Bioelectrochemical systems (BESs) have attracted wide attention owing to their utility in sustainable biotechnology processes, such as microbial fuel cells and electrofermentation systems. In BESs, electrochemically active bacteria (EAB) form biofilms on electrode surfaces, thereby serving as effective catalysts for the interconversion between chemical and electric energy.
KEYWORDS: cyclic di-GMP, diguanylate cyclase, biofilm, electrochemically active bacteria, flow cell
ABSTRACT
In many bacteria, cyclic diguanosine monophosphate (c-di-GMP), synthesized by diguanylate cyclase (DGC), serves as a second messenger involved in the regulation of biofilm formation. Although studies have suggested that c-di-GMP also regulates the formation of electrochemically active biofilms (EABFs) by Shewanella oneidensis MR-1, DGCs involved in this process remained to be identified. Here, we report that the SO_1646 gene, hereafter named dgcS, is upregulated under medium flow conditions in electrochemical flow cells (EFCs), and its product (DgcS) functions as a major DGC in MR-1. In vitro assays demonstrated that purified DgcS catalyzed the synthesis of c-di-GMP from GTP. Comparisons of intracellular c-di-GMP levels in the wild-type strain and a dgcS deletion mutant (ΔdgcS mutant) showed that production of c-di-GMP was markedly reduced in the ΔdgcS mutant when cells were grown in batch cultures and on electrodes in EFCs. Cultivation of the ΔdgcS mutant in EFCs also revealed that the loss of DgcS resulted in impaired biofilm formation and decreased current generation. These findings demonstrate that MR-1 uses DgcS to synthesize c-di-GMP under medium flow conditions, thereby activating biofilm formation on electrodes.
IMPORTANCE Bioelectrochemical systems (BESs) have attracted wide attention owing to their utility in sustainable biotechnology processes, such as microbial fuel cells and electrofermentation systems. In BESs, electrochemically active bacteria (EAB) form biofilms on electrode surfaces, thereby serving as effective catalysts for the interconversion between chemical and electric energy. It is therefore important to understand mechanisms for the formation of biofilm by EAB grown on electrodes. Here, we show that a model EAB, S. oneidensis MR-1, expresses DgcS as a major DGC, thereby activating the formation of biofilms on electrodes via c-di-GMP-dependent signal transduction cascades. The findings presented herein provide the molecular basis for improving electrochemical interactions between EAB and electrodes in BESs. The results also offer molecular insights into how Shewanella regulates biofilm formation on solid surfaces in the natural environment.
INTRODUCTION
Bioelectrochemical systems (BESs) are engineered systems that utilize electrochemical interactions between electrochemically active bacteria (EAB) and electrodes (1). Potential applications of BESs include microbial fuel cells (MFCs) and electrofermentation systems, and these are expected to be used for electricity generation from wastewater and bioproduction of value-added chemicals, respectively (2–4). In BESs, EAB form biofilms during growth on electrode surfaces, thereby functioning as effective catalysts for the interconversion between electric and chemical energy (5). It is also known that the biofilms facilitate the long-range transfer of electrons from EAB cells to electrodes by association with self-secreted redox-active compounds, such as cytochromes (6, 7) and flavins (8). The roles of biofilms formed by EAB (i.e., electrochemically active biofilms [EABFs]) are considered particularly important in practical BESs that are assumed to be operated under a continuous flow of electrolytes (e.g., wastewater and culture medium), since planktonic microbes are washed out from these systems (9). It is therefore expected that understanding the mechanisms behind EABF formation would provide insights into the development of high-performance BESs.
Extensive microscopic and electrochemical measurements have been performed to characterize EABFs formed by model EAB, such as Geobacter sulfurreducens PCA (10–12) and Shewanella oneidensis MR-1 (13–15). However, limited information is available on how EAB regulate EABF formation on electrode surfaces. To explore genes involved in the EABF formation by S. oneidensis MR-1, we previously cultivated this strain in electrochemical flow cells (EFCs) and examined the transcriptional profiles of the electrode-associated cells grown in the presence and absence of the flow of medium (16). It was revealed that under the medium flow conditions, MR-1 significantly upregulated the expression of a putative diguanylate cyclase (DGC) gene, SO_1646, as well as other genes related to cell motility and signal transduction (16). DGCs catalyze the synthesis of cyclic diguanosine monophosphate (c-di-GMP), a key second messenger involved in the regulation of biofilm formation in bacteria (17). It is therefore suggested that MR-1 expresses SO_1646 under the flow of medium, thereby facilitating the formation of biofilms on electrodes via c-di-GMP-dependent signal transduction cascades.
The concentration of c-di-GMP in bacterial cells depends on the activities of two enzymes, DGC and phosphodiesterase (PDE) (18). DGCs contain GGDEF domains that catalyze the synthesis of c-di-GMP from two GTP molecules, while PDEs contain EAL or HD-GYP domains that catalyze the hydrolysis of c-di-GMP into 5′-phosphoguanylyl-(3′–5′)-guanosine (pGpG) or two GMP molecules (19). Bifunctional proteins that have both DGC and PDE activities are also present (20, 21). The genome of MR-1 encodes 51 GGDEF domain-containing proteins, including 20 hybrid proteins containing both GGDEF and EAL domains (22). Although the physiological functions of these GGDEF domain-containing proteins are largely unknown, a previous study reported that the overexpression of a DGC gene that originated from Escherichia coli in MR-1 resulted in enhanced biofilm formation on electrodes and increased current generation in MFCs (23). It has also been reported that the disruption of a PDE gene, pdeB, in MR-1 facilitates biofilm formation on glass surfaces in aerobically operated flow cells (24). These observations suggest that c-di-GMP plays important roles in the regulation of biofilm formation by MR-1, although roles of its endogenous DGCs in the formation of electrode-associated biofilms remain to be elucidated.
Based on the above-mentioned knowledge, we hypothesized that MR-1 regulates biofilm formation on electrodes via c-di-GMP-dependent signal transduction cascades, in which the gene product of SO_1646, hereafter named DgcS (for diguanylate cyclase in S. oneidensis), plays a key role. To address this hypothesis, in vitro and in vivo experiments were conducted in this study to investigate the involvement of DgcS in c-di-GMP synthesis and EABF formation by MR-1.
RESULTS
Transcriptional regulation of dgcS.
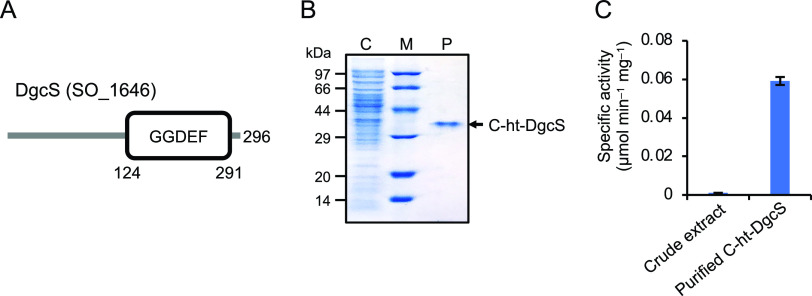
Our previous transcriptome analyses for MR-1 cells grown in EFCs found that the expression of the SO_1646 gene (dgcS) was upregulated in the presence of medium flow (16). To verify this result, we performed quantitative reverse transcription (RT)-PCR analysis to compare the expression levels of this gene in MR-1 cells grown in EFCs operated under medium flow and nonflow conditions (see Fig. S1 in the supplemental material). The results confirmed that the presence of medium flow significantly upregulated the expression of dgcS, supporting the hypothesis that this gene is involved in the formation of electrode-associated biofilms by MR-1 cells exposed to medium flow. Given that a σ54-dependent promoter-like sequence is present at the upstream region of dgcS (25) and that the downstream tilS (SO_1645) gene is arranged in the opposite direction, it is suggested that dgcS is transcribed as a single gene from the upstream promoter. It is known that the activities of many DGC proteins are controlled by N-terminal sensor domains, such as PAS, GAF, CHASE, and HAMP domains (26). However, in silico domain prediction using the Simple Modular Architecture Research Tool (SMART) (27) showed that the DgcS protein contains no known domain other than a C-terminal GGDEF domain (Fig. 1A). Taken together, these findings suggest that transcriptional regulation plays an important role in controlling the activity of DgcS, although the involvement of posttranscriptional regulation cannot be excluded.
FIG 1.
Enzymatic characterization of DgcS. (A) Predicted domain structure of DgcS. (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of C-ht-DgcS protein samples. Protein samples (5 μg) were analyzed on 12.5% SDS-polyacrylamide gels. Lanes: C, E. coli BL21(DE3) (pET-C-ht-dgcS) crude extract; M, molecular weight marker; P; purified C-ht-DgcS. (C) Specific DGC activity of E. coli BL21(DE3) (pET-C-ht-dgcS) crude extract and purified C-ht-DgcS. Results are expressed as μmol c-di-GMP produced min−1 mg−1 protein. Error bars represent standard deviations calculated from the results of three independent experiments.
In vitro enzyme assay for DgcS.
To examine if DgcS is a DGC, we conducted in vitro assays using purified DgcS and examined the synthesis of c-di-GMP from GTP. C-terminally histidine-tagged DgcS (C-ht-DgcS) was expressed in recombinant Escherichia coli cells and purified by nickel affinity chromatography (Fig. 1B). The purified protein was incubated in a reaction mixture containing GTP, and the consumption of GTP and the production of c-di-GMP were analyzed by high-performance liquid chromatography (HPLC). Control experiments using crude (nonpurified) extracts from the C-ht-dgcS-expressing E. coli cells were also performed. The HPLC analysis revealed that synthesis of c-di-GMP, along with GTP consumption (data not shown), occurred in the presence of purified DgcS. The specific activity of purified DgcS was 53 times higher than that of the crude extract (Fig. 1C), demonstrating that DgcS functions as a DGC that catalyzes the synthesis of c-di-GMP from GTP.
Contribution of DgcS to intracellular c-di-GMP synthesis.
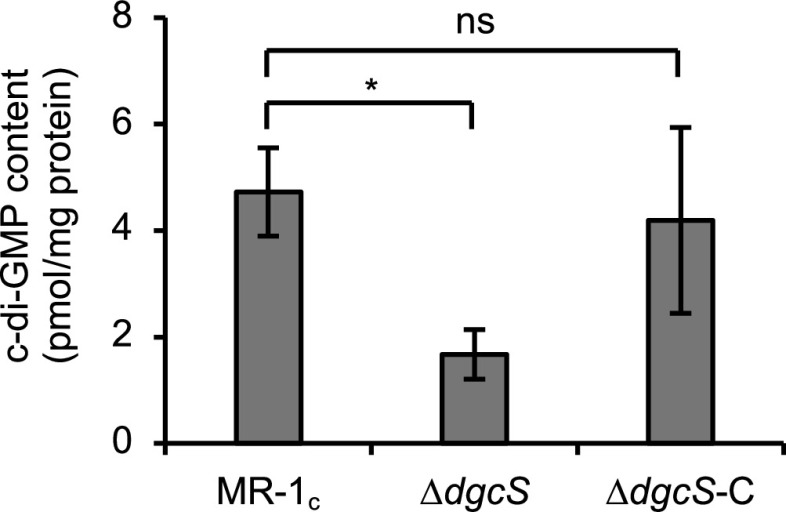
To evaluate the contribution of DgcS to in vivo c-di-GMP synthesis, a dgcS deletion mutant of MR-1 (ΔdgcS mutant) and its complemented strain (ΔdgcS-C strain) were constructed (Table 1). A plasmid expressing an anaerobic fluorescent protein (AFP) (evoglow-Bs2; evocatal GmbH, Düsseldorf, Germany) (28) had been introduced into these strains for subsequent biofilm observation in EFCs. When these mutant strains were cultivated in test tubes containing lactate minimal medium (LMM), they showed growth similar to that of a wild-type control strain (MR-1c) (Table 1) (Fig. S2), suggesting that the loss of DgcS does not affect the growth of MR-1 in batch cultures. These strains were cultivated in LMM until the stationary growth phase and analyzed for intracellular c-di-GMP contents using liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Fig. 2). Analyses revealed that the c-di-GMP content in the ΔdgcS mutant was approximately 35% of that in MR-1c and that this decrease was restored in the ΔdgcS-C strain. These results demonstrated that among multiple GGDEF domain-containing proteins encoded in the genome, DgcS served as a major DGC that affected global c-di-GMP levels in MR-1 cells.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli | ||
| WM6026 | Donor strain for conjugation; lacIq rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 (rhaBAD)568 rph-1 att-lambda::pAE12 Δ(oriR6K-cat::Frt5) Δ(endA)::Frt uidA(ΔMluI)::pir(wt) attHK::pJK1006-Δ1/2 (ΔoriR6K-cat::Frt5 ΔtrfA::Frt) | William Metcalf, University of Illinois |
| BL21(DE3) | F– ompT hsdSB(rB– mB–) gal(λcI 857 ind1 sam7 nin5 lacUV5-T7 gene 1) dcm(DE3) | Novagen |
| Shewanella oneidensis | ||
| MR-1 | Wild type | ATCC |
| MR-1c | MR-1 harboring pMElacZ-AFP and pBBR1MCS-2; Gmr Kmr | This study |
| ΔdgcS mutant | In-frame SO_1646 deletion mutant of MR-1c; Gmr Kmr | This study |
| ΔdgcS-C mutant | ΔdgcS mutant harboring pBBR-dgcS instead of pBBR1MCS-2; Gmr Kmr | This study |
| Plasmids | ||
| pMElacZ-AFP | Broad-host-range plasmid expressing AFP | 16 |
| pSMV-10 | 9.1-kb mobilizable suicide vector; oriR6 K mobRP4 sacB Kmr Gmr | 42 |
| pSMV-dgcS | 1.6-kb fusion PCR fragment containing ΔSO_1646 cloned into the SpeI site of pSMV-10 | This study |
| pBBR1MCS-2 | Broad-host-range vector, lacZ promoter; Kmr | 45 |
| pBBR-dgcS | pBBR1MCS-2-based plasmid expressing dgcS | This study |
| pET-28a(+) | Expression vector; Kmr | Novagen |
| pET-C-ht-dgcS | pET-28a(+)-based plasmid expressing C-ht-dgcS | This study |
| pUC18 | Cloning vector; Apr | TaKaRa |
FIG 2.
Intracellular c-di-GMP contents in S. oneidensis derivatives. Cells were grown under aerobic conditions and harvested at the stationary growth phase. Error bars represent standard deviations calculated from the results of three independent experiments. Asterisks indicate a statistically significant difference (P < 0.05; one-way ANOVA followed by LSD test; ns, not significant).
Involvement of DgcS in biofilm formation on electrodes.
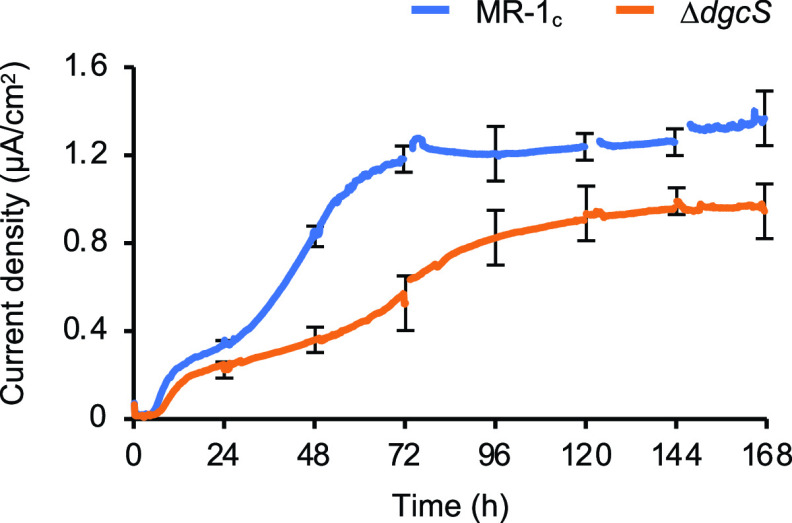
To examine the involvement of DgcS in EABF formation and current generation in BESs, EFCs were inoculated with MR-1c or the ΔdgcS mutant and operated under a medium flow condition at a working electrode (WE) potential of +0.4 V (versus the standard hydrogen electrode [SHE]) until the current generation by both strains became stable (for 168 h). Measurement of currents (Fig. 3) revealed that the initial increase in current generation by the ΔdgcS mutant was slower than that for MR-1c and that the maximum current density achieved by the ΔdgcS mutant (0.97 μA/cm2) was approximately 71% of that for MR-1c (1.36 μA/cm2). These results demonstrate that the loss of DgcS results in decreased current generation in EFCs.
FIG 3.
Time courses of current generation by MR-1c and the ΔdgcS mutant in EFC. Results and error bars represent the means and standard deviations, respectively, calculated from the results of three independent experiments.
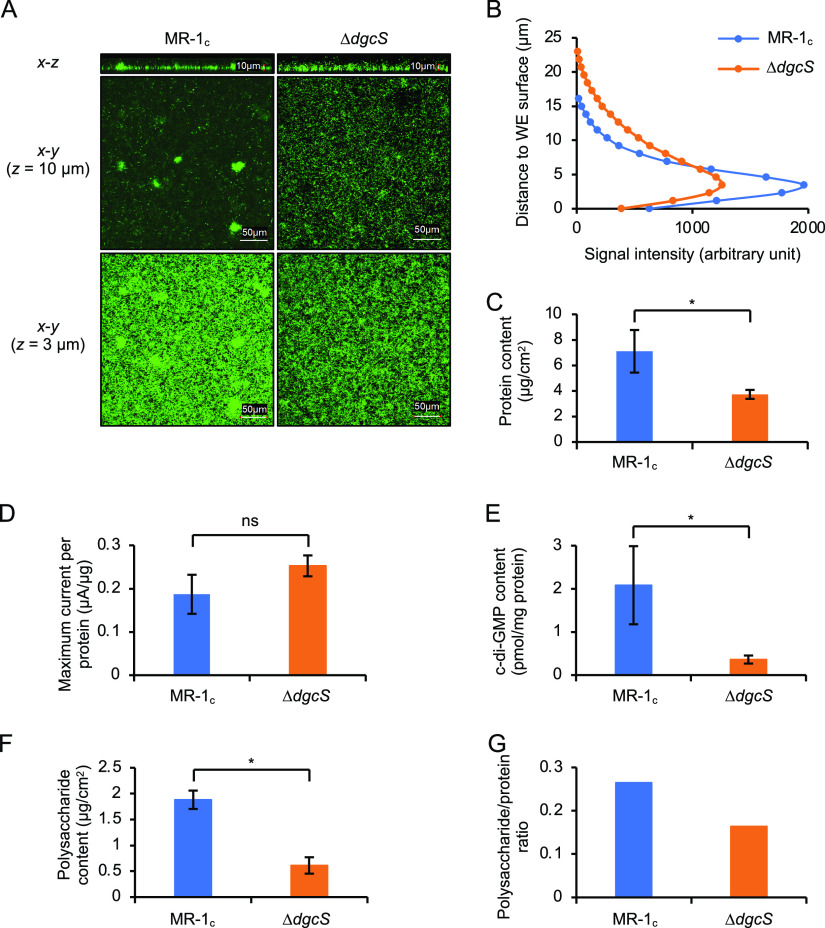
Figure 4A shows typical confocal laser scanning microscopy (CLSM) images for WE-associated and planktonic cells in EFCs when the growth of WE-associated cells reached a steady state (168 h after the start of operation). As observed in previous studies, MR-1c formed a thin, flat biofilm on the WE surface (Fig. 4A). However, the ΔdgcS mutant appeared to form a thicker and sparser biofilm than MR-1c, as seen in the side (x-z) view of Fig. 4A. In the ΔdgcS mutant-inoculated EFC, the number of cells in the lower zone (at 3 μm from the WE surface; z = 3 μm) was smaller than that in the MR-1c-inoculated EFC, while the number of cells in the upper zone (z = 10 μm) was larger (Fig. 4A). This trend was confirmed by analyzing the vertical distribution of fluorescent intensities in the CLSM images (Fig. 4B). To quantify the biomass attached to WE, WE-associated MR-1c and ΔdgcS cells were collected from EFCs operated for 168 h, and their protein contents were determined (Fig. 4C). The analysis revealed that the protein content in ΔdgcS biofilm was significantly lower than that in MR-1c biofilm. These results demonstrate that the ΔdgcS mutant exhibits an impaired ability to form biofilms in EFCs. When the maximum current densities achieved by the ΔdgcS mutant and MR-1c were normalized by their protein content in WE-associated biofilms, the normalized values were not significantly different between these strains (Fig. 4D), suggesting that the loss of DgcS does not affect current generation per cell. We therefore concluded that the decreased current generation observed for the ΔdgcS mutant (Fig. 3) was attributable to the impaired ability of this mutant to physically associate with WE. We also determined the intracellular c-di-GMP contents in WE-associated MR-1c and ΔdgcS cells. The results confirmed that the ΔdgcS mutant contained a lower level of c-di-GMP than did MR-1c (Fig. 4E), although, for unknown reasons, the c-di-GMP contents in the WE-associated cells were significantly lower than those in aerobically grown cells (Fig. 2). Taken together, these findings indicate that DgcS facilitated biofilm formation on electrodes by activating c-di-GMP-dependent signal transduction.
FIG 4.
Characterization of the ΔdgcS mutant in EFC. (A) Representative CLSM images of MR-1c and the ΔdgcS mutant grown in EFC. Vertical (x-z) and horizontal (x-y) images were obtained from EFCs operated for 168 h. Horizontal (x-y) images were obtained at heights of 3 μm and 10 μm from the WE surface (z = 3 μm and 10 μm, respectively). (B) Vertical distribution of AFP signal intensities in CLSM images obtained from EFCs operated for 168 h. (C to G) Protein contents (C), maximum current density normalized to protein contents (D), intracellular c-di-GMP contents (E), polysaccharide contents (F), and ratios of polysaccharide contents to protein contents (G) in WE-associated MR-1c and ΔdgcS biofilms. Cells were grown in EFCs for 168 h. In all graphs, asterisks indicate a statistically significant difference (P < 0.05; Student's t test; ns, not significant). Error bars represent standard deviations calculated from the results of three independent experiments.
It is well known that c-di-GMP-dependent signal transduction cascades regulate the synthesis of extracellular polysaccharides, thereby modulating the ability of bacterial cells to associate with solid surfaces (29). We therefore determined the extracellular polysaccharide content of WE-associated biofilms formed by the ΔdgcS mutant and MR-1c to examine how DgcS facilitates biofilm formation on electrodes. As shown in Fig. 4F, the ΔdgcS biofilm contained a lower level of polysaccharides than the MR-1c biofilm. Notably, when polysaccharide contents were normalized by protein contents in WE-associated biofilms, the polysaccharide/protein ratio was lower in the ΔdgcS mutant than in MR-1c (Fig. 4G), indicating that the ΔdgcS mutant produced less extracellular polysaccharides per cell than MR-1c. These results indicate that c-di-GMP production catalyzed by DgcS activated the synthesis of extracellular polysaccharides, thereby facilitating the association of MR-1 cells with electrodes.
DISCUSSION
Here, we found that MR-1 activates biofilm formation on electrodes via the DgcS-dependent c-di-GMP signaling cascades. Our results indicate that DgcS catalyzes the production of c-di-GMP, which induces the synthesis of extracellular polysaccharides and facilitates the association of MR-1 cells with electrode surfaces (Fig. 1 and 4). Notably, a recent study reported that an elevated c-di-GMP level in MR-1 cells by the overexpression of a heterologous DGC gene resulted in increased expression of c-type cytochrome genes involved in extracellular electron transfer (EET) (30). In the present study, however, we observed that current generation normalized by the biomass of WE-associated cells was not significantly different between the wild type and the ΔdgcS mutant (Fig. 4D), suggesting that the loss of DgcS does not affect the EET activity of MR-1 cells. We consider that the intracellular level of c-di-GMP in ΔdgcS is sufficient for the expression of EET-related genes, while it is lower than the threshold level at which the biosynthesis of extracellular polysaccharides is promoted.
In Pseudomonas aeruginosa, c-di-GMP regulates biofilm formation by binding to a σ54-dependent transcriptional regulator, FleQ, that activates the expression of biofilm-related genes, including those involved in the biosynthesis of extracellular polysaccharides (31, 32). A homolog of FleQ, referred to as FlrA in MR-1, is found in many members of the genus Shewanella, suggesting that similar mechanisms are also present in members of this genus. In support of this idea, a recent study has reported that a spontaneous MR-1 mutant in which the flrA gene is disrupted by an insertion sequence exhibited impaired biofilm formation on electrodes (33). Previous studies have also shown that FlrA in Shewanella regulates the expression of genes for the synthesis of flagella (34) and an adhesin protein, BpfA (35). It is therefore possible that these biofilm-related factors other than extracellular polysaccharides are also involved in EABF formation by MR-1. It is also known that c-di-GMP directly binds to polysaccharide biosynthetic enzymes, such as the glycosyltransferase BcsA, thereby regulating biofilm formation at the posttranslational level (36). Furthermore, previous studies have suggested that the MxdA protein containing a degenerate GGDEF (NVDEF) motif functions as a c-di-GMP receptor and interacts with the MxdB glycosyltransferase, thereby activating the biosynthesis of extracellular polysaccharides in MR-1 (37, 38). These studies provide information for predicting downstream processes of a DgcS-dependent c-di-GMP signaling cascade, and further studies will be conducted to investigate the involvement of these biofilm-related factors in EABF formation by MR-1.
The genome of MR-1 encodes 51 GGDEF domain-containing proteins, of which only a few have been functionally characterized. For instance, a recent study has reported that the GGDEF-containing proteins PdgA and PdgB play essential roles in the formation of pellicles, floating biofilms formed at the air-liquid interface (38). Although enzymatic activities of these proteins have not been examined in vitro, evidence obtained from in vivo experiments has suggested that they serve as DGCs (38). It is noteworthy that the study also suggests that PdgA and PdgB specifically function for pellicle biogenesis but not for biofilm formation on solid surfaces (38). These observations, together with the findings in the present study, suggest that MR-1 utilizes multiple DGCs to specifically regulate the formation of solid surface-associated biofilms and pellicles according to environmental conditions. In relation to this idea, recent studies have provided evidence suggesting that local signaling based on direct interaction between DGCs and c-di-GMP receptor proteins regulates specific cellular processes (39, 40). In contrast to these observations, the present study demonstrates that the loss of DgcS results in a significant decrease in the overall intracellular level of c-di-GMP in MR-1 grown in EFCs (Fig. 4E). We therefore suggest that DgcS globally activates c-di-GMP-dependent signal transduction pathways that are involved in biofilm formation on electrodes. It is also notable that the decrease in the c-di-GMP content was also observed in ΔdgcS cells grown under batch culture conditions, where most cells were planktonic (Fig. 2). One possible explanation for this result is that DgcS is activated in response to hydrodynamic signals, such as changes in shear force (41), given that cells are exposed to fluid (medium) flows in both EFCs and shaking cultures. However, considering that medium flow can cause changes in many environmental factors, such as shifts in local nutrient concentrations, identification of an environmental factor(s) that functionalizes DgcS requires additional investigation.
We previously reported that MR-1 uses the extracytoplasmic function (ECF) sigma factor encoded by SO_3096 to activate the expression of many genes, including the dsbD genes involved in cytochrome c maturation, under medium flow conditions (16). It is not likely, however, that this ECF sigma factor is involved in the transcriptional regulation of dgcS, because the disruption of SO_3096 did not significantly alter the transcription of dgcS (16). We therefore consider that there may exist an unknown transcription regulator(s) that activates the expression of this gene. We expect that the development of a comprehensive understanding of the DgcS-mediated signaling cascades will provide vital information in regard to EABF formation and current generation by S. oneidensis MR-1.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. oneidensis strains were cultivated at 30°C in lysogeny broth (LB) medium or lactate minimal medium (LMM) containing 15 mM dl-lactate as the sole carbon and energy source (15). For aerobic cultivation, LB medium or LMM in test tubes was inoculated with an S. oneidensis strain and incubated by reciprocal shaking at 180 rpm. The optical density at 600 nm (OD600) was measured using a mini photo 518R photometer (Taitec, Tokyo, Japan). E. coli strains were cultivated in LB medium at 37°C. The E. coli mating strain (WM6026) required 100 μg/ml 2,6-diaminopimelic acid for growth. When necessary, 50 μg/ml kanamycin (Km), 15 μg/ml gentamicin (Gm), or 100 μg/ml ampicillin (Ap) was added to culture media. Agar plates contained 1.6% Bacto agar (Difco, Franklin Lakes, NJ).
Operation of EFCs and CLSM of biofilms.
The setup and operation of EFCs were performed according to methods described previously (16). Briefly, a three-electrode EFC comprising a graphite plate WE (68 by 140 mm; IG-70; Toyo Tanso, Osaka, Japan), an Ag/AgCl reference electrode (HX-R5; Hokuto Denko, Tokyo, Japan), and a titanium mesh counter electrode (25 by 25 mm, 100 mesh, 8 pieces; Nilaco, Tokyo, Japan) was filled with LMM and inoculated with an S. oneidensis strain. The electrodes were connected to a potentiostat (VMP3; Bio-Logic Science Instruments, Seyssinet-Pariset, France), and current was measured at a WE potential of +0.4 V (versus SHE). For EFC operation under medium flow conditions, LMM was continuously supplied from a reservoir tank to the EFC using a peristaltic pump at a constant linear velocity of 5.5 mm min−1 (a flow rate of 1.38 ml min−1). The medium in the reservoir tank was constantly purged with high-purity nitrogen gas to maintain anaerobic conditions. During operation, the EFC was put in an incubator at 30°C. CLSM was performed using an FV1200 (BX61WI configuration) microscope system (Olympus, Tokyo, Japan) according to a method described previously (16). Biofilms expressing AFP were observed at an excitation wavelength of 473 nm.
RNA extraction and quantitative RT-PCR.
RNA extraction from WE-attached MR-1 cells and subsequent quantitative RT-PCR were performed according to methods described previously (16). Briefly, EFCs inoculated with MR-1 cells were operated for 72 h under the medium flow condition or for 48 h under the nonflow condition until current generation became stable along with the growth of WE-associated cells. Total RNA was extracted from the WE-attached cells using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA) and was subsequently purified using an RNeasy minikit and an RNase-free DNase set (Qiagen, Valencia, CA). The quantity and quality of the purified RNA were checked using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific) and an Agilent 2100 bioanalyzer with RNA 6000 pico reagents and RNA pico chips (Agilent Technologies, Santa Clara, CA). For quantitative RT-PCR, cDNA was synthesized from total RNA using Superscript III reverse transcriptase (Thermo Fisher Scientific) and random primers (Thermo Fisher Scientific). Quantitative PCR was performed using the Applied Biosystems StepOnePlus real-time PCR system (Thermo Fisher Scientific), Power SYBR green PCR master mix (Thermo Fisher Scientific), and the specific primers listed in Table S1 in the supplemental material. The specificity of the quantitative PCR was verified by dissociation curve analysis. Expression levels of target genes were normalized based on expression levels of the 16S rRNA gene.
Construction of plasmids and mutants.
The in-frame disruption of the dgcS gene in MR-1 was performed using a two-step homologous recombination method with the suicide plasmid pSMV-10 (42) (Table 1) as described previously (43). Briefly, a 1.6-kb fusion product, consisting of upstream and downstream sequences of the dgcS (SO_1646) gene joined by an 18-bp linker sequence, was constructed by overlap extension PCR (44) using primers listed in Table S1 in the supplemental material and Phusion high-fidelity DNA polymerase (New England Biolabs, Beverly, MA). The amplified fusion product was ligated into the SpeI site of pSMV-10, and the resultant plasmid, pSMV-dgcS (Table 1), was introduced into MR-1 by filter mating with E. coli WM6026. Double-crossover mutants in which the dgcS gene was disrupted in-frame were isolated on LB plates containing 10% (wt/vol) sucrose, and one representative strain was selected and transformed with the AFP-expressing plasmid pMElacZ-AFP (15) and the control vector pBBR1MCS-2 (45) (Table 1) by electroporation (46) and filter mating, respectively. The resultant mutant strain was designated the ΔdgcS mutant (Table 1). The control MR-1 strain transformed with pMElacZ-AFP and pBBR1MCS-2 was designated MR-1c (Table 1).
To construct the dgcS-expressing plasmid pBBR-dgcS (Table 1), the DNA fragment consisting of the dgcS gene and its 500-bp upstream region was PCR amplified using primers listed in Table S1. The amplified product was subcloned into the SmaI site of pUC18 (TaKaRa, Tokyo, Japan), and sequencing confirmed that no error occurred in the PCR amplification. The insert was digested with NsiI and KpnI and cloned into the corresponding sites of pBBRMCS-2 (45) (Table 1) to yield pBBR-dgcS. The ΔdgcS derivative transformed with pBBR-dgcS instead of pBBR1MCS-2 was designated the ΔdgcS-C strain (Table 1).
Purification of DgcS.
To construct a plasmid expressing C-ht-DgcS, the dgcS gene was amplified from the total DNA of MR-1 using primers listed in Table S1. The amplified product was digested with NcoI and HindIII and cloned between the corresponding sites of the histidine tag expression vector pET-28a(+) (Novagen, Madison, WI). The resultant plasmid, pET-C-ht-dgcS (Table 1), was introduced into E. coli BL21(DE3). Cells carrying the plasmid were grown in 300-ml baffled Erlenmeyer flasks containing 100 ml 2× YT medium supplemented with Km at 37°C. Isopropyl-1-thio-β-d-galactopyranoside (IPTG) was added to a final concentration of 0.1 mM when the OD600 reached 0.5. After further cultivation for 16 h, the cells were harvested by centrifugation and suspended in 20 mM sodium phosphate buffer (pH 7.4) containing 0.5 M NaCl and 20 mM imidazole. The cell suspension was subjected to ultrasonication with an S4000 sonicator (Misonix, Farmingdale, NY) and centrifuged at 1,000 × g for 10 min to remove cell debris. C-ht-DgcS contained in the supernatant was purified using a His GraviTrap column (Cytiva, Marlborough, MA) according to the manufacturer’s instructions. The protein concentration of the extracted solution was determined using a Micro BCA (bicinchoninic acid) protein assay kit (Thermo Fisher Scientific). The purity of the protein samples was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining according to the standard procedure.
In vitro DGC activity assay.
For the DGC activity assay, 50 μg of purified C-ht-DgcS was mixed with 200 nmol GTP in 600 μl reaction buffer (50 mM Tris-HCl, 300 mM NaCl, and 20 mM MgCl2; pH 8.0). The reaction mixture was incubated at 30°C for 15 min, and the reaction was stopped by heating at 95°C for 10 min. The consumption of GTP and the production of c-di-GMP were analyzed using an HPLC system (1100 series; Agilent Technologies) equipped with a Zorbax SB-Aq column (4.6 mm by 150 mm, 5 μm; Agilent Technologies) as described previously (47). Briefly, the reaction mixture was diluted with an HPLC mobile phase consisting of 20 mM triethyl ammonium bicarbonate buffer and 9% (vol/vol) methanol and was neutralized to pH 7.0 using acetic acid. The sample solution was filtered using a membrane filter unit (0.20-μm pore size, DISMIC-25HP; Advantec, Tokyo, Japan) and injected into the HPLC column. Isocratic elution of GTP and c-di-GMP was achieved by chromatography using the above-mentioned mobile phase at an elution rate of 0.2 ml/min. Quantitative analysis was performed using a UV detector (Agilent Technologies) set at 243 nm and commercially available c-di-GMP (Sigma-Aldrich, St. Louis, MO) as a standard.
Extraction and quantification of intracellular c-di-GMP.
Intracellular c-di-GMP in S. oneidensis strains was extracted and quantified according to methods described previously (48) with modifications. S. oneidensis cells were grown under aerobic conditions in test tubes containing LMM until the stationary growth phase or under anaerobic medium flow conditions in EFCs until the growth of WE-associated cells reached a steady state. The aerobically grown or WE-associated cells were suspended in 300 μl of an ice-cold extraction solvent consisting of acetonitrile-methanol-water (40:40:20 [vol/vol/vol]) containing 50 pmol cyclic 3′,5′-xanthosine monophosphate (cXMP) as an internal standard. The cell suspension was incubated at 4°C for 15 min and then heated to 95°C for 10 min. After cooling, the cell suspension was centrifuged at 20,000 × g for 5 min to separate the insoluble (pellet) and soluble (supernatant) fractions. The extraction procedure from the insoluble fraction was repeated twice, omitting the heat treatment, using 200 μl of the extraction solvent without cXMP. The solvent of the combined supernatants (700 μl) was evaporated under vacuum using a centrifugal concentrator (VC-15S; Taitec).
Analyses of extracts were conducted by liquid chromatography-electrospray ionization‐tandem mass spectrometry in positive ionization mode [LC-ESI(+)‐MS/MS] by using a Waters 2690 separation module (Waters Corp., Milford, MA) in‐line with a Waters 2998 photodiode array detector that was interfaced with a Quattro Ultima triple-stage quadrupole mass spectrometer (Waters‐Micromass, Manchester, UK). Sample aliquots of 20 μl were analyzed on a Cosmosil 5C18-AR-II column (4.6 mm inside diameter by 250 mm; Shimadzu, Kyoto, Japan) that was in-line with a Security Guard cartridge system precolumn fitted with a wide-pore C18 cartridge (Phenomenex, Torrance, CA, USA). Compounds were eluted at a flow rate of 0.3 ml min−1 via a gradient that began at a ratio of 97% water to 3% methanol, which was changed to a ratio of 25% water to 75% methanol over a period of 30 min. Conditions were returned to a ratio of 97% water to 3% methanol and held through 60 min. Sample components were delivered to the mass spectrometer by electrospray, whereby nitrogen gas was used as the nebulizing gas. The ion source temperature was 130°C, the desolvation temperature was 360°C, and the cone voltage was operated at 35 V. Nitrogen gas was also used as the desolvation gas, at 600 liters/h, and the cone gas, at 60 liters/h. Argon gas (99.9999%) was used as the collision cell gas. MS analyses were conducted in single reaction monitoring (SRM) mode by monitoring c-di-GMP and cXMP at the m/z transitions 691 > 152 and 347 > 153, respectively. The collision cell energies utilized for these transitions were 61 eV and 29 eV, respectively. The c-di-GMP content of cells was normalized based on their total protein content determined using a Micro BCA protein assay kit.
Quantification of extracellular polysaccharides and proteins in biofilms.
WE-associated biofilms were recovered from EFCs operated for 168 h. Polysaccharides and proteins in the biofilms were determined using a phenol-sulfuric acid method (49) and a Micro BCA protein assay kit, respectively, as described previously (15).
Statistical and in silico analysis.
Data were statistically evaluated using Student's t test or one-way analysis of variance (ANOVA), followed by Fisher’s least significant difference (LSD) test using js-STAR 2012 (http://www.kisnet.or.jp/nappa/software/star/). The domain structure of DgcS was predicted using the SMART program (27).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by JSPS KAKENHI grant numbers 15H01753 (to K.W.) and 18K05399 (to A.K.).
A.M. carried out the majority of the experimental work and drafted the manuscript. R.K. supported EFC operation and CLSM observation. R.A.K. performed LC-MS/MS analyses and manuscript editing. A.K. designed the study and drafted the manuscript. K.W. supervised the study and performed manuscript editing. All authors read and approved the final manuscript.
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Rabaey K, Angenent L, Schroder U, Keller J (ed). 2009. Bioelectrochemical systems: from extracellular electron transfer to biotechnological application. International Water Association, London, United Kingdom. [Google Scholar]
- 2.Watanabe K. 2008. Recent developments in microbial fuel cell technologies for sustainable bioenergy. J Biosci Bioeng 106:528–536. 10.1263/jbb.106.528. [DOI] [PubMed] [Google Scholar]
- 3.Hirose A, Kouzuma A, Watanabe K. 2019. Towards development of electrogenetics using electrochemically active bacteria. Biotechnol Adv 37:107351. 10.1016/j.biotechadv.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Rabaey K, Rozendal RA. 2010. Microbial electrosynthesis—revisiting the electrical route for microbial production. Nat Rev Microbiol 8:706–716. 10.1038/nrmicro2422. [DOI] [PubMed] [Google Scholar]
- 5.Borole AP, Reguera G, Ringeisen B, Wang Z-W, Feng Y, Kim BH. 2011. Electroactive biofilms: current status and future research needs. Energy Environ Sci 4:4813–4813. 10.1039/c1ee02511b. [DOI] [Google Scholar]
- 6.Rollefson JB, Stephen CS, Tien M, Bond DR. 2011. Identification of an extracellular polysaccharide network essential for cytochrome anchoring and biofilm formation in Geobacter sulfurreducens. J Bacteriol 193:1023–1033. 10.1128/JB.01092-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue K, Leang C, Franks AE, Woodard TL, Nevin KP, Lovley DR. 2011. Specific localization of the c-type cytochrome OmcZ at the anode surface in current-producing biofilms of Geobacter sulfurreducens. Environ Microbiol Rep 3:211–217. 10.1111/j.1758-2229.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 8.Choi S, Kim B, Chang IS. 2018. Tracking of Shewanella oneidensis MR-1 biofilm formation of a microbial electrochemical system via differential pulse voltammetry. Bioresour Technol 254:357–361. 10.1016/j.biortech.2018.01.047. [DOI] [PubMed] [Google Scholar]
- 9.Miyahara M, Hashimoto K, Watanabe K. 2013. Use of cassette-electrode microbial fuel cell for wastewater treatment. J Biosci Bioeng 115:176–181. 10.1016/j.jbiosc.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Marsili E, Rollefson JB, Baron DB, Hozalski RM, Bond DR. 2008. Microbial biofilm voltammetry: direct electrochemical characterization of catalytic electrode-attached biofilms. Appl Environ Microbiol 74:7329–7337. 10.1128/AEM.00177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stockl M, Teubner NC, Holtmann D, Mangold KM, Sand W. 2019. Extracellular polymeric substances from Geobacter sulfurreducens biofilms in microbial fuel cells. ACS Appl Mater Interfaces 11:8961–8968. 10.1021/acsami.8b14340. [DOI] [PubMed] [Google Scholar]
- 12.Yang G, Huang L, Yu Z, Liu X, Chen S, Zeng J, Zhou S, Zhuang L. 2019. Anode potentials regulate Geobacter biofilms: new insights from the composition and spatial structure of extracellular polymeric substances. Water Res 159:294–301. 10.1016/j.watres.2019.05.027. [DOI] [PubMed] [Google Scholar]
- 13.Kouzuma A, Meng XY, Kimura N, Hashimoto K, Watanabe K. 2010. Disruption of the putative cell surface polysaccharide biosynthesis gene SO3177 in Shewanella oneidensis MR-1 enhances adhesion to electrodes and current generation in microbial fuel cells. Appl Environ Microbiol 76:4151–4157. 10.1128/AEM.00117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto D, Coradin T, Laberty-Robert C. 2018. Effect of anode polarization on biofilm formation and electron transfer in Shewanella oneidensis/graphite felt microbial fuel cells. Bioelectrochemistry 120:1–9. 10.1016/j.bioelechem.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Kitayama M, Koga R, Kasai T, Kouzuma A, Watanabe K. 2017. Structures, compositions, and activities of live Shewanella biofilms formed on graphite electrodes in electrochemical flow cells. Appl Environ Microbiol 83:166–172. 10.1128/AEM.00903-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koga R, Matsumoto A, Kouzuma A, Watanabe K. 2020. Identification of an extracytoplasmic function sigma factor that facilitates c-type cytochrome maturation and current generation under electrolyte-flow conditions in Shewanella oneidensis MR-1. Environ Microbiol 22:3671–3684. 10.1111/1462-2920.15131. [DOI] [PubMed] [Google Scholar]
- 17.Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15:271–284. 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 18.Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40:385–407. 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 19.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bharati BK, Mukherjee R, Chatterji D. 2018. Substrate-induced domain movement in a bifunctional protein, DcpA, regulates cyclic di-GMP turnover: functional implications of a highly conserved motif. J Biol Chem 293:14065–14079. 10.1074/jbc.RA118.003917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang F, Wang Y, Cen C, Fu L, Wang Y. 2020. A tandem GGDEF-EAL domain protein-regulated c-di-GMP signal contributes to spoilage-related activities of Shewanella baltica OS155. Appl Microbiol Biotechnol 104:2205–2216. 10.1007/s00253-020-10357-w. [DOI] [PubMed] [Google Scholar]
- 22.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, Rodionov DA, Rodrigues JL, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM. 2008. Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6:592–603. 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 23.Liu T, Yu YY, Deng XP, Ng CK, Cao B, Wang JY, Rice SA, Kjelleberg S, Song H. 2015. Enhanced Shewanella biofilm promotes bioelectricity generation. Biotechnol Bioeng 112:2051–2059. 10.1002/bit.25624. [DOI] [PubMed] [Google Scholar]
- 24.Chao L, Rakshe S, Leff M, Spormann AM. 2013. PdeB, a cyclic Di-GMP-specific phosphodiesterase that regulates Shewanella oneidensis MR-1 motility and biofilm formation. J Bacteriol 195:3827–3833. 10.1128/JB.00498-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi M, Gao T, Ju L, Yao Y, Gao H. 2014. Effects of FlrBC on flagellar biosynthesis of Shewanella oneidensis. Mol Microbiol 93:1269–1283. 10.1111/mmi.12731. [DOI] [PubMed] [Google Scholar]
- 26.Hengge R, Grundling A, Jenal U, Ryan R, Yildiz F. 2016. Bacterial signal transduction by cyclic di-GMP and other nucleotide second messengers. J Bacteriol 198:15–26. 10.1128/JB.00331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496. 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drepper T, Eggert T, Circolone F, Heck A, Krauss U, Guterl JK, Wendorff M, Losi A, Gartner W, Jaeger KE. 2007. Reporter proteins for in vivo fluorescence without oxygen. Nat Biotechnol 25:443–445. 10.1038/nbt1293. [DOI] [PubMed] [Google Scholar]
- 29.Maunders E, Welch M. 2017. Matrix exopolysaccharides; the sticky side of biofilm formation. FEMS Microbiol Lett 364:fnx120. 10.1093/femsle/fnx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng CK, Xu J, Cai Z, Yang L, Thompson IP, Huang WE, Cao B. 2020. Elevated intracellular cyclic-di-GMP level in Shewanella oneidensis increases expression of c-type cytochromes. Microb Biotechnol 13:1904–1916. 10.1111/1751-7915.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuyama BY, Krasteva PV, Baraquet C, Harwood CS, Sondermann H, Navarro MV. 2016. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 113:E209–E218. 10.1073/pnas.1523148113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng L, Min D, Liu DF, Zhu TT, Wang KL, Yu HQ. 2020. Deteriorated biofilm-forming capacity and electroactivity of Shewanella oneidnsis MR-1 induced by insertion sequence (IS) elements. Biosens Bioelectron 156:112136. 10.1016/j.bios.2020.112136. [DOI] [PubMed] [Google Scholar]
- 34.Gao T, Shi M, Gao H. 2018. Partially reciprocal replacement of FlrA and FlrC in regulation of Shewanella oneidensis flagellar biosynthesis. J Bacteriol 200:e00796-17. 10.1128/JB.00796-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng YY, Wu C, Wu JY, Jia HL, Wang MY, Wang HY, Zou SM, Sun RR, Jia R, Xiao YZ. 2017. FlrA represses transcription of the biofilm-associated bpfA operon in Shewanella putrefaciens. Appl Environ Microbiol 83:e02410-16. 10.1128/AEM.02410-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan JL, McNamara JT, Zimmer J. 2014. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21:489–496. 10.1038/nsmb.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rakshe S, Leff M, Spormann AM. 2011. Indirect modulation of the intracellular c-Di-GMP level in Shewanella oneidensis MR-1 by MxdA. Appl Environ Microbiol 77:2196–2198. 10.1128/AEM.01985-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gambari C, Boyeldieu A, Armitano J, Méjean V, Jourlin-Castelli C. 2019. Control of pellicle biogenesis involves the diguanylate cyclases PdgA and PdgB, the c-di-GMP binding protein MxdA and the chemotaxis response regulator CheY3 in Shewanella oneidensis. Environ Microbiol 21:81–97. 10.1111/1462-2920.14424. [DOI] [PubMed] [Google Scholar]
- 39.Dahlstrom KM, Giglio KM, Collins AJ, Sondermann H, O'Toole GA. 2015. Contribution of physical interactions to signaling specificity between a diguanylate cyclase and its effector. mBio 6:e01978-15. 10.1128/mBio.01978-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarenko O, Klauck G, Wilke FM, Pfiffer V, Richter AM, Herbst S, Kaever V, Hengge R. 2017. More than enzymes that make or break cyclic Di-GMP-local signaling in the interactome of GGDEF/EAL domain proteins of Escherichia coli. mBio 8:e01639-17. 10.1128/mBio.01639-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanfilippo JE, Lorestani A, Koch MD, Bratton BP, Siryaporn A, Stone HA, Gitai Z. 2019. Microfluidic-based transcriptomics reveal force-independent bacterial rheosensing. Nat Microbiol 4:1274–1281. 10.1038/s41564-019-0455-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saltikov CW, Newman DK. 2003. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci U S A 100:10983–10988. 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirose A, Kasai T, Aoki M, Umemura T, Watanabe K, Kouzuma A. 2018. Electrochemically active bacteria sense electrode potentials for regulating catabolic pathways. Nat Commun 9:1083. 10.1038/s41467-018-03416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki Y, Kouzuma A, Watanabe K. 2020. CRISPR/Cas9-mediated genome editing of Shewanella oneidensis MR-1 using a broad host-range pBBR1-based plasmid. J Gen Appl Microbiol 66:41–45. 10.2323/jgam.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 45.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 46.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 47.Sauer K (ed). 2017. C-di-GMP signaling: methods and protocols. Humana Press, Totowa, NJ. [Google Scholar]
- 48.Spangler C, Bohm A, Jenal U, Seifert R, Kaever V. 2010. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J Microbiol Methods 81:226–231. 10.1016/j.mimet.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 49.Dubois M, Gilles KA, Hamilton JK, Rebers P, Smith F. 1956. Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356. 10.1021/ac60111a017. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.