significantly earlier during infection with Sheila SmithT. These molecular patterns of potential virulence factors strengthen the hypothesis of a slower and constant growth of Sawtooth, compared to the rapid and more destructive growth of Sheila SmithT. Indeed, smaller amounts of Sawtooth and the same pattern of bacterium/cell ratios were detected in vivo. This could explain why so few Sawtooth transcripts were identified in vivo. A low growth rate was previously hypothesized to play a role in the propagation of R. prowazekii (20). As a confirmation of Sawtooth’s slow growth in vivo, granulomas were identified exclusively in Sawtooth-infected spleen samples and in no other tissue from animals infected with any other isolate, suggesting that relatively slow growth of this isolate allowed a robust host response. The important equilibrium between host response and bacterial growth is supported additionally by the recent confirmation of fatal RMSF in a human host caused by an Hlp strain of R. rickettsii (13). Guinea pigs infected with the first described Hlp strains exhibited milder disease than those infected with a classically virulent strain of R. rickettsii, including longer incubation periods, shorter febrile intervals, diminished scrotal pathology, and, most notably, the absence of fatal outcomes in the Hlp-infected animals (21). In this context, it is plausible that even subtle changes in host immunity can affect the outcome of infection with mildly virulent strains of. R. rickettsii.
The data from this investigation, obtained under highly controlled settings with standardized inocula and collected during the log phase of bacterial replication, illuminate quantitative phenotypic and transcriptional differences among geographically distinct R. rickettsii isolates within the classically recognized animal model of infection. Collectively, this information supports previously recognized qualitative assessments of strain virulence and suggests a critical and previously underappreciated balance between bacterial growth and host immune response that leverages strain pathogenicity. Finally, these data indicate that an overall assessment of strain virulence based on geographical origin should be undertaken in the context of regional host factors that influence the immune response to infections with R. rickettsii.
MATERIALS AND METHODS
Ethics statement.
All animals were maintained in a facility fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International. Care and husbandry of the animals were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (22). The procedures of the study were approved by the Centers for Disease Control and Prevention Institutional Animal Care and Use Committee under protocol 2834LEVGUIC and monitored by a veterinarian stationed on site, following the U.S. Animal Welfare Act and all associated regulations.
Inocula and animal host.
All isolates used in the study—Iowa, Sawtooth, Gila, Sheila SmithT, Costa Rica, and Taiaçu—are included in the CDC Rickettsial Isolate Reference Collection (CRIRC) as described in Table 2. These isolates were selected by geographical origin, previous genotype characterization (13), and previous use in vitro or in vivo evaluations of virulence, when available (8–11). All isolates were cultured in African green monkey (Cercopithecus aethiops) kidney cells (Vero E6) at 34°C in a 5% CO2. To prepare the inocula, the bacteria were initially grown in 150-cm2 flasks of confluent Vero E6 cells until they reached the log phase of replication. Cells were then harvested and processed as described previously (23, 24) with a few modifications. Briefly, cells were detached using sterile 3-mm glass beads, transferred to 50-ml centrifuge tubes and centrifuged at 4°C and 12.000 × g for 20 min to resuspend pellets in cold Snyder-1 medium (25, 26). The suspension was transferred to a 20-ml syringe, passed through a 20-gauge needle 10 times, and filtered through a 2-μm syringe filter. The number of viable bacteria was determined by counting the bacteria stained with a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes) in a Petroff-Hausser bacterial counting chamber (Hausser Scientific) and examined using a Zeiss microscope (45) in order to standardize all inocula at 107 viable bacteria. This inoculum was selected based on data acquired from an initial pilot study using Sheila SmithT which showed systemic tissue distribution and significant morbidity in guinea pigs at 7 days postinfection (data not shown). To standardize the volume of inoculum, the Rickettsia suspension was then diluted in Snyder-1 (1:1). Groups of 10 (9 in the case of Costa Rica) 6- to 9-week-old pathogen-free male Hartley guinea pigs (Cavia porcellus) weighing 400 to 500 g (Charles River Laboratories) were intraperitoneally (i.p.) inoculated with an isolate. Negative-control animals received noninfected Vero cells in a volume equivalent to that of the inoculum given to the infected groups.
Rickettsia rickettsii infection and sample collection.
Animals were followed for 6 days postinoculation. Clinical signs, including body temperature, scrotal edema (presence/absence), and footpad dermatitis (presence/absence), were recorded daily (46), while body weight was measured on days 1 and 6 of the experiment. Six days after inoculation, animals were euthanized and necropsied. Three samples of each spleen, liver, lung, and testis were collected (i) in RNAlater solution (Thermo Fisher Scientific) for nucleic acid extraction, (ii) in 10% formalin for histopathology processing, and (iii) in sterile sucrose-phosphate-glutamic acid (SPG) buffer for bacterial isolation. The spleen weight and final body weight were used for calculations as described previously (47).
Histopathology and immunohistochemistry.
Representative portions of spleens, livers, lungs, and testes were fixed in 10% neutral buffered formalin for 48 h and then embedded in paraffin. Three-micrometer sections were cut from paraffin blocks, stained with hematoxylin-eosin, and also stained by an immunoalkaline phosphatase technique with a polyclonal rabbit anti-R. rickettsii antibody diluted at 1/500 and counterstained with hematoxylin (48).
DNA and RNA extraction.
Simultaneous extraction of genomic DNA (gDNA) and total RNA was performed individually on all samples using the Qiagen AllPrep DNA/RNA minikit according to the manufacturer’s instructions. A 30-mg sample of each tissue was disrupted and homogenized using sterile 5-mm stainless steel beads (Qiagen) on a TissueLyser II system (Qiagen) for two cycles of 1 min at 30 Hz. Extracted gDNA was stored at −20°C until needed for quantitative real-time PCR (qPCR), while the total RNA was stored at −80°C until required for transcription assays.
In vitro infection.
For in vitro growth analysis, R. rickettsii Sawtooth and Sheila SmithT were each cultured in a T25 flask of confluent Vero E6 cells at 34°C in the presence of 5% CO2 for 6 days, at a multiplicity of infection (MOI) of 3. Samples were collected daily to evaluate infection progression via acridine orange (Becton Dickinson) staining and qPCR as described below. Simultaneous extraction of DNA and RNA was performed, as described above, in all biological triplicates at all time points.
Quantification assays.
For the Rickettsia-specific qPCR, the rickettsial 50S ribosomal protein L16 was targeted as described previously (49). For the guinea pig-specific qPCR, the C. porcellus beta-actin gene (actB) was targeted using 0.8 μM concentrations each of the forward primer 5′-CCTGTATGCCTCTGGCCGCA-3′ and reverse primer 5′-GGACTTCGAGCAGGAGATGG-3′ in addition to a 40 nM concentration of the fluorescence-labeled probe 5′-(Quasar 670)ACTGTGCCCATCTACGAG-3′ as described previously (50) for mouse actin with a few modifications on the forward and reverse primer sequences to match the C. porcellus sequence, generating a 260-bp amplicon. The reaction conditions were a 15-min incubation at 95°C, followed by 55 cycles of 95°C for 15 s, 53°C for 15 s, and 72°C for 30 s. For the Vero cell-specific qPCR, the C. aethiops (AB004047) beta-actin was targeted using 0.8 μM concentrations of the forward primer 5′-TGAAGTGTGACGTGGACATCCATA-3′ and the reverse primer 5′-AGGATGCAGAAGGAGATTACTGCC-3′ in addition to a 40 nM concentration of the fluorescence-labeled probe 5′-(Quasar 670)TGGCACCACCATGTACCCTGGCATTGCT-3′, generating a 98-bp amplicon, also described previously (50), with a single modification on the forward primer sequence to match the C. aethiops sequence available in NCBI. In order to perform an absolute quantification, control plasmids were created for all targets described above by cloning each amplicon into a pCR 4-TOPO TA vector system (Thermo Fisher Scientific), followed by sequencing to confirm the plasmid construction. All reactions were performed with QuantiTec Probe PCR master mix (Qiagen) using a CFX96 real-time system (Bio-Rad), while the analysis was conducted with CFX Manager software. Three technical replicates were analyzed for each sample.
Rickettsia rickettsii isolation.
Samples of testes collected during necropsies from all isolate-infected animals were kept at −80°C in sterile SPG buffer until needed. After thawing, 50 mg of each tissue was individually disrupted and homogenized using a Covidien Precision disposable tissue grinder system in 0.5 ml supplemented minimal essential medium (MEM) (5% fetal bovine serum, 0.1 mM MEM nonessential amino acids, 2 mM l-glutamine, 10 mM sodium pyruvate, 10 mM HEPES buffer, 10 U/ml penicillin G, and 10 μg/ml streptomycin sulfate). The inoculum was placed on a confluent monolayer of Vero E6 cells in a T25 tissue culture flask and incubated at 34°C in a 5% CO2-in-air atmosphere. Cultures were monitored weekly for rickettsial infection by acridine orange (Becton Dickinson) staining and real-time PCR, described below.
RT-qPCR.
The transcriptional profile of 11 selected R. rickettsii genes was determined by quantitative reverse transcription-PCR (RT-qPCR). Some have been previously associated with rickettsial virulence in vitro and in vertebrate or invertebrate hosts (19, 40–44, 51, 52). They are the genes encoding the 190-kDa cell surface antigen, surface cell antigen Sca2, outer membrane protein B (cell surface antigen sca5), preprotein translocase subunit SecA, preprotein translocase subunit SecE, VirB8 protein, VirB10 protein, type IV secretion system ATPase VirB11, Arp2/3 complex activation protein, patatin b1 precursor, and hypothetical protein/ankyrin repeats (RARP-1). Primers used for the experiment (Table 3) were designed at conserved regions among 10 R. rickettsii genomes (Sheila SmithT, Iowa, Arizona, Colombia, Hlp#2, Hino, Hauke, Morgan, R, and Brazil) available in the National Center for Biotechnology Information (NCBI) database using Primer3 (53) or as described previously (51). Two hundred nanograms of DNA-free RNA was used as the template for reverse transcription (RT) in cDNA using SuperScript III reverse transcriptase (Life Technologies) according to the manufacturer’s protocol. To determine the efficiency of each primer (0.8 μM each), standard curves were generated using six serial dilutions of a unique gDNA sample composed of a pool of gDNA from the biological samples. The curves were tested at annealing temperatures of 60°C, 61°C, and 62°C. Primers presenting efficiencies between 92 and 100% at a specific tested temperature were included in the analysis. PCRs were run as follows: 95°C for 15 min, followed by 40 cycles of 94°C for 15 s, 60 to 62°C for 30 s, and 72°C for 30 s, followed by a melting curve. QuantiTect SYBR green PCR master mix (Qiagen) was used. Reactions were performed using a CFX96 real-time system (Bio-Rad), while the analysis was conducted with CFX Manager software. Three technical replicates were analyzed for each sample. R. rickettsii gDNA was used for calculation of the standard curve. qPCR-derived fold change values are expressed using the equation described previously (54).
TABLE 3.
Primers used in gene expression analysis
| Gene ID | Annotation | Sequence (5′–3′) |
Annealing temp (°C) | Referencea | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| A1G_06990 | 190-kDa cell surface antigen | ACCAGCCATTAACTGACCTGT | GACGTGTTATTGCAACGACACT | 60 | This work |
| RRR_00670 | Surface cell antigen Sca2 | GCATCAGCCCCGATGGTTA | GAGGGGTATGGATTAGCGGC | 61 | This work |
| A1G_06030 | Outer membrane protein B (cell surface antigen sca5) | AGTAGTACCGCCGCCTAAAA | TGGTGCAGGATTACAAGGAA | 60 | 51 |
| A1G_04855 | Preprotein translocase subunit SecA | TACCCCGTCCTGCCATATTA | AAACGCCAAATTTCATGAGC | 61 | 51 |
| A1G_01005 | Preprotein translocase subunit SecE | TGTTGCTTCAACGTTAGTAGTGG | GCCGATATTAAGCAAAAGCTG | 61 | 51 |
| A1G_02210 | VirB8 protein | GGCATATCAGGTGATGTATTGG | ACTTTTGAATCACTGGCAAAAA | 60 | 51 |
| A1G_02230 | VirB10 protein | CGCCGGTCTTACCTCCTACT | TGCATCGCTCTCAACTAACG | 61 | 51 |
| A1G_02235 | Type IV secretion system ATPase VirB11 | GAGGCCTGTTTGCGTTTAAG | TGAACCGGGATGACCTGTAT | 60 | 51 |
| RrIowa_1080 | Arp2/3 complex activation protein | TGCAACCGGTTTTCCTGTAT | GGAAAATCAACCACGTCCTTC | 61 | 51 |
| A1G_05085 | Patatin b1 precursor | CGATTTTACTGGAGGGACCA | TTGCGCTGCACTGAATAAAG | 61 | 51 |
| A1G_01760 | Hypothetical protein/ankyrin repeats (RARP-1) | GCCGCTTATACCCGTTGTTG | ATTTGGAGAGCTTGCCGGAG | 60 | This work |
Where the design description can be found.
Statistical analysis.
To compare percent change in body weight and spleen indexes among isolates, a standard one-way fixed-effects model was fitted with the factor isolate, using generalized least squares (equivalent to maximum likelihood). Different variance parameters for each isolate were included to account for potential differences in variability by isolates, and the model was fitted with generalized least squares. These two models were compared considering both the Akaike information criterion (AIC), for which smaller values indicate a better fit, and the likelihood ratio test. Ninety-five percent confidence intervals (CI) were computed for model parameters, and all pairwise differences of estimated means (95% CIs) were computed using Tukey’s method to adjust for multiple comparisons and incorporated heterogeneity if indicated. Standard model diagnostics were used, including graphical evaluation of homoscedasticity and normality of the residuals.
For bacterial load analysis, the multivariate data were recorded as log(qPCR value) or as log(qPCR value + 1) for qPCR values of 0, presented as pairwise plots (55). Multiple imputation (56) was applied as a standard approach for modeling in the presence of missing data, assuming the Gaussian distributions for the log(qPCR) data together with linear models. We used four completed data sets based on standard diagnostics. To evaluate differences in bacterial loads among isolates, multivariate analysis of variance (MANOVA) was used to compare the multivariate means (i.e., mean vectors). As heterogeneity among isolates was anticipated, methods of standard MANOVA were used that have been adapted to this case (57). These methods were implemented in the R package MANOVA.RM. For comparison of the mean vectors for multivariate responses, the sum of the isolate pairwise differences of the means over the multivariate measurements [log(qPCR)] were used for the four tissues. Simultaneous 95% CIs for these isolates’ pairwise differences were computed using the parametric bootstrap by computing appropriate quantile (q*) of the estimated difference statistics, as described in reference 57. To employ this approach while accounting for the uncertainty associated with multiple imputation, we averaged , which represents the computed quantiles from the analysis of each multiple imputation completed data set. Further, we calculated a revised value for the quantile from the method of Friedrich and Pauly adapted to include Rubin’s rules by multiplying by the ratio of the t value with degrees of freedom computed using Rubin’s rules to the “plain” t value with degrees of freedom computed assuming no adjustments for simultaneous inference (so using, generically, n + m − 2). This ratio of t values captures the adjustment to the standard pairwise t comparison made by the multiple imputation adjustment using Rubin’s Rules, and multiplying it by by the method described in reference 57 preserves the simultaneous and distribution-free nature of their approach.
We compared the proportion of reisolated samples among the isolates using Fisher’s exact followed by pairwise comparisons adjusted for multiple comparisons using the Holm procedure (58), as implemented in the RVAideMemoire package in R.
To compare the number of bacteria per host cell, we fitted a linear model to the within-sample differences between the logarithms of the loads [log(Rickettsia) − log(actin)], including day and isolate as covariates for the in vitro studies comparing Sheila SmithT (SS) to Sawtooth (SAW) and also for the in vivo study comparing Sheila SmithT to Sawtooth for livers, lungs, and testes. Standard diagnostics were performed to evaluate model assumptions. Linear functions of the model coefficients were used to estimate the effect of isolate by day (in vitro) and tissue (in vivo), and the corresponding simultaneous 95% CIs were computed using methods detailed previously (58). Resulting estimates and CIs were exponentiated to provide inference for the ratio of the ratios of concentrations, i.e., the SS Rickettsia-to-actin ratio to the SAW Rickettsia-to-actin ratio, as a means of summarizing the differential growth for the isolates.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Levin and the CDC’s Medical Entomology Laboratory personnel for technical assistance in animal studies and Kim Slater for assistance in the in vitro experiments. We are also grateful to Andrea C. Fogaça from University of Sao Paulo, Brazil, for providing the Taiaçu isolate used in this study.
M.F.B.M.G., S.K., and C.P., conceived the hypothesis. M.F.B.M.G. designed and developed the project. M.F.B.M.G., J.H., and S.K. carried out the in vivo experiments. J.R. and C.P. carried out the histological analysis. B.B. performed the statistical analyzes. M.F.B.M.G., S.K., and C.P. wrote the manuscript. All authors reviewed and revised the final version of the manuscript.
The research reported here was supported in part by an appointment of M.F.B.M.G. to the Research Participation Program at the Centers for Disease Control and Prevention administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the CDC.
The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Alvarez-Hernandez G, Roldan JFG, Milan NSH, Lash RR, Behravesh CB, Paddock CD. 2017. Rocky Mountain spotted fever in Mexico: past, present, and future. Lancet Infect Dis 17:e189–e196. doi: 10.1016/S1473-3099(17)30173-1. [DOI] [PubMed] [Google Scholar]
- 2.Drexler NA, Yaglom H, Casal M, Fierro M, Kriner P, Murphy B, Kjemtrup A, Paddock CD. 2017. Fatal Rocky Mountain spotted fever along the United States-Mexico Border, 2013–2016. Emerg Infect Dis 23:1621–1626. doi: 10.3201/eid2310.170309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drexler NA, Dahlgren FS, Heitman KN, Massung RF, Paddock CD, Behravesh CB. 2016. National surveillance of spotted fever group rickettsioses in the United States, 2008–2012. Am J Trop Med Hyg 94:26–34. doi: 10.4269/ajtmh.15-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg R, Lindsey NP, Fischer M, Gregory CJ, Hinckley AF, Mead PS, Paz-Bailey G, Waterman SH, Drexler NA, Kersh GJ, Hooks H, Partridge SK, Visser SN, Beard CB, Petersen LR. 2018. Vital signs: trends in reported vectorborne disease cases - United States and territories, 2004–2016. MMWR Morb Mortal Wkly Rep 67:496–501. doi: 10.15585/mmwr.mm6717e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, Abdad MY, Stenos J, Bitam I, Fournier PE, Raoult D. 2013. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev 26:657–702. doi: 10.1128/CMR.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Oliveira SV, Guimaraes JN, Reckziegel GC, Neves BM, Araujo-Vilges KM, Fonseca LX, Pinna FV, Pereira SV, de Caldas EP, Gazeta GS, Gurgel-Goncalves R. 2016. An update on the epidemiological situation of spotted fever in Brazil. J Venom Anim Toxins Incl Trop Dis 22:22. doi: 10.1186/s40409-016-0077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angerami RN, Resende MR, Feltrin AF, Katz G, Nascimento EM, Stucchi RS, Silva LJ. 2006. Brazilian spotted fever: a case series from an endemic area in southeastern Brazil: clinical aspects. Ann N Y Acad Sci 1078:252–254. doi: 10.1196/annals.1374.044. [DOI] [PubMed] [Google Scholar]
- 8.Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Porcella SF, Hackstadt T. 2008. Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect Immun 76:542–550. doi: 10.1128/IAI.00952-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark TR, Noriea NF, Bublitz DC, Ellison DW, Martens C, Lutter EI, Hackstadt T. 2015. Comparative genome sequencing of Rickettsia rickettsii strains that differ in virulence. Infect Immun 83:1568–1576. doi: 10.1128/IAI.03140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eremeeva ME, Dasch GA, Silverman DJ. 2001. Quantitative analyses of variations in the injury of endothelial cells elicited by 11 isolates of Rickettsia rickettsii. Clin Diagn Lab Immunol 8:788–796. doi: 10.1128/CDLI.8.4.788-796.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anacker RL, Philip RN, Williams JC, List RH, Mann RE. 1984. Biochemical and immunochemical analysis of Rickettsia rickettsii strains of various degrees of virulence. Infect Immun 44:559–564. doi: 10.1128/IAI.44.3.559-564.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karpathy SE, Dasch GA, Eremeeva ME. 2007. Molecular typing of isolates of Rickettsia rickettsii by use of DNA sequencing of variable intergenic regions. J Clin Microbiol 45:2545–2553. doi: 10.1128/JCM.00367-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paddock CD, Denison AM, Lash RR, Liu L, Bollweg BC, Dahlgren FS, Kanamura CT, Angerami RN, Pereira dos Santos FC, Brasil Martines R, Karpathy SE. 2014. Phylogeography of Rickettsia rickettsii genotypes associated with fatal Rocky Mountain spotted fever. Am J Trop Med Hyg 91:589–597. doi: 10.4269/ajtmh.14-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price WH. 1953. The epidemiology of Rocky Mountain spotted fever. I. The characterization of strain virulence of Rickettsia rickettsii. Am J Hyg 58:248–268. doi: 10.1093/oxfordjournals.aje.a119604. [DOI] [PubMed] [Google Scholar]
- 15.Bell EJ, Pickens EG. 1953. A toxic substance associated with the rickettsias of the spotted fever group. J Immunol 70:461–472. [PubMed] [Google Scholar]
- 16.Cox HR. 1941. Cultivation of rickettsiae of the Rocky Mountain spotted fever, typhus and Q fever groups in the embryonic tissues of developing chicks. Science 94:399–403. doi: 10.1126/science.94.2444.399. [DOI] [PubMed] [Google Scholar]
- 17.Esteves E, Fongsaran C, Langohr IM, Riley SP, Labruna MB, Daffre S, Fogaça AC, Macaluso KR. 2020. Comparative Analysis of Infection by Rickettsia rickettsii Sheila Smith and Taiaçu strains in a murine model. Pathogens 9:744. doi: 10.3390/pathogens9090744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burgdorfer W. 1963. Investigation of “transovarial transmission” of Rickettsia rickettsii in the wood tick, Dermacentor andersoni. Exp Parasitol 14:152–159. doi: 10.1016/0014-4894(63)90019-5. [DOI] [Google Scholar]
- 19.Uchiyama T, Kawano H, Kusuhara Y. 2006. The major outer membrane protein rOmpB of spotted fever group rickettsiae functions in the rickettsial adherence to and invasion of Vero cells. Microbes Infect 8:801–809. doi: 10.1016/j.micinf.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Winkler HH. 1995. Rickettsia prowazekii, ribosomes and slow growth. Trends Microbiol 3:196–198. doi: 10.1016/s0966-842x(00)88920-9. [DOI] [PubMed] [Google Scholar]
- 21.Parker RR, Pickens EG, Lackman DB, Belle EJ, Thraikill FB. 1951. Isolation and characterization of Rocky Mountain spotted fever rickettsiae from the rabbit tick Haemaphysalis leporis-palustris Packard. Public Health Rep 66:455–463. doi: 10.2307/4587691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 23.Weiss E. 1973. Growth and physiology of rickettsiae. Bacteriol Rev 37:259–283. doi: 10.1128/BR.37.3.259-283.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petchampai N, Sunyakumthorn P, Guillotte ML, Thepparit C, Kearney MT, Mulenga A, Azad AF, Macaluso KR. 2014. Molecular and functional characterization of vacuolar-ATPase from the American dog tick Dermacentor variabilis. Insect Mol Biol 23:42–51. doi: 10.1111/imb.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bovarnick MR, Miller JC, Snyder JC. 1950. The influence of certain salts, amino acids, sugars, and proteins on the stability of rickettsiae. J Bacteriol 59:509–522. doi: 10.1128/JB.59.4.509-522.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson EB, Smadel JE. 1951. Immunization against scrub typhus. II. Preparation of lyophilized living vaccine. Am J Hyg 53:326–331. doi: 10.1093/oxfordjournals.aje.a119457. [DOI] [PubMed] [Google Scholar]
- 27.Stokes JV, Walker DH, Varela-Stokes AS. 2020. The guinea pig model for tick-borne spotted fever rickettsioses: a second look. Ticks Tick Borne Dis 11:101538. doi: 10.1016/j.ttbdis.2020.101538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spencer RR, Parker RR. 1923. Rocky Mountain spotted fever: infectivity of fasting and recently fed ticks. Public Health Rep 38:333–339. doi: 10.2307/4576667.19314866 [DOI] [Google Scholar]
- 29.Price WH, Emerson H, Preston C. 1961. The transmission of low virulent strains of Rickettsia rickettsii. Pathol Microbiol (Basel) 24(Suppl):132–139. doi: 10.1159/000161251. [DOI] [PubMed] [Google Scholar]
- 30.Blanton LS, Wilson NM, Quade BR, Walker DH. 2019. Susceptibility of Rickettsia rickettsii to tigecycline in a cell culture assay and animal model for Rocky Mountain spotted fever. Am J Trop Med Hyg 101:1091–1095. doi: 10.4269/ajtmh.19-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin ML, Ford SL, Hartzer K, Krapiunaya L, Stanley H, Snellgrove AN. 2020. Minimal duration of tick attachment sufficient for transmission of infectious Rickettsia rickettsii (Rickettsiales: Rickettsiaceae) by its primary vector Dermacentor variabilis (Acari: Ixodidae): duration of rickettsial reactivation in the vector revisited. J Med Entomol 57:585–594. doi: 10.1093/jme/tjz191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin ML, Schumacher LBM, Snellgrove A. 2018. Effects of Rickettsia amblyommatis infection on the vector competence of Amblyomma americanum ticks for Rickettsia rickettsii. Vector Borne Zoonotic Dis 18:579–587. doi: 10.1089/vbz.2018.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eremeeva ME, Dasch GA. 2009. Closing the gaps between genotype and phenotype in Rickettsia rickettsii. Ann N Y Acad Sci 1166:12–26. doi: 10.1111/j.1749-6632.2009.04526.x. [DOI] [PubMed] [Google Scholar]
- 34.Hun LHL, Fuentes L, Vargas M. 1991. Tres nuevos casos de Fiebres Manchadas de las Monta-as Rocosas en Costa Rica. Rev Costarricense Ciencias Medicas 12:51–56. [Google Scholar]
- 35.Pinter A, Labruna MB. 2006. Isolation of Rickettsia rickettsii and Rickettsia bellii in cell culture from the tick Amblyomma aureolatum in Brazil. Ann N Y Acad Sci 1078:523–529. doi: 10.1196/annals.1374.103. [DOI] [PubMed] [Google Scholar]
- 36.Noriea NF, Clark TR, Hackstadt T. 2015. Targeted knockout of the Rickettsia rickettsii OmpA surface antigen does not diminish virulence in a mammalian model system. mBio 6:e00323-15. doi: 10.1128/mBio.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker DH, Yu XJ. 2005. Progress in rickettsial genome analysis from pioneering of Rickettsia prowazekii to the recent Rickettsia typhi. Ann N Y Acad Sci 1063:13–25. doi: 10.1196/annals.1355.003. [DOI] [PubMed] [Google Scholar]
- 38.Pivkin IV, Peng Z, Karniadakis GE, Buffet PA, Dao M, Suresh S. 2016. Biomechanics of red blood cells in human spleen and consequences for physiology and disease. Proc Natl Acad Sci U S A 113:7804–7809. doi: 10.1073/pnas.1606751113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapila V, Wehrle CJ, Tuma F. 2020. Physiology, spleen. StatPearls, Treasure Island, FL. [PubMed] [Google Scholar]
- 40.Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. 2010. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun 78:2240–2247. doi: 10.1128/IAI.00100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed SCO, Lamason RL, Risca VI, Abernathy E, Welch MD. 2014. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr Biol 24:98–103. doi: 10.1016/j.cub.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaur SJ, Rahman MS, Ammerman NC, Beier-Sexton M, Ceraul SM, Gillespie JJ, Azad AF. 2012. TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi. J Bacteriol 194:4920–4932. doi: 10.1128/JB.00793-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gillespie JJ, Kaur SJ, Rahman MS, Rennoll-Bankert K, Sears KT, Beier-Sexton M, Azad AF. 2015. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev 39:47–80. doi: 10.1111/1574-6976.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Walker DH. 1998. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb Pathog 24:289–298. doi: 10.1006/mpat.1997.0197. [DOI] [PubMed] [Google Scholar]
- 45.Kurtti TJ, Simser JA, Baldridge GD, Palmer AT, Munderloh UG. 2005. Factors influencing in vitro infectivity and growth of Rickettsia peacockii (Rickettsiales: Rickettsiaceae), an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni (Acari, Ixodidae). J Invertebr Pathol 90:177–186. doi: 10.1016/j.jip.2005.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker DH, Harrison A, Henderson F, Murphy FA. 1977. Identification of Rickettsia rickettsii in a guinea pig model by immunofluorescent and electron microscopic techniques. Am J Pathol 86:343–358. [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Carrillo C. 1977. Relationship between bodyweight and spleen size in guinea-pigs. Lab Anim 11:175–180. doi: 10.1258/002367777780936594. [DOI] [PubMed] [Google Scholar]
- 48.Paddock CD, Greer PW, Ferebee TL, Singleton J, Jr, McKechnie DB, Treadwell TA, Krebs JW, Clarke MJ, Holman RC, Olson JG, Childs JE, Zaki SR. 1999. Hidden mortality attributable to Rocky Mountain spotted fever: immunohistochemical detection of fatal, serologically unconfirmed disease. J Infect Dis 179:1469–1476. doi: 10.1086/314776. [DOI] [PubMed] [Google Scholar]
- 49.Kato CY, Chung IH, Robinson LK, Austin AL, Dasch GA, Massung RF. 2013. Assessment of real-time PCR assay for detection of Rickettsia spp. and Rickettsia rickettsii in banked clinical samples. J Clin Microbiol 51:314–317. doi: 10.1128/JCM.01723-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riley SP, Fish AI, Garza DA, Banajee KH, Harris EK, del Piero F, Martinez JJ. 2016. Nonselective persistence of a Rickettsia conorii extrachromosomal plasmid during mammalian infection. Infect Immun 84:790–797. doi: 10.1128/IAI.01205-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galletti MF, Fujita A, Nishiyama MY, Jr, Malossi CD, Pinter A, Soares JF, Daffre S, Labruna MB, Fogaca AC. 2013. Natural blood feeding and temperature shift modulate the global transcriptional profile of Rickettsia rickettsii infecting its tick vector. PLoS One 8:e77388. doi: 10.1371/journal.pone.0077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galletti MF, Fujita A, Rosa RD, Martins LA, Soares HS, Labruna MB, Daffre S, Fogaca AC. 2016. Virulence genes of Rickettsia rickettsii are differentially modulated by either temperature upshift or blood-feeding in tick midgut and salivary glands. Parasit Vectors 9:331. doi: 10.1186/s13071-016-1581-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koressaar T, Lepamets M, Kaplinski L, Raime K, Andreson R, Remm M. 2018. Primer3_masker: integrating masking of template sequence with primer design software. Bioinformatics 34:1937–1938. doi: 10.1093/bioinformatics/bty036. [DOI] [PubMed] [Google Scholar]
- 54.Riley SP, Pruneau L, Martinez JJ. 2017. Evaluation of changes to the Rickettsia rickettsii transcriptome during mammalian infection. PLoS One 12:e0182290. doi: 10.1371/journal.pone.0182290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.National Institute of Standards and Technology. 2012. NIST/SEMATECH e-Handbook of Statistical Methods.
- 56.Little RJA, Rubin DB. 2002. Statistical analysis with missing data, 2nd ed. Wiley, Hoboken, NJ. [Google Scholar]
- 57.Friedrich S, Pauly M. 2018. MATS: inference for potentially singular and heteroscedastic MANOVA. J Multivariate Analysis 165:166–179. doi: 10.1016/j.jmva.2017.12.008. [DOI] [Google Scholar]
- 58.Bretz F, Hothorn T, Westfall P. 2010. Multiple comparisons using R. CRC Press, Boca Raton, FL. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.