Lp is a major cause of nosocomial and community-acquired pneumonia. Lp is found in water systems, including hot water distribution systems. Heat treatment is a method of disinfection often used to limit the presence of Lp in such systems; however, the benefit is usually short term, as Lp is able to quickly recolonize these systems.
KEYWORDS: Legionella pneumophila, tail-specific protease, small regulatory RNA, thermal stress, cis-encoded sRNA, Legionnaires’ disease, amoeba, CpxR
ABSTRACT
Legionella pneumophila (Lp) is an inhabitant of natural and human-made water systems, where it replicates within amoebae and ciliates and survives within biofilms. When Lp-contaminated aerosols are breathed in, Lp can enter the lungs and may infect human alveolar macrophages, causing severe pneumonia known as Legionnaires’ disease. Lp is often found in hot water distribution systems (HWDS), which are linked to nosocomial outbreaks. Heat treatment is used to disinfect HWDS and reduce the concentration of Lp. However, Lp is often able to recolonize these water systems, indicating an efficient heat shock response. Tail-specific proteases (Tsp) are typically periplasmic proteases implicated in degrading aberrant proteins in the periplasm and important for surviving thermal stress. In Lp Philadelphia-1, Tsp is encoded by the lpg0499 gene. In this paper, we show that Tsp is important for surviving thermal stress in water and for optimal infection of amoeba when a shift in temperature occurs during intracellular growth. We also demonstrate that Tsp is expressed in the postexponential phase but repressed in the exponential phase and that the cis-encoded small regulatory RNA Lpr17 shows the opposite expression, suggesting that it represses translation of tsp. In addition, our results show that tsp is regulated by CpxR, a major regulator in Lp, in an Lpr17-independent manner. Deletion of CpxR also reduced the ability of Lp to survive heat shock. In conclusion, our study shows that Tsp is likely an important factor for the survival and growth of Lp in water systems.
IMPORTANCE Lp is a major cause of nosocomial and community-acquired pneumonia. Lp is found in water systems, including hot water distribution systems. Heat treatment is a method of disinfection often used to limit the presence of Lp in such systems; however, the benefit is usually short term, as Lp is able to quickly recolonize these systems. Presumably, Lp responds efficiently to thermal stress, but so far, not much is known about the genes involved. In this paper, we show that the Tsp and the two-component system CpxRA are required for resistance to thermal stress when Lp is free in water and when it is inside host cells. Our study identifies critical systems for the survival of Lp in its natural environment under thermal stress.
INTRODUCTION
Legionnaires’ disease (LD) is a severe form of pneumonia in humans caused by the Gram-negative bacterium Legionella pneumophila (Lp) (1). Lp is often responsible for nosocomial and community-acquired pneumonia (1). In the environment, Lp can be found in natural and human-made aquatic environments, such as cooling towers and water distribution systems, where it can replicate within phagocytic protozoa (2–4). Notably, Lp replicates within Vermamoeba vermiformis (formerly Hartmannella vermiformis), a thermotolerant amoeba commonly found in both natural and human-made water systems (5, 6). V. vermiformis protects Lp from predation, competition, and various disinfection methods such as heat treatments, potentially contributing to nosocomial infections (3, 5, 6). In addition, intracellular growth of Lp within amoeba increases its pathogenicity and facilitates the establishment of infection in humans (3). The main virulence factor of Lp is the type IVb secretion system called Icm/Dot, which translocates more than 300 effectors inside the host cells (7). These effectors are responsible for stopping the maturation of the phagosome and creating a specialized vacuole where Lp can grow (7).
The preferred growth temperature of Lp is between 25°C and 42°C, though it has been found in water systems at temperatures below 20°C or above 60°C (8). Its ability to live in a wide range of temperatures allows Lp to colonize hot water distribution systems (HWDS) (9). The higher temperature found within HWDS also reduces the microbial diversity, therefore making it easier for Lp to compete in these environments (10, 11). A method used to limit the proliferation of Lp in water systems is to maintain the temperature at the outlet above 55°C (8, 9). When a system is heavily colonized by Lp, it can be treated by superheat and flush, also referred to as pasteurization, consisting of increasing the temperature at the heater to about 75°C in order to provide a temperature of at least 65°C at the outlets (12).
An increase in temperature, even by a few degrees, can cause proteins to unfold, which leads to protein aggregates that can be lethal, as these aggregates accumulate in the cells (13, 14). Protein misfolding and aggregation result in an imbalance of protein homeostasis, which triggers the heat shock response (14). The canonical heat shock response is composed of two heat shock regulons, the σH and σE heat shock regulons (14). This response results in the production of chaperones and proteases to refold or destroy misfolded proteins and aggregates (14). In Lp, the two-component system LetA/S is the only regulatory system known to be important for surviving thermal stress (15).
Protein degradation by proteases is an important cellular function, as it allows the removal of aberrant proteins, the regulation of intracellular protein concentration; produces active molecules from precursors such as the alpha-toxin from Staphylococcus aureus or the heat-stable toxin produced by enterotoxigenic Escherichia coli (ETEC); and allows the cell to recycle amino acids during starvation (16, 17). Carboxyl-terminal proteases (CTPs) are serine proteases conserved in most Gram-negative bacteria (18–20). They can also be found in Archaea, Gram-positive bacteria, eukaryotes, viruses, as well as in organelles such as chloroplasts (20–22). CTPs are mostly located in the periplasm, though some are located in the cytoplasm, while others are secreted in the extracellular environment (19). CTPs are involved in several different processes in Gram-negative bacteria. E. coli’s CTP, called tail-specific protease (Tsp) and sometimes Prc, was the first bacterial CTP characterized and is involved in regulating peptidoglycan assembly (19). The assembly of the peptidoglycan layer plays a role in the bacteria’s size and shape (23). This important process is dependent on penicillin-binding proteins (PBP) (20, 24). Tsp recognizes a sequence of amino acids located on the C-terminal end of the precursor of PBP-3, resulting in its activation (25, 26). As a result, the E. coli’s tsp mutant is sensitive to various antibiotics and thermal and osmotic stress, and it displays a filamentous morphology (25, 27). Overproduction of Tsp is detrimental to cell growth (25). The susceptibility to thermal stress is dependent on osmolarity. These phenotypes are attributed to increased permeability of the outer membrane in the tsp mutant (25, 28). The tsp mutant and the wild type (WT) expressed similar levels of GroEL and DnaK, two heat shock proteins, when exposed to thermal stress in high-osmolarity buffer (25). However, when the strains were in low-osmolarity buffer and exposed to thermal stress, the tsp mutant showed decreased expression of DnaK and almost no expression of GroEL (25). E. coli’s Tsp also targets MepS, a peptidoglycan hydrolase which cleaves peptide cross-links between the glycan chains of the peptidoglycan layer (29, 30). Tsp is also able to degrade misfolded proteins in the periplasm, suggesting a role in maintaining protein homeostasis in the periplasm (26, 31, 32).
Pseudomonas aeruginosa codes two CTPs, called Prc and CtpA (19, 33). Prc is a homologue of E. coli’s Tsp and has been shown to degrade a mutant form of the anti-sigma factor MucA, preventing development of mucoidy (33–35). CtpA is involved in the proper function of the type 3 secretion system (T3SS), required for cytotoxicity in cultured cells and virulence in the animal model of acute pneumonia (19, 33). The activity of CtpA is dependent on LcbA, an outer membrane lipoprotein with tetratricopeptide repeat (TPR) motifs (36). CtpA degrades four peptidoglycan hydrolases, including MepM and PA4404, belonging to the M23 peptidase family, and PA1198 and PA1199, belonging to the NlpC/P60 peptidase family (36).
The lpg0499 gene in Lp encodes a CTP named Tsp, most homologous to the P. aeruginosa CtpA (37). The tsp gene is upregulated 54-fold in the postexponential (PE) phase compared to the exponential (E) phase (38). The tsp gene was also strongly upregulated during infection of Acanthamoeba castellanii and THP-1 (38, 39). tsp is regulated by the Legionella quorum-sensing system, an important activator of genes required in the transmissive phase (40). Transcriptomic analysis of a letS mutant in water revealed that tsp is downregulated in the mutant (15). In addition, the expression of tsp was downregulated in a cpxR mutant grown to the PE phase, suggesting that CpxR is also required for expression of tsp in the PE phase (41). CpxR regulates the expression of several Icm/Dot effectors and is required for growth in A. castellanii (41–44). A previous study showed that a tsp mutant in the Lp serogroup 1 strain 130b did not have a growth defect in Acanthamoeba castellanii, THP-1 macrophages, or low-salt chemically defined media at 42°C (37). A small regulatory RNA (sRNA) named Lpr17 (Lppnc0140 in strain Paris) is encoded complementary to lpg0499 (45, 46). sRNAs are short RNA molecules involved in posttranscriptional regulation of genes required for virulence and response to various conditions such as sugar metabolism, iron homeostasis, and biofilm formation (47, 48). Base-pairing sRNAs are the most common type of sRNA, and they act by hybridizing to their target mRNA. Lpr17 is a cis-encoded sRNA. Such sRNAs pair perfectly with their target due to their being encoded on the complementary strand of their target (47). Lpr17 is therefore likely to control tsp expression.
In this paper, we have investigated the role of the Tsp of Lp in the resistance to thermal stress in water and during infection of amoeba. In addition, we have studied the regulatory function of the cis-encoded sRNA Lpr17 as well as confirmed the role of CpxR in the regulation of Tsp.
RESULTS
Tsp is important for L. pneumophila to survive thermal stress.
Since tail-specific proteases have been implicated in managing thermal stress in other bacteria, the importance of Tsp for the survival of Lp after a heat shock at 55°C was tested (Fig. 1). The strains were suspended in the freshwater artificial medium Fraquil and incubated for 24 h at room temperature prior to the temperature stress. Incubation in Fraquil induces a specific transcriptomic program related to starvation and long-term survival in water (49, 50). After 15 min at 55°C, the CFU count of the Δtsp mutant decreased by 10,000-fold, while the CFU count of the WT and the complemented strain decreased by only 10-fold. Thirty minutes after thermal stress, the CFU count of the Δtsp mutant was 100 times lower than the CFU count of the WT and complemented strain; however, this was not statistically significant (P > 0.05).
FIG 1.
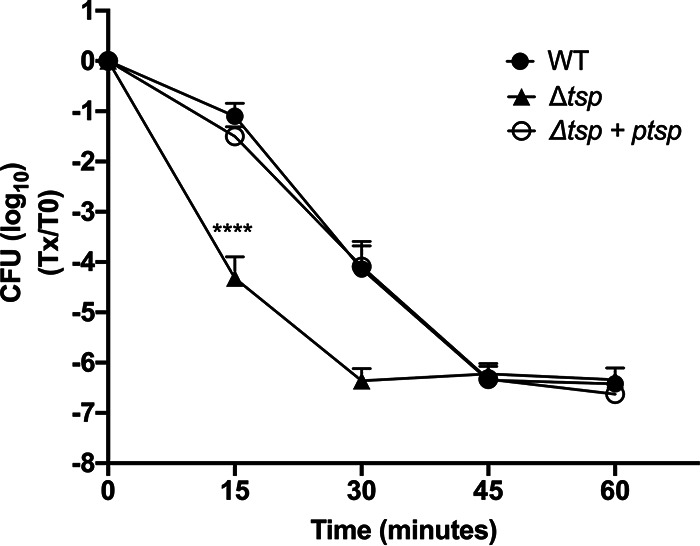
Tsp is required for Lp to survive a thermal stress in water. The WT, tsp mutant (Δtsp), and complemented strain (Δtsp + ptsp) were suspended in Fraquil for 24 h and then subjected to thermal stress at 55°C. The survival of the strains was measured by CFU counts every 15 min from 0 to 60 min. The data shown represent the average of three independent biological replicates with standard deviation. A two-way analysis of variance (ANOVA) with a Tukey correction for multiple comparison was used to determine statistical difference (****, P < 0.0001).
Tsp is important for intracellular multiplication in V. vermiformis following a temperature shift.
Given the susceptibility of the Δtsp mutant to thermal stress, we hypothesized that the ability of the mutant to grow inside amoeba could be compromised if a temperature shift occurs, which is likely to happen in HWDS. The ability of the Δtsp mutant to replicate within V. vermiformis was then investigated (Fig. 2). The infection was therefore carried out at 3 different temperatures in parallel as follows: room temperature for 5 days, 37°C for 5 days, and room temperature for 2 days followed by 37°C for 3 days (referred to in the manuscript as a temperature shift). The temperature shift would simulate the change in temperature encountered in HWDS, albeit to a lower degree, and would test the importance of Tsp for survival under these conditions. At room temperature, none of the strains tested were able to replicate intracellularly (Fig. 2A). At 37°C, all strains were able to replicate intracellularly as well as the WT, with the exception of the dotA mutant, the negative control (Fig. 2C), confirming what was previously reported (37). However, when the temperature was shifted from 25°C to 37°C 2 days after the start of the infection, the yield of intracellular bacteria of the Δtsp mutant at the end of the experiment was significantly lower than that of the WT and the complemented strain (Fig. 2B).
FIG 2.
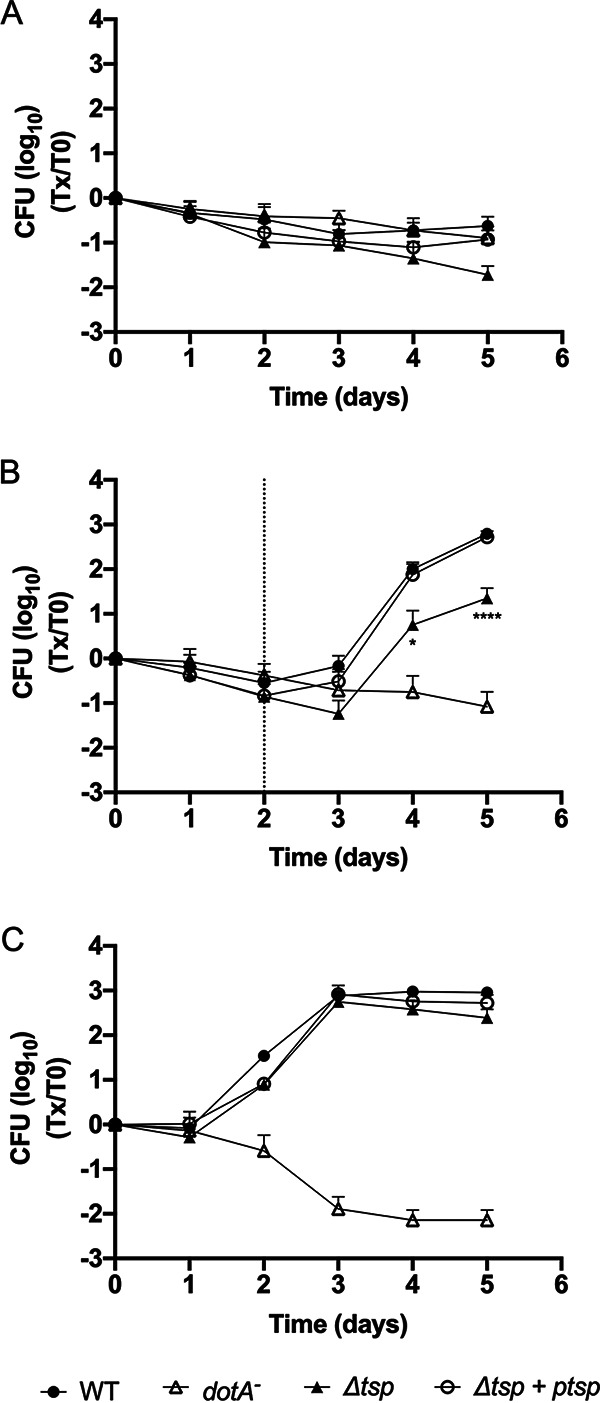
Tsp is required for intracellular multiplication in V. vermiformis during a temperature shift. Infection of the host cell V. vermiformis was carried out at room temperature for 5 days (A), at room temperature for 2 days and then at 37°C for 3 days (B), and at 37°C for 5 days (C). Host cells were infected with the WT, tsp mutant (Δtsp), complemented strain (Δtsp + ptsp), and the dotA mutant, a negative control, suspended in Fraquil. The dotted line in panel B represents the day on which the shift in temperature occurred. The data represent average and standard deviation of 6 biological replicates. A two-way analysis of variance (ANOVA) with a Tukey correction for multiple comparison was used to determine if the results were statistically different. The statistical significance shown compares the WT and tsp mutant (*, P < 0.05; ****, P < 0.0001).
Lpr17 is expressed in the E phase.
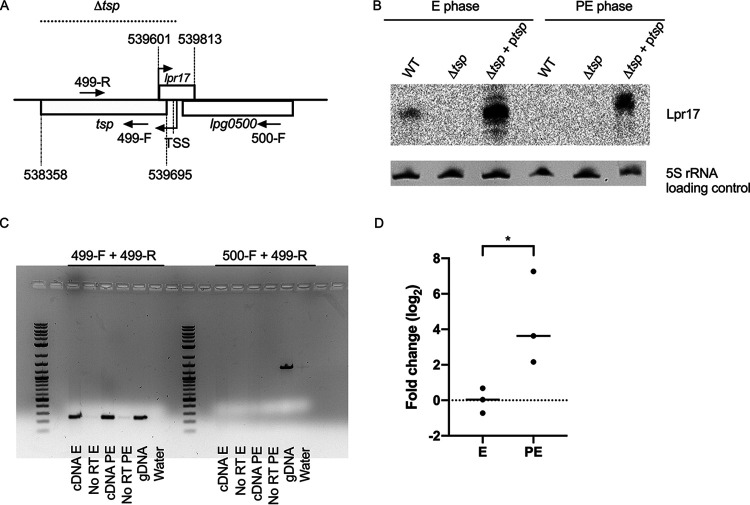
Next, we sought to investigate the regulation of Tsp. The small regulatory RNA Lpr17 is encoded antisense to the 5′ end and promoter region of the tsp gene and the 3′ end of lpg0500, which codes for a peptidase of the M23/M37 family (Fig. 3A). cis-Encoded sRNAs tend to regulate the gene encoded on the complementary strand (47). Therefore, the expression of the Lpr17 sRNA was analyzed by Northern blotting in the WT, tsp mutant (Δtsp), and complemented strain (Δtsp + ptsp) (Fig. 3B). To make the mutant, the tsp gene was replaced with a kanamycin resistance cassette, and, in the process, a section of the Lpr17 coding region, including its transcription start site (TSS), was also removed. The complementation in trans was done by cloning the full-length tsp with 441 bp upstream of the ATG to include the putative promoter on a plasmid. This fragment also contains the full-length lpr17 gene. Lpr17 was only detected in the E phase by Northern blotting. The expression of Lpr17 is stronger in the complemented strain, probably caused by the copy number of the plasmid used for complementation. In the complemented strain, Lpr17 was also expressed in the PE phase, although much less than in the E phase.
FIG 3.
cis-Encoded Lpr17 sRNA is expressed in the E phase, while tsp is expressed in both the E and PE phases. (A) tsp is encoded downstream of lpg0500, and the Lpr17 sRNA is encoded on the complementary strand of tsp overlapping with its TSS and the promoter region. The coordinates of tsp and lpr17 in the Philadelphia-1 genome are indicated. The dotted line indicates the portion of the genome that was replaced with a kanamycin resistance cassette in the Δtsp mutant. (B) Northern blotting was used to investigate the expression of Lpr17 in the WT, tsp mutant (Δtsp), and complemented strain (Δtsp + ptsp) grown to exponential phase (E) and postexponential phase (PE) in AYE broth. 5S rRNA was used as a loading control. (C) An RT-PCR was performed on cDNA from the WT strain grown to the E and PE phases in AYE broth to determine if tsp is encoded on a polycistronic operon with the upstream gene lpg0500. RNA from the E and PE phases that was not reverse transcribed (No RT) as well as water were used as a negative control; genomic DNA (gDNA) of the WT strain served as a positive control. (D) RT-qPCR was used to investigate the expression of tsp in the WT grown in AYE broth to the PE phase compared to the E phase. One microgram of RNA was used for reverse transcription. The Ct values of tsp were normalized against 16S rRNA. An unpaired t test was used to access statistical significance (*, P < 0.05).
tsp is transcribed in the E and PE phases.
Reverse transcriptase PCR (RT-PCR) was used to investigate when the tsp gene is transcribed and if the tsp and lpg0500 genes are transcribed as part of a polycistronic mRNA (Fig. 3C). Amplification with a forward primer (499-R) and a reverse primer (499-F) within the tsp gene revealed that the gene is transcribed in both the E and PE phases. The 499-R primer, along with a forward primer within lpg0500 (500-F), showed that the 2 genes are not expressed as polycistronic mRNA. Our RT-PCR results suggest that tsp is slightly more expressed in the PE phase than the E phase. Using RT-quantitative PCR (RT-qPCR), tsp is 4.3-fold more expressed in the PE phase than the E phase (Fig. 3D).
Tsp is expressed in the PE phase.
In order to detect the expression of Tsp, the protein was tagged with a hexahistidine tag and cloned onto pXDC39, a derivative of pMMB207c lacking the Ptac promoter and lacIq (51). This plasmid also encodes a full-length copy of lpr17. The expression of Tsp and Lpr17 was investigated by Western blotting and Northern blotting, respectively, in the WT and the tsp mutant harboring the Tsp-His construct (ptsp-His) in the E and PE phases (Fig. 4). The WT and the tsp mutant containing an empty vector served as controls. As expected, the Lpr17 sRNA is more expressed in the E phase than the PE phase (Fig. 4A), similar to what was seen in the complemented strain (Fig. 3B). In contrast, the Tsp protein is detected only in the PE phase (Fig. 4B).
FIG 4.
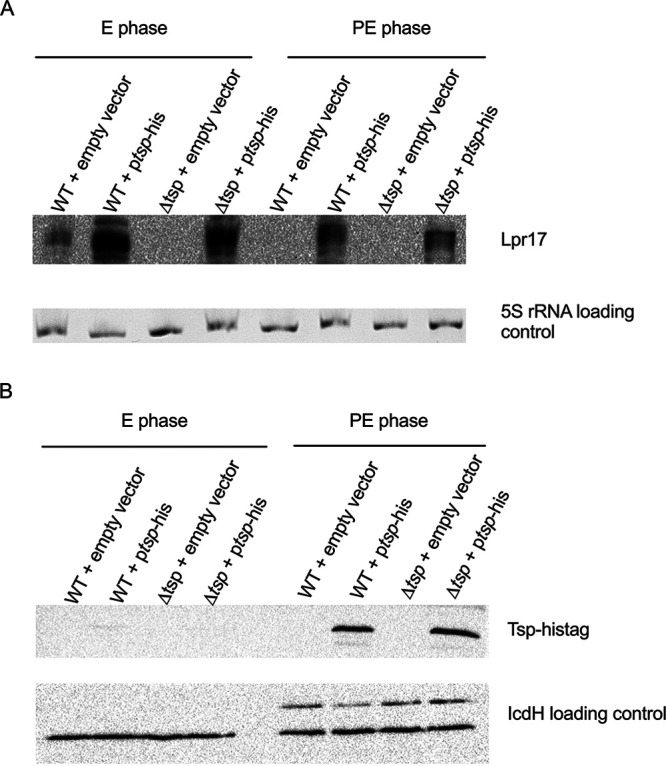
Tsp is expressed in the PE phase. Tsp was tagged with a hexahistidine tag and cloned in plasmid pXDC39 under its own promoter and transferred to the WT (WT + ptsp-His) and tsp mutant (Δtsp + ptsp-His) strains. The strains were grown to the E and PE phases in AYE broth, and expression of the Lpr17 was analyzed by Northern blotting (A), while the expression of the Tsp was analyzed by Western blotting (B). The strains with the empty vector (WT + empty vector; Δtsp + empty vector) served as negative controls. 5S rRNA and IcdH serve as the loading control for Northern blotting and Western blotting, respectively.
Lpr17 is repressed during thermal stress.
Lpr17 is expressed in an opposite manner to Tsp: the sRNA is expressed in the E phase, while the protein is expressed only in the PE phase (Fig. 4). However, tsp seems to be transcribed in both the E and PE phases (Fig. 3C), and the relatively small increase in expression of the transcript between the E and PE phases is insufficient to explain this observation. This suggests that Lpr17 blocks expression of Tsp. Since Tsp is important for survival at 55°C, we hypothesize that Lpr17 should be repressed under this condition as well. Therefore, the expression of Lpr17 during thermal stress was analyzed by Northern blotting (Fig. 5). The strains were grown to the E phase in order to induce expression of Lpr17 and then subjected to thermal stress at 55°C. As seen in Fig. 5, Lpr17 sRNA was strongly repressed after 15 min at 55°C in the WT and complemented strains. This could be due to transcriptional repression or increased sensitivity of the sRNA to RNases or both. Lpr17 is repressed whenever Tsp is produced or needed and supports the hypothesis that Lpr17 blocks translation of Tsp. Further experiments will be needed to confirm this possibility.
FIG 5.
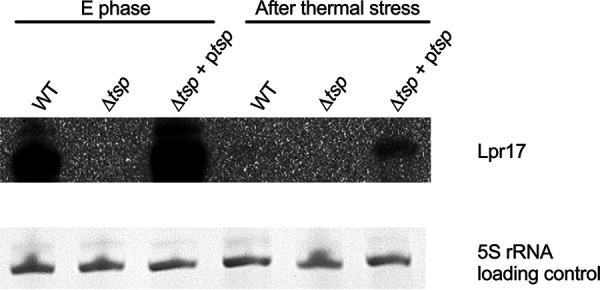
Lpr17 is repressed following thermal stress. The WT, tsp mutant (Δtsp), and complemented strain (Δtsp + ptsp) were grown to E phase in AYE broth and then subjected to heat shock at 55°C for 15 min. Samples for RNA extraction were taken before (E phase) and after thermal stress for Northern blot analysis.
CpxR regulates tsp independently of Lpr17.
A previous transcriptomic study has identified CpxR as a potential regulator of tsp in Lp (41) since the tsp gene was downregulated in the ΔcpxR mutant in the PE phase (41). In order to confirm that the downregulation observed also affects Tsp protein levels, the ptsp-his plasmid was electroporated in the ΔcpxR mutant, and the expression of Tsp in the ΔcpxR mutant was analyzed by Western blotting (Fig. 6A). In the PE phase, Tsp is not expressed in the absence of CpxR. Since the Lpr17 sRNA seems to negatively regulate the expression of Tsp, we hypothesized that the deletion of CpxR may cause overexpression of Lpr17, which would inhibit Tsp expression. The expression of Lpr17 was therefore analyzed by Northern blotting (Fig. 6B). The expression level of Lpr17 in the PE phase in the ΔcpxR mutant is similar to the WT, indicating that the regulation of Tsp by CpxR is Lpr17 independent.
FIG 6.
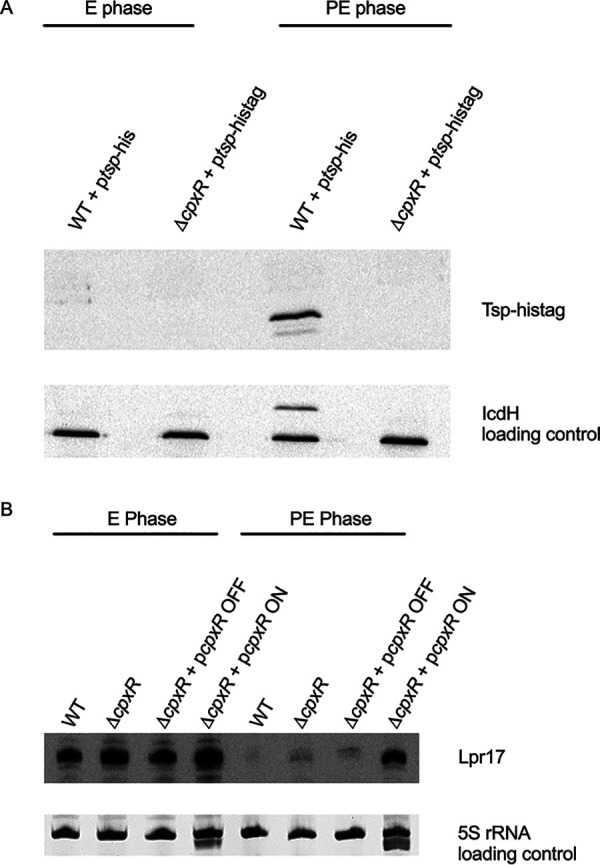
CpxR regulates Tsp in an Lpr17-independent manner. The WT and cpxR mutant ΔcpxR containing ptsp-His were grown to the E and PE phases and analyzed by Western blotting (A) and Northern blotting (B). 5S rRNA and IcdH were used as a loading control for Northern blotting and Western blotting, respectively.
The response regulator CpxR is important for surviving thermal stress.
Since the cpxR mutant produces no Tsp in the PE phase, the ability of the cpxR mutant to survive thermal stress was investigated (Fig. 7). The cpxR mutation was complemented in trans by cloning the ΔcpxR gene under the control of a Ptac promoter in the vector pMMB207c, since cpxR is the fourth gene in a polycistronic mRNA. The cpxR mutant is unable to survive thermal stress compared to the WT and shows a much faster death than the wild type. Basal expression from the vector without addition of isopropyl-β-d-thiogalactopyranoside (IPTG) was enough to complement the mutation (ΔcpxR + pcpxR). To ensure the complementation of the ΔcpxR mutant was due to the cpxR gene cloned, the cpxR mutant with the empty vector pMMB207c (ΔcpxR + pMMB207c) was also tested and showed a similar survival defect to the ΔcpxR mutant. The results were only significantly different after 15 min (P = 0.001). The CFU counts for the three replicates at the later time points showed large variation, and the difference was not considered significantly different.
FIG 7.
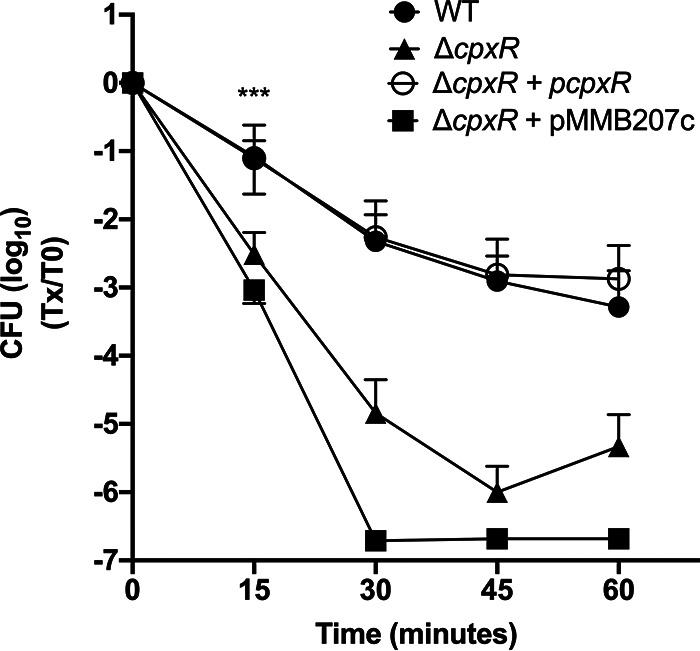
CpxR is required for Lp to survive a thermal stress. The WT, cpxR mutant (ΔcpxR), cpxR mutant with the empty vector (ΔcpxR + pMMB207c), and complemented strain (ΔcpxR + pcpxR) were suspended in Fraquil for 24 h and then subjected to thermal stress at 55°C. The survival of the strains was measured by CFU counts every 15 min from 0 to 60 min. The data shown represent the average of three independent replicate with standard deviation. A two-way ANOVA with a Tukey correction for multiple comparison was used to determine if the results were significantly different. The statistical difference shown in Fig. 7 compares the WT and tsp mutant; ***, P < 0.005.
DISCUSSION
Similar to tsp mutants in other species, the tsp mutant of Lp is sensitive to thermal stress. The tsp mutant shows increased susceptibility to 55°C for 15 min (Fig. 1). The tsp mutant is also sensitive to changes in temperatures. During infection of V. vermiformis, shifting the temperature from 25°C to 37°C impacts the ability of the tsp mutant to reach a similar level to the WT at the end of the experiment (Fig. 2B). The tsp mutant is unable to reach the same yield achieved at 37°C without a temperature shift (Fig. 2C). The yield of the tsp mutant is similar to the WT when the temperature is maintained at 37°C throughout the infection (Fig. 2C). This phenotype could be the result of higher mortality of the tsp mutant within cells during the temperature shift, which would reduce the effective infection dose, or a reduced ability to grow intracellularly following the shift. Nevertheless, the ability of Lp to replicate within V. vermiformis despite shifts in temperature is likely important for its persistence in hot water distribution systems. Sporadic use of HWDS is likely to generate sections of lower temperature, such as in dead ends, that momentarily get warmer when the hot water flows through the system. In addition, increasing the temperature of the system is often used to disinfect hot water distribution systems (8). The amoeba cysts are able to survive treatments up to 80°C, and the ability of Lp to survive and replicate efficiently once the temperature decreases is important for recolonization of the water system (3).
Possibly, the absence of Tsp causes a disturbance in the cell membrane, resulting in increased sensitivity to thermal stress. This reasoning was previously suggested to explain decreased resistance to thermal stress of tsp mutants in Borrelia burgdorferi, Escherichia coli, and Staphylococcus aureus (20, 25, 52). Alternatively, accumulation of misfolded proteins in the tsp mutant could explain the inability to cope with thermal stress. It has previously been shown in other bacteria that Tsp degrades misfolded proteins that accumulate in the periplasm (26, 31, 32). Degradation of misfolded proteins increases the pool of amino acids available for de novo protein synthesis. Possibly, the accumulated misfolded proteins are not degraded in the absence of Tsp, leading to a decrease in the pool of amino acids (13, 14). This might explain the reduced yield of the tsp mutant during infection of amoeba following the temperature shift.
The tsp mutant is still able to replicate within amoeba to some extent following the temperature shift, but the yield, compared to the WT, is lower than when infection is carried out at a constant temperature of 37°C (Fig. 2). This suggests that in the absence of Tsp, other proteases are able to degrade misfolded proteins, thereby relieving the stress. The presence of an N-terminal secretion signal in Tsp was found using two bioinformatics software programs, SignalP and Phobius (53–55). This is consistent with tsp from other species that also have an N-terminal secretion signal required for transport to the periplasm (19). This suggests that the Tsp of Lp is located in the periplasm and could therefore degrade misfolded and aggregated proteins in the periplasm. The expression of alternate periplasmic proteases could explain the reduced yield compared to the WT observed following the temperature shift. In E. coli, the DegP periplasmic protease is important for surviving elevated temperatures and is responsible for degrading misfolded proteins and aggregated proteins in the periplasm (56–60). DegQ is a homolog of DegP that is also found in the periplasm and is responsible for degrading denatured proteins (61–63). In Legionella, the DegP homologue is required for surviving thermal stress but is not required for infection of amoeba (64). The infection was done at a constant temperature, and therefore, it is unknown if DegP plays a role during amoeba infection if a temperature change occurs (64). Legionella codes for a DegQ homologue; however, its role in surviving thermal stress has not been investigated (65, 66). In the absence of tsp, it is possible that these proteases also contribute to removing the aggregated and misfolded proteins at the temperature that occurs during amoeba infection. However, subjecting the strain to 55°C might be too big of a stress for the other proteases to compensate for the lack of tsp.
The CtpA of P. aeruginosa cleaves MepM, belonging to the M23 peptidase family. It is interesting to note that tsp is encoded downstream of lpg0500, which codes for an M23/M37 family peptidase. Lp codes for two other peptidases belonging to the M23/M37 family, lpg0567 and lpg0825, the former being a homologue of MepM of P. aeruginosa. In addition, Lp codes for at least 10 TPR proteins, the same type of protein as the CtpA partner LcbA, suggesting one or some of these TPR proteins might be required for the activity of the Tsp of Lp, similar to what has been reported in P. aeruginosa (67).
cis-Encoded sRNAs, such as Lpr17, typically regulate genes coded on the complementary strand (47). The expression patterns of Lpr17 and Tsp are opposite from each other, where the sRNA is expressed only in the E phase (Fig. 3B), while the Tsp protein is expressed only in the PE phase (Fig. 4B). This pattern of expression suggests a negative regulation of Tsp by Lpr17. This is further supported by the complete repression of Lpr17 during thermal stress (Fig. 5). Furthermore, RT-PCR on cDNA extracted from the E and PE phases (Fig. 3C) shows the presence of the tsp mRNA transcript in both phases, whereas the protein is detected only in the PE phase. Taken together, our results suggest that the Lpr17 sRNA inhibits translation of the protein, possibly by blocking the binding of ribosome to the ribosome binding site (RBS) without initiating degradation of the transcript (Fig. 3A). This is reminiscent of several cis-encoded sRNAs that overlap the 5′ untranslated region of their target gene (68). Determining the exact mechanism would require additional experiments that are beyond the scope of the present study.
In E. coli, the CpxR/A two-component system regulates genes involved in dealing with envelope stress and misfolded proteins in the periplasm (69). In E. coli, CpxR upregulates the DegP periplasmic protease (70–72). In Lp, CpxR does not regulate DegP and DegQ in the E and PE phases, but seems essential for expression of Tsp (41). Therefore, the inability of the cpxR mutant to tolerate thermal stress could rely on the absence of Tsp; however, complementation of cpxR defect by expressing tsp in trans was unsuccessful, suggesting that CpxR regulates other determinants of thermal stress resistance. It is unclear if CpxR directly binds to the promoter region or if the regulation of tsp is indirect. Additional experiments are required to investigate the exact mechanism.
In conclusion, we have demonstrated that Tsp is necessary for survival of thermal stress in water and during intracellular growth. Such situations are probably encountered by Lp in its normal environment. Therefore, Tsp is likely to be a critical genetic determinant for survival and growth in water systems. Our results show that tsp is likely regulated by a complex network consisting minimally of Lpr17 and CpxR. Finally, we have determined that CpxR is an important regulator of thermal stress tolerance in Lp.
MATERIALS AND METHODS
Bacterial strains and media.
Table 1 describes the bacterial strains used in this study. The WT Lp strain used in this study is KS79, a ΔcomR mutant of the JR32 strain, which allows the strain to be constitutively competent (73). JR32 is a Philadelphia-1 derivative that is salt sensitive, streptomycin resistant, and restriction negative (74). Lp strains were cultured on CYE agar [N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered charcoal yeast extract] supplemented with 0.25 mg·ml−1 of l-cysteine and 0.4 mg·ml−1 of ferric pyrophosphate (75). Strains were cultured in AYE broth, which is CYE lacking charcoal and agar. If required, media were supplemented with 25 μg·ml−1 of kanamycin and 5 μg·ml−1 of chloramphenicol (75).
TABLE 1.
Strains, mutants, and plasmids used in this study
| Strain, mutant, or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Legionella pneumophila Philadelphia-1 strains and mutants | ||
| JR32 | Philadelphia-1 derivative; Smr; r−; m+ | 74 |
| KS79 | JR32 ΔcomR | 73 |
| dotA mutant | JR32 dotA::Tn903dIIlacZ | 74 |
| Δtsp (SPF365) | KS79 tsp::Km; Kmr | This study |
| Δtsp + ptsp (SPF403) | Kmr, Cmr | This study |
| KS79 + pXDC39 empty vector (SPF132) | Cmr | This study |
| KS79 + ptsp-His (SPF451) | Cmr | This study |
| Δtsp + pXDC39 empty vector (SPF469) | Kmr, Cmr | This study |
| Δtsp + ptsp-His (SPF452) | Kmr, Cmr | This study |
| Plasmids | ||
| pSF6 | pGEMT-easy-rrnb | 77 |
| pXDC39 | pMMB207c ΔPtac, ΔlacIq, Cmr | Xavier Charpentier |
| ptsp (pSF113) | pXDC39 containing tsp; Cmr | This study |
| ptsp-his (pSF129) | pXDC39 containing tsp-His; Cmr | This study |
Deletion of tsp and complementation of the mutant.
The tsp gene was replaced with a kanamycin resistance cassette by allelic exchange, as previously described, to construct the deletion mutant strain (76). PCR primers are described in Table 2. A 1-kb fragment upstream of tsp was amplified using primers Lpg499-UF and Lpg499-UR. A 1-kb fragment downstream of tsp was amplified using primers Lpg499-DF and Lpg499-DR. The kanamycin cassette was amplified from the pSF6 plasmid (77) using primers Lpg499-Km-F and Lpg499-Km-R. Each 1-kb fragment was purified using a gel extraction kit (Qiagen) and ligated by PCR using primers Lpg499-DR and Lpg499-UF to generate a 3-kb fragment that was purified by a gel extraction kit (Qiagen). The purified 3-kb fragment was introduced into the KS79 strain by natural transformation (78), and the recombinants were selected on CYE agar supplemented with kanamycin. PCR was used to confirm the allelic exchange, and the Δtsp mutant strain was named SPF365.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′) | Source |
|---|---|---|
| Lpg499-UF | ATAAACGGACTGTGCTAAACCAAGAGCTG | This study |
| Lpg499-UR | GTCTAGCTATCGCCATGTAAGCGATCTCCTCAGATGC | This study |
| Lpg499-DF | GCTGAAGATCAGTTGGGTTGACCAATTAGTCACTCC | This study |
| Lpg499-DR | TATCTATGCTCCGTTTTCCAGTCATTCAGC | This study |
| Lpg499-Km-F | GCATCTGAGGAGATCGCTTACATGGCGATAGCTAGAC | This study |
| Lpg499-Km-R | GGAGTGACTAATTGGTCAACCCAACTGATCTTCAGC | This study |
| Lpg499-F1 | TTCGAGCTCTAATTCAAAGCGGCAAACCGTTCAC | This study |
| Lpg499-R1 | CGACTCTAGATTATCTGTTAGCTAACGCCATTCCTTCC | This study |
| pXDC39-scr-R | AAACAGCCAAGCTTGCATGC | This study |
| 499-F | ATGCTAACCGGCCTTGATCC | This study |
| 499-R | CCGCGCATTAAATTAACAGC | This study |
| 500-F | TGCAAGCTCAGCAGGAAATG | This study |
| Lpr17-5O | TCCAACAAAACGAGCAAATCGC | This study |
| Lpr17-5I | TTGTCTTAAAGCTTAGCCGCTGGC | This study |
| Lpr17-3O | TGGTTTCTTCAGCCGAAAACGC | This study |
| Lpr17-3I | ATAGCGATCTCCTCAGATGCAAGG | This study |
| 5′RACE Adapter | GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA | This study |
| Poly(dT) | GCGAGCACAGAATTAATACGACTCACTATAGGT12VN | This study |
| 5′RACE-Outer | GCTGATGGCGATGAATGAACACTG | This study |
| 5′RACE-Inner | CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG | This study |
| 3′RACE-Outer | GCGAGCACAGAATTAATACGACT | This study |
| 3′RACE-Inner | CGCGGATCCGAATTAATACGACTCACTATAGG | This study |
| Lpg499-his-F | ATCCTCTAGAATCGCAAGTGTAGGCCATACCGGTGG | This study |
| Lpg499-his-R | ATGCCTGCAGTTAATGGTGATGGTGATGGTGTCTGTTAGCTAACGCCATTCCTTCC | This study |
| 499-qPCR-F | TCTGAAGCAGAGGCAGAACC | This study |
| 499-qPCR-R | CAATGACAAGGCAGGTAAGC | This study |
The tsp mutation was complemented in trans by amplifying the tsp gene with its native promoter from the KS79 WT genomic DNA using primers Lpg499-F1 and Lpg499-R1. The Lpg499-F1 primer is located 441 nucleotides upstream of the ATG of tsp, and the amplicon includes both the tsp and lpr17 genes. The amplicon was cloned in the pXDC39 plasmid vector using the restriction enzymes SacI (New England Biolabs) and XbaI (New England Biolabs). The restriction digestion was done at 37°C for 1 h, column purified (Qiagen), and ligated for 2 h at room temperature using T4 DNA ligase (New England Biolabs). The ligation product was transformed into E. coli DH5α, and the transformants were selected on LB agar containing 25 μg/ml of chloramphenicol. Cloning was confirmed by PCR using primers Lpg499-F1 and pXDC39-scr-R. The recombinant plasmid (ptsp) was extracted and electroporated into the Δtsp mutant strain. The transformants were selected on CYE agar containing 5 μg·ml−1 of chloramphenicol and then patched on CYE agar containing 25 μg·ml−1 of kanamycin and 5 μg·ml−1 of chloramphenicol. Screening was done by PCR using primers Lpg499-F1 and pXDC39-scr-R, and the resulting strain was called SPF403.
Thermal stress.
Lp strains were grown on CYE agar for 3 days at 37°C. A suspension was prepared from a few colonies in Fraquil, an artificial freshwater medium (79, 80), at an optical density at 600 nm (OD600) of 1.0. The suspension was washed three times with Fraquil to remove any trace of nutrients. The strains were incubated at 25°C for 24 h in 25-cm2 cell culture flasks (Sarstedt). The strains were then diluted 1:10 in Fraquil, and 1 ml was distributed in 13-ml tubes (Sarstedt) and incubated in a 55°C water bath. Individual tubes were used for each time point. Tubes were removed from the water bath for CFU counts on CYE agar to determine the survival of the strains. CFU counts were done at 0, 15, 30, 45, and 60 min.
Vermamoeba vermiformis culture and infection.
V. vermiformis cells were grown at room temperature in 75-cm2 cell culture flasks (Sarstedt) in modified PYNFH medium (ATCC medium 1034) and passaged when confluence was reached. Modified PYNFH contains, per liter, 10 g of peptone, 10 g of yeast extract, 1 g of yeast nucleic acid, 15 mg of folic acid, 1 mg of hemin, 100 ml of heat-inactivated fetal bovine serum, and 20 ml of buffer solution (18.1 g of KH2PO4 and 25 g of Na2HPO4 per liter). The amoebas were centrifuged at 200 × g for 10 min, the supernatant was discarded, and cells were resuspended in fresh modified PYNFH medium and incubated for 3 days prior to infection. On the day the infection was to be carried out, the cells were centrifuged at 200 × g for 10 min and resuspended at a concentration of 5.0 × 105 cells/ml in modified PYNFH lacking fetal bovine serum (FBS) and the buffer solution. Legionella is able to grow in modified PYNFH containing FBS and the buffer solution; therefore, it was omitted during the infection to ensure that the growth observed is due to intracellular multiplication. One milliliter of cells was seeded in each well of the 24-well plate (Sarstedt).
Bacterial strains were grown on CYE agar with antibiotics for 3 days at 37°C. On the day of the infection, the bacteria were suspended in Fraquil at an OD600 of 0.1 and then diluted 1:10 in Fraquil. Five microliters of the 1:10 dilution were added to each well in triplicate in order to have a starting multiplicity of infection (MOI) of 0.1. The infection was carried out at room temperature for 5 days, at 37°C for 5 days, at 25°C for 2 days, and then at 37°C for an additional 3 days. CFU counts were performed on a daily basis.
RNA extraction.
TRIzol (Thermo Fisher Scientific) was used for RNA extraction according to the manufacturer’s protocol. Briefly, 200 μl of chloroform (Thermo Fisher Scientific) was added to the bacterial pellet suspended in TRIzol, shaken, and incubated for 3 min at room temperature. The sample was added to a Phase Lock Gel Heavy 2-ml tube (VWR) and centrifuged at 12,000 × g for 15 min at room temperature. Approximately 600 μl of the supernatant was transferred to a microtube containing 600 μl of 100% isopropanol (Thermo Fisher Scientific) and 5 μg of glycogen (Thermo Fisher Scientific). Following incubation at room temperature for 10 min, the tubes were centrifuged at 17,000 × g for 10 min at 4°C. The supernatant was removed, and 1 ml of 75% ice-cold ethanol (Greenfield Global) was added to the RNA pellet. The tubes were centrifuged at 17,000 × g for 10 min at 4°C, the supernatant was removed, and the pellet was air-dried and resuspended in nuclease-free water (Thermo Fisher Scientific). The RNA was quantified using a UV spectrophotometer (Thermo Fisher Scientific).
Northern blotting.
The expression of the Lpr17 sRNA was examined by Northern blotting as previously described (15). The strains were grown on CYE agar with required antibiotics for 3 days at 37°C, inoculated in AYE broth with required antibiotics, and grown at 37°C with shaking at 250 rpm. Aliquots of 10 ml of exponential-phase culture (OD600, 0.5 to 1.0) and 5 ml of PE phase culture (OD600 > 3) were centrifuged at 4,000 × g for 10 min, the supernatant was removed, and the bacterial pellet was resuspended in 1 ml of TRIzol reagent. RNA extraction was carried out as mentioned above. Five micrograms of RNA were loaded on a 6% Tris-borate-EDTA-urea polyacrylamide gel, and the samples were migrated at 180 mV. The RNA was transferred to a positively charged nylon membrane (Thermo Fisher Scientific) using a semidry gel blotting system (Bio-Rad) for 20 min at 200 mA. The membrane was prehybridized in ULTRAhyb-Oligo hybridization buffer (Thermo Fisher Scientific) for 1 h at 37°C in a rotating chamber. We added 5 nM 5′ biotinylated Lpr10 probe (Integrated DNA Technologies) to the prehybridization buffer, and the membrane was incubated overnight at 37°C in the rotating chamber. The membrane was twice washed with 2 × SSC (0.15 M NaCl and 0.015 M sodium citrate) and 0.5% SDS (Bio-Rad) for 30 min at 37°C. The probes were detected with the chemiluminescent nucleic acid detection module (Thermo Fisher Scientific).
RT-PCR.
RNA extracted from the KS79 WT strain grown to the E and PE phases was treated with DNase (Thermo Fisher Scientific). One microgram of DNase treated RNA was reverse transcribed with Protoscript II (New England Biolabs) and a no-reverse transcriptase control (no RT) was included by replacing the reverse transcriptase by nuclease-free water. The PCR was performed on cDNA, no-RT control, WT gDNA, and a no-template control (water) using primers described in Table 2. The amplicon was analyzed on a 1% agarose gel.
Quantitative PCR.
RNA was extracted and cDNA synthesized as described above. Following cDNA synthesis, qPCR was done with primers 499-qPCR-F and 499-qPCR-R. qPCR was performed using iTaq universal SYBR green supermix (Bio-Rad) according to the manufacturer’s protocol. The efficiency of the primer pairs was determined using dilution series of gDNA. Threshold cycle (Ct) values were normalized to the 16S rRNA as described previously (81).
Cloning of Tsp with hexahistidine tag.
The hexahistidine tag was added to tsp by PCR. tsp, along with its native promoter, was amplified using primers Lpg499-his-F and Lpg499-his-R, the latter containing a hexahistidine tag. The amplicon was cloned in the pXDC39 plasmid vector using XbaI (New England Biolabs) and PstI (New England Biolabs). The restriction digestion was carried out at 37°C for 1 h, followed by column purification (Qiagen) and ligation at room temperature for 2 h using T4 DNA ligase (New England Biolabs). The ligation product was transformed into E. coli DH5α, and the transformants were selected on LB agar supplemented with 25 μg·ml−1 of chloramphenicol. The cloning was confirmed by PCR using primers Lpg499-his-F and pXDC39-scr-R. The plasmid (ptsp-his) was extracted and electroporated into KS79, and the tsp mutant and transformants were selected on CYE containing 5 μg·ml−1 of chloramphenicol. The KS79 ptsp-His transformants were patched on CYE containing 5 μg·ml−1 of chloramphenicol, while the tsp mutant ptsp-His transformants were patched on CYE containing 25 μg·ml−1 of kanamycin and 5 μg·ml−1 of chloramphenicol. Screening was done by PCR using primers Lpg499-his-F and pXDC39-scr-R. The pXDC39 empty vector was also electroporated in the KS79 WT and the tsp mutant, which serve as controls.
Western blotting.
Strains were grown to the E and PE phases in AYE broth. Aliquots of 1.5 ml were centrifuged at 17,000 × g for 3 min, the supernatant was decanted, and the cell pellet was resuspended in 200 μl of 1× sample buffer (10% glycerol, 62.5 mM Tris-HCl, pH 6.8, 2.5% SDS, 0.002% bromophenol blue, and 5% β-mercaptoethanol). The samples were boiled for 5 min and sonicated for 15 min in an ice-cold water bath using ultrasonic cleaner (Cole-Palmer). Samples were then centrifuged for 15 min at 17,000 × g at 4°C. The supernatant was transferred to a new tube and stored at −20°C until it was ready to use. The protein samples were standardized by adjusting the final OD600 to 0.5 by diluting the samples with 1× sample buffer. We loaded 15 μl of the standardized protein sample on a 12.5% polyacrylamide gel, and samples were migrated at 100 V. The proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) at 16 V for 24 h. The membrane was blocked for 30 min with a 5% milk protein solution. Tsp-His was detected with the anti-6× His tag antibody (Thermo Fisher Scientific), which is already conjugated with horseradish peroxidase (HRP) and does not require a secondary antibody for detection. The blot was incubated with this antibody at room temperature for the following 2 h. The IcdH antibody (Sigma) was used as a loading control. In this case, the blot was incubated with the IcdH primary antibody at room temperature for 2 h and with the secondary antibody (anti-rabbit HRP; Sigma) at room temperature for 30 min. Antibodies were detected using ECL Prime Western blotting detection reagents (GE Healthcare).
ACKNOWLEDGMENTS
The pXDC39 plasmid is a kind gift from Xavier Charpentier.
This study was supported by Canadian Institute of Health Research open operating grant number 142208 to S.P.F. J.S. was supported by a FRQNT doctoral scholarship and a CRIPA scholarship supported by the Fonds de recherche du Québec-Nature et technologies n°RS-170946.
REFERENCES
- 1.Swanson MS, Hammer BK. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu Rev Microbiol 54:567–613. doi: 10.1146/annurev.micro.54.1.567. [DOI] [PubMed] [Google Scholar]
- 2.Wadowsky RM, Wilson TM, Kapp NJ, West AJ, Kuchta JM, States SJ, Dowling JN, Yee RB. 1991. Multiplication of Legionella spp. in tap water containing Hartmannella vermiformis. Appl Environ Microbiol 57:1950–1955. doi: 10.1128/AEM.57.7.1950-1955.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storey MV, Winiecka-Krusnell J, Ashbolt NJ, Stenstrom TA. 2004. The efficacy of heat and chlorine treatment against thermotolerant Acanthamoebae and Legionellae. Scand J Infect Dis 36:656–662. doi: 10.1080/00365540410020785. [DOI] [PubMed] [Google Scholar]
- 4.Taylor M, Ross K, Bentham R. 2009. Legionella, protozoa, and biofilms: interactions within complex microbial systems. Microb Ecol 58:538–547. doi: 10.1007/s00248-009-9514-z. [DOI] [PubMed] [Google Scholar]
- 5.Kuchta JM, Navratil JS, Shepherd ME, Wadowsky RM, Dowling JN, States SJ, Yee RB. 1993. Impact of chlorine and heat on the survival of Hartmannella vermiformis and subsequent growth of Legionella pneumophila. Appl Environ Microbiol 59:4096–4100. doi: 10.1128/AEM.59.12.4096-4100.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delafont V, Rodier MH, Maisonneuve E, Cateau E. 2018. Vermamoeba vermiformis: a free-living amoeba of interest. Microb Ecol 76:991–1001. doi: 10.1007/s00248-018-1199-8. [DOI] [PubMed] [Google Scholar]
- 7.Ensminger AW. 2016. Legionella pneumophila, armed to the hilt: justifying the largest arsenal of effectors in the bacterial world. Curr Opin Microbiol 29:74–80. doi: 10.1016/j.mib.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Bedard E, Fey S, Charron D, Lalancette C, Cantin P, Dolce P, Laferriere C, Deziel E, Prevost M. 2015. Temperature diagnostic to identify high risk areas and optimize Legionella pneumophila surveillance in hot water distribution systems. Water Res 71:244–256. doi: 10.1016/j.watres.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Borella P, Guerrieri E, Marchesi I, Bondi M, Messi P. 2005. Water ecology of Legionella and protozoan: environmental and public health perspectives. Biotechnol Annu Rev 11:355–380. doi: 10.1016/S1387-2656(05)11011-4. [DOI] [PubMed] [Google Scholar]
- 10.Henne K, Kahlisch L, Hofle MG, Brettar I. 2013. Seasonal dynamics of bacterial community structure and composition in cold and hot drinking water derived from surface water reservoirs. Water Res 47:5614–5630. doi: 10.1016/j.watres.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 11.Lesnik R, Brettar I, Hofle MG. 2016. Legionella species diversity and dynamics from surface reservoir to tap water: from cold adaptation to thermophily. ISME J 10:1064–1080. doi: 10.1038/ismej.2015.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whiley H, Bentham R, Brown MH. 2017. Legionella persistence in manufactured water systems: pasteurization potentially selecting for thermal tolerance. Front Microbiol 8:1330. doi: 10.3389/fmicb.2017.01330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter K, Haslbeck M, Buchner J. 2010. The heat shock response: life on the verge of death. Mol Cell 40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Schumann W. 2016. Regulation of bacterial heat shock stimulons. Cell Stress Chaperones 21:959–968. doi: 10.1007/s12192-016-0727-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendis N, McBride P, Saoud J, Mani T, Faucher SP. 2018. The LetA/S two-component system regulates transcriptomic changes that are essential for the culturability of Legionella pneumophila in water. Sci Rep 8:6764. doi: 10.1038/s41598-018-24263-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt CK, Meysick KC, O'Brien AD. 1999. Bacterial toxins: friends or foes? Emerg Infect Dis 5:224–234. doi: 10.3201/eid0502.990206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beebe KD, Shin J, Peng J, Chaudhury C, Khera J, Pei D. 2000. Substrate recognition through a PDZ domain in tail-specific protease. Biochemistry 39:3149–3155. doi: 10.1021/bi992709s. [DOI] [PubMed] [Google Scholar]
- 18.Rawlings ND, Barrett AJ, Bateman A. 2010. MEROPS: the peptidase database. Nucleic Acids Res 38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoge R, Laschinski M, Jaeger KE, Wilhelm S, Rosenau F. 2011. The subcellular localization of a C-terminal processing protease in Pseudomonas aeruginosa. FEMS Microbiol Lett 316:23–30. doi: 10.1111/j.1574-6968.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- 20.Carroll RK, Rivera FE, Cavaco CK, Johnson GM, Martin D, Shaw LN. 2014. The lone S41 family C-terminal processing protease in Staphylococcus aureus is localized to the cell wall and contributes to virulence. Microbiology (Reading) 160:1737–1748. doi: 10.1099/mic.0.079798-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandara AB, Sriranganathan N, Schurig GG, Boyle SM. 2005. Carboxyl-terminal protease regulates Brucella suis morphology in culture and persistence in macrophages and mice. J Bacteriol 187:5767–5775. doi: 10.1128/JB.187.16.5767-5775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldman AR, Lee J, Delmas B, Paetzel M. 2006. Crystal structure of a novel viral protease with a serine/lysine catalytic dyad mechanism. J Mol Biol 358:1378–1389. doi: 10.1016/j.jmb.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 23.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waxman DJ, Strominger JL. 1983. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu Rev Biochem 52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 25.Hara H, Yamamoto Y, Higashitani A, Suzuki H, Nishimura Y. 1991. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol 173:4799–4813. doi: 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 27.Seoane A, Sabbaj A, McMurry LM, Levy SB. 1992. Multiple antibiotic susceptibility associated with inactivation of the prc gene. J Bacteriol 174:7844–7847. doi: 10.1128/jb.174.23.7844-7847.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang CY, Wang SW, Huang WC, Kim KS, Chang NS, Wang YH, Wu MH, Teng CH. 2012. Prc contributes to Escherichia coli evasion of classical complement-mediated serum killing. Infect Immun 80:3399–3409. doi: 10.1128/IAI.00321-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh SK, Parveen S, SaiSree L, Reddy M. 2015. Regulated proteolysis of a cross-link-specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proc Natl Acad Sci U S A 112:10956–10961. doi: 10.1073/pnas.1507760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh SK, SaiSree L, Amrutha RN, Reddy M. 2012. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol 86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- 31.Keiler KC, Silber KR, Downard KM, Papayannopoulos IA, Biemann K, Sauer RT. 1995. C-terminal specific protein degradation: activity and substrate specificity of the Tsp protease. Protein Sci 4:1507–1515. doi: 10.1002/pro.5560040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silber KR, Keiler KC, Sauer RT. 1992. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc Natl Acad Sci U S A 89:295–299. doi: 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo J, Darwin AJ. 2013. The Pseudomonas aeruginosa periplasmic protease CtpA can affect systems that impact its ability to mount both acute and chronic infections. Infect Immun 81:4561–4570. doi: 10.1128/IAI.01035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiling SA, Jansen JA, Henley BJ, Singh S, Chattin C, Chandler M, Rowen DW. 2005. Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology (Reading) 151:2251–2261. doi: 10.1099/mic.0.27772-0. [DOI] [PubMed] [Google Scholar]
- 35.Sautter R, Ramos D, Schneper L, Ciofu O, Wassermann T, Koh CL, Heydorn A, Hentzer M, Hoiby N, Kharazmi A, Molin S, Devries CA, Ohman DE, Mathee K. 2012. A complex multilevel attack on Pseudomonas aeruginosa algT/U expression and algT/U activity results in the loss of alginate production. Gene 498:242–253. doi: 10.1016/j.gene.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivastava D, Seo J, Rimal B, Kim SJ, Zhen S, Darwin AJ. 2018. A proteolytic complex targets multiple cell wall hydrolases in Pseudomonas aeruginosa. mBio 9:e00972-18. doi: 10.1128/mBio.00972-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence A, K Nicholls S, H Stansfield S, M Huston W. 2014. Characterization of the tail-specific protease (Tsp) from Legionella. J Gen Appl Microbiol 60:95–100. doi: 10.2323/jgam.60.95. [DOI] [PubMed] [Google Scholar]
- 38.Bruggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, Kunst F, Steinert M, Heuner K, Coppee JY, Buchrieser C. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol 8:1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- 39.Faucher SP, Friedlander G, Livny J, Margalit H, Shuman HA. 2010. Legionella pneumophila 6S RNA optimizes intracellular multiplication. Proc Natl Acad Sci U S A 107:7533–7538. doi: 10.1073/pnas.0911764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiaden A, Spirig T, Carranza P, Bruggemann H, Riedel K, Eberl L, Buchrieser C, Hilbi H. 2008. Synergistic contribution of the Legionella pneumophila lqs genes to pathogen-host interactions. J Bacteriol 190:7532–7547. doi: 10.1128/JB.01002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanner JR, Li L, Faucher SP, Brassinga AK. 2016. The CpxRA two-component system contributes to Legionella pneumophila virulence. Mol Microbiol 100:1017–1038. doi: 10.1111/mmi.13365. [DOI] [PubMed] [Google Scholar]
- 42.Gal-Mor O, Segal G. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J Bacteriol 185:4908–4919. doi: 10.1128/jb.185.16.4908-4919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altman E, Segal G. 2008. The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J Bacteriol 190:1985–1996. doi: 10.1128/JB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feldheim YS, Zusman T, Speiser Y, Segal G. 2016. The Legionella pneumophila CpxRA two‐component regulatory system: new insights into CpxR’s function as a dual regulator and its connection to the effectors regulatory network. Mol Microbiol 99:1059–1079. doi: 10.1111/mmi.13290. [DOI] [PubMed] [Google Scholar]
- 45.Weissenmayer BA, Prendergast JG, Lohan AJ, Loftus BJ. 2011. Sequencing illustrates the transcriptional response of Legionella pneumophila during infection and identifies seventy novel small non-coding RNAs. PLoS One 6:e17570. doi: 10.1371/journal.pone.0017570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sahr T, Rusniok C, Dervins-Ravault D, Sismeiro O, Coppee JY, Buchrieser C. 2012. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol 9:503–519. doi: 10.4161/rna.20270. [DOI] [PubMed] [Google Scholar]
- 47.Apura P, Domingues S, Viegas SC, Arraiano CM. 2019. Reprogramming bacteria with RNA regulators. Biochem Soc Trans 47:1279–1289. doi: 10.1042/BST20190173. [DOI] [PubMed] [Google Scholar]
- 48.Desgranges E, Caldelari I, Marzi S, Lalaouna D. 2020. Navigation through the twists and turns of RNA sequencing technologies: application to bacterial regulatory RNAs. Biochim Biophys Acta Gene Regul Mech 1863:194506. doi: 10.1016/j.bbagrm.2020.194506. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Mendis N, Trigui H, Faucher SP. 2015. Transcriptomic changes of Legionella pneumophila in water. BMC Genomics 16:637. doi: 10.1186/s12864-015-1869-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trigui H, Dudyk P, Oh J, Hong JI, Faucher SP. 2015. A regulatory feedback loop between RpoS and SpoT supports the survival of Legionella pneumophila in water. Appl Environ Microbiol 81:918–928. doi: 10.1128/AEM.03132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frey J, Bagdasarian MM, Bagdasarian M. 1992. Replication and copy number control of the broad-host-range plasmid RSF1010. Gene 113:101–106. doi: 10.1016/0378-1119(92)90675-f. [DOI] [PubMed] [Google Scholar]
- 52.Kumru OS, Bunikis I, Sorokina I, Bergstrom S, Zuckert WR. 2011. Specificity and role of the Borrelia burgdorferi CtpA protease in outer membrane protein processing. J Bacteriol 193:5759–5765. doi: 10.1128/JB.05622-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 54.Kall L, Krogh A, Sonnhammer EL. 2007. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res 35:W429–32. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 56.Strauch KL, Beckwith J. 1988. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc Natl Acad Sci U S A 85:1576–1580. doi: 10.1073/pnas.85.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lipinska B, Fayet O, Baird L, Georgopoulos C. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J Bacteriol 171:1574–1584. doi: 10.1128/jb.171.3.1574-1584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strauch KL, Johnson K, Beckwith J. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol 171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seol JH, Woo SK, Jung EM, Yoo SJ, Lee CS, Kim KJ, Tanaka K, Ichihara A, Ha DB, Chung CH. 1991. Protease do is essential for survival of Escherichia coli at high temperatures: its identity with the htrA gene product. Biochem Biophys Res Commun 176:730–736. doi: 10.1016/s0006-291x(05)80245-1. [DOI] [PubMed] [Google Scholar]
- 60.Laskowska E, Kuczyńska-Wiśnik D, Skórko-Glonek J, Taylor A. 1996. Degradation by proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol Microbiol 22:555–571. doi: 10.1046/j.1365-2958.1996.1231493.x. [DOI] [PubMed] [Google Scholar]
- 61.Waller PR, Sauer RT. 1996. Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J Bacteriol 178:1146–1153. doi: 10.1128/jb.178.4.1146-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolmar H, Waller PR, Sauer RT. 1996. The DegP and DegQ periplasmic endoproteases of Escherichia coli: specificity for cleavage sites and substrate conformation. J Bacteriol 178:5925–5929. doi: 10.1128/jb.178.20.5925-5929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malet H, Canellas F, Sawa J, Yan J, Thalassinos K, Ehrmann M, Clausen T, Saibil HR. 2012. Newly folded substrates inside the molecular cage of the HtrA chaperone DegQ. Nat Struct Mol Biol 19:152–157. doi: 10.1038/nsmb.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pedersen LL, Radulic M, Doric M, Abu Kwaik Y. 2001. HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Infect Immun 69:2569–2579. doi: 10.1128/IAI.69.4.2569-2579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wrase R, Scott H, Hilgenfeld R, Hansen G. 2011. The Legionella HtrA homologue DegQ is a self-compartmentizing protease that forms large 12-meric assemblies. Proc Natl Acad Sci U S A 108:10490–10495. doi: 10.1073/pnas.1101084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schubert A, Wrase R, Hilgenfeld R, Hansen G. 2015. Structures of DegQ from Legionella pneumophila define distinct ON and OFF states. J Mol Biol 427:2840–2851. doi: 10.1016/j.jmb.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 67.Bandyopadhyay P, Sumer EU, Jayakumar D, Liu S, Xiao H, Steinman HM. 2012. Implication of proteins containing tetratricopeptide repeats in conditional virulence phenotypes of Legionella pneumophila. J Bacteriol 194:3579–3588. doi: 10.1128/JB.00399-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ellis MJ, Trussler RS, Haniford DB. 2015. A cis-encoded sRNA, Hfq and mRNA secondary structure act independently to suppress IS200 transposition. Nucleic Acids Res 43:6511–6527. doi: 10.1093/nar/gkv584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Wulf P, Kwon O, Lin EC. 1999. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J Bacteriol 181:6772–6778. doi: 10.1128/JB.181.21.6772-6778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev 9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 71.Pogliano J, Lynch AS, Belin D, Lin EC, Beckwith J. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev 11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 72.Dartigalongue C, Missiakas D, Raina S. 2001. Characterization of the Escherichia coli sigma E regulon. J Biol Chem 276:20866–20875. doi: 10.1074/jbc.M100464200. [DOI] [PubMed] [Google Scholar]
- 73.de Felipe KS, Glover RT, Charpentier X, Anderson OR, Reyes M, Pericone CD, Shuman HA. 2008. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog 4:e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61:5361–5373. doi: 10.1128/IAI.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feeley JC, Gibson RJ, Gorman GW, Langford NC, Rasheed JK, Mackel DC, Baine WB. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol 10:437–441. doi: 10.1128/JCM.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hovel-Miner G, Pampou S, Faucher SP, Clarke M, Morozova I, Morozov P, Russo JJ, Shuman HA, Kalachikov S. 2009. SigmaS controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J Bacteriol 191:2461–2473. doi: 10.1128/JB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Faucher SP, Mueller CA, Shuman HA. 2011. Legionella Pneumophila transcriptome during intracellular multiplication in human macrophages. Front Microbiol 2:60. doi: 10.3389/fmicb.2011.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sexton JA, Vogel JP. 2004. Regulation of hypercompetence in Legionella pneumophila. J Bacteriol 186:3814–3825. doi: 10.1128/JB.186.12.3814-3825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morel FMM, Westall JC, Rueter JG, Chaplick JP. 1975. Description of the algal growth media "Aquil" and "Fraquil." Technical Report 16. Water Quality Laboratory, Ralph Parsons Laboratory for Water Resources and Hydrodynamics, Massachusetts Institute of Technology, Cambridge, MA. [Google Scholar]
- 80.Mendis N, McBride P, Faucher SP. 2015. Short-term and long-term survival and virulence of Legionella pneumophila in the defined freshwater medium Fraquil. PLoS One 10:e0139277. doi: 10.1371/journal.pone.0139277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]