The exploration of deep marine sediments has unearthed many new lineages of microbes. The finding of this novel phylum of Asgard archaea is important, since understanding the diversity and evolution of Asgard archaea may inform also about the evolution of eukaryotic cells. The comparison of metabolic potentials of the Asgard archaea can help inform about selective pressures the lineages have faced during evolution.
KEYWORDS: Asgard, subsurface, metagenomics
ABSTRACT
The Asgard superphylum is a deeply branching monophyletic group of Archaea, recently described as some of the closest relatives of the eukaryotic ancestor. The wide application of genomic analyses from metagenome sequencing has established six distinct phyla, whose genomes encode diverse metabolic capacities and which play important biogeochemical and ecological roles in marine sediments. Here, we describe two metagenome-assembled genomes (MAGs) recovered from deep marine sediments off the Costa Rica margin, defining a novel lineage phylogenetically married to “Candidatus Thorarchaeota”; as such, we propose the name “Sifarchaeota” for this phylum. The two Sifarchaeota MAGs encode an anaerobic pathway for methylotrophy enabling the utilization of C1 to C3 compounds (methanol and methylamines) to synthesize acetyl coenzyme A (acetyl-CoA). The MAGs showed a remarkable saccharolytic capabilities compared to other Asgard lineages and encoded diverse classes of carbohydrate active enzymes (CAZymes) targeting different mono-, di-, and oligosaccharides. Comparative genomic analysis based on the full metabolic profiles of different Asgard lineages revealed the close relation between Sifarchaeota and “Candidatus Odinarchaeota” MAGs, which suggested similar metabolic potentials and ecological roles. Furthermore, we identified multiple HGT events from different bacterial donors within Sifarchaeota MAGs, which hypothetically expanded Sifarchaeota capacities for substrate utilization, energy production, and niche adaptation.
IMPORTANCE The exploration of deep marine sediments has unearthed many new lineages of microbes. The finding of this novel phylum of Asgard archaea is important, since understanding the diversity and evolution of Asgard archaea may inform also about the evolution of eukaryotic cells. The comparison of metabolic potentials of the Asgard archaea can help inform about selective pressures the lineages have faced during evolution.
INTRODUCTION
Deep marine sediments are the home of many poorly described archaeal lineages, most of which are yet uncultured (1, 2). Recently, the discovery of Asgard archaea in benthic environments has generated great interest in novel lineages from marine sediments. Additionally, greater attention is being directed toward studying Asgard archaea in order to better understand eukaryotic evolution, as this superphylum represents the archaeal group most closely related to eukaryotes and their genomes encode multiple homologs of eukaryotic proteins (3, 4). Currently, there are 6 phyla proposed to be part of the Asgard superphylum: “Candidatus Lokiarchaeota,” “Candidatus Thorarchaeota,” “Candidatus Odinarchaeota,” “Candidatus Heimdallarchaeota,” “Candidatus Helarchaeota,” and Gerdarchaeota (4–6). The number of novel lineages yet to be found is at this point unknown. This raises the need for more genome-resolved metagenome surveys to recover genomes of these archaeal lineages, decipher their metabolic capacities, and place them in the context of microbial ecology. Previous studies targeting deep sediment from the Costa Rica margin subseafloor have shown the presence of abundant and diverse archaeal communities (7, 8). Among these archaeal lineages, members of the Asgard superphylum were highly abundant at multiple depths, making up 17% of the archaeal communities present (7, 8). With newly sequenced metagenomes from similar Costa Rica margin subseafloor sediments, we employed genome-resolved metagenomics to describe the metabolic potential of the genomes belonging to a new Asgard phylum. Phylogenomic analysis placed the sequences of the new Asgard genomes as a sister clade to the sequences of “Candidatus Thorarchaeota,” and we propose the name “Sifarchaeota” to describe the new Asgard phylum. Comparative analysis showed distinct differences between Sifarchaeota and previously reported Asgard archaea in terms of substrate utilization, energy production and niche adaptation strategies. Finally, we detected multiple horizontal gene transfer (HGT) events from different bacterial donors that likely expanded the substrate utilization, energy production, and secondary metabolite production capacities of the Sifarchaeota.
RESULTS
MAG construction and phylogenomic analysis.
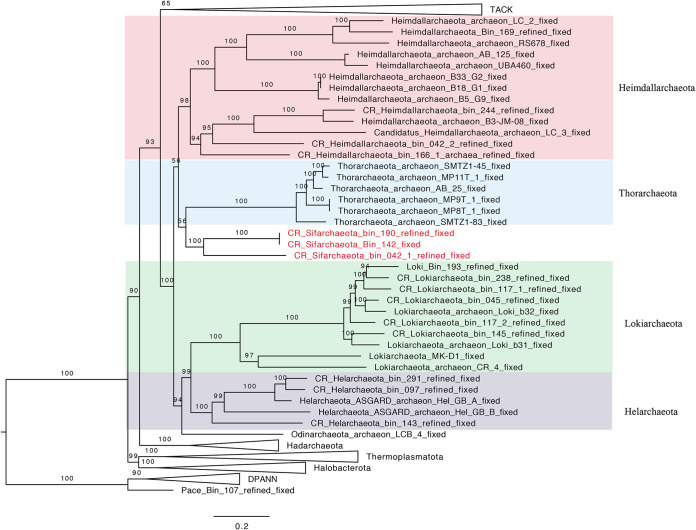
We reconstructed 3 medium-quality draft metagenome-assembled genomes (MAGs) belonging to a potentially novel Asgard phylum with moderate completion levels (67 to 80%) and very low contamination levels (0.93 to 1.9%) (Table 1) (9). Asgard archaea made up on average 5.6% of the total microbial community. The full composition of the microbial community was described by Zhao and Biddle (10). Upon genome binning and refinement, we used GTDB-tk (11) to classify the recovered archaeal MAGs using a set of 122 archaeon-specific marker proteins, three of which were suggested to be novel Asgard archaea outside the six established Asgard phyla. The taxonomic affiliations of these potentially novel Asgard MAGs were confirmed using a phylogenomic tree with 16 ribosomal proteins, and it showed that both MAGs clustered together and formed a sister lineage to MAGs belonging to the phylum “Candidatus Thorarchaeota” (Fig. 1). To further confirm the unique positions of this candidate novel Asgard lineage, we calculated the average nucleotide identities (ANI) and average amino acid identities (AAI) between the 3 novel Asgard MAGs and other MAGs representing the other Asgard lineages, including “Candidatus Lokiarchaeota,” “Candidatus Thorarchaeota,” “Candidatus Heimdallarchaeota,” and “Candidatus Odinarchaeota”. On average, the 3 novel Asgard MAGs showed low AAI values compared to the other Asgard MAGs (<50%) (see Tables S1 and S2 in the supplemental material).
TABLE 1.
Details of the Sifarchaeota MAGs analyzed in this studya
| Bin ID | Completion (%) | Redundancy (%) | Strain heterogeneity (%) | No. of contigs |
|---|---|---|---|---|
| 042_1 | 67.76 | 0.93 | 0 | 301 |
| 190 | 74.4 | 1.9 | 0 | 786 |
| 142 | 81.4 | 1.9 | 0 | 300 |
All samples were from the Costa Rica margin.
FIG 1.
Maximum-likelihood phylogenetic tree of archaeal genomes based on 16 concatenated ribosomal proteins. This tree was inferred using IQ-TREE v1.6.10 applying ModelFinder (-m option) to select the best-fit evolutionary model, which is LG+R8 model and 1,000 ultrafast bootstraps. The Sifarchaeota MAGs recovered in this study are highlighted in red. Lineages of the Asgard superphylum are expanded, while the other lineages were collapsed, if possible. The bar shows estimated sequence substitutions per residue.
Two of the new Asgard MAGs, bins 190 and 142, showed a very high similarity, with an AAI of 98.81% and an ANI of 99.52%; therefore, we focused all our subsequent analysis on two MAGs, bins 042 and 142, since they were the most complete and unique MAGs in this study.
Description of the taxa.
Although the two Sifarchaeota MAGs, bin 042 and bin 142, shared many metabolic similarities, there are significant phylogenetic distances between the two MAGs (Fig. 1) and the genomic differences as described using ANI and AAI values (see Tables S1 and S2) that prevent them from being described as one type strain. Therefore, we used two type strains to describe the two putative Sifarchaeota lineages. The genome designated bin 042 represents “Candidatus Sifarchaeotum marinoarchaea (marinoarchaea, from a marine environment). The genome designated bin 142 represents “Candidatus Sifarchaeotum subterraneus” (subterraneus, subsurface). Based on these genera, we further propose a new Asgard phylum, named Sifarchaeota phylum nov.
Metabolic reconstruction of Sifarchaeota MAGs showed remarkable saccharolytic capacities and potential anaerobic methylotrophy.
Previous reports showed that Asgard genomes encode proteins with a wide range of protein and fatty acid degradation capacities (12). So far, most of the known Asgard genomes showed limited saccharolytic capacities, emphasized by their low genomic densities of carbohydrate-active enzymes (CAZymes) and sugar transporters. However, Sifarchaeota showed a high abundance and diversity of CAZymes encoded by their MAGs specifically targeting sugars varying in complexity from low (C1 to C3) to moderate (C4 to C6), including mono-, di-, and oligosaccharides (see Fig. S1 and Tables S3 and S5; key metabolic pathways are shown in Fig. 2). Interestingly, CAZyme-based analysis revealed the presence of different glycoside hydrolase families, including cellulases and endoglucanases (GH5 and GH9), cyclomaltodextrinase (GH13), α-glucosidase (GH63), β-1,4-mannooligosaccharide phosphorylase (GH130), and β-l-arabinofuranosidase (GH142), targeting cellobiose, maltose, and maltooligosaccharides, alpha-glucosaccharides, glucose/mannose, and arabinosaccharides, respectively. We identified dedicated sugar transporters mediating the transfer of these sugars inside the Sifarchaeota cells, where the degradation and fermentation processes take place (see Table S6). Metabolic reconstruction of Sifarchaeota MAGs predicted an anaerobic and heterotrophic lifestyle, with multiple anaerobic respiration capabilities, emphasized by the presence of various fermentation pathways capable of producing different fermentative products, including acetate, acetoin, and butanediol, under strictly anaerobic conditions as well as the potential capabilities to anaerobically respire sulfate to sulfite and nitrite to ammonia. Notably, the degradation capacities for proteins, peptides, and amino acids as well as amino acid metabolism are limited in Sifarchaeota compared to the other benthic archaea. Hence, Sifarchaeota may make up for the short supply of fixed nitrogen by reducing nitrite to ammonia and encoding amidases to extract fixed nitrogen from nitrogen-containing amides like formamide.
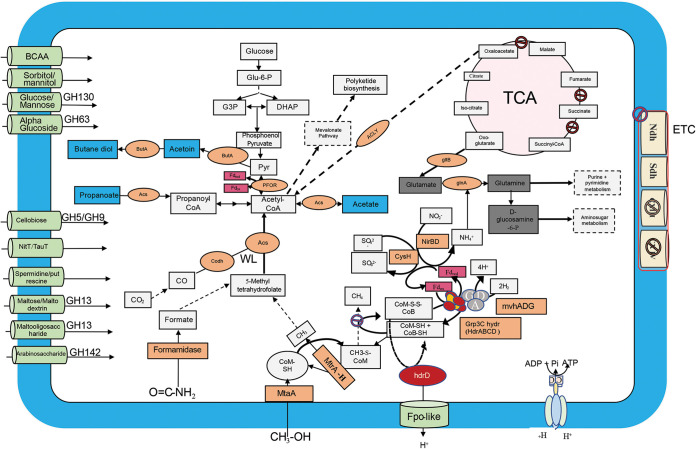
FIG 2.
Metabolic reconstruction of the key metabolic pathways encoded by the Sifarchaeota MAGs. Central metabolic pathways are shown in gray boxes, carbon fixation pathways (WL and reverse tricarboxylic acid [rTCA] cycles) are shown in pink, electron transport chain (ETC) proteins are shown in yellow, fermentation products are shown in blue boxes, enzymes and enzyme complexes are shown in orange circles, energy carriers are shown in red, and metabolite and amino acid transporters are shown in light green.
Sifarchaeota MAGs showed the capacity to utilize various C1 compounds, including formate and methanol. For example, methanol is metabolized using an anaerobic methylotrophy pathway described in reference 8, suggesting the potential widespread presence of this pathway among benthic marine archaea to metabolize methylated compounds. This potential anaerobic methylotrophic capacity was inferred by detecting an incomplete methylotrophic methanogenesis pathway, where the key genes encoding the methyl coenzyme reductase complex were completely absent. In addition, genes for an incomplete Wood-Ljungdahl (WL) pathway were identified, where only the genes of the carbonyl branch were present and the genes of the methyl branch were completely absent. Detecting this set of genes in Sifarchaeota MAGs suggests the presence of anaerobic methylotrophic capability, enabling Sifarchaeota to recycle methyl groups within methanol and other methylated compounds. Then, these methyl groups are transferred to a tetrahydrofolate complex, replacing the function of the methyl branch of the WL pathway and ultimately producing acetyl coenzyme A (acetyl-CoA).
Comparative genomic analyses between Asgard lineages show diverse metabolic features and lifestyle patterns.
To understand the key metabolic differences between the Asgard lineages, we conducted comprehensive genome-centric analyses. As described in Materials and Methods, we parsed 13 different MAGs (2 MAGs obtained from this study and 11 publicly available MAGs) belonging to 5 Asgard phyla (“Candidatus Lokiarchaeota,” “Candidatus Thorarchaeota,” “Candidatus Heimdallarchaeota,” “Candidatus Odinarchaeota” and Sifarchaeota) against the KOfam database using a hidden Markov model (HMM) search tool (Table S7). The analysis output focused on two goals: (i) exploring the range of lifestyle diversity within the Asgard superphylum and (ii) comparing the metabolic capabilities of different Asgard lineages.
Overall, the comparative analyses grouped the Asgard superphylum into 4 distinct clusters (Fig. 3) based on their whole metabolic profiles. Notably, all the MAGs within the Asgard superphylum suggested similar anaerobic and heterotrophic lifestyles, where various fermentation pathways were present as well as the absence of oxidative phosphorylation- and oxygen tolerance-related genes. Here, we describe the clusters and the features that drive their clustering.
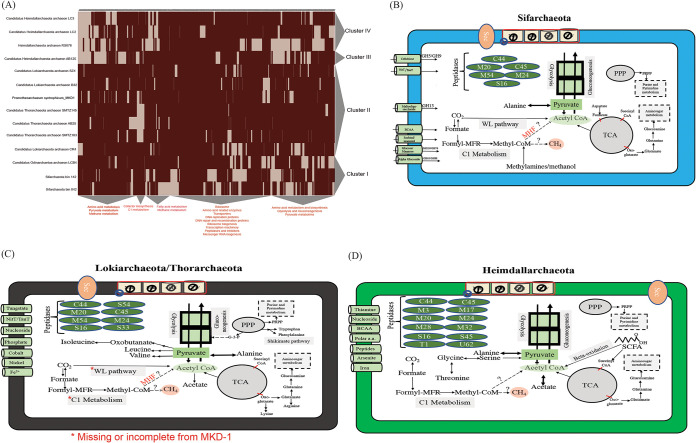
FIG 3.
Comparative analysis between MAGs representing different Asgard members. (A) Heat map clustering the different Asgard MAGs (x axis) based on their total metabolic profiles as predicted by the KOfam database (y axis). The clustering was performed using Euclidean distance and complete linkage methods. (B) Metabolic model illustrating the key metabolic features identified in the Sifarchaeota cluster. (C) Metabolic model illustrating the key metabolic features identified in the cluster comprising “Candidatus Lokiarchaeota” and “Candidatus Thorarchaeota.” (D) Metabolic model illustrating the key metabolic features identified in “Candidatus Heimdallarchaeota” clusters.
The first cluster grouped the 2 Sifarchaeota MAGs (bins 42 and 142), obtained in this study, with “Candidatus Odinarchaeota” LCB4. Sifarchaeota MAGs shared multiple metabolic similarities with the “Candidatus Odinarchaeota” MAGs, including limited amino acid and fatty acid metabolic potentials and evident saccharolytic activities, emphasized by a high density of CAZyme-encoding genes targeting different mono-, di-, and oligosaccharides. We could only identify genes encoding the oxidative branch of the pentose phosphate pathway, producing phosphoribosyl pyrophosphate (PRPP), which eventually channeled to the purine and pyrimidine metabolic pathways to be used for nucleotide and nucleic acid biosynthesis. MAGs within this cluster encoded a large number of proteins involved in C1 metabolism, including formate, methylamines, and methanol. Genes encoding formate dehydrogenase and methanol- and methylamine-specific corrinoid protein-coenzyme M methyltransferases were present, which suggests the potential capability of both lineages to utilize formate, methylamines, and methanol as carbon sources, respectively.
The second cluster grouped “Candidatus Lokiarchaeota” and “Candidatus Thorarchaeota” MAGs. Unlike the previous cluster, MAGs belonging to this cluster are characterized by their protein- and peptide-degrading capabilities, emphasized by the presence of high numbers of protease- and peptidase-encoding genes, ranging in density from 126 to 193 proteins/megabase (Mb) and belonging to different families of serine and metalloproteases. Unlike other Asgard groups, “Candidatus Thorarchaeota” and “Candidatus Lokiarchaeota” showed the capacities to synthesize different amino acids, including nonpolar amino acids (e.g., isoleucine, leucine, valine, and alanine), aromatic amino acids (e.g., tryptophan and phenylalanine) via the shikimate pathway, and charged amino acids (e.g., glutamate and lysine). Similar to other Asgard archaea, MAGs belonging to “Candidatus Thorarchaeota” and “Candidatus Lokiarchaeota” encoded proteins mediating the metabolism of C1 compounds (e.g., formate); however, the absence of methyltransferase-encoding genes excluded the use of methylated compounds as one of the potential substrates. Interestingly, MK-D1 fell within the cluster including “Candidatus Lokiarchaeota” and “Candidatus Thorarchaeota,” which is characterized by protein- and peptide-degrading capabilities. However, many of the key genes of the WL pathway and methane metabolism were absent in MK-D1. Considering that these pathways are key features of almost all Asgard archaea, this loss might be due to the effect of long-term lab cultivation and MK-D1’s dependence on a limited variety of resources (13).
Finally, the third and fourth clusters included different “Candidatus Heimdallarchaeota” MAGs. The separation between members of “Candidatus Heimdallarchaeota” based on their metabolic profiles suggests the presence of fundamental metabolic differences between the members of “Candidatus Heimdallarchaeota” and supports the previous findings that described this phylum as a polyphyletic group (3, 6). This raises the need for wider sampling efforts targeting “Candidatus Heimdallarchaeota” genomes to fully resolve their phylogenetic position and evolutionary history. However, in this study we grouped all the “Candidatus Heimdallarchaeota” MAGs and treated them as one phylum, and we designed a model that highlights the difference between them and the other Asgard lineages. Notably, all “Candidatus Heimdallarchaeota” MAGs showed the capacity to utilize proteins and short-chain fatty acids as carbon sources, while polysaccharide degradation was less supported. Similar to “Candidatus Lokiarchaeota” and “Candidatus Thorarchaeota” MAGs, “Candidatus Heimdallarchaeota” MAGs encoded a high number of peptidases with coding densities of 110 to 210 proteins/Mb and belonging to diverse families of proteases and peptidases, including serine peptidases, metallopeptidases, cysteine peptidases, and threonine peptidases (Table S4). “Candidatus Heimdallarchaeota” MAGs showed the capacity to metabolize and synthesize nonpolar amino acids (e.g., alanine, glycine, and threonine). Among all the Asgard MAGs included in this analysis, only “Candidatus Heimdallarchaeota” MAGs encoded enzymes mediating the beta-oxidation pathway, including acyl-CoA dehydrogenase, enoyl-CoA hydratase, and hydroxyacyl-CoA dehydrogenase, suggesting their potential to use short-chain fatty acids (SCFA) as carbon and energy sources. Similar to all other Asgard archaea, “Candidatus Heimdallarchaeota” encoded an incomplete WL pathway, which could have a role in metabolizing C1 compounds like formate and formaldehyde.
Due to the limited access to the surrounding microbial community composition and environmental conditions as well as the underrepresentation of the some of the lineages (only 1 MAG from “Candidatus Odinarchaeota”), we could not assess the exact reasons for these diverse substrate preferences between the Asgard phyla (e.g., polysaccharides in Sifarchaeota, proteins in “Candidatus Thorarchaeota” and “Candidatus Lokiarchaeota,” and SCFAs in “Candidatus Heimdallarchaeota”). At this point, we are unable to determine whether these findings could be generalized for all the members within each phylum or whether they are limited to the lineages/MAGs included in the study.
Role of HGT in enhancing Sifarchaeota metabolic capacities and niche adaptations.
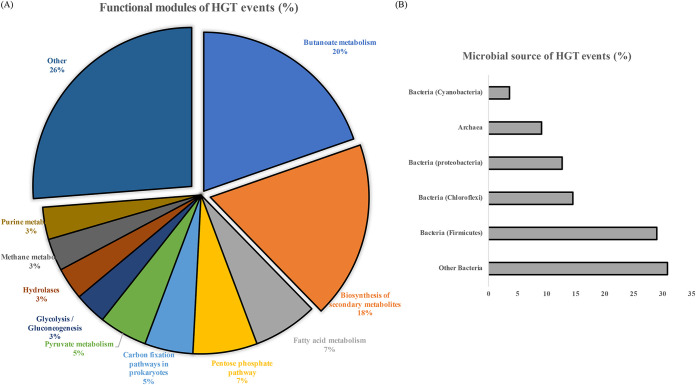
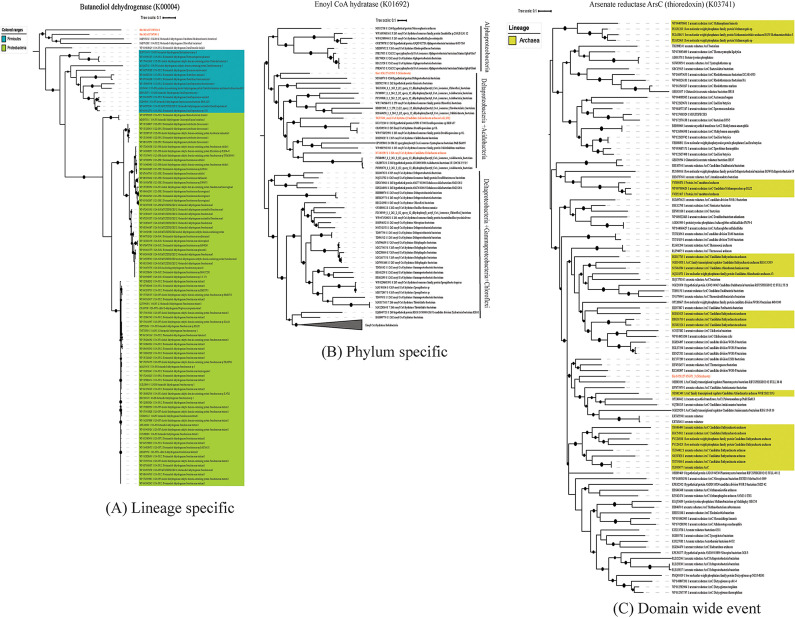
We investigated the role of HGT in expanding the metabolic capacities, substrate utilization, and niche adaptation of the Sifarchaeota phylum (Fig. 4). We traced the origin of each of the HGT events and identified the extent of the spread of each of the genes within the Asgard superphylum (Fig. S2). We successfully identified a total of 65 HGT events in the Sifarchaeota, 12 (0.65% of the total proteins) and 53 (1.34% of the total proteins) events in bins 42 and 142, respectively. Most likely, the majority of the HGT events identified were lineage specific (58 and 89.2%), and only a few similar events were detected in other Asgard lineages (7 and 11.8%). Also, we identified the majority of the potential donors of the horizontally transferred genes, of which (∼90%) were of bacterial origin. The major bacterial contributors of the horizontally transferred genes were Firmicutes (∼30%), Chloroflexi (∼15%), Proteobacteria (∼13%), Cyanobacteria (5%), and other bacterial lineages (∼30%). Only a small fraction (∼10%) of the horizontally transferred genes were of archaeal origin outside the Asgard superphylum (Fig. 5; also, see Trees S1 to S3 and Tables S8 and S9 in the supplemental material).
FIG 4.
Summary of HGT events detected in the Sifarchaeota MAGs. (A) Different functional modules of horizontally transferred genes. (B) Major donors of the horizontally transferred genes.
FIG 5.
Examples of horizontally transferred genes in Sifarchaeota MAGs. (A) Lineage-specific HGT event. Maximum-likelihood phylogenetic tree of butanediol dehydrogenase gene sequences. Shaded areas correspond to the potential source organisms. The tree was constructed on the basis of butanediol dehydrogenase gene sequences using FastTree. Reference sequences were obtained using AnnoTree (K00004). (B) Phylum-specific HGT event. Maximum-likelihood phylogenetic tree of enoyl-CoA hydratase gene sequences. The tree was constructed on the basis of enoyl-CoA hydratase gene sequences using FastTree. Reference sequences were obtained using AnnoTree (K01692). (C) Domain-wide HGT event. Maximum-likelihood phylogenetic tree of arsenate reductase (ArsC) thioredoxin gene sequences. The tree was constructed on the basis of ArsC thioredoxin gene sequences using FastTree. Reference sequences were obtained using AnnoTree (K03741).
We classified HGT events based on how widespread the transferred genes were among the different Asgard phyla, as well as other archaeal lineages. Accordingly, the HGT events were classified into lineage-specific, phylum-specific, and domain-wide events. In the lineage-specific events, the closest relatives of the transferred genes were bacteria; the genes were exclusively found in Sifarchaeota MAGs, and no orthologs were found in other Asgard or any other archaeal phyla (e.g., butanediol dehydrogenase). In the phylum-specific events, the closest relatives of the transferred genes were bacteria and the genes were found in multiple Asgard phyla (e.g., enoyl-CoA hydratase). In the domain-wide events, the closest relatives of the transferred genes were bacteria and the genes were found in different archaeal phyla (e.g., arsenate reductase arsC thioredoxin) (Fig. 5). In general, the functional annotation of the transferred genes showed that these genes are involved in augmenting the Sifarchaeota metabolic repertoire. The majority of the transferred genes fall within two metabolic modules: butanoate metabolism and biosynthesis of secondary metabolites. In butanoate metabolism, the majority of transferred genes encoded butane diol dehydrogenases and glutaconate-CoA transferase subunits mediating the key steps in pyruvate and acetate formation from butane diol and hydroxyglutaryl-CoA, respectively.
The majority of the genes involved in the biosynthesis of secondary metabolites were part of the porphyrin and chlorophyll metabolism and terpenoid biosynthesis (e.g., anaerobic magnesium-protoporphyrin IX monomethyl ester cyclase and 1,4-dihydroxy-2-naphthoate polyprenyltransferase). Interestingly, multiple genes involved in niche adaptation may be acquired from the surrounding bacterial communities. These genes include those encoding formamidase and MtaA/CmuA family methyltransferase which potentially enable Sifarchaeota to utilize amide-containing compounds and methylated compounds as nitrogen and carbon sources, respectively. A gene for arsenate reductase was potentially acquired from a candidate division KSB1 bacterium, potentially allowing Sifarchaeota to use arsenate-containing compounds as final electron acceptors.
DISCUSSION
In this study, we recovered three MAGs from deep Costa Rica sediments belonging to a new Asgard phylum, forming a sister clade to MAGs belonging to “Candidatus Thorarchaeota.” This population was not seen during previous studies of the Costa Rica margin (7, 8). We propose the name Sifarchaeota for this phylum. Putative collective metabolic profiles of the Sifarchaeota MAGs showed remarkable differences in lifestyle and niche adaptations compared to the other Asgard members. We predict a saccharolytic, anaerobic, and heterotrophic lifestyle with limited amino acid and fatty acid metabolism, whereas most of the Asgard archaea identified before this study were identified as having peptide degradation and short-chain fatty acid oxidation capacities (3, 5, 12). We detected genes encoding incomplete methanogenesis pathways coupled with the carbonyl branch of the WL pathway, suggesting the capability of Sifarchaeota to perform anaerobic methylotrophy, enabling the utilization of various methylated compounds (e.g., methanol and methylamines). The widespread presence of anaerobic methylotrophy in multiple benthic archaea highlights the importance of this pathway as an effective strategy to utilize various methylated compounds commonly encountered in marine sediment niches (8, 14). On the other hand, Sifarchaeota MAGs shared potential biogeochemical functions with other Asgard archaea, including the presence of nitrite reductase (nirBD) genes, putatively enabling Sifarchaeota members to reduce nitrite to ammonia, as well as genes encoding sulfate adenylyltransferase (sat) and phosphoadenosine phosphosulfate reductase (cysH), signifying their putative capability to perform assimilatory sulfate reduction and sulfate activation. These shared characteristics between Asgard genomes confirm the significant roles of different Asgard lineages in nitrogen and sulfur biogeochemical cycles in marine sediment environments.
We gauged the role of HGT in shaping the evolution of Sifarchaeota. Our analysis suggests that HGT events either added novel genes to the Sifarchaeota pangenome that impart new functions, e.g., butanoate metabolism and biosynthesis of secondary metabolites, or enabled the utilization of alternative nonorganic compounds as electron sources, e.g., arsenate reductase. Moreover, we explored the range of donors of horizontally transferred genes, and we concluded that HGT is not limited to a specific phylogenetic group and is probably acquired from the surrounding bacterial communities normally present in deep marine sediments. Finally, 91% of HGT events are Sifarchaeota lineage specific and probably took place relatively recently during the course of evolution, after the diversification of Sifarchaeota from the other Asgard lineages. We determined that only 9% of the events happened prior to the full diversification of Sifarchaeota from other Asgard archaea, as well as other archaeal lineages. This could be a plausible explanation for the presence of shared functions of nonarchaeal origin between most Asgard lineages.
MATERIALS AND METHODS
Sample collection.
Samples were collected during International Ocean Drilling Program (IODP) Expedition 334 at site U1379B on the Costa Rica margin. Details of the site location and sampling methods were described previously (7, 15). Microbiology samples (whole-round cores) were collected on board and frozen immediately at −80°C. They were shipped to the Gulf Core Repository (College Station, TX) on dry ice and stored at −80°C until shipping to the Biddle lab (Lewes, DE) on dry ice and further storage at −80°C. Metagenome sequencing data were generated from four silty clay sediment horizons (2H-1, 2H-2, 2H-5A, and 2H-5B) at a depth interval of 2 to 9 m below the sea floor (mbsf), within the sulfate reduction zone.
DNA extraction, library construction, and sequencing.
DNA for metagenomic sequencing was extracted from ∼7 g sediment (∼0.7 g sediment in 10 individual lysis tubes) using the PowerSoil DNA isolation kit (Qiagen) following the manufacturer’s instructions, except for the following minor modification: the lysing tubes were incubated in a water bath of 60°C for 15 min prior to bead beating on the MP machine at the highest speed (grade of 6) for 45 s. The DNA extracts were iteratively eluted from the 10 spin columns into a final volume of 100 μl of double-distilled H2O for further analysis. Metagenomic libraries were prepared and sequenced (150-bp paired-end reads) on an Illumina NextSeq 500 sequencer at the Genome Sequencing & Genotype Center at the University of Delaware.
Assembly and genome binning.
The raw sequencing data were processed with Trimmomatic v.0.36 (16) to remove Illumina adapters and low-quality reads (SLIDINGWINDOW:10:25). The quality-controlled reads from the eight samples were de novo coassembled into contigs using Megahit v.1.1.2 (17) with the k-mer length varying from 27 to 117. Contigs longer than 1,000 bp were automatically binned using MaxBin2 (18) and Metabat2 (19), and the best-quality ones were selected using DAS_Tool (20) with the default parameters. The resulting MAGs were quality assessed using CheckM (21) and taxonomically classified using GTDBTk v1.3.0 (11) using the default parameters. Genome bins of >50% completeness were manually refined using gbtools (22) based on the GC content, taxonomic assignments, and differential coverages in different samples. Coverages of contigs in each sample were determined by mapping trimmed reads onto the contigs using BBMap v.37.61 (23). Taxonomy of contigs was assigned according to the taxonomy of the single-copy marker genes in contigs identified using a script modified from blobology (24) and classified by BLASTn. rRNA gene sequences in contigs were identified using Barrnap (Seeman 2015, GitHub; https://github.com/tseemann/barrnap), and classified using VSEARCH with the SILVA 132 release (25) as the reference.
To improve the quality of the two novel Asgard archaeal MAGs, we recruited quality-controlled reads using BBMap from 2H-2, because the highest genome coverages of these two MAGs were detected in this particular sample. The recruited reads were then reassembled using SPAdes v.3.12.0 (26) using default parameters. After removal of contigs shorter than 1 kb, the resulting scaffolds were visualized and rebinned manually using gbtools (22) as described above. The quality of the resulting Asgard archaea genomes were checked using CheckM v.1.0.7 (21) with the lineage_wf option.
Concatenated ribosomal protein phylogeny.
To determine the phylogenetic affiliations of the two Sifarchaeota MAGs in the domain Archaea, we performed a thorough phylogenomic analysis based on the concatenation of 16 ribosomal proteins (L2, L3, L4, L5, L6, L14, L15, L16, L18, L22, L24, S3, S8, S10, S17, and S19). Reference genomes were selected from all the major archaeal phyla (3 to 5 for each) included in the GTDB database (27), except for the Asgard superphylum, for which all available genomes were included. Ribosomal protein sequences were detected in Anvi’o (28) using the respective HMM profiles, aligned using MUSCLE v.3.8.31 (29), and concatenated. The maximum-likelihood phylogenetic tree was reconstructed using IQ-Tree (v1.6.6) (30) (located on the CIPRES web server [31]) with LG+R8 as the best-fit substitution model selected by ModelFinder (32), and single-branch location was tested using 1,000 ultrafast bootstraps and approximate Bayesian computation (33). In addition to phylogenomic analysis, we calculated the average nucleotide identity (ANI) using FastANI (34) and average amino acid identity (AAI) using CompareM (https://github.com/dparks1134/CompareM) with default settings between these novel Asgard MAGs and other publicly available ones (i.e., those included in the GTDB database), to further explore the novelty of these MAGs.
Metabolic reconstruction.
Amino acid sequences encoded by the Sifarchaeota MAGs were predicted using Prodigal v2.6.3 (35) applying the default parameters and using translation table 11. The resulting amino acid sequences were screened using the HMMsearch tool (36) against custom HMM databases (37) representing the key genes for specific metabolic pathways to understand the potential metabolic capacities of Sifarchaeota and their ecological roles. The presence/absence profiles of the metabolic pathways and their completion levels were further assessed through querying the predicted amino acids against KEGG database using BlastKoala tool (38). Carbohydrate-active enzymes encoded by the Sifarchaeota MAGs were analyzed using the dbCAN-fam-HMMs (v6) database (39). Proteases, peptidases, and peptidase inhibitors encoded by the MAGS were detected with the USEARCH-ublast tool (40) against the MEROPS database v12.1 (41). Finally, the predicted amino acid sequences were queried against the TCDB database (42) using the USEARCH-ublast tool (40) to identify potential transporters.
Genome-centric comparative analysis for the Asgard superphylum.
We compared the total metabolic profiles of Sifarchaeota MAGs with representatives from other Asgard lineages, including (“Candidatus Odinarchaeota,” “Candidatus Thorarchaeota,” “Candidatus Lokiarchaeota” and “Candidatus Heimdallarchaeota”) in addition to the only available Asgard culture representative, “Candidatus Prometheoarchaeum syntrophicum” strain MK-D1, to identify the key metabolic differences between the phyla within the Asgard superphylum and understand their role in their potential ecological roles in their native habitats. We queried the representative MAGs of each of the Asgard lineages against the KOfam database (KEGG release 94.1) with the KofamKOALA web tool (43) and using an E value of 10−3. Then, the Asgard MAGs were clustered based on the presence/absence profiles of the identified KOfams, shared between at least 3 genomes, using the web-based tool clustergrammer (44) and applying the Euclidean distance and average linkage type.
HGT analysis.
HGT events were detected by querying predicted Sifarchaeota amino acids against the KEGG database (KEGG release 94.1) via the GhostKoala web interface (38). Proteins with KEGG ontology (KO) annotation, non-Asgard hits, and a bit score of >100 were considered potential HGT candidate proteins. Then, the candidate set of proteins were queried against nonredundant (nr) and UniProtKB/Swiss-Prot databases, and all proteins showing Asgard hits as one of the top hits were removed from any further analysis. Finally, each candidate protein was aligned to reference set of proteins collected via AnnoTree (45) using the corresponding KO entry, and the HGT events were confirmed through creating approximately maximum-likelihood phylogenetic tree for each candidate protein using FastTree v2.1 (46). An outline for the HGT detection pipeline is illustrated in Fig. S1. HGT events were further confirmed via reconstructing likelihood trees for the potential HGT proteins using IQ tree software. In the IQ trees, we applied the ModelFinder module with 1,000 bootstrap replicates, and single-branch position was tested using the approximate Bayes test (Trees S1 to S3).
Data availability.
All sequencing data used in this study are available in the NCBI Short Reads Archive under the project number PRJNA599172. In particular, the raw metagenomic sequencing data are available in the NCBI database under the BioSample numbers SAMN13740702 to SAMN13740710. The three MAGs discussed in this study are available under the accession numbers SAMN16874501 to SAMN16874503.
Supplementary Material
ACKNOWLEDGMENTS
We thank the shipboard scientists and crew of Integrated Ocean Drilling Program (IODP) Expedition 334 for collecting the sediments and the shore-based curators of the Gulf Coast Repository for their faithful stewardship of precious frozen subsurface samples. We also thank our high-performance computation cluster administrator, Karol Miaskiewicz, for his tireless work.
This work was supported by a W. M. Keck Foundation award to J.F.B.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Teske A, Sørensen KB. 2008. Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J 2:3–18. 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- 2.Solden L, Lloyd K, Wrighton K. 2016. The bright side of microbial dark matter: lessons learned from the uncultivated majority. Curr Opin Microbiol 31:217–226. 10.1016/j.mib.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 3.Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521:173–179. 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, Nunoura T, Banfield JF, Schramm A, Baker BJ, Spang A, Ettema TJG. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541:353–358. 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 5.Seitz KW, Dombrowski N, Eme L, Spang A, Lombard J, Sieber JR, Teske AP, Ettema TJG, Baker BJ. 2019. Asgard archaea capable of anaerobic hydrocarbon cycling. Nat Commun 10:1822. 10.1038/s41467-019-09364-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai M, Liu Y, Yin X, Zhou Z, Friedrich MW, Richter-Heitmann T, Nimzyk R, Kulkarni A, Wang X, Li W, Pan J, Yang Y, Gu J-D, Li M. 2020. Diverse Asgard archaea including the novel phylum Gerdarchaeota participate in organic matter degradation. Sci China Life Sci 63:886–897. 10.1007/s11427-020-1679-1. [DOI] [PubMed] [Google Scholar]
- 7.Martino A, Rhodes ME, León-Zayas R, Valente IE, Biddle JF, House CH. 2019. Microbial diversity in sub-seafloor sediments from the Costa Rica margin. Geosciences 9:218. 10.3390/geosciences9050218. [DOI] [Google Scholar]
- 8.Farag IF, Biddle JF, Zhao R, Martino AJ, House CH, León-Zayas RI. 2020. Metabolic potentials of archaeal lineages resolved from metagenomes of deep Costa Rica sediments. ISME J 14:1345–1358. 10.1038/s41396-020-0615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glöckner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu W-T, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Lapidus A, Meyer F, Yilmaz P, Parks DH, Eren AM, Genome Standards Consortium, et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao R, Biddle J. 2021. Helarchaeota and co-occurring sulfate-reducing bacteria in subseafloor sediments from the Costa Rica margin. bioRxiv 10.1101/2021.01.19.427333. [DOI] [PMC free article] [PubMed]
- 11.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927. 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacLeod F, Kindler GS, Wong HL, Chen R, Burns BP. 2019. Asgard archaea: diversity, function, and evolutionary implications in a range of microbiomes. AIMS Microbiol 5:48–61. 10.3934/microbiol.2019.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, Matsui Y, Miyazaki M, Murata K, Saito Y, Sakai S, Song C, Tasumi E, Yamanaka Y, Yamaguchi T, Kamagata Y, Tamaki H, Takai K. 2020. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577:519–525. 10.1038/s41586-019-1916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anda VD, Chen L-X, Dombrowski N, Hua Z, Jiang H-C, Banfield J, Li W-J, Baker B. 2020. Brockarchaeota, a novel archaeal lineage capable of methylotrophy. Research Square 10.21203/rs.3.rs-39998/v1. [DOI] [Google Scholar]
- 15.Vannucchi P, Ujiie K, Stroncik N, IODP Exp. 334 Scientific Party. 2013. IODP Expedition 334: an investigation of the sedimentary record, fluid flow and state of stress on top of the seismogenic zone of an erosive subduction margin. Sci Drilling 15:23–30. 10.5194/sd-15-23-2013. [DOI] [Google Scholar]
- 16.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y-W, Tang Y-H, Tringe SG, Simmons BA, Singer SW. 2014. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2:26. 10.1186/2049-2618-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, Banfield JF. 2018. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol 3:836–843. 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seah BKB, Gruber-Vodicka HR. 2015. gbtools: interactive visualization of metagenome bins in R. Front Microbiol 6:1451. 10.3389/fmicb.2015.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. No. LBNL-7065E. Ernest Orlando Lawrence Berkeley National Laboratory, Berkeley, CA. [Google Scholar]
- 24.Kumar S, Jones M, Koutsovoulos G, Clarke M, Blaxter M. 2013. Blobology: exploring raw genome data for contaminants, symbionts and parasites using taxon-annotated GC-coverage plots. Front Genet 4:237. 10.3389/fgene.2013.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 28.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ’omics data. PeerJ 3:e1319. 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA. 10.1109/GCE.2010.5676129. [DOI] [Google Scholar]
- 32.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson LS, Eddy SR, Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431. 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beckmann S, Farag IF, Zhao R, Christman GD, Prouty NG, Biddle JF. 2021. Expanding the repertoire of electron acceptors for the anaerobic oxidation of methane in carbonates in the Atlantic and Pacific Ocean. ISME J 10.1038/s41396-021-00918-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 41.Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. 2018. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res 46:D624–D632. 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saier MH, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. 2016. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res 44:D372–D379. 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H. 2020. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36:2251–2252. 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernandez NF, Gundersen GW, Rahman A, Grimes ML, Rikova K, Hornbeck P, Ma'ayan A. 2017. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci Data 4:170151. 10.1038/sdata.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendler K, Chen H, Parks DH, Lobb B, Hug LA, Doxey AC. 2019. AnnoTree: visualization and exploration of a functionally annotated microbial tree of life. Nucleic Acids Res 47:4442–4448. 10.1093/nar/gkz246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data used in this study are available in the NCBI Short Reads Archive under the project number PRJNA599172. In particular, the raw metagenomic sequencing data are available in the NCBI database under the BioSample numbers SAMN13740702 to SAMN13740710. The three MAGs discussed in this study are available under the accession numbers SAMN16874501 to SAMN16874503.